Signal transduction networks allow cells to perceive changes in the extracellular environment and to mount an appropriate response. Mitogen-activated protein kinase (MAPK) cascades are among the most thoroughly studied of signal transduction systems and have been shown to participate in a diverse array of cellular programs, including cell differentiation, cell movement, cell division, and cell death. A key question in studies of this cascade is, how does a ubiquitously activated regulatory enzume generate a specific and biologically appropriate cellular response? In this review we describe recent findings that provide insight into ways that the regulation, structure, and localization of MAPKs and the participation of adapters and scaffolds can help determine biological outcomes.
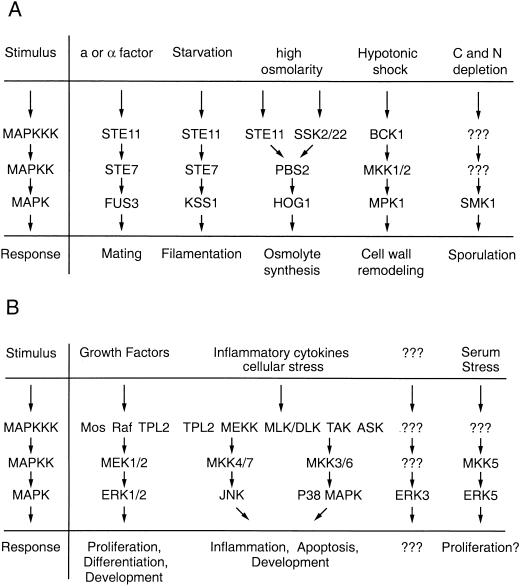
MAPK cascades are evolutionarily conserved in all eucaryotes and play a key role in the regulation of gene expression as well as cytoplasmic activities. They typically are organized in a three-kinase architecture consisting of a MAPK, a MAPK activator (MEK, MKK, or MAPK kinase), and a MEK activator (MEK kinase [MEKK] or MAPK kinase kinase). Transmission of signals is achieved by sequential phosphorylation and activation of the components specific to a respective cascade. In the yeast Saccharomyces cerevisiae, five MAPK modules have been described; they regulate mating, filamentation, high-osmolarity responses, cell wall remodeling, and sporulation (Fig. 1A) (reviewed in references 56 and 77). In mammalian systems five distinguishable MAPK modules have been identified so far (Fig. 1B). These include the extracellular signal-regulated kinase 1 and 2 (ERK1/2) cascade, which preferentially regulates cell growth and differentiation, as well as the c-Jun N-terminal kinase (JNK) and p38 MAPK cascades, which function mainly in stress responses like inflammation and apoptosis (reviewed in references 57, 74, and 103). Moreover, MAPK pathways control several developmental programs, such as morphogenesis and spatial patterning in Dictyostelium amoebae (17, 45), eye development in Drosophila melanogaster (124), vulva induction in Caenorhabditis elegans (113), and T-cell development in mammals (31).
FIG. 1.
Schematic overview of MAPK modules. (A) In S. cerevisiae, five MAPK modules regulate mating, filamentation, high-osmolarity responses, cell wall remodeling, and sporulation. (B) Mammalian MAPK modules regulate cell growth, differentiation, stress responses, and development. Abbreviations: MAPKK, MAPK kinase; MAPKKK, MAPK kinase kinase.
Individual MAPK modules generally can signal independently from each other, and this specificity is manifested in distinct physiologic responses. This is most obvious when studying MAPK signaling in S. cerevisiae. Here a particular extracellular event characteristically activates a specific MAPK module and initiates a unique cellular program (reviewed in references 56 and 77). For example, stimulation of cells with pheromone leads to the activation of the pheromone response pathway (STE11, STE7, and FUS3) (Fig. 2), which ultimately results in cell cycle arrest and the induction of mating-specific genes. However, related MAPKs whose modules share some components with the pheromone response pathway are not affected by pheromone stimulation but are activated only in response to the appropriate stimulus. For example, under conditions of high osmolarity Ste11 can lead to activation of Hog1 but does not induce mating-specific genes. Conversely, conditions that activate the filamentation pathway (which utilizes STE11 and STE7) induce only genes that regulate filamentous growth without triggering pheromone responses or responses to high osmolarity. These observations suggest that yeast cells have developed efficient mechanisms to generate pathway specificity and to successfully suppress cross talk, even when individual components participate in more than one signaling pathway.
FIG. 2.
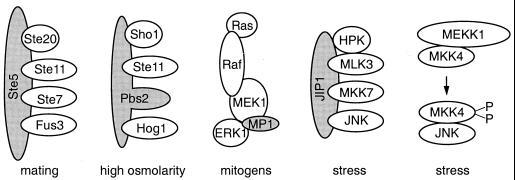
Scaffold and adapter molecules in MAPK pathways. MAPK scaffolds and adapters (gray shading) are thought to promote the formation of oligomeric protein complexes with components that function in a specific MAPK module. Scaffolds have been identified in S. cerevisiae (Ste5 and Pbs2) and mammals (MP1 and JIP1). MKK4 may have scaffolding properties comparable to those of PBS2.
In metazoan cells the problem is more complex because each cell is simultaneously exposed to multiple extracellular signals and must integrate these inputs to choose an appropriate response. Thus, the biological context of a signal plays a determinative role in the way that MAPK activation is interpreted. For example, although ERKs generally regulate cell growth and cell differentiation and JNKs participate in a stress response, this is not always the case and in certain cell types activation of JNKs can induce proliferation (110). This indicates that in mammalian systems physiologic responses associated with a certain MAPK module can be cell type specific. Moreover, in PC12 cells, transient stimulation of the ERK cascade leads to proliferation whereas sustained stimulation leads to differentiation, as measured by neurite outgrowth (81). Thus, activation of the ERK cascade can lead to contrasting physiological responses in the same cellular context, suggesting that signal specificity is also determined by regulatory mechanisms other than the selective activation of a MAPK module.
In this short review, we outline recent advances in understanding of this signaling system that help to explain how MAPK cascades are regulated and how specificity can be generated. Because of the power of yeast genetics, understanding of MAPK signaling in S. cerevisiae is at an advanced level, and thus many examples that utilize this organism are given. However, analogous mechanisms appear to be operative in metazoans as well. We discuss in turn the role of enzyme-substrate interactions, scaffolding proteins, subcellular targeting and localization, temporal regulation, and signal integration in determining the biological outcome of MAPK activation.
ENZYME-SUBSTRATE INTERACTIONS
Since MAPK pathways form a cascade of kinases, each downstream kinase serves as a substrate for the upstream activator. Thus, direct enzyme-substrate interactions play a critical role in the transmission of signals and offer a potential platform for generating specificity. Indeed, in vitro and in vivo certain upstream kinases exhibit substrate specificity and preferentially signal to a subset of possible effectors. For example, members of the Raf family specifically bind to and activate MEKs (28, 68, 93, 94, 118) but not MKKs in the stress pathways (83). The interaction of MEKs with Raf is dependent on a proline-rich sequence unique to MEKs and not found in other MKKs. Deletion of this proline-rich sequence ablates the ability of MEK to bind to Raf and greatly diminishes the ability of Raf to activate MEK (10). A-Raf preferentially activates MEK1, whereas c-Raf-1 seems to activate both MEK1 and MEK2 equally well (130); full-length B-Raf binds to both MEK1 and MEK2 but activates MEK1 better than MEK2, whereas another MEK activator that may be a splicing isoform of B-Raf displays the opposite specificity (102). These results indicate that Raf family members can interact differentially with the substrates MEK1 and MEK2. However, much of this specificity is a consequence of quantitative differences in protein-protein affinities and can be lost when the partners are overexpressed (10). Likewise, MEKK1, a MAPK kinase kinase of the stress pathway, appears to be specifically coupled to unique sets of activators and effectors (141), although specificity can be lost when MEKK1 is N-terminally truncated or overexpressed (133, 134). MEKs and ERKs also can bind directly to each other (although weakly) (43), and as expected, MEKs activate ERKs but not JNKs.
