Abstract
Genetically encoded fluorescent indicators of neuronal activity are ultimately developed to dissect functions of neuronal ensembles during behavior in living animals. Recent development of near-infrared shifted calcium and voltage indicators moved us closer to this goal and enabled crosstalk-free combination with blue light-controlled optogenetic tools for all-optical control and readout. Here I discuss designs of recent near-infrared and far-red calcium and voltage indicators, compare their properties and performance, and overview their applications to spectral multiplexing and in vivo imaging. I also provide perspectives for further development.
Keywords: Near-infrared, far-red, GECI, GEVI, GECO, iGECI, miRFP, SomArchon, all-optical electrophysiology, optogenetics, calcium indicator, voltage indicator
Introduction.
Imaging neuronal activity in cells and living animals can move us closer to understanding of the brain. Fluorescent indicators for calcium changes, voltage dynamics, and neurotransmitter release combined with advanced imaging technologies already became state-of-the-art in modern neuroscience 1–3. Here I focus on near-infrared (NIR) and far-red genetically encoded indicators of neuronal activity with excitation peaks above 590 nm and emission above 640 nm. These probes can be excited with 610–640 nm lasers and emit close to or within the NIR window of tissue transparency (~650 nm - 950 nm), where the combined absorption of water and hemoglobin is minimal allowing deep tissue penetration 4. In addition to penetration depth, NIR light can allow minimally invasive large-scale recordings in vivo, since it has low scattering and induces minimal autofluorescence 5–8. Further, NIR light has minimal toxicity and influence on cellular processes in neurons, in contrast to blue light that was shown to increase expression of genes that are normally regulated through neuronal activity 9. NIR fluorescent probes also provide opportunities for crosstalk-free combinations with optogenetics and biosensors in the visible spectral range 10. Moreover, NIR indicators are the only options to be used in single-photon imaging mode in retinal neurons sensing visible light 11. While far-red light (here ~590–640 nm) yields to NIR light in tissue penetration, light scattering and autofluorescence, it outperforms green-red light in these characteristics 12, 13.
Here I discuss both fully genetically encoded indicators and chemigenetic or hybrid indicators, which rely on fluorescence of synthetic fluorogenic dyes but use genetically encoded dye binding proteins, such as HaloTag and SNAP-tag 14. A use of synthetic dyes makes chemigenetic indicators brighter than FP-based ones 15. Modern dyes, such as Janelia Fluor series of dyes, can easily penetrate brain-blood barrier and can be applied in vivo 16, 17. Although fluorogenic dyes are developed to be not fluorescent in solution in unbound state and fluorescent when bound to its respective protein, there is still an issue of background fluorescence18.
Recently progress in NIR and far-red probe development became possible with (i) engineering of new fluorescent proteins, such as monomeric bacteriophytochrome-derived fluorescent proteins miRFPs 19–21 and mIFP 22, (ii) a development of advanced fluorogenic synthetic dyes to be used in chemigenetic indicators 15–17 and new circular permutant dye-binding modules 23, (iii) application of advanced directed molecular evolution approaches resulting in substantial improvement in indicator performance 24.
As more researchers are interested in using NIR calcium and voltage indicators to solve problems in neuroscience, I aim to provide an overview by succinctly discussing available tools, their designs, advantages and limitations, imaging technologies to combine with, and demonstrated applications. I also provide a perspective on the development of new and improved indicators and their applications to advanced imaging technologies.
Genetically encoded calcium indicators (GECI).
Design and performance.
GECIs are the most widely used indicators for neuronal activity, as calcium increase reliably serves as a proxy for action potentials in neurons. Currently there are four types of NIR and far-red calcium indicators: (i) intensiometric NIR-GECO series based on bacterial phytochrome NIR FP mIFP 25, 26, (ii) NIR FRET iGECI based on miRFP670-miRFP720 FRET pair27, (iii) intensiometric far-red FR-GECOs based on circularly permuted GFP-like FPs 28, (iv) chemigenetic HCaMP1 intensiometric indicators based on circular permuted HaloTag23.
NIR GECIs incorporate monomeric FPs engineered from bacterial phytochrome photoreceptors. These FPs utilize biliverdin IXα (BV) as a chromophore. BV is naturally present in eukaryotic cells as an intermediate of heme metabolism 29. For BV-binding FPs or biosensors, the effective brightness in cells do not always correlate with molecular brightness 5. While early FPs, such as IFP1.4 have very low brightness in cells due to low affinity and specificity for BV binding, modern FPs, such as (m)iRFP series 7, 19, 20, were successfully optimized for efficient binding of endogenous BV.
The first NIR indicator NIR-GECO1 was developed as an intensiometric GECI 26. While widely used GFP-like intensiometric GECIs of GCaMP3–6 series 30–33 are based on circular permutant versions of EGFP, this design could not be adapted to NIR FPs. It was not possible to obtain a circular permutant of mIFP, since its N- and C-termini are distantly located. Therefore, NIR-GECOs were developed using an insertion of calcium-sensing module (calmodulin and its binding peptide RS20) into the loop near the chromophore pocket (Fig. 1a).
Figure 1. Near-infrared and far-red GECIs and their representative applications.
