Abstract
Background and aims
SARS-CoV-2 antibody assays are relevant in managing the COVID-19 pandemic, providing valuable data on the immunization status of the population. However, current serology tests are highly variable, due to their different characteristics and to the lack of reference materials. The aim of the World Health Organization (WHO) first International Standard (IS) for anti-SARS-CoV-2 immunoglobulin is to harmonize humoral immune response assessment after natural infection or vaccination, and recommend reporting the results for binding activity in Binding Antibody Units (BAU).
Materials and methods
This study analyzed six commercial quantitative anti-SARS-CoV-2 S-protein assays in a head-to-head comparison, using the manufacturers' conversion factors for the WHO IS to obtain BAU/mL values.
Results
Our data showed good alignment up to 1000 BAU/mL, then began to disperse, exhibiting some discrepancies. Moreover, correlations among methods varied with Cohen’s Kappa ranging from 0.580 to 1.00, with the lowest agreement values for kits using different target antigens or different antibody isotypes, making it clear that the laboratory report should include this information. Values expressed as BAU/ml showed a reduced between-assays variability compared to AU/ml (median coefficients of variation 0.38 and 0.68, respectively; p < 0.001).
Conclusion
On the basis of these data at present anti-SARS CoV-2 serological assays’ results are not interchangeable, and, more importantly, individual immune monitoring should be performed with the same method.
Keywords: COVID-19 Serological Testing, anti-SARS-CoV-2 antibodies, WHO international standard, Harmonization, Serological tests, COVID-19
1. Introduction
SARS-CoV-2 antibody assays have been and continue to be essential in managing the COVID-19 pandemic [1], [2], [3].
Since the beginning, the US Food and Drug Administration (FDA) has issued Emergency Use Authorizations (EUA) for hundreds of serological assays to support COVID-19 diagnosis [4].
As of 12 April 2021, there are at least 655 immunoassays for antibodies that are either commercially available or in development, according to the Foundation for Innovative New Diagnostics (FIND) (https://www.finddx.org/covid-19/pipeline/)[5], a World Health Organization (WHO) collaborating center for Laboratory Strengthening and Diagnostic Technology Evaluation. Simultaneously, in September 2020 the WHO issued the target product profiles for COVID-19 diagnostics, indicating 95–97% and 98–99% as acceptable criteria for sensitivity and specificity, respectively [6].
Nowadays, antibody testing may provide valuable data on: 1) surveillance and epidemiological assessment to evaluate the immunization status of the population [7], [8]; 2) diagnosis of suspected cases with negative viral RNA test to increased diagnostic sensitivity for COVID‐19 patients [9], [10], [11]; 3) selection of hyperimmune plasma for convalescent plasma therapy [12], [13]; 4) therapeutic antibody development and evaluation [14], [15]; 5) assessment of COVID-19 vaccines immune responses and durability [16], [17]. Regarding vaccine application, Gudbjartsson et al. highlighted the utility of antibody assays as a highly cost-effective tool for a vaccination strategy to promote public health [18]. Although this new role has not yet been fully validated scientifically, as we do not know how long persisting antibodies confer protection, it is increasingly clear that antibody assays are needed to monitor and check responses to vaccines to support the interpretation of the clinical outcomes.
For this purpose, standardized anti-SARS-CoV-2 quantitative and neutralizing assays are pivotal in assessing immune responses to vaccines [19], and it will become essential if a correlation of protection level could be identified with an antibody titre.
Among the serological assays, those targeting the spike protein, against the Receptor Binding Domain (RBD), the Subunit 1 (S1) or the full Spike (S), have played an important role in vaccine campaigns. However, current anti-S protein assays are highly variable in clinical practice [20] according to the different neutralizing activities, possibly due to a lack of reference materials. The Centers for Disease Control and Prevention (CDC) recently highlighted the need for standardized SARS-CoV-2 quantitative IgG and neutralization assays [21]. Improvements in antibody assay comparability are anticipated with the recent introduction of the WHO first International Standard (IS) for anti-SARS-CoV-2 immunoglobulin (NIBSC code 20/136), with the aim of harmonizing immune response assessment after natural infection or vaccination [22].
The WHO IS is a pool of eleven human plasma from convalescent patients and was established in December 2020 by the WHO Expert Committee on Biological Standardization with an arbitrary assigned unitage of 1000 International Unit per mL (IU/mL) for neutralization activity [23], [24]; the role of the IS in binding antibody assays was discussed. Not enough clinical data were available to assign an International Unit for binding activity and it was suggested to report the results in Binding Antibody Units (BAU) to assist in comparing assays that detect the same class of immunoglobulins with the same specificity (e.g., anti-receptor-binding domain IgG, anti-N IgM, etc).
In the present work, we aimed to compare six commercial quantitative anti-S protein serological assays, all with the manufacturers' conversion factor for the WHO IS for anti-SARS-CoV-2 immunoglobulin, in a head-to-head comparison to verify in “real life” whether the WHO IS effectively harmonizes laboratory testing.
