Significance
Resistance to therapeutics is a challenge when treating cancer patients. Cancer can become resistant to therapies in distinct, unpredictable ways. Therefore, it is important to understand how resistance occurs to enable development of second-line or combination treatment strategies. This study develops cells that are resistant to inhibitors of protein arginine methyltransferase 5 (PRMT5), which is up-regulated in many cancers and being targeted in current clinical trials. We show that resistant lung adenocarcinoma cells are now sensitive to the widely used chemotherapy, paclitaxel, and identify a single gene, Stmn2, that is responsible for both the resistance to inhibitors of PRMT5 and the collateral sensitivity to paclitaxel. Our data suggest that combined treatment with PRMT5 inhibitors and taxanes could be a success.
Keywords: PRMT5i resistance, STMN2, collateral sensitivity
Abstract
Epigenetic regulators play key roles in cancer and are increasingly being targeted for treatment. However, for many, little is known about mechanisms of resistance to the inhibition of these regulators. We have generated a model of resistance to inhibitors of protein arginine methyltransferase 5 (PRMT5). This study was conducted in KrasG12D;Tp53-null lung adenocarcinoma (LUAD) cell lines. Resistance to PRMT5 inhibitors (PRMT5i) arose rapidly, and barcoding experiments showed that this resulted from a drug-induced transcriptional state switch, not selection of a preexisting population. This resistant state is both stable and conserved across variants arising from distinct LUAD lines. Moreover, it brought with it vulnerabilities to other chemotherapeutics, especially the taxane paclitaxel. This paclitaxel sensitivity depended on the presence of stathmin 2 (STMN2), a microtubule regulator that is specifically expressed in the resistant state. Remarkably, STMN2 was also essential for resistance to PRMT5 inhibition. Thus, a single gene is required for both acquisition of resistance to PRMT5i and collateral sensitivity to paclitaxel in our LUAD cells. Accordingly, the combination of PRMT5i and paclitaxel yielded potent and synergistic killing of the murine LUAD cells. Importantly, the synergy between PRMT5i and paclitaxel also extended to human cancer cell lines. Finally, analysis of The Cancer Genome Atlas patient data showed that high STMN2 levels correlate with complete regression of tumors in response to taxane treatment. Collectively, this study reveals a recurring mechanism of PRMT5i resistance in LUAD and identifies collateral sensitivities that have potential clinical relevance.
Resistance to targeted chemotherapies is an important clinical problem. Development of drug-resistant cell lines can provide key insight into the origins and mechanisms of resistance, informing better treatment strategies. The first targeted therapies were developed against kinases that promote oncogenic signaling. These early kinase inhibitors were remarkably successful in a subset of patients, but resistance typically emerged. A common mechanism of resistance for many early generation kinase inhibitors was the presence of point mutations within the target protein that impaired drug binding (1). At a broader level, it is now clear that resistance to targeted therapies can result from either inhibitor-specific mechanisms, such as mutations that impact drug binding or up-regulation of bypass pathways, or inhibitor-agnostic mechanisms (1). Numerous studies found that these resistant variants typically existed within the tumor prior to therapy, due to high mutational burden and tumor heterogeneity, and drug treatment simply drove their enrichment within the reemerging tumors (1). This forged the prevailing view that resistance to targeted therapies develops primarily through selection of preexisting cells with genetic mutations.
