Abstract
Efficient cleavage of both forms of eukaryotic initiation factor 4G (eIF4G-1 and eIF4G-2) has been achieved in HeLa cells by incubation with hybrid proteins containing poliovirus 2Apro. Entry of these proteins into cells is promoted by adenovirus particles. Substantial levels of ongoing translation on preexisting cellular mRNAs still continue for several hours after eIF4G degradation. Treatment of control HeLa cells with hypertonic medium causes an inhibition of translation that is reversed upon restoration of cells to normal medium. Protein synthesis is not restored in cells lacking intact eIF4G after hypertonic treatment. Notably, induction of synthesis of heat shock proteins still occurs in cells pretreated with poliovirus 2Apro, suggesting that transcription and translation of these mRNAs takes place even in the presence of cleaved eIF4G. Finally, the synthesis of luciferase was examined in a HeLa cell line bearing the luciferase gene under control of a tetracycline-regulated promoter. Transcription of the luciferase gene and transport of the mRNA to the cytoplasm occurs at control levels in eIF4G-deficient cells. However, luciferase synthesis is strongly inhibited in these cells. These findings indicate that intact eIF4G is necessary for the translation of mRNAs not engaged in translation with the exception of heat shock mRNAs but is not necessary for the translation of mRNAs that are being translated.
The initiation of translation in eukaryotes is a complex process that requires the functioning of a number of initiation factors in addition to the mRNA and the 40S ribosomal subunit (32, 57). Among those factors, eukaryotic translation initiation factor 4F* (eIF4F*) is involved in the early steps of mRNA recognition, facilitating the interaction of the mRNA with eIF3 and the small ribosomal subunit (58, 66). eIF4F* is a protein complex formed by the 25-kDa cap-binding protein eIF4E, eIF4A, a 50-kDa protein with helicase activity, and p220, also designated eIF4G (23, 58, 66). Recently described is a homologue of eIF4G, named eIF4G-2, that interacts with eIF4E, eIF4A, and eIF3 as well (21). A second homologue, PAIP-1, binds to the poly(A)-binding protein, providing a link between the 5′ and 3′ ends of mRNAs. PAIP-1 shows homology to the central region of mammalian eIF4G and interacts with the initiation factor eIF4A (12).
The role proposed for eIF4F* during translation is to recognize and attach to the cap structure present in the majority of eukaryotic mRNAs (62) in order to unwind the secondary structure of the untranslated 5′ region of mRNA (35, 59). This cap recognition step is accomplished by the eIF4E subunit and is required for the functioning of other initiation factors, including eIF4B, which stimulates helicase activity present in eIF4F* (29, 67). The RNA-unwinding capacity of the eIF4F* complex is higher than that found with eIF4A (67). The coordinate functioning of eIF4F* and eIF4B, together with eIF3 and the 40S ribosomal subunit containing eIF2–Met-tRNA–GTP, finally leads to the formation of the 43S initiation complex at the AUG initiation codon of the mRNA to form the 48S complex.
However, the cap recognition step is not absolutely required for an mRNA to be translated; artificially uncapped mRNAs are also translated both in intact cells and in cell-free systems, albeit with reduced efficiency (60, 70). The translatability of artificially uncapped mRNAs implies that either eIF4F* is not essential for mRNA translation or eIF4F* also participates in protein synthesis directed by uncapped mRNAs through a still undefined mechanism that would not involve cap recognition. Additional evidence that cap recognition is not an absolute requirement for translation comes from the finding that picornavirus mRNAs are naturally uncapped. These mRNAs are efficiently translated both in vivo and in cell-free systems (65). Elegant experiments demonstrated that the translation of picornavirus mRNAs follows a particular and efficient mechanism of initiation termed internal initiation (3, 53, 54). Intact eIF4G or the C-terminal moiety of this factor participates in the translation of naturally uncapped mRNAs, such as picornavirus RNAs (33, 51, 56). Addition of this factor to cell-free systems clearly stimulates translation of these mRNAs (4, 72). Moreover, inactivation of eIF4G blocks the translation of both artificially uncapped mRNA or picornavirus RNAs (48, 52), suggesting that eIF4F* participates in translation even in the absence of a cap structure in the mRNA.
In addition to uncapped mRNAs, certain other cellular mRNAs may not depend on the usual cap recognition step during the initiation of translation (42). This is the case for some heat shock mRNAs even though they contain a typical cap structure at the 5′ end (30, 41, 63). Poliovirus-infected cells still synthesize some heat shock proteins after the shutoff of cellular translation (45), owing to the fact that these mRNAs contain a leader sequence that participates in translation independently of eIF4F* (5, 14, 30, 39).
The infection of cells by poliovirus leads to the efficient and rapid inhibition of ongoing cellular translation (6, 9). Cleavage of initiation factor eIF4G by the poliovirus protease 2Apro has been proposed as the cause of this shutoff phenomenon (15). In fact, addition of picornavirus 2Apro to cell-free systems leads to eIF4G proteolysis and to the inhibition of cellular mRNA translation (34, 36, 48, 50). Apart from eIF4G proteolysis, 2Apro may have other cellular substrates involved in different cell functions. Notably, the synthesis of proteins from mRNAs containing the picornavirus leader sequence is usually stimulated by the respective picornavirus protease after eIF4G cleavage (24). Thus, picornavirus proteases, including poliovirus 2Apro, are useful tools for analyzing the exact function of eIF4G during translation (28). We have devised a method for introducing the poliovirus 2Apro into HeLa cells that leads to the efficient proteolysis of eIF4G (46) and used it to investigate the requirement of eIF4G for gene expression in intact human cells. Our findings indicate that eIF4G is not necessary for each initiation event of translation but rather may be necessary to bring the mRNA to the protein-synthesizing machinery.
MATERIALS AND METHODS
Cell cultures and viruses.
Dulbecco modified Eagle’s medium supplemented with 10% newborn calf serum was used for growth and maintenance of HeLa cell cultures. HeLa clone X1/5 cells express luciferase in a tetracycline-dependent manner (20). Chicken adenovirus (CELO virus) was obtained as described previously (11). The CsCl-banded virus was resuspended in 40% glycerol–150 mM NaCl–20 mM HEPES (pH 7.4) at 1012 virus particles/ml (11). CELO virus was generously provided by M. Cotten (Research Institute of Molecular Pathology, Vienna, Austria).
Purification of the fusion proteins MBP–β-Gal-α, MBP-2A, and MBP–PE-III+2A.
Escherichia coli DH5 cells were transformed either with pMal-c2, pMal-c.2A (48), or pMal-PE-III+2A (46). Addition of 1 mM isopropyl-β-d-thiogalactopyranoside induces the expression of genes encoding the fusion proteins maltose-binding protein (MBP)–β-galactosidase-α (β-Gal-α), MBP-2Apro (MBP-2A), and MBP-PE-III+2A (see Results for descriptions). These proteins were purified by column chromatography with an amylose resin (New England BioLabs) as described elsewhere (48).
Analysis of proteins by polyacrylamide gel electrophoresis (PAGE).
At the times indicated, cell monolayers were incubated for 1 h in methionine-free medium containing 20 μCi of [35S]methionine (1,000 Ci/mmol; Amersham) per ml. The monolayers were washed with phosphate-buffered saline (PBS) and solubilized in 100 μl of extraction buffer (10 mM Tris-HCl [pH 8.5], 150 mM NaCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.5% Nonidet P-40, 10 mM phenylmethylsulfonyl fluoride), and protein content was determined by the Bio-Rad protein assay. Aliquots containing equivalent amounts of protein were loaded on sodium dodecyl sulfate (SDS)–15% polyacrylamide gels. Fluorography and autoradiography of the gels were performed as described elsewhere (26).
