Abstract
Background
Biliary atresia (BA) is the most common obstructive cholangiopathy in neonates, often progressing to end-stage cirrhosis. BA pathogenesis is believed to be multifactorial, but the genetic contribution, especially for nonsyndromic BA (common form: > 85%) remains poorly defined.
Methods
We conducted whole exome sequencing on 89 nonsyndromic BA trios to identify rare variants contributing to BA etiology. Functional evaluation using patients’ liver biopsies, human cell and zebrafish models were performed. Clinical impact on respiratory system was assessed with clinical evaluation, nasal nitric oxide (nNO), high speed video analysis and transmission electron microscopy.
Findings
We detected rare, deleterious de novo or biallelic variants in liver-expressed ciliary genes in 31.5% (28/89) of the BA patients. Burden test revealed 2.6-fold (odds ratio (OR) [95% confidence intervals (CI)]= 2.58 [1.15–6.07], adjusted p = 0.034) over-representation of rare, deleterious mutations in liver-expressed ciliary gene set in patients compared to controls. Functional analyses further demonstrated absence of cilia in the BA livers with KIF3B and TTC17 mutations, and knockdown of PCNT, KIF3B and TTC17 in human control fibroblasts and cholangiocytes resulted in reduced number of cilia. Additionally, CRISPR/Cas9-engineered zebrafish knockouts of KIF3B, PCNT and TTC17 displayed reduced biliary flow. Abnormally low level of nNO was detected in 80% (8/10) of BA patients carrying deleterious ciliary mutations, implicating the intrinsic ciliary defects.
Interpretation
Our findings support strong genetic susceptibility for nonsyndromic BA. Ciliary gene mutations leading to cholangiocyte cilia malformation and dysfunction could be a key biological mechanism in BA pathogenesis.
Funding
The study is supported by General Research Fund, HMRF Commissioned Paediatric Research at HKCH and Li Ka Shing Faculty of Medicine Enhanced New Staff Start-up Fund.
Keywords: Biliary atresia, Whole exome sequencing, Rare variants, Cilia dysfunction
Research in context.
Evidence before this study
Biliary atresia (BA) is a rare, complex hepatobiliary disorder that usually presents during the first few weeks of life. Surgery is the only treatment, yet the outcome is notoriously poor, with less than 60% of patients having long term survival with native liver. Genetics and gene expression studies suggested that the genetic vulnerability of the patient is critical for BA development; however, the genetic etiology of BA remains largely unexplored. Thus far, only one single gene has been linked to syndromic form of BA while a few common variants were associated with increased risk of nonsyndromic BA.
Added value of this study
Using trio-based whole exome sequencing, this study for the first time demonstrated rare variant contribution to nonsyndromic BA, such that rare, de novo or biallelic deleterious variants in liver-expressed ciliary genes was associated with a significant two-fold increased risk of BA. This finding suggests that deleterious ciliary mutations may underlie the cholangiocyte cilia abnormalities observed in livers of nonsyndromic BA patients. Depletion of candidate ciliary genes can induce BA-like phenotype of reduced bile flow in zebrafish models, implying that ciliary malformation and dysfunction could be a novel disease mechanism of BA. The low nasal nitric oxide levels detected in most of the BA patients carrying ciliary mutations suggests a genetic predisposition to multisystem ciliary abnormalities.
Implication of all the available evidence
This study provides novel evidence that genetic factors play a major role in a substantial proportion of nonsyndromic BA cases, and a strong genetic basis for the previously reported cholangiocyte ciliary abnormalities. Rare deleterious ciliary mutations predisposed to higher risk of nonsyndromic BA. The predisposition to ciliary dysfunction may act beyond the hepatobiliary system and genetic screening for deleterious mutations in the cilia gene set would allow patient stratification to further assess the pleiotropic clinical features and to improve clinical management. As reflected by the high genetic heterogeneity of ciliary mutations uncovered in this study, the nonsyndromic BA is unlikely to have a single genetic etiology. Patient stratification aided by transcriptomic and immune profiling may help leverage statistical power and elucidate the polygenic etiology of BA.
Alt-text: Unlabelled box
1. Introduction
Biliary Atresia (BA) is a major cause of neonatal cholestasis, characterized by progressive fibrosclerosing and inflammatory obliteration of the biliary system during the first few weeks of life. BA is rare and varies widely in incidence among populations, being most common in East Asia (1 in 5000 live births in Asians, compared to 1 in 18,000 live births in Caucasians) [1]. The occurrence of BA is mostly sporadic, though a limited number of familial cases have been reported previously [2]. The only current treatment is the Kasai portoenterostomy to restore bile flow. Yet a high proportion of patients, even when bile flow is reestablished, still develop progressive inflammation and sclerosis in the intrahepatic biliary tree, leading to secondary liver cirrhosis. For these patients and those with failed portoenterostomy, liver transplantation is the only treatment option. In neonates, BA is the most common indication for liver transplantation.
BA can be classified into two major groups. The syndromic form, found in 5–20% of BA cases, is characterized by the coexistence of other major congenital anomalies, most commonly laterality defects and splenic malformation. This could be attributed to defective morphogenesis of the bile duct during embryonic development and chromosomal anomalies are a possible genetic origin [3,4]. The nonsyndromic form of BA accounts for over 80% of cases, often presenting with later onset of pale stools and jaundice compared to the syndromic form. Nonsyndromic BA is thought to be caused by a combination of environmental insults and genetic predisposition [5]. It is hypothesized that a still unknown exogenous factor, possibly viral infection (e.g. cytomegalovirus) or toxins (e.g. biliatresone), meets the innate immune system of a genetically predisposed individual and thereby induces an uncontrollable and potentially self-limiting immune response, which manifests as liver fibrosis and atresia of the bile ducts [6,7]. To explore the genetic contribution to BA, we conducted the first genome-wide association studies on nonsyndromic BA and identified ADD3 as a susceptibility gene, showing that common genetic variants could alter disease risk by affecting ADD3 expression levels in liver [8,9]. Since then, there has been growing evidence that common genetic variations contribute to BA risk, with the identification of the additional susceptibility loci GPC [10], EFEMP1 [11] and ARF6 [12]. Recent whole exome sequencing (WES) study on syndromic patients with biliary atresia splenic malformation (BASM) uncovered potentially damaging rare variants in polycystic kidney disease 1 like 1 (PKD1L1) involved in laterality determination and ciliary function, which might account for the syndromic presentation of BASM [13]. Yet, convincing evidence that genetic factors act beyond common variant genetic susceptibility in the pathogenesis of nonsyndromic BA remains to be found [14].
We hypothesized that rare coding variants predispose not only to syndromic but also to nonsyndromic BA. Here, we report a trio-based WES study on 89 BA trios (affected proband and unaffected parents), which utilizes the inheritance models to discover a set of liver-expressed ciliary genes enriched with rare, deleterious de novo or biallelic mutations in nonsyndromic BA patients. Through functional characterization of candidate ciliary genes in patients’ liver biopsies, human cholangiocytes and fibroblast cells as well as zebrafish models, we demonstrated their functional impact on ciliary abnormalities and their relevance to BA phenotype. Lastly, we assessed their clinical impact on patients carrying the rare deleterious mutations with nasal nitric oxide (nNO) test, respiratory evaluation, high speed video analysis and transmission electron microscopy (TEM).
2. Methods
2.1. Human subjects
A total of 91 unrelated nonsyndromic BA patients and their unaffected parents from the Southeast Asian population participated in the study, of which 45 trios were recruited from Queen Mary Hospital, Hong Kong and 46 trios from the National Hospital of Pediatrics in Vietnam. BA was diagnosed by hepatobiliary scintigraphy and operative cholangiography. After sequencing, we excluded two BA trios due to problematic biological relatedness and sample contamination, which resulted in 89 trios for subsequent genetic analysis. Of these 89 BA patients analyzed, 38.2% of the subjects (n = 34) were male and 88.8% (n = 79) had undergone Kasai hepatoportoenterostomy at the average age of 2.2 months (range: 1–7 months).
2.2. Ethics
The study protocol was approved by the Institutional Review Board of the University of Hong Kong - Hospital Authority Hong Kong West Cluster (UW 05–282 T/945). Informed consent, or informed parental consent for those under 18 years old, was obtained from all participants. Liver biopsies of non-BA controls were taken during operations with full informed consent from parents or patients, and the study was approved by Hong Kong West Cluster-Hong Kong University Cluster Research Ethics Committee/Institutional Review Board (UW 16–052).
2.3. WES and bioinformatics analysis
Genomic DNA was extracted from blood samples for both BA cases and their unaffected parents. We then performed WES on DNA using the xGen Exome Research Panel v1.0 (xGen Lockdown®Probes) (83 BA trios) or TruSeq Exome Enrichment Kit v1.0 (8 BA trios) for exome enrichment, sequenced using the Illumina HiSeq 2000 platform at the Centre for PanorOmic Sciences, University of Hong Kong. Sequence reads were aligned by BWA [15] and processed according to the Genome Analysis Toolkit (GATK) best practice [16] version 3.4 for calling single nucleotide variants (SNVs) and small INDELs. Quality control was performed using PLINK [17] and KGGseq [18] (Supplementary Methods).