2. Materials and methods
Eighty-eight consecutive serum samples (F:M ratio 1.4:1, mean age = 57.0 ± 14.0 years) screened for anti-S protein antibodies at the Clinical Chemistry Laboratory of “Tor Vergata” University Hospital (Rome, Italy), were tested using six different commercial kits widely used worldwide for anti-SARS-CoV-2 IgG or total antibodies (tAb) detection. Our study cohort reasonably consisted of a mixed population (vaccinated subjects, recovered patients from COVID-19, and healthy individuals) and no longitudinal samples were included. All assays were performed according to the manufacturers’ cutoff and instructions.
The characteristics of the different serological tests evaluated are listed in Table 1 , including detailed information about targeted antigens. The linear dynamic ranges have been extended conforming to the manufacturers’instructions.
Table 1.
Characteristics of anti-SARS-CoV-2 serological assays: target antigen, method, immunoglobulin class detected, cut-off value in arbitrary units (AU/mL), BAU conversion factor, cut-off value in BAU/mL and dynamic range.
| MANUFACTURER | KIT ASSAY | TARGET ANTIGEN | METHOD | IMMUNOGLOBULIN DETECTED | CUT-OFF (AU/mL) | CONVERSION FACTOR (BAU/mL) |
CUT-OFF (BAU/mL) | DYNAMIC RANGE* (BAU/mL) |
|---|---|---|---|---|---|---|---|---|
| Mindray |
SARS-CoV-2 S-RBD IgG |
S-RBD | CLIA | IgG | 10 | 1.216 | 12.16 | 3.65–1216 |
| Roche | Elecsys | S-RBD | ECLIA | IgA, IgM, IgG | 0.80 | 1.288 | 0.823 | 0.40–243 |
| anti-SARS-CoV-2 S | ||||||||
| Snibe | Maglumi | S-RBD | CLIA | IgG | 1 | 4.33 | 4.33 | 0.78–433 |
|
SARS-CoV-2 S-RBD IgG | ||||||||
| DiaSorin |
LIAISON SARS CoV-2 |
Trimeric S | CLIA | IgG | 13 | 2.6 | 33.8 | 4.8–2080 |
| Trimeric S IgG | ||||||||
| Thermo Fisher | EliA SARS-CoV-2-Sp1 IgG | S1 | FEIA | IgG | 7 (7–10 borderline) |
4 | 28 (28–40 borderline) |
2.8–816 |
| Euroimmun | Anti-SARS-CoV-2 | S1 | ELISA | IgG | 8 (8–11 borderline) |
3.2 | 25.6 (25.6–35.2 borderline) |
3.2–384 |
| QuantiVac ELISA IgG |
CLIA, chemiluminescence immunoassay; ECLIA, electrochemiluminescence immunoassay; ELISA, enzyme-linked immunosorbent assay; FEIA, fluorescence enzyme immunoassay.
The study was performed according to local ethical approval protocol no. R.S.44.20. Informed consent was obtained from all subjects enrolled in the study. The study was in accordance with the Helsinki Declaration, as revised in 2013.
3. Maglumi SARS-CoV-2 S-RBD IgG
The Maglumi SARS-CoV-2 S-RBD IgG (Snibe S-RBD IgG) is an indirect chemiluminescence immunoassay (CLIA) for the in vitro quantitative determination of IgG antibodies to SARS-CoV-2 S-RBD protein, performed by fully automated Maglumi 800 analytical system (Snibe Diagnostic, Shenzhen, China). The samples, buffer solution and magnetic beads coated with S-RBD recombinant antigen are mixed thoroughly. After settling in a magnetic field, the supernatant is decanted and a wash cycle is performed. Then, anti-human IgG antibodies labelled with amino-butyl-ethyl-isoluminol (ABEI) are added and incubated to form immune-complexes. After a second precipitation in a magnetic field, the supernatant is decanted and another wash cycle is performed. A starter reagent is added to initiate a chemiluminescent reaction, producing a light signal measured by a photomultiplier as Relative Light Units (RLUs) which is proportional to the concentration of anti-SARS-CoV-2 S-RBD IgG present in the sample.
The cut-off value in arbitrary units (AU)/mL, the conversion factor to obtain BAU/mL, the cut-off value in BAU/mL and the linearity range in AU/mL are respectively: 1, 4.33, 4.33 and 0.18–100, as declared by the manufacturer.
Samples with values over 100 AU/mL (433 BAU/mL) were diluted and measured 1:10 or 1:20 (if necessary), allowing extension of the dynamic range of analysis to 2000 AU/mL (8660 BAU/mL).
The manufacturer states intra- and inter- assay precision between 1% and 4%, with a clinical specificity of 99,6% (95% CI: 98,7%–100%) and a cumulative sensitivity of 100% (95% CI: 99,9%–100%), calculated at 15 days or more after the first positive PCR.