Immunoblot assays against eIF4G.
Western blot analysis of eIF4G was carried out with SDS–7.5% polyacrylamide gels. Polyclonal eIF4G antibodies were obtained from rabbits immunized with synthetic peptides (2, 18). Aliquots containing equivalent amounts of protein were separated by PAGE on SDS–7.5% polyacrylamide gels. Proteins were transferred overnight to a nitrocellulose membrane (Trans-blot transfer medium; Bio-Rad) at 200 mA in a transfer buffer (25 mM Tris-HCl [pH 8.3], 90 mM glycine, 20% methanol, 0.1% SDS). After incubation with 5% nonfat dry milk in PBS and with human anti-eIF4G rabbit polyclonal antibodies, the immunoreacted bands were visualized with peroxidase-coupled secondary antibodies (Pierce) and enhanced chemiluminescence (ECL kit; Amersham) (2, 47). The peptides used to raise polyclonal antibodies against eIF4G correspond to amino acids 35 to 55 and 995 to 1020 of eIF4G (accession no. Q04637). These two peptides have a low homology with the new homologue of eIF4G, named eIF4G-2 (21). Specific antibodies kindly provided by N. Sonenberg (McGill University, Montreal, Quebec, Canada) against eIF4G-2 were used to detect the cleavage of this protein after incubation with CELO virus and hybrid proteins.
Luciferase assays.
After 1 h of labeling with [35S]methionine, monolayers of HeLa clone X1/5 cells were washed with PBS before lysis in 25 mM glycylglycine (pH 7.8)–1 mM DTT–0.5% Triton X-100 for 1 min at room temperature. Aliquots (10 μl) of the lysate were mixed with 190 μl of 25 mM glycylglycine (pH 7.8)–5 mM ATP–15 mM MgSO4–1 mM DTT–100 μg of bovine serum albumin per ml and assayed for luciferase activity in a Monolight 2010 (Analytical Luminescence Laboratory). d-Luciferin (Boehringer Mannheim) was used at 0.33 mM.
RNase protection assay.
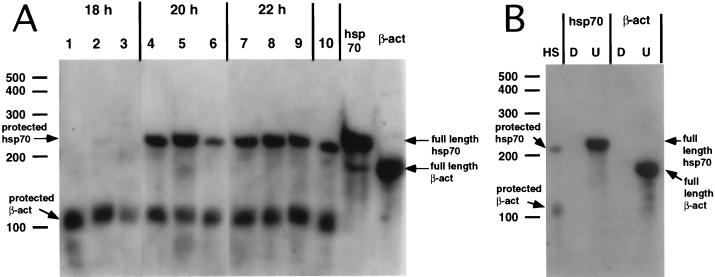
Detection of hsp70, luciferase, and β-actin mRNAs was carried out by RNase protection assay. A fragment of 240 bp that corresponds to nucleotides (nt) 1981 to 2220 of the human hsp70 gene (accession no. M11717) and a fragment of 327 bp corresponding to nt 749 to 1076 of the luciferase gene (accession number M15077) were amplified by PCR and subcloned into pcDNA3 (Invitrogen) and pBluescript KS (Stratagene), respectively. The sequences of both inserts were determined by sequence analysis. After digestion of each plasmid, T7 RNA polymerase was used to synthesize antisense probes as indicated by the manufacturer (Ambion). β-Actin antisense probe (protects a band of 127 nt) was synthesized as specified by Ambion. The sizes of full-length β-actin, hsp70, and luciferase probes are 188, 256, and 493 nt, respectively.
Total cytoplasmic RNA was extracted as described elsewhere (17). As a control at each time point, a fraction of the cells from the same dish was used to detect eIF4G (heat-shocked HeLa cells and HeLa clone X1/5 cells) and luciferase activity (HeLa clone X1/5 cells). The concentration of RNA was quantitated by measuring the absorbance at 260 nm. Total RNA was hybridized overnight at 42°C to psoralen-biotin-labeled antisense RNA probes. After hybridization, RNA samples were digested with a mixture of RNases A and T1, analyzed by electrophoresis on a 5% polyacrylamide–8 M urea gel, and transferred to a positively charged nylon membrane (Ambion). A chemiluminescence detection kit was used to detect the protected RNA fragments (Ambion). Due to the low level of detection of luciferase protected bands (Fig. 6), these protected bands for luciferase and β-actin were quantified by densitometric analysis.
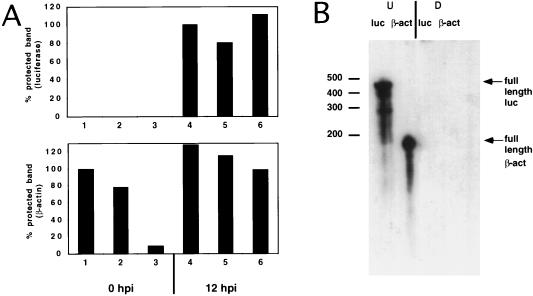
FIG. 6.
Effect of eIF4G cleavage on luciferase and β-actin mRNA levels. HeLa clone X1/5 cells grown in 60-mm-diameter dishes were incubated with purification buffer (lanes 1 and 4), with 900 μg of MBP–β-Gal-α and 48 μl of CELO virus (lanes 2 and 5), or with 900 μg of MBP-2A and 48 μl of CELO virus (lanes 3 and 6) in the presence of tetracycline (20 ng/ml) for 15 h. Total cytoplasmic RNA was extracted at 0 (lanes 1 to 3) and 12 (lanes 4 to 6) h postinduction (hpi) (0 and 12 h after tetracycline removal). Twenty-microgram aliquots of RNA were hybridized to β-actin and luciferase antisense probes. (A) RNase protection assay showing the densitometric quantification of protected bands for luciferase mRNAs (upper graph) and for β-actin mRNAs (lower graph). (B) Twenty micrograms of yeast RNA was hybridized with either a luciferase (luc) or a β-actin (β-act) antisense probe, and RNase mixture was added (D [digested]) or not (U [undigested full-length probe]).
RESULTS
Entry of hybrid proteins containing poliovirus 2Apro into HeLa cells and cleavage of eIF4G.
Picornavirus proteases like poliovirus 2Apro promote the cleavage of eIF4G (also known as eIF4G-1 or eIF4GI) between residues Arg485 and Gly486 to yield two polypeptides of approximately 100 to 130 kDa (33, 34, 64). Several polypeptides corresponding to the N-terminal of eIF4G are apparent after cleavage, reflecting heterogeneity of the factor in this region (77). The fact that poliovirus 2Apro proteolytically degrades eIF4G directly or indirectly in a cascade fashion (15) makes this protease a valuable tool for investigating the exact functioning of this factor in gene expression. The approach that we used to inactivate the function of eIF4G in intact human cells was to introduce the protein 2Apro directly into cells (46). To this end, poliovirus 2Apro was obtained as the MBP-2A hybrid protein. In addition, we engineered a Pseudomonas exotoxin (PE) gene in which the active domain of the toxin was replaced by 2Apro. As a result, MBP–PE-III+2A was produced and purified on amylose columns. Recently, we showed that these hybrid proteins bearing MBP are easily purified and cleave eIF4G in cell-free systems (48). Moreover, a hybrid protein bearing the receptor binding domain of PE and 2Apro entered into cells in the presence of animal virus particles (46). However, it was not known if a hybrid protein containing 2Apro devoid of receptor binding activity could still be translocated to the cytoplasm by the virus particles.