2.4. Prioritization of rare, damaging de novo and biallelic mutations in liver-expressed genes
De novo and biallelic variants were first identified using KGGseq. A variant is defined as de novo if it is absent in both parents but present in the affected child, while biallelic variants (whether homozygous or compound heterozygous) refer to variants present in both the paternally-inherited and the maternally-inherited copies of the same gene in the affected child. Sanger validation was performed to validate the de novo variants. Variants with the same genotype observed in healthy parents as well as the BA probands were excluded (Supplementary Methods). These variants were then annotated in KGGseq with the relevant RefSeq gene features, population allele frequencies in public databases, in silico deleteriousness predictions, and known disease associations from the OMIM and ClinVar databases. To prioritize variants of BA association potential, we focused on protein-altering variants that were: (i) rare, (ii) predicted to be functionally damaging and (iii) located in genes expressed in liver or biliary tissues. To define rare variants, population minor allele frequency (MAF; Supplementary Methods) thresholds of < 0.005 for de novo, < 0.01 for compound heterozygous variants, and < 0.05 for homozygous variants were used. Functionally damaging variants included all protein-truncating variants and all missense or inframe coding variants that were predicted by SIFT as “deleterious”, by PolyPhen2 as “probably damaging” or that had a CADD score ≥ 20. Candidate gene mRNA and protein expression in liver or bile duct tissue were checked using the human tissue expression database in EMBL-EBI Expression Atlas (www.ebi.ac.uk) and our in-house BA liver organoid expression database [19]. The resulting rare, damaging biallelic variants were visually checked using Integrative Genomics Viewer (IGV). Sanger validation result of selected ciliary de novo mutations and IGV plots for those rare, damaging liver expressed biallelic variants in ciliary genes were included as Supplementary Materials, Figs. S1–S3.
2.5. Functional enrichment analysis
Potential disease pathways and molecular mechanisms underlying BA pathogenesis were identified by functional enrichment test of the liver expressed genes with rare, damaging variants against the gene ontology (GO) database, using g:Profiler. Overrepresented functional terms were defined by adjusted p-value < 0.05 [20] after multiple testing correction using the default g:SCS algorithm.
2.6. Ciliary gene set
The list of ciliary genes was derived by combining: (i) SYSCILIA gold standard cilia gene list (303 genes) [21] and (ii) genes with the annotation “cilium” in the GO human database, searched for using AmiGO (791 genes) [22]. The resulting list contained 864 genes involved in ciliary functions with gene products localized within and/or outside the ciliary compartment (Supplementary Materials, Table S1). One-tailed hypergeometric test was performed to evaluate any over-representation of ciliary genes in the set of liver-expressed genes.
2.7. Gene set burden test
We compared the mutation burden of rare, deleterious de novo or homozygous variants between 81 (which passed quality check out of 83) BA trios sequenced by xGen Exome Research Panel v1.0 capture kit and 148 ethnicity-matched control trios with non-hepatic congenital condition (8 BA trios enriched by the TruSeq Exome Kit excluded, to minimize any technical variation in sequencing data). Whole genome sequence data of the control trios (mean sequencing depth 39.1x) were processed using identical procedures and software versions to those of the BA variant calling. Quality metrics for sequencing reads and variants in the exonic regions of both cohorts can be found in Supplementary Materials, Table S2. Four gene sets were subjected to burden testing: (a) all protein coding genes exome-wide, (b) all selected ciliary genes in our ciliary gene set (864 genes), (c) liver expressed ciliary genes (586 genes), and (d) non-liver expressed ciliary genes (278 genes). Burden tests were implemented using ProxECAT, a method formulated by the ratio of the counts of functional to non-functional variants, where the counts of non-functional variants are used as a proxy for the confounding effect that may arise from technical variations (e.g. sequencing techniques) in the case-control data [23]. In each gene set, we tested for the difference in the mutation burden of rare, damaging de novo and homozygous variants as previously defined, while using the rare, synonymous variants in the corresponding gene set as the non-functional proxy (Supplementary Methods). Unless otherwise specified, all statistical analyses in this study were implemented using the R package.
To assess if the BA phenotype was associated with rare, damaging variants, in each of the four gene sets we carried out logistic regression using Firth's method on the case-control status. We controlled for any background variation effects by including an individual's count of rare, synonymous variants in the gene set as covariate.
For multiple testing correction, we used the Benjamini-Hochberg method and obtained the FDR-adjusted p-value, using the p.adjust function in R. An FDR-adjusted P< 0.05 over the four gene sets was considered to be study-wide significant.
2.8. Immunofluorescence staining of liver sections
For BA patients with ciliary gene mutations of interest (BA634C: KIF3B and BA650C: TTC17), we examined for the expression of ciliary proteins, acetylated α-Tubulin (1/1000) and pericentrin (PCNT; 1/200), by immunostaining of liver biopsy specimens obtained for diagnostic histopathology from the Kasai procedure or liver transplantation. A total of 11 and 12 bile ducts were examined in liver sections of BA650C and BA634C patients, respectively. Non-tumour liver biopsy specimens from hepatoblastoma (HB) subjects and liver biopsy specimens from choledochal cyst (CC) subjects were used as non-BA controls. Liver biopsies of non-BA controls were taken during operations with full informed consent from parents or patients, and the study was approved by Hong Kong West Cluster-Hong Kong University Cluster Research Ethics Committee/Institutional Review Board (UW 16-052). Liver biopsies of BA patients with no mutation detected in any of the ciliary genes were included as non-mutant BA. Collectively, 83 and 65 bile ducts were examined in non-BA control and non-mutant BA liver sections, respectively. Sections were also immuno-stained for CK19 (1/300) plus PTCH1 (1/50) or GLI1 (1/100) to assay for Sonic Hh signalling in BA and control bile ducts. For details, see Supplementary Methods.
2.9. Cell culture and transfection
Human foreskin fibroblast cells (BJ, CRL-2522; ATCC) were cultured in DMEM with 10% fetal bovine serum (FBS) and penicillin (100 IU/mL) and streptomycin (100 µg/mL) (Invitrogen). Human intra-hepatic cholangiocytes were derived from liver biopsy via the generation of bile duct organoids following our previously published protocol [19]. Human cholagniocytes were cultured as a monolayer on cover slip coated with Matrigel (356231; Corning Biocoat) in organoid medium until ready for short interfering RNAs (siRNA) transfection. The siRNAs specifically target to PCNT, KIF3B, TTC17 and control-scrambled siRNA were purchased from Santa Cruz Biotechnology. The fibroblast cells and human cholangiocytes were transfected with siRNAs via Lipofectamine® RNAiMAX reagent (Life technologies) according to the manufacturer's instructions. Fibroblast cells were grown to 80% confluence on the day of transfection and starved of serum for 48 h. Human cholangiocytes were grown to 80% confluence on the day of transfection in organoid medium.
2.10. Scanning electron microscopy
For cilia imaging by scanning electron microscopy (SEM) of human cholangiocytes, at 48 h post siRNA transfection, the culture medium was removed and the cholangiocytes were fixed in 4% PFA/PBS (paraformaldehyde in phosphate buffered saline) for 16 h, dehydrated in a series of graded ethanol until 100% (EM grade) and processed for SEM analysis by Electron Microscopy Unit (EMU) of the Queen Mary Hospital, Hong Kong following standard protocols. Samples were examined on Hitachi S-4800FEG (Tokyo, Japan) and LEO 1530 FEG (Georgia Tech, Georgia Institute of Technology, Atlanta, USA) scanning electron microscopes. At least 50 cells in each siRNA transfected culture were examined for the presence of cilia and the percentage of ciliated cells in each transfected cultures were quantified. Each siRNA transfection were performed in duplicate.
2.11. Real-time PCR
RNA was isolated from BJ cells with RNeasy Mini Kit (Qiagen) and reverse-transcribed with PrimeScript™RT reagent Kit with gDNA Eraser (Takara). Samples were assayed with iTaq™ Universal SYBR Green Supermix (Bio-Rad). Real-time PCR was performed with ViiA 7 (Applied Biosystems). Relative expression levels of PCNT, KIF3B and TTC17 in transfected cells were determined using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as internal reference and 2–ΔΔCt method. The relative fold change of expression between scrambled and PCNT, KIF3B and TTC17 siRNA transfected cells were calculated taking the relative expression level of PCNT, KIF3B and TTC17 in scrambled siRNA transfected cells arbitrarily as one, and expressed as mean ± standard deviation (SD).
2.12. Immunofluorescence staining of cells
The cells were fixed with 4% PFA and incubated with anti-acetylated-α-tubulin (1/1000) and anti-PCNT (1/200) antibodies. Alexa Fluor® 488-conjugated goat anti-rabbit IgG and Alexa Fluor® 594 goat anti-mouse IgG secondary antibodies (1/300, Invitrogen) were used. Cells were then counterstained with DAPI. Confocal images were captured with the Carl Zeiss LSM 880 microscope. Image processing was performed with the ZEN 2.3 software. Images were analyzed with Image J.
