Abstract
Large T antigens from polyomaviruses are multifunctional proteins with roles in transcriptional regulation, viral DNA replication, and cellular transformation. They have been shown to enhance the activity of various cellular transcription factors. In the case of the POU protein Tst-1/Oct6/SCIP, this enhancement involves a direct physical interaction between the POU domain of the transcription factor and the amino-terminal region of large T antigen. Here we have analyzed the structural requirements for synergistic interaction between the two proteins in greater detail. Tst-1/Oct6/SCIP and the related POU protein Brn-1 were both capable of direct physical interaction with large T antigen. Nevertheless, only Tst-1/Oct6/SCIP functioned synergistically with large T antigen. This differential behavior was due to differences in the amino-terminal regions of the proteins, as evident from chimeras between Tst-1/Oct6/SCIP and Brn-1. Synergy was specifically observed for constructs containing the amino-terminal region of Tst-1/Oct6/SCIP. Large T antigen, on the other hand, functioned synergistically with Tst-1/Oct6/SCIP only when the integrity of its J-domain-containing amino terminus was maintained. Mutations that disrupted the J domain concomitantly abolished the ability to enhance the function of Tst-1/Oct6/SCIP. The J domain of T antigen was also responsible for the physical interaction with Tst-1/Oct6/SCIP and could be replaced in this property by other J domains. Intriguingly, a heterologous J domain from a human DnaJ protein partially substituted for the amino terminus of T antigen even with regard to the synergistic enhancement of Tst-1/Oct6/SCIP function. Given the general role of J domains, we propose chaperone activity as the underlying mechanism for synergy between Tst-1/Oct6/SCIP and large T antigens.
POU proteins form a large family of transcriptional regulators with numerous functions in determination, specification, and differentiation events during development (42). Whereas all POU proteins have significant homology within their composite DNA-binding domain, the so-called POU domain (19), similarity outside the POU domain is confined to members within a given class, of which there are at least six (42, 52). Currently, there are four known members of class III in mammals, all of which are encoded by intronless genes (16). These are Brn-1, Brn-2, Tst-1/Oct6/SCIP, and Brn-4. Deletions in these genes have been shown to interfere with specific developmental processes, leading to defects in peripheral myelination (3, 21) or the endocrine hypothalamus (35, 44) or to sensorineural deafness (10). These morphologically detectable defects mark the cell types in which the respective POU proteins are uniquely expressed. The spatially restricted, clearly defined phenotype observed in knockout animals contrasts with the widespread, strongly overlapping expression of class III POU proteins during development (1, 18). This overlap probably allows POU proteins to function redundantly in cell types in which they are coexpressed.
Oligodendrocytes, which are the myelin-forming glial cells of the central nervous system, might represent such a case. All class III POU proteins except Brn-4 are expressed in cells of the oligodendrocyte lineage (45), and these cells seem to be largely unaffected by inactivation of either Brn-2 or Tst-1/Oct6/SCIP (3, 21, 35, 44). So far, no cellular gene that is dependent for its oligodendrocytic expression on class III POU proteins has been identified. However, we have previously shown that both the early and late promoters of the human papovavirus JC virus can be activated by Tst-1/Oct6/SCIP (53). Tst-1/Oct6/SCIP-dependent activation of viral gene expression might contribute to the observed tropism of JC virus for glial cells: lytic infection of JC virus can be observed only in oligodendrocytes, the destruction of which results in the demyelinating disease progressive multifocal leukoencephalopathy (31). Similar to the case for other POU proteins, transcriptional activity of Tst-1/Oct6/SCIP can be significantly enhanced by synergistic interaction with other transcription factors or coactivators, some of which are glia specific (27, 28, 33, 47). Interestingly, synergistic enhancement of Tst-1/Oct6/SCIP function could also be observed for large T antigen, the early gene product of JC virus (38). This synergy might be a crucial factor for the role of Tst-1/Oct6/SCIP in the establishment of lytic JC virus infection in oligodendrocytes. At the molecular level, synergistic interaction was paralleled by direct physical contact between the amino-terminal 82 residues of large T antigen and the POU domain of Tst-1/Oct6/SCIP (38). These initial studies already indicated that although necessary, the interaction with the POU domain was not sufficient for synergy. In addition, synergy required the amino-terminal region of Tst-1/Oct6/SCIP. Given the high sequence conservation of the amino termini of large T antigens from various papovaviruses (36), it was not surprising that large T antigen from simian virus 40 (SV40) could efficiently replace JC viral large T antigen in synergy with Tst-1/Oct6/SCIP (38).
SV40 large T antigen has been previously shown to activate a whole range of viral and cellular promoters in a manner that requires large T antigen to interact through protein-protein interactions with both the basal transcription complex and transcription factors bound to the respective promoters (13, 14, 41). This function of SV40 large T antigen has been attributed, at least in part, to its ability to augment or replace a function of the TATA-binding protein (TBP)-associated factor (TAF) TAFII250 (8) and to stabilize the TBP-TFIIa complex on certain TATA elements (9).
How the synergistic interaction between Tst-1/Oct6/SCIP and large T antigen relates to these known mechanisms of large-T-antigen-dependent transcriptional activation is unknown. Here, we have started to dissect the functional interaction between Tst-1/Oct6/SCIP and large T antigen on the molecular level, and we unexpectedly discovered an important role for the J domain of T antigen.
MATERIALS AND METHODS
Plasmids.
The POU domains of wild-type Tst-1/Oct6/SCIP, mutant Tst-1/Oct6/SCIP mt (53), and Brn-1 (45) were generated as PCR fragments and cloned into pGEX-KG to produce glutathione S-transferase (GST) fusions. pGEX-Tst-1 expressed amino acids 96 to 448 of Tst-1 (53).
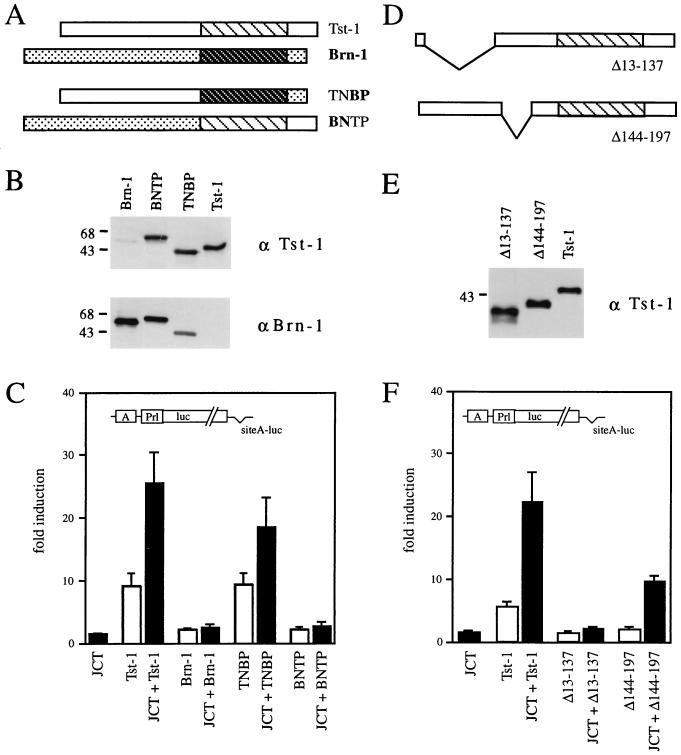
The luciferase reporter plasmids containing the early promoter of JC virus (JCearly-luc), the functional POU protein binding site from this promoter (siteA-luc), or estrogen response elements (2xERE-luc) have been described, as have the cytomegalovirus (CMV)-driven expression plasmids for Tst-1/Oct6/SCIP and Brn-1 (39, 45, 53). The Δ13-137 and Δ144-197 mutants of Tst-1/Oct6/SCIP were obtained by deleting either an internal ApaI fragment or a SmaI/PvuII fragment. Chimeras between Tst-1/Oct6/SCIP and Brn-1 were generated by introducing silent SmaI and EcoRV sites immediately in front of the POU domains of Tst-1/Oct6/SCIP and Brn-1, respectively. By using these sites, amino-terminal domains were switched with respect to the POU domain and carboxy terminus. The resulting chimeras, TNBP and BNTP (see Fig. 3A), were also inserted into CMV expression vectors.
