Summary
Earthquakes occur thousands of times every day around the world. They are naturally destructive seismic events and often result in soil liquefaction. Soil microbiota plays a vital role in soil environments and may serve as an effective indicator to assess soil liquefaction after earthquakes. This study aimed to detect the microbial community abundance and composition in soil samples of different depths. Soil samples were collected in Southern Taiwan immediately after the 2010 earthquake. Their physical characteristics were determined, and their microbial communities were analyzed through 16S amplicon sequencing. The results revealed that Nitrospirae phylum dominated in the liquefied layer. In particular, the genus HB118, dominant in the liquefied layer, was not detected at other soil depths or in the expelled liquefied soil. This finding not only provides valuable insights into changes in microbial community composition at different soil depths after earthquakes but also suggests a useful indicator for monitoring liquefied soil.
Subject areas: Earth sciences, Soil science, Biogeochemistry, Seismology, Microbiology
Graphical abstract
Highlights
-
•
This study characterized the microbial composition of liquefied soil after an earthquake
-
•
Most abundant phylum Nitrospirae found in liquefied soil if 3 most abundant phyla removed
-
•
HB118 spp is correlated with liquefied soil
-
•
We set up the alternative monitoring methods of soil liquefaction after seismic events
Earth sciences; Soil science; Biogeochemistry; Seismology; Microbiology
Introduction
Soil liquefaction is one of the major geohazards induced by earthquakes and is a complicated phenomenon that is influenced by many factors; moreover, it is undetectable at the soil surface. Soil liquefaction occurs when the soil fails owing to the loss of shear strength and increase in pore water pressure (Dobry and Abdoun, 2017; Lashkari et al., 2017; Segata et al., 2011). It usually develops when (1) the soil particles are cohesionless and loose, (2) the soil becomes uncompacted and the particle size ranges from fine to coarse, and (3) sufficient, long-duration ground shaking occurs (Castiglia et al., 2020; Zhang and Wang, 2020). When the pore water pressure is over the limit, the water and some of the sandy soil can erupt out of the ground along the cracks in the stratum, and sandblasting occurs. Following an earthquake, the excess pore water pressure dissipates, and the density of the soil structure increases.
After liquefaction, volumetric strain causes ground surface subsidence phenomena (e.g., the settlement and tilting of residential buildings). In recent years, microbial communities have played increasingly important roles in various fields, including agricultural ecosystems (Chen et al., 2020), water safety (Hou et al., 2018), biogeochemical processes (Bardgett et al., 2008), and drinking water safety after earthquakes, such as Haiti earthquake of 2010 (Roy et al., 2018) and Nepal earthquake of 2015 (Uprety et al., 2017). Despite the extensive presence of soil microbial communities throughout soil profiles, the compositions of soil microbial communities after seismic events have not been reported.
Taiwan is located between the Eurasian and Philippine Sea plates and is one of the most seismically active regions in the world. Investigating the rupture behavior of earthquakes is important in the region of Taiwan, where many destructive earthquakes have occurred in the past few decades (Hwang et al., 2020). In 2010, the JiaSian earthquake struck Southern Taiwan; this earthquake was the most powerful earthquake in the region since 1900. Although the earthquake did not cause any deaths, 96 people were injured. Soil liquefaction occurred after the JiaSian earthquake. Therefore, we explored the characteristics of not only different soil depth profiles from the surface to 10 m underground but also the liquefied soil. It is important to identify liquefaction-prone areas; however, direct analyses of the microbial communities in seismic soil and soil liquefaction are lacking.
To our knowledge, this is the first seismic event study to examine bacterial abundance as it relates to soil depth, especially in deeper soil profiles. In this study, we investigated the soil depth distribution of bacterial phyla using a data set of 16S amplicons from different depths within soil and from liquefied soil that were sequenced on the Illumina MiSeq platform. We provide insights into the direct relationship between microbial communities and seismic events that could assist in detecting soil microbes that may cause soil liquefaction after earthquakes.
Results and discussion
Soil characteristics
The geological survey results from Hsin Hwa site (HH01) are shown in Table 1. The location of the sampling site was shown in Figure 1. According to the results of the geological drilling report, the in situ groundwater level was at 1.4 m depth. There were four soil layers, and their distribution is as follows.
Table 1.
Geological survey results for the Hsin Hwa area
| Depth (m) | 0–3.4 m | 3.4 m–8.1 m | 8.1 m–20.5 m | 20.05 m–35 m |
| SPT-N | 2–5 | 11–14 | 3–7 | 15–64 |
| Soil type | Brown and yellow silty clay interbedded with thin layers of fine sand | Brown and gray sandy silt interbedded with thin layers of clay | Gray silty clay and nonplastic silty sand | Gray and silt |
| USCS | CL, ML | SM | CL, CL-ML | SM, ML, CL-ML |
| ω (%) | 20.6–23.7 | 18.7–22.4 | 21.6–39.7 | 19.4–24.1 |
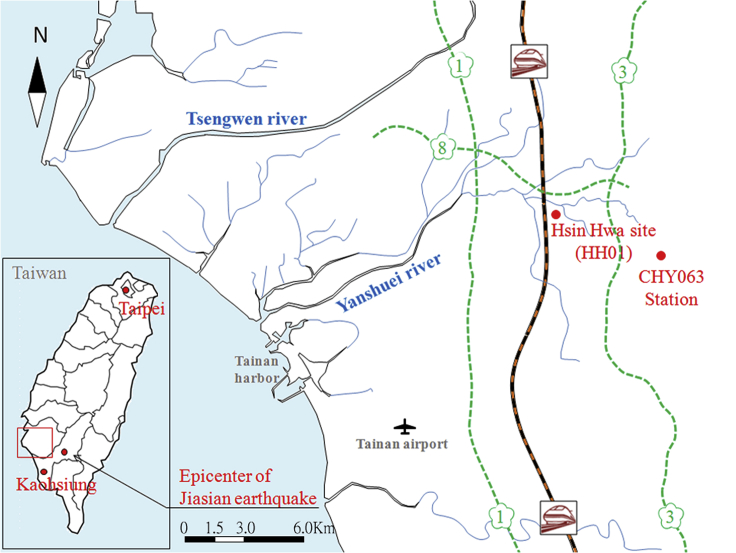
Figure 1.