2.13. CRISPR/Cas9 gene knockout in zebrafish model
We used wildtype zebrafish and transgenic zebrafish (Danio rerio) lines (um14Tg[Tg(tp1-MmHbb:EGFP)um14] (ZL1950; Zebrafish International Resource Center (ZIRC), 5274 University of Oregon, Eugene, OR 97403–5274, USA)) as animal models to investigate the in vivo association between candidate ciliary genes and the BA phenotype, by CRISPR/Cas9 gene knockout. The um14Tg[Tg(tp1-MmHbb:EGFP)um14] transgenic fish expressed strong green fluorescent protein signal in bile ducts and were used to investigate the biliary development in zebrafish [24], [25], [26], [27]. Bile flow was assessed in 51–57 independently injected embryos per wildtype or gene-knockout test group, by measuring the ability to process N-([6-(2,4-dinitro-phenyl)amino]hexanoyl)-1-palmitoyl-2-BODIPY-FL-pentanoyl-sn-glycerol-3-phosphoethanolamine (PED6) [28], a fluorescent lipid reporter for examination of biliary function. We cultured gRNA-injected and un-injected embryos until 5 days post-fertilization, after which they were released to swim in a 5 µM solution of PED6 for 2 h. During this period, images using an Olympus SZX7 fluorescent microscope with fixed fluorescence intensity were taken at the same magnification with identical brightness and contrast settings at 30 min intervals. We analysed the microscopic images using ImageJ [29]. The total green fluorescence in the gall bladder was calculated as integrated density (area × mean fluorescence intensity). Comparison of PED6 uptake between each mutant embryo group and wild type was performed using t-test. P < 0.05 was considered to be statistically significant.
2.14. Measurements of nNO, ciliary beat frequency, beat pattern and ultrastructure
Ten BA patients identified to have rare, damaging ciliary mutations could be recalled and agreed to further clinical evaluation. We performed tidal breathing nNO (TB-nNO) test using NIOX® (Nitric Oxide Monitoring System) MINO hand-held electrochemical device sampling at flow rates of 2 ml/s (MINO2) or 5 ml/s (MINO5) on these patients. Nasal NO concentrations were measured with non-velum closure techniques in parts per billion (ppb) and were compared against the device-specific ranges previously published for healthy and primary ciliary dyskinesia (PCD) subjects [30]. Ciliated epithelial samples were obtained by brushing the inferior nasal turbinate. Ciliary beat frequency (CBF) and beat pattern were analyzed using high speed video microscopy (HSVM) while ciliary ultrastructure were assessed using TEM as described previously [31] (See Supplementary Methods). The CBF, beat pattern, and ultrastructure of the ciliated epithelium and ciliary axonemes of the BA patients were compared against the normal age-related healthy reference ranges established on Chinese children and adults [31].
2.15. Role of funding source
Funders had no role in study design, data collection, data analyses, interpretation, or writing of the manuscript.
3. Results
WES was performed on 91 Southeast Asian nonsyndromic BA trios, each with one affected patient plus their two unaffected parents. A total of 89 BA trios passed quality controls and were subject to genetic analysis. The mean and median sequencing depths were 31.9x and 28x, respectively.
3.1. Most BA cases carry rare, damaging de novo or biallelic (RDL) variants in liver expressed genes
To identify potentially pathogenic variants, we filtered Rare, Damaging protein-altering variants present in the BA proband but not in their unaffected parents and in genes expressed in Liver/biliary tissues (hereafter known as RDL variants). Among 92% of the BA subjects (n = 82), we identified 45 de novo RDL variants, of which 8 were predicted to cause loss-of-function (6 stopgain and 2 frameshift), and 233 biallelic (112 homozygous and 121 compound heterozygous) RDL variants, distributed in 239 genes. Of these 239 genes, none had been linked to any form of BA in previous studies nor case reports [14]. Only PKHD1 that was found to have a biallelic variant in one of our BA patient was screened in a previous study of perinatal BA patients, but no pathogenic mutation was found [32]. Three genes had multiple RDL variants observed in > 2 BA patients but all of which are predicted as being tolerant to missense mutations according to the constraint score (missense z-score: TTN, -1.10; PLEC, -2.57; USH2A, -2.47).
3.2. Ciliary genes are enriched for RDL variants in BA patients
To gain insights into the major biological or molecular functions associated with these 239 RDL genes, we performed functional enrichment analysis using g:Profiler. We detected seven highly related GO biological processes that were significantly enriched with RDL variants (adjusted P < 0.05; Table 1), namely cytoskeleton organization, cilium organization, microtubule cytoskeleton organization, microtubule-based process, cilium assembly, spindle organization, and organelle assembly, covering ∼20% (= 49/239) of the RDL genes and half of (n = 26) which are ciliary genes. Similar enrichment of GO molecular function terms were observed. In fact, microtubule is an integral part of both cytoskeleton and cilia. It is a core component of the cilium axenome and is essential in cilia assembly and in the regulation of ciliogenesis. Specially, among the eight genes with protein truncating de novo mutations that are considered as most pathogenic, around 37.5% (n = 3; KIF3B, PCNT, and SPEF2) were ciliary genes (Supplementary Materials, Table S3). Strikingly, 31.5% (28/89) of the BA subjects carried at least one RDL variant in ciliary genes. The genetic profiles of these RDL ciliary variants in BA patients were given in Supplementary Materials, Table S4. To further delineate if the enrichment of cilia-related GO terms could be biased simply due to the selection of genes expressed in liver, we performed hypergeometric test but did not find overrepresentation of ciliary genes in liver-expressed gene set (P = 1.0). This implies that the GO enrichment is specific to genes identified to contain rare, de novo or biallelic deleterious variants in BA patients. These findings corroborated with previous report of association of PKD1L1, also a ciliary gene, with syndromic BA-BASM, leading us to hypothesize that the cilium is the key organelle affected by the genetic mutations observed in nonsyndromic BA subjects [13].
Table 1.
Results of gene set enrichment analysis on 239 genes with RDL variants. List of overrepresented GO biological process and molecular function with adjusted p-value < 0.05.
| Term description | GO term ID | Gene count | P |
|---|---|---|---|
| GO biological process | |||
| Cytoskeleton organization a,b,c | GO:0007010 | 39 | 1.09 × 10−3 |
| Cilium organization a,b,c | GO:0044782 | 18 | 3.22 × 10−3 |
| Microtubule cytoskeleton organization a,b | GO:0000226 | 22 | 4.23 × 10−3 |
| Microtubule-based process a,b | GO:0007017 | 26 | 5.84 × 10−3 |
| Cilium assembly a,b | GO:0060271 | 17 | 7.24 × 10−3 |
| Spindle organization a,b | GO:0007051 | 11 | 2.08 × 10−2 |
| Organelle assembly a,b,c | GO:0060271 | 26 | 3.38 × 10−2 |
| GO molecular Function | |||
| Cytoskeletal protein binding a | GO:0008092 | 34 | 4.09 × 10−5 |
| Cell adhesion molecule binding | GO:0050839 | 20 | 2.82 × 10−3 |
| Protein-containing complex binding a | GO:0044877 | 31 | 1.68 × 10−2 |
| Ankyrin binding | GO:0030506 | 4 | 4.50 × 10−2 |
denotes the enriched GO term containing KIF3B.
denotes the enriched GO term containing PCNT.
denotes the enriched GO term containing TTC17.
3.3. Excess of rare deleterious variants in ciliary genes compared to controls
To further confirm the excess of ciliary mutations in nonsyndromic BA patients, we performed gene set burden tests for rare, de novo and homozygous variants relative to 148 population control trios by ProxECAT and logistic regression while adjusted for counts of rare synonymous variants. Considering all protein coding genes, no enrichment of these rare deleterious mutations was found in patients (odds ratio (OR) [95% CI]=1.12 [0.93–1.35], false discovery rate (FDR) adjusted P [P-adjusted]= 0.241; ProxECAT: P-adjusted= 0.484), demonstrating the comparability of the case-control data (Table 2). In contrast, when we considered only the liver expressed ciliary genes, both tests consistently detected case-control differences and the presence of an RDL ciliary variant increased risk of BA by 2.6-fold (OR [95% CI]= 2.58 [1.15–6.07], P-adjusted = 0.034; ProxECAT: P-adjusted = 0.048). Results for non-liver expressed ciliary genes were mixed and the inconclusive result is probably due to the sparse variant counts in that gene set (around 5 synonymous/nonsynonymous variants in the whole control cohort, Table 2) and/or the difference in statistical power between the two methods. Irrespective of the gene expression in liver tissues, excess burden was observed in patients for all ciliary genes by both methods (OR [95% CI]= 3.24 [1.63–6.67], P-adjusted = 0.030; ProxECAT: P-adjusted = 0.048). Further stratification of the patients by ancestries (Chinese versus Vietnamese) and by sex detected no difference between groups, implicating the robustness of the enrichment (Supplementary Materials, Table S5).
Table 2.
