Abstract
Background/purpose
Previous studies have suggested that there is a mutual antagonism between caries and periodontitis. This research aimed to investigate the ecological connection and bacterial interaction of these two diseases.
Materials and methods
We profiled and analyzed the salivary microbiota from 124 individuals (including 38 caries patients, 34 periodontitis patients, 15 comorbid diseases patients, and 37 healthy controls) by using 16 S rRNA gene sequencing and bioinformatics approaches, and also quantified their salivary bacteria loads via quantitative real-time PCR. The putative biological functions of the salivary microbiome of the different groups were predicted by PICRUSt.
Results
We observed that both the total bacteria loads and the overall microbial richness in the saliva of the periodontitis group were higher than that in the healthy group. The principal coordinate analysis (PCoA) showed that the caries, periodontitis and healthy groups were separated from each other, and that the samples from comorbid diseases were located at the overlap of caries and periodontitis groups. Using LEfSe analysis, 20 differentially abundant genera were identified as potential biomarkers. These genera also performed complicated interactions among the four groups. Additionally, the PICRUSt analysis indicated caries-related and periodontitis-related functions (e.g., carbohydrate metabolism and bacteria proliferation) respectively.
Conclusion
We disclosed the significant differences in the salivary bacterial community under caries, periodontitis and comorbid diseases. The periodontitis group was marked by the increased complexity of the salivary microbiota. The result may have vital clinical significance to the screening and early treatment of caries-active and periodontitis-active individuals.
Keywords: Microbiota, Saliva, 16S rRNA gene, Dental caries, Periodontitis
Introduction
Dental caries and periodontitis are the two most common infectious and inflammatory diseases in the oral cavity, both of which are initiated by the oral microbiome and can progressively lead to the destructed integrity of dental and maxillofacial system. The polymicrobial etiologies and clinical manifestations are widely different in caries and periodontitis; therefore, researchers usually discuss caries and periodontal disease separately and may ignore the relationship between these two diseases. However, in clinical practices, we found that the patients prone to caries usually have better periodontal conditions, while patients who suffer from periodontitis usually have lower incidence of caries, which is quite an interesting phenomenon. Several studies have tried to investigate the connection and relationship between these two diseases. On one hand, some researches revealed a reverse relationship between the occurrence of dental caries and periodontal disease.1 After the systematic treatment of periodontal disease, the prevalence of dental caries could be increased.2 Besides, in vitro co-cultured experiments revealed that cariogenic bacteria and periodontal pathogens had antagonistic effect on each other.3 On the other hand, there were also some epidemiological studies indicating that dental caries and periodontitis occurred together.4 Until now, the relationship between caries and periodontitis has still remained unclear.
With the birth of metagenomics sequencing technology and the advanced understanding of bacterial communities, the dysbiosis of human microbiota has been implicated in the pathogenesis and development of various diseases. Therefore, clarifying the oral microbiota is also a critical part to study oral diseases. In the oral cavity, teeth and oral mucosa are surrounded by the saliva. As saliva acquisition is noninvasive, fast, safe, easy to transport and store and inexpensive, it is an important medium for studying biomarkers associated with oral health and disease.5 Therefore, we hypothesized that the diverse susceptibilities to caries and periodontitis are associated with changes of the salivary microbial communities. Despite accumulated evidence suggesting the structural and taxonomic shift of the salivary microbiota in a variety of oral diseases, limited research is comprehensively concerned with the ecological connection and bacterial interaction in the saliva of patients with caries, periodontitis, comorbid diseases (those with both caries and periodontitis) and healthy controls.
In this study, we presented the salivary bacterial profiles in patients with caries, periodontitis, comorbid diseases and healthy controls by using 16 S rRNA gene sequencing and performed comparisons of the overall community diversity, composition and function across groups with respect to different oral statues. The aim of this study was to investigate the discrepancy and relationship of the oral microbiome to caries and periodontitis, which may provide potentials for the prediction, screening and treatment strategies of these two common oral diseases.
Materials and methods
Study population
The participants in the present study were chosen from patients of the Affiliated Stomatology Hospital of Tongji University during years 2014–2016. A total of 124 adults meeting the inclusion and exclusion criteria were enrolled, which consisted of 38 dental caries-active patients, 34 periodontitis patients, 15 comorbid disease patients (those with both caries and periodontitis), and 37 healthy controls. All participants provided written informed consent before participating in the study; and the study protocols were approved by the ethics committee of faculty of medicine for human studies, school of medicine, Tongji University (ethical reference number: 2010-085; approval time: year 2010).
We performed the clinical examination according to the recommendations of the WHO (2013).6 Caries was evaluated according to criteria defined by the National Institute of Dental and Craniofacial Research (NIDCR; USA) for caries diagnosis and recording. The DFT (decayed and filled teeth) index was adopted to measure the caries status of individuals and thus to define caries-active patients (DFT ≥ 6 and untreated caries lesions ≥3).7 Periodontitis were diagnosed from clinical exams and radiographic findings, which showed deep pockets, attachment loss and bone destruction.8 The CAL (clinical attachment level), PD (pocket depth) and BOP(bleeding on probing) were adopted to measure the periodontal status and thus to define the periodontitis individuals.9 Two professional dentists who were trained and calibrated for the evaluation and sampling performed all dental examinations. Inclusion criteria were as follows: (1) caries-active patients (CA): DFT ≥ 6 (untreated caries lesions ≥ 3), PD < 3 mm and CAL < 2 (all the remaining teeth), BOP < 20%; (2) periodontitis patients (P): minimum two teeth with PD ≥ 6 mm, minimum two teeth with CAL ≥ 5 mm, BOP≥ 25%, DFT = 0; (3) patients with both caries and periodontitis (CAP): DFT ≥ 6 (untreated caries lesions ≥ 3), minimum two teeth with PD ≥ 6 mm, minimum two teeth with CAL ≥ 5 mm, BOP ≥ 25%; (4) healthy controls (H): no caries lesions, PD < 3 mm and CAL < 2 mm (all the remaining teeth), BOP<20%. Exclusion criteria of participants were as follows: (1) having other oral diseases (e.g., salivary gland disease); (2) having removable prosthesis; (3) presence of any systemic disease or cancer; (4) taking antibiotics or antifungal agents within the last 1 month prior to sampling; (5) receiving periodontal treatment within the last 1 year prior to participation; (6) smokers; (7) pregnant or breast-feeding females.