FIG. 3.
Requirement for the amino-terminal domain of Tst-1/Oct6/SCIP. (A and D) Schematic representation of the TNBP and BNTP chimeras and the Tst-1 mutants Δ13-137 and Δ144-197 in relation to Tst-1/Oct6/SCIP and Brn-1. (B and E) Expression of TNBP and BNTP chimeric proteins and the Tst-1 mutants Δ13-137 and Δ144-197 in transfected cells, controlled by Western blot analyses of CV-1 whole-cell extracts with polyclonal antisera against Tst-1/Oct6/SCIP (α Tst-1) and Brn-1 (α Brn-1). Numbers on the left indicate sizes of molecular mass markers in kilodaltons. (C and F) Synergy of TNBP and BNTP chimeras as well as the Tst-1 mutants Δ13-137 and Δ144-197 with JC viral T antigen. The luciferase reporter plasmid siteA-luc was cotransfected with pCMV/Tst-1, pCMV/Brn-1, pCMV/TNBP, pCMV/BNTP, pCMV/Tst-1Δ13-137, pCMV/Tst-1Δ144-197, pRSV/JCT, or combinations thereof in U138 cells as indicated. Luciferase activities were determined in three independent experiments, each performed in duplicate. Values from transfections with luciferase reporter and empty expression plasmids were arbitrarily set to 1. Data from all other transfections are presented as fold induction above this level. Error bars indicate standard deviations.
Large T antigen of JC virus and the human estrogen receptor were expressed under the control of the Rous sarcoma virus (RSV) long terminal repeat and have been described before (38, 39). RSV-driven expression vectors for SV40 T antigen (pRSV-B-neo) and the dl1135, 5110, and 3213 mutants were obtained from J. Pipas, University of Pittsburgh, Pittsburgh, Pa. (48). The SV40 K67P T-antigen mutant, in which lysine at position 67 was replaced by proline, was generated from pRSV-B-neo by site-directed mutagenesis. PCR fragments corresponding to amino acids 1 to 82 of SV40 T antigen and the dl1135, 5110, and K67P mutants were inserted between the EcoRI and SalI sites of pGEX-KG in a continuous reading frame with GST. GST fusion vectors for amino acids 1 to 65 and 66 to 82 of wild-type SV40 T antigen were constructed in an analogous manner.
Plasmids pSG5-HSJ1-T and pSG5-HSJ1-HQ were obtained from J. A. DeCaprio, Dana-Farber Cancer Institute, Boston, Mass. (50, 54). The coding sequence for DnaJ from Escherichia coli (a gift from B. Bukau, Freiburg, Germany) was inserted into pCMV as an EcoRI/XbaI fragment after addition of a eukaryotic translation initiation consensus sequence.
Cell culture, transfections, and luciferase assays.
CV1 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS). U138 human glioblastoma cells were propagated in RPMI medium supplemented with 10% FCS. Two days before transfection, cells were plated at a density of 5 × 105 per 60-mm-diameter plate (1.5 × 106 per 100-mm-diameter plate) in Dulbecco’s modified Eagle’s medium with 10% FCS. Cells were transfected by the calcium phosphate technique. For extract preparations (100-mm-diameter plates), 10 μg of expression plasmids was used. For luciferase assays (60 mm-diameter plates), 2 μg of luciferase reporter plasmid was transfected with 0.2 μg of expression plasmids for POU proteins, T-antigen variants, or human estrogen receptor or with 2 μg of DnaJ. The total amount of plasmid was kept constant. At 4 h posttransfection, cells were treated for 1 min with 30% (vol/vol) glycerol in phosphate-buffered saline and placed in fresh medium. Where indicated, estrogen was added at a final concentration of 10−6 M. Cells were harvested after 48 h. Extracts were assayed for luciferase activity as described previously (53).
Nuclear and whole-cell extracts, proteins, and antisera.
Protein extracts were prepared from transiently transfected CV1 cells as described previously (47). GST fusion proteins were produced in bacteria and purified by standard procedures (38). JC viral large T antigen was produced in baculovirus-infected Sf9 cells and purified by affinity chromatography (40). Purified DnaJ protein and a corresponding antiserum were obtained from Stressgen (Victoria, Canada). Polyclonal rabbit antisera against Tst-1/Oct6/SCIP, Brn-1, and JC viral T antigen have been described previously (38, 46).
Protein-protein interaction and Western blotting.
Purified GST, GST–Tst-1, GST–Tst-1 Pou, GST–Tst-1 Pou mt, or GST–Brn-1 Pou was immobilized on glutathione-agarose beads (Sigma). Resins carrying comparable amounts of GST or its fusion derivatives were incubated for 2 h at 4°C with T antigen purified from baculovirus-infected Sf9 cells or nuclear extracts in 100 mM NaCl–20 mM HEPES (pH 7.8)–2 mM EDTA–0.2% Nonidet P-40–5 mM dithiothreitol–10% glycerol–1 μg of leupeptin per ml–1 μg of aprotinin per ml. The same buffer supplemented with 0.2% nonfat dry milk was used in incubations of Tst-1/Oct6/SCIP-containing nuclear extracts with resins carrying fusions between GST and SV40 T antigen. After extensive washing, resin-bound proteins were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels, transferred to nitrocellulose membranes, and detected by Western blotting. Western blotting was performed with polyclonal antisera, horseradish peroxidase-coupled secondary antibodies, and the ECL detection system (Amersham).
RESULTS
Papovaviral T antigen physically interacts with both Tst-1/Oct6/SCIP and the related class III POU-domain protein Brn-1.
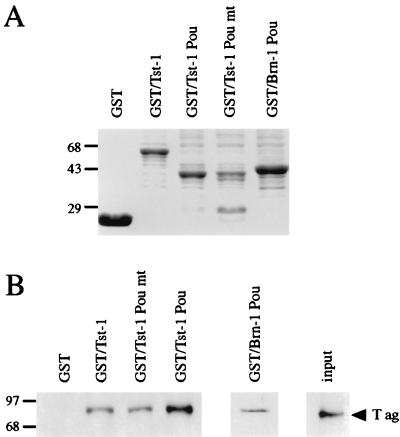
An important prerequisite for the synergistic interaction of large T antigen and Tst-1/Oct6/SCIP is the physical interaction between the two proteins. This interaction is mediated by the amino-terminal 82 amino acids of large T antigen and the POU domain of Tst-1/Oct6/SCIP (38). The POU domain is the most conserved region in all POU proteins (52), and the POU domain of Tst-1/Oct6/SCIP resembles very closely the POU domain of the related Brn-1 protein (45). Given the high degree of similarity between these two POU domains, it seemed possible that interaction with large T antigen was not restricted to Tst-1/Oct6/SCIP but could also be observed with Brn-1. To address this issue, we performed GST pulldown assays in which JC viral large T antigen purified from baculovirus-infected Sf9 insect cells was assessed for its ability to bind to a variety of immobilized GST fusion proteins (Fig. 1A). JC viral large T antigen interacted not only with the POU domain of Tst-1/Oct6/SCIP but also with the POU domain of Brn-1 both in the absence and presence of ethidium bromide (Fig. 1B and data not shown). Large-T-antigen binding was observed for both the wild-type POU domain and a mutant version in which alteration of two conserved amino acids within helix 3 of the POU homeodomain led to a severe reduction of DNA binding (53). These results confirm the existence of a bona fide protein-protein interaction of T antigen with Tst-1/Oct6/SCIP and provide evidence for a similar interaction with Brn-1.