How enzyme substrate specificity is generated in MAPK cascades is possibly best understood for the MAPKs themselves. Substrate specificity was originally determined by sequencing the phosphorylation site of in vitro MAPK substrates and subsequently by peptide selection. These studies revealed that MAPKs are proline-directed serine/threonine kinases that recognize a proline at the +1 position of a potential substrate (with ERKs, but not JNKs or p38 MAPKs, having a secondary preference for prolines at the −2 position) (18, 35, 111).
The three-dimensional structures of both inactive (143) and active (8) ERK2 have been solved by X-ray crystallography and greatly improve our understanding of how these enzyme substrate interactions are regulated. Upon activation by dual phosphorylation on Thr183 and Tyr185, the activation lip of ERK2 undergoes a conformational change, thereby creating a surface pocket that is specific for proline. Interestingly, this proline-specific pocket is occupied by the side chain of Arg192 when ERK2 is in an inactive state. Therefore, access of proline to the catalytic specificity pocket is blocked when the enzyme is not activated, and a productive enzyme-substrate interaction cannot take place (8).
Sequences in the C-terminal domain of MAPKs that are outside of the proline-specific pocket are likely to be involved in additional enzyme-substrate interactions and may have some targeting function. These sequences correspond to what is termed an extended substrate-binding groove in cyclic AMP (cAMP)-dependent protein kinase (66). Interestingly, substrate specificity in stress-activated JNK2 is determined by a region that spans residues 208 to 230 and that resides within the predicted substrate-binding groove (61). This region is necessary for both binding to and phosphorylation of the substrate c-Jun by JNK2. JNK1, a MAPK that interacts poorly with c-Jun, can be redirected to c-Jun by replacing the predicted substrate binding region of JNK1 with corresponding sequences from JNK2 (61), suggesting that distinct regions participate in the recognition of MAPK substrates.
In contrast to the proline-specific pocket, the structure of this C-terminal substrate-binding groove in ERK2 is not significantly affected by the state of ERK2 activation (8). Thus, it is likely that protein-protein interactions mediated by this domain are independent of enzyme activation and that enzyme-substrate complexes are formed prior to activation. This would allow inactive MAPKs to regulate substrates, either through direct binding or by restricting access of the substrate to kinases with related specificity. A mechanism like this explains a series of otherwise puzzling observations in yeast where kinase-deficient point mutants of the MAPKs Kss1 and Fus3 inhibit signaling in a pathway-specific fashion (2, 21, 78). Kinase-deficient Kss1 selectively blocks filamentation and inhibits the activation of transcription factor Ste12, whereas kinase-deficient Fus3 blocks mating. However, in a MAPK deletion null mutant this specificity is lost and each MAPK can functionally complement the other. For example, in the complete absence of Fus3 (but not in the presence of kinase-deficient Fus3) Kss1 can substitute for Fus3 and can regulate mating. These conceptually important findings from studies in yeast, which show that inactive kinases can regulate signaling pathways, are likely also to be applicable to mammalian systems.
Additional specificity-determining domains in MAPKs have been identified in a study using chimeric ERK1-p38 MAPK molecules (7). Consistent with previous findings, Brunet and Pouyssegur demonstrate that C-terminal regions, including the phosphorylation lip, play a role in substrate recognition. Interestingly, a small region of 40 residues in the N-terminal domain of p38 MAPK determines responsiveness to particular agonists and thus may be involved in directing p38 MAPK to a specific activator. A chimera consisting of the N-terminal regions of p38 MAPK and C-terminal regions of ERK1 was able to receive stress signals and to redirect them into mitogenic responses, suggesting a modular structure for MAPK with distinct domains responsible for substrate recognition and agonist specificity.
Various MAPKs can recognize substrates differentially, as demonstrated in a recent study that examined the interactions of ERKs, JNKs, and p38 MAPKs with transcription factor Elk-1 (135). Although all three MAPK family members can phosphorylate Elk-1 in vitro and in vivo, they interact with Elk-1 through clearly distinct mechanisms. Both ERKs and JNKs associate with Elk-1 through the so-called D domain of Elk-1 (residues 310 to 334) but require a different set of residues within this domain. In contrast, p38 MAPK does not need the D domain for functional interaction with Elk-1, suggesting that this MAPK utilizes a different mechanism to select ELK-1 as a substrate.
Taken together, these findings indicate that enzyme specificity of MAPKs is determined by a combination of the intrinsic specificity of the catalytic region for serines or threonines with a proline at the +1 position, as well as domains that determine stable binding to the substrate. As more in vivo substrates of MAPKs are identified, it will be possible to determine which of these factors is the predominant determinant of signaling specificity.
SCAFFOLDS AND ADAPTERS
Several lines of evidence suggest that enzymatic activation of kinases in a given MAPK cascade may not be sufficient for successful propagation of a specific signal. For example, even though a constitutively active fragment of MEKK1, when overexpressed in cells, can phosphorylate and activate MEKs, under these conditions activated MEK is unable to signal further downstream, resulting in a poor activation of ERKs (133). In addition, NIH 3T3 cells grown in suspension fail to activate MEKs and ERKs after growth factor stimulation, although c-Raf-1 can still be activated (100). Finally, mutationally activated MEK that lacks the proline-rich sequence between kinase domains 9 and 10 exhibits ample kinase activity in vitro but fails to activate ERKs in vivo and to transform cells (10, 25). These observations led us and others to predict the existence of additional factors in mammalian MAPK cascades that facilitate optimal information flow. The above-reported defects in signaling could be explained by inappropriate targeting of affected components so that functional enzyme-substrate interactions would not take place. Targeting could be regulated by a mechanism that directly affected enzyme-substrate affinities or by putative accessory molecules like adapters or scaffolds involved in facilitating the assembly of enzyme-substrate complexes.
In MAPK pathways in S. cerevisiae, scaffolding functions have been identified for Ste5, a protein that is essential in the mating pathway, and for Pbs2, a MAPK activator that regulates the high-osmolarity response (reviewed in reference 128). Ste5 selectively associates with Ste11, Ste7, and Fus3 (14, 80, 98) and apparently functions by facilitating the formation of an oligomeric protein complex (Fig. 2). Because this complex would increase the local concentration of the components involved in the mating response, the efficiency of enzyme-substrate interactions should be greatly enhanced. Moreover, by tethering components of a specific cascade into an oligomeric protein complex and by excluding from this complex related components that operate in parallel cascades, illegitimate cross-interactions with related MAPK modules may be minimized (136). Thus, scaffolds like Ste5 may insulate a given MAPK module from related pathways, thereby increasing the specificity of signaling. Similar scaffolding properties were suggested for the MKK homologue Pbs2, which associates with the Pbs2 activator Ste11, the MAPK Hog1, and a putative osmosensor, Sho1 (97). Interestingly, here the predicted scaffolding function is located in domains of Pbs2, a kinase in the nodule, suggesting that both accessory molecules and components of MAPK cascades can serve as scaffolds.