The location of the sampling site
The first layer, which was silty clay interbedded with thin layers of fine sand, was classified as clay of low plasticity (CL) and low plastic silt (ML) in the Unified Soil Classification System (USCS), a soil classification system used in engineering and geology. Its distribution range was ground surface elevation (GL) of 0.0 ∼ −3.0 m, and its SPT-N value was approximately 4. The SPT-N value means the N value of the standard penetration test which mainly indicates the degree of soil hardness or compactness. Its value is about 1–50. When N value is smaller, the soil layer is softer. The harder soil layer has a larger N value. Generally speaking, when the SPT-N value is larger than 50, it means that the soil layer is hard and will not liquefy. The second layer, which was sandy silt interbedded with thin layers of clay, was classified as silty sand (SM) in the USCS. Its distribution range was GL −3.0 ∼ −9.0 m, and its SPT-N value is approximately 13. The third layer, which was silty clay and nonplastic silty sand, was classified as CL and CL-ML in the USCS. Its distribution range was located at GL −9.0 ∼ −20.0 m, and its SPT-N value was approximately 6. The fourth layer, which was sandy silt, was classified as SM in the USCS system. Its distribution range was below GL −20.0 m, and its SPT-N value was approximately 15.
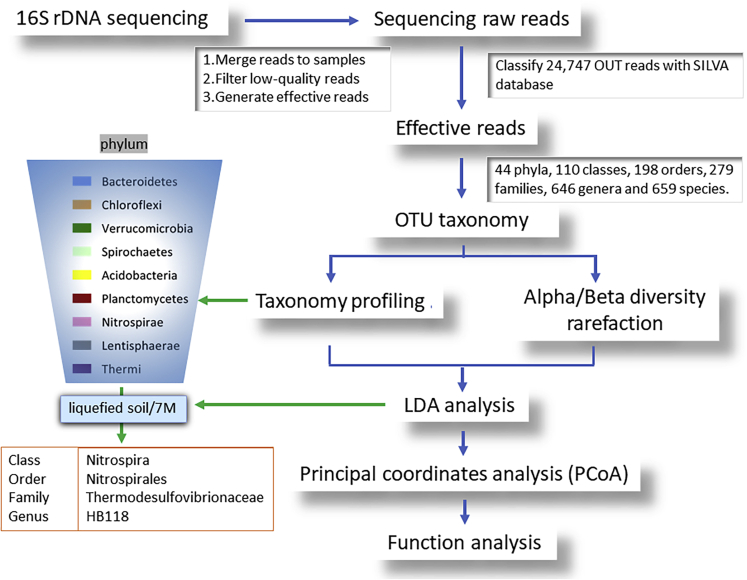
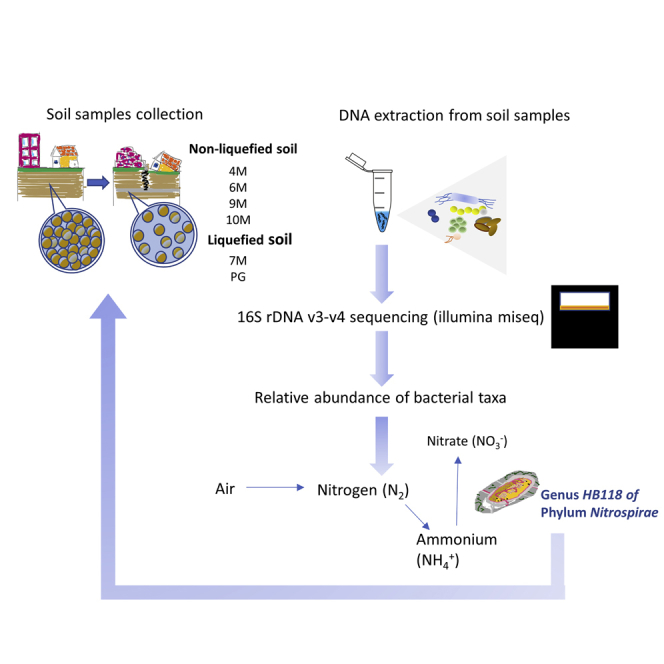
In general, the SM that was within 20 m below the surface and under the groundwater level was easily liquefied. In soil liquefaction research, dynamic triaxial tests were usually used to investigate soil liquefaction engineering properties. The dynamic triaxial test apparatus used in this research was designed by Chan et al. (Chan et al., 1976). In the test, the initial liquefaction was set as the double amplitude (DA) of axial strain exceeding 5%. This meant that the soil specimen was considered to be liquefied when the DA reached 5%, and the test was ended at that point. In accordance with the results of the dynamic triaxial test, the soil layer located between GL −6.0 m ∼ and 8.0 m has high potential for soil liquefaction. The basic physical properties of the soil in the Hsin Hwa area are shown in Table 2. We characterized the microbial community composition of liquefied soil after an earthquake, and our bioinformatics analysis pipeline is briefly shown in Figure 2.
Table 2.
The basic physical properties of soils in the Hsin Hwa area
| Depth (m) | 5M | 7M | 9M | 10M |
| Effective confining pressure (kPa) | 50 | 75 | 100 | 105 |
| Fines content, Fc (%) | 15.06–48.59 | 4.94–19.06 | 3.3–7.7 | 26.8–29.1 |
| Initial void ratio, e | 0.620–0.699 | 0.663–0.805 | 0.861–0.886 | 0.690–0.765 |
| Specific gravity, Gs | 2.60 | 2.61 | 2.62 | 2.62 |
| Moist unit weight, γm (g/cm3) | 1.85–2.03 | 1.71–1.96 | 1.85–1.88 | 1.94–2.03 |
| Dry unit weight, γd (g/cm3) | 1.53–1.61 | 1.44–1.56 | 1.39–1.40 | 1.48–1.55 |
| Moisture content, ω(%) | 21.1–23.9 | 17.0–28.9 | 33.49 | 30.69 |
| Liquid limit, LL (%) | 25.36 | 24.54 | 15.41 | 18.76 |
| Plastic limit, PL (%) | NP | NP | NP | NP |
| USCS | SM | SM | SM | SM |
Figure 2.