Indeed, animal viruses promote the entry of proteins from the medium to the cytosol of cultured cells (8, 10). Adenoviruses are particularly effective in this activity (10, 19). Chicken adenoviruses (e.g., CELO virus) that produce an abortive infection in human cells have been used successfully to introduce large DNA molecules into cells (11). Therefore, we made use of CELO virus to internalize the hybrid molecules containing poliovirus 2Apro. Addition of CELO virus or each one of the hybrid molecules separately to the culture medium has no effect on eIF4G in HeLa cells (results not shown). However, the simultaneous presence of MBP-2A or MBP–PE-III+2A plus CELO virus leads to cleavage of eIF4G-1 to an extent similar to that observed in poliovirus-infected cells (Fig. 1A). These results indicate that irrespective of the presence of cellular receptors for the hybrid protein, CELO virus efficiently promotes the internalization of the protein into cells. The hybrid molecule appears in the cytoplasm in an active form, as shown by the generation of the eIF4G cleavage products (Fig. 1A). Using a polyclonal anti-2Apro antibody, we could not detect the hybrid proteins delivered by CELO virus into the cytosol, although this antibody detected the 2Apro synthesized during a poliovirus infection (data not shown). Therefore, the amount of 2Apro delivered by CELO virus is below the level of 2Apro present in poliovirus-infected cells but is enough to cleave eIF4G.
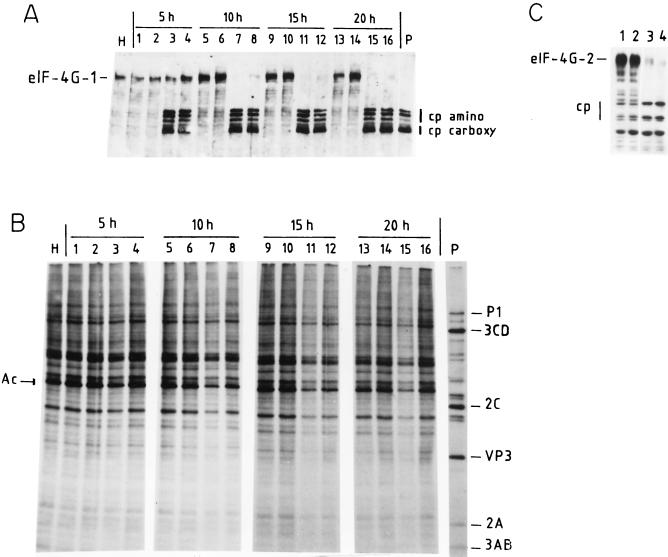
FIG. 1.
Time course of protein synthesis and eIF4G cleavage in HeLa cells incubated with MBP-2A or MBP–PE-III+2A plus CELO virus. HeLa cells grown in 24-well dishes were incubated with column purification buffer (lanes 1, 5, 9, and 13), with 100 μg of MBP–β-Gal-α and 10 μl of CELO virus (lanes 2, 6, 10, and 14), with 100 μg of MBP-2A and 10 μl of CELO virus (lanes 3, 7, 11, and 15), or with 100 μg of MBP–PE-III+2A and 10 μl of CELO virus (lanes 4, 8, 12, and 16). [35S]methionine was added to the medium 1 h before each time point, and cells were incubated for 1 h. Cells were harvested at 5 h (lanes 1 to 4), 10 h (lanes 5 to 8), 15 h (lanes 9 to 12), and 20 h (lanes 13 to 16). Cell extracts were separated by SDS-PAGE. (A) Western blot analysis with anti-eIF4G polyclonal antibodies. Intact eIF4G-1 and the amino-terminal (cp amino) and carboxy-terminal (cp carboxy) fragments of eIF4G are indicated. H, extract from mock-infected HeLa cells; P, extract from poliovirus-infected HeLa cells. (B) Labeled proteins were analyzed as described in Materials and Methods. The migration of some poliovirus proteins is indicated. Ac, cellular actin. (C) Cleavage of eIF4G-2 by hybrid proteins and CELO virus, determined by Western blot analysis using specific antibodies against human eIF4G-2. The intact protein and the cleavage products are shown. Lane 1, incubation with column purification buffer; lane 2, incubation with 100 μg of MBP–β-Gal-α and 10 μl of CELO virus; lane 3, 40 μg of MBP-2A and 10 μl of CELO virus; lane 4, 60 μg of MBP-2A and CELO virus. Cells were harvested 15 h after incubation, and the proteins were analyzed as described in Materials and Methods.
Effects of eIF4G cleavage on ongoing cellular translation and in cells treated with hypertonic medium.
Having observed that both MBP-2A and MBP-PE-III+2A fusion proteins cleaved eIF4G in HeLa cells, we tested the capacity of the cells to synthesize proteins under these conditions. Figure 1 shows both the kinetics of eIF4G cleavage by the hybrid proteins and the translation capacity of human cells upon various treatments. After 5 h of incubation with both CELO virus and either one of the hybrid molecules, about 50% of eIF4G is cleaved (Fig. 1A) although no effect on cellular translation is observed, as determined by the incorporation of [35S]methionine into proteins (Fig. 1B). After 10 h of incubation, eIF4G is almost completely degraded by the hybrid toxins, particularly when MBP-2A is assayed. Notably, substantial levels of cellular protein synthesis are observed under these circumstances. The same observation applies after 15 and 20 h of incubation. Densitometric analysis indicates a 65% inhibition of protein synthesis in cells treated with MBP-2A (Fig. 1B, lanes 7, 11, and 15) compared with control cells treated with MBP–β-Gal-α and a 55% inhibition after treatment with MBP–PE-III+2A (lanes 8 and 12). Therefore, we conclude that ongoing cellular translation is not totally abrogated in cells in which significant degradation of eIF4G has occurred.
Recently, a second form of eIF4G has been described (21). Analysis of poliovirus-infected cells indicate that this second form of eIF4G, named eIF4G-2 (or eIF4GII), is less sensitive to 2Apro and its cleavage correlates with the shutoff of cellular protein synthesis (22). Therefore, the cleavage of eIF4G-2 by the hybrid proteins plus CELO virus was also assayed (Fig. 1C). Extensive cleavage of eIF4G-2 clearly occurs after 15 h of incubation. Thus, this approach leads to cleavage of both forms of eIF4G in HeLa cells. In subsequent experiments cells were incubated with the hybrid proteins and CELO virus for approximately 15 h to ensure the extensive cleavage of both eIF4G-1 and eIF4G-2.