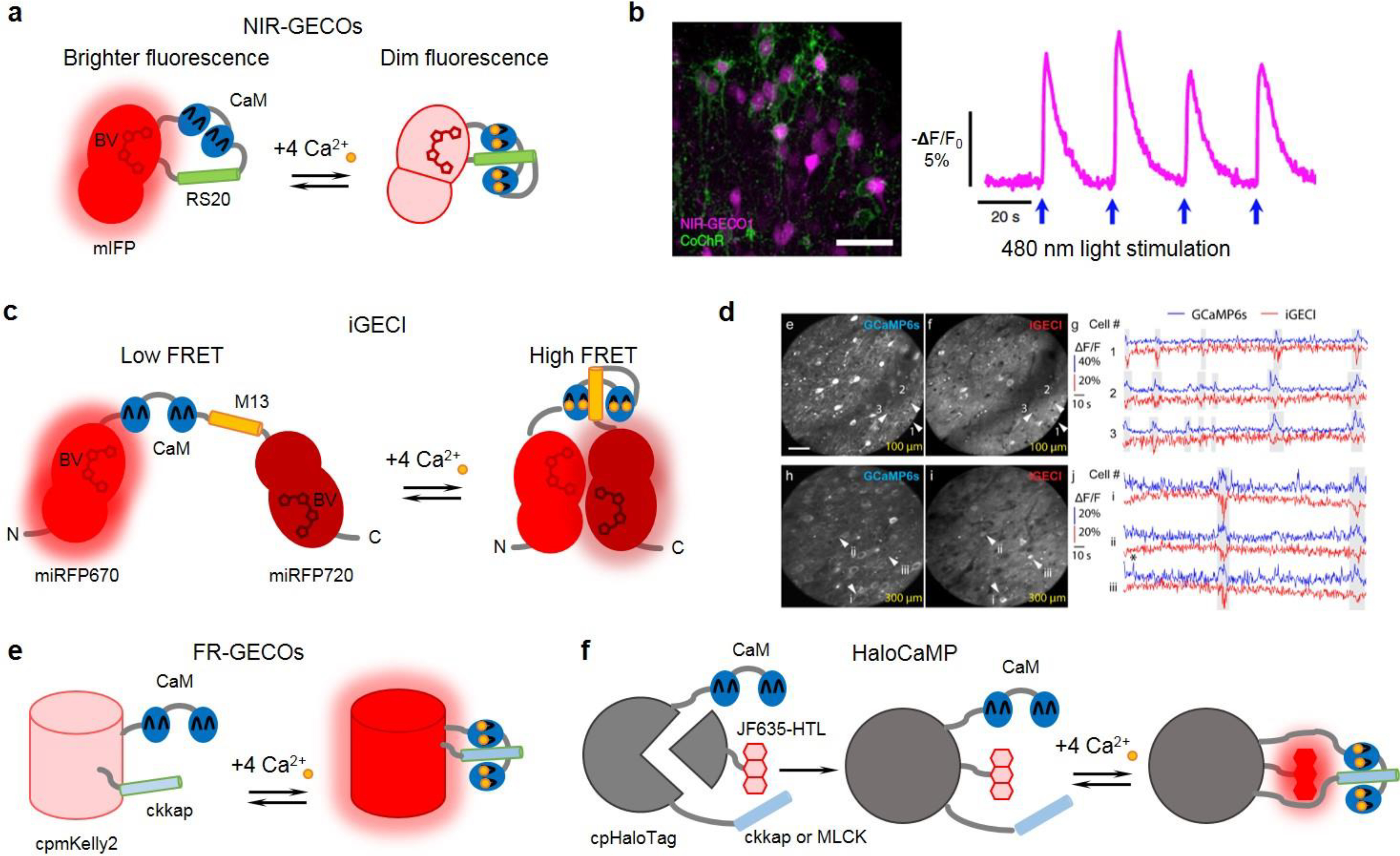
(a) Schematics of NIR-GECO series of indicators. (b) All-optical control and readout of neuronal activity. (left) Confocal images of neurons co-expressing NIR-GECO1 (magenta pseudocolor) and CoChR-mTagBFP2-Kv2.2motif (green pseudocolor) in a brain section. (right) NIR-GECO1 responses to CoChR activation. Adapted, with permission, from26 (c) Schematics of iGECI FRET based indicator. (d) Two-photon imaging of spontaneous neuronal activity with iGECI in vivo. (left) Two-photon fluorescence images of neurons in the mouse primary visual cortex at 100 μm (top) and 300 μm (bottom) below dura. Scale bar: 50 μm. (right) Example ΔF/F calcium transients for neurons co-expressing GCaMP6s and iGECI at 100 μm (top, white arrowheads) and 300 μm (bottom, white arrowheads). In all experiments, no exogenous BV was supplied. Adapted, with permission, from27. (e) Schematics of FR-GECO series of indicators. (f) Schematics of HaloCaMP1 series of indicators. (a, c, e, f) CaM: calmodulin; RS20, M13, ckkap or MLCK: CaM binding peptides; BV: biliverdin; cpmKelly2 and cpHaloTag: circular permutant mKelly2 FP and HaloTag; JF635-HTL: synthetic rhodamine dye
NIR-GECO1 has inverted signal response (also called negative response), i.e. its fluorescence reduces upon calcium binding. It was characterized in mammalian cells, cultured neurons, and tested in vivo. It was shown to be functional and reliably responding to single action potential stimulation in neurons. Still, it yields to GCaMP3–6 indicators in brightness, dynamic range, and rise and decay kinetics (Table 1). The limitation of NIR-GECO1 is its low brightness in vivo. While strong stimulation allowed mesoscale imaging with NIR-GECO1, green GCaMP6s exhibited 10-fold greater fluorescence changes in the same conditions26. Low brightness of NIR-GECO1 originates from its design: calcium-binding disturbs the chromophore pocket and, thus, binding of BV. Therefore, more than 80% of NIR-GECO1 molecules in mammalian cells do not contain the chromophore and interfere with calcium detection by buffering calcium ions26. While the BV chromophore can be supplied in vitro, it is not practical in vivo, since it does not penetrate the blood-brain barrier. Attempted co-expression of heme oxygenase, which is an enzyme that converts heme to BV, did not help much25. It can be explained by tight binding of BV to this enzyme and quick processing of BV, an intermediate metabolite, into bilirubin by biliverdin reductase29.
Table 1.
Characteristics of far-red and NIR GECIs in comparison with widely used GCaMP6s.
| GECI name | Ex/Em (nm) | Molecular brightness vs. EGFP, % (1) | pKa apo, pKa sat, | Photostability t0.5, s (2) | Effective brightness, % (3) | Dynamic range (ΔF/F), fold | Hill coefficients n1 and n2 | Rise time, s (4) | Decay time, s (4) | Kd1 and Kd2, nM | ΔF/F, % per 1 AP | Ref | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| w/o Ca2+ | with Ca2+ | HeLa cells | mouse neurons | w/o BV | with BV | in neurons | in vivo | |||||||||
| iGECI | Donor 640/670 | 37 | 6.0 | 4.5, 4.5 | 1795 | 1735 | 46 | 100 | 6× | 2.5 and 0.90 | 0.70 | 14 | 15 and 890 | −5.7 (−12.9 with BV) | ∼ −5–20 | 27 |
| Acceptor 702/720 | ||||||||||||||||
| NIR-GECO1 | 678/704 | 12 | 1.0 | 5.1, 4.9 | 100 (2) | 134 (2) | 2.4 (6) | 20 (6) | 8× | 0.99 | 1.5 (6) | 4.0 (6) | 215 885 |
∼ −4.5 (∼ −10 with BV (6)) | n.a. | 26 |
| NIR-GECO2G | 678/704 | 13 | 1.3 | 5.3, 4.8 | n.a. | ∼130 (5) | ∼5 (5) | n.a. | n.a. | 0.78 | ∼1.2 | ∼3 | 480 | ∼ −16 | n.a. | 25 |
| NIR-GECO2 | 678/704 | 12 | 0.75 | 5.3, 4.8 | n.a. | ∼130 (5) | ∼2 (5) | n.a. | n.a. | 0.94 | ∼1.3 | ∼3.5 | 331 | ∼ −17 | n.a. | 25 |
| FR-GECO1a | 596/642 | 3.2 | 27 | 8.9, 8.0 | n.a. | n.a. | n.a. | n.a. | 6× | 1.5 | 0.008 | 0.3 | 29 | ∼ 10 | n.a. | 28 |
| FR-GECO1c | 596/646 | 1.1 | 28 | 9.5, 7.9 | n.a. | n.a. | n.a. | n.a. | 18× | 2.0 | 0.001 | 0.16 | 83 | ∼ 33 | n.a. | 28 |
| HaloCaMP1a | 640–642/653–655 | n.a. | 223 | n.a. | n.a. | n.a. | n.a. | n.a. | 5× | n.a. | n.a. | n.a. | 190 | 2 | n.a. | 23 |
| HaloCaMP1b | 640–642/653–655 | n.a. | 134 | n.a. | n.a. | n.a. | n.a. | n.a. | 9× | n.a. | n.a. | n.a. | 43 | 8 | n.a. | 23 |
| GCaM6s | 497/515 | ∼5 | 123 | 9.8, 6.0 | n.a. | n.a. | n.a. | n.a. | 60× | 2.9 | 0.48 | 1.8 | 144 | 28 | 23 | 32 |
Molecular brightness is determined by extinction coefficient multiplied by quantum yield.