The illustration of the bioinformatics analysis pipeline
Alpha diversity of the different soil depths
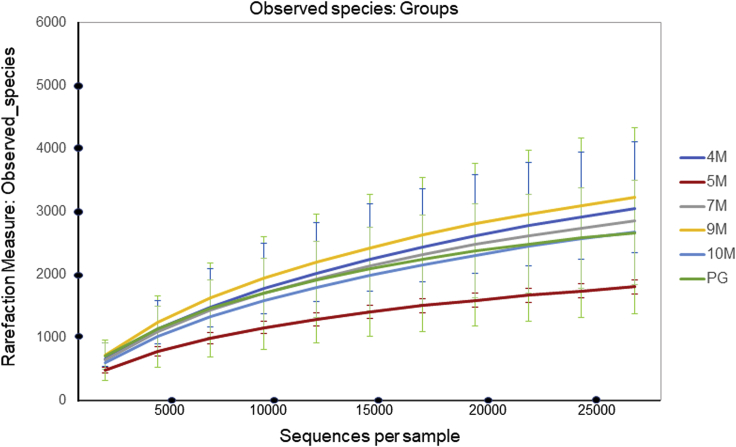
All amplified rarefaction curves increased from 0 to 10,000 sequences but tended to plateau when they reached 20,000 sequencing reads, which indicated that the diversity and richness in our study were sufficient to characterize the species in every sample (Figure 3). The bacterial community richness and diversity at different depths were shown (Table 3). Chao1 and abundance-based coverage estimator (ACE) were used as the richness indexes for estimating the richness from the abundance data. The Simpson and Shannon indexes allowed us to monitor the diversity indexes in each individual sample. There were no significant differences in the Shannon and Simpson indexes among the different soil depths. However, the 5M and PG layers had slightly lower Chao1 and ACE values than the other soil layers, which meant that the 5M and PG layers had lower bacterial community richness. PG samples, the liquefied soil in this study, were obtained when soil layer under the ground surface was liquefied and ejected out along the cracks to ground surface.
Figure 3.
Alpha diversity, measured by observed species
4M–10M indicate the depth of the soil collected. The soil sample used in this study, No. PG, is a sample of the liquefied soil expelled onto the ground after soil liquefaction. Data were expressed as mean ± standard error.
Table 3.
OTU richness and diversity indexes of different soil depth profiles
| Group | Chao1 | Shannon | Simpson | ACE |
|---|---|---|---|---|
| 4M | 4784.601 | 8.150 | 0.969 | 5079.479 |
| 5M | 2373.743 | 7.093 | 0.956 | 2488.49 |
| 7M | 4087.754 | 8.064 | 0.974 | 4342.043 |
| 9M | 4499.078 | 8.257 | 0.970 | 4835.748 |
| 10M | 4131.202 | 7.921 | 0.976 | 4397.975 |
| PG | 3283.383 | 8.596 | 0.979 | 3533.861 |
Microbial composition by soil depth and in liquefied soil
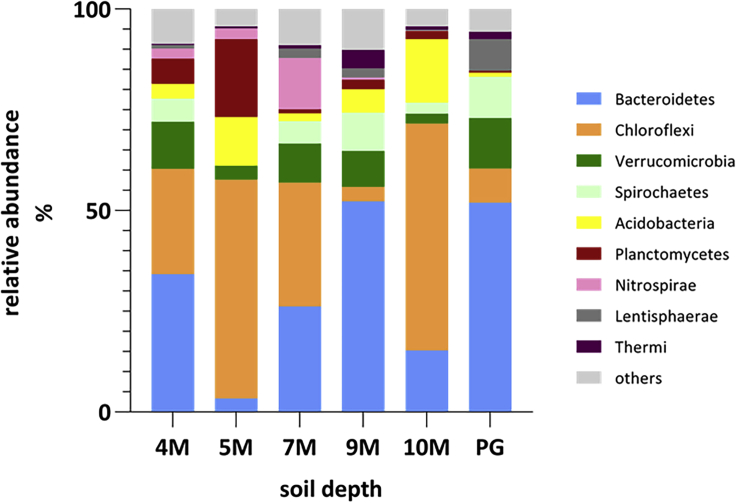
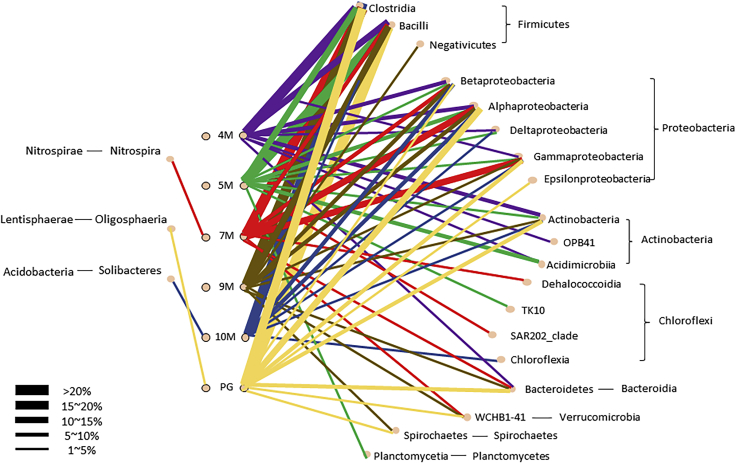
To assess whether the microbial community composition changed at different soil depths after earthquakes, three samples were collected along a soil depth gradient. From a total of 24,747 Operational taxonomic units (OTUs), only a few reads were classified as archaea. After removing singletons (OTUs consisting of a maximum of one sequence read), 99.96% of the sequence reads in 18 soil samples were classified as bacteria and assigned to 44 phyla, 110 classes, 198 orders, 279 families, 646 genera, and 659 species. In general, the dominant phyla in the soil bacterial communities were Proteobacteria, Acidobacteria, Actinobacteria, Verrucomicrobia, Bacteroides, Chloroflexi, Planctomycetes, and Gemmatimonadetes (Asano et al., 2020; Janssen, 2006). Table S1 showed the top 3 abundant bacterial phyla in our study, and the ranked order of the predominant phyla was Firmicutes (59.8%), followed by Proteobacteria (21.34%), Actinobacteria (6.27%), Bacteroidetes (4.21%), Chloroflexi (3.31%), Verrucomicrobia (1.12%), Spirochaetes (0.80%), Acidobacteria (0.67%), Planctomycetes (0.54%), Nitrospirae (0.47%), and Lentisphaerae (0.37%). The remaining phyla represented 0.9% of the total samples (Figure 4A; see also Figure S1). Previous studies have shown that bacterial phyla change in relative abundance with soil depth. Firmicutes