Mutation burden of rare, damaging de novo and homozygous variants in 4 gene sets: (a) all protein coding genes, (b) all ciliary genes (n = 864), (c) liver expressed ciliary genes (n = 586), and (d) non-liver expressed ciliary genes (n = 278), in 81 BA trios compared to 148 ethnicity-matched control trios.
| All genes |
Ciliary genes |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Overall |
Liver expressed |
Non-liver expressed |
|||||||
| BA | Control | BA | Control | BA | Control | BA | Control | ||
| Per-person rare mutation count (mean) | |||||||||
| Synonymous (S) | 2.19 | 2.24 | 0.14 | 0.15 | 0.08 | 0.11 | 0.06 | 0.03 | |
| Damaging non-synonymous (dNS) | 2.25 | 2.01 | 0.29 | 0.10 | 0.18 | 0.07 | 0.11 | 0.03 | |
| dNS/S | 1.03 | 0.90 | 2.09 | 0.68 | 2.33 | 0.59 | 1.80 | 1.00 | |
| ProxECAT | |||||||||
| Pa | 0.364 | 0.021 | 0.024 | 0.484 | |||||
| FDR-adjusted Pb | 0.484 | 0.048 | 0.048 | 0.484 | |||||
| Logistic regression | |||||||||
| Odds ratio (95% CI) c | 1.12 (0.93-1.35) | 3.24 (1.63-6.67) | 2.58 (1.15-6.07) | 3.36 (1.16-10.70) | |||||
| Pa | 0.241 | 0.007 | 0.021 | 0.026 | |||||
| FDR-adjusted Pb | 0.241 | 0.030 | 0.034 | 0.034 | |||||
P-value of gene set burden test (< 0.05 in bold) before multiple comparison adjustment.
FDR-adjusted p-value (< 0.05 in bold) of the gene set burden test derived by Benjamini-Hochberg method.
Odds ratio from logistic regression on the case-control status (in bold: both P and FDR-adjusted P < 0.05). CI, confidence interval.
3.4. High level of locus heterogeneity in nonsyndromic BA involving diverse ciliary functions
For our nonsyndromic BA patients, instead of having excess mutations in a single or a few major ciliary genes, there is high level of locus heterogeneity in which multiple occurrence of these RDL variants per ciliary gene was sparse (only DNAH8, PKD1 and USH2A with RDL variants in > 1 BA cases; Table 3 and Supplementary Materials, Table S4). A broad spectrum of ciliary genes with diverse localizations and ciliary functions were involved (Table 3). For example, PCNT located at the basal body that forms the pericentriolar material, a nucleation site for microtubules, is involved in cilia assembly. The GLI1 transcription factor on the axoneme mediates the Hedgehog (Hh) pathway while KIF3B is an essential anterograde intraflagellar transport (IFT) motor driver for cilia assembly and maintenance. On the other hand, TTC17 localizes outside the ciliary compartment and is involved in actin organization and pre-ciliogenesis. To evaluate the potential impact of the ciliary mutations on cilia structure and BA phenotypes, we selected three ciliary genes affected by de novo mutations in our BA probands—KIF3B, PCNT and TTC17—as the representatives of key ciliary localizations and functions in cilia assembly for functional characterization.
Table 3.
List of 26 ciliary genes with RDL variants in the BA probands categorized by ciliary localization and functions.
| Gene | Description | Localization / functional category | Ciliopathy association a | Inheritance b [> 1 case] |
|---|---|---|---|---|
| KIF3B | kinesin family member 3B | IFT-kinesin | De novo | |
| DNAH8 | dynein axonemal heavy chain 8 | Outer dynein arm | Spermatogenesis defects | Homo [2] |
| BBS9 | Bardet-Biedl syndrome 9 | BBSome-IFT-associated, basal body | Bardet-Biedl syndrome 9 | Homo |
| GLI1 | GLI family zinc finger 1 | Axoneme (tip), transcription factor | CompHet | |
| MYO15A | myosin XVA | Axoneme (tip) | CompHet | |
| TEKT4 | tektin 4 | Axoneme | Asthenozoospermia (M); subfertility (M) | Homo |
| PKD1 | polycystin 1, transient receptor potential channel interacting | Axoneme, membrane - signaling | ADPKD | CompHet [2] |
| PKHD1 | PKHD1 ciliary IPT domain containing fibrocystin/polyductin | Axoneme, basal body - signaling | ARPKD | CompHet |
| TTLL3 | tubulin tyrosine ligase like 3 | Axoneme modification | CompHet | |
| PCNT | pericentrin | Centrosome | Microcephalic osteodysplastic primordial dwarfism, type II | De novo |
| HAUS1 | HAUS augmin like complex subunit 1 | Centrosome | Homo | |
| CLASP1 | cytoplasmic linker associated protein 1 | Centrosome; microtubule plus-end | Homo | |
| CEP131 | centrosomal protein 131 | Centriolar Satellite | De novo | |
| PCM1 | pericentriolar material 1 | Centriolar Satellite | CompHet | |
| SPAG17 | sperm associated antigen 17 | Central Pair | CompHet | |
| SPEF2 | sperm flagellar 2 | Central Pair | Spermatogenesis defects (M); PCD (M) | De novo |
| USH2A | usherin | Basal body | Usher syndrome; retinitis pigmentosa | CompHet [3] |
| DCTN1 | dynactin subunit 1 | Subdistal appendage, basal foot | ALS; Perry syndrome; neuropathy, distal hereditary motor, type VIIB | CompHet |
| TTC17 | tetratricopeptide repeat domain 17 | Actin filament polymerization | De novo | |
| DHRS3 | dehydrogenase/reductase 3 | Membrane | Homo | |
| PROM1 | prominin 1 | Membrane, endoplasmic reticulum | Homo | |
| NOTCH1 | notch receptor 1 | Signalling | CompHet | |
| PKHD1L1 | PKHD1 like 1 | Signalling | CompHet | |
| SYNE2 | spectrin repeat containing nuclear envelope protein 2 | Trafficking, actin remodeling | Emery-dreifuss muscular dystrophy 5 | Homo, CompHet |
| EXOC6 | exocyst complex component 6 | Exocytosis - vesicle docking | Homo | |
| LAMA5 | laminin subunit alpha 5 | Extracellular - integrin binding | CompHet |
Abbreviation: (M), phenotypes specific to mice; ADPKD, Autosomal dominant polycystic kidney disease; ALS, Amyotrophic lateral sclerosis; ARPKD, autosomal recessive polycystic kidney disease; BBSome, Bardet-Biedl syndrome complex; IFT, intraflagellar transport; PCD, primary cilia dyskinesia.
Mutation type: Homo, homozygous; CompHet, compound heterozygous. Number in brackets indicates case count (> 1) of RDL variants.
3.5. Abnormal cilia in the bile ducts of BA patients
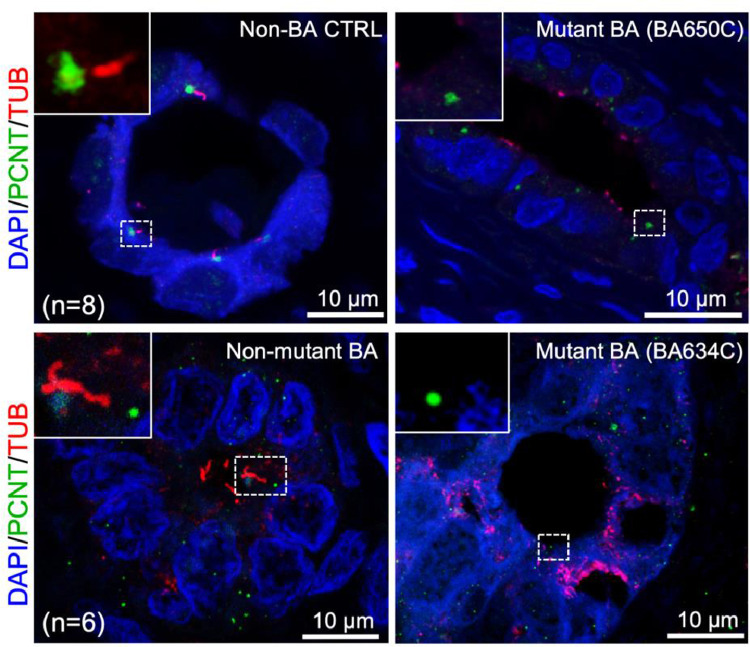
We first examined if the ciliary mutations correlated with cilia formation in the bile duct of the corresponding BA patients. We examined the cilia marker co-immunofluorescence staining of the BA livers for patients carrying de novo stopgain and missense RDL variants in KIF3B (BA634C) and TTC17 (BA650C), respectively compared to the liver biopsies of non-BA controls (total n=8; including biopsies of non-tumor liver of hepatoblastoma (HB; n = 4), liver of choledochal cysts (CC; n = 4)), and non-mutant BA subjects without ciliary mutation (n = 6) (Fig. 1). In contrast to an average of 3.6 ([S.E.M.] ± 0.1) and 3.2 ([S.E.M.] ± 0.1) cilia detected per bile duct in non-BA control and non-mutant BA livers, respectively, cilium axoneme were absent in all bile ducts (n = 11 and 12) of the both mutant BA subjects. BA634C has two de novo ultra-rare stopgain RDL variants not present in any public databases, one in KIF3B and another in SPEF2. SPEF2 is known to function in motile cilia, especially for sperm development and tracheal cilia beating, since depletion in SPEF2 causes male infertility and primary ciliary dyskinesia (PCD) in mice [33]. Therefore the absence of primary cilia in cholangiocytes was more likely to be attributed to the KIF3B mutation. The liver tissue sample of the BA subject with PCNT mutation was not available, but a previous study reported that PCNT depletion results in loss of primary cilia in human epithelial cells [34]. Given the importance of primary cilia in Hh signaling pathway in cholangiocytes, we further examined the expression of sonic Hh markers, PTCH1 and GLI1, in the bile ducts of these two BA subjects and compared the expression to those of non-BA control (HB, n = 3; CC, n = 3) and non-mutant BA (n = 9) subjects. Dysregulation of PTCH1 and GLI1 were observed in both mutant BA subjects (Supplementary Materials, Fig. S4), suggesting correlation between abnormal cilia and aberrant Hh regulation in mutant BA patients.