FIG. 1.
Physical interaction of JC viral large T antigen with POU domains. (A) GST or GST fusions were immobilized on glutathione-agarose resins, purified, and analyzed on a Coomassie blue-stained SDS-polyacrylamide gel. GST fusions were prepared from the following proteins: wild-type Tst-1/Oct6/SCIP (Tst-1), the isolated POU domain of Tst-1/Oct6/SCIP (Tst-1 Pou), a mutant Tst-1/Oct6/SCIP with tryptophane-to-cysteine and phenylalanine-to-serine mutations in helix 3 of the POU homeodomain (Tst-1 Pou mt), and the isolated POU domain of Brn-1 (Brn-1 POU). (B) Purified large T antigen (T ag) was incubated with glutathione-agarose resins carrying GST or GST fusions. Resin-bound T antigen was detected after SDS-polyacrylamide gel electrophoresis by Western blotting with an anti-JC viral T-antigen antiserum. One-fifth of the amount of T antigen incubated with the resins is shown for comparison (input). The sizes of molecular mass markers are indicated in kilodaltons.
Papovaviral T antigen synergistically functions with Tst-1/Oct6/SCIP but not with Brn-1.
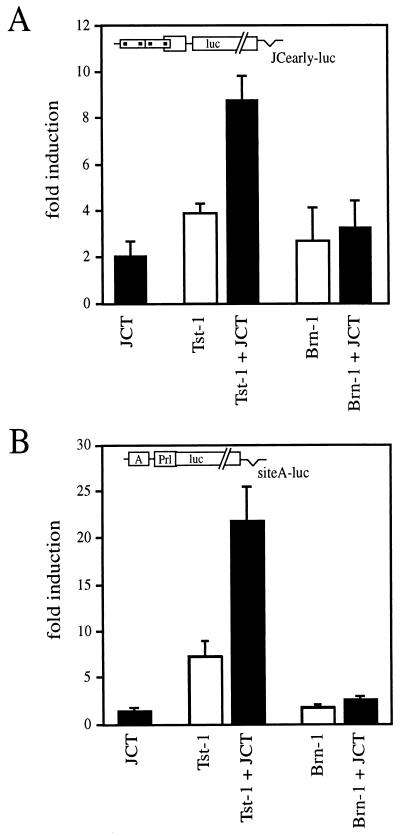
Next we asked whether the physical interaction between JC viral large T antigen and Brn-1 would be sufficient to warrant functional interaction between the two proteins in transient transfections. The early promoter of JC virus was activated approximately fourfold by ectopically expressed Tst-1/Oct6/SCIP and approximately threefold by Brn-1 (Fig. 2A). When combined with JC viral large T antigen, promoter activation by Tst-1/Oct6/SCIP increased to approximately ninefold, whereas the combination of large T antigen and Brn-1 was not any more active than Brn-1 alone. Similar results were obtained when a simple promoter was used instead of the JC viral early promoter. This simple promoter consisted of a TATA box and an adjacent AT-rich binding site for class III POU proteins taken from the JC viral promoter (45, 53). The activity of this simple promoter was stimulated 7-fold by Tst-1/Oct6/SCIP and less than 2-fold by JC viral large T antigen but 22-fold by the combination of Tst-1/Oct6/SCIP and large T antigen (Fig. 2B). When Brn-1 was cotransfected with the luciferase promoter instead of Tst-1/Oct6/SCIP, a twofold stimulation of the simple promoter was observed. Promoter activity was not further increased in the presence of both Brn-1 and large T antigen. Taken together, the results from our transient transfections indicate that physical interaction between Brn-1 and large T antigen is not sufficient for functional synergy.
FIG. 2.
Functional comparison between Tst-1/Oct6/SCIP and Brn-1. Luciferase reporter plasmids pJCearly-luc (A) and siteA-luc (B) were cotransfected with pCMV/Tst-1, pCMV/Brn-1, pRSV/JCT, or combinations thereof in U138 cells as indicated. Luciferase activities were determined in three independent experiments, each performed in duplicate. Values from transfections with luciferase reporter and empty expression plasmids were arbitrarily set to 1. Data from all other transfections are presented as fold induction above this level. Error bars indicate standard deviations.
Synergistic activation requires the amino-terminal region of Tst-1/Oct6/SCIP.
To analyze the requirements for functional synergy in greater detail, we performed domain swaps between Tst-1/Oct6/SCIP and Brn-1 (Fig. 3A). In the resulting chimeras, regions amino terminal to the POU domains were exchanged, such that the BNTP mutant contained amino terminal sequences of Brn-1 in front of the POU domain and carboxy-terminal sequences of Tst-1/Oct6/SCIP. TNBP, on the other hand, consisted of amino-terminal Tst-1/Oct6/SCIP sequences preceding the POU domain and carboxy-terminal sequences of Brn-1. The chimeric nature of these proteins was also evident from Western blot analyses (Fig. 3B). The BNTP and TNBP proteins were recognized equally well by antiserum directed against either Brn-1 or Tst-1/Oct6/SCIP, whereas Brn-1 and Tst-1/Oct6/SCIP were preferentially recognized by those antibodies that were specifically directed against the respective protein. When tested in transient transfections for activation of a POU-responsive simple promoter construct, TNBP behaved very similarly to Tst-1/Oct6/SCIP (Fig. 3C). Alone, TNBP activated the simple promoter construct ninefold. In combination with JC viral large T antigen, TNBP stimulated reporter gene expression approximately 19-fold, whereas T antigen alone only caused a minor 1.5-fold activation. BNTP, on the other hand, was capable of only a twofold activation of the same promoter construct. This rate was not further increased by the addition of JC viral large T antigen, despite the fact that the POU domain of BNTP interacts with T antigen (see Fig. 1B). Thus, our results indicate a role of the amino-terminal region of Tst-1/Oct6/SCIP in functional synergy with large T antigen, and show that the amino-terminal region of the related Brn-1 cannot substitute in this function.
To more closely define the synergy domain within the amino-terminal region of Tst-1/Oct6/SCIP, we generated two mutants in which either amino acids 13 to 137 (Δ13-137) or amino acids 144 to 197 (Δ144-197) were missing (Fig. 3D). The first 146 amino acids of Tst-1/Oct6/SCIP had previously been shown to contain the protein’s transactivation domain (32, 33). This transactivation domain is therefore absent in the Δ13-137 mutant but present in the Δ144-197 mutant, which targets a region between the transactivation domain and the DNA-binding POU domain. Both Tst-1/Oct6/SCIP mutants localized to the nucleus and were produced in comparable amounts in transfected cells (Fig. 3E and data not shown). Nevertheless, only the Δ144-197 mutant exhibited functional synergy with JC large viral T antigen, with activation rates for the simple promoter construct increasing from a mere 2-fold to a considerable 10-fold in the presence of T antigen (Fig. 3F). No significant promoter activation was obtained for T antigen alone, Δ13-137, or a combination of both. We conclude that the transactivation domain is the structure within the amino-terminal region of Tst-1/Oct6/SCIP involved in functional synergy with T antigen. Interestingly, this region has little sequence similarity to the corresponding region in Brn-1 (16), thus offering T antigen a possibility of distinguishing between the two class III POU proteins.