Recently, there has been a growing appreciation of the importance of epigenetic regulation in cancer. Mutation, amplification, or gene expression changes of epigenetic regulators can drive cellular plasticity and tumor heterogeneity, which contributes to cancer development and progression (1). Epigenetic changes can also yield resistance by enabling gene expression changes that decrease drug sensitivity (2). These nongenetic resistance variants may preexist within the tumor population, either as stable variants or in a preresistant state that is stabilized by the presence of drug (3). Recently, a few studies have reported resistance to targeted therapies against B-RAF, MEK, and BET in which the drug-induced formation of stable, nongenetic resistant states, sometimes called Lamarckian induction (4–12). In the case of BET inhibitors, induced resistance has been observed in various different tumor types (5, 13), suggesting that this is the preferred resistance mechanism. In contrast, for other epigenetic regulators, including HDAC (14) and EZH2 (15), inhibitor resistance results from a variety of preexisting mechanisms, more similar to resistance arising from other targeted therapies. Thus, it is an open question whether Lamarckian induction is a common mechanism of resistance for inhibitors of epigenetic regulators or whether BET inhibitors are an exceptional case.
Regardless of the mechanism by which resistance arises, it is of great clinical importance to understand whether, and how, the resistant state alters the response to other therapeutic treatments by creating collateral resistance or collateral sensitivity to another drug. A classic example of collateral resistance is up-regulation of efflux pumps, which can yield broad resistance to many drugs and have dire clinical consequences (1). Collateral sensitivities are of particular clinical interest because they offer opportunities to treat relapsed tumors or to be used together with the first inhibitor as up-front therapy. Collateral sensitivities have been extensively pursued in the context of antibiotic resistance (16), but there are far fewer examples in cancer research.
Given the documented importance of epigenetic regulation in cancer development, numerous companies are developing small-molecule inhibitors to target these regulators. Many of these therapies are still in clinical trials, and thus it remains an open question whether they will be effective and if resistance will arise. We have pursued this question for PRMT5 inhibitors (PRMT5i). PRMT5 is a type II arginine methyltransferase, which catalyzes the symmetric dimethylation of arginine (SDMA) on a wide variety of targets including histones, transcription factors, splicing factors, translation factors, and signaling regulators (17). PRMT5 is highly expressed in embryonic stem cells, where it is essential for proliferation and self-renewal (18). PRMT5 levels are low in postmitotic tissues but up-regulated in a wide variety of tumor types (17). EPZ015666 is the first-in-class PRMT5i and acts by binding the substrate binding pocket (19). It is highly selective for PRMT5, versus other PRMTs, but has low potency (19). Its higher potency derivative, GSK3326595, is currently in clinical trials (20, 21). Many other PRMT5is have been developed (21–30), of which JNJ64619178, PF-06939999, and PRT543, are also in clinical trials (31–33). Notably, some work through distinct mechanisms from EPZ015666 and GSK3326595, for example JNJ64619178 targets the SAM binding domain (21). Importantly, cell line studies show that PRMT5i are effective in a wide variety of tumors (34). However, even within a tumor type, the responses vary greatly, and the determinants are not understood. Thus, there is strong rationale to identify potential mechanisms of PRMT5i response and resistance.
We previously discovered that PRMT5 inhibition suppresses glioblastoma, including being effective in K-ras and Tp53 (KP) mutant lines (35). We chose to extend our analyses of PRMT5i to lung adenocarcinomas (LUAD) because these tumors are often KP mutant and lack effective treatments, show a significant correlation between high PRMT5 expression and lower survival (17, 36), have reduced proliferation from PRMT5 knockdown (37), and are included in the current PRMT5i clinical trials (20, 31). We find that murine KP mutant LUAD lines initially respond to PRMT5i, but resistance emerges rapidly via Lamarckian induction, which is conserved in all of our independently generated resistant lines. Importantly, the resistant state brings collateral sensitivities to several standard-of-care chemotherapies, particularly paclitaxel. Remarkably, we found that one gene in the resistance signature, Stmn2, is necessary for both PRMT5i resistance and paclitaxel sensitivity. Moreover, these findings have clinical relevance, as we detected a highly significant association between high levels of human STMN2 and complete regression of tumors treated with taxanes in The Cancer Genome Atlas (TCGA) patient data and found that the combination of PRMT5i and paclitaxel is potent and synergistic in murine and human cell lines.