4. Mindray SARS-CoV-2 S-RBD IgG
The Mindray SARS-CoV-2 S-RBD IgG (Mindray S-RBD IgG) is a two-step CLIA for quantitative determination of SARS-CoV-2 S-RBD IgG in human serum or plasma, performed on the fully automated Mindray CL 1200i analytical system (Mindray Bio-Medical Electronic Co Ltd, Shenzhen, China). In the first step, sample, sample treatment solution and paramagnetic microparticles coated with SARS-CoV-2 S-RBD antigen are added to a reaction vessel. After incubation, SARS-CoV-2 S-RBD IgG antibodies present in the sample bind to S-RBD coated on microparticles. Afterwards, microparticles are magnetically captured while other unbound substances are removed by washing. In the second step, diluent solution and alkaline phosphatase (ALP) labeled anti-human IgG monoclonal antibodies are added to the reaction vessel to form a structure with the microparticles captured S-RBD IgG antibodies. Then, a substrate solution is added with a resulting chemiluminescent reaction measured as RLUs by a photomultiplier. The amount of anti-S-RBD IgG antibodies present in the sample is proportional to the RLUs generated during the reaction. The cut-off value in AU/mL, the conversion factor to obtain BAU/mL, the cut-off value in BAU/mL and the linearity range in AU/mL are respectively: 10, 0.8229, 8.229, 3.0–1000, as declared by the manufacturer. Samples with values over 1000 AU/mL (822.9 BAU/mL) were diluted and measured 1:10, allowing extension of the dynamic range of analysis to 10,000 AU/mL (8229 BAU/mL). The manufacturer states intra- and inter-assay precision between 1.7% and 4.08%, with a clinical specificity of 99.6% (95% CI: 99.3%–99.8%) and a cumulative sensitivity of 99.6% (95% CI 98.9%-99.8%), calculated at 14 days or more, after the first positive PCR.
5. Elecsys Anti-SARS-CoV-2 S
The Elecsys anti-SARS-CoV-2 S (Roche S-RBD tAb) is an electrochemiluminescence immunoassay (ECLIA) for the in vitro quantitative determination of total antibodies (IgG/IgA/IgM) to the SARS-CoV-2 S-RBD protein in human serum and plasma, performed by fully automated Roche Cobas E602 analyzer (Roche Diagnostics GmbH, Mannheim, Germany). The assay is a three-step test process that uses a recombinant protein representing the RBD of the S antigen in a double-antigen assay format, which favours detection of high affinity antibodies against SARS-CoV-2. Patient samples are incubated with a mix of biotinylated and ruthenylated RBD antigens to form double antigen immune-complexes. After the addition of streptavidin-coated microparticles, DAGS complexes bind to the solid phase. The reagent mixture is then transferred to the measuring cell, where the microparticles are magnetically captured. Electrochemiluminescence is induced by applying a voltage and measured with a photomultiplier. The signal yield increases with the antibody titre.The cut-off value in arbitrary units (AU/mL), the conversion factor to obtain BAU/mL, the cut-off value in BAU/mL and the linearity range in AU/mL are respectively: 0.8, 1.0288, 0.823 and 0.40–250, as declared by the manufacturer.
Samples with values over 250 AU/mL (257.2 BAU/mL) were diluted and measured 1:50, allowing the extension of the dynamic range of analysis to 12,550 AU/mL (12911 BAU/mL). The manufacturer states intra- and inter-assay precision between 1.4% and 2.4%, with a clinical specificity of 99.9% (95% CI: 99.9%–100%) and a cumulative sensitivity of 98,8% (95% CI: 98.1%–99.3%), calculated after 14 days or more from symptoms onset.
6. EliA SARS-CoV-2-Sp1 IgG
The EliA SARS-CoV-2-Sp1 IgG (Thermo Fisher S1 IgG) is a fluoroenzyme-immunoassay (FEIA) for the in vitro quantitative measurement of IgG antibodies in human serum and plasma directed to the SARS-CoV-2 spike 1 protein, performed on the Phadia 250 instrument (Thermo Fisher, Uppsala, Sweden).
The EliA SARS-CoV-2-Sp1 IgG wells are coated with recombinant SARS-CoV-2 S1 protein.
After washing non-bound antibodies, enzyme-labelled anti-human IgG are added to form antibody-conjugate complexes incubated with a development solution. After stopping the reaction, the fluorescence in the reaction mixture is measured.
The cut-off value in AU/mL, the conversion factor to obtain BAU/mL, the cut-off value in BAU/mL and the linearity range in AU/mL are respectively: 7.0, 4.0, 28 and 0.7–204, as declared by the manufacturer.
Samples with values above 204 AU/mL (816 BAU/mL) were diluted and measured 1:2 allowing the extension of the dynamic range up to 408 U/mL (1632 BAU/mL).