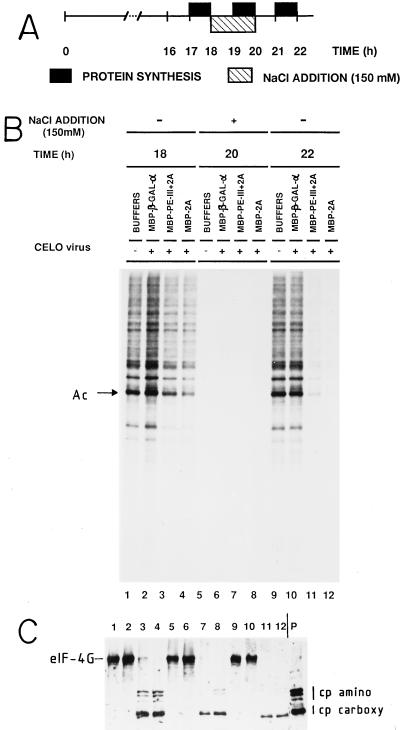
To test if initiation of translation can occur on mRNAs removed from the protein-synthesizing machinery in cells where eIF4G degradation has occurred, the experiment shown in Fig. 2 was carried out. Runoff of polysomes takes place in mammalian cells incubated in hypertonic media (49, 61). The salt excess blocks the initiation of translation, while elongation still occurs, leading to stripped mRNAs (61). This effect is fully reversible upon removal of excess salt and incubation in normal medium (Fig. 2). HeLa cells in which eIF4G was cleaved by incubation with either of the hybrid proteins plus CELO virus synthesize proteins at about 50% of the level observed in control cells after 18 h of incubation. Under hypertonic conditions, protein synthesis is essentially blocked regardless whether cells were treated with 2Apro (Fig. 2B, 20 h). Upon restoration of isotonic conditions, protein synthesis returned to normal in control cultures. However, translation was not restored in cells in which eIF4G had been cleaved as a result of 2Apro activity (Fig. 2B and C, 22 h). These results indicate that cells containing eIF4G proteolyzed by the method described in this work support significant levels of protein synthesis for several hours (Fig. 1) but are unable to initiate de novo the translation of mRNAs which have been released from the protein-synthesizing machinery and are not engaged in translation (Fig. 2). The cleavage products of eIF4G, especially the N-terminal fragments, are weakly detected during and after hypertonic treatment, perhaps as a result of their degradation. It may be that even though eIF4G is efficiently cleaved under the conditions used for Fig. 1 and 2, the cleaved products of eIF4G remain attached to the mRNA and participate as such in ongoing cellular translation, while these fragments cannot support de novo initiation once they are stripped from mRNAs by hypertonic treatment.
FIG. 2.
Effect of eIF4G cleavage on the reinitiation of protein synthesis after exposure to hypertonic medium. HeLa cells grown in 24-well dishes were incubated with column purification buffer (lanes 1, 5, and 9), with 100 μg of MBP–β-Gal-α and 10 μl of CELO virus (lanes 2, 6, and 10), with 100 μg of MBP–PE-III+2A and 10 μl of CELO virus (lanes 3, 7, and 11), or with 100 μg of MBP-2A and 10 μl of CELO virus (lanes 4, 8, and 12) for 16 h. From 18 to 20 h, the concentration of NaCl in the medium was increased to 300 mM. At the times indicated, cell monolayers were labeled with [35S]methionine for 1 h. (A) Schematic representation of the protocol. (B) Labeled proteins analyzed by SDS-PAGE. (C) Western blot analysis using the anti-eIF4G polyclonal antibodies. P, extract from poliovirus-infected HeLa cells. Intact eIF4G and the amino-terminal (cp amino) and carboxy-terminal (cp carboxy) fragments of eIF4G are indicated. Ac, actin.
Translation of heat shock mRNAs in HeLa cells containing cleaved eIF4G.
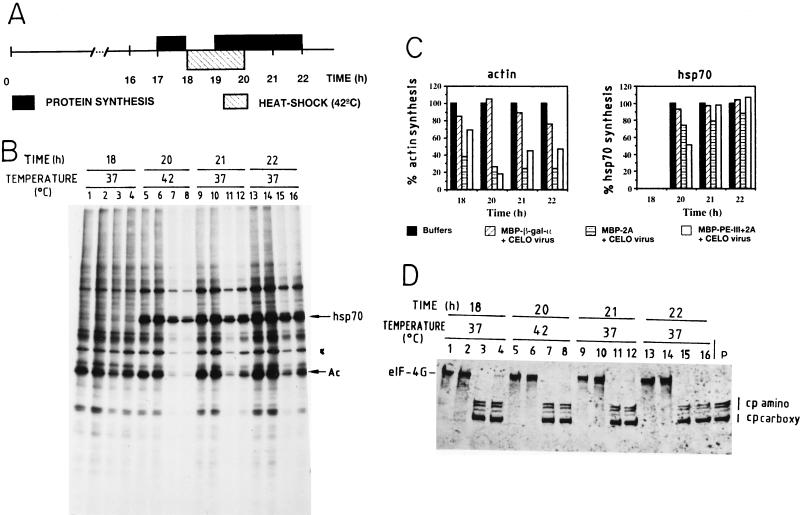
Both prokaryotic and eukaryotic cells trigger the expression of a number of genes when challenged with a variety of stress conditions (37). Thus, human cells incubated at supraoptimal temperatures induce the transcription of the so-called heat shock genes, followed by the subsequent translation of the mRNAs synthesized, giving rise to the heat shock proteins (14). Translation of heat shock mRNAs has special requirements for initiation factors compared to other cellular mRNAs (14, 63). Thus, poliovirus-infected HeLa cells still translate the heat shock mRNAs under conditions where the shutoff of host protein synthesis has taken place (45). The mechanism of translation of these mRNAs has been shown to be cap independent, not requiring the functioning of eIF4F* (5, 30, 39, 79). Therefore, it was of interest to test the translation pattern in heat-shocked cells defective in eIF4G. To this end, HeLa cells were incubated for 16 h with each of the hybrid toxins and CELO virus (Fig. 3A) to ensure that eIF4G has been efficiently proteolyzed (Fig. 3D). In good agreement with the results presented in Fig. 1, substantial levels of ongoing protein synthesis are detected from 17 to 18 h (Fig. 3B and C, 18 h), even in cells where almost no intact eIF4G is observed. Incubation of cells at 42°C induces the appearance of heat shock proteins (Fig. 3B, 20 h). Notably, HeLa cells synthesize heat shock proteins even when eIF4G has been cleaved, while the translation of other cellular mRNAs, including actin, is clearly defective. HeLa cells incubated at 42°C for 2 h still continue to translate most cellular mRNAs, both at 42°C and after restoration to the physiological temperature. However, cells where eIF4G has been cleaved do not translate most cellular mRNAs (Fig. 3B and C), although increasing amounts of actin synthesis are detected. The mRNA levels were analyzed by RNase protection assay from cells treated as described for Fig. 3A. Each RNA sample was hybridized to antisense β-actin and hsp70 probes (Fig. 4B). A band corresponding to the protected hsp70 mRNA was detected during and after treatment at 42°C (Fig. 4A, lanes 4 to 9). Initially the level of hsp70 mRNAs was lower in cells containing cleaved eIF4G than in control cells during the heat shock (Fig. 4, lanes 4 to 6), which correlates with reduced levels of hsp70 protein synthesis (Fig. 3B and C, 20 h). However, after the heat shock, the levels of hsp70 mRNAs were similar regardless of the state of eIF4G. We found a reduced amount of β-actin mRNA before the heat shock treatment followed by a recovery after several hours, when eIF4G was cleared. These differences are not due to differences in the amount of sample added, because RNA content was also analyzed by ethidium bromide staining. It may be possible that delivery of 2Apro to cells when CELO virus is present causes a reduction in mRNAs levels, and once the protein and CELO virus are removed the concentration of 2Apro decreases and the levels of mRNAs are reconstituted.
FIG. 3.