Results
Inhibition of PRMT5 in KP LUAD Cells Causes Proliferation Defects and Induces Apoptosis.
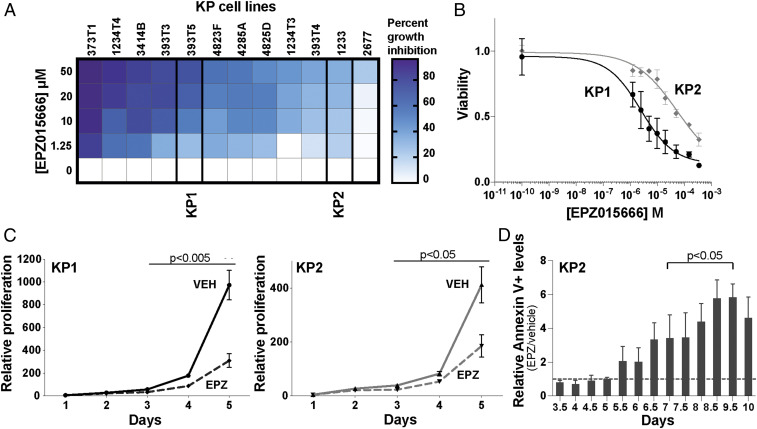
To analyze the response to PRMT5 inhibition in KP mutant LUAD, we used a panel of 12 murine cell lines generated from independent tumors arising in an autochthonous KP mutant mouse model. These lines showed cell-to-cell variation in morphology, suggesting that they retained heterogeneity. We treated these murine LUAD lines for 5 d with vehicle control (DMSO) and a range of EPZ015666 concentrations (1.25 to 50 µM) that are typical for this first-in-class PRMT5i. Quantification of live cells showed that EPZ015666 treatment yielded growth inhibition in every line but with significant variation in sensitivity (Fig. 1A). We selected a relatively sensitive line (KP1) and a relatively resistant line (KP2) for further characterization and validated their differential sensitivities over an extended dose range (Fig. 1B).
Fig. 1.
Inhibition of PRMT5 in KP LUAD cells causes proliferation defects and induces apoptosis. (A) Percent growth inhibition of 12 KP cell lines after treatment with EPZ015666 (EPZ) was quantified after 5 d (n = 3 replicates/condition). (B) Graph shows the response of KP1 (black) and KP2 (gray) to EPZ015666 after 5-d treatment (n = 5 replicates/condition). Viability is relative to DMSO control. (C) Analysis of relative proliferation of KP1 and KP2 cells (n = 3 replicates/condition) treated with vehicle control (KP1: black; KP2: gray) or 10 µM EPZ015666 (dashed). Significance between vehicle- and EPZ-treated cells was determined using Welch’s t test with the Holm–Sidak correction. (D) Relative Annexin-V levels, comparing KP2 cells treated with 10 µM EPZ015666 versus vehicle control at the indicated time points (n = 3 replicates/condition). The dotted line indicates equal levels of apoptosis between the two conditions. Significance was determined using Welch’s t test with the Holm–Sidak correction: (day 7 to day 9.5: adjusted p < 0.03).
To determine whether EPZ015666 was acting on target, we treated the KP1 cells with two different PRMT5 small interfering RNAs (siRNAs) and found that these shifted the EPZ015666 dose–response curve toward increased sensitivity in accordance with the degree of PRMT5 knockdown (SI Appendix, Fig. S1A). We next wanted to determine what concentrations of EPZ015666 were able to inhibit PRMT5 activity. When we treated the KP1 cells for 5 d with vehicle control, 0.1, 1, 5, or 10 µM EPZ015666, and immunoblotted whole-cell lysates for pan-SDMA, we found that PRMT5 inhibition increases across this range and is only effective at 5 and 10 µM (SI Appendix, Fig. S1B). We decided to analyze the biological consequences of PRMT5 inhibition, selecting 10 µM EPZ015666 as this corresponds to 50% growth inhibition of KP1 after 5 d (SI Appendix, Fig. S1B).