Synergistic activation requires the J domain of T antigen.
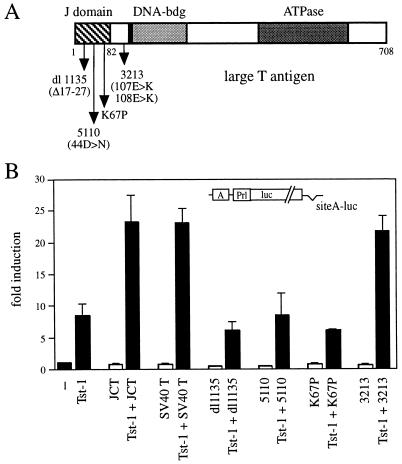
We had previously mapped the region responsible for both physical and functional interaction to the amino-terminal 82 residues of large T antigen, which the protein shares with the other early viral gene product, small t antigen (38). The same region exhibits substantial homology to a domain which is conserved in all prokaryotic and eukaryotic members of the DnaJ family of chaperones (25). Even more intriguing, recent analyses of SV40 large-T-antigen mutants indicated that this part of large T antigen indeed functions as a J domain (48, 54). We had previously shown that large T antigens from JC virus and the related SV40 not only are highly homologous in their amino-terminal domain but also functioned interchangeably when tested for their influence on the transcriptional activity of Tst-1/Oct6/SCIP (38). Therefore, we were able to test mutations proven to interfere with J-domain function of SV40 large T antigen (48) for their effect on synergy with Tst-1/Oct6/SCIP (Fig. 4). The dl1135 mutant carries a deletion of 10 amino acids (Δ17-27) in the cr1 motif of large T antigen, and the 5110 mutant is characterized by an aspartate-to-asparagine substitution at position 44 which changes a hexapeptide motif from HPDKGG to HPNKGG. This hexapeptide motif is conserved among all polyomaviruses and contains in its first half the HPD hallmark signature of all known J domains (23), which in case of T antigen is involved in hsc70 binding (4). In our transfection experiments, neither of these two mutants was capable of enhancing the activity of Tst-1/Oct6/SCIP on the simple promoter construct. Whereas the presence of wild-type JC virus or SV40 large T antigen increased Tst-1/Oct6/SCIP activity such that a 23-fold induction of the test promoter was observed instead of the 9-fold induction for Tst-1/Oct6/SCIP alone, the dl1135 and 5110 mutants even slightly decreased the activity of Tst-1/Oct6/SCIP. The same loss of functional synergy was also observed upon lysine-to-proline substitution at position 67 of SV40 large T antigen (K67P in Fig. 4). This lysine residue is situated in helix 4 of the J domain and is conserved between J domains but has so far not been evaluated for its importance in J-domain function.
FIG. 4.
Requirement for J-domain function of large T antigen. (A) Schematic representation of SV40 dl1135, 5110, K67P, and 3213 large-T-antigen mutants. DNA-bdg, DNA-binding domain. (B) The luciferase reporter plasmid siteA-luc was cotransfected in U138 cells with pCMV/Tst-1, pRSV/JCT, or pRSV-B-neo (SV40T) and its mutants (dl1135, 5110, K67P, and 3213) in the indicated combinations. Luciferase activities were determined in three independent experiments, each performed in duplicate. Values from transfections with luciferase reporter and empty expression plasmids were arbitrarily set to 1. Data from all other transfections are presented as fold induction above this level. Error bars indicate standard deviations.
The failure to functionally interact with Tst-1/Oct6/SCIP was specific for mutations within the J domain, as evident from the behavior of the large-T-antigen 3213 mutant. This mutant carries two amino acid substitutions within the cr2 region of large T antigen, which is adjacent to but not within the J domain. As a result of this mutation, the 3213 mutant is no longer able to interact with the Rb protein (48). However, when analyzed for its ability to functionally interact with Tst-1/Oct6/SCIP, the 3213 mutant exhibited the same activity as wild-type large T antigen. Our results therefore indicate that the synergy with Tst-1/Oct6/SCIP involves the J-domain function of large T antigen.
Different amino acids of the J domain of T antigen are involved in physical interaction and functional synergy with Tst-1/Oct6/SCIP.
Next we analyzed whether the same large-T-antigen mutations that interfered with functional interaction also affected the physical interaction with Tst-1/Oct6/SCIP. CV-1 extracts containing wild-type SV40 large T antigen or the dl1135, 5110, and 3213 mutants were passed over columns that carried a GST–Tst-1 fusion protein or GST alone. Analogous experiments were performed with 35S-labeled small t antigens with wild-type or mutant sequence obtained by in vitro transcription and translation. None of the large T or small t antigens bound to the GST column to significant levels, whereas all of them were retained on the GST–Tst-1 column (data not shown).
We also generated GST fusion proteins containing amino acids 1 to 82 of wild-type T antigen or the corresponding regions of the dl1135 and 5110 mutants (Fig. 5B). When used as resins in GST pulldown assays, both mutants interacted as effectively with Tst-1/Oct6/SCIP as the wild type (Fig. 5A). Thus, it has to be concluded that neither the dl1135 nor the 5110 mutation affects the physical interaction between T antigen and Tst-1/Oct6/SCIP, despite their severe impact on synergistic function.
FIG. 5.
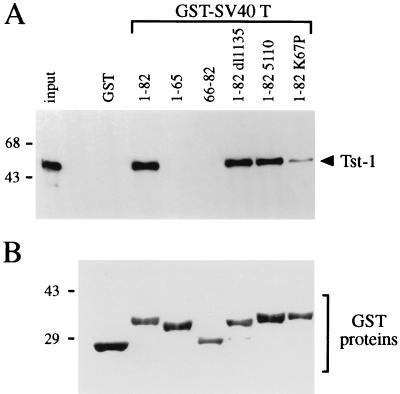
Requirement for physical interactions between the J domain of large T antigen and Tst-1/Oct6/SCIP. (A) CV1 nuclear extract containing Tst-1/Oct6/SCIP was incubated with glutathione-agarose resins carrying GST or GST fusions with SV40 T antigen (GST-SV40 T). The GST fusions contained the following T-antigen sequences: amino acids 1 to 82, 1 to 65, and 66 to 82 of wild-type T antigen (1-82, 1-65, and 66-82, respectively) and amino acids 1 to 82 of the dl1135 mutant (1-82 dl1135), of the 5110 mutant (1-82 5110), and of the K67P mutant (1-82 K67P). Resin-bound Tst-1/Oct6/SCIP was detected after SDS-polyacrylamide gel electrophoresis by Western blotting with an anti-Tst-1 antiserum. One-fifth of the amount of Tst-1/Oct6/SCIP incubated with the resins is shown for comparison (input). (B) Coomassie blue-stained SDS-polyacrylamide gel of GST proteins. The sizes of molecular mass markers are indicated in kilodaltons.