Fig. 1.
Absence of cilia staining in the bile ducts of BA livers of the two patients harboring LOF mutations in the ciliary genes. Co-immunofluorescence staining for α-Tubulin (TUB; red) and Pericentrin (PCNT; green) was performed on liver sections of Non-BA control (Non-BA CTRL; top left), BA patients with no ciliary gene mutation (Non-mutant BA; bottom left) and BA patients with ciliary gene mutation (Mutant BA; BA650C and BA634C; top and bottom right). α-Tubulin immuno-reactivity was co-localised with PCNT at the luminal surface of the bile ducts of Non-BA CTRL and Non-mutant BA livers. In contrast, only immuno-staining for PCNT were detected but expression of α-Tubulin were absent in BA650C and BA634C patients’ bile ducts. Nuclei were stained with DAPI. Representative photos were shown for comparison. Highlighted regions were magnified as shown in inset. Number of patients examined in each group was indicated as “n”. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
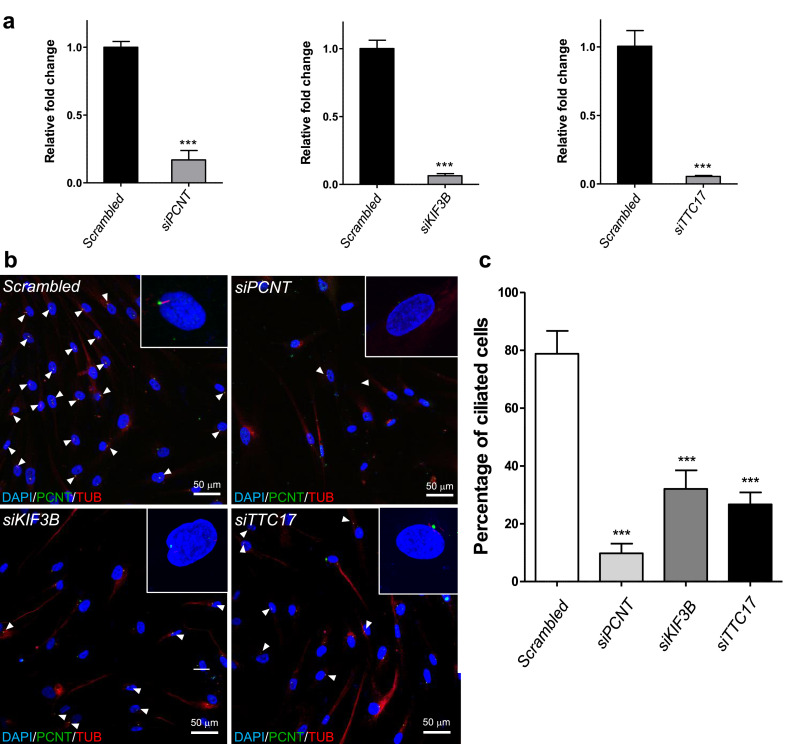
3.6. Gene knockdown resulted in reduction in number of cilia in human fibroblast and cholangiocyte cells
To directly assess the impact of loss of function of these ciliary genes, we further knocked down KIF3B, PCNT and TTC17 by siRNA transfection in human control cholangiocyte and fibroblast cells. A significant reduction in the number of ciliated human fibroblast cells was observed compared to controls transfected with non-specific scrambled siRNA (Fig. 2). Effect of siRNA knockdown on cilia of human cholangiocytes were also investigated by SEM analysis. Under SEM examination, primary cilia appeared as a long and thick protrusion from the cholangiocyte cell surface as compared to the surrounding short and thin microvilli (Supplementary Materials, Fig. S5). Primary cilia could be identified in 40-50% of cells in scrambled siRNA transfected cultures (Supplementary Materials, Fig. S5a). In contrast, cilia were either undetected or detected at lower percentages in KIF3B, PCNT and TTC17 siRNA transfected cultures (0-2% in KIF3B siRNA transfected cultures; 0% in PCNT siRNA transfected cultures; 10–12% in TTC17 siRNA transfected cultures) (Supplementary Materials, Fig. S5b,c and data not shown). This reduction in the number of ciliated human cholangiocyte cells compared to controls transfected with non-specific scrambled siRNA (Supplementary Materials, Fig. S5) further confirmed the effect of abnormal cilia formation caused by the depletion of the selected ciliary genes.
Fig. 2.
Knockdown of PCNT, KIF3B and TTC17 resulted in reduced number of cilia in cells. (a) Examination of the knockdown efficiency by siRNAs in human fibroblasts by real-time quantitative PCR assay using GAPDH as internal controls. Three independent experiments were performed and data represented as means ± standard deviation (SD). *** p < 0.0001, two-sided t-test. Cells transfected with non-specific scrambled siRNA were employed as negative controls and relative expression levels of PCNT, KIF3B and TTC17 to GAPDH in scrambled siRNA transfected cells were arbitrarily regarded as 1. (b) Effect of PCNT, KIF3B and TTC17 knockdown on cilia. Cells were transfected with siRNAs and serum-starved for 48 h, then immuno-stained for acetylated-α-tubulin (TUB; red) and pericentrin (PCNT, green). Nuclei were stained with DAPI (blue). Representative images of each of the siRNA transfection were shown. Ciliated cells (arrowheads) were abundant in scrambled siRNA transfected cells, whereas ciliated cells were scarce in siPCNT, siKIF3B and siTTC17 transfected cells. Ciliated cell and non-ciliated cells were magnified as shown as inset. (c) Percentage of ciliated cells in scrambled siRNA, siPCNT, siKIF3B and siTTC17 transfected cells was determined by counting ciliated cells and total number of cells. Two independent transfection experiments were performed for each siRNA, and over 100 cells were counted in each experiment. Data was shown as means±SD. *** p < 0.0001, one-way ANOVA, two-sided test. All scale bars = 50 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
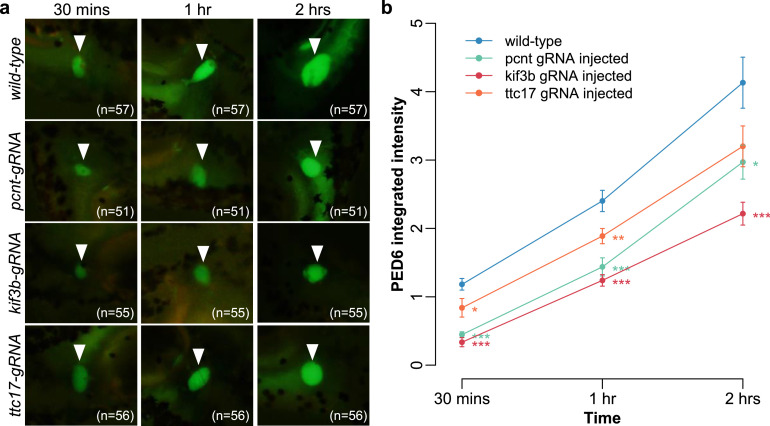
3.7. Ciliary gene knockout leads to impaired biliary function in zebrafish
We next assessed if ciliary gene depletion could cause BA phenotype in vivo using zebrafish model. Knockout of pcnt, kif3b and ttc17 was performed by CRISPR/Cas9-mediated genome editing. Introduction of INDEL in pcnt, kif3b and ttc17 genes was confirmed by T7E1 assay and reduced expression of pcnt, kif3b and ttc17 in injected embryo was demonstrated by semi-quantitative RT-PCR (Supplementary Materials, Fig. S6). The biliary development of un-injected (Cas9 protein alone) and pcnt-, kif3b- and ttc17- gRNA injected 5-dpf um14Tg[Tg(tp1-MmHbb:EGFP)um14] larvae were studied by examining the bile duct green fluorescence signal intensity under fluorescence microscope. Zebrafish larvae with the knockout of pcnt, kif3b and ttc17 showed comparable biliary green fluorescence signal as control larvae, which suggested that knockout of these genes did not cause gross abnormal bile duct development in zebrafish (Supplementary Materials, Fig. S7). To further examine the impact of the knockout of pcnt, kif3b and ttc17 on bile flow from liver to the gall bladder in zebrafish, we performed PED6 bile flow assay in pcnt, kif3b and ttc17 knockout non-transgenic fish. Knockout of all three genes resulted in defective bile flow in fish embryos, as measured by significantly lower integrated density of PED6 in the gall bladder in the embryo mutant groups compared to the control group (Fig. 3). This demonstrated that BA-like phenotype of impaired bile blow in zebrafish model can be ascribed to disrupted function of PCNT, KIF3B and TTC17.