Synthesis of heat shock proteins in cells containing cleaved eIF4G. HeLa cells grown in 24-well dishes were incubated with purification buffer (lanes 1, 5, 9, and 13), with 100 μg of MBP–β-Gal-α and 10 μl of CELO virus (lanes 2, 6, 10, and 14), with 100 μg of MBP-2A and 10 μl of CELO virus (lanes 3, 7, 11, and 15), or with 100 μg of MBP–PE-III+2A and 10 μl of CELO virus (lanes 4, 8, 12, and 16) for 16 h. The cells were incubated at 42°C (heat shock) for 2 h (between 18 and 20 h). Protein synthesis was estimated by [35S]methionine incorporation before (17 to 18 h), during (19 to 20 h), and after (20 to 21 and 21 to 22 h) heat shock. (A) Schematic representation of the protocol. (B) Labeled proteins analyzed by SDS-PAGE as described in Materials and Methods. Position of migration of hsp70 and actin (Ac) are indicated. (C) Densitometric analysis of actin and hsp70 bands from the autoradiogram shown in panel B. (D) Western blot analysis with anti eIF4G polyclonal antibodies. P, extract from poliovirus-infected HeLa cells. Positions of amino-terminal (cp amino) and carboxy-terminal (cp terminal) fragments are shown.
FIG. 4.
Detection of β-actin and hsp70 mRNAs by RNase protection assay. HeLa cells grown in 35-mm-diameter dishes were incubated with purification buffer (lanes 1, 4, and 7), with 400 μg of MBP–β-gal-α and 24 μl of CELO virus (lanes 2, 5, and 8), or with 400 μg of MBP-2A and 24 μl of CELO virus (lanes 3, 6, and 9) for 16 h. The cells were incubated at 42°C (heat shock) for 2 h (between 18 and 20 h). Total RNA was extracted before (18 h), during (20 h), and after (22 h) heat shock. Five-microgram aliquots of total RNA were hybridized to β-actin and hsp70 antisense probes as described in Materials and Methods. Lane 10, total RNA of HeLa cells heat shocked for 2 h at 42°C hybridized with β-actin and hsp70 antisense probes. hsp70 and β-act lanes, full-length undigested hsp70 and β-actin antisense probes hybridized with 5 μg of yeast RNA. (A) RNase protection assay showing the protected bands for hsp70 and β-actin (arrows on the left) and full-length hsp70 and β-actin probes (arrows on the right). (B) HS, protected bands obtained after hybridizing 5 μg of total RNA of heat-shocked HeLa cells with hsp70 and β-actin probes. Five micrograms of yeast RNA was hybridized to each antisense probe and treated with RNase mixture (D [digested]) or not treated (U [undigested full-length probe]).
These findings indicate that translation of heat shock mRNAs is not dependent on the integrity of eIF4G in cultured human cells. Alternatively, it may be that translation of these mRNAs requires only very little eIF4G or that the cleaved eIF4G products participate in their translation. Moreover, the shutdown of translation of the rest of cellular mRNAs is clearly evident in cells containing no detectable eIF4G. This result provides an internal control to show that translation of most cellular mRNAs is blocked, while heat shock protein synthesis occurs at control levels in cells containing no detectable intact eIF4G.
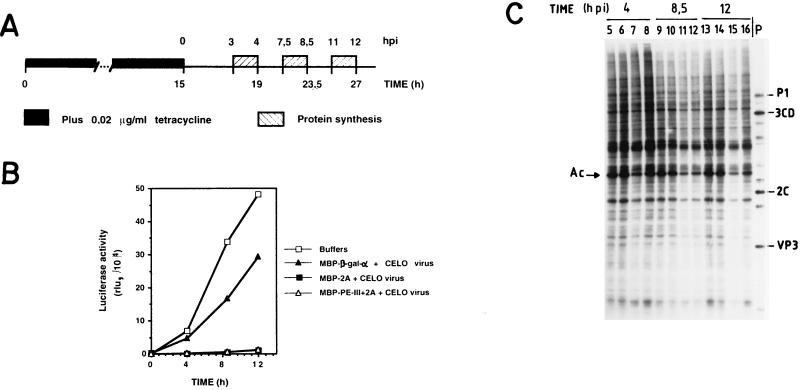
Inducible synthesis of luciferase in HeLa cells and effect of eIF4G cleavage by poliovirus 2Apro.
We then decided to test the effects of eIF4G cleavage on the expression of a newly synthesized cellular mRNA. To this end, we took advantage of HeLa cell lines bearing an integrated luciferase gene whose expression is controlled by tetracycline (20). This cell line offers an elegant model system with which to test the requirement for eIF4G during gene expression. Initially, we assayed the optimal conditions for luciferase repression and for its induction upon removal of the antibiotic. We found that as little as 20 ng of tetracycline per ml strongly repressed luciferase expression, while treatment with this concentration of tetracycline is fully reversible upon extensive washing, leading to the concomitant induction of luciferase synthesis (results not shown). Once we determined the amount of tetracycline needed to repress the expression of luciferase, the experiment shown in Fig. 5A was conducted. HeLa clone X1/5 cells were treated with hybrid proteins and CELO virus in order to cleave eIF4G in the presence of tetracycline. After 15 h of treatment, cells were washed and luciferase expression was analyzed at different time points after tetracycline removal (Fig. 5B). One hour before each time point, [35S]methionine was added to the medium to estimate ongoing protein synthesis (Fig. 5C). Figure 5B shows that virtually no luciferase activity is detected when HeLa clone X 1/5 cells are incubated with tetracycline and thus are repressed (time zero), but significant synthesis of luciferase appears after 4, 8.5, and 12 h of induction. This induction is partially blocked in cells treated with both CELO virus and the control hybrid protein MBP–β-Gal-α. Strikingly, luciferase synthesis is almost completely inhibited in cells incubated with the hybrid proteins bearing the poliovirus 2Apro plus CELO virus. The eIF4G present in these cells was extensively cleaved by either of the hybrid proteins (results not shown). It was important to analyze the level of protein synthesis in this HeLa cell line under conditions where luciferase synthesis was so strongly inhibited. In agreement with the results shown in Fig. 1, ongoing translation is affected much less than luciferase synthesis in cells containing cleaved eIF4G (Fig. 5C). Therefore, HeLa cells containing almost no detectable intact eIF4G, where luciferase synthesis is strongly hampered, are still able to support significant levels of translation of preexisting mRNAs over a period of several hours. These findings clearly illustrate the differential dependence on eIF4G for translation of the newly made luciferase mRNA compared to preexisting cellular mRNAs already engaged in translation in the same cells.
FIG. 5.
Effect of eIF4G cleavage on the inducible expression of luciferase. (A) Schematic representation of the protocol followed. HeLa clone X1/5 cells cultured in the presence of tetracycline (20 ng/ml) were incubated with purification buffer or with purified proteins in the presence of CELO virus for 15 h. Afterwards, cells were washed and tetracycline-free medium was added to induce luciferase gene expression (time zero). (B) At the indicated times postinduction (pi) (4, 8.5, and 12 h after tetracycline removal), luciferase activity (relative luciferase units [rlu]) was determined as described in Materials and Methods. (C) [35S]methionine was added 1 h before each time point, the medium was incubated for 1 h, and protein synthesis was analyzed; HeLa clone X1/5 cells were incubated with purification buffer (lanes 5, 9, and 13), with 100 μg of MBP–β-gal-α and 10 μl of CELO virus (lanes 6, 10, and 14), with 100 μg of MBP-2A and 10 μl of CELO virus (lanes 7, 11, and 15), or with 100 μg of MBP–PE-III+2A and 10 μl of CELO virus (lanes 8, 12, and 16). pi, postinduction; P, extract from poliovirus-infected HeLa cells; Ac, actin.