Sample collection
Oral sampling was performed 2 h after meals, drinks, and tooth brushing. After a gentle rinsing with water, 1.5 mL unstimulated saliva samples were collected using sterile tubes, then frozen at −20 °C immediately and subsequently stored at −80 °C until further analysis.
DNA isolation, PCR amplification and 16S rRNA gene sequencing
The genomic DNA was isolated using the QIAamp DNA micro Kit (Qiagen, Valencia, CA, USA), following the manufacturer's instructions.
Then the PCR amplifications and 16 S rRNA gene amplicon sequencing libraries were prepared. The universal PCR primers for the V3–V4 hypervariable regions of 16 S rRNA gene were ultimately designed as follows: forward primer (341 F: 5′-CCTAYGGGRBGCASCAG-3′) and reverse primer (806 R: 5′-GGACTACNNGGGTWTCTAAT-3′). Unique barcodes were attached to the primers in order to allow multiplex deep sequencing. Pools of amplicons from all samples were sequenced on the Illumina HiSeq 2500 sequencing platform (Illumina, San Diego, CA, USA).
Total bacteria load quantification in saliva
Quantitative real-time PCRs (qPCRs) were performed by using the Applied Biosystems QuantStudio™ 3 Real-Time PCR Instrument (Applied Biosystems, Singapore). Two pairs of universal-specific primers for qPCR targeting 16 S rRNA gene and β-actin were designed. β-actin was considered as an internal control. The primers for 16 S rRNA gene were CGGTAATACGGAGGGTGCAA (16 S–F) and CACCTGCATGCGCTTTACG (16 S-R).10 The primers for β-actin designed according to NCBI were as follows: TCATGAAGTGTGACGTGGACATC (β-Actin-F) and CAGGAGGAGCAATGATCTTGATCT (β-Actin-R).
The relative ratio of bacteria to human genome in each sample was calculated using the 2-DCt method to compare the cycle threshold (Ct).11 One-way ANOVA was used to identify the differences among the four groups.
Sequencing data processing and statistical analysis
The Trimmomatic12 and QIIME software (version 1.8)13 were applied for sequences processing and analysis. OTUs (Operational taxonomic units) were picked using an open-reference strategy against the Greengenes database with 97% identity. Microbial taxonomy was assigned by using RDP classifier.14
The estimations of alpha and beta diversities were done by QIIME. Alpha diversity was carried out to indicate the microbiota quantities with Shannon index and Chao 1 index. Beta diversity was calculated with the UniFrac distances. The Kruskal–Wallis rank sum test with a post hoc Dunn's test was performed to compare the microbial diversities among the four groups. For the detection of differential taxa, linear discriminant analysis effect size (LEfSe)15 was used, which is an algorithm for high-dimensional biomarker discovery and characterizing the genomic differences between two or more biological conditions. The ecological connection and interaction were further investigated by performing Pearson's correlations among the differential genera using Rhea pipeline.16 Finally, the microbial functions were predicted by PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states),17 and LEfSe analysis was carried out to detect inner function differentials among groups.
Result
Participant characteristics
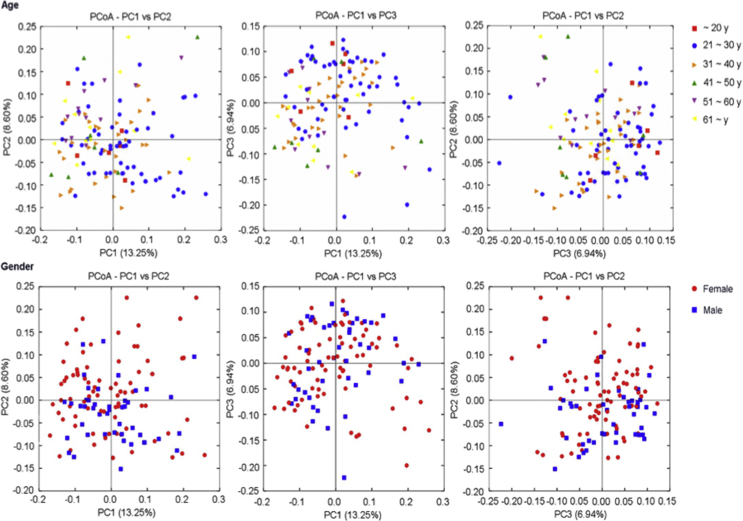
The demographic and clinical characteristics of the 124 subjects were shown in Table 1. Of these participants, 38 (30.65%) belonged to caries-active group (CA), 34 (27.42%) belonged to periodontitis group (P), 15 (12.10%) belonged to CAP group (patients with both caries and periodontitis), and 37 (29.83%) belonged to healthy controls (H). The PCoA plots (Supplementary Fig. 1) based on gender and age indicated they had no obvious influence on grouping.