Determined in27, using 605/30 nm excitation and 647 nm longpass emission filters at 14 mW/cm2 light power density at the back aperture of the lens, and normalized to absorption efficiency of the biosensors at 605 nm.
Determined in transiently transfected live HeLa cells. Effective (a.k.a. cellular) brightness of iGECI with 25 μM of exogenous BV was assumed to be 100%.
Determined for 1–10 electrical pulses in cultured neurons, without adding exogenous BV.
n.a., not available
Intensiometic indicators NIR-GECO2 and NIR-GECO2G were recently developed by applying directed molecular evolution to NIR-GECO125. NIR-GECO2 has higher dynamic range, but lower brightness (Table 1). NIR-GECO2G demonstrated both slightly increased brightness and dynamic range, compared to NIR-GECO1 (Table 1). NIR-GECO2G was found to have the highest dynamic range of response and brightness among NIR-GECO variants when expressed in acute brain slices25. Ineffective BV binding also limits a use of improved NIR-GECOs in living mammals. NIR-GECO1 was applied to imaging in Caenorhabditis elegans (with co-expressed heme oxygenase-1 (HO1) to increase the conversion of heme into BV) and in the olfactory bulb of Xenopus laevis tadpoles. All NIR-GECOs are characterized by relatively poor photostability (Table 1). This characteristic is practically important, since it limits the maximum acquisition rate of calcium recording. In wide-field imaging experiments, continuous illumination with 631/28 nm excitation light of 2.8 mW/mm2 (100 ms exposure time were used) resulted in 50% bleaching in neurons in 30 min25. Imaging at lower frequencies decreased photobleaching: at acquisition rate of 2 Hz with 100 ms exposure time (conditions recommended by the authors) the sensors retained 68–85% of fluorescence25.
Another type of intensiometric NIR indicator was developed from a small GAF-FP protein 34, 35. While this small sensor is an interesting achievement in GECI engineering, its very low brightness in mammalian cells and high dependence on BV prevented its use in neurons.
Recently developed NIR FRET based iGECI is the first NIR calcium indicator that demonstrated satisfactory performance in neuronal cultures, brain slices and in single-cell in vivo imaging27. This FRET indicator incorporates undisturbed optimized FPs miRFP67019 and miRFP72021 as the donor and the acceptor, correspondingly. Calcium binding causes the conformational change that translates into the FRET change between FPs (Fig. 1c). Directed molecular evolution was applied to optimize the linkers between the FPs and the calcium-sensing module. iGECI demonstrated the dynamic range of response comparable to that of NIR-GECO1 (Table 1). The effective brightness of iGECI was almost 20-fold higher than that of NIR-GECO1 without external BV and 5-fold higher with BV supply. iGECI was shown to be considerably more photostable than NIR-GECO1 in neurons. In neuronal cultures, iGECI demonstrated response amplitudes similar to those of NIR-GECO1 for small number of pulses. A limitation of iGECIs is its slow decay kinetics, whereas its rise kinetics is faster than that of NIR-GECO1 (Table 1).
Although iGECI is a ratiometric indicator, the fluorescence in the “FRET” channel (donor excitation, acceptor emission) changes only slightly, compared to the “donor” channel (donor excitation, donor emission). This effect is caused by relatively wide spectra of the miRFPs resulting in a compensation of the acceptor fluorescence increase by the donor fluorescence decrease in the “FRET” channel27. Therefore, iGECI can be used as an intensiometric inverse response (or negative response) indicator by monitoring changes in the “donor” channel only. Its relatively stable fluorescence in the “FRET” channel can be used for correction of motion- and laser-induced fluctuations27.
New monomeric far-red FPs mKelly1 and mKelly236 were used to design far-red GFP-like GECIs named FR-GECO1a and FR-GECO1c (excitation peaks at 596 nm, emission peaks at 642–646 nm) 28. These indicators are composed of circular permuted versions of mKelly2 that change their fluorescence upon the interaction between calmodulin and its binding ckkap peptide attached to the new N- and C-termini (Fig. 1e). This design is similar to that of GCaMP series of indicators: the conformational changes near the chromophore of a circular permuted FP result in the chromophore transition between the fluorescent anionic and the nonfluorescent protonated forms. FR-GECOs increase their fluorescence upon calcium binding (positive fluorescence response to calcium). These sensors have not been applied in vivo yet, although their functionality in cultured neurons was demonstrated. Both FR-GECOs showed relatively high responses to single action potentials, fast kinetics, and the ability to be imaged by widely used 633 nm laser. FR-GECOs were found to be bright under two-photon illumination at 1050–1200 nm that opens new possibilities for their applications. A limitation of FR-GECOs is their photoactivation by blue light, similar to red R-GECO series of indicators30, 31, 37, that makes them incompatible for spectral multiplexing.
Bright far-red chemigenetic calcium indicators were recently developed using JF635-HTL dye and a re-engineered circular permutant HaloTag named cpHaloTag23 (Fig. 1f). Janelia Fluor rhodamine analogues, including JF635, exist mostly in non-fluorescent lactone form in aqueous solutions. Upon binding to HaloTag, the dye switches to the zwitterionic state and its fluorescence increases more than 100-fold. Observation that the synthetic rhodamine dyes partially undergo a conformational change into the fluorescent ionic conformation when they bind to HaloTag, led to a development of new indicators that modulate conformational changes in the bound dye through conformational changes in the circular permuted cpHaloTag. The latter are induced by the interaction of calmodulin with its binding peptide attached to the new N- and C-termini in close proximity to the dye. These GECIs named HaloCaMP1a and 1b bound to JF635-HTL were validated in neurons in vitro and demonstrated high brightness and decent response (Table 1).
Applications in spectral multiplexing.
NIR GECIs can be combined with blue-light controlled optogenetic tools (OTs) without a crosstalk. While red GECIs based on mApple and mRuby FPs were also combined with channelrhodopsin OTs30, there may be a cross-talk since action spectra of channelrhodopsins are wide and illumination at 530–560 nm needed for red GECI imaging may result in detectable currents38. To avoid this, the intensity of the excitation light used for imaging of red indicators should be carefully selected31, 38. Also, mApple-based red GECIs were reported to be photoactivated by blue light leading to artifacts30.