Because many of the enzymatic components of the mammalian MAPK cascades bind directly to each other, it was not clear whether these signaling modules would contain scaffolds that were separate proteins. Rather, it seemed likely that, as with Pbs2, protein-protein interaction domains intrinsic to components of a respective MAPK module would direct the formation of oligomeric signaling complexes. This is especially plausible for the ERK cascade, where Raf family members interact with Ras-GTP through interactions between a well-defined effector domain on Ras and Ras-binding domains on Raf (85, 119, 122, 126, 144), MEKs bind to Raf family members (10, 93, 130), and ERKs bind to N-terminal regions of MEK (43). Similar interactions have been reported for the JNK activator MKK4, which seems to organize a MAPK module consisting of MEKK1, MKK4, and JNK1 (131). Theoretically, these protein-protein interactions should be sufficient to facilitate the formation of an oligomeric signaling complex, making accessory scaffolding molecules like yeast Ste5 unnecessary in higher eucaryotic systems.
However, the identification of JNK interacting protein 1 (JIP1) and MEK partner 1 (MP1), two scaffold-like molecules that function in mammalian MAPK modules, suggests that scaffolds may have a more universal role in the generation of specificity than previously anticipated. JIP1 operates in the JNK pathway and selectively binds to JNK, JNK activator MKK7, and MKK7 activators mixed-lineage kinase 3 (MLK3) and dual leucine zipper-bearing kinase (DLK) (127). Overexpression of JIP1 can enhance the activation of JNK in the presence of ectopically expressed MKK7 (but not MKK4) and MLK3, suggesting that JIP1 is specific in regard to both binding and enzymatic activation of this MAPK module. MP1 functions in the ERK cascade and has properties somewhat different from those of JIP1. This small protein (14 kDa) selectively associates with ERK1 and ERK activator MEK1 but fails to detectably interact with the close family members ERK2 and MEK2 (105). Moreover, physical interactions of MP1 with MEK1 activators like c-Raf-1 (equivalent to the JIP1-MLK3 interaction) have not been detected so far. In vitro, MP1 enhances the activation of MEK by Raf, and when overexpressed in cells, MP1 can selectively enhance the activation of ERK1 but not ERK2, suggesting that MP1 helps to discriminate between these two MAPK isoforms. Since both JIP1 and MP1 bind selectively to a subset of components of a MAPK module and specifically enhance signaling that is routed through these components, they exhibit properties of scaffolding proteins. A scaffolding function was also suggested for Ksr (kinase suppressor of Ras), which can physically associate with Raf, MEK, and ERK (132, 139); however, the precise role of Ksr is incompletely understood, and whether this protein functions as a scaffold in MAPK signaling remains to be seen.
Interestingly, the known MAPK scaffolds (Ste5, Pbs2, JIP1, and MP1) show no obvious similarity in amino acid sequence. This raises the possibility that these molecules did not originate from a common ancestor but have evolved independently. It is likely that more scaffolding molecules that operate at different steps in MAPK modules will be identified in the near future. In particular, for the ERK cascade, we predict additional small proteins like MP1 that associate with family members of ERK1 and MEK1 and that possibly connect these components to the upstream activators Raf, Mos, and TPL2.
Considering that many players in MAPK pathways physically interact with both their activators and effectors, there arises the question of why nature evolved a variety of distinct scaffolding proteins that form additional contacts to components of MAPK cascades. Why, for example, do we need MP1 to bridge ERK1 and MEK1 if ERK1 is capable of directly interacting with MEK1 (43)? The answer to this question may lie in the potential of such complexes to achieve a high degree of regulatory flexibility. It is likely that signaling complexes form dynamic structures with constant assembly and disassembly of components. Because several sites of relatively weak contact within a complex can act cooperatively, components exhibiting such a binding behavior can establish disproportionately stable associations. However, selective disruption of some of the interactions may result in the loss of synergistic binding and the subsequent dissociation of a binding partner. Therefore, stable association and disassociation could be effectively regulated by controlling a few relatively weak protein-protein interactions. It is plausible that conformational changes induced by activating (or feedback) phosphorylation in these kinase cascades are sufficient to mediate these effects.
SPATIAL ORGANIZATION OF MAPK MODULES
A growing number of reports indicate that MAPKs can be targeted to specific sites within a cell. For example, ERKs associate with the cytoskeleton (101) and a small but constitutively active pool of ERK1 copurifies with tubulin, whereas ERK2 copurifies with microtubule-associated protein 2 (87). Moreover, active JNK is localized to punctate structures along microtubules together with JNK kinase kinase MLK2 (90), suggesting that both MAPKs and upstream activators can be recruited to particular cellular locations. Finally, active ERK is found at the kinetochores, asters, and midbody during certain stages of mitosis (109, 142) and associates with centromeric protein CENP-E (142), a finding that is particularly interesting because ERKs (51, 121) and p38 MAPKs (114) have been implicated in checkpoint controls during M phase.
At present, targeting mechanisms for MAPK pathway components are poorly understood and several strategies for targeting are possible. For example, substrates or upstream activators with a restricted subcellular distribution could facilitate the recruitment of the respective kinases by direct physical interactions. To illustrate, membrane-associated Ras binds to Raf and can recruit this kinase to the membrane (69). Similarly, CENP-E that colocalizes with active ERK at the kinetochores may target ERKs to this cellular location (142). Comparable mechanisms could also be in effect for other MAPK substrates, such as transcription factors, that can often directly bind to their upstream activators (2, 52, 60). Alternatively, targeting could be regulated by so far unknown anchoring molecules that operate in a manner similar to A-kinase-anchoring proteins. These molecular anchors can recruit protein kinase A (PKA) to specific cellular sites (65, 72, 95, 106). In addition, subcellular distribution of a given component could be affected by internal targeting sequences, as suggested for ERK activators MEK1 and MEK2. Both contain an intrinsic nuclear export signal (NES) that regulates the distribution of MEKs between the nucleus and the cytoplasm (41, 42, 58, 125).
MAPKs can undergo a pronounced nuclear translocation following stimulation by agonists (12, 49, 70), a process that is best understood for MAPKs ERK1 and ERK2. Here nuclear translocation involves at least three distinct regulatory steps, including cytoplasmic retention of ERKs by MEK (43), phosphorylation and subsequent dimerization of ERKs (63), and active transport of ERK dimers across the nuclear membrane (63).
ERKs are cytoplasmic in quiescent cells and associate with MEK (43). Because MEK has a NES and is actively exported from the nucleus, it may serve as a cytoplasmic anchor for ERKs under these conditions. This idea is supported by the finding that MEK mutants with dysfunctional NESs were unable to retain ERKs in the cytoplasm, resulting in a uniform distribution of ERKs and MEKs in the cytoplasm and the nucleus (43). Serum stimulation disrupts the ERK-MEK interaction by an unknown mechanism allowing ERKs to translocate to the nuclear compartment (43).
Nuclear translocation of ERKs is facilitated by phosphorylation of ERKs at the sites of activation, but it does not require ERK kinase activity (63). Interestingly, phosphorylation of yeast MAPK Hog1 also induces transient nuclear translocation in the absence of kinase activity (38), suggesting that this mechanism may be common to several MAPKs. Phosphorylated ERKs dimerize, and this seems to be important for nuclear translocation, because a dimerization-deficient mutant of ERK, although otherwise functionally intact, cannot translocate to the nucleus (63).