According to the manufacturer, intra- and inter-assay precision data ranges between 2.1% and 3.1%; clinical specificity is 99.7% (95% CI: 98.4%-100%) and clinical sensitivity is 100% (95% CI: 99.5%-100%), calculated at 8 days or more, after the first positive PCR.
7. LIAISON SARS-CoV-2 Trimeric S IgG
The LIAISON SARS-CoV-2 Trimeric S IgG (Diasorin TrimericS IgG) is a CLIA for the detection of IgG antibodies to SARS-CoV-2 in human serum and plasma samples. The principal components of the test are paramagnetic particles (solid phase) coated with recombinant trimeric SARS-CoV-2 S protein and a conjugate reagent containing an anti-human IgG mouse monoclonal antibody linked to an isoluminol derivative (isoluminol-antibody conjugate). During the first incubation SARS-CoV-2 IgG antibodies bind to the solid phase. Unbound material is then removed with a wash cycle. Subsequently, the starter reagents are added, and a flash chemiluminescence reaction is induced. The light signal is measured by a photomultiplier as RLUs and is proportional to the amount of antibodies to SARS CoV-2 present. The assay is performed by the LIAISON analyzer (DiaSorin, Stillwater, USA).
The cut-off value in AU/mL, the conversion factor to obtain BAU/mL, the cut-off value in BAU/mL and the linearity range in AU/mL are respectively 13.0, 2.6, 33.8, 1.84–800, as declared by the manufacturer.
Samples with values above 800 AU/mL (2080 BAU/mL) were not allowed to be diluted at the date of the experiment (March 20,2021).
According to the manufacturer, intra- and inter-assay precision ranges between 3.6% and 9%; clinical specificity is 99.5% (95% CI: 99.0%–99.7%) and clinical sensitivity is 98.7% (95% CI: 94.5%–99.6%), calculated at 15 days or more, after the first positive PCR.
8. Anti-SARS-CoV-2 QuantiVac ELISA IgG
The anti-SARS-CoV-2 QuantiVac enzyme immunosorbent assay (ELISA) IgG (Euroimmun S1 IgG) is a commercially available quantitative immunoassay test system (Euroimmun, Lübeck, Germany). The QuantiVac ELISA detects IgG antibodies from serum, plasma, or dried blood spot (DBS) using the S1 domain of the recombinant SARS-CoV-2 spike protein expressed in the HEK 293 human cell line. In the first step, diluted samples are incubated in the wells coated with S1. The specific IgG antibodies bind to the S1 antigens and are detected after a second incubation step, performed using an enzyme-labelled anti-human IgG to catalyze a color reaction. The absorbancies are measured with a standard microplate reader at a wavelength of 450 nm. A standard curve obtained by the calibrators optical densities is used to assess the concentration of antibodies in the samples.
The cut-off value in AU/mL, the conversion factor to obtain BAU/mL, the cut-off value in BAU/mL and the linearity range in AU/mL are respectively 8.0, 3.2, 25.6, 6–120, as declared by the manufacturer.
Samples with values above 120 AU/mL (384 BAU/mL) were diluted and measured 1:10 allowing the extension of the dynamic range up to 1200 AU/mL (3840 BAU/mL).
According to the manufacturer, intra- and inter-assay precision ranges between 6.3% and 9.1%; clinical specificity is 99.8% and clinical sensitivity is 90.3%, calculated at 21 days or more, after symptoms onset.
8.1. Statistical analysis
Analyses were performed using MedCalc 19.2.0 (MedCalc Software Ltd, Ostend, Belgium). The level of agreement between the two assays was evaluated by Cohen’s kappa coefficient (κ), assuming a substantial agreement with a κ value of: 0.01 – 0.20 slight agreement; 0.21 – 0.40 fair agreement; 0.41 – 0.60 moderate agreement; 0.61 – 0.80 substantial agreement; 0.81 – 1.00 almost perfect or perfect agreement. The Pearson correlation coefficient was used to compare the six commercial quantitative anti-S protein serological assays. In addition, to assess variability, between-assays coefficients of variation (CV) were calculated and compared between kit’s arbitrary unit (AU)/mL and BAU/mL using the Wilkoxon Signed Rank test. CV calculation and correlation analyses were performed only on values comprised inside the linearity range of the method (46 samples included). Finally an intraclass correlation coefficient (ICC) was used to compare results < 1000 and greater than 1000 BAU of the different assays. ICC values < 0.5 are indicative of poor reliability, values between 0.5 and 0.75 indicate moderate reliability, values between 0.75 and 0.9 indicate good reliability, and values greater than 0.90 indicate excellent reliabilityIn all analyses, a p-value of < 0.05 was considered as statistically significant.