Possibilities for spectral multiplexing were explored for all NIR GECIs. NIR-GECOs and iGECI were used to record neuronal activities induced by optogenetic actuators CheRiff or CoChR in crosstalk-free experiments25–27 (Fig. 1b). iGECI was used for detection of single spike responses induced by light-stimulated ChR2 in acute brain slices. NIR-GECO2G was tested for recording of optogenetically-induced calcium changes in iPSC-derived cardiomyocytes25. Further, NIR-GECO1 was applied to spectral multiplexing with red Pink Flamindo biosensor for cAMP and cyan-yellow FRET AKAR4 biosensor for protein kinase A (PKA) in MIN6 β-cells26. In this experiment, four-color (cyan, yellow, red, near-infrared) imaging allowed tracing of synchronous oscillations in calcium changes, cAMP, and PKA activities induced pharmacologically.
Applications in deep-tissue and in vivo imaging.
Mesoscale imaging in the mouse brain was performed with NIR-GECO1 and iGECI26, 27. For iGECI, mesoscale level imaging was combined with photoacoustic detection of blood oxygenation using a hybrid photoacoustic-fluorescence microscopy. Simultaneous detection of neuronal activity and local hemodynamics allowed visualization of neurovascular coupling in the brain.
As discussed above, poor binding of BV limits NIR-GECO1 applications in vivo. While BV supply and co-expression of heme oxygenase are not practical to increase BV levels in vivo, another successful approach was recently reported. It was shown that knock-out of biliverdin reductase A, the enzyme that converts BV to bilirubin, allows to increase performance of bacterial phytochrome derived FPs and NIR-GECO1 sensor39 The knock-out mice did not show gross abnormalities. However, they might have health issues, as it was found that bilirubin protects cells from oxidative stress40, 41. In neurons, low concentrations of bilirubin were shown to have neuroprotective effect by scavenging superoxide, whereas at high concentrations bilirubin is toxic41. I believe that BV concentration can be increased without affecting bilirubin levels and associated side effects by obtaining transgenic animals stably expressing BV binding biosensors. As BV is an intermediate of heme metabolism, its extra amount should be quickly absorbed by a BV binding protein without affecting the whole heme metabolism pathway.
While two-photon microscopy is a well-established technique for in vivo imaging of single cells at depths, single-photon single-objective light-sheet microscopy techniques, such as single objective-based scanned oblique plane illumination (SOPi)42 and Swept, Confocally-Aligned Planar Excitation (SCAPE) microscopy43 are emerging for imaging at high-speed and large-scale at depths. iGECI was imaged by single objective-based scanned oblique plane illumination (SOPi) in brain slices27. SOPi imaging of iGECI demonstrated that a functional response of this NIR indicator did not decline with depth, in contrast to GCaMP6s. Still, these technologies do not allow signal-to-noise (SNR, can be defined as the maximum fluorescence change during a response divided by the standard deviation of the baseline) high enough for in vivo imaging in mammals.
iGECI was applied to two-photon microscopy imaging of neuronal activity in the visual cortex of living mice at subcellular resolution27. Co-expressed GCaMP6s was imaged as an internal control of iGECI functionality. Both indicators were excited with 900 nm laser, since this wavelength was found to be optimal for their two-photon excitation. 900 nm two-photon excitation corresponds to the so called Soret band, which is a minor absorption peak at ~400 nm common to bacterial phytochrome-derived FPs44, 45. iGECI could detect visually-evoked neuronal activity in response to drifting grating visual stimuli. While recording spontaneous activity, correlated calcium transients from iGECI and GCaMP6s were observed, which reached up to 25% of dynamic range of response (Fig. 1d). Importantly, ΔF/F was similar at two depths (100 μm and 300 μm) for iGECI.
Genetically-encoded voltage sensors (GEVIs).
Design and performance.
Compared to GECIs capable of detection action potentials, GEVIs also allow imaging of depolarizing and hyperpolarizing subthreshold potentials underlying information processing in neurons. Voltage imaging is more challenging than calcium46, 47. While calcium changes occur at tens of milliseconds timescale, changes in electrical potential have sub-millisecond dynamics. Shorter acquisition time results in intrinsically lower signals to be detected. In addition, voltage indicators localized to the plasma membrane have lower total signal per neuron than calcium indicators localized in the cytoplasm. Therefore, signal-to-noise (SNR, can be defined as the maximum fluorescence change during a response divided by the standard deviation of the baseline) becomes critical in GEVI imaging, in addition to indicator’s dynamic range of response (sensitivity) and kinetics. SNR depends on the experimental conditions: the wavelength of excitation light, the light intensity, and the acquisition rate.
NIR and far-red GEVIs can be classified by their voltage sensing domains: GEVIs based on microbial rhodopsins, and GEVIs based on voltage sensing domains (VSD) from voltage sensitive phosphatases. Far-red GEVIs based on microbial rhodopsins either (i) use an intrinsic ability of microbial rhodopsin derived from Archaerhodopsin 3 (Arch) to emit light in response to voltage changes24, 48–51, or (ii) use so-called electrochromic FRET from the HaloTag-bound synthetic fluorogenic dye to the nearby covalently linked rhodopsin that changes its absorbance in response to voltage changes52. There are two types of far-red and NIR VSD-based GEVIs: (i) NIR FRET GEVI nirButterfly that uses miRFP670-miRFP720 FRET pair53, (ii) chemigenetic GEVIs that use modulation of fluorescence of a dye bound to the circular permuted HaloTag (“ASAP”-like HASAP1 and “ArcLight”-like HArcLight1 variants)23.
Microbial rhodopsin GEVIs are characterized by relatively fast kinetics of voltage response compatible with the kinetics of voltage changes in neurons (Fig. 2a). They are also characterized by the high dynamic range, although they suffer from the low brightness54 (Table 2). Therefore, their imaging requires high excitation light intensities to achieve SNR sufficient for detection of subthreshold voltage changes in vitro (usually >10-fold the noise level55). Most rhodopsin-based GEVIs have a mutation that abolishes light-driven currents48. Early rhodopsin-based GEVIs had low signal and poor membrane localization48, whereas each generation of modern GEVIs are becoming better in terms of their membrane localization and brightness. Rhodopsin-based GEVIs were found to perform best, compared to all other types of GEVIs, in in vitro conditions54. In contrast, their low brightness complicates their use in vivo, where light scattering and light from out-of-focus cells interfere.
Figure 2. Near-infrared and far-red GEVIs and their representative applications.