Since ERK dimers exceed the maximal size for passive diffusion, it is likely that these dimers are actively transported across the nuclear membrane. However, ERKs lack an intrinsic nuclear localization signal (NLS), and it is unclear how the nuclear import machinery recognizes ERKs. One possibility is that ERK dimers interact with a third partner that provides the NLS. Alternatively, ERKs could contain a discontinuous NLS that is recognized only after dimerization of two ERK monomers (63). Genetic evidence from S. cerevisiae supports the notion that MAPKs are actively transported across the nuclear membrane. Nuclear import of Hog1 requires the activity of the small GTP-binding protein Ran-GSP1 (38), which plays an important role in nuclear transport (19, 82, 86). Moreover, the activity of the novel importin β homologue NMD5 that presumably functions in targeting Hog1 to the nuclear pore complex is essential for nuclear translocation of Hog1 (38). Interestingly, nuclear export of dephosphorylated Hog1 requires the function of the NES receptor XPO1-CRM1 (38, 112), suggesting that unphosphorylated MAPKs can be exported actively from the nuclear compartment.
MAPKs can also regulate subcellular distribution of downstream effectors, thereby affecting the signaling properties of these proteins. This is especially apparent for Ca2+-activated transcription factor NFAT4, which requires dephosphorylation of two residues (Ser163 and Ser165) in order to translocate to the nucleus. Interestingly, these sites can be phosphorylated by JNK, and JNK activation leads to nuclear exclusion of NFAT4 (15), indicating that JNK activation can counteract NFAT4 signaling. Similarly, phosphorylation of nuclear MAPK-activated protein (MAPKAP) kinase 2 by p38 MAPK results in exclusion of both p38 MAPK and its substrate MAPKAP kinase 2 from the nuclear compartment (4, 33).
TEMPORAL ORGANIZATION OF MAPK MODULES
Temporal organization of MAPK activities can play an important role in the generation of specific biologic responses. This is especially apparent in PC12 cells, where ERK activation can initiate two opposing biologic programs, cell growth and cell differentiation. Sustained ERK activation followed by nuclear translocation of ERKs leads to cellular differentiation, as measured by neurite outgrowth (81). In contrast, cell growth can be induced by transient activation of ERKs (81). Interestingly, sustained activation of ERKs in fibroblasts leads to growth and transformation (24). Thus, the biologic response to these variations in the timing of kinase activation is cell type dependent.
Control of the persistence of signaling in PC12 cells has been examined by Stork and coworkers (137), who reported that MAPK activation in response to nerve growth factor is controlled by both Ras and Ras-related GTP-binding protein Rap1. Whereas Ras is responsible for the early activation of ERKs, Rap1 mediates the persistent activation that is required to induce neuronal differentiation. Thus, the ability of a cell or agonist to make use of these two GTP-binding proteins could determine the timing of a MAPK signal. However, these interpretations have been challenged by recent reports from Bos and colleagues (145).
Whether activation of MAPKs is sustained or transient depends also on the induction of feedback inhibition pathways. These include activation of phosphatases, notably members of the large family of dual-specificity phosphatases that inactivate MAPKs (reviewed in reference 74). These phosphatases display unique substrate specificities for MAPK family members. For example, MAPK phosphatase 3 (MKP-3) selectively inactivates ERKs (89) and M3/6 is restricted to stress-activated MAPKs (89). However, MKP-1 and MKP-2 dephosphorylate ERK, JNK, and p38 MAPK to various degrees (16), suggesting that some dual-specificity phosphatases have overlapping substrates. Moreover, many dual-specificity phosphatases are confined to specific subcellular compartments. For example, MKP-1 is exclusively localized to the nucleus (6, 73) whereas MKP-3 is cytoplasmic (50, 88). A nuclear phosphatase such as MKP-1 would selectively turn off MAPK-induced changes in gene expression while allowing continued functioning of cytoplasmic MAPKs.
Serine/threonine phosphatases such as PP1 and PP2A also can regulate MAPK cascades, presumably by dephosphorylation of activated MEK and Raf. Indeed, inhibition of these phosphatases with okadaic acid leads to constitutive activation of MAPKs (1, 9, 55). However, whether these phosphatases are regulated or serve only as a basal inactivation mechanism is unknown.
Feedback phosphorylations also are likely to play a role in determining the persistence of a MAPK signal. MEK (10, 37, 79), Raf (108, 117), and SOS (23, 32) have all been shown to be subject to feedback phosphorylation, but the role of these phosphorylations in regulating signaling is still uncertain.
SPECIFICITY BY CROSS TALK AND SIGNAL INTEGRATION
Mammalian MAPK modules are intertwined in a complex network of signaling cascades, and the induction of a particular biologic response often requires more than one input signal. In order to generate an appropriate output signal, coordination of input pathways and subsequent integration of signals are necessary. To achieve this, MAPK modules have evolved strategies to communicate with other pathways. This cross talk can affect signaling properties and information flow and can modify the specificity of the pathways involved.
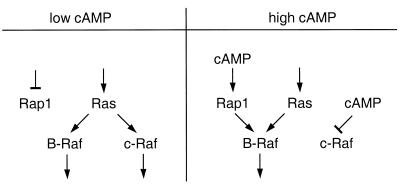
To illustrate, elevated levels of cAMP that induce the activation of PKA can determine which Raf isoform will be engaged in a cell to stimulate MEKs. In Rat-1 fibroblast cells, the activation of c-Raf-1 can be selectively inhibited with high levels of intracellular cAMP (22, 107, 129), possibly by a direct phosphorylation of c-Raf-1 by PKA (129). In contrast, in PC12 cells B-Raf can be activated by elevated levels of cAMP in a Ras-independent manner (34, 39, 138), and in LNCaP prostate cancer cells cAMP potentiates rather than inhibits the activation of MAPK in response to epidermal growth factor (13). This activation is mediated by the cAMP-responsive small GTPase Rap1 (30), which directly signals to the B-Raf isoform (120, 137). Therefore, high levels of cAMP and subsequent activation of PKA can change the routing of information, resulting in selective activation of B-Raf and the inhibition of c-Raf-1 (Fig. 3). This suggests that cAMP could function as a molecular switch to determine isoform specificity in the mitogenic MAPK cascade. Similarly, MEK1 may be regulated by the cell cycle machinery, since phosphorylation of MEK1 (but not MEK2) by cyclin-dependent kinase 2 can reduce kinase activity of this MEK (104).
FIG. 3.
cAMP can act as a molecular switch to generate isoform specificity. In the absence of cAMP, both B-Raf and c-Raf-1 can be activated by p21ras. High levels of cAMP inhibit activation of c-Raf-1 and activate the small GTP-binding protein Rap1, which then can selectively activate B-Raf in a Ras-independent manner.
MAPK modules can affect the properties of other signaling pathways. As mentioned previously, JNK can counteract the activation of transcription factor NFAT4 by direct phosphorylation (15). ERKs can communicate with estrogen signals by phosphorylating the estrogen receptor at serine 118 (62). This phosphorylation enhances estrogen-induced transcriptional activity of the estrogen receptor, suggesting that estrogen binding and growth factor stimulation of ERKs can cooperate to maximally activate the estrogen receptor. ERKs also interact with alpha/beta interferon-mediated activation of the JAK-STAT signaling cascade, and dominant-negative ERK2 can inhibit transcriptional activation of an interferon-responsive reporter gene (26).