We then decided to investigate whether transcription of the luciferase gene takes place in cells deficient in eIF4G. To this end, cells were ruptured, the nuclei were removed, and RNA was extracted from the cytoplasmic fraction. Total RNA was extracted at 0 and 12 h after induction of luciferase expression in control cells and in cells previously treated with the hybrid protein MBP-2A plus CELO virus. Twenty micrograms of total RNA was hybridized to β-actin and luciferase antisense probes (Fig. 6A). No luciferase mRNA is detected in uninduced cells (time zero), whereas this mRNA is detected after 12 h of induction (Fig. 6A, upper panel). As a control, the same samples were hybridized with a probe to detect β-actin mRNAs (Fig. 6A, lower panel). Addition of the hybrid molecules plus CELO virus causes a reduction of β-actin mRNA levels compared to CELO virus alone at 0 h postinduction (0 h corresponds to hybrid proteins and CELO virus removal), and again a recovery is detected several hours postinduction (Fig. 6A, lower panel). These results indicate that there may be small differences in the amounts of luciferase mRNAs, but these differences do not explain the inhibition of luciferase mRNA translation when eIF4G is cleaved with hybrid proteins and CELO virus. Therefore, eIF4G integrity is necessary for the synthesis of luciferase, but transcription and transport of the luciferase mRNA to the cytoplasm are much less affected by eIF4G cleavage.
DISCUSSION
Poliovirus infection of HeLa cells leads to a rapid inhibition of host protein synthesis, while poliovirus RNA translation continues for a few hours (9, 65). The proposed model that explains poliovirus inhibition of host cell protein synthesis is based on the cleavage of the translation initiation factor eIF4G (15). We found that the CELO virus induced permeabilization of mammalian cells to hybrid proteins containing poliovirus 2Apro yields the complete cleavage of eIF4G-1 and eIF4G-2 (also known as eIF4GI and eIF4GII, respectively). The approach used in the present work to unveil the function of eIF4G offers the advantage that extensive cleavage of eIF4G is achieved in human cells, opening the possibility to test the effects of factor inactivation on ongoing cellular translation. Previous approaches to test the action of picornavirus 2Apro on gene expression in cultured cells focused on the effect of this protease on a reporter gene (13, 69). Due to the limited number of cells expressing the picornavirus 2Apro, ongoing cellular translation was not assayed in these studies. Although the transient expression of 2Apro with the recombinant vaccinia virus bearing the T7 RNA polymerase allowed a high efficiency of eIF4G cleavage, only vaccinia virus protein expression was analyzed due to the inhibition of cellular protein synthesis provoked by vaccinia virus infection (1, 2, 18).
Our present findings show that ongoing cellular protein synthesis takes place at substantial levels in cells containing cleaved eIF4G. We also found that there was a clear inhibition of de novo initiation of translation after hypertonic treatment in cells that contain cleaved eIF4G. Cleavage of eIF4G divides this protein in two functional domains: the N-terminal domain, which binds to eIF4E, and the C-terminal domain, which interacts with eIF4A and eIF3 (33, 40). The N-terminal domain, which recognizes the cap structure of the mRNA, could be dispensable when preexisting mRNAs are being translated, while the C-terminal domain could allow the binding of preexisting mRNAs to the ribosomal subunit and their translation. Therefore, the eIF4G cleaved products may be able to support new rounds of initiation events on mRNAs already engaged in translation. When these preexisting mRNAs are stripped from the protein synthesis machinery, intact eIF4G would be required to promote de novo initiation on these mRNAs. The discovery of new homologues of eIF4G also opens the possibility that one of these proteins is involved in the translation of preexisting mRNAs. Our experiments have analyzed the effect on translation caused by eIF4G-1 and eIF4G-2 cleavage, although we do not know if other homologues of eIF4G such as PAIP-1 are cleaved by treatment with the hybrid proteins and CELO virus.
With respect to protein synthesis on newly synthesized mRNAs, we have found that eIF4G cleavage strongly blocks the translation of newly synthesized mRNAs. An exception to this rule is constituted by heat shock mRNAs which also bear a cap structure at the 5′ end (14, 63). This finding is consistent with previous results demonstrating the ability of heat shock mRNAs to be translated in conditions of limiting eIF4G (5, 30, 68), suggesting that initiation factors involved in the binding of the cap structure and recruitment of the mRNA to the ribosome are not essential for heat shock mRNA translation.
Thus, these findings suggest a major importance of the integrity of eIF4G during the presentation of an mRNA to the translation machinery, but it seems to be dispensable in each further round of translation of an mRNA. These results also seem to exclude the possibility that some residual intact eIF4G in our 2Apro-permeabilized cells was responsible for maintaining the cellular translation.
The working model that eIF4G as part of eIF4F* is involved in the very first initiation event and is not required for further rounds of initiation is consistent with a number of results obtained both for cell-free systems and in intact cells. Thus, eIF4F* is not absolutely required for in vitro translation of mRNAs, and the eIF4F* dependence is modulated by several factors, including mRNA concentration (4, 23, 48, 67, 72). Moreover, eIF4F* stimulates the translation of both capped and uncapped mRNAs, indicating that this factor plays a pivotal role in the presentation of exogenously added mRNAs to the protein-synthesizing machinery regardless of the presence of a cap structure at the 5′ end. However, the cap structure would facilitate the interaction of most mRNAs with eIF4F* through eIF4E binding to the cap structure. Both natural and artificially uncapped mRNAs still interact with eIF4F* and require this factor for efficient translation (48, 52, 56). The experiments with poliovirus-infected HeLa cells, where no strict correlation exists between the shutoff of host translation and eIF4G degradation (7, 27, 55), are easily explained if eIF4G and hence eIF4F* are involved in the coupling of newly synthesized mRNAs to the protein-synthesizing machinery. Finally, the cleavage of eIF4G in Xenopus oocytes by recombinant coxsackievirus B4 protease 2A led to a decrease of protein synthesis of only 35% (31).
Recent findings indicate that the mRNA may adopt a circular structure, and this circularization could facilitate sequential rounds of translation (25, 71). Thus, the 5′ end and the poly(A) tail of mRNAs, facilitated by some polypeptides, may interact with each other to signal the availability of these mRNAs for translation. In fact, eIF4G in yeast interacts directly with the poly(A)-binding protein (PABP) (71), and PAIP, a homologue of eIF4G in mammals, binds directly to PABP (12). These results help to explain the influence of both 5′ and 3′ ends of mRNA on the initiation of protein synthesis (28, 44, 70). Perhaps the circularization of mRNAs allows their translation even in the presence of cleaved eIF4G, or possibly some other homologue of eIF4G can support the translation of mRNAs already engaged in translation. In fact, PAIP does not bind to the cap-binding protein eIF4E but can bind to the helicase protein eIF4A and to PABP.