Table 1.
Characteristics of the patients in the study.
| Group | Healthy control | Caries-active | Periodontitis | Caries-active and Periodontitis |
|---|---|---|---|---|
| Num. of subjects | 37 | 38 | 34 | 15 |
| Age (mean ± sd) | 27.95 ± 7.67 y | 30.61 ± 9.19 y | 43.79 ± 14.17 y | 47.33 ± 15.20 y |
| Gender (F/M) | 17 F/20 M | 29 F/9 M | 21 F/13 M | 12 F/3 M |
| DFT | 0.027 ± 0.16 | 10.53 ± 4.23 | 0.49 ± 0.74 | 8.67 ± 2.67 |
| DT | 0 | 9.11 ± 4.09 | 0.057 ± 0.24 | 6.93 ± 2.46 |
| PD (mm) | 2.23 ± 0.55 | 2.34 ± 0.47 | 7.20 ± 0.90 | 7.43 ± 0.73 |
| CAL(mm) | 1.81 ± 0.43 | 1.83 ± 0.52 | 6.89 ± 1.39 | 7.19 ± 0.83 |
| BOP(%) | 4.50 ± 2.73 | 6.07 ± 3.09 | 84.39 ± 7.07 | 81.75 ± 7.69 |
DFT(decayed-filled tooth), DT(decayed tooth), PD(probing depth), CAL(clinical attachment loss), BOP(bleeding on probing).
Relative bacterial density in saliva of the four groups
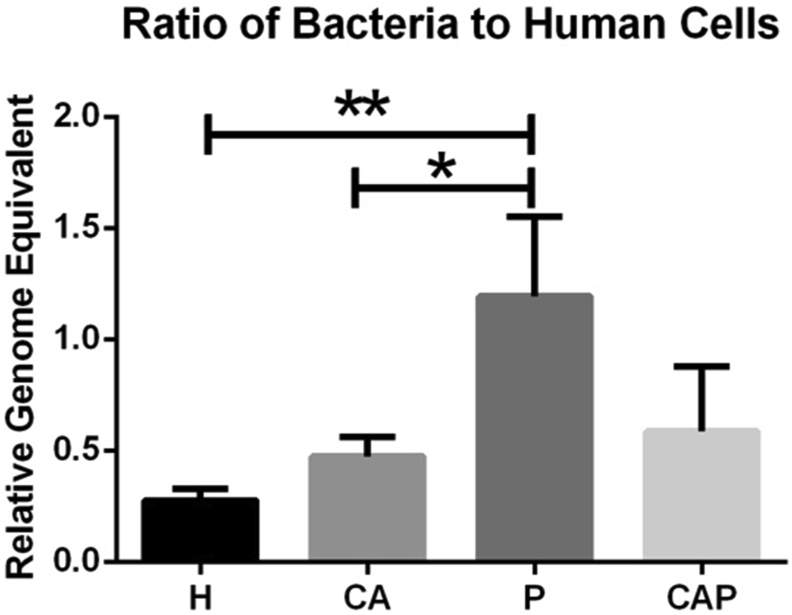
To obtain the general microbial information of the four groups, we first determined the relative bacterial density of each group by comparing the ratio of 16s rRNA gene value/β-actin11 (Fig. 1). The relative bacterial density was higher in the caries-active, periodontitis, and comorbid diseases groups than that in the healthy group, whereas a significant higher density was found in the periodontitis compared with the caries-active group (P < 0.05) and healthy controls (P < 0.05).
Figure 1.
Relative bacterial density in saliva of the four groups (CA: caries-active group, P: periodontitis group, CAP: caries-active and periodontitis group, H: healthy control group). The 16s RNA gene value/β-actin ratio of each specimen was calculated. Group P had a significantly higher density than H and CA (P < 0.05).
Distinct bacterial diversity with respect to different oral statuses
124 saliva samples were sequenced and a total of 4,436,374 high-quality sequences were obtained with an average of 35,777 sequences per sample. We then clustered the 16 S rRNA gene sequences into OTUs and ultimately detected 24 phyla, 48 classes, 85 orders, 139 families, 215 genera.
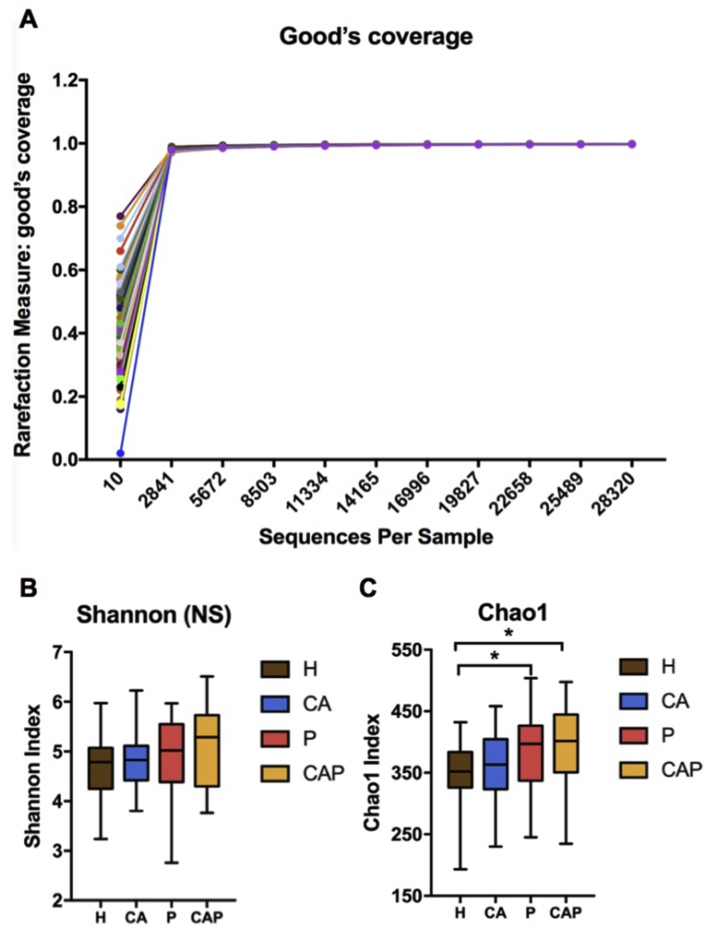
As for the Rarefaction curve, Good's coverage estimator for each sample almost approached 100% (>99.7%), indicating that the sequencing depth in this study was enough to reflect the majority of bacterial characteristics of the samples (Fig. 2A). The alpha diversity was measured by Shannon (Fig. 2B) and Chao 1 indexes (Fig. 2C). P and CAP groups had higher Chao1 index compared with healthy controls, suggesting a higher bacterial richness in individuals suffering from periodontitis (P < 0.05, Fig. 2C). However, the Shannon index considering both bacterial richness and evenness showed no significant differences among the four groups.