Signaling properties of MAPK modules can also be modified by closely related MAPK cascades. For example, the small G proteins Rac1 and Cdc42, which normally activate the JNK module, can cooperate with Raf to activate the ERK cascade (40). This cooperative effect seems to require the Rac- and Cdc42-activated kinase p21-activated kinase 1 (PAK1), which can phosphorylate MEK1 in a proline-rich region previously implicated in signaling (10, 25, 59), thereby increasing MEK’s affinity for c-Raf-1 (40). Interestingly, PAK3, a member of the same kinase family, was recently reported to enhance c-Raf-1 activity by direct phosphorylation at serine 338 (64), further supporting the concept of cross-cascade communication. In the case of MEKK1, a more indirect cross talk network that can induce gene expression of MKP-1 occurs, (5), thereby possibly attenuating signaling of heterologous MAPK cascades.
Interestingly, downstream of MAPK modules signals can converge; for example, both ERK and p38 MAPK can activate the downstream kinases MAPK signal-integrating kinase 1 (MNK1) (44, 123) and mitogen- and stress-activated protein kinase 1 (MSK1) (27). Similarly, MAPKAP kinase 3 (also known as 3pK) can be targeted by ERK, JNK, and p38 MAPK (75). In addition, many transcription factors are regulated by more than one MAPK pathway. For example, ERK (47), JNK (11, 48), and p38 MAPK (99) can activate transcription factor Elk-1. Other transcription factors interact with a subset of MAPKs, as demonstrated for ATF2, which is regulated by JNK (53) and p38 MAPK (99), or c-Jun, which is targeted by JNK (29, 84) and in some cellular contexts also by ERK (71, 96). This raises the question of why cells first put a lot of effort into the generation of a specific signal if in the end many pathways converge on the same downstream targets. In other words, how can specificity of signals be maintained when several pathways utilize the same target protein as an endpoint of signaling? Although understanding of this issue is incomplete, several mechanisms have so far been identified.
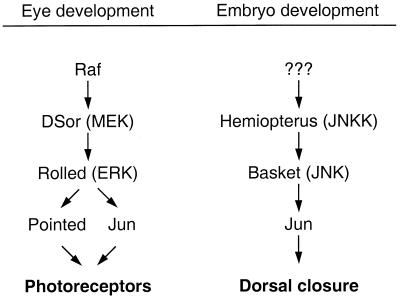
First, MAPK modules may be linked to certain downstream effectors in a cell-type-specific manner. This was suggested, for example, in a recent study demonstrating that the Drosophila MAPK Rolled (ERK homologue) can activate Jun in eye development, whereas Basket (JNK homologue) signals to Jun during dorsal closure (Fig. 4) (67, 96, 116). Similarly, in PC 12 cells ERKs but not JNKs activate c-Jun to induce neurite outgrowth (71). However, in other cell lines c-Jun is selectively regulated by JNK (83, 84), suggesting that additional cell-type-specific factors can determine which MAPK module is linked to c-Jun.
FIG. 4.
Jun can be targeted by two MAPK modules in Drosophila. During eye development, Jun is activated by the ERK cascade and cooperates with Pointed to regulate differentiation of photoreceptors. During embryonal development Jun is activated by the JNK pathway to regulate dorsal closure. JNKK, JNK kinase.
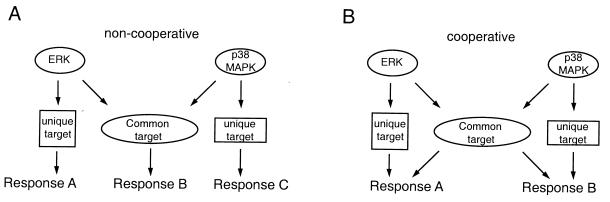
Second, common targets of MAPK modules may be involved in the regulation of general cellular functions, such as the regulation of translation, that are required for virtually all transitions in cell state (Fig. 5A). This notion is supported by the observation that MNK1, a kinase that is a common substrate of ERK and p38 MAPK, is able to phosphorylate eukaryotic translation initiation factor 4E at a physiologically relevant site, thereby possibly affecting initiation of translation (123).
FIG. 5.
MAPK modules can induce specific responses by signaling to unique and common downstream targets. Unique and common substrates can induce separate responses (A) or cooperate to induce a coordinated response (B).
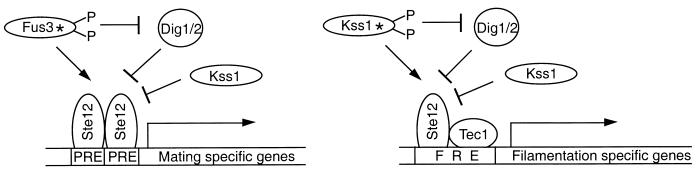
Finally, common downstream targets of MAPKs also could cooperate with input signals unique to a specific MAPK module to generate discrete biologic responses (Fig. 5B). This is presumably the case for S. cerevisiae transcription factor Ste12, which is regulated by both Fus3 and Kss1 to induce genes specific to mating or filamentous growth. Induction of mating-specific genes requires binding of Ste12 to pheromone response elements in pheromone-inducible promoters (54, 77, 140) and may be enhanced by a second transcription factor, Mcm1 (36, 91). In contrast, during filamentous growth, Ste12 binds cooperatively with transcription factor Tec1 to an enhancer element called FRE (filamentation and invasion response element) to induce genes that regulate filamentous growth (Fig. 6) (46, 76, 77). Therefore, by regulating the availability of transcription factor Tec1, a cell may be able to influence whether activation of Ste12 leads to mating or to filamentous growth. Moreover, additional regulatory elements participate in the control of Ste12 activity. For example, inactive Kss1 can cooperate with Dig1/2 to repress transcriptional activity of Ste12. This repression can be overcome by phosphorylation of Dig1/2 by MAPKs Fus3 and Kss1 (2, 3, 20, 115).
FIG. 6.
Activation of Ste12 by Fus3 or Kss1 induces distinct biologic responses. Mating-specific genes can be induced when Ste12 binds to pheromone response elements (PRE). Induction of filamentation-specific genes requires cooperative binding of Ste12 and Tec1 to a specific filamentation response element (FRE). Additional signals other than MAPK activity may be necessary to regulate Tec1 or to target Ste12 to specific response elements. Inactive Kss1 can cooperate with Dig1/2 to repress Ste12 activity. Derepression of Ste12 requires phosphorylation of Dig1/2 by MAPK Fus3 or Kss1.
CONCLUSIONS
MAPK cascades were initially believed to operate predominantly as linear signaling pathways that directly link a specific input signal to a specific biologic response. However, it now seems that communication takes place through a complex network of signaling cascades and that information flows not only from and to the outside world but also laterally from one pathway to another. By integrating several different signals, organisms can mount appropriate responses to an enormous variety of challenges and signals while making use of a minimal number of novel components.
The way regulatory signals are interpreted will depend on the other signals that are also received, just as middle C on a piano sounds different when played as part of a C major or an A minor chord. In a more general way, cells may generate a diversity of activities with a limited number of signaling pathways, much as a composer can create music with just 12 keys. Activation of signaling cascades in various combinations would correspond to the different sounds an orchestra creates by varying harmony, rhythm, and instrumentation. The challenge now is to learn how the various pathways are coordinated and integrated to orchestrate richly diverse biological responses.
ACKNOWLEDGMENTS
We thank Andy Catling and other members of the T. Parsons, M. Weber, and S. Parsons laboratories for many helpful discussions and Maja Zecevic, Charu Sharma, and Dan Gioeli for critical comments on the manuscript.
This work was supported by USPHS/NIH grants CA39076, CA76500, and GM47332 and a gift from CaPCURE.