9. Results
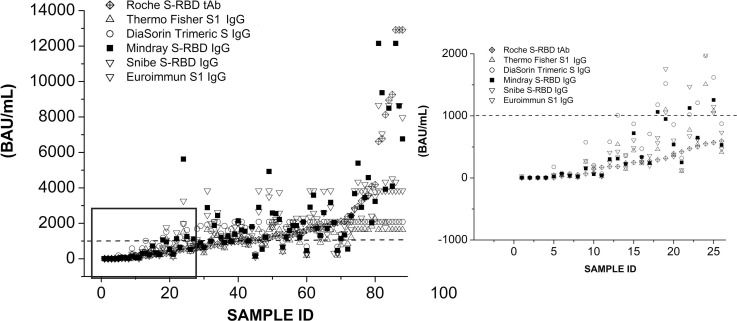
In our study, all samples have been tested with six commercial anti-SARS-CoV-2 S protein antibody assays and values were compared, expressing the results in both kit’s arbitrary unit (AU)/mL and BAU/mL (Supplemental Data Table S1). Table 1 illustrates the main features of each serological assay related to its target antigen, method, immunoglobulin class detected, cut-off (AU/mL), conversion factor, cut-off (BAU/mL) and dynamic range, according to the manufacturer’s instructions. In Fig. 1 all data, detected by the serological assays in BAU/mL, were gradually sorted from the lowest to the highest value and have been shown as scatter-plot. Since Roche method has the highest linearity range (0–12911 BAU/mL), and is able to detect total antibodies to the SARS-CoV-2 S-RBD protein, all values for each data set have been analyzed following the Roche results trend. Data have the best linear fit for values under 1000 BAU/mL and they scattered over 1000 BAU/mL showing some discrepancies. In fact, the enlargement of Fig. 1 shows a good alignment up to 1000 BAU/mL (ICC = 0.71) which then begins to disperse, becoming extremely evident over 1000 BAU/mL (ICC = 0.48). Moreover, it should also be considered that some serological assays reached a plateau, represented by the upper value of their linearity range allowed.
Fig. 1.
Scatter-plot for each serological assay in BAU/mL. Data were sorted from the lowest to the highest value. Dotted line represents the value of 1000 BAU/mL. On the right an enlargement of the figure shows a good alignment up to about 1000 BAU/mL. Samples who have reached a plateau, represented by their maximum linearity range, have been excluded.
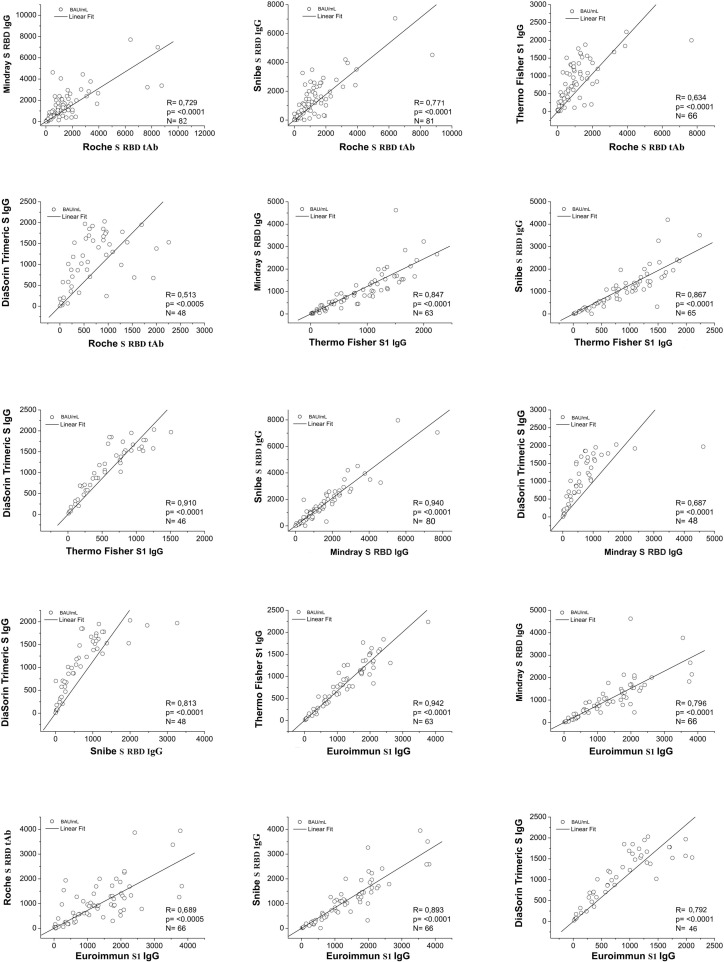
In Table 2 a correlation matrix shows the correlation coefficients for the serological assays. In each cell the correlation factors (R), the significance levels (p) and the number of the samples (n) have been reported. In particular, the lowest correlation coefficient value was obtained from Roche S-RBD tAb -DiaSorin Trimeric S IgG comparison (R = 0.513); the highest correlation coefficient value was obtained from Euroimmun S1 IgG-Thermo Fisher S1 IgG comparison (R = 0.942). Interestingly, on the one hand the highest correlation coefficients were obtained comparing methods with the same target antigen and isotype (S1 IgG for Euroimmun-Thermo Fisher, R = 0.942; S-RBD IgG for Snibe-Mindray, R = 0.940); on the other hand a different target antigen and/or antibody isotype gave a moderate correlation coefficient (S-RBD tAb for Roche and Trimeric S IgG for DiaSorin, R = 0.513). All data were statistically significant (p < 0.001).