Figure 2.
Alpha diversity analysis of caries-active (CA), periodontitis (P), caries-active and periodontitis (CAP) and healthy controls (H). (A) Rarefaction curves of all samples. The rarefaction curves approached asymptotes for each sample when approximately 5000 sequences were extracted and Good's coverage estimator of each group came up to 99.7%, indicating that the sequencing depth was sufficient. (B) Shannon index and (C) Chao 1 index of each group. When 28,320 sequences were randomly subsampled to obtain equal numbers of sequences from each dataset, the diversity of the four communities had no significant differences based on Shannon index. However, significant differences between P and H, CAP and H were observed under Chao1 index (∗: P value < 0.05).
Changes in the microbial composition of different oral statuses
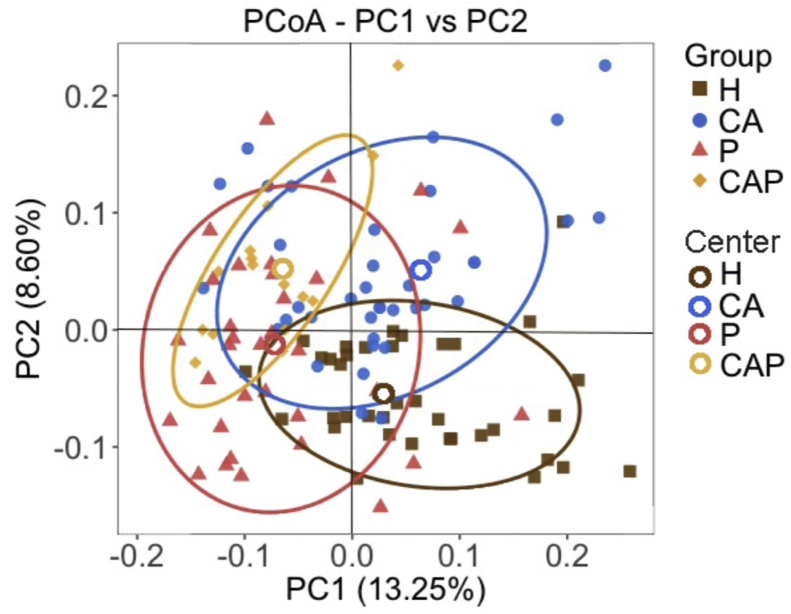
We then further compared the microbial composition among the four groups. The principal coordinate analysis (PCoA) plot based on the unweighted UniFrac distances showed a decent separation of the CA, P and H groups (Fig. 3), suggesting different bacterial structure of caries, periodontitis and healthy individuals. Moreover, the patients from the CAP group were located at the overlap of CA and P groups, which was quite reasonable.
Figure 3.
Principal coordinate analysis (PCoA) among caries-active (CA, blue), periodontitis (P, red), caries-active and periodontitis (CAP, yellow) and healthy controls (H, brown). The PCoA plot showed a reasonable separation of samples from caries-active (blue), periodontitis (red) and healthy controls (brown). The samples from CAP (yellow) were located at the overlap of caries-active and periodontitis groups and were clearly separated from the healthy controls (brown).