REFERENCES
- 1.Amaral M C, Casillas A M, Nel A E. Contrasting effects of two tumor promoters, phorbol myristate acetate and okadaic acid, on T-cell responses and activation of p42 MAP-kinase/ERK-2. Immunology. 1993;79:24–31. [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell L, Cook J G, Voora D, Baggott D M, Martinez A R, Thorner J. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell L, Cook J G, Zhu-Shimoni J X, Voora D, Thorner J. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc Natl Acad Sci USA. 1998;95:15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benlevy R, Hooper S, Wilson R, Paterson H F, Marshall C J. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr Biol. 1998;8:1049–1057. doi: 10.1016/s0960-9822(98)70442-7. [DOI] [PubMed] [Google Scholar]
- 5.Bokemeyer D, Sorokin A, Yan M, Ahn N G, Templeton D J, Dunn M J. Induction of mitogen-activated protein kinase phosphatase 1 by the stress-activated protein kinase signaling pathway but not by extracellular signal-regulated kinase in fibroblasts. J Biol Chem. 1996;271:639–642. doi: 10.1074/jbc.271.2.639. [DOI] [PubMed] [Google Scholar]
- 6.Brondello J M, McKenzie F R, Sun H, Tonks N K, Pouyssegur J. Constitutive MAP kinase phosphatase (MKP-1) expression blocks G1 specific gene transcription and S-phase entry in fibroblasts. Oncogene. 1995;10:1895–1904. [PubMed] [Google Scholar]
- 7.Brunet A, Pouyssegur J. Identification of MAP kinase domains by redirecting stress signals into growth factor responses. Science. 1996;272:1652–1655. doi: 10.1126/science.272.5268.1652. [DOI] [PubMed] [Google Scholar]
- 8.Canagarajah B J, Khokhlatchev A, Cobb M H, Goldsmith E J. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 9.Casillas A M, Amaral K, Chegini-Farahani S, Nel A E. Okadaic acid activates p42 mitogen-activated protein kinase (MAP kinase; ERK-2) in B-lymphocytes but inhibits rather than augments cellular proliferation: contrast with phorbol 12-myristate 13-acetate. Biochem J. 1993;290:545–550. doi: 10.1042/bj2900545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catling A D, Schaeffer H J, Reuter C W, Reddy G R, Weber M J. A proline-rich sequence unique to MEK1 and MEK2 is required for Raf binding and regulates MEK function. Mol Cell Biol. 1995;15:5214–5225. doi: 10.1128/mcb.15.10.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavigelli M, Dolfi F, Claret F X, Karin M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R H, Sarnecki C, Blenis J. Nuclear localization and regulation of ERK- and RSK-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, Cho R W, Stork P J, Weber M J. Elevation of cyclic adenosine 3′,5′-monophosphate (cAMP) potentiates activation of mitogen-activated protein kinase (MAP kinase) by growth factors in LNCaP prostate cancer cells. Cancer Res. 1999;59:213–218. [PubMed] [Google Scholar]
- 14.Choi K Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 15.Chow C W, Rincon M, Cavanagh J, Dickens M, Davis R J. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 16.Chu Y, Solski P A, Khosravi-Far R, Der C J, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem. 1996;271:6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 17.Chung C Y, Reddy T K, Zhou K M, Firtel R A. A novel, putative MEK kinase controls developmental timing and spatial patterning in Dictyostelium and is regulated by ubiquitin-mediated protein degradation. Genes Dev. 1998;12:3564–3578. doi: 10.1101/gad.12.22.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark-Lewis I, Sanghera J S, Pelech S L. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 19.Cole C N, Hammell C M. Nucleocytoplasmic transport: driving and directing transport. Curr Biol. 1998;8:R368–R372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- 20.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 21.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 22.Cook S J, McCormick F. Inhibition by cAMP of Ras-dependent activation of Raf. Science. 1993;262:1069–1072. doi: 10.1126/science.7694367. [DOI] [PubMed] [Google Scholar]
- 23.Corbalan-Garcia S, Yang S S, Degenhardt K R, Bar-Sagi D. Identification of the mitogen-activated protein kinase phosphorylation sites on human Sos1 that regulate interaction with Grb2. Mol Cell Biol. 1996;16:5674–5682. doi: 10.1128/mcb.16.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 25.Dang A, Frost J A, Cobb M H. The MEK1 proline-rich insert is required for efficient activation of the mitogen-activated protein kinases ERK1 and ERK2 in mammalian cells. J Biol Chem. 1998;273:19909–19913. doi: 10.1074/jbc.273.31.19909. [DOI] [PubMed] [Google Scholar]
- 26.David M, Petricoin E, Benjamin C, Pine R, Weber M J, Larner A C. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 27.Deak M, Clifton A D, Lucocq L M, Alessi D R. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dent P, Haser W, Haystead T A, Vincent L A, Roberts T M, Sturgill T W. Activation of mitogen-activated protein kinase kinase by v-Raf in NIH 3T3 cells and in vitro. Science. 1992;257:1404–1407. doi: 10.1126/science.1326789. [DOI] [PubMed] [Google Scholar]
- 29.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 30.de Rooij J, Zwartkruis F J T, Verheijen M H, Cool R H, Nijman S M, Wittinghofer A, Bos J L. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 31.Dong C, Yang D, Wysk M, Whitmarsh A J, Davis R J, Flavell R A. Defective T cell differentiation in the absence of Jnk1. Science. 1998;282:2092–2095. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 32.Douville E, Downward J. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene. 1997;15:373–383. doi: 10.1038/sj.onc.1201214. [DOI] [PubMed] [Google Scholar]
- 33.Engel K, Kotlyarov A, Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 1998;17:3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erhardt P, Troppmair J, Rapp U R, Cooper G M. Differential regulation of Raf-1 and B-Raf and Ras-dependent activation of mitogen-activated protein kinase by cyclic AMP in PC12 cells. Mol Cell Biol. 1995;15:5524–5530. doi: 10.1128/mcb.15.10.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erickson A K, Payne D M, Martino P A, Rossomando A J, Shabanowitz J, Weber M J, Hunt D F, Sturgill T W. Identification by mass spectrometry of threonine 97 in bovine myelin basic protein as a specific phosphorylation site for mitogen-activated protein kinase. J Biol Chem. 1990;265:19728–19735. [PubMed] [Google Scholar]
- 36.Errede B, Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989;3:1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- 37.Errede B, Ge Q Y. Feedback regulation of MAP kinase signal pathways. Philos Trans R Soc Lond B Biol Sci. 1996;351:143–148. doi: 10.1098/rstb.1996.0010. [DOI] [PubMed] [Google Scholar]
- 38.Ferrigno P, Posas F, Koepp D, Saito H, Silver P A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frodin M, Peraldi P, Van Obberghen E. Cyclic AMP activates the mitogen-activated protein kinase cascade in PC12 cells. J Biol Chem. 1994;269:6207–6214. [PubMed] [Google Scholar]
- 40.Frost J A, Steen H, Shapiro P, Lewis T, Ahn N, Shaw P E, Cobb M H. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 1997;16:6426–6438. doi: 10.1093/emboj/16.21.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda M, Gotoh I, Adachi M, Gotoh Y, Nishida E. A novel regulatory mechanism in the mitogen-activated protein (MAP) kinase cascade. Role of nuclear export signal of MAP kinase kinase. J Biol Chem. 1997;272:32642–32648. doi: 10.1074/jbc.272.51.32642. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 43.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901–1908. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaskins C, Clark A M, Aubry L, Segall J E, Firtel R A. The Dictyostelium MAP kinase ERK2 regulates multiple, independent developmental pathways. Genes Dev. 1996;10:118–128. doi: 10.1101/gad.10.1.118. [DOI] [PubMed] [Google Scholar]