Table 2.
Correlation matrix for the six serological assays; the Pearson’s coefficient of correlation (r) and the number of samples (n) are indicated in each box. All data were statistically significant with p<0.001.
 |
Furthermore, the correlation data have been represented in Fig. 2 . The linear fit of the correlation rate between two assays, is shown by a straight line and the analysis was performed only considering the detectable values, excluding all samples outside the linearity range of each method.
Fig. 2.
Two-test pair correlation data for the serological assays. The straight line represents the linear fit; the coefficient of correlation (R), the number of samples (n) and the level of significance (p) are also indicated.
Lastly, Table 3 highlights the values (k) obtained through Cohen's Kappa analysis. All the k values agreed significantly (p < 0.001) ranging from substantial (0.711) to perfect (1.00), and with the only exception of the pairwise agreement between Snibe S RBD IgG and Thermo Fisher S1 IgG which revealed a moderate agreement (k = 0.580). When between-assays variability was assessed, the difference between the median value of the BAU/mL CV (0.38; IQR: 0.23) and the median value of the (AU)/mL CV (0.68; IQR: 0.26) was statistically significant (p < 0.001).
Table 3.
Pairwise Cohen’s Kappa agreement among the six serological assays (95% confidence intervals and level of significance). All data were statistically significant with p < 0.001.
| DiaSorin Trimeric S IgG |
Snibe S RBD IgG |
Roche S RBD tAb |
Mindray S RBD IgG |
Euroimmun S1 IgG |
|
|---|---|---|---|---|---|
| Thermo Fisher S1 IgG |
k = 0.822 95 %CI = 0.5809;1.00 |
k = 0.580 95 %CI = 0.215;0.945 |
k = 0.711 95 %CI = 0.4023;1.00 |
k = 0.711 95 %CI = 0.4023;1.00 |
k = 0.822 95 %CI = 0.5809;1.00 |
| DiaSorin Trimeric S IgG |
k = 0.739 95 %CI = 0.3936;1.00 |
k = 0.883 95 %CI = 0.6566;1.00 |
k = 0.883 95 %CI = 0.6566;1.00 |
k = 1.00 | |
| Snibe S RBD IgG |
k = 0.851 95 %CI = 0.6138;1.00 |
k = 0.851 95 %CI = 0.565;1.00 |
k = 0.739 95 %CI = 0.3936;1.00 |
||
| Roche S RBD tAb |
k = 1.00 |
k = 0.883 95 %CI = 0.6566; 1.00 |
|||
| Mindray S RBD IgG |
k = 0.833 95 %CI = 0.6566; 1.00 |
10. Discussion
Immunity to SARS-CoV-2 is provided through the contribution of both B cells and T cells, but even if immunological memory can be maintained over time in the absence of measurable levels of serum antibodies, serological assays could be used as surrogates to determine the protection of infection and vaccine-induced immunity to SARS-CoV-2. In a clinical laboratory they also increasingly represent an important element for long-term strategies, as for the recent hypothesis of supplemental vaccine doses [25].
Since S protein constitutes the primary target of all current vaccines, serological assays are increasingly geared towards mainly using this protein or parts of it; also the S protein is likely to be the target of neutralizing antibodies, particularly the RBD. Nevertheless, it is still unclear exactly which antigen(s) would be preferable for the antibody assays in the disease and vaccine monitoring. Analogously to what happens for vaccines, a specific assay might in fact not work well, and starts producing more false negatives, because it no longer binds to the target peptides as they have mutated in the emerging virus variants. In addition, it is evident from the literature that variability among different assays exist, due to the different method architecture, antigen to target, test antibody quality, and isotype of antibodies to be detected [26], [27]. Several studies have been recently published on this topic comparing SARS-CoV-2 serological assays focusing mainly on the the sensitivities of the different kits at different time intervals from symptoms’ onset and therefore evaluating the best sensitive method for COVID-19 instead of antibodies levels [28], [29], [30], [31].
In a 2020 collaborative study, the use of the WHO International Standard showed a reduction in the results reported from different laboratories worldwide, of 2000-fold for ELISA methods and more than 50 times for neutralization assays [32].
The authors have clearly underlined in the document that despite the strong harmonisation observed in the collaborative study results, even between binding assays targeting different viral antigens, that may not reflect what happens in a diagnostic laboratory. Indeed, the study samples were mainly pools of samples collected before May 2020 at similar time points post symptomps. More data are required and until then, as included in the WHO instructions for use, the results expressed in BAU/mL have to be antigen specific and serological assay targeting different antigens (e.g. RBD versus S) are not necessarily harmonised, even after recalculation in BAU/mL.