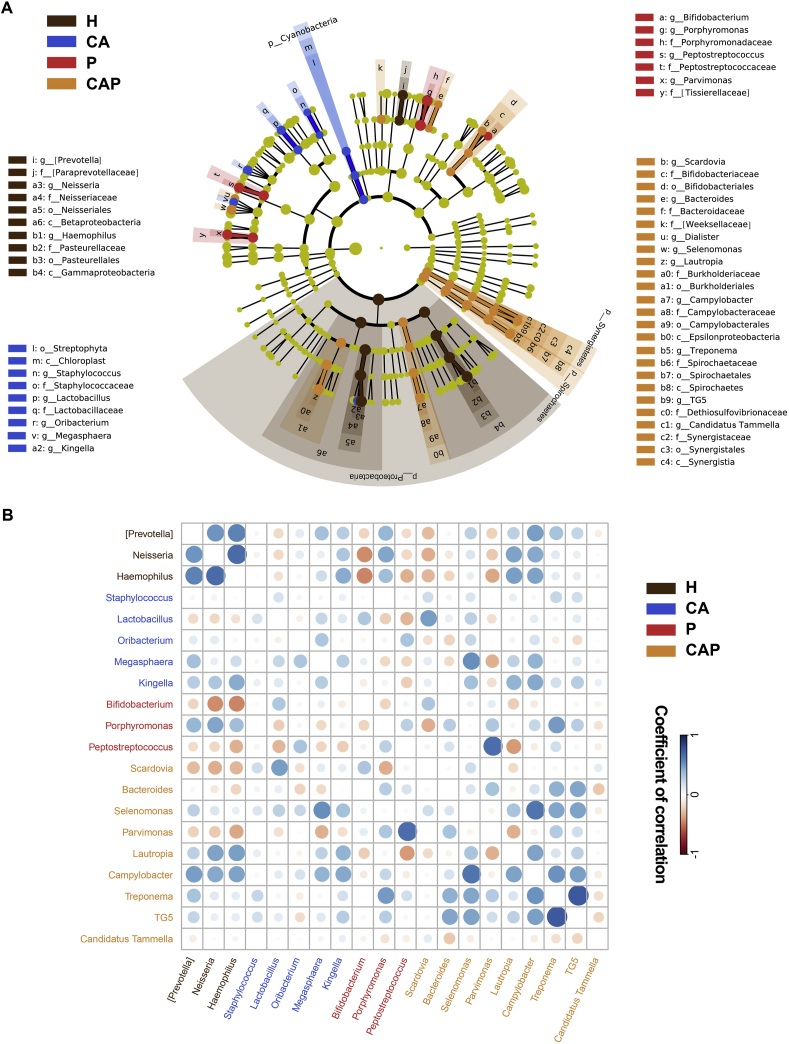
The results showed taxonomic distributions of the phyla and the most five predominant phyla included Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes and Fusobacteria. The composition of the microbial communities among the four groups was compared at different levels using LEfSe analysis (Fig. 4A). At the phylum level, Cyanobacteria was enriched in the caries-active group, and Synergistetes and Spirochaetes were more abundant in the comorbid diseases group. At the genus level, 20 differentially abundant genera were identified as potential biomarkers. The caries-active group was enriched with Staphylococcus, Lactobacillus, Oribacterium, Megasphaera and Kingella; the periodontitis group was enriched with Bifidobacterium, Porphyromonas and Peptostreptococcus; and comorbid diseases were marked by the higher Scardovia, Bacteroides, Selenomonas, Parvimonas, Lautropia, Campylobacter, Treponema, TG5 and Candidatus Tammella. Nevertheless, the [Prevotella], Neisseria and Haemophilus genera exhibited higher abundances in the healthy group.
Figure 4.
The differential taxa at different taxonomic levels (phylum, class, order, family, and genus) of the caries-active (CA, blue), periodontitis (P, red), caries-active and periodontitis (CAP, yellow) saliva in comparison with healthy controls (H, brown). (A) The significant taxa were tested by LEfSe (LDA score > 2.0) and showed using the cladogram. The cladogram graphically represents the discovered biomarkers in a taxonomic tree specified by the hierarchical feature names. (B) Pearson's correlations of differentially abundant genera among the four microbial communities. The colors of the circles represent the directions of the correlations, and their sizes correspond to the significance of the P values (the larger the circle, the smaller the P value and thus, the higher the significance).
The ecological connection and interaction were further investigated by performing Pearson's correlations among the differential genera. As shown in Fig. 4B, the enriched Lactobacillus in caries was negatively correlated with abundant Porphyromonas and Peptostreptococcus and positively correlated with abundant Bifidobacterium in periodontitis; the enriched Peptostreptococcus in the periodontitis group was negatively correlated with higher Kingella, Megasphaera and positively correlated with higher Oribacterium in caries-active group. Furthermore, the respective enriched genera in caries and periodontitis showed positive correlations with most enriched genera in comorbid diseases. In addition, Neisseria and Haemophilus were positively correlated with each other in the healthy group and were negatively correlated with Peptostreptococcus and Bifidobacterium in the periodontitis and Parvimonas and Scardovia in comorbid diseases. Collectively, these results showed great changes and complex connections in the salivary microbiota with regard to caries and periodontitis.