Poliovirus 2Apro has been implicated in several processes during poliovirus infection. As a protease is involved in the cleavage of viral protein precursors (73) as well as in the cleavage of cellular proteins, such as eIF4G-1 and eIF4G-2 (16, 21, 22). 2Apro also stimulates the translation of poliovirus mRNA through a mechanism that requires the poliovirus internal ribosomal entry site located at the 5′ untranslated region (24, 75, 80). Finally 2Apro may play a role in viral replication (38, 43, 78). Initial experiments to determine the action of 2Apro on gene expression by transfection with a plasmid encoding a reporter gene suggested that poliovirus 2Apro blocked transcription more powerfully than translation (13). The analysis of mRNAs in cells treated with the hybrid proteins and CELO virus showed a reduced presence of β-actin mRNAs at early time points after hybrid protein and CELO virus removal, although this treatment did not prevent the transcription of new mRNAs like hsp70 and luciferase mRNAs and the recovery of β-actin mRNA levels. We cannot discard the possibility that a higher level of 2Apro is delivered to the cells while CELO virus is present, causing also an inhibition of transcription. Once the protein and virus are removed, the level of 2Apro decreases and the levels of mRNAs are reconstituted. Recently, it has been found that protease 2Apro cleaves also the TATA-binding protein, although this cleavage did not affect the transcriptional activity of this protein in vitro (76). These authors suggested that the effect of 2Apro on transcription could be due to a secondary effect caused by inhibition of translation, although direct cleavage of a protein involved in transcription cannot be excluded. In fact, the expression of 2Apro mutants by recombinant vaccinia virus VT7 indicates that the cleavage of eIF4G can be separated from the inhibition of transcription, suggesting that 2Apro interferes with cellular gene expression at both transcriptional and translational levels (74). The CELO-mediated delivery of 2Apro to mammalian cells is a helpful system for analyzing the role of eIF4G in cellular translation, but it may be less useful for elucidating targets of 2Apro that potentially interfere with transcription. The development of new systems that mimic the levels of 2Apro present in the cytoplasm of poliovirus-infected cells could help to elucidate the effect of 2Apro on transcription.
ACKNOWLEDGMENTS
The expert technical assistance of M. A. Sanz is acknowledged. E. Feduchi, J. M. Sierra, and M. G. Rush are acknowledged for their help and for critical reading of the manuscript. We thank M. Cotten (Research Institute of Molecular Pathology, Vienna, Austria) for kindly providing CELO virus. H. Bujard (Zentrum für Molekulare Biologie, Heidelberg, Germany) is acknowledged for providing the HeLa cell line X1/5. N. Sonenberg is acknowledged for providing specific antibodies against human eIF4G-2.
Plan Nacional project PB94-0148 and the institutional grant to the CBM of Fundación Ramón Areces are acknowledged for financial support. I.N. is a holder of a Gobierno Vasco fellowship.
REFERENCES
- 1.Aldabe R, Feduchi E, Novoa I, Carrasco L. Expression of poliovirus 2Apro in mammalian cells: effects on translation. FEBS Lett. 1995;377:1–5. doi: 10.1016/0014-5793(95)01269-9. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe R, Feduchi E, Novoa I, Carrasco L. Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: Effects on vaccinia virus. Biochem Biophys Res Commun. 1995;215:928–936. doi: 10.1006/bbrc.1995.2553. [DOI] [PubMed] [Google Scholar]
- 3.Alexander L, Lu H H, Wimmer E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony D D, Merrick W C. Eukaryotic initiation factor (eIF)-4F. Implications for a role in internal initiation of translation. J Biol Chem. 1991;266:10218–10226. [PubMed] [Google Scholar]
- 5.Barnes C A, MacKenzie M M, Johnston G C, Singer R A. Efficient translation of an SSA1-derived heat-shock mRNA in yeast cells limited for cap-binding protein and eIF-4F. Mol Gen Genet. 1995;246:619–627. doi: 10.1007/BF00298969. [DOI] [PubMed] [Google Scholar]
- 6.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonneau A M, Sonenberg N. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco L. Entry of animal viruses and macromolecules into cells. FEBS Lett. 1994;350:152–154. doi: 10.1016/0014-5793(94)00780-2. [DOI] [PubMed] [Google Scholar]
- 9.Carrasco L. Picornavirus inhibitors. Pharmacol Ther. 1994;64:215–290. doi: 10.1016/0163-7258(94)90040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrasco L. Modification of membrane permeability by animal viruses. Adv Virus Res. 1995;45:61–112. doi: 10.1016/S0065-3527(08)60058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotten M, Wagner E, Zatloukal K, Birnstiel M L. Chicken adenovirus (CELO virus) particles augment receptor-mediated DNA delivery to mammalian cells and yield exceptional levels of stable transformants. J Virol. 1993;67:3777–3785. doi: 10.1128/jvi.67.7.3777-3785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig A W B, Haghighat A, Yu A T K, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 13.Davies M V, Pelletier J, Meerovitch K, Sonenberg N, Kaufman R J. The effect of poliovirus proteinase 2Apro on cellular metabolism. J Biol Chem. 1991;266:14714–14720. [PubMed] [Google Scholar]
- 14.Duncan R F. Translational control during heat shock. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 271–293. [Google Scholar]
- 15.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 16.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 17.Favaloro J, Treisman R, Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65:718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- 18.Feduchi E, Aldabe R, Novoa I, Carrasco L. Effects of poliovirus 2Apro on vaccinia virus gene expression. Eur J Biochem. 1995;234:849–854. doi: 10.1111/j.1432-1033.1995.849_a.x. [DOI] [PubMed] [Google Scholar]
- 19.Fernández-Puentes C, Carrasco L. Viral infection permeabilizes mammalian cells to protein toxins. Cell. 1980;20:769–775. doi: 10.1016/0092-8674(80)90323-2. [DOI] [PubMed] [Google Scholar]
- 20.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grifo J A, Tahara S M, Morgan M A, Shatkin A J, Merrick W C. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258:5804–5810. [PubMed] [Google Scholar]
- 24.Hambidge S J, Sarnow P. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc Natl Acad Sci USA. 1992;89:10272–10276. doi: 10.1073/pnas.89.21.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentze M W. Translation—eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 26.Irurzun A, Perez L, Carrasco L. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology. 1992;191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- 27.Irurzun A, Sanchez-Palomino S, Novoa I, Carrasco L. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. J Virol. 1995;69:7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson R J, Hunt S L, Reynolds J E, Kaminski A. Cap-Dependent and cap-independent translation: operational distinctions and mechanistics interpretations. Curr Top Microbiol Immunol. 1996;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 29.Jaramillo M, Dever T E, Merrick W C, Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol Cell Biol. 1991;11:5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joshi-Barve S, De Benedetti A, Rhoads R E. Preferential translation of heat shock mRNAs in HeLa cells deficient in protein synthesis initiation factors eIF-4E and eIF-4gamma. J Biol Chem. 1992;267:21038–21043. [PubMed] [Google Scholar]