was the most dominant phylum across all samples. Proteobacteria was the second most abundant phylum, and its abundance peaked slightly in the 7M layer. Proteobacteria was found in a study of soil that had undergone long-term fertilization and was the dominant phylum in the deep soil (Li et al., 2014). The relative abundance of Actinobacteria decreased with soil depth but slightly increased in the PG layer. Actinobacteria was known for its cellulose degradation capability and abundance in terrestrial and aquatic environments (Servin et al., 2008). Actinobacteria were economically important to humans because agriculture and forests depend on their contributions to soil systems. They helped to digest the organic material in dead organisms so that the nutrients can be taken up by plants. Some soil Actinobacteria can fix nitrogen for plants and in turn utilizing carbon compounds such as sugars and plaint origin. Among the most abundance phylum Actinobacteria, class OPB41 was observed in 4M layer. Phylum Chloroflexi accounted for 16% of the bacterial community in the 5M and 7M layers, respectively. The abundance of Bacteroidetes showed no correlation with soil depth in our study. Bacteroidetes were usually found in surface soils owing to the availability of organic carbon (Fierer et al., 2007). Verrucomicrobia and Spirochaetes did not show clear changes in abundance across profiles. The relative abundance of Planctomycetes increased slightly in the 5M layer, but they were rarely detected in the 7M layer. Planctomycetia was significantly correlated to potential nitrification rate with the increment of N fertilization rates (Liao et al., 2020). It has been reported that Planctomycetes derive their energy for growth from the conversion of ammonium and nitrite into dinitrogen gas in the complete absence of oxygen, thereby regulating the nitrogen cycle (Delmont et al., 2018). The previous study reported that sediment Planctomycetes transitioned to aquatic environments, where they induce new freshwater-specific clades (Andrei et al., 2019). Besides, Planctomycetia of phylum Planctomycetes class TK10 of phylum Chloroflexi was unique in 5M. TK10 was also recorded to be enriched in sludge-amended soils (Rutgersson et al., 2020). Class Solibacteres of phylum Acidobacteria was found distinct than other soil layers. Although more than 12,000 distinct phylotypes and more than 6,500 species OTUs had been reported in phylum Acidobacteria so far, it was described by only 56 cultivable species belonging to 28 genera (Dedysh and Yilmaz, 2018). It was observed in the 10M layer; however, it had limited information about their biological traits. Phylum Lentisphaerae, class Oligosphaeria is unique in the PG layer. The class Oligosphaeria is gram-negative bacteria and mostly obtained from uncultivated bacteria retrieved mainly from mesophilic anaerobic digester sludges and landfill leachate (Qiu et al., 2013).
Figure 4.
The most abundant bacterial phyla in the soil samples, with the 3 most abundant phyla removed
Candidate microbes are relatively more abundant in liquefied soil
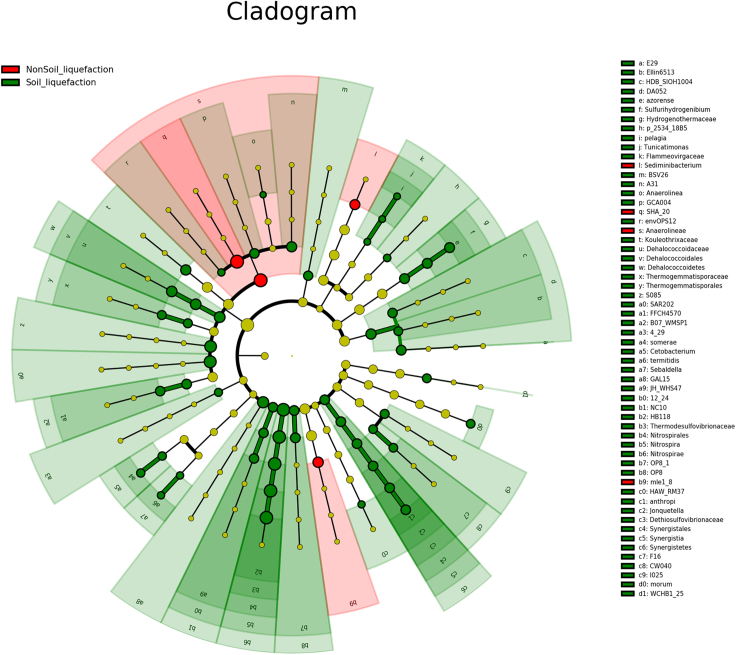
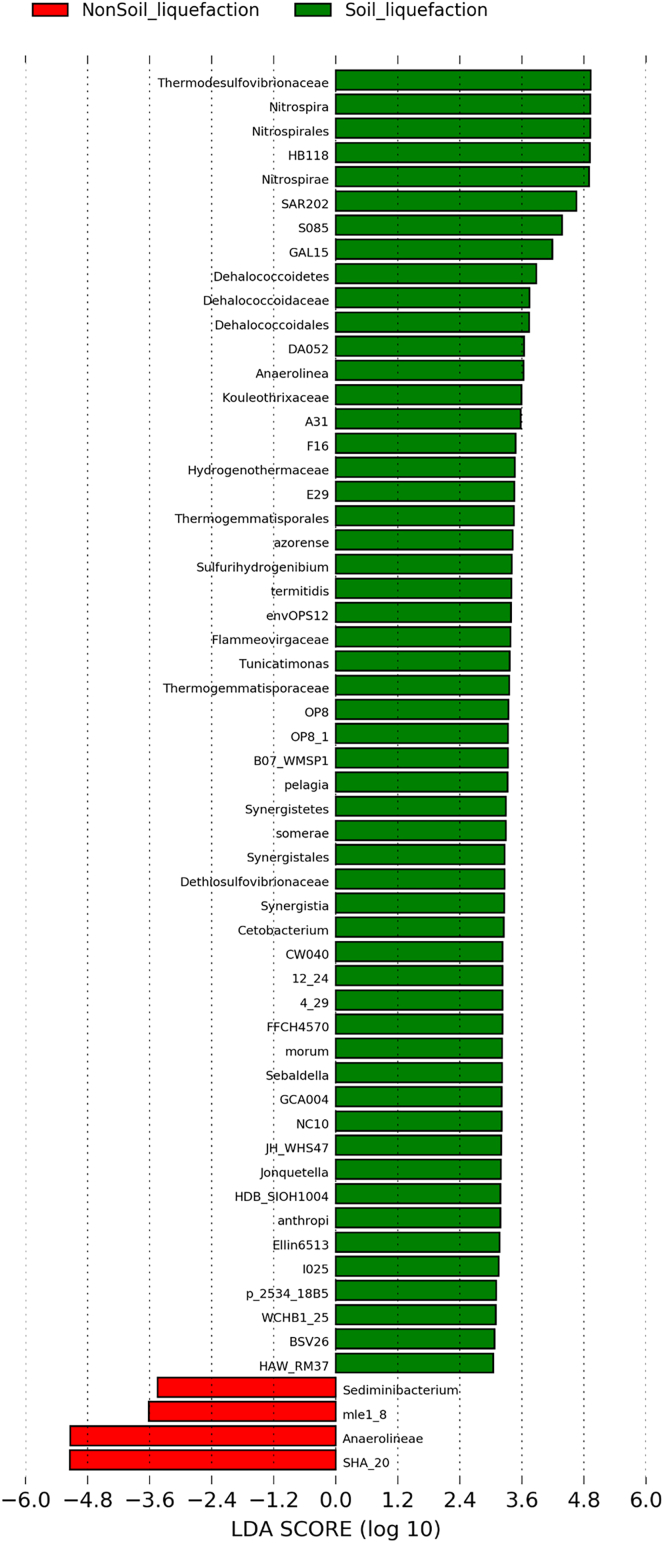