The J domain of T antigen is predicted to contain four α-helical segments (23). The dl1135 mutation disrupts helix 1 and helix 2, and the 5110 mutation is localized to an exposed loop between helix 2 and helix 3. To map the region involved in physical interaction with Tst-1/Oct6/SCIP, we generated a GST fusion protein which contained only the first 65 amino acids and lacked helix 4 of the J domain (GST-SV40 T 1-65 in Fig. 5). This mutant failed to interact with Tst-1/Oct6/SCIP in GST pulldown assays (Fig. 5A). To analyze whether helix 4 alone is sufficient for interaction with Tst-1/Oct6/SCIP, we also tested a fusion between GST and amino acids 66 to 82 of SV40 T antigen (GST-SV40 T 66-82 in Fig. 5). Again, this fusion did not interact with Tst-1/Oct6/SCIP in pulldown assays (Fig. 5A). These results indicate that helix 4 of the J domain is not sufficient for interaction but is certainly an important determinant for Tst-1/Oct6/SCIP binding.
Of particular interest were the interaction studies performed with a GST fusion carrying amino acids 1 to 82 of the K67P mutant. Contrary to the results with the dl1135 and 5110 mutants, this mutant exhibited only weak residual binding to Tst-1/Oct6/SCIP in GST pulldown assays (Fig. 5A). Our results thus indicate that physical interaction with Tst-1/Oct6/SCIP and J-domain activity are mediated by the same region of T antigen but are separate functions, as they are differentially affected by single amino acid substitutions within this region.
The POU domain of Tst-1/Oct6/SCIP specifically interacts with J domains.
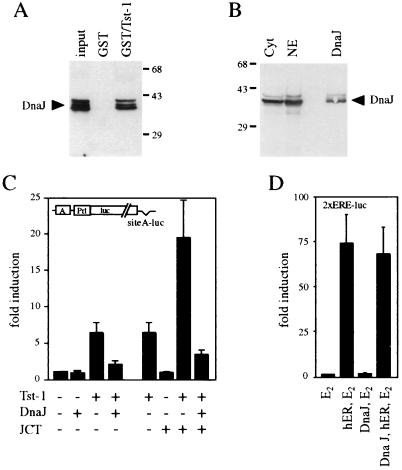
Given the fact that the J domain of large T antigen was necessary for physical and functional interaction with Tst-1/Oct6/SCIP, we wanted to know whether other J-domain proteins could substitute for large T antigen. We chose the bacterial DnaJ chaperone, which contains the prototypic J domain. First, we tested whether purified DnaJ protein would interact with the POU domain of Tst-1/Oct6/SCIP in either GST pulldown or coimmunoprecipitation experiments (Fig. 6A and data not shown). This was indeed the case.
FIG. 6.
Effects of DnaJ on synergy between Tst-1/Oct6/SCIP and large T antigen. (A) Purified DnaJ was incubated with glutathione-agarose resins carrying GST or GST/Tst-1. Resin-bound DnaJ was detected after SDS-polyacrylamide gel electrophoresis by Western blotting with a polyclonal antiserum against DnaJ. (B) Nuclear (NE) and cytosolic (Cyt) extracts from CV-1 cells ectopically expressing DnaJ were analyzed by Western blotting with the DnaJ antiserum. Numbers on the right in panel A and on the left in panel B indicate sizes of molecular mass markers in kilodaltons. (C) The luciferase reporter plasmid siteA-luc was cotransfected with pCMV/Tst-1, pCMV/DnaJ, pRSV/JCT, or combinations thereof in U138 cells as indicated. (D) The estrogen-responsive luciferase reporter 2xERE-luc was cotransfected with pRSV/hER, pCMV/DnaJ, or both in estrogen (E2)-stimulated U138 cells as indicated. Luciferase activities for panels C and D were determined in three independent experiments, each performed in duplicate. Values from transfections with luciferase reporter and empty expression plasmids were arbitrarily set to 1. Data from all other transfections are presented as fold induction above this level. Error bars indicate standard deviations.
To evaluate the capacity for functional interaction with Tst-1/Oct6/SCIP, we next expressed DnaJ in eukaryotic cells and analyzed its cellular distribution. As evident from Western blot analysis, approximately equal amounts of DnaJ were present in both cytosolic and nuclear extracts (Fig. 6B). The presence in the nucleus allowed us to test the influence of DnaJ on Tst-1/Oct6/SCIP function in cotransfection experiments. As already observed for large T antigen, DnaJ on its own did not have any significant influence on the activity of the test promoter construct (Fig. 6C). When combined with Tst-1/Oct6/SCIP, however, the presence of DnaJ became visible. Contrary to the case for large T antigen, DnaJ did not cause a synergistic stimulation of Tst-1/Oct6/SCIP activity but instead led to an almost complete inhibition of Tst-1/Oct6/SCIP. Furthermore, DnaJ interfered with the synergistic function of large T antigen. Thus, triple transfections of Tst-1/Oct6/SCIP, large T antigen, and DnaJ led to activation rates of the test promoter that were significantly below activation rates obtained with Tst-1/Oct6/SCIP alone. This strong inhibitory effect of DnaJ was not an unspecific one, as the presence of DnaJ did not interfere with the activation of an estrogen-responsive simple promoter construct by liganded estrogen receptor (Fig. 6D). Taken together, the results indicate that DnaJ and large T antigen can both physically interact with Tst-1/Oct6/SCIP. However, this interaction leads to functional inactivation of Tst-1/Oct6/SCIP in the case of DnaJ, whereas it leads to functional synergy in the case of large T antigen.
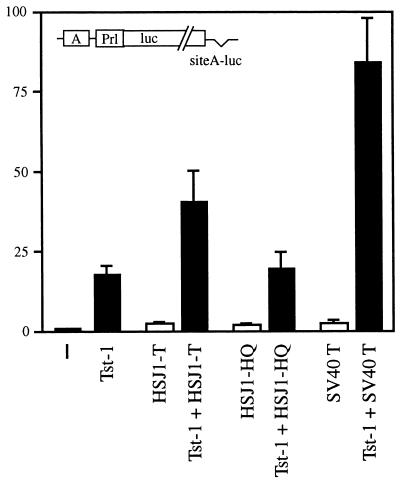
This difference could be due either to intrinsic differences in the J domains of DnaJ and T antigen or to the different contexts in which the two J domains are found. To distinguish between these possibilities, we performed transient transfections with a chimera in which the J domain of human HSJ1 was fused to amino acids 83 to 708 of SV40 T antigen, thus replacing the homologous J domain by a heterologous one (50, 54). In transfections with a POU-responsive luciferase reporter, this chimera did not influence reporter gene expression on its own (HSJ1-T in Fig. 7). When combined with Tst-1/Oct6/SCIP, however, it significantly stimulated expression rates above the level obtained with Tst-1/Oct6/SCIP alone, although induction rates were only half as high as those observed after cotransfection of Tst-1/Oct6/SCIP and wild-type SV40 large T antigen. This effect depended on a functional J domain, as inactivation of the J domain from HSJ1 abolished cooperativity between Tst-1/Oct6/SCIP and the chimera completely (HSJ1-HQ in Fig. 7). Thus, we conclude that heterologous J domains can synergistically enhance Tst-1/Oct6/SCIP function in the context of large T antigen.
FIG. 7.
Effects of a heterologous J domain on synergy between Tst-1/Oct6/SCIP and large T antigen. The luciferase reporter plasmid siteA-luc was cotransfected in U138 cells with pCMV/Tst-1, pSG5-HSJ1-T, or pSG5-HSJ1-HQ in the indicated combinations. Luciferase activities were determined in three independent experiments, each performed in duplicate. Values from transfections with luciferase reporter and empty expression plasmids were arbitrarily set to 1. Data from all other transfections are presented as fold induction above this level. Error bars indicate standard deviations.