- 31.Keiper B D, Rhoads R E. Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res. 1997;25:395–402. doi: 10.1093/nar/25.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Regulation of translation in eukaryotic systems. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- 33.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases—implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 34.Lamphear B J, Yan R, Yang F, Waters D, Liebig H-D, Klump H, Kuechler E, Skern T, Rhoads R E. Mapping the cleavage site in protein synthesis initiation factor eIF-4gamma of the 2A proteases from human coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 35.Lawson T G, Ray B K, Dodds J T, Grifo J A, Abramson R D, Merrick W C, Betsch D F, Weith H L, Thach R E. Influence of 5′ proximal secondary structure on the translational efficiency of eukaryotic mRNAs and on their interaction with initiation factors. J Biol Chem. 1986;261:13979–13989. [PubMed] [Google Scholar]
- 36.Liebig H-D, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R, Kuechler E, Skern T. Purification of two picornaviral 2A proteinases: interaction with eIF-4gamma and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 37.Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 38.Lu H H, Li X Y, Cuconati A, Wimmer E. Analysis of picornavirus 2Apro proteins: separation of proteinase from translation and replication functions. J Virol. 1995;69:7445–7452. doi: 10.1128/jvi.69.12.7445-7452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macejak D, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 40.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maroto F G, Sierra J M. Translational control in heat-shocked Drosophila embryos. Evidence for the inactivation of initiation factor(s) involved in the recognition of mRNA cap structure. J Biol Chem. 1988;263:15720–15725. [PubMed] [Google Scholar]
- 42.McBratney S, Chen C Y, Sarnow P. Internal initiation of translation. Curr Opin Cell Biol. 1993;5:961–965. doi: 10.1016/0955-0674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 43.Molla A, Paul A V, Schmid M, Jang S K, Wimmer E. Studies on dicistronic polioviruses implicate viral proteinase 2Apro in RNA replication. Virology. 1993;196:739–747. doi: 10.1006/viro.1993.1531. [DOI] [PubMed] [Google Scholar]
- 44.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muñoz A, Alonso M A, Carrasco L. Synthesis of heat-shock proteins in HeLa cells: inhibition by virus infection. Virology. 1984;137:150–159. doi: 10.1016/0042-6822(84)90018-7. [DOI] [PubMed] [Google Scholar]
- 46.Novoa I, Cotten M, Carrasco L. Hybrid proteins between Pseudomonas aeruginosa exotoxin A and poliovirus 2Apro cleave p220 in HeLa cells. J Virol. 1996;70:3319–3324. doi: 10.1128/jvi.70.5.3319-3324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novoa I, Feduchi E, Carrasco L. Hybrid proteins between Pseudomonas exotoxin A and poliovirus 2Apro. FEBS Lett. 1994;355:45–48. doi: 10.1016/0014-5793(94)01157-5. [DOI] [PubMed] [Google Scholar]
- 48.Novoa I, Martínez-Abarca F, Fortes P, Ortín J, Carrasco L. Cleavage of p220 by purified poliovirus 2Apro in cell-free systems. Effects on translation of capped and uncapped mRNAs. Biochemistry. 1997;36:7802–7809. doi: 10.1021/bi9631172. [DOI] [PubMed] [Google Scholar]
- 49.Nuss D L, Oppermann H, Koch G. Selective blockage of initiation of host protein synthesis in RNA-virus-infected cells. Proc Natl Acad Sci USA. 1975;72:1258–1262. doi: 10.1073/pnas.72.4.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohlmann T, Rau M, Morley S J, Pain V M. Proteolytic cleavage of initiation factor eIF-4gamma in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 52.Pause A, Méthot N, Svitkin Y, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pelletier J, Kaplan G, Racaniello V R, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 55.Perez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 56.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhoads R E. Regulation of eukaryotic protein synthesis by initiation factors. J Biol Chem. 1993;268:3017–3020. [PubMed] [Google Scholar]
- 58.Rhoads R E, Joshi B, Minich W B. Participation of initiation factors in the recruitment of mRNA to ribosomes. Biochimie. 1994;76:831–838. doi: 10.1016/0300-9084(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 59.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell P J, Hambidge S J, Kirkegaard K. Direct introduction and transient expression of capped and non-capped RNA in Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4949–4953. doi: 10.1093/nar/19.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saborio J L, Pong S S, Koch G. Selective and reversible inhibition of initiation of protein synthesis in mammalian cells. J Mol Biol. 1974;85:195–211. doi: 10.1016/0022-2836(74)90360-x. [DOI] [PubMed] [Google Scholar]
- 62.Shatkin A J. mRNA cap binding proteins: essential factors for initiating translation. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 63.Sierra J M, Zapata J M. Translational regulation of the heat shock response. Mol Biol Rep. 1994;19:211–220. doi: 10.1007/BF00986963. [DOI] [PubMed] [Google Scholar]
- 64.Sommergruber W, Ahorn H, Klump H, Seipelt J, Zoephel A, Fessl F, Krystek E, Blaas D, Kuechler E, Liebig H-D, Skern T. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4gamma via a common recognition motif. Virology. 1994;198:741–745. doi: 10.1006/viro.1994.1089. [DOI] [PubMed] [Google Scholar]
- 65.Sonenberg N. Poliovirus translation. Curr Top Microbiol Immunol. 1990;161:23–47. doi: 10.1007/978-3-642-75602-3_2. [DOI] [PubMed] [Google Scholar]
- 66.Sonenberg N. Regulation of translation and cell growth by eIF-4E. Biochimie. 1994;76:839–846. doi: 10.1016/0300-9084(94)90185-6. [DOI] [PubMed] [Google Scholar]
- 67.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 68.Song H-J, Gallie D R, Duncan R F. m7GpppG cap dependence for efficient translation of Drosophila 70-kDa heat-shock-protein (Hsp70) mRNA. Eur J Biochem. 1995;232:778–788. [PubMed] [Google Scholar]
- 69.Sun X H, Baltimore D. Human immunodeficiency virus tat-activated expression of poliovirus protein 2A inhibits mRNA translation. Proc Natl Acad Sci USA. 1989;86:2143–2146. doi: 10.1073/pnas.86.7.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tarun S Z, Jr, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 71.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas A A, ter Haar E, Wellink J, Voorma H O. Cowpea mosaic virus middle component RNA contains a sequence that allows internal binding of ribosomes and that requires eukaryotic initiation factor 4F for optimal translation. J Virol. 1991;65:2953–2959. doi: 10.1128/jvi.65.6.2953-2959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toyoda H, Nicklin M J, Murray M G, Anderson C W, Dunn J J, Studier F W, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 74.Ventoso I, Barco A, Carrasco L. Mutational analysis of poliovirus 2Apro. Distinct inhibitory functions of 2Apro on translation and transcription. J Biol Chem. 1998;273:27960–27967. doi: 10.1074/jbc.273.43.27960. [DOI] [PubMed] [Google Scholar]
- 75.Ventoso I, Carrasco L. A poliovirus 2Apro mutant unable to cleave 3CD shows inefficient viral protein synthesis and transactivation defects. J Virol. 1995;69:6280–6288. doi: 10.1128/jvi.69.10.6280-6288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yalamanchili P, Banerjee R, Dasgupta A. Poliovirus-encoded protease 2Apro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J Virol. 1997;71:6881–6886. doi: 10.1128/jvi.71.9.6881-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan R, Rychlik W, Etchison D, Rhoads R E. Amino acid sequence of the human protein synthesis initiation factor eIF-4gamma. J Biol Chem. 1992;267:23226–23231. [PubMed] [Google Scholar]
- 78.Yu S F, Benton P, Bovee M, Sessions J, Lloyd R E. Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. J Virol. 1995;69:247–252. doi: 10.1128/jvi.69.1.247-252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zapata J M, Maroto F G, Sierra J M. Inactivation of mRNA cap-binding protein complex in Drosophila melanogaster embryos under heat shock. J Biol Chem. 1991;266:16007–16014. [PubMed] [Google Scholar]
- 80.Ziegler E, Borman A M, Deliat F G, Liebig H D, Jugovic D, Kean K M, Skern T, Kuechler E. Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology. 1995;213:549–557. doi: 10.1016/s0042-6822(95)90001-2. [DOI] [PubMed] [Google Scholar]