To determine the impact of PRMT5 inhibition, we first assessed cell proliferation using a fluorescent label retention assay and observed impairment in both KP1 and KP2 lines that achieved significance after 3 d of treatment (KP1: adjusted p = 0.0044, KP2: adjusted p = 0.0270; Fig. 1C). We then screened for apoptosis. Measuring cleaved caspase 3 and 7 activity revealed a significant drug-induced induction of apoptosis in both KP1 and KP2 cells at day 6 of treatment (SI Appendix, Fig. S1C). We then assessed the kinetics of apoptosis induction through analysis of Annexin-V staining of KP2 cells (Fig. 1D). This revealed increased levels in the drug-treated cells, versus controls, which was first apparent at day 5.5 and achieved significance at day 7 (Fig. 1D). Thus, PRMT5 inhibition of LUAD lines causes proliferation defects and apoptosis. Notably, although significant apoptosis continued beyond day 7, we observed rare, dispersed healthy cells that subsequently expanded into small colonies under the continuous presence of EPZ015666. This suggested that a subset of the KP LUAD cells displayed resistance to PRMT5 inhibition.
PRMT5i-Resistant KP LUAD Cells Arise via Acquired Resistance.
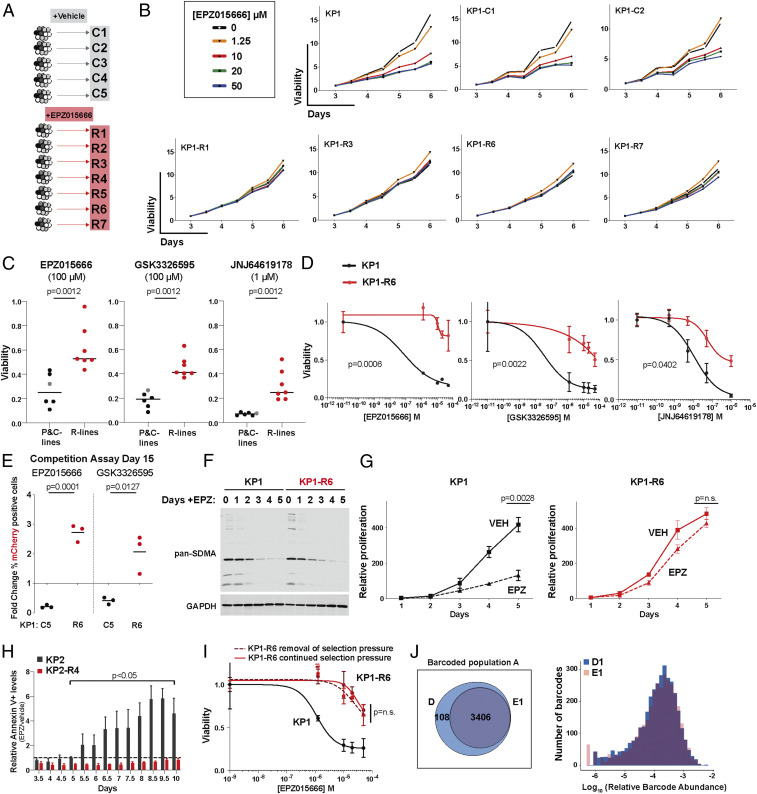
Given the detection of surviving cells, we set out to generate PRMT5i resistant variants of our KP1 and KP2 lines. As schematized (Fig. 2A), we plated multiple independent aliquots of 4 × 104 cells for both KP1 and KP2 and treated these with either vehicle (DMSO; n = 5) or 10 µM EPZ015666 (n = 7). Each aliquot was maintained in vehicle or EPZ015666 for 12 d, splitting and replating 4 × 104 cells every 4 d, and then allowed to expand in the same treatment conditions, to enable selection of viable, proliferating populations. We then tested the drug response of the selected populations by treating them with a range of EPZ015666 concentrations (0 to 50 µM) and measuring viability between 3 and 6 d of treatment (Fig. 2B and SI Appendix, Fig. S2 A and B). The vehicle-selected populations all responded in a comparable manner to their relevant parental (KP1 or KP2) line, while each of the drug-selected populations were significantly more resistant, showing little or no impairment in the presence of 10, 50, or even 100 µM EPZ015666 (Fig. 2 B and C and SI Appendix, Fig. S2 A and B). Based on these observations, we named these control (C) and resistant (R) lines, respectively.