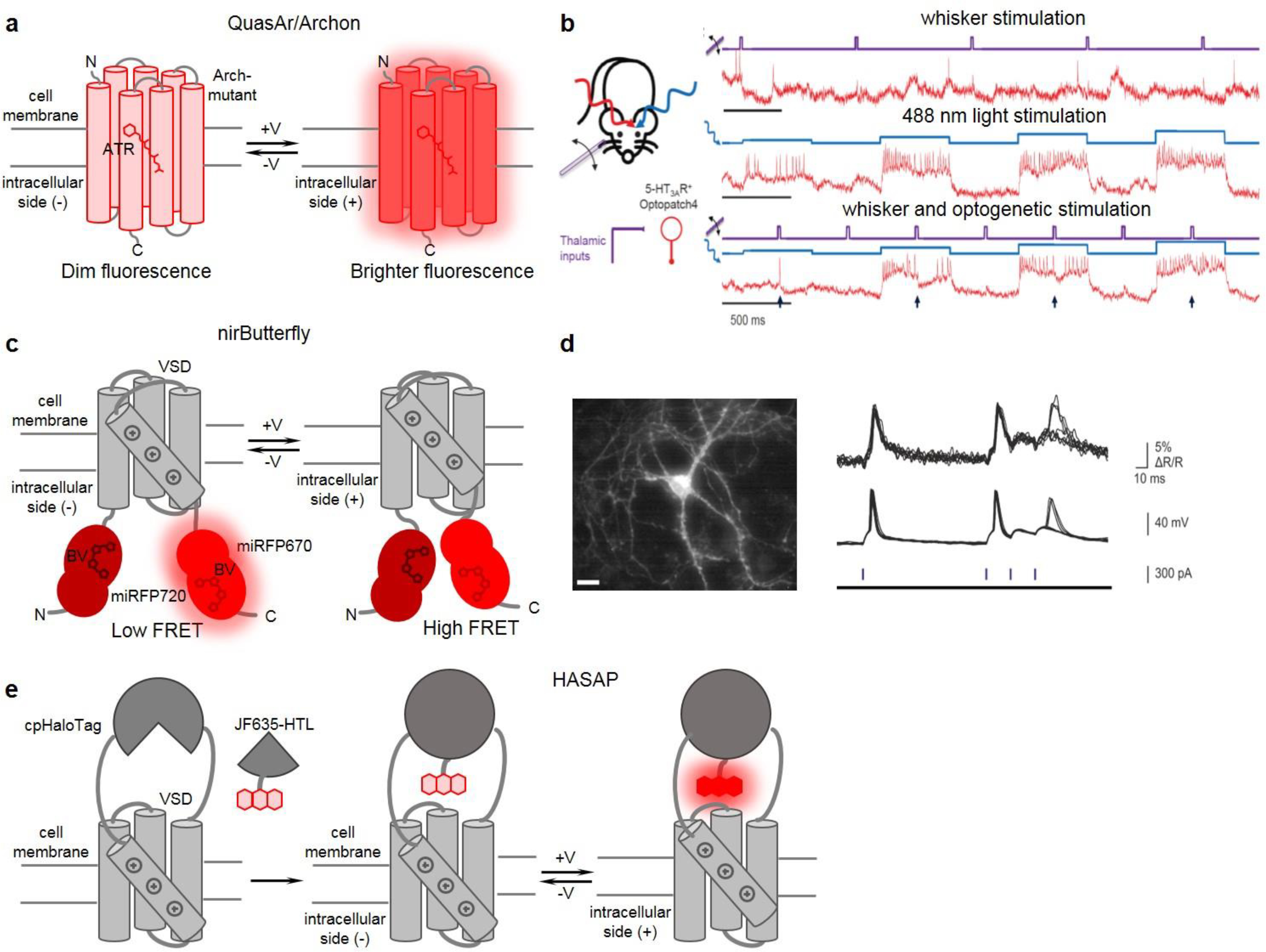
(a) Schematics of rhodopsin-based GEVIs of Archon/QuasAr series. (b) All-optical electrophysiology applied to optical dissection of neuronal excitation and inhibition in vivo. (left) Schematics of applied stimuli (whisker stimuli and single-cell optogenetic stimuli with 488 nm) and recording of SomArchon GEVI activity with 639 nm light. (right) Recordings from a single neuron showing SomArchon response to whisker stimuli (top), optogenetic stimuli (middle), and their combination (bottom). Whisker stimuli are in violet, optogenetic stimuli are in blue, and SomArchon GEVI responses are in red. Adapted, with permission, from57. (c) Schematics of nirButterfly FRET GEVIs. (d) nirButterfly in all-optical electrophysiology in vitro. (left) Fluorescence image of a neuron expressing nirButterfly. (right) nirButterfly response to CheRiff photocurrent-triggered action potentials and subthreshold depolarization. Optical signal (top), microelectrode recording (middle), pulses of 488 nm light (bottom). Adapted, with permission, from53. (e) Schematics of HASAP indicator as an example of a chemigenetic voltage indicator. (a, c, e) Arch-mutant: GEVI derived as Archaerhodopsin 3 mutant; ATR: all-trans retinal; BV: biliverdin; VSD: voltage sensing domains from voltage sensitive phosphatases; cpHaloTag: circular permutant HaloTag; JF635-HTL: synthetic rhodamine dye.
Table 2.
Characteristics and performance of far-red and NIR GEVIs.
| GEVI name | Ex/Em (nm) | Molecular brightness vs. EGFP, % (1) | Effective brightness vs. QuasAr2, % (2) | ΔF/F, % | SNR in in brain slice(3) | On kinetics, ms (4) | Off kinetics, ms (4) | Ref | ||
|---|---|---|---|---|---|---|---|---|---|---|
| per 100 mV | per 1 AP | In brain slice(3) | ||||||||
| nirButterfly | Donor 640/670 | 37(5) | n.a. | −9–16 | −4–8 | n.a. | n.a. | 2.2 (single exponential fit) | 2.1 (single exponential fit) | 53 |
| Acceptor 702/720 | ||||||||||
| QuasAr1 | 580–590/∼715 | ∼1.5(6) | ∼3–4 | n.a. | 21 | n.a. | n.a. | 0.05 (94)/3.2 | 0.07 (88)/1.9 | 49 |
| QuasAr2 | 580–590/∼715 | ∼0.75(6) | 1 | 39 | 48 | 15 | 8.5 (8 W/mm2at 1 kHz) | 0.9 (67)/11(8); 2.3 (half-time) (8) | 1.6 (76)/20(8); 0.6 (half-time) (8) | 49 |
| QuasAr3 | 580–590/∼715 | n.a. | n.a. | n.a. | n.a. | 14 | 6.4 (1.5 W/mm2 at 0.5 kHz) | 1.2 (77)/10 | 0.87 (91)/9 | 50 |
| paQuasAr3 | 580–590/∼715; 488 nm for activation | n.a. | n.a. | n.a. | n.a. | 23 | 28 (1.5 W/mm2 at 0.5 kHz) | 0.9 (57)/15 | 0.93 (79)/15 | 50 |
| Archer1(9) | 580–590/∼715 | n.a. | 1.03 | 34 (85 original) | 25–40 | n.a. | n.a. | 0.6 (68)/33(8) | 1.1 (77)/87(8) | 51 |
| Archon1 (Som-Archon) | 580–590/∼715 | n.a. | 2.78 | 43 | 30 | 26–53 | 20–37(1.5 W/mm2 at 0.5–1 kHz) | 0.6 (88)/8.1(8); 3.6 (half-time) (8) | 1.1 (88)/13(8); 1.7 (half-time) (8) | 24,56 |
| Archon2 | 580–590/∼715 | n.a. | 8.01 | 19 | 18 | n.a. | n.a. | 0.06 (70)/6.7(8) | 0.17 (92)/7.0(8) | 56 |
| Voltron635 | 640–642/653–655 | 134(7) | n.a. | −7 | −3 | n.a. | n.a. | n.a. | n.a. | 52 |
| HASAP | 640–642/653–655 | >100 | n.a. | n.a. | ∼4–6 | n.a. | n.a. | n.a. | n.a. | 23 |
| HArcLight | 640–642/653–655 | >100 | n.a. | n.a. | ∼3–4 | n.a. | n.a. | n.a. | n.a. | 23 |
Molecular brightness is determined by extinction coefficient multiplied by quantum yield.