Harmonization and standardization are related but different concepts. Harmonizing in laboratory testing considers all phases, from pre-analytical to post-analytical, while the standardization is a specific process, referred as assay calibrator and calibration development [33]. To this matter, each manufacturer should indicate the complete methodological traceability chain and lot-to-lot variation in terms of measurement uncertainty, according to the EN ISO 17511:2020 (en) [34].
However, when the first WHO 20/136 IS became available, none of the manufacturers adopted the new standardization on the existing assays, but through experimental tests provided a conversion factor (AU/mL to BAU/mL) to create a harmonization.
In the current study we performed a head-to-head comparison of six commercially available anti-SARS-CoV-2 serological tests, confirming the effect of the WHO IS in reducing the results variability (Supplemental Data Table S1). The main limitation of the the study is related to the sample size reduced in some statistical analysis because only detectable values could be taken into account. Moreover, the highest dispersions observed by the comparison of curves for values greater than 1000 AU/mL may be explained by the different dynamic ranges among methods, whereby some of them had reached a plateau. In detail, when comparing assays with various measuring ranges, it is important to consider the specific concentrations assigned to the calibrators.
Furthermore, the intrinsic variability of each specimen should be considered. In fact, due to differing binding characteristics of the antibodies in patients’ sera, not all the samples can be diluted linearly within the measuring range.
We analyzed serological assays in pairs and we found in some cases a very good correlation (i.e. Euroimmun S1 IgG vs Thermo Fisher IgG: ρ = 0.942) and in other cases significant differences (TriS DiaSorin IgG vs Roche S-RBD tAb: ρ = 0.513). Interestingly, since the lowest agreement values were found when kits using different target antigens or different antibody isotypes were compared, these discrepancies might be due mainly to the different target antigens used for the assays (S1, S-RBD, trimeric S) and/or to the immunoglobin classes detected (IgG or total IgG/IgA/IgM), as well as the detection methods based on several technologies (fluorescence, chemiluminescence, electrochemiluminescence). A similar finding was observed by Van Elsande et al., where the agreement between the two IgG anti-S assays (0.915) was higher than between an anti-N assay and anti-S assay (0.839–0.901) [35]. This further makes clear why it is advisable that the laboratory report should include the information on the target antigen and immunoglobulin isotype of the assay kit.
The average agreement among methods for positive/negative results assessed by the Cohen’s Kappa was almost perfect (κ = 0.820), better than the average of correlation coefficients (ρ = 0.786).
Hence, this could signify that today the immunological response to the virus should be interpreted mainly in terms of positive/negative. Then, only after the harmonization and standardization processes have moved on, would it make sense to compare antibody levels in patients or establish a precise threshold for immunity, even when tested by different manufacturer’s assays.
At present anti-SARS CoV-2 serological assays’ results are not interchangeable and individual immune monitoring should be performed using the same method.
The expression in BAU/mL may allow: 1) a first important step in the harmonization process, reducing differences among reported values compared to the expression in AU/mL; 2) a common language to report SARS-CoV-2 quantitative antibody results.
The WHO, the CDC and the European Commission’s Joint Research Centre (JRC) have worked and are still working, in synergy with manufactures, to further improve harmonization and to reach the standardization of SARS-CoV-2 quantitative antibodies and of the neutralization assays.
CRediT authorship contribution statement
Maria Infantino: Conceptualization, Investigation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. Massimo Pieri: Conceptualization, Investigation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. Marzia Nuccetelli: Conceptualization, Investigation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. Valentina Grossi: Formal analysis, Methodology, Visualization. Barbara Lari: Formal analysis, Methodology, Visualization. Flaminia Tomassetti: Formal analysis, Methodology, Visualization. Graziella Calugi: Formal analysis, Methodology, Visualization. Silvia Pancani: Formal analysis, Methodology, Visualization. Maurizio Benucci: Formal analysis, Methodology, Visualization. Patrizia Casprini: Conceptualization, Visualization, Supervision, Conceptualization, Visualization, Supervision. Mariangela Manfredi: Conceptualization, Visualization, Supervision. Sergio Bernardini: Conceptualization, Visualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to thank Giada Mattiuzzo and Mark Page (National Institute for Biological Standards and Control, UK) for their critical reading of the manuscript and valuable comments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.108095.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Krammer F., Simon V. Serology assays to manage covid-19. Science. 2020;368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 2.Infantino M., Damiani A., Gobbi F.L., Grossi V., Lari B., Macchia D., et al. Serological Assays for SARS-CoV-2 Infectious Disease: Benefits, Limitations and Perspectives. Isr Med Assoc J. 2020;22:203–210. [PubMed] [Google Scholar]
- 3.Nuccetelli M., Pieri M., Grelli S., Ciotti M., Miano R., Andreoni M., et al. SARS-CoV-2 infection serology: a useful tool to overcome lockdown? Cell Death Discov. 2020;6:38. doi: 10.1038/s41420-020-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.US Food and Drug Administration. Coronavirus disease 2019 (COVID-19) emergency use authorizations for medical devices: in vitro diagnostics EUAs. 2020. Available at: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas. Accessed 28 September 2020.