Fig. 2.
PRMT5i-resistant KP lung adenocarcinoma cells arise via acquired resistance. (A) Schematic of the generation of R cell lines, selected with 10 µM EPZ015666 (EPZ), versus C lines, selected in 0.01% DMSO. (B) Relative growth of parental (KP1), representative control (KP1-C1, C2), and resistant (KP1-R1, R3, R6, R7) lines at indicated times and doses of EPZ015666 treatment (n = 3 replicates/condition). Growth is normalized to day 3. (C) Parental KP1 (gray), C (black; n = 5), and R (red; n = 7) lines were grown for 6 d in vehicle, 100 µM EPZ015666, 100 µM GSK3326595, or 1 µM JNJ64619178. Viability was measured with resazurin and is relative to DMSO control. For significance calculations (Mann–Whitney U test), the parental and C lines were collectively compared to the R lines. (D) Graph shows the response of KP1 (black) versus KP1-R6 (red) to the indicated PRMT5i after a 5-d treatment (n = 3 replicates/condition). Viability is relative to DMSO control. Significance was determined using Student’s t test. (E) mCherry-labeled C or R cell lines were mixed with unlabeled parental cells in a 10:90 ratio and treated with vehicle control or the indicated PRMT5i for 15 d. Graph shows the fold change in the mCherry-labeled cells with drug treatment, normalized to the DMSO control. Significance determined by Student’s t test. (F) Western blot for pan-SDMA in lysates of KP1 and KP1-R6 at indicated time points of treatment with 10 µM EPZ015666. (G) Relative proliferation of KP1 (black) and KP1-R6 (red) grown in vehicle (solid) or 10 µM EPZ015666 (dashed) for 5 d (n = 3 replicates/condition). Significance determined by Welch’s t test with the Holm–Sidak correction. (H) Relative Annexin-V levels for KP2 (gray) and KP2-R4 (red) after treatment with 10 µM EPZ015666 (n = 3 replicates/condition). Significance determined by Welch’s t test with the Holm–Sidak correction. (I) Graph shows the EPZ015666 response of KP1 parental cells (black) versus KP1-R6 variants that, prior to treatment, had been grow for 10 passages in the presence (red; continued selection pressure) or absence (red dashed; removal of selection pressure) of 10 µM EPZ015666. Viability and significance as in D. (J) Representation (Venn diagram) and relative abundance (graph) of barcodes in the KP1 parental cells (population A) after selection of surviving cells with 10 µM EPZ015666 (representative sample E1) versus DMSO vehicle (D).