Diverse functions in the microbial community of different oral statuses
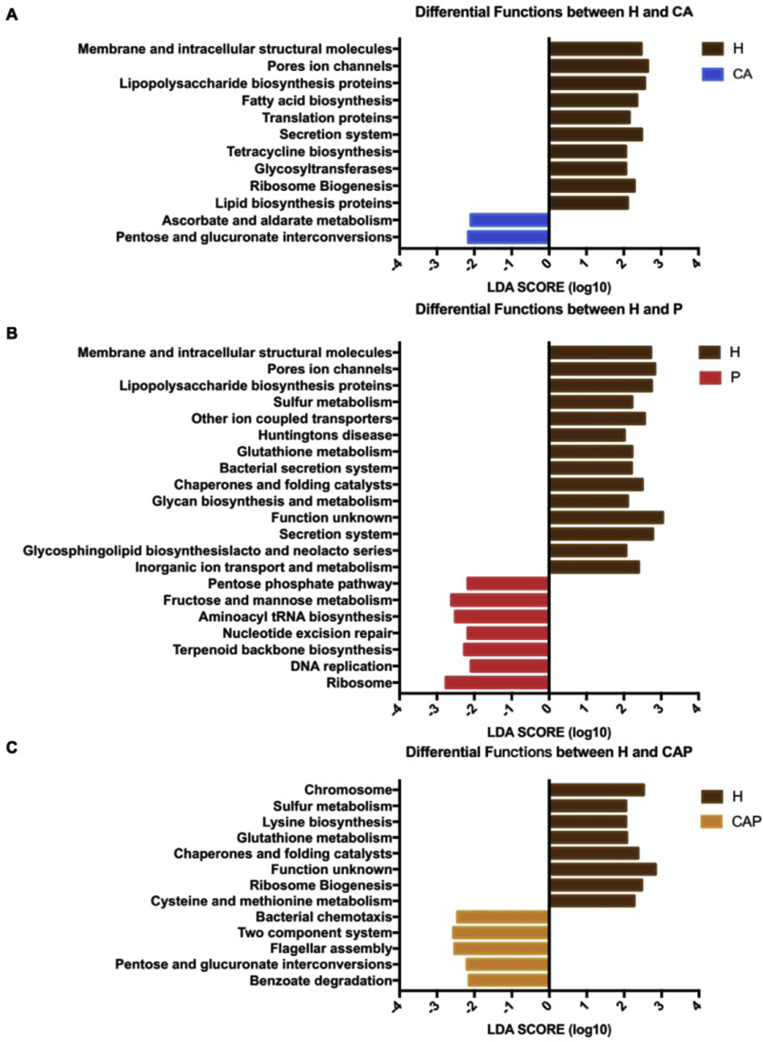
The dysbosis of microbial function is also an important reflection of diseases. Therefore, we also performed putative biological functions of the four oral statuses using the PICRUSt algorithm (Fig. 5) to comprehensively illustrate the microbial differences among the salivary ecologies. For the caries-active group, an enrichment of carbohydrate metabolism (ascorbate and aldarate metabolism, pentose and glucuronate interconversions) has been observed when compared with the healthy group. For the periodontitis group, genes involved in carbohydrate metabolism (fructose and mannose metabolism, pentose phosphate pathway), translation (ribosome, aminoacyl-tRNA biosynthesis), replication and repair (nucleotide excision repair, DNA replication) were significantly more abundant compared with healthy controls. Conversely, amino acid metabolism (glutathione metabolism), energy metabolism (sulfur metabolism), membrane transport (bacterial secretion system) were significantly decreased in periodontitis when compared with healthy controls. In the CAP group, the enrichment of carbohydrate metabolism (pentose and glucuronate interconversions), cell motility (bacterial chemotaxis, flagellar assembly) and signal transduction (two-component system) were observed. Taken together, the present study demonstrated not only compositional changes but also functional divergences among the individuals with caries, periodontitis, comorbid diseases and healthy controls.
Figure 5.
Functional changes of the caries-active (CA), periodontitis (P), caries-active and periodontitis (CAP) salivary microbiomes in comparison with healthy controls (H). Microbial functions pathways were predicted using PICRUSt algorithm and LEfSe analysis was carried out to detect differential functions among groups. Significantly differential functions between (A) caries-active group and healthy controls, (B) periodontitis group and healthy controls, (C) CAP and healthy controls were showed (LDA score > 2.0).
Discussion
The ecological connection and bacterial interaction of caries and periodontitis have drawn attention from dentists for a long time, but it has still remained unclear until now. The present study targeted on salivary microbiota in the caries-active, periodontitis, comorbid patients and healthy controls, which aimed to investigate the discrepancy and relationship of the oral microbiome to caries and periodontitis. In the process of case collection, we observed an antagonism trend of caries and periodontitis. So the number of comorbid disease patients group in this study was small.
We observed that both the total bacteria loads and the overall microbial richness in the saliva of the periodontitis group were higher than that in the healthy group, suggesting that periodontitis is characterized by an increase in the complexity of the oral microbiome. In addition, CAP group also had a higher richness compared with healthy controls. It might probably due to the local immune dysfunction and the increased gingival crevicular fluid in periodontitis, then the availability nutrients increase and it is good for bacteria growth.18
The principal coordinate analysis (PCoA) showed that caries-active, periodontitis and healthy groups were separated from each other, suggesting the structural difference of their salivary microbial communities. It was also showed that each of these three groups had its own unique salivary bacterial profiles. The result is in accordance with Belstrom D.19, 20, 21, 22 who reported that patients with periodontitis, dental caries and healthy controls all had a unique saliva bacterial profile different from each other using Human Oral Microbe Identification Microarray (HOMIM) method. They also found that bacterial gene expression and the relative abundance of specific oral bacterial species in saliva were different in caries and periodontitis by metagenomics and metatranscriptomics.23 Besides, another study had detected seven bacterial that allows discrimination of healthy and caries-affected individuals by the human oral metaproteome.24
Using LEfSe analysis, 20 differentially abundant genera were identified as potential biomarkers. The caries-active group had a higher abundance of Staphylococcus, Lactobacillus, Oribacterium, Megasphaera and Kingella in the genus level, which might be considered as screening indicators of caries. Kingella is best known as the fifth member of the HACEK group of fastidious Gram-negative bacteria and could cause a series of inflammatory diseases, such as septic arthritis, osteomyelitis, spondylodiscitis, bacteremia and endocarditis, and less frequently lower respiratory tract infections and meningitis.25
Periodontitis group had a higher abundance of Bifidobacterium, Porphyromonas and Peptostreptococcus. The Porphyromonas is a Gram-negative, non-spore-forming, obligately anaerobic genus. Porphyromonas gingivalis is one of the classic periodontal pathogen.26 Peptostreptococcus is a gram-positive anaerobe and can cause brain, liver, breast, and lung abscesses, as well as generalized necrotizing soft tissue infections. Peptostreptococcus magnus was usually regarded as the oral pathogens (gingival and teeth).27 A previous study analyzed bacterial communities in the subgingival paper point samples from individuals with gingivitis, periodontitis and no sign of disease using 454 FLX Titanium pyrosequencing.28 The results showed that high levels of Porphyromonas, Fusobacterium, Rothia, Filifactor, and Treponema genera were detected in periodontitis subject. There are some differences compared to our results and it may be due to different sampling sites.