Fig. 3.
Knockout of pcnt, kif3b and ttc17 resulted in defective bile flow in mutant zebrafish. (a) Representative epifluorescence images and (b) line plot of pcnt (n = 51), kif3b (n = 55) and ttc17 (n = 56) mutant embryo groups at 5 days post-fertilization showing significantly lower accumulated PED6 integrated density in gallbladder (arrowhead) after incubation for 30 min (left), 1 h (middle) and 2 h (right) compared to wild type (n = 57). Data expressed as mean ± standard error of the mean (SEM). Results of pairwise t-test: ***, P < 0.001; **, P < 0.01, *, P < 0.05.
3.8. Impaired respiratory ciliary function in majority of patients with RDL variants
Clinically, disorders of cilia dysfunction often present with complex multisystem involvement. Although nonmotile ciliopathies are clinically distinct from motile ones (e.g. PCD), they sometimes display common clinical features, including hepatobiliary and kidney diseases, due to the common involvement of ciliary processes and underlying signaling defects between the motile and primary cilia [35]. Prior to this WES, we came across a syndromic BA patient with dextrocardia and situs invertus. The patient was suspected to have PCD and displayed high proportion of disarranged respiratory cilia upon respiratory ciliary test. Thus, we attempted to use a simple non-invasive nNO test, a well-established first line screening test for PCD, on those BA patients with RDL variants to indirectly assess ciliary dysfunction clinically [36]. PCD patients with abnormal ciliary structure and/or function have exceptionally low nNO levels. NO biosynthesis depends on nitric oxide synthases (NOS) and NOS activity relies on normal ciliary function [37,38]. Production and release of NO may depend on the activation of cilia in response to fluid flow-generated shear stress and thus measures of NO concentration may indirectly reflect ciliary function and signaling [39]. Surprisingly, as shown in Table 4, eight out of the 10 BA patients with ciliary mutations have abnormally low nNO levels. Four patients have nNO levels in the range suggestive for PCD while the other four patients have low nNO levels outside the normal range of healthy subjects. While motile respiratory cilia ultrastructure abnormalities detected by TEM were within the normal range (Supplementary Materials, Fig. S8) and the average ciliary beat frequencies (CBF) and beat pattern detected using HSVM also appeared normal for all 10 BA patients, detailed side-view visual examinations revealed that three patients have their CBF recorded at the lower end (<=7.5 Hz) in a proportion of edges (1–3 in 10). Representative high speed videos of a BA patient with abnormal CBF and a healthy subject were included in Supplementary Materials, video S1 and S2. Altogether, the low nNO levels and abnormal CBF for a subset of respiratory motile cilia suggested subtle impaired ciliary function in BA patients with ciliary mutations and highlighted the plausible multiorgan involvement in this patient subgroup.
Table 4.
Analysis of ciliary beat frequency (CBF) and nasal nitric oxide (nNO) levels for 10 BA patients with rare deleterious ciliary mutations.
| Ciliary mutation |
HSVMc |
nNO test |
||||||
|---|---|---|---|---|---|---|---|---|
| Subject ID | Inheritance modea | Gene | Amino acid changeb | Testing Age(years) | Mean CBF (Hz) | Number of edges (out of 10) with visual CBF <=7.5 | Hand-held analyzer | nNO concentrations (ppb)d |
| BA171C | CompHet | MYO15A | p.V2872M/p.I3421M | 18 | 11.7 | 0 | MINO5 | 341 |
| BA230C | CompHet | NOTCH1 | p.G1195R/p.T1035S | 13 | 10.7 | 0 | MINO5 | 185 |
| CompHet | PKD1 | p.R3046C/p.S1679R | ||||||
| BA307C | Homo | PROM1 | p.S281R | 11 | 9.5 | 3 | MINO2 | 152 |
| BA596C | CompHet | USH2A | p.R1870W/p.T3014N | 8 | 11.4 | 0 | MINO2 | 853 |
| Homo | DNAH8 | p.E972V | ||||||
| BA611C | CompHet | USH2A | p.C729*+ p.S333P /p.A3438T | 7 | 9.4 | 0 | MINO2 | 168 |
| BA613C | CompHet | PKD1 | p.S699L/p.R2075C | 7 | 11.5 | 1 | MINO2 | 25 |
| BA634C | De novo | KIF3B | p.R153* | 7 | 9.2 | 2 | MINO2 | 27 |
| De novo | SPEF2 | p.Y473* | ||||||
| BA636C | Homo | CLASP1 | p.S617P | 6 | 9.5 | 0 | MINO2 | 29 |
| BA642C | CompHet | PCM1 | p.T1174A/p.T1513S | 5 | 11.3 | 0 | MINO2 | 102 |
| BA645C | Homo | DNAH8 | p.E972V | 5 | 12.8 | 0 | MINO2 | 16 |
CompHet: Compound heterozygous; Homo: Homozygous
Amino acid changes for compound heterozygous variant are ordered by paternal/maternal inheritance
HSVM: high speed video-microscopy; The ranges (5th - 95th percentiles) of CBF are (i) 6.3 – 13.5 and (ii) 5.5 – 11.9 for healthy subjects with age (i) under 18 and (ii) above 18, respectively as reported in Lee et al.[31].
Ppb:parts per billion; BA patients with nNO levels lower than the 1 standard error (SEM) of healthy subjects are bolded and those within the range of 1 SEM of primary ciliary dyskinesia (PCD) patients are italicized. The ranges (±1 SEM) of nNO levels are (i) 4 – 72 and (ii) 317 – 363 for (i) PCD patients and (ii) healthy subjects, respectively using hand-held analyzer MINO5 and (iii) 51 – 97 and (iv) 693 – 811 for (iii) PCD patients and (iv) healthy subjects, respectively using hand-held analyzer MINO2.
4. Discussion
BA clinically presents with obstruction, inflammation and fibrosis in the biliary tree and is widely considered as multifactorial in origin, involving poorly defined environmental and genetic factors. In this largest WES study on nonsyndromic BA trios to date, we found an excess burden of rare, deleterious mutations carried by BA patients, in a wide spectrum of liver expressed ciliary genes that play different roles in ciliary function and ciliogenesis. Cilia were found absent in the liver of our BA patients with KIF3B or TTC17 mutations and the corresponding zebrafish mutants (including PCNT) exhibited impaired biliary function. We further demonstrated reduction of cilia formation caused by the depletion of these genes independent of the genetic background of the patients, which were consistent with other experimental studies [34,[40], [41], [42]]. Clinical evaluation by nNO test revealed abnormally low level of nNO in a majority (80%) of BA patients with ciliary mutations, implicating intrinsic ciliary defects and highlighting a subset of patients with potential pleiotropic phenotypes. Based on these findings, we suggest that genetic factors play a major role in a substantial proportion of nonsyndromic BA cases. Our findings also implicate defective ciliary structure and function as one of the underlying mechanisms of BA pathogenesis.
We hypothesize that ciliary gene mutations can lead to the development of BA phenotypes through two interconnected biological mechanisms. First, it was suggested that defective cholangiocyte cilia structure and function can in itself cause dysregulation of the Hh pathway, which promotes dysfunctional tissue repair and leads to hepatic inflammation and fibrogenesis [43]. The primary cilium is indispensable in the Hh signaling pathway both physiologically and biologically, interacting with various components at different points of the signaling cascade to regulate liver regeneration and repair. The Hh ligand receptors and transcription factors depend on primary cilia for activation, mediation and suppression, and the core signaling components are localized to cilia, thus requiring IFT for their trafficking to the functional sites for regulatory activities [44]. However, there have been growing evidence that non-canonical pathway under the absence of cilia was also involved in liver pathophysiology progressed from chronic liver injury [44,45] and tumorigenesis [46]. Secondly, it is likely that defective cilia would compromise the protective function of immature neonatal cholangiocytes against bile acid insults, leading to chronic liver injury, which can also trigger Hh signalling. In different tissue types, aberrant or absent cilia, or mutations in IFT proteins were shown to cause Hh loss- or gain-of-function phenotypes [46,47]. Indeed, shorter, misoriented, or less abundant cholangiocyte cilia were commonly observed in several studies of both syndromic and nonsyndromic BA patients [48], [49], [50], meanwhile Hh activity was shown to be associated with jaundice-free survival of BA patients in another study [51]. Our current study is the first to suggest genetic defects might underlie these observations in nonsyndromic patients. We showed convincing evidence that at least some of the nonsyndromic patients with rare damaging mutations in ciliary genes have no primary cilia in their bile ducts and concurrently with deregulated Hh signaling. Elucidating the molecular mechanisms by which the defective primary cilium dysregulate Hh signaling may provide a better understanding of the disease pathogenesis and offer potential targets for alternative therapies.