- 5.Finddx.org. SARS-COV-2 diagnostic pipeline. 2020. Available from: https://www.finddx.org/covid-19/pipeline (accessed12 April 2021).
- 6.World Health Organization. COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.0.1; 2020. Available https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1[Accessed 29 April 2021].
- 7.Eckerle I., Meyer B. SARS-CoV-2 seroprevalence in COVID-19 hotspots. Lancet. 2020;396:514–515. doi: 10.1016/S0140-6736(20)31482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M., et al. ENE-COVID Study Group. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin Infect Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marovich M., Mascola J.R., Monoclonal C.MS. Antibodies for Prevention and Treatment of COVID-19. JAMA. 2020;324:131–132. doi: 10.1001/jama.2020.10245. [DOI] [PubMed] [Google Scholar]
- 15.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 16.Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samrat S.K., Tharappel A.M., Li Z., Li H. Prospect of SARS-CoV-2 spike protein: Potential role in vaccine and therapeutic development. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gundlapalli AV, Salerno RM, Brooks JT, Averhoff F, Petersen LR, McDonald LC, et al; CDC COVID-19 Response. SARS-CoV-2 Serologic Assay Needs for the Next Phase of the US COVID-19 Pandemic Response. Open Forum Infect Dis. 2020;8:ofaa555. [DOI] [PMC free article] [PubMed]
- 20.U.S. Food and Drug Administration. Coronavirus Disease 2019 (COVID-19) Emergency Use Authorization for Medical Devices: In Vitro Diagnostics EUAs 2020; FDA: Silver Spring, MD, USA, 2020. [PubMed]
- 21.Gundlapalli A.V., Salerno R.M., Brooks J.T., Averhoff F., Petersen L.R., McDonald L.C., et al. SARS-CoV-2 Serologic Assay Needs for the Next Phase of the US COVID-19 Pandemic Response. Open forum infectious diseases. 2021;8(1):ofaa555. doi: 10.1093/ofid/ofaa555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. First WHO International Standard for anti-SARS-CoV-2 immunoglobulin. 2021. https://www.who.int/groups/expertcommittee-on-biological-standardization (accessed March 8, 2021). [DOI] [PMC free article] [PubMed]
- 23.https://www.who.int/publications/m/item/ECBS-Executive-Summary.IF.IK.TW-15_Dec_2020.
- 24.WHO Expert Committee on Biological Standardization, seventy-first report. Geneva: World Health Organization; 2021 (WHO Technical Report Series, No. 1030).
- 25.Kim D.S., Rowland-Jones S., Gea-Mallorquí E. Will SARS-CoV-2 Infection Elicit Long-Lasting Protective or Sterilising Immunity? Implications for Vaccine Strategies (2020) Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.571481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Post N., Eddy D., Huntley C., van Schalkwyk M.C.I., Shrotri M., Leeman D., et al. Antibody response to SARS-CoV-2 infection in humans: A systematic review. PLoS One. 2020;15 doi: 10.1371/journal.pone.0244126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370 doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coste A.T., Jaton K., Papadimitriou-Olivgeris M., Greub G., Croxatto A. Comparison of SARS-CoV-2 serological tests with different antigen targets. J Clin Virol. 2021 Jan;134 doi: 10.1016/j.jcv.2020.104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brochot E., Demey B., Handala L., François C., Duverlie G., Castelain S. Comparison of different serological assays for SARS-CoV-2 in real life. J Clin Virol. 2020;130 doi: 10.1016/j.jcv.2020.104569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James J., Rhodes S., Ross C.S., Skinner P., Smith S.P., Shipley R., et al. Comparison of Serological Assays for the Detection of SARS-CoV-2 Antibodies. Viruses. 2021;13:713. doi: 10.3390/v13040713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol. 2020;38:1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.https://www.who.int/publications/m/item/WHO-BS-2020.2403.
- 33.Tate J.R., Johnson R., Barth J., Panteghini M. Harmonization of laboratory testing - Current achievements and future strategies. Clin Chim Acta. 2014;432:4–7. doi: 10.1016/j.cca.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 34.ISO 15189:2012E 2012(en). Available from: https://www.iso.org/ standard/69984.html).
- 35.Van Elslande J., Decru B., Jonckheere S., Van Wijngaerden E., Houben E., Vandecandelaere P., et al. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin Microbiol Infect. 2020;26:1557.e1–1557.e7. doi: 10.1016/j.cmi.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.