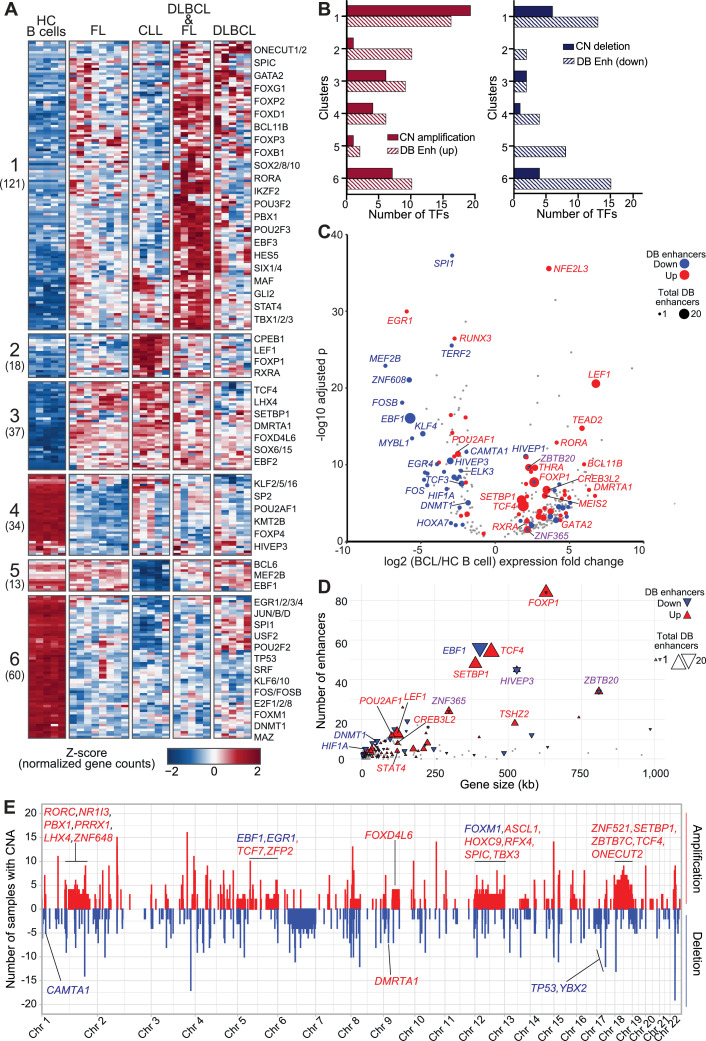
Fig. 5.
BCL-altered transcription factor expression profiles associated with genome copy number alterations and differentially bound enhancers. A) Heatmap shows normalized gene count z-scores (RNA-seq) for transcription factors (TF) differentially expressed (adjusted p-value < 0.01, absolute log2 fold change > 1, n = 283) in at least one BCL subtype compared to healthy control (HC) B-cells. Six clusters of distinct expression profiles were detected; the number of genes is shown beneath each cluster label. B) Bar chart show the number of TFs with copy number alterations and differentially bound (DB) enhancers in each cluster. (Left) Copy number (CN) amplified, DB increased H3K27ac in BCL (DB up); (Right) CN deletion, DB decreased H3K27ac in BCL (DB down). C) Volcano plot shows log2 fold change and -log10 adjusted p value for expression of TFs (as in A) in BCL versus HC B cells. TF genes with overlapping DB enhancers are highlighted by color (red = increased, blue = decreased, grey = unchanged level of histone acetylation) and size (corresponding to the number of DB enhancers). Color of the gene label corresponds to the status of the overlapping DB enhancers (red = all increased, blue = all decreased, purple = both increased and decreased DB enhancers overlap). D) XY plot shows the number of total enhancers overlapping each TF (Y axis) by TF gene size in kilobases (kb) (X axis). Size and colors as in (C). E) Whole genome frequency plot of number of BCL samples with copy number amplification (red) or deletion (blue) (100kb bins). TFs (from A) with at least 3 samples with overlapping CN alterations (CNA) are labeled on the upper panel if they overlap an amplified region and on the lower panel for deleted regions. Gene label color indicates if expression is increased (red) or decreased (blue) in BCL relative to HC B cells.