One of the interesting findings is the low nNO levels in 80% of BA patients with ciliary mutations. Although primary cilia (cholangiocyte cilia) are structurally and functionally distinct from motile cilia (respiratory cilia), the similarity in cilium–centrosome complex and the shared signaling pathways (e.g. Hh signaling) across tissues with ciliated cells may account for the involvement of multiple organ systems in ciliopathies generally. Genetic predisposition to dysregulation of liver-expressed ciliary genes may thereby exert a functional impact beyond the hepatobiliary system. Such predisposition may not be necessarily sufficient for clinical manifestation as a ciliary defect in nonsyndromic BA patients, rather milder or subclinical phenotypes may reside and other modifiers (genetic or environmental factors) can play a pathogenic role in triggering ciliary dysfunction. This finding suggested a further follow up and thorough clinical assessment of this patient subgroup for any pleiotropic clinical features to establish genotype-phenotype correlation and to improve clinical management and disease outcome.
Familial aggregation of BA is rare with low recurrence in siblings, even among monozygotic twins [52]. Mendelian form of inheritance of rare causal variants with high penetrance is therefore unlikely to account for the majority of BA etiology. Unlike syndromic BA with laterality defects that can be caused by high penetrant ciliary mutations similar to other ciliopathies, we hypothesize that RDL ciliary mutations confer a strong genetic predisposition to risk of nonsyndromic BA and provide a sensitized genetic background such that other genetic and/or environmental factors may act as modifiers of penetrance or triggers for the disease manifestation. The synergistic effect of genetic modifier(s) has been demonstrated in classical zebrafish model of BA in which knockdown of both Egfr and Arf6 of the same signaling pathway resulted in a more severe biliary defect compared to minimal or no defects in intrahepatic biliary structure when knockdown each gene alone [12]. Inherited common genetic variations in BA susceptibility genes, e.g. ADD3 and GPC1, may further contribute to the genetic susceptibility and disease expressivity through perturbation of interconnected biological pathways including Hh signaling [8,10,53]. Compared to the multiple occurrence of rare damaging biallelic variants in a single gene, PKD1L1, in syndromic patients with BASM, the scarce multiple occurrences observed for individual variants or genes in nonsyndromic BA patients has important implications on future genetic analyses of this patient subgroup. Unlike the syndromic form, the nonsyndromic BA is unlikely to have a single genetic etiology. Instead of focusing on mutational burden on single gene-level, the genetic heterogeneity of nonsyndromic BA calls for a gene set-based enrichment analysis aggregating genetic effects across multiple genes in pathways or biological meaningful gene sets and possibly a more lenient damaging criteria to detect the strong predisposing effect. Examining mainly the top 0.1% damaging novel variants transmitted in a Mendelian fashion might partially account for the lack of genetic association for a recently published WES study on nonsyndromic BA [54].
The genetic contributions to cholangiocyte ciliary malformation and ciliary protein dysfunction appear to be heterogeneous, demonstrated by the diverse spectrum of liver expressed ciliary genes identified to carry RDL variants in BA cases in this study. We found almost a third of BA subjects with rare, damaging, liver expressed ciliary gene variants, whilst a previous descriptive histopathological study observed cilia abnormalities in 86% of BA subjects [49]. The considerable gap could be attributed to the ciliary gene list we applied is relatively conserved, including only genes from SYSCILIA gold standard and GO databases for a list of confident ciliary genes, but not the candidate genes in other databases where stronger evidence of their ciliary nature are yet to be established. It could also be that the genetic variations in the non-coding regions contribute to cilia abnormality. Further genetic studies using whole genome sequencing along with further functional studies to delineate the complex effects that perturbation of cilia structure and function have on the Hh signaling pathway in cholangiocytes in relation to the BA phenotype, are warranted.
To conclude, our findings indicate that genetic factors have a more direct role in nonsyndromic BA pathogenesis than previously thought. The excess burden of mutations in liver expressed ciliary genes, conferring absence of cilia in some patients’ bile duct, implicate that malformed cilia or reduced ciliary function in the vulnerable neonatal biliary system can lead to aberrant Hh signaling pathway. Especially when exposed to bile acids, the ciliary abnormalities may result in biliary fibrosis, inflammation, and eventually chronic liver injury. Elucidation of disease mechanisms of BA may help in the development of preventative or/and therapeutic strategies to improve the clinical outcome.
Contributors
W.Y.L. performed the genetics analyses. W.Y.L. and J.S.H contributed to bioinformatics data processing. M.T.S. prepared the samples, performed Sanger sequencing and functional studies. H.Y. generated the human cholangiocytes and performed the functional studies. P.H.Y.C., J.M.N., F.Y., C.W.D.L., D.N.N, P.A.H.N., and S.L.L. recruited the subjects, collected the samples and clinical data. H.M.M, D.J. and C.O. provided insightful comments and suggestions. W.Y.L. and C.S.M.T. wrote the manuscript and verified the genetic data. P.C.S., M.M.G.B., V.C.H.L., C.S.M.T. and P.K.H.T. designed the study and supervised the project. All authors read and approved the final version of the manuscript.
Data sharing statement
The data of this study are available within the article and in the Supplementary Information, or from the authors upon request.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
We extend our gratitude to all the patients and their families who participated in the study, the physicians who referred and treated the patients, and other members of our laboratories for their valuable contributions over the years. Authors thank the technical staff of the Electron Microscope Unit, Queen Mary Hospital, Hong Kong for their expert assistance in the scanning and transmission electron microscopy studies.
This work was supported by research grants from the Hong Kong General Research Fund (Grant Nos. 17107314 and 766112 to M.M.G.B., 17105119 to V.C.H.L. and 17109918 to P.K.H.T.) and Health and Medical Research Fund Commissioned Paediatric Research at Hong Kong Children's Hospital (Grant No. PR-HKU-1 to P.K.H.T.). P.H.Y.C. was supported by The University of Hong Kong Li Ka Shing Faculty of Medicine Enhanced New Staff Start-up Fund. C.S.M.T. was supported by Theme-based Research scheme (T12C-714/14-R). P.K.H.T., C.S.M.T. and W.Y.L were supported by Dr. Li Dak-Sum Research Fund.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103530.
Contributor Information
Vincent Chi-Hang Lui, Email: vchlui@hku.hk.
Paul Kwong-Hang Tam, Email: paultam@hku.hk.
Appendix. Supplementary materials
References
- 1.Jimenez-Rivera C., Jolin-Dahel K.S., Fortinsky K.J., Gozdyra P., Benchimol EI. International incidence and outcomes of biliary atresia. J Pediatr Gastroenterol Nutr. 2013;56(4):344–354. doi: 10.1097/MPG.0b013e318282a913. [DOI] [PubMed] [Google Scholar]
- 2.Smith B.M., Laberge J.M., Schreiber R., Weber A.M., Blanchard H. Familial biliary atresia in three siblings including twins. J Pediatr Surg. 1991;26(11):1331–1333. doi: 10.1016/0022-3468(91)90613-x. [DOI] [PubMed] [Google Scholar]
- 3.Silveira T.R., Salzano F.M., Howard E.R., Mowat AP. Congenital structural abnormalities in biliary atresia: evidence for etiopathogenic heterogeneity and therapeutic implications. Acta Paediatr Scand. 1991;80(12):1192–1199. doi: 10.1111/j.1651-2227.1991.tb11808.x. [DOI] [PubMed] [Google Scholar]
- 4.Tam P.K.H., Chung P.H.Y., St Peter S.D., Gayer C.P., Ford H.R., Tam G.C.H. Advances in paediatric gastroenterology. Lancet North Am Ed. 2017;390(10099):1072–1082. doi: 10.1016/S0140-6736(17)32284-5. [DOI] [PubMed] [Google Scholar]
- 5.Mack C.L., Sokol RJ. Unraveling the pathogenesis and etiology of biliary atresia. Pediatr Res. 2005;57(7):87–94. doi: 10.1203/01.PDR.0000159569.57354.47. [DOI] [PubMed] [Google Scholar]
- 6.Bezerra J.A., Wells R.G., Mack C.L., Karpen S.J., Hoofnagle J.H., Doo E. Biliary atresia: clinical and research challenges for the twenty-first century. Hepatology. 2018;68(3):1163–1173. doi: 10.1002/hep.29905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kilgore A., Mack C.L. Update on investigations pertaining to the pathogenesis of biliary atresia. Pediatr Surg Int. 2017;33(12):1233–1241. doi: 10.1007/s00383-017-4172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Barceló M.M., Yeung M.Y., Miao X.P., Tang C.S., Cheng G., Chen G. Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Hum Mol Genet. 2010;19(14):2917–2925. doi: 10.1093/hmg/ddq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng G., Tang C.S., Wong E.H., Cheng W.W., So M.T., Miao X. Common genetic variants regulating ADD3 gene expression alter biliary atresia risk. J Hepatol. 2013;59(6):1285–1291. doi: 10.1016/j.jhep.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Cui S., Leyva–Vega M., Tsai E.A., EauClaire S.F., Glessner J.T., Hakonarson H. Evidence from human and zebrafish that GPC1 is a biliary atresia susceptibility gene. Gastroenterology. 2013;144(5):1107–1115.e3. doi: 10.1053/j.gastro.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Gilbert M.A., Grochowski C.M., McEldrew D., Llewellyn J., Waisbourd-Zinman O. A genome-wide association study identifies a susceptibility locus for biliary atresia on 2p16.1 within the gene EFEMP1. PLoS Genet. 2018;14(8) doi: 10.1371/journal.pgen.1007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ningappa M., So J., Glessner J., Ashokkumar C., Ranganathan S., Min J. The role of ARF6 in biliary atresia. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0138381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berauer J.P., Mezina A.I., Okou D.T., Sabo A., Muzny D.M., Gibbs R.A. Identification of polycystic kidney disease 1 like 1 gene variants in children with biliary atresia splenic malformation syndrome. Hepatology. 2019;70(3):899–910. doi: 10.1002/hep.30515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girard M., Panasyuk G. Genetics in biliary atresia. Curr Opin Gastroenterol. 2019;35(2):73–81. doi: 10.1097/MOG.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 15.H. Li Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:13033997v1 [q-bioGN].