The unique bacterial phyla identified in this study were Nitrospirae (2.23%) and GAL15 (0.31%) (Table S2). The relative abundances of Nitrospirae and GAL15 were significantly higher in the 7M layer, which was the liquefied soil layer, than in any other layer. Nitrospirae contains only one class, Nitrospira, which was significantly higher in the 7M layer (p < 0.05) (Figure 5). Phylum Lentisphaerae, class Oligosphaeria and Phylum Acidobacteria, class Solibacteres were not significant different, respectively, in the PG and 10M layers (p > 0.05). Examination of differences in microbiota composition using linear discriminant analysis effect size (LEfSe) was done to compare the estimated samples of soil liquefaction (7M) and nonsoil liquefaction (4M, 5M, 9M and 10M). The cladograms showed the soil liquefaction microbial communities were diverse compared with those in nonsoil liquefaction. The results indicated differences in the phylogenetic distributions of the microbiotas of two groups (Figure 6). In our study, family Thermodesulfovibrionaceae was enriched in liquefied soil, followed by abundance of Nitrospira, Nitrospirales, and genus level HB118 (Figure 7). Nonliquefied soil microbiota was more enriched for order level SHA 20 and class level Anaerolineae (linear discriminant analysis [LDA] score [log10] > 3, p < 0.05).
Figure 5.
Connection diagram between different soil layers and relative proportion of bacterial class
Phylum Nitrospirae, class Nitrospira is only present in 7M, whereas Phylum Lentisphaerae, class Oligosphaeria and Phylum Acidobacteria, class Solibacteres are present respectively in PG and 10M without a significant difference (p > 0.05).
Figure 6.
Cladogram using the Linear discriminant analysis Effect size (LEfSe) method indicating the phylogenetic distribution
Cladogram using the linear discriminant analysis effect size (LEfSe) method indicating the phylogenetic distribution of microbiota associated with liquefied and nonliquefied soil samples A cladogram for differentially distributed taxa (p < 0.05, LDA > 3) between 16S rDNA regions in liquefied soil samples (green, n = 3) and nonliquefied soil samples (red, n = 12). The brightness of each dot is proportional to its effect size, and the nodes of taxa which were not significantly differentially represented were colored yellow.
Figure 7.
Linear discriminant analysis (LDA) effect size (LEfSe) analysis revealed significant bacterial differences in liquefied soil and nonliquefied soil
Taxa were indicated with a positive LDA score (green), and taxa enriched in nonliquefied soil have a negative score (red). (LDA score [log10] > 3, p < 0.05).
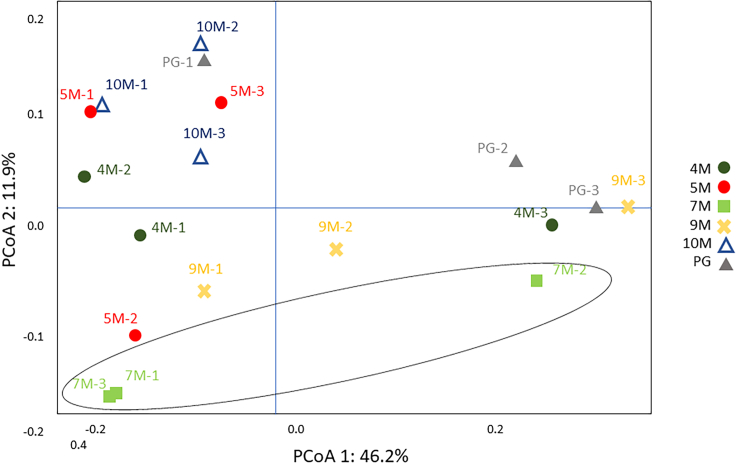
The Principal coordinate analysis (PCoA) results showed that the 7M layer was quite different from the other layers, and the two dimensions, PCoA1 and PCoA2, explained 46.2% and 11.9% of the variation, respectively (Figure 8). The Nitrospirae phylum is known to play a pivotal role in nitrification. The nitrite oxidizing process involves the oxidation of ammonia via nitrite to nitrate and is considered to be a two-step process catalyzed by chemolithoautotrophic microorganisms oxidizing either ammonia or nitrite (Daims et al., 2015; Daims and Wagner, 2018). Nitrospirae have been detected predominantly in marine, in postearthquake environments (Baskaran et al., 2020; Uprety et al., 2017) in transitional areas from desert to oasis (Li et al., 2015), and from noncultivated soil to cultivated soil.
Figure 8.
