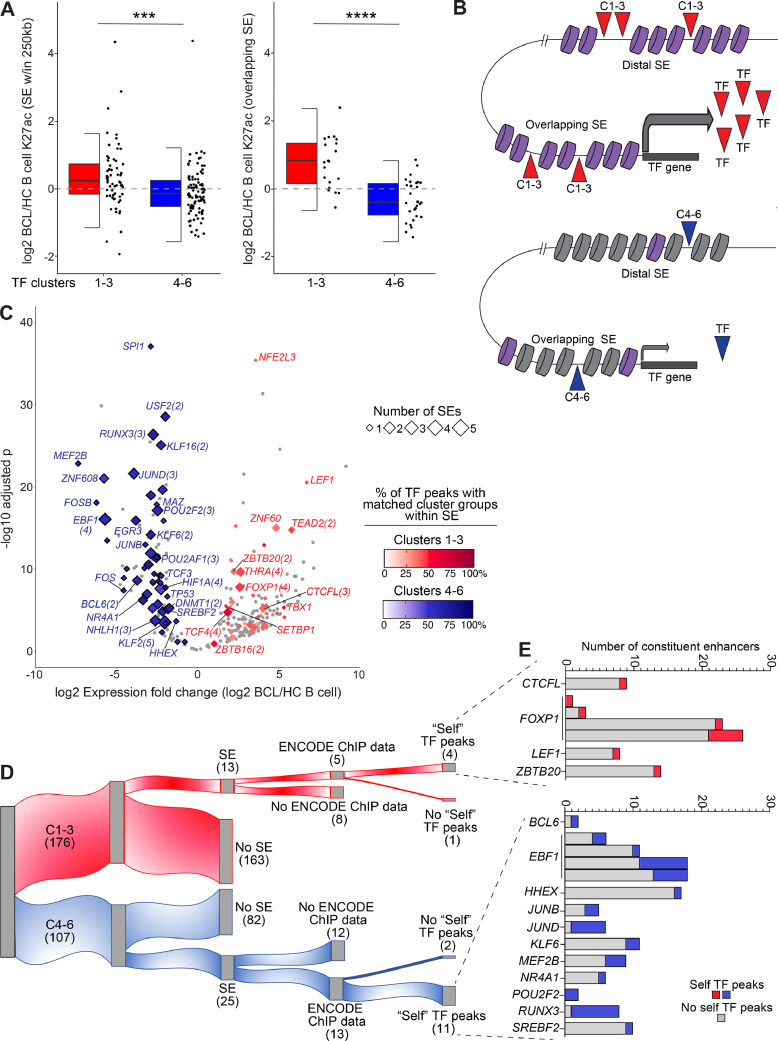
Fig. 6.
BCL-altered transcription factors have super-enhancer powered transcriptional feedback loops. A) Half box/dot-plot shows the log2 fold change of K27ac in BCL versus healthy control (HC) B cells for super-enhancers (SE) within 250kb (left) or overlapping (right) the differentially expressed TF genes (red: clusters 1-3, upregulated in BCL, blue: clusters 4-6, downregulated in BCL, from Fig. 5). Wilcoxon test *** p < 0.001; **** p < 0.0001. B) Cartoon depicts distal and overlapping SEs with high (purple) or low (grey) levels of H3K27ac regulating the expression of a TF gene from one of clusters 1-3 (red) or 4-6 (blue). TFs from the same (matching) cluster groups (1-3/4-6) bind within the SEs and control TF expression. C) Volcano plot shows log2 fold change and -log10 adjusted p value for expression of TFs (from Fig. 5) in BCL versus HC B cells. TF genes with SE within 250kb are highlighted by diamonds, with size indicating number of SEs and color corresponding to the percentage of matching TF cluster group peaks within the SE(s), red = TF gene in Clusters 1-3, blue = TF gene in Clusters 4-6 (ENCODE 3 TF ChIPseq). Gene labels include the number of SEs if greater than one. D) Alluvial plot segregates TFs with overlapping SEs and ENCODE 3 ChIPseq data. “Self” TF peaks: ChIPseq binding peaks for a TF were found in the SE overlapping the gene encoding that TF, an indication of TF self-regulation. E) For 15 TFs with “self” TF peaks in overlapping super-enhancers from (D), bar graphs show the number of constituent enhancers with (red/blue) or without (grey) binding peaks of the “self” TF. Red = TF in Clusters 1-3, blue = TF in Clusters 4-6.