- 16.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011:43. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M.X., Gui H.S., Kwan J.S., Bao S.Y., Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of mendelian diseases. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babu R.O., Lui V.C.H., Chen Y., Yiu R.S.W., Ye Y., Niu B. Beta-amyloid deposition around hepatic bile ducts is a novel pathobiological and diagnostic feature of biliary atresia. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:Profiler – a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(suppl_2):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Dam T.J., Wheway G., Slaats G.G., Huynen M.A., Giles R.H., Group SS. The syscilia gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013;2(1):7. doi: 10.1186/2046-2530-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. (Oxford, England) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendricks A.E., Billups S.C., Pike H.N.C., Farooqi I.S., Zeggini E., Santorico S.A. ProxECAT: proxy external controls association test. a new case-control gene region association test using allele frequencies from public controls. PLoS Genet. 2018;14(10) doi: 10.1371/journal.pgen.1007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung I.D., Bagnat M., Ma T.P., Datta A., Evason K., Moore J.C. Regulation of intrahepatic biliary duct morphogenesis by claudin 15-like b. Dev Biol. 2012;361(1):68–78. doi: 10.1016/j.ydbio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garnaas M.K., Cutting C.C., Meyers A., Kelsey P.B., Harris J.M., North T.E. Rargb regulates organ laterality in a zebrafish model of right atrial isomerism. Dev Biol. 2012;372(2):178–189. doi: 10.1016/j.ydbio.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaub M., Nussbaum J., Verkade H., Ober E.A., Stainier D.Y., Sakaguchi T.F. Mutation of zebrafish snapc4 is associated with loss of the intrahepatic biliary network. Dev Biol. 2012;363(1):128–137. doi: 10.1016/j.ydbio.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins B.J., Gong W., Pack M. A novel keratin18 promoter that drives reporter gene expression in the intrahepatic and extrahepatic biliary system allows isolation of cell-type specific transcripts from zebrafish liver. Gene Expr Patterns. 2014;14(2):62–68. doi: 10.1016/j.gep.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews R.P., EauClaire S.F., Mugnier M., Lorent K., Cui S., Ross M.M. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53(3):905–914. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider C.A., Rasband W.S., Eliceiri KW. NIH image to imageJ: 25 years of image analysis. Nat Methods. 2012;9:671. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marthin J.K., Nielsen KG. Hand-held tidal breathing nasal nitric oxide measurement-a promising targeted case-finding tool for the diagnosis of primary ciliary dyskinesia. PLoS One. 2013;8(2):e57262. doi: 10.1371/journal.pone.0057262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S.L., O'Callaghan C., Lau Y.L., Lee CD. Functional analysis and evaluation of respiratory cilia in healthy Chinese children. Respir Res. 2020;21(1):259. doi: 10.1186/s12931-020-01506-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartley J.L., O'Callaghan C., Rossetti S., Consugar M., Ward C.J., Kelly D.A. Investigation of primary cilia in the pathogenesis of biliary atresia. J Pediatr Gastroenterol Nutr. 2011;52(4):485–488. doi: 10.1097/MPG.0b013e318200eb6f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sironen A., Kotaja N., Mulhern H., Wyatt T.A., Sisson J.H., Pavlik J.A. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol Reprod. 2011;85(4):690–701. doi: 10.1095/biolreprod.111.091132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurczyk A., Gromley A., Redick S., Agustin J.S., Witman G., Pazour G.J. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol. 2004;166(5):637–643. doi: 10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobin J.L., Beales PL. The nonmotile ciliopathies. Genet Med. 2009;11(6):386–402. doi: 10.1097/GIM.0b013e3181a02882. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro A.J., Davis S.D., Polineni D., Manion M., Rosenfeld M., Dell S.D. Diagnosis of primary ciliary dyskinesia. An official American thoracic society clinical practice guideline. Am J Respir Crit Care Med. 2018;197(12):e24–e39. doi: 10.1164/rccm.201805-0819ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narang I., Ersu R., Wilson N.M., Bush A. Nitric oxide in chronic airway inflammation in children: diagnostic use and pathophysiological significance. Thorax. 2002;57(7):586–589. doi: 10.1136/thorax.57.7.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker W.T., Jackson C.L., Lackie P.M., Hogg C., Lucas JS. Nitric oxide in primary ciliary dyskinesia. Eur Respir J. 2012;40(4):1024–1032. doi: 10.1183/09031936.00176111. [DOI] [PubMed] [Google Scholar]
- 39.Saternos H.C., AbouAlaiwi WA. Signaling interplay between primary cilia and nitric oxide: a mini review. Nitric Oxide. 2018;80:108–112. doi: 10.1016/j.niox.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bontems F., Fish R.J., Borlat I., Lembo F., Chocu S., Chalmel F. C2orf62 and TTC17 are involved in actin organization and ciliogenesis in zebrafish and human. PLoS One. 2014;9(1):e86476. doi: 10.1371/journal.pone.0086476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95(6):829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 42.Engelke M.F., Waas B., Kearns S.E., Suber A., Boss A., Allen B.L. Acute inhibition of heterotrimeric kinesin-2 function reveals mechanisms of intraflagellar transport in mammalian cilia. Curr Biol. 2019;29(7):1137–1148.e4. doi: 10.1016/j.cub.2019.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omenetti A., Diehl AM. Hedgehog signaling in cholangiocytes. Curr Opin Gastroenterol. 2011;27(3):268–275. doi: 10.1097/MOG.0b013e32834550b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machado M.V., Diehl AM. Hedgehog signaling in liver pathophysiology. J Hepatol. 2018;68(3):550–562. doi: 10.1016/j.jhep.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grzelak C.A., Martelotto L.G., Sigglekow N.D., Patkunanathan B., Ajami K., Calabro S.R. The intrahepatic signaling niche of hedgehog is defined by primary cilia positive cells during chronic liver injury. J Hepatol. 2014;60(1):143–151. doi: 10.1016/j.jhep.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Wong S.Y., Seol A.D., So P.L., Ermilov A.N., Bichakjian C.K., Epstein E.H. Primary cilia can both mediate and suppress hedgehog pathway–dependent tumorigenesis. Nat Med. 2009;15(9):1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huangfu D., Anderson KV. Cilia and hedgehog responsiveness in the mouse. Proc Natl Acad Sci U. S. A. 2005;102(32):11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu A.S., Russo P.A., Wells RG. Cholangiocyte cilia are abnormal in syndromic and non-syndromic biliary atresia. Mod Pathol. 2012;25:751. doi: 10.1038/modpathol.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frassetto R., Parolini F., Marceddu S., Satta G., Papacciuoli V., Pinna M.A. Intrahepatic bile duct primary cilia in biliary atresia. Hepatol Res. 2018;48(8):664–674. doi: 10.1111/hepr.13060. [DOI] [PubMed] [Google Scholar]
- 50.Karjoo S., Hand N.J., Loarca L., Russo P.A., Friedman J.R., Wells RG. Extrahepatic cholangiocyte cilia are abnormal in biliary atresia. J Pediatr Gastroenterol Nutr. 2013;57(1):96–101. doi: 10.1097/MPG.0b013e318296e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jung H.Y., Jing J., Lee K.B., Jang JJ. Sonic hedgehog (SHH) and glioblastoma-2 (Gli-2) expressions are associated with poor jaundice-free survival in biliary atresia. J Pediatr Surg. 2015;50(3):371–376. doi: 10.1016/j.jpedsurg.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 52.Xu X., Zhan J. Biliary atresia in twins: a systematic review and meta-analysis. Pediatr Surg Int. 2020;36(8):953–958. doi: 10.1007/s00383-020-04690-4. [DOI] [PubMed] [Google Scholar]
- 53.Tang V., Cofer Z.C., Cui S., Sapp V., Loomes K.M., Matthews RP. Loss of a candidate biliary atresia susceptibility gene, add3a, causes biliary developmental defects in zebrafish. J Pediatr Gastroenterol Nutr. 2016;63(5):524–530. doi: 10.1097/MPG.0000000000001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajagopalan R., Tsai E.A., Grochowski C.M., Kelly S.M., Loomes K.M., Spinner N.B. Exome sequencing in individuals with isolated biliary atresia. Sci Rep. 2020;10(1):2709. doi: 10.1038/s41598-020-59379-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.