Principal coordinate analysis (PCoA) of the samples from the SILVA database with weighted UniFrac distance
Characteristic functional profiles of the dominant bacterial phylum
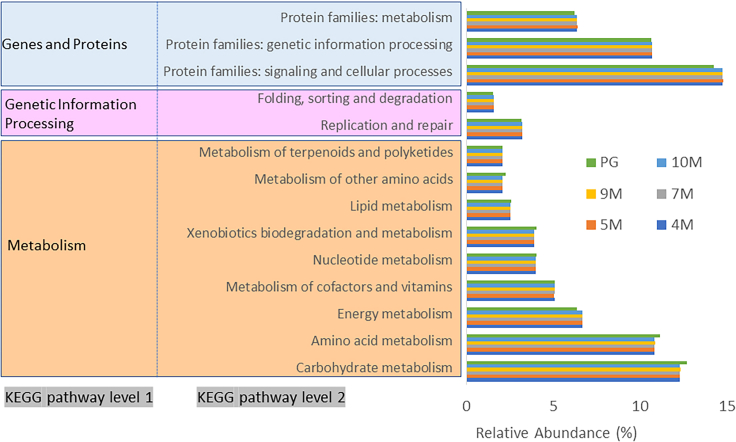
On the basis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, the functional behavior of the microbes present in the different soil layer samples was determined and is shown in Figure S2. Some pathways, such as protein families: signaling and cellular processes, carbohydrate metabolism, amino acid metabolism, protein families: genetic information processing, energy metabolism, and protein families: metabolism, were predominantly enriched at every depth. However, there were no significant differences between soil layers. The percentage of OTUs vs. pathways was plotted using PICRUSt software (Figure 9). Environmental information processing and membrane-transported signal transduction are highly enriched in microbiomes (Sun et al., 2020). Signal transduction is similar to the cascade when chemical or physical signals are transmitted through a cell as a series of molecular events (Kleerebezem et al., 1997). These systems regulate physiological processes based on environmental changes and enable microbiomes to adapt to earthquakes. Carbohydrate metabolism is responsible for the formation, breakdown, and conversion of carbohydrates in every organism. The main carbohydrate compound is glucose, which participates in multiple metabolic pathways. The most dominant phyla in our study, Firmicutes, Proteobacteria, and Actinobacteria, are also responsible for multiple N cycling processes; their contributions to different processes are different, and their relative abundances may determine the strength of nitrification (Delmont et al., 2018; Gtari et al., 2012; Jeong and Bae, 2021; Nishihara et al., 2018). The uptake of various N forms, such as ammonia, ammonium salts, nitrite, nitrate, and other inorganic nitrogen forms, from the soil by the bacterial community to produce proteins and nitrogen-containing substances activates amino acid metabolism.
Figure 9.
The relative abundance of the top 10 pathways from the KEGG enrichment analysis at different soil depths predicted with PICRUSt
Relationships between the phylum Nitrospirae and soil liquefaction
In geotechnical engineering studies, different types of microbially mediated processes, such as combined biodesaturation, biocementation, and bioclogging methods, were used to mitigate soil liquefaction hazards (Haryati et al., 2018; Ivanov et al., 2019). Biocementation is a newly developed method that applies microbiological activity to improve the engineering properties of soils (Mujah et al., 2016) and is based on the simultaneous production of nitrogen gas by denitrifying anaerobic bacteria. Soil desaturation involves injecting water-saturated soils with insoluble gas bubbles to mitigate soil liquefaction during earthquakes (Ivanov et al., 2019). The relative abundance of nitrifying bacteria, Nitrospirae, observed in liquefying soil at 7M is in accordance with the results of a previous biotechnological study, further confirming that Nitrospirae tend to be present when liquefaction occurs. However, in the PG layer, no Nitrospirae were detected.
The family Nitrospiraceae consists of the genera Nitrospira, Leptospirillum, Candidatus Magnetobacterium, Candidatus Magnetoovum, and Thermodesulfovibrio. The genus Thermodesulfovibrio represents 89% reads of the phylum Nitrospirae and was by far the most abundant family in this study after removal of the 3 most dominant bacteria. Thermodesulfovibrio is an obligate anaerobic thermophilic genus that plays an important role in sulfate reduction and growth on limited organic substrates with lactate and pyruvate (Adjeroud et al., 2020; Frank et al., 2016). Sulfate-reducing bacteria are very common in aquatic and terrestrial environments that become anoxic owing to microbial decomposition processes (Madigan and Martinko, 2005). The growth temperature range for these bacteria is between 40 and 70°C, and the acceptable pH generally ranges from 6.5 to 8.5 (Daims, 2014). Among the Thermodesulfovibrio members, HB118 represents 99% of the genus and is the most abundant species in the family. HB118 can be found in different soil environments (Kumar et al., 2017) and has been reported to be an important contributor to soil phosphorus metabolism (Torres-Cortes et al., 2015). Thus, we speculate that HB118 has a very strong relationship to liquefaction. This is the first study that sets up a potential microbial biomarker as an alternative model to detect the seismic soil liquefaction.
Conclusions
Soil liquefaction after seismic events is a substantial threat to both properties and human lives. We had the extraordinary opportunity to obtain a deep soil profile after the 2010 JiaSian earthquake and to determine the microbiota of one significant species in the liquefied soil. In this study, we provide valuable insights that identify useful markers for the soil liquefaction monitoring. Microbial communities are strongly influenced by soil properties, which subsequently affect microbial diversity and function in soil. This relationship gives scientific researchers and industry a tool for monitoring microbial activities after seismic events.