Experimentally measured in56.
Experimentally measured in24 for soma-targeted GEVIs.
Kinetics evaluated by bi-exponential fitting (fast, slow), the value in parentheses represents the % of magnitude in the fast component, measured in HEK293 cells, unless otherwise noted.
Estimated based on the molecular brightness of the donor.
Estimated based on reported quantum yield and extinction coefficient of the bacterial rhodopsin48, 49.
Estimated based on the brightness of JF635 bound to HaloTag17.
Measured in neuronal cultures in56.
For Archer1, some photocurrents were observed in56.
n.a., not available
A use of directed molecular evolution with advanced screening strategies resulted in a recent development of the best performing rhodopsin GEVIs. paQuasAr3 GEVI derived from QuasAr2 is significantly brighter than its precursor but requires blue illumination for photoactivation to achieve that maximum brightness50 (Table 2). Archon1 GEVI was derived from QuasAr2 using a directed molecular evolution with robotic picking of cells expressing GEVI mutants with several enhanced properties (brightness, sensitivity, membrane localization)56. Selection for efficient membrane localization allowed to improve SNR by reducing background fluorescence caused by improperly trafficked molecules.
Addition of a soma-targeting sequence to rhodopsin-based GEVIs resulted in a substantial increase of SNR in vivo24, 50. Soma targeting allowed localization of signals from individual neurons in dense populations and reduction of contaminating signals from neuropil that includes neurites of adjacent cells and microvasculature. Soma-targeted paQuasAr3 and SomArchon are the first rhodopsin-based GEVIs that were successfully applied to recording of neuronal populations in vivo24, 50, 57.
Chemigenetic FRET-based GEVI Voltron uses FRET from the HaloTag-bound dye to the linked rhodopsin, whose absorbance changes in response to voltage (electrochromic effect, eFRET)52. Compared to rhodopsin-based GEVIs, eFRET-based GEVIs are substantially brighter but have lower dynamic range of response58. Unfortunately, absorbance spectra of current rhodopsins do not allow efficient FRET from far-red and NIR FPs and synthetic dyes. For example, Voltron chemigenetic GEVI works best with the yellow dye JF525 due to better spectral overlap. Voltron525 bound to JF525 produced more than 10% dynamic range of response in single-trial action potentials induced by current injections, while Voltron635 bound to JF635 demonstrated 3% changes in the same conditions52 (Table 2).
nirButterfly GEVI is based on FRET changes between the miRFP670 donor and the miRFP720 acceptor attached to the N- and C-termini of VSD53 (Fig. 2c). This sensor is characterized by relatively high brightness, although its dynamic range and kinetics yield to opsin-based GEVIs (Table 2). nirButterfly showed good performance in neuronal culture (Fig. 2d). It was not tested in vivo. Most possibly, its relatively low dynamic range of response is a limitation for in vivo applications.
Far-red chemigenetic voltage indicators were engineered using the circular-permuted HaloTag bound to the JF635-HTL dye, i.e. similar fluorescent module as in the chemigenetic calcium indicators HaloCaMP1a/1b23 (Fig. 2e). The sensors named HASAP1 and HArcLight1 demonstrated small but decent responses to spikes in neuronal culture (ΔF/F ~4–6% for HASAP1, ~3–4% for HArcLight1) (Table 2).
Applications in spectral multiplexing.
Far-red rhodopsin-based GEVIs were long envisioned as ideal tools for electrode-free all-optical electrophysiology49 that allows crosstalk-free control and readout of neuronal activity. All-optical electrophysiology was considered as a technique for streamline screening of electrophysiological properties of iPSC-derived neurons59. However, the power of this technique is the most striking in in vivo applications. Crosstalk-free combination of far-red and NIR GEVIs with spectrally matching opsin tools for optogenetic activation is suitable for noninvasive activation and recording of electric potentials of neurons with light at fast enough frequencies. This should allow testing various hypotheses of how specific genetically defined neuronal ensembles in the brain integrate and process input signals. Recent improved GEVIs, such as SomArchon, in combination with spectrally matching opsin tools, such as CheRiff, and advanced imaging techniques achieving high SNR made all-optical electrophysiology applicable to in vivo studies as described below.
Applications in deep-tissue and in vivo imaging.
Voltage imaging of neuronal populations in vivo can be used to test correlation and coherence of subthreshold neuronal activities. Single-photon wide field imaging of SomArchon was demonstrated24. High excitation light powers of 1.6–4 W/mm2 apparently did not induced overheating and allowed imaging at depths of 50–150 μm from the surface. A simultaneous monitoring of voltage changes in neuronal populations of ~10 neurons in several brain regions (cortex, hippocampus, striatum) was achieved in head-fixed living mice. This imaging was combined with the optogenetic control. By monitoring positive and negative responses induced by locomotion, bi-directional modulation of striatal neurons was detected. This technique was also used to find out how spikes relate to subthreshold theta-oscillations in individual neurons in hippocampus vs local field potential theta-oscillations. It was shown that neurons tend to be phase-locked to their own subthreshold theta-oscillations.
Although wide-field imaging was demonstrated, it is obviously challenging, since SNR is relatively low. Therefore, patterned illumination to the membrane region was introduced to enhance SNR50, 57. Two-photon structural imaging was used to support patterned single-photon excitation targeting individual cell bodies for imaging of far-red soma-targeted paQuasAr3, which also needs blue light for activation50. Although demonstrated in combination with CheRiff in all-optical electrophysiology, paQuasAr3 is suboptimal for this application. Light controlling CheRiff activated this GEVI and CheRiff activation can occur during blue light photoactivation of paQuasAr3. Population imaging of paQuasAr3 in vivo was used for detection of behavior-induced changes in neuronal excitability in the hippocampus. Optogenetic control enabled to distinguish between excitatory and inhibitory inputs to membrane potentials.