DISCUSSION
Papovaviral large T antigens are multifunctional proteins with diverse roles in DNA replication, transcriptional regulation, virion assembly, cellular transformation, and tumorigenesis. Thus, it cannot be surprising that there exists a multitude of functionally important, partially overlapping domains within large T antigen which are required for one function or another (11). We are interested in studying the influence of JC viral large T antigen on transcriptional processes and in analyzing the underlying mechanisms. In particular, we would like to understand how JC viral large T antigen synergistically stimulates the transcriptional activity of the glial transcription factor Tst-1/Oct6/SCIP (38), which is a candidate protein for ensuring the glia specificity of viral gene expression, and consequently the tropism of this human papovavirus for glial cells, by stimulating both the viral early and late gene promoters (47, 53).
As reported recently, some glial cell types express more than one class III POU protein. Cells of the oligodendrocyte lineage, which are the ones that are lytically infected by JC virus during progressive multifocal leukoencephalopathy, express Brn-1 and Brn-2 in addition to Tst-1/Oct6/SCIP (45). Both Brn-1 and Brn-2 are closely related to Tst-1/Oct6/SCIP and are members of POU protein class III (42, 52). Furthermore, Brn-1 and Tst-1/Oct6/SCIP behave very similarly in comparative DNA binding studies (45). We have previously shown that a direct interaction between the POU domain of Tst-1/Oct6/SCIP and large T antigen is an important prerequisite for the synergy between the two proteins (38). Here, we wanted to determine whether the POU domain of Brn-1 was also able to bind to large T antigen. Given the high degree of sequence similarity between the POU domains of Brn-1 and Tst-1/Oct6/SCIP, it was indeed not surprising that Brn-1 interacted with large T antigen in a manner identical to that of Tst-1/Oct6/SCIP.
Similar binding of related POU proteins to a mutual coactivator has been described before in the case of OBF-1/Bob1/OCA-B (15, 30, 49). This B-cell-specific coactivator associates equally well with the POU domain of either Oct-1 or Oct-2. Association with either POU domain leads to activation of suitable target promoters, such as the immunoglobulin heavy-chain and κ light-chain promoters. Although it was necessary, interaction of OBF-1/Bob1/OCA-B with the POU domain was not sufficient for immunoglobulin promoter activation; the Oct-1 or Oct-2 activation domain was additionally required for functional synergy (30).
In our case, the situation is somewhat different. Although Tst-1/Oct6/SCIP and Brn-1 bound large T antigen equally well, only Tst-1/Oct6/SCIP exhibited functional synergy with the viral early protein. The interaction between large T antigen and Brn-1, on the other hand, was nonproductive. As far as synergy with large T antigen is concerned, Tst-1/Oct6/SCIP and Brn-1 did not function interchangeably, thus giving at least one example in which closely related and partially coexpressed proteins are not functionally redundant.
Similarly to what has been described for OBF-1/Bob1/OCA-B and Oct-1/Oct-2, synergistic promoter activation by Tst-1/Oct6/SCIP and large T antigen required the presence of the amino-terminal part of Tst-1/Oct6/SCIP (38). When the amino-terminal region of Tst-1/Oct6/SCIP was replaced by the amino-terminal region of Brn-1, synergy with large T antigen was lost. This clearly shows that the amino-terminal region of Tst-1/Oct6/SCIP is an important determinant for synergy and that it cannot be replaced by regions from Brn-1.
The amino terminus of Tst-1/Oct6/SCIP contains the protein’s transactivation domain between amino acids 1 and 146 (32, 33, 45). Using deletion mutants of Tst-1/Oct6/SCIP, we were able to show that it is this transactivation domain and not the adjacent amino-terminal region which is required for synergy. The amino-terminal part of Brn-1 also contains the transactivation domain (45). However, the transactivation domains of Tst-1/Oct6/SCIP and Brn-1 are localized within different parts of the amino terminus, with amino acids 1 to 146 containing the transactivation function of Tst-1/Oct6/SCIP and amino acids 119 to 237 containing the transactivation function of Brn-1 (45). Importantly, no strong homology to amino acids 1 to 146 of Tst-1/Oct6/SCIP is present in Brn-1, thus offering an explanation of why functional synergy with T antigen is restricted to Tst-1/Oct6/SCIP.
We were also able to show that the capacity to cooperate with T antigen was conferred upon the POU domain of Brn-1 by transfer of the amino-terminal region of Tst-1/Oct6/SCIP. Thus, it seems that the transactivation domain of Tst-1/Oct6/SCIP is the most decisive parameter for the specificity of the observed synergy with T antigen. A simple model for the synergy between Tst-1/Oct6/SCIP and large T antigen could therefore invoke a set of protein-protein interactions between the transactivation domain of Tst-1/Oct6/SCIP and general cofactors or components of the basal transcription machinery and a second set of complementary interactions involving the POU-domain-bound T antigen and the transcription apparatus. Such a model has been suggested for the function of OBF-1/Bob1/OCA-B (29). However, as discussed below, our results are also compatible with alternative models. These models would predict that large T antigen somehow alters the state of the transactivation domain when recruited to Tst-1/Oct6/SCIP via the POU domain, thereby allowing the transactivation domain to engage in productive interactions with the basal transcription complex.
We have shown in the past that synergy between Tst-1/Oct6/SCIP and JC viral large T antigen required amino acids 1 to 82 of large T antigen, the same region that was also involved in a direct protein-protein interaction with the POU domain of Tst-1/Oct6/SCIP (38). Although JC viral large T antigen is a known DNA-binding protein, this property was not required for synergistic enhancement of Tst-1/Oct6/SCIP function. Similar requirements have also been reported for the activation of many viral and cellular promoters by large T antigen of SV40. Here too, promoter activation was dependent on binding of a transcription factor but not on binding of SV40 large T antigen to the promoter (5, 12, 56). Instead, direct interaction between SV40 large T antigen and the transcription factor was essential (2, 14). As in the case of Tst-1/Oct6/SCIP and JC viral large T antigen, synergistic activation not only was obtained in the context of the natural promoter but was reproduced in the context of simple promoters which consisted only of a transcription factor binding site and an adjacent TATA box or initiator element (13, 14, 41). In recent years, SV40 large T antigen has also been shown to interact with TBP and numerous TAFs, leading to the assumption that large T antigen mediates contacts between transcription factors and the basal transcription machinery and thus performs a TAF-like function (2, 8, 22, 55). In line with this, SV40 large T antigen was capable of rescuing a temperature-sensitive defect in TAFII250 (8). In addition, large T antigen was recently shown to influence the rate of transcription by stabilizing the TBP-TFIIa complex on certain TATA elements (9). Where tested, small t antigen, the other early papovaviral gene product, was unable to replace SV40 large T antigen in any of these functions despite the fact that the two proteins have a common domain corresponding to amino acids 1 to 82 (2, 8, 22, 55). This contrasts with our finding that JC viral small t antigen could efficiently replace large T antigen in both direct protein-protein interaction and functional synergy with Tst-1/Oct6/SCIP (38). Thus, it is unlikely that the observed functional interaction between JC viral large T antigen and Tst-1/Oct6/SCIP can be fully explained by the above-mentioned general transcriptional activities of large T antigen.
Amino acids 1 to 82 of papovaviral large T antigens resemble a J domain, which is the evolutionarily conserved region of all known members of the DnaJ family of molecular chaperones (6, 23, 25). This J domain consists of four α-helical segments, and nuclear magnetic resonance spectroscopy indicates the presence of a finger-like structure formed by helix 2, helix 3, and the connecting invariant loop which carries the highly conserved HPD sequence (20, 37, 51). Ample evidence from the study of hybrid proteins and from biochemical analysis of large T antigen suggests that the J domain is functional and has important roles in SV40-mediated cell transformation (24, 48, 50, 54). This J domain is likewise present in large T antigen and small t antigen.