- 46.Gavrias V, Andrianopoulos A, Gimeno C J, Timberlake W E. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 47.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gille H, Strahl T, Shaw P E. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr Biol. 1995;5:1191–1200. doi: 10.1016/s0960-9822(95)00235-1. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez F A, Seth A, Raden D L, Bowman D S, Fay F S, Davis R J. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. J Cell Biol. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Groom L A, Sneddon A A, Alessi D R, Dowd S, Keyse S M. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15:3621–3632. [PMC free article] [PubMed] [Google Scholar]
- 51.Guadagno T M, Ferrell J E. Requirement for MAPK activation for normal mitotic progression in Xenopus egg extracts. Science. 1998;282:1312–1315. doi: 10.1126/science.282.5392.1312. [DOI] [PubMed] [Google Scholar]
- 52.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Derijard B, Davis R J. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta S, Campbell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 54.Hagen D C, McCaffrey G, Sprague G J. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2952–2961. doi: 10.1128/mcb.11.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haystead T A, Weiel J E, Litchfield D W, Tsukitani Y, Fischer E H, Krebs E G. Okadaic acid mimics the action of insulin in stimulating protein kinase activity in isolated adipocytes. The role of protein phosphatase 2a in attenuation of the signal. J Biol Chem. 1990;265:16571–16580. [PubMed] [Google Scholar]
- 56.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 57.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 58.Jaaro H, Rubinfeld H, Hanoch T, Seger R. Nuclear translocation of mitogen-activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc Natl Acad Sci USA. 1997;94:3742–3747. doi: 10.1073/pnas.94.8.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jelinek T, Catling A D, Reuter C W, Moodie S A, Wolfman A, Weber M J. RAS and RAF-1 form a signalling complex with MEK-1 but not MEK-2. Mol Cell Biol. 1994;14:8212–8218. doi: 10.1128/mcb.14.12.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 61.Kallunki T, Su B, Tsigelny I, Sluss H K, Derijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 62.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 63.Khokhlatchev A V, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb M H. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 64.King A J, Sun H Y, Diaz B, Barnard D, Miao W Y, Bagrodia S, Marshall M S. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature. 1998;396:180–183. doi: 10.1038/24184. [DOI] [PubMed] [Google Scholar]
- 65.Klauck T M, Faux M C, Labudda K, Langeberg L K, Jaken S, Scott J D. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 66.Knighton D R, Zheng J H, Ten E F, Ashford V A, Xuong N H, Taylor S S, Sowadski J M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 67.Kockel L, Zeitlinger J, Staszewski L M, Mlodzik M, Bohmann D. Jun in Drosophila development: redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes Dev. 1997;11:1748–1758. doi: 10.1101/gad.11.13.1748. [DOI] [PubMed] [Google Scholar]
- 68.Kyriakis J M, App H, Zhang X F, Banerjee P, Brautigan D L, Rapp U R, Avruch J. Raf-1 activates MAP kinase-kinase. Nature. 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 69.Leevers S J, Paterson H F, Marshall C J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 70.Lenormand P, Sardet C, Pages G, Allemain G, Brunet A, Pouyssegur J. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leppa S, Saffrich R, Ansorge W, Bohmann D. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 1998;17:4404–4413. doi: 10.1093/emboj/17.15.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lester L B, Scott J D. Anchoring and scaffold proteins for kinases and phosphatases. Recent Prog Horm Res. 1997;52:409–429. [PubMed] [Google Scholar]
- 73.Lewis T, Groom L A, Sneddon A A, Smythe C, Keyse S M. XCL100, an inducible nuclear MAP kinase phosphatase from Xenopus laevis: its role in MAP kinase inactivation in differentiated cells and its expression during early development. J Cell Sci. 1995;108:2885–2896. doi: 10.1242/jcs.108.8.2885. [DOI] [PubMed] [Google Scholar]
- 74.Lewis T S, Shapiro P S, Ahn N G. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 75.Ludwig S, Engel K, Hoffmeyer A, Sithanandam G, Neufeld B, Palm D, Gaestel M, Rapp U R. 3pK, a novel mitogen-activated protein (MAP) kinase-activated protein kinase, is targeted by three MAP kinase pathways. Mol Cell Biol. 1996;16:6687–6697. doi: 10.1128/mcb.16.12.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 77.Madhani H D, Fink G R. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 78.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 79.Mansour S J, Resing K A, Candi J M, Hermann A S, Gloor J W, Herskind K R, Wartmann M, Davis R J, Ahn N G. Mitogen-activated protein (MAP) kinase phosphorylation of MAP kinase kinase: determination of phosphorylation sites by mass spectrometry and site-directed mutagenesis. J Biochem. 1994;116:304–314. doi: 10.1093/oxfordjournals.jbchem.a124524. [DOI] [PubMed] [Google Scholar]
- 80.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 82.Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 83.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 84.Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moodie S A, Willumsen B M, Weber M J, Wolfman A. Complexes of Ras · GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 86.Moore M S. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 87.Morishima-Kawashima M, Kosik K S. The pool of MAP kinase associated with microtubules is small but constitutively active. Mol Biol Cell. 1996;7:893–905. doi: 10.1091/mbc.7.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muda M, Boschert U, Dickinson R, Martinou J C, Martinou I, Camps M, Schlegel W, Arkinstall S. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J Biol Chem. 1996;271:4319–4326. doi: 10.1074/jbc.271.8.4319. [DOI] [PubMed] [Google Scholar]
- 89.Muda M, Theodosiou A, Rodrigues N, Boschert U, Camps M, Gillieron C, Davies K, Ashworth A, Arkinstall S. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J Biol Chem. 1996;271:27205–27208. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- 90.Nagata K, Puls A, Futter C, Aspenstrom P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oehlen L J, McKinney J D, Cross F R. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol Cell Biol. 1996;16:2830–2837. doi: 10.1128/mcb.16.6.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Papin C, Denouel-Galy A, Laugier D, Calothy G, Eychene A. Modulation of kinase activity and oncogenic properties by alternative splicing reveals a novel regulatory mechanism for B-Raf. J Biol Chem. 1998;273:24939–24947. doi: 10.1074/jbc.273.38.24939. [DOI] [PubMed] [Google Scholar]
- 93.Papin C, Denouel A, Calothy G, Eychene A. Identification of signalling proteins interacting with B-Raf in the yeast two-hybrid system. Oncogene. 1996;12:2213–2221. [PubMed] [Google Scholar]
- 94.Papin C, Eychene A, Brunet A, Pages G, Pouyssegur J, Calothy G, Barnier J V. B-Raf protein isoforms interact with and phosphorylate MEK-1 on serine residues 218 and 222. Oncogene. 1995;10:1647–1651. [PubMed] [Google Scholar]
- 95.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 96.Peverali F A, Isaksson A, Papavassiliou A A, Plastina P, Staszewski L M, Mlodzik M, Bohmann D. Phosphorylation of Drosophila Jun by the MAP kinase rolled regulates photoreceptor differentiation. EMBO J. 1996;15:3943–3950. [PMC free article] [PubMed] [Google Scholar]
- 97.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 98.Printen J A, Sprague G F., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raingeaud J, Whitmarsh A J, Barrett T, Derijard B, Davis R J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Renshaw M W, Ren X D, Schwartz M A. Growth factor activation of MAP kinase requires cell adhesion. EMBO J. 1997;16:5592–5599. doi: 10.1093/emboj/16.18.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reszka A A, Seger R, Diltz C D, Krebs E G, Fischer E H. Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc Natl Acad Sci USA. 1995;92:8881–8885. doi: 10.1073/pnas.92.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reuter, C. W., H. J. Schaeffer, A. Eychène, C. Papin, and M. J. Weber. Differential activation of MEK-1 and MEK-2 and the role of B-Raf isoforms. Submitted for publication.