All-optical electrophysiology was developed further for in vivo use with SomArchon (Fig. 2b). Here patterned illumination was used to enhance SNR by reducing optical crosstalk from scattered and out-of-focus light57. Holographic patterned illumination for GEVI imaging and for opsin activation was restricted to cell membranes and reportedly significantly improved SNR of optically detected spikes. Further, the activity-based image segmentation was applied to remove residual out-of-focus crosstalk. The technique was applied to imaging of interneurons in superficial L1 neocortical layer. L1 consists of inhibitory neurons and was considered as a hub responsible for gating sensory inputs to the underlying cortex60. Optogenetic control revealed lateral inhibition within L1 and confirmed a hypothesis that L1 neuronal network functions as a high-pass filter for excitatory inputs. Although rhodopsin-based GEVIs have been applied to population voltage imaging in vivo, these GEVIs do not work in two-photon illumination regimen, that would allow imaging of neurons in L2/3 and deeper layers.
Summary and Perspectives.
To summarize, NIR and far-red calcium and voltage indicators combined with advanced imaging technologies opened up new possibilities for crosstalk free all-optical control and readout of neuronal activity and multiplexing with biosensors of the visible range. While, red-shifted opsins have recently become available61–63 for combination with green indicators in alternative all-optical setups, NIR indicators seem to be the most versatile choice, since they can be combined with a variety of well-studied blue-green optogenetic tools and visible biosensors. In vivo all-optical control and readout can be now used to directly test hypotheses of how neuronal circuits process information during behavior.
The NIR and far-red fluorescent modules used in GEVIs and GECIs can be adapted to development of NIR and far-red biosensors for neurotransmitters and neuromodulators 64. Such sensors would allow more possibilities for multiplexing: combination with green GCaMP6–732, 65 and red biosensors, such as red cpmApple-based dopamine biosensor66, for simultaneous monitoring of multiple neurotransmitters and neuronal activity, and combination with optogenetics. The most promising seems a use of cpHaloTag bound to JF635. This module works similar to cpEGFP and cpmApple modules in intensiometric biosensor and should be interchangeable in biosensors for glutamate67, GABA68, acetylcholine69, norepinephrine70, and dopamine66, 71.
Improved NIR biosensors can be developed using recently developed FPs, such as a small (2-fold smaller than miRFPs, 1.6-fold smaller than EGFP) and stable miRFP670nano72. This FP should be amenable to circular permutations as its N- and C-termini are located closely. The circular permuted miRFP670nano can be used to design new intensiometric indicators and ratiometric FRET indicators with better performance. For development of improved NIR GEVIs, it is possible that red-shifted proton-pumping rhodopsins can be found using structure-based genome mining63. Such red-shifted opsins would allow efficient eFRET from far-red, NIR FPs, and fluorogenic dyes like JF635.
Current indicators discussed in this review can be improved for better performance in vivo. Specifically, appending a soma targeting sequence to iGECI and obtaining a transgenic mouse stably expressing iGECI for upregulation of the endogenous BV level without affecting heme metabolism should increase SNR and create the best performing NIR GECI to date.
All-optical control and readout of neuronal activity are reaching a stage when they can be used to generate new data on how the brain works, beyond proof-of-principle demonstrations57, 63. While patterned illumination increased SNR in a single-photon GEVI imaging, this technique is limited in imaging depth (below ~230 μm with a reasonable density of labeled neurons)57. Approaches reducing light scattering like adaptive optics73, graded index lens endoscopes74 and prism-based imaging75 should allow deeper single-cell single-photon imaging of GEVIs and GECIs. For imaging and light control of single cells deeper in the brain, two-photon imaging and optogenetics are promising63, 76. While the current rhodopsin-based GEVIs do not respond to voltage under two-photon illumination77, green GFP-based GEVI ASAP3, which was also produced using advanced molecular evolution, was successfully used in fast two-photon in vivo imaging78. NIR iGECI already was shown to be functional in two-photon in vivo imaging. However, a brighter version that produces enough signal at ~1200 nm two-photon excitation peak of (m)iRFPs45 is required to be paired with optogenetics in two-photon setup. For GEVI engineering, it is challenging to propose a strategy for a search of rhodopsins that change fluorescence in response to two-photon excitation in genome databases. However, these rhodopsins may exist, since it was possible to find opsins that produce large currents in two-photon optogenetics63, 79.
Highlights.
Near-infrared shifted indicators are well suited for spectral multiplexing.
NIR calcium indicator iGECI demonstrated satisfactory in vivo performance.
SomArchon voltage indicator allowed all-optical electrophysiology in vivo.
Patterned illumination is used in voltage in vivo imaging for best results.
Chemigenetic indicators based on cpHaloTag are promising bright probes.
Acknowledgments
I thank Konstantin Rumyantsev (Moscow Clinical Scientific Center, Moscow, Russia) for help with the figure drafts and Mikhail Baloban (Albert Einstein College of Medicine, USA) for useful discussions. This work was supported by the grants EY030705 and GM122567.