We show here that mutations within the J domain of large T antigen also interfere with the ability to enhance the function of Tst-1/Oct6/SCIP. These mutations are localized within helix 1 (dl1135) and the invariant HPD sequence (5110) of the J domain and have previously been shown to disrupt J-domain function (48). A third T-antigen variant with a mutation of the Rb binding pocket that is situated close to but not within the J domain (3213) failed to interfere with synergistic activation, pointing to the specificity of the observed effect. Of equal importance, replacement of amino acids 1 to 82 of large T antigen by a heterologous J domain left synergy with Tst-1/Oct6/SCIP largely intact, clearly indicating that J-domain function of the amino terminus of T antigen mediates synergy with Tst-1/Oct6/SCIP.
The importance of the J domain was also evident from transfections carried out in the presence of DnaJ from E. coli. DnaJ interfered both with Tst-1/Oct6/SCIP-dependent promoter activation and with T-antigen-dependent enhancement of Tst-1/Oct6/SCIP function, whereas it did not have any effect on the transcriptional activity of the unrelated estrogen receptor. This result is best explained by the inhibitory activity of regions from DnaJ situated outside the J domain. After all, DnaJ is a complex molecule that contains a number of functional domains, including a glycine- and-phenylalanine-rich module and a zinc finger in addition to its J domain (7). Similar to T antigen, DnaJ would engage Tst-1/Oct6/SCIP in a complex via its J domain, but sequences outside the J domain would not be compatible with a function of this complex in transcriptional activation. As DnaJ targets the same region of Tst-1/Oct6/SCIP that is recognized by T antigen, it would also interfere with T-antigen function by competition, as observed in our experiments. Thus, J domains function in a context-dependent manner, with both large T and small t antigens, but not DnaJ, being permissive for synergistic function. This hypothesis is also supported by the fact that the J domain of human HSJ1 is functional in the context of large T antigen.
Interestingly, neither the dl1135 nor the 5110 T-antigen mutant, which interfered with J-domain function and transcriptional synergy, had any effect on the ability of T antigen to physically interact with Tst-1/Oct6/SCIP. Binding of Tst-1/Oct6/SCIP was instead lost when amino acids corresponding to helix 4 of the J domain were deleted. Although helix 4 alone was also unable to interact with Tst-1/Oct6/SCIP, it contains important determinants for Tst-1/Oct6/SCIP binding, as evident from the K67P mutation within helix 4 that reduced interaction between T antigen and Tst-1/Oct6/SCIP to a minimal level. The altered residue is conserved between J domains, making it likely that the corresponding lysine within the J domains of DnaJ and HSJ1 also participates in the physical interaction with Tst-1/Oct6/SCIP. The fact that replacement of single amino acids within the first 82 amino acids of T antigen differentially affects J-domain function and Tst-1/Oct6/SCIP binding furthermore indicates that they are separate activities of a single domain.
The J domain has also been implicated in the T-antigen-dependent inactivation of Rb family proteins through alterations of phosphorylation status, enhanced degradation, and dissociation of the Rb-E2F complex (17, 48, 50, 54). The J domain also interacts with the hsc70 family of chaperones (43). This interaction involves the HPD motif of the J domain and is therefore missing in the 5110 mutant (4). A model has been proposed in which the J domain directs hsc70 or related proteins to Rb proteins which are part of multiprotein complexes. One such multiprotein complex contains E2F and DP proteins in addition to Rb. It is assumed that this multiprotein complex is then rearranged or disassembled, possibly by participation of hsc70 or related proteins in a T-antigen-dependent manner such that Rb is released from the complex and engaged in a new complex with T antigen (48, 54).
It is tempting to invoke a similar mechanism to explain the observed synergy between Tst-1/Oct6/SCIP and T antigen. It is possible that the amino-terminal domain of Tst-1/Oct6/SCIP is in a conformation that does not allow its interaction with general cofactors or components of the basal machinery. It could even be envisaged that the amino-terminal domain of Tst-1/Oct6/SCIP is normally engaged in some intramolecular interactions, which curtail its function. In accord with such a hypothesis, it is quite obvious from in vitro DNA binding studies, that the Tst-1/Oct6/SCIP holoprotein binds to DNA less avidly than the isolated POU domain. It would then be the function of the J domain, and possibly of the associated hsc70 chaperone, to change the conformation of the amino-terminal region such that intramolecular contacts are dissolved and productive intermolecular interactions are now allowed. In that respect, it is important to note that simultaneous interaction of Tst-1/Oct6/SCIP and hsc70 with T antigen seems possible, as the 5110 mutation of T antigen, which affects hsc70 binding (4), has no influence on binding of Tst-1/Oct6/SCIP. Other T-antigen regions outside the J domain could establish additional contacts with the transcription machinery but are not essential, as they can be replaced by small-t-antigen sequences which exhibit no apparent homology outside the J domain.
In another scenario, the amino-terminal region of Tst-1/Oct6/SCIP could be complexed with corepressors or coactivators. Such a possibility is supported by the fact that the amino-terminal domain of Tst-1/Oct6/SCIP mediated both repression and activation in tissue culture experiments (33, 34). In this case the J domain of T antigen could be instrumental in disassembling the repressive complex and instead assembling coactivators on the transactivation domain of Tst-1/Oct6/SCIP. At present, it is unclear what these coactivators could be, as all interaction partners of Tst-1/Oct6/SCIP which have been identified so far interact with the POU domain rather than with the amino-terminal transactivation domain (28, 38). It should be noted, however, that there exists a functional synergy between Tst-1/Oct6/SCIP and the recently identified HMG-box protein Sox10, which cannot be mimicked by Brn-1 (26, 27). It is tempting to speculate that this synergy could involve interactions with the amino-terminal region of Tst-1/Oct6/SCIP. Future studies will help to distinguish between these possibilities.
ACKNOWLEDGMENTS
We thank J. Pipas, J. A. DeCaprio, and B. Bukau for the gift of plasmids.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (We 1326/5-2) and the Wilhelm Sander-Stiftung (93.066.2) to M.W.