Patients in CAP group were related to the high proportions of the genera Scardovia, Bacteroides, Selenomonas, Parvimonas, Lautropia, Campylobacter, Treponema, TG5 and Candidatus Tammella. Scardovia, one of the aciduric microbial taxa,29 was recognized as cariogenic bacteria involved in the later stage of severe early childhood caries. It has been reported that the detection of Scardovia wiggsiae and Streptococcus mutans improves the positive predictive value and the specificity of early childhood caries test.30 Selenomonas is a gram-negative anaerobe and it was reported more abundant in patients with gingivitis and periodontitis.31 Some researchers believe that due to the virulence factor of Selenomonas sputigena and its key role in agglomeration and plaque maturation, it might be an important periodontal pathogen.32 Parvimonas micra is the only species in Parvimonas genus and is a gram-positive anaerobic coccus that is most commonly isolated from plaque in chronic periodontitis patients.33 It can cause bleeding of the gum, retraction, alveolar bone loss, and tooth movement and it is the common bacteria of mixed anaerobic infections.34
Meanwhile, the higher Neisseria and Haemophilus in the healthy group suggested a protective role against caries and periodontitis. Neisseria is a large genus of bacteria that usually colonize the mucosal surfaces. Most species of this genus are believed to be commensal or nonpathogenic. The higher relative abundance of Neisseria and Haemophilus were also recorded in saliva samples from the healthy group compared with the caries group in the previous study of Belstrom D.35
In all, the result showed that each group has its own specific high abundance bacteria, which may have vital clinical significance to the screening and early treatment of caries-active and periodontitis-active individuals. Further studies are warranted to evaluate if screening of salivary microbial activity of specific oral bacterial species can identify periodontitis and dental caries at preclinical stages.
The correlation analysis showed that the enriched Lactobacillus in caries was negatively correlated with abundant Porphyromonas and Peptostreptococcus and positively correlated with abundant Bifidobacterium in periodontitis; the enriched Peptostreptococcus in the periodontitis group was negatively correlated with higher Kingella, Megasphaera and positively correlated with higher Oribacterium in caries-active group; the respective enriched genera in caries and periodontitis showed positive correlations with most enriched genera in comorbid diseases. Besides, the CAP group were located in the overlap of CA and P groups according to the PCoA result. These indicated that caries-active related bacteria and periodontitis related bacteria could co-exist and have complex interactions. It may partially explain the phenomenon that after periodontal therapy, prevalence of dental caries increased.1 The results reminded us that a stricter caries preventive program should be performed during initial periodontal therapy.
To identify how an alteration of salivary microbiota influenced caries and periodontitis, the microbial function was predicted by PICRUSt analysis.17 Notably, the functions associated with carbohydrate metabolism were abundant in the caries-active group, which might be due to the active carbohydrate metabolism during the development of caries. In addition, Ribosome, Aminoacyl-tRNA biosynthesis, Nucleotide excision repair and DNA replication, belonging to genetic information processing, were highly represented in the periodontitis group, which suggested that the functions related to bacteria proliferation were higher in the periodontitis. This may partly explain the higher diversity and density in periodontitis and CAP groups.
In conclusion, there were significant divergences in salivary microbial community under different disease conditions. The periodontitis group was marked by the increased complexity of the salivary microbiota. Each group has its own specific high abundance bacteria. Furthermore, the caries-associated bacteria and periodontitis-associated bacteria can co-exist. These results may provide potentials for the prediction, screening and treatment strategies of these two common oral diseases.
Declaration of competing interest
The authors declare that they have no conflicts of interest relevant to this article.
Acknowledgements
This work was financially supported by the Natural Science Foundation of China (81870764), and the Fundamental Research Funds for the Central Universities (1504219038).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jds.2020.12.002.
Contributor Information
Ruixin Zhu, Email: rxzhu@tongji.edu.cn.
Yuan He, Email: drheyuan@tongji.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Supplementary Figure 1.