Footnotes
Competing interests
The author declares no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lin MZ & Schnitzer MJ Genetically encoded indicators of neuronal activity. Nat Neurosci 19, 1142–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W, Kim CK & Ting AY Molecular tools for imaging and recording neuronal activity. Nat Chem Biol 15, 101–110 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Luo L, Callaway EM & Svoboda K Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 98, 865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissleder R A clearer vision for in vivo imaging. Nat Biotechnol 19, 316–317 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Shcherbakova DM, Baloban M & Verkhusha VV Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr Opin Chem Biol 27, 52–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luker KE et al. Comparative study reveals better far-red fluorescent protein for whole body imaging. Sci Rep 5, 10332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shcherbakova DM & Verkhusha VV Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods 10, 751–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frangioni JV In vivo near-infrared fluorescence imaging. Curr Opin Chem Biol 7, 626–634 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Tyssowski KM & Gray JM Blue Light Increases Neuronal Activity-Regulated Gene Expression in the Absence of Optogenetic Proteins. eNeuro 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shcherbakova DM, Stepanenko OV, Turoverov KK & Verkhusha VV Near-Infrared Fluorescent Proteins: Multiplexing and Optogenetics across Scales. Trends Biotechnol 36, 1230–1243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fyk-Kolodziej B, Hellmer CB & Ichinose T Marking cells with infrared fluorescent proteins to preserve photoresponsiveness in the retina. Biotechniques 57, 245–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu J et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods 11, 572–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morozova KS et al. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J 99, L13–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoelzel CA & Zhang X Visualizing and Manipulating Biological Processes by Using HaloTag and SNAP-Tag Technologies. Chembiochem 21, 1935–1946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 12, 244–250, 243 p following 250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm JB et al. A general method to optimize and functionalize red-shifted rhodamine dyes. Nat Methods 17, 815–821 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm JB et al. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods 14, 987–994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm JB, Brown TA, English BP, Lionnet T & Lavis LD Synthesis of Janelia Fluor HaloTag and SNAP-Tag Ligands and Their Use in Cellular Imaging Experiments. Methods Mol Biol 1663, 179–188 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Shcherbakova DM et al. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun 7, 12405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matlashov ME et al. A set of monomeric near-infrared fluorescent proteins for multicolor imaging across scales. Nat Commun 11, 239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shcherbakova DM, Cox Cammer N, Huisman TM, Verkhusha VV & Hodgson L Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET. Nat Chem Biol 14, 591–600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Methods 12, 763–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deo C et al. The HaloTag as a general scaffold for far-red tunable chemigenetic indicators. Nat chemical biology 17, 718–723 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Piatkevich KD et al. Population imaging of neural activity in awake behaving mice. Nature 574, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian Y et al. Improved genetically encoded near-infrared fluorescent calcium ion indicators for in vivo imaging. PLoS Biol 18, e3000965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian Y et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat Methods 16, 171–174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shemetov AA et al. A near-infrared genetically encoded calcium indicator for in vivo imaging. Nat Biotechnol 39, 368–377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalangin R et al. Far-red fluorescent genetically encoded calcium ion indicators. bioRxiv, 2020.2011.2012.380089 (2020). [Google Scholar]
- 29.Shemetov AA, Oliinyk OS & Verkhusha VV How to Increase Brightness of Near-Infrared Fluorescent Proteins in Mammalian Cells. Cell chemical biology 24, 758–766 e753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dana H et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akerboom J et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in molecular neuroscience 6, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen TW et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian L et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6, 875–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subach OM, Barykina NV, Anokhin KV, Piatkevich KD & Subach FV Near-Infrared Genetically Encoded Positive Calcium Indicator Based on GAF-FP Bacterial Phytochrome. Int J Mol Sci 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rumyantsev KA et al. Minimal domain of bacterial phytochrome required for chromophore binding and fluorescence. Sci Rep 5, 18348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wannier TM et al. Monomerization of far-red fluorescent proteins. Proceedings of the National Academy of Sciences 115, E11294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J et al. Improved orange and red Ca(2)+/− indicators and photophysical considerations for optogenetic applications. ACS chemical neuroscience 4, 963–972 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klapoetke NC et al. Independent optical excitation of distinct neural populations. Nat Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobachi K et al. Biliverdin Reductase-A Deficiency Brighten and Sensitize Biliverdin-binding Chromoproteins. Cell Struct Funct 45, 131–141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W et al. Absence of the biliverdin reductase-a gene is associated with increased endogenous oxidative stress. Free Radic Biol Med 115, 156–165 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Vasavda C et al. Bilirubin Links Heme Metabolism to Neuroprotection by Scavenging Superoxide. Cell chemical biology 26, 1450–1460.e1457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar M, Kishore S, Nasenbeny J, McLean DL & Kozorovitskiy Y Integrated one- and two-photon scanned oblique plane illumination (SOPi) microscopy for rapid volumetric imaging. Optics Express 26, 13027–13041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouchard MB et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat Photonics 9, 113–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shcherbakova DM et al. Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins. Chem Biol 22, 1540–1551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piatkevich KD et al. Near-Infrared Fluorescent Proteins Engineered from Bacterial Phytochromes in Neuroimaging. Biophys J 113, 2299–2309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piatkevich KD, Murdock MH & Subach FV Advances in Engineering and Application of Optogenetic Indicators for Neuroscience. Applied Sciences 9, 562 (2019). [Google Scholar]
- 47.Knöpfel T & Song C Optical voltage imaging in neurons: moving from technology development to practical tool. Nat Rev Neurosci 20, 719–727 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D & Cohen AE Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods 9, 90–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hochbaum DR et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods 11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adam Y et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flytzanis NC et al. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat Commun 5, 4894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdelfattah AS et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Monakhov MV et al. Screening and Cellular Characterization of Genetically Encoded Voltage Indicators Based on Near-Infrared Fluorescent Proteins. ACS chemical neuroscience 11, 3523–3531 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Bando Y, Sakamoto M, Kim S, Ayzenshtat I & Yuste R Comparative Evaluation of Genetically Encoded Voltage Indicators. Cell Rep 26, 802–813 e804 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong Y The evolving capabilities of rhodopsin-based genetically encoded voltage indicators. Curr Opin Chem Biol 27, 84–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piatkevich KD et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat Chem Biol 14, 352–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan LZ et al. All-Optical Electrophysiology Reveals the Role of Lateral Inhibition in Sensory Processing in Cortical Layer 1. Cell 180, 521–535 e518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong Y et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiskinis E et al. All-Optical Electrophysiology for High-Throughput Functional Characterization of a Human iPSC-Derived Motor Neuron Model of ALS. Stem cell reports 10, 1991–2004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang X, Wang G, Lee AJ, Stornetta RL & Zhu JJ The organization of two new cortical interneuronal circuits. Nat Neurosci 16, 210–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broser M et al. NeoR, a near-infrared absorbing rhodopsin. Nat Communications 11, 5682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Govorunova EG et al. RubyACRs, nonalgal anion channelrhodopsins with highly red-shifted absorption. Proceedings of the National Academy of Sciences 117, 22833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshel JH et al. Cortical layer–specific critical dynamics triggering perception. Science 365, eaaw5202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leopold AV, Shcherbakova DM & Verkhusha VV Fluorescent Biosensors for Neurotransmission and Neuromodulation: Engineering and Applications. Frontiers in cellular neuroscience 13, 474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dana H et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16, 649–657 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Patriarchi T et al. An expanded palette of dopamine sensors for multiplex imaging in vivo. Nat Methods 17, 1147–1155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marvin JS et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods 10, 162–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marvin JS et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat Methods 16, 763–770 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Jing M et al. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat Biotechnol 36, 726–737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng J et al. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 102, 745–761 e748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun F et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174, 481–496 e419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oliinyk OS, Shemetov AA, Pletnev S, Shcherbakova DM & Verkhusha VV Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nat Commun 10, 279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji N Adaptive optical fluorescence microscopy. Nat Methods 14, 374–380 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Jung JC, Mehta AD, Aksay E, Stepnoski R & Schnitzer MJ In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J Neurophysiol 92, 3121–3133 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andermann ML et al. Chronic cellular imaging of entire cortical columns in awake mice using microprisms. Neuron 80, 900–913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Forli A et al. Two-Photon Bidirectional Control and Imaging of Neuronal Excitability with High Spatial Resolution In Vivo. Cell Rep 22, 3087–3098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brinks D, Klein AJ & Cohen AE Two-Photon Lifetime Imaging of Voltage Indicating Proteins as a Probe of Absolute Membrane Voltage. Biophys J 109, 914–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Villette V et al. Ultrafast Two-Photon Imaging of a High-Gain Voltage Indicator in Awake Behaving Mice. Cell 179, 1590–1608 e1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prakash R et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods 9, 1171–1179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]