Limitations of the study
The composition of bacterial communities can vary owing to many factors, such as pH, temperature, moisture, and pressure. Soil sample collection cannot always be performed in a timely manner, and the cost of geological drilling is high. Meanwhile, it was very hard to get good control data in general because earthquakes cannot be predictable. Thus, it was difficult to detect a normal soil profile. The discrepancies between our soil profile study and previous studies may be because of the type of soil material we considered. Our findings described earlier are exciting and promising for future studies. Despite the considerable interesting and positive results obtained so far, there is still some room for improvement in our study methods.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Different soil layers | This paper | NA |
| Chemicals, peptides, and recombinant proteins | ||
| KAPA HiFi PCR Kit | KapaBiosystems | KK2103 |
| DNeasy PowerSoil Kit | QIAGEN | Cat. No.: 12888 |
| QIAquick PCR Purification Kit | QIAGEN | Cat. No.: 28104 |
| AMPure XP beads | Beckman Coulter, Inc. | Item No: A63880 |
| MinEluteGel Extraction Kit | QIAGEN | Cat. No.: 28604 |
| Critical commercial assays | ||
| Celero DNA-Seq | NuGEN | 0360-24 |
| Deposited data | ||
| Raw and analyzed data | This paper | PRJNA704838 |
| SILVA reference database (release version 128) | Quast et al., 2013 | https://www.arb-silva.de/ |
| Oligonucleotides | ||
| Primer 341F: CCTACGGGNGGCWGCAG | This paper | NA |
| Primer 805R: GACTACHVGGGTATCTAATCC | This paper | NA |
| Software and algorithms | ||
| Mothur | Schloss et al., 2009 | https://mothur.org/wiki/mothur_manual/ |
| UCHIME | Edgar et al., 2011 | http://drive5.com/uchime |
| QIIME | Caporaso et al., 2010 | http://qiime.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be filled up by the lead contact, Yi Chiung, Hsu (syic@ncu.edu.tw).
Materials availability
This work did not generate new unique reagents.
Experimental model and subject details
Any animals, human subjects, plants, microbe strains, cell lines, primary cell cultures were not used in the study.
Acknowledgments
We would like to thank Dr. Jia-Jyun Dong for reviewing the article. This study was supported by grants from the Ministry of Science and Technology (MOST 108-2314-B-008-00 and MOST 109-2622-E-008-023).
Author contributions
Conceptualization, Y.C.H., Y.T.C, and J.W.C.; Methodology, Y.C.H.; Formal Analysis, Y.C.H and S.H.L.; Investigation, Y.C.H., J.W.C., C.C.C. W.F.L., and Y.T.C.; Data Curation, Y.C.H., Y.T.C., and S.H.L.; Writing – Original Draft, Y.T.C, M.H.C., and Y.C.H.; Writing – Review and Editing, Y.C.H., Y.T.C., and M.H.C.; Funding Acquisition, Y.C.H.; Supervision, Y.C.H.
Declaration of interests
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102984.
Supplemental information
Data and code availability
The data are available upon request by contacting Lead Contact, Yi Chiung, Hsu (syic@ncu.edu.tw). No new code was generated during this study. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adjeroud M., Escuder-Rodríguez J.-J., González-Siso M.-I., Kecha M. Metagenomic investigation of bacterial and archaeal diversity of Hammam Essalihine hot spring from Khenchela, Algeria. Geomicrobiol. J. 2020;37:804–817. [Google Scholar]
- Andrei A.S., Salcher M.M., Mehrshad M., Rychtecky P., Znachor P., Ghai R. Niche-directed evolution modulates genome architecture in freshwater Planctomycetes. ISME J. 2019;13:1056–1071. doi: 10.1038/s41396-018-0332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano R., Hayakawa A., Fukushima J., Nakai Y., Shimura Y., Abe M., Inamoto T. Changes in bacterial communities in seawater-flooded soil in the four years after the 2011 Tohoku Tsunami in Japan. J. Mar. Sci. Eng. 2020;8:76. [Google Scholar]
- Bardgett R.D., Freeman C., Ostle N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008;2:805–814. doi: 10.1038/ismej.2008.58. [DOI] [PubMed] [Google Scholar]
- Baskaran V., Patil P.K., Antony M.L., Avunje S., Nagaraju V.T., Ghate S.D., Nathamuni S., Dineshkumar N., Alavandi S.V., Vijayan K.K. Microbial community profiling of ammonia and nitrite oxidizing bacterial enrichments from brackishwater ecosystems for mitigating nitrogen species. Sci. Rep. 2020;10:5201. doi: 10.1038/s41598-020-62183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglia M., Fierro T., Santucci de Magistris F. Pipeline performances under earthquake-induced soil liquefaction: State of the art on real observations, model tests, and numerical simulations. Shock Vib. 2020;2020:1–20. [Google Scholar]
- Chan C.K., ASCE M., Mulilis J.P. Pneumatic sinusoidal loading system. J. Geotech. Geoenviron. Eng. 1976;102:277–282. [Google Scholar]
- Chen Q.L., Ding J., Zhu Y.G., He J.Z., Hu H.W. Soil bacterial taxonomic diversity is critical to maintaining the plant productivity. Environ. Int. 2020;140:105766. doi: 10.1016/j.envint.2020.105766. [DOI] [PubMed] [Google Scholar]
- Daims H. The Prokaryotes. 2014. The family Nitrospiraceae; pp. 733–749. [Google Scholar]
- Daims H., Lebedeva E.V., Pjevac P., Han P., Herbold C., Albertsen M., Jehmlich N., Palatinszky M., Vierheilig J., Bulaev A. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Wagner M. Nitrospira. Trends Microbiol. 2018;26:462–463. doi: 10.1016/j.tim.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Dedysh S.N., Yilmaz P. Refining the taxonomic structure of the phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018;68:3796–3806. doi: 10.1099/ijsem.0.003062. [DOI] [PubMed] [Google Scholar]
- Delmont T.O., Quince C., Shaiber A., Esen O.C., Lee S.T., Rappe M.S., McLellan S.L., Lucker S., Eren A.M. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 2018;3:804–813. doi: 10.1038/s41564-018-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobry R., Abdoun T. Recent findings on liquefaction triggering in clean and silty sands during earthquakes. J. Geotech. Geoenviron. Eng. 2017;143 040170771–040170719. [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Bradford M.A., Jackson R.B. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Frank Y.A., Kadnikov V.V., Lukina A.P., Banks D., Beletsky A.V., Mardanov A.V., Sen'kina E.I., Avakyan M.R., Karnachuk O.V., Ravin N.V. Characterization and genome analysis of the first facultatively alkaliphilic Thermodesulfovibrio isolated from the deep terrestrial subsurface. Front. Microbiol. 2016;7:2000. doi: 10.3389/fmicb.2016.02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gtari M., Ghodhbane-Gtari F., Nouioui I., Beauchemin N., Tisa L.S. Phylogenetic perspectives of nitrogen-fixing actinobacteria. Arch. Microbiol. 2012;194:3–11. doi: 10.1007/s00203-011-0733-6. [DOI] [PubMed] [Google Scholar]