Principal coordinates analysis (PCoA) of all 124 microbial communities according to age and gender. Different color represented different age, while female marked in red and male marked in blue
References
- 1.Iwano Y., Sugano N., Matsumoto K. Salivary microbial levels in relation to periodontal status and caries development. J Periodontal Res. 2010;45:165–169. doi: 10.1111/j.1600-0765.2009.01213.x. [DOI] [PubMed] [Google Scholar]
- 2.De Soete M., Dekeyser C., Pauwels M., Teughels W., van Steenberghe D., Quirynen M. Increase in cariogenic bacteria after initial periodontal therapy. J Dent Res. 2005;84:48–53. doi: 10.1177/154405910508400108. [DOI] [PubMed] [Google Scholar]
- 3.Kozlovsky A., Wolff A., Saminsky M., Mazor Y., Venezia E., Bar-Ness Greenstein R. Effect of Aggregatibacter actinomycetemcomitans from aggressive periodontitis patients on Streptococcus mutans. Oral Dis. 2015;21:955–961. doi: 10.1111/odi.12362. [DOI] [PubMed] [Google Scholar]
- 4.Mattila P.T., Niskanen M.C., Vehkalahti M.M., Nordblad A., Knuuttila M.L. Prevalence and simultaneous occurrence of periodontitis and dental caries. J Clin Periodontol. 2010;37:962–967. doi: 10.1111/j.1600-051X.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 5.Yoshizawa J.M., Schafer C.A., Schafer J.J., Farrell J.J., Paster B.J., Wong D.T. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev. 2013;26:781–791. doi: 10.1128/CMR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization . World Health Organization; Geneva, Switzerland: 2013. Oral health surveys: basic methods. [Google Scholar]
- 7.Yang F., Zeng X., Ning K. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J. 2012;6:1–10. doi: 10.1038/ismej.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiebe C.B., Putnins E.E. The periodontal disease classification system of the American academy of periodontology--an update. J Can Dent Assoc. 2000;66:594–597. [PubMed] [Google Scholar]
- 9.Eke P.I., Dye B.A., Wei L. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 – 2012. J Periodontol. 2015;86:611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C.-c., Pang B., Jiang X.-g., Kan B. Selection of reference genes for gene expression analysis in Vibrio cholerae. Chin J Zoonoses. 2014:433–438. [Google Scholar]
- 11.He Y., Gong D., Shi C., Shao F., Shi J., Fei J. Dysbiosis of oral buccal mucosa microbiota in patients with oral lichen planus. Oral Dis. 2017;23:674–682. doi: 10.1111/odi.12657. [DOI] [PubMed] [Google Scholar]
- 12.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso J.G., Kuczynski J., Stombaugh J. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata N., Izard J., Waldron L. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagkouvardos I., Fischer S., Kumar N., Clavel T. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. Peer J. 2017;5 doi: 10.7717/peerj.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langille M.G., Zaneveld J., Caporaso J.G. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mira A., Simon-Soro A., Curtis M.A. Role of microbial communities in the pathogenesis of periodontal diseases and caries. J Clin Periodontol. 2017;44(Suppl 18):S23–S38. doi: 10.1111/jcpe.12671. [DOI] [PubMed] [Google Scholar]
- 19.Belstrom D., Fiehn N.E., Nielsen C.H. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol. 2014;41:104–112. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- 20.Belstrom D., Fiehn N.E., Nielsen C.H. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study. Caries Res. 2014;48:368–375. doi: 10.1159/000357502. [DOI] [PubMed] [Google Scholar]
- 21.Belstrom D., Jersie-Christensen R.R., Lyon D. Metaproteomics of saliva identifies human protein markers specific for individuals with periodontitis and dental caries compared to orally healthy controls. Peer J. 2016;4 doi: 10.7717/peerj.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belstrom D., Fiehn N.E., Nielsen C.H. Differentiation of salivary bacterial profiles of subjects with periodontitis and dental caries. J Oral Microbiol. 2015;7:27429. doi: 10.3402/jom.v7.27429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belstrom D., Constancias F., Liu Y. Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. Npj Biofilms Microbi. 2017;3:8. doi: 10.1038/s41522-017-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belda-Ferre P., Williamson J., Simon-Soro A., Artacho A., Jensen O.N., Mira A. The human oral metaproteome reveals potential biomarkers for caries disease. Proteomics. 2015;15:3497–3507. doi: 10.1002/pmic.201400600. [DOI] [PubMed] [Google Scholar]
- 25.Yagupsky P. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect Dis. 2004;4:358–367. doi: 10.1016/S1473-3099(04)01046-1. [DOI] [PubMed] [Google Scholar]
- 26.Costalonga M., Herzberg M.C. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas-Jaimes J., Diaz-Tello A., Bazán C.C., Kerrigan M. Subdural empyema caused by Peptostreptococcus sp.: a complication of acute pharyngitis. Rev Inst Med Trop SP. 2017;59:e83. doi: 10.1590/S1678-9946201759083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park O.J., Yi H., Jeon J.H. Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. J Dent Res. 2015;94:921–927. doi: 10.1177/0022034515583531. [DOI] [PubMed] [Google Scholar]
- 29.Henne K., Rheinberg A., Melzer-Krick B., Conrads G. Aciduric microbial taxa including Scardovia wiggsiae and Bifidobacterium spp. in caries and caries free subjects. Anaerobe. 2015;35:60–65. doi: 10.1016/j.anaerobe.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Vacharaksa A., Suvansopee P., Opaswanich N., Sukarawan W. PCR detection of Scardovia wiggsiae in combination with Streptococcus mutans for early childhood caries-risk prediction. Eur J Oral Sci. 2015;123:312–318. doi: 10.1111/eos.12208. [DOI] [PubMed] [Google Scholar]
- 31.Hespell R.B., Paster B.J., Dewhirst F.E. The genus Selenomonas. Prokaryotes. 2006;4:982–990. [Google Scholar]
- 32.Hiranmayi K.V., Sirisha K., Ramoji Rao M.V., Sudhakar P. Novel pathogens in periodontal microbiology. J Pharm Bioallied Sci. 2017;9:155–163. doi: 10.4103/jpbs.JPBS_288_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar P.S. Smoking and the subgingival ecosystem. a pathogen-enriched community. Future Microbiol. 2012;7:917–919. doi: 10.2217/fmb.12.71. [DOI] [PubMed] [Google Scholar]
- 34.Ang M.Y., Dymock D., Tan J.L. Genome sequence of Parvimonas micra strain A293, isolated from an abdominal abscess from a patient in the United Kingdom. Genome Announc. 2013;1 doi: 10.1128/genomeA.01025-13. e01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belstrom D., Holmstrup P., Fiehn N.E. Salivary microbiota in individuals with different levels of caries experience. J Oral Microbiol. 2017;9:1270614. doi: 10.1080/20002297.2016.1270614. [DOI] [PMC free article] [PubMed] [Google Scholar]