- 103.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 104.Rossomando A J, Dent P, Sturgill T W, Marshak D R. Mitogen-activated protein kinase kinase 1 (MKK1) is negatively regulated by threonine phosphorylation. Mol Cell Biol. 1994;14:1594–1602. doi: 10.1128/mcb.14.3.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schaeffer H J, Catling A D, Eblen S T, Collier L S, Krauss A, Weber M J. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 106.Scott J D. Dissection of protein kinase and phosphatase targeting interactions. Soc Gen Physiol Ser. 1997;52:227–239. [PubMed] [Google Scholar]
- 107.Sevetson B R, Kong X, Lawrence J J. Increasing cAMP attenuates activation of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:10305–10309. doi: 10.1073/pnas.90.21.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shapiro P S, Ahn N G. Feedback regulation of Raf-1 and mitogen-activated protein kinase (MAP) kinase kinases 1 and 2 by MAP kinase phosphatase-1 (MKP-1) J Biol Chem. 1998;273:1788–1793. doi: 10.1074/jbc.273.3.1788. [DOI] [PubMed] [Google Scholar]
- 109.Shapiro P S, Vaisberg E, Hunt A J, Tolwinski N S, Whalen A M, Mcintosh J R, Ahn N G. Activation of the MKK/ERK pathway during somatic cell mitosis—direct interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J Cell Biol. 1998;142:1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith A, Ramos-Morales F, Ashworth A, Collins M. A role for JNK/SAPK in proliferation, but not apoptosis, of IL-3-dependent cells. Curr Biol. 1997;7:893–896. doi: 10.1016/s0960-9822(06)00380-0. [DOI] [PubMed] [Google Scholar]
- 111.Songyang Z, Lu K P, Kwon Y T, Tsai L-H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hunter T, Cantley L C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 113.Sundaram M, Han M. Control and integration of cell signaling pathways during C. elegans vulval development. Bioessays. 1996;18:473–480. doi: 10.1002/bies.950180609. [DOI] [PubMed] [Google Scholar]
- 114.Takenaka K, Moriguchi T, Nishida E. Activation of the protein kinase p38 in the spindle assembly checkpoint and mitotic arrest. Science. 1998;280:599–602. doi: 10.1126/science.280.5363.599. [DOI] [PubMed] [Google Scholar]
- 115.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 116.Treier M, Bohmann D, Mlodzik M. JUN cooperates with the ETS domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell. 1995;83:753–760. doi: 10.1016/0092-8674(95)90188-4. [DOI] [PubMed] [Google Scholar]
- 117.Ueki K, Matsuda S, Tobe K, Gotoh Y, Tamemoto H, Yachi M, Akanuma Y, Yazaki Y, Nishida E, Kadowaki T. Feedback regulation of mitogen-activated protein kinase kinase kinase activity of c-Raf-1 by insulin and phorbol ester stimulation. J Biol Chem. 1994;269:15756–15761. [PubMed] [Google Scholar]
- 118.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 120.Vossler M R, Yao H, York R D, Pan M G, Rim C S, Stork P J. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 121.Wang X M, Zhai Y, Ferrell J J. A role for mitogen-activated protein kinase in the spindle assembly checkpoint in XTC cells. J Cell Biol. 1997;137:433–443. doi: 10.1083/jcb.137.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Warne P H, Viciana P R, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 123.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases MNK1 and MNK2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wassarman D A, Therrien M, Rubin G M. The Ras signaling pathway in Drosophila. Curr Opin Genet Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 125.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 126.White M A, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M H. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 127.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 128.Whitmarsh A J, Davis R J. Structural organization of MAP kinase signaling molecules by scaffold proteins in yeast and mammals. Trends Biochem Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 129.Wu J, Dent P, Jelinek T, Wolfman A, Weber M J, Sturgill T W. Inhibition of the EGF-activated MAP kinase signaling pathway by adenosine 3′,5′-monophosphate. Science. 1993;262:1065–1069. doi: 10.1126/science.7694366. [DOI] [PubMed] [Google Scholar]
- 130.Wu X, Noh S J, Zhou G, Dixon J E, Guan K L. Selective activation of MEK1 but not MEK2 by A-Raf from epidermal growth factor-stimulated Hela cells. J Biol Chem. 1996;271:3265–3271. doi: 10.1074/jbc.271.6.3265. [DOI] [PubMed] [Google Scholar]
- 131.Xia Y, Wu Z G, Su B, Murray B, Karin M. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 1998;12:3369–3381. doi: 10.1101/gad.12.21.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xing H, Kornfeld K, Muslin A J. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 133.Xu S, Robbins D, Frost J, Dang A, Lange-Carter C, Cobb M H. MEKK1 phosphorylates MEK1 and MEK2 but does not cause activation of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1995;92:6808–6812. doi: 10.1073/pnas.92.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 135.Yang S H, Whitmarsh A J, Davis R J, Sharrocks A D. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yashar B, Irie K, Printen J A, Stevenson B J, Sprague G F, Jr, Matsumoto K, Errede B. Yeast MEK-dependent signal transduction: response thresholds and parameters affecting fidelity. Mol Cell Biol. 1995;15:6545–6553. doi: 10.1128/mcb.15.12.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.York R D, Yao H, Dillon T, Ellig C L, Eckert S P, McCleskey E W, Stork P J. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- 138.Young S W, Dickens M, Tavare J M. Differentiation of PC12 cells in response to a cAMP analogue is accompanied by sustained activation of mitogen-activated protein kinase. Comparison with the effects of insulin, growth factors and phorbol esters. FEBS Lett. 1994;338:212–216. doi: 10.1016/0014-5793(94)80367-6. [DOI] [PubMed] [Google Scholar]
- 139.Yu W, Fantl W J, Harrowe G, Williams L T. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1998;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- 140.Yuan Y L, Fields S. Properties of the DNA-binding domain of the Saccharomyces cerevisiae STE12 protein. Mol Cell Biol. 1991;11:5910–5918. doi: 10.1128/mcb.11.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yujiri T, Sather S, Fanger G R, Johnson G L. Role of MEKK1 in cell survival and activation of JNK and ERK pathways defined by targeted disruption. Science. 1998;282:1911–1914. doi: 10.1126/science.282.5395.1911. [DOI] [PubMed] [Google Scholar]
- 142.Zecevic M, Catling A D, Eblen S T, Renzi L, Hittle J C, Yen T J, Gorbsky G J, Weber M J. Active MAP kinase in mitosis—localization at kinetochores and association with the motor protein CENP-E. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang F, Strand A, Robbins D, Cobb M H, Goldsmith E J. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 144.Zhang X F, Settleman J, Kyriakis J M, Takeuchi-Suzuki E, Elledge S J, Marshall M S, Bruder J T, Rapp U R, Avruch J. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 145.Zwartkuis F J T, Wolthius R M F, Nabben N M J M, Franke B, Bos J L. Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signaling. EMBO J. 1998;17:5905–5912. doi: 10.1093/emboj/17.20.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]