- Haryati Y., Zango M.U., Kassim K.A., Mohammed A.S., Nor Zurairahetty M.Y., Izni Syahrizal I. Bio-desaturation and bio-sealing techniques for mitigation of soil liquefaction: A review. MATEC Web Conf. 2018;250:1–13. [Google Scholar]
- Hou L., Zhou Q., Wu Q., Gu Q., Sun M., Zhang J. Spatiotemporal changes in bacterial community and microbial activity in a full-scale drinking water treatment plant. Sci. Total Environ. 2018;625:449–459. doi: 10.1016/j.scitotenv.2017.12.301. [DOI] [PubMed] [Google Scholar]
- Hwang R.-D., Ho C.-Y., Lin T.-W., Chang W.-Y., Huang Y.-L., Lin C.-Y., Lin C.-Y. Relationship between seismic moment and source duration for seismogenic earthquakes in Taiwan: Implications for the product of static stress drop and the cube of rupture velocity. Pure Appl. Geophys. 2020;177:3191–3203. [Google Scholar]
- Ivanov V., Stabnikov V., Stabnikova O., Kawasaki S. Environmental safety and biosafety in construction biotechnology. World J. Microbiol. Biotechnol. 2019;35:26. doi: 10.1007/s11274-019-2598-9. [DOI] [PubMed] [Google Scholar]
- Janssen P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D., Bae H. Insight into functionally active bacteria in nitrification following Na(+) and Mg(2+) exposure based on 16S rDNA and 16S rRNA sequencing. Sci. Total Environ. 2021;758:143592. doi: 10.1016/j.scitotenv.2020.143592. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M., Quadri L.E., Kuipers O.P., de Vos W.M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- Kumar R., Verma H., Haider S., Bajaj A., Sood U., Ponnusamy K., Nagar S., Shakarad M.N., Negi R.K., Singh Y. Comparative genomic analysis reveals habitat-specific genes and regulatory hubs within the genus Novosphingobium. mSystems. 2017;2:e00020-17. doi: 10.1128/mSystems.00020-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Yan K., Tang L., Jia Z., Li Y. Change in deep soil microbial communities due to long-term fertilization. Soil Biol. Biochem. 2014;75:264–272. [Google Scholar]
- Lashkari A., Karimi A., Fakharian K., Kaviani-Hamedani F. Prediction of undrained behavior of isotropically and anisotropically consolidated Firoozkuh sand: Instability and flow liquefaction. Int. J. Geomech. 2017;17 040170831–40170817. [Google Scholar]
- Li C.H., Tang L.S., Jia Z.J., Li Y. Profile changes in the soil microbial community when desert becomes oasis. PLoS One. 2015;10:e0139626. doi: 10.1371/journal.pone.0139626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Qin F., Wang K., Zhang Y., Hao X., Chen W., Huang Q. Long-term chemical fertilization-driving changes in soil autotrophic microbial community depresses soil CO2 fixation in a Mollisol. Sci. Total Environ. 2020;748:141317. doi: 10.1016/j.scitotenv.2020.141317. [DOI] [PubMed] [Google Scholar]
- Madigan M.T., Martinko J.M. Brock biology of microorganisms. Int. Microbiol. 2005;9:149–152. [Google Scholar]
- Mujah D., Shahin M.A., Cheng L. State-of-the-Art review of biocementation by microbially induced calcite precipitation (MICP) for soil stabilization. Geomicrobiol. J. 2016;34:524–537. [Google Scholar]
- Nishihara A., Thiel V., Matsuura K., McGlynn S.E., Haruta S. Phylogenetic diversity of nitrogenase reductase genes and possible nitrogen-fixing bacteria in thermophilic chemosynthetic microbial communities in Nakabusa hot springs. Microbes Environ. 2018;33:357–365. doi: 10.1264/jsme2.ME18030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y.L., Muramatsu M., Hanada S., Kamagata Y., Guo R.B., Sekiguchi Y. Oligosphaera ethanolica gen. nov., sp. nov., an anaerobic, carbohydrate-fermenting bacterium isolated from methanogenic sludge, and description of Oligosphaeria classis nov. in the phylum Lentisphaerae. Int. J. Syst. Evol. Microbiol. 2013;63:533–539. doi: 10.1099/ijs.0.039545-0. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gks1219. D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M.A., Arnaud J.M., Jasmin P.M., Hamner S., Hasan N.A., Colwell R.R., Ford T.E. A metagenomic approach to evaluating surface water quality in Haiti. Int. J. Environ. Res. Public Health. 2018;15:1–16. doi: 10.3390/ijerph15102211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutgersson C., Ebmeyer S., Lassen S.B., Karkman A., Fick J., Kristiansson E., Brandt K.K., Flach C.F., Larsson D.G.J. Long-term application of Swedish sewage sludge on farmland does not cause clear changes in the soil bacterial resistome. Environ. Int. 2020;137:105339. doi: 10.1016/j.envint.2019.105339. [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin J.A., Herbold C.W., Skophammer R.G., Lake J.A. Evidence excluding the root of the tree of life from the actinobacteria. Mol. Biol. Evol. 2008;25:1–4. doi: 10.1093/molbev/msm249. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang L., Pei J., Huang L.F. Regulatory relationship between quality variation and environment of Cistanche deserticola in three ecotypes based on soil microbiome analysis. Sci. Rep. 2020;10:6662. doi: 10.1038/s41598-020-63607-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cortes G., Ghignone S., Bonfante P., Schussler A. Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: Transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7785–7790. doi: 10.1073/pnas.1501540112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprety S., Hong P.Y., Sadik N., Dangol B., Adhikari R., Jutla A., Shisler J.L., Degnan P., Nguyen T.H. The effect of the 2015 earthquake on the bacterial community compositions in water in Nepal. Front. Microbiol. 2017;8:2380. doi: 10.3389/fmicb.2017.02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang Y. An ensemble method to improve prediction of earthquake-induced soil liquefaction: A multi-dataset study. Neural Comput. Appl. 2020;33:1533–1546. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available upon request by contacting Lead Contact, Yi Chiung, Hsu (syic@ncu.edu.tw). No new code was generated during this study. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.