REFERENCES
- 1.Alvarez B G, Rosenfeld M G, Swanson L W. Model of forebrain regionalization based on spatiotemporal patterns of POU-III homeobox gene expression, birthdates, and morphological features. J Comp Neurol. 1995;355:237–295. doi: 10.1002/cne.903550207. [DOI] [PubMed] [Google Scholar]
- 2.Berger L, Smith D B, Davidson I, Hwang J-J, Fanning E, Wildeman A G. Interaction between T antigen and TEA domain of the factor TEF-1 derepresses simian virus 40 late promoter in vitro: identification of T-antigen domains important for transcriptional control. J Virol. 1996;70:1203–1212. doi: 10.1128/jvi.70.2.1203-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermingham J R, Scherer S S, O’Connell S, Arroyo E, Kalla K A, Powell F L, Rosenfeld M G. Tst-1/Oct6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 4.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 5.Casaz P, Sundseth R, Hansen U. trans activation of the simian virus 40 late promoter by large T antigen requires binding sites for the cellular transcription factor TEF-1. J Virol. 1991;65:6535–6543. doi: 10.1128/jvi.65.12.6535-6543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheetham M E, Brion J P, Anderton B H. Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem J. 1992;284:469–476. doi: 10.1042/bj2840469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheetham M E, Caplan A J. Structure, function, and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 9.Damania B, Lieberman P, Alwine J C. Simian virus 40 large T antigen stabilizes the TATA-binding protein–TFIIA complex on the TATA element. Mol Cell Biol. 1998;18:3926–3935. doi: 10.1128/mcb.18.7.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kok Y, van der Maarel S M, Bitner-Glindzicz M, Huber I, Monaco A P, Malcolm S, Pembrey M E, Ropers H H, Cremers F P. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- 11.Fanning E. Simian virus 40 large T antigen: the puzzle, the pieces, and the emerging picture. J Virol. 1992;66:1289–1293. doi: 10.1128/jvi.66.3.1289-1293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo G J, Gruda M C, Manupello J R, Alwine J C. Activity of simian DNA-binding factors is altered in the presence of simian virus 40 (SV40) early proteins: characterization of factors binding to elements involved in activation of the SV40 late promoter. J Virol. 1990;64:173–184. doi: 10.1128/jvi.64.1.173-184.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilinger G, Alwine J C. Transcriptional activation by simina virus 40 large T antigen: requirement for simple promoter structures containing either TAT or initiator elements with variable upstream factor binding sites. J Virol. 1993;67:6682–6688. doi: 10.1128/jvi.67.11.6682-6688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruda M, Zabolotny J, Xiao J, Davidson I, Alwine J. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens C M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 16.Hara Y, Rovescalli A C, Kim Y, Nirenberg M. Structure and evolution of four POU domain genes expressed in mouse brain. Proc Natl Acad Sci USA. 1992;89:3280–3284. doi: 10.1073/pnas.89.8.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris K F, Christensen J B, Radany E H, Imperiale M J. Novel mechanisms of E2F induction by BK virus large T antigen: requirement of both the pRB-binding and the J domain. Mol Cell Biol. 1998;18:1746–1756. doi: 10.1128/mcb.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Treacy M N, Simmons D M, Ingraham H A, Swanson L W, Rosenfeld M G. Expression of a large family of POU domain regulatory genes in mammalian brain development. Nature. 1989;340:35–42. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- 19.Herr W, Cleary M A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 20.Hill R B, Flanagan J M, Prestegard J H. 1H and 15N magnetic resonance assignments, secondary structure, and tertiary fold of Escherichia coli DnaJ(1-78) Biochemistry. 1995;34:5587–5596. doi: 10.1021/bi00016a033. [DOI] [PubMed] [Google Scholar]
- 21.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 22.Johnston S D, Yu X-M, Mertz J E. The major transcriptional transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J Virol. 1996;70:1191–1202. doi: 10.1128/jvi.70.2.1191-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley W L. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 24.Kelley W L, Georgopoulos C. The T/t common exon of SV40, JCV, and BK polyomavirus T antigens can functionally replace the J-domain of Escherichia coli Dna J molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley W L, Landy S J. Chaperone power in a virus? Trends Biochem Sci. 1994;19:277–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 26.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and Sox proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 27.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leger H, Sock E, Renner K, Grummt F, Wegner M. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol Cell Biol. 1995;15:3738–3747. doi: 10.1128/mcb.15.7.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Y, Ge H, Stevens S, Xiao H, Roeder R G. Coactivation by OCA-B: definition of critical regions and synergism with general cofactors. Mol Cell Biol. 1998;18:3803–3810. doi: 10.1128/mcb.18.7.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Y, Roeder R G. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Major E O, Amemiya K, Tornatore C S, Houff S A, Berger J R. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijer D, Graus A, Grosveld G. Mapping the transactivation domain of the Oct-6 POU transcription factor. Nucleic Acids Res. 1992;20:2241–2247. doi: 10.1093/nar/20.9.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monuki S E, Kuhn R, Lemke G. Cell-specific action and mutable structure of a transcription factor effector domain. Proc Natl Acad Sci USA. 1993;90:9978–9982. doi: 10.1073/pnas.90.21.9978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monuki S E, Kuhn R, Lemke G. Repression of the myelin PO gene by the POU transcription factor SCIP. Mech Dev. 1993;42:15–32. doi: 10.1016/0925-4773(93)90095-f. [DOI] [PubMed] [Google Scholar]
- 35.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, Shiba K, Noda T. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 36.Pipas J M. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian Y Q, Patel D, Hartl F-U, McColl D J. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol. 1996;260:224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- 38.Renner K, Leger H, Wegner M. The POU-domain protein Tst-1 and papovaviral T-antigen function synergistically to stimulate glia-specific gene expression of JC virus. Proc Natl Acad Sci USA. 1994;91:6433–6437. doi: 10.1073/pnas.91.14.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renner K, Sock E, Bermingham J R, Wegner M. Expression of the gene for the POU domain transcription factor Tst-1/Oct6 is regulated by an estrogen-dependent enhancer. Nucleic Acids Res. 1996;24:4552–4557. doi: 10.1093/nar/24.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renner K, Sock E, Gerber J-K, Wegner M. T antigen of human papovavirus JC stimulates transcription of the POU domain factor Tst-1/Oct6/SCIP. DNA Cell Biol. 1996;15:1057–1062. doi: 10.1089/dna.1996.15.1057. [DOI] [PubMed] [Google Scholar]
- 41.Rice P W, Cole C N. Efficient transcriptional activation of many simple modular promoters by simian virus 40 large T antigen. J Virol. 1993;67:6689–6697. doi: 10.1128/jvi.67.11.6689-6697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan A K, Rosenfeld M G. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- 43.Sawai E T, Butel J S. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J Virol. 1989;63:3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schonemann M D, Ryan A K, Mcevilly R J, Oconnell S M, Arias C A, Kalla K A, Li P, Sawchenko P E, Rosenfeld M G. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 1995;9:3122–3135. doi: 10.1101/gad.9.24.3122. [DOI] [PubMed] [Google Scholar]
- 45.Schreiber J, Enderich J, Sock E, Schmidt C, Richter-Landsberg C, Wegner M. Redundancy of class III POU proteins in the oligodendrocyte lineage. J Biol Chem. 1997;272:32286–32293. doi: 10.1074/jbc.272.51.32286. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber J, Sock E, Wegner M. The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain. Proc Natl Acad Sci USA. 1997;94:4739–4744. doi: 10.1073/pnas.94.9.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sock E, Renner K, Feist D, Leger H, Wegner M. Functional comparison of progressive multifocal leukoencephalopathy-type and archetype strains of JC virus. J Virol. 1996;70:1512–1520. doi: 10.1128/jvi.70.3.1512-1520.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 50.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szyperski T, Pellechia M, Wall D, Georgopoulos C, Wüthrich K. NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc Natl Acad Sci USA. 1994;91:11343–11347. doi: 10.1073/pnas.91.24.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wegner M, Drolet D W, Rosenfeld M G. POU-domain proteins: structure and function of developmental regulators. Curr Opin Cell Biol. 1993;5:488–498. doi: 10.1016/0955-0674(93)90015-i. [DOI] [PubMed] [Google Scholar]
- 53.Wegner M, Drolet D W, Rosenfeld M G. Regulation of JC virus by the POU-domain transcription factor Tst-1, implications for progressive multifocal leukoencephalopathy. Proc Natl Acad Sci USA. 1993;90:4743–4747. doi: 10.1073/pnas.90.10.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhai W, Tuan J, Comai L. SV40 large T antigen binds to the TBP-TAFI complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]
- 56.Zhu J, Rice P W, Chamberlain M, Cole C N. Mapping the transcriptional activation function of simian virus 40 large T antigen. J Virol. 1991;65:2778–2790. doi: 10.1128/jvi.65.6.2778-2790.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]