We next asked whether the resistance phenotype of the R lines was specific to EPZ015666 or whether it extended to two other PRMT5i currently in clinical trials: the EPZ015666-derivative GSK3326595 and the more-potent inhibitor JNJ64619178, which inhibits PRMT5 through a distinct mechanism. We found that JNJ64619178 was a more-potent inhibitor of our parental KP1 line than EPZ015666 or GSK3326595 and is effective in the nanomolar range (SI Appendix, Fig. S2C). We selected concentrations of these drugs that were highly effective at targeting the parental KP1 cells: 100 µM EPZ015666, 100 µM GSK3326595, and 1 µM JNJ64619178 (SI Appendix, Fig. S2C). In crystal violet staining assays, these drug doses resulted in similar dramatic loss of viable KP1 parental cells or KP1-C lines but were all significantly less effective (p = 0.0012) at eliminating the KP1-R lines (Fig. 2C and SI Appendix, Fig. S2D). Dose–response curve experiments for a representative R line, KP1-R6, confirmed that the IC50 values were dramatically shifted for all three drugs (from >40-fold to >300-fold), compared to the KP1 parental control (Fig. 2D and SI Appendix, Fig. S2E). We also directly compared the viability of representative R and C lines to their parental controls by conducting competition assays in which fluorescently labeled R (KP1-R6, KP1-R7, and KP2-R1) or C (KP1-C4, KP1-C5, and KP2-C1) lines were mixed in a 10:90 ratio with the relevant, unlabeled parental line and then cultured in DMSO, 10 µM EPZ015666, or 10 µM GSK3326595. After 15 d of coculture, treatment with EPZ015666 or GSK3326595 did not result in significant enrichment of the three C populations compared to the DMSO control (Fig. 2E and SI Appendix, Fig. S2F). In stark contrast, the three R populations all showed significant drug-induced enrichment, in some cases reaching 90% of the population (Fig. 2E and SI Appendix, Fig. S2F). We did observe some variation in the ability of the C-labeled lines to compete against the parental line in vehicle control, but the profound enrichment of the R lines in the presence of PRMT5i greatly exceeded this variability. Collectively, the reduced sensitivity to PRMT5 inhibition, and the ability to outcompete the parental cells, suggests that our R lines are broadly resistant to different PRMT5 inhibitors.
Having validated the resistance properties of our R lines, we next probed the nature of this resistance. The simplest explanations were that the basal level of PRMT5 activity was increased or that PRMT5 was no longer inhibited in the R lines. To address these possibilities, we treated KP1-R6 and the parental KP1 cells with 10 µM EPZ015666 for 5 d and then immunoblotted whole-cell lysates for SDMA marks, using the pan-SDMA antibody. Remarkably, the KP1-R6 and parental KP1 cells showed no apparent difference in the levels or pattern of SDMA-modified proteins before drug treatment (Fig. 2F). Moreover, PRMT5 inhibition reduced the SDMA levels to the same extent, and with similar kinetics, in both contexts (Fig. 2F). Thus, the resistance phenotype cannot be explained by a gross difference in PRMT5 basal activity or a bypass of effective inhibition.
We next considered the cellular consequences of PRMT5 inhibition. Our data show that PRMT5 inhibition impairs proliferation and induces apoptosis in the parental LUAD lines. Given this, we hypothesized that the resistant populations bypass the cellular consequences of PRMT5 inhibition by altering their proliferative capacity. However, direct comparison of representative R lines with parental controls showed that the R and C lines had the same, or higher, proliferative capacity as the parental lines in the absence of PRMT5i (SI Appendix, Fig. S2 G and H). We then examined the effect of drug treatment on proliferation. Strikingly, while 10 µM EPZ015666 profoundly suppressed the proliferation of the KP1 parental cells (p = 0.0006), it did not significantly reduce the proliferation rate of representative KP1-R lines KP1-R6 (Fig. 2G) and KP1-R7 (SI Appendix, Fig. S2I). Consistent with the differential proliferative response of parental and R cells, the EPZ015666-induced apoptosis occurring in parental cells was completely rescued in a corresponding R line (Fig. 2H). Given these observations, we conclude that PRMT5 inhibition still efficiently suppresses PRMT5 activity and SDMA marks in the R cells, but this fails to trigger the proliferative impairment and consequent apoptosis that is a hallmark of the parental response to PRMT5 inhibition.
We next asked whether resistance to PRMT5 inhibition was stable (independent of drug) or transient (requiring the continued presence of drug for maintenance). For this, we cultured two representative KP1-R lines with or without EPZ015666 for 10 passages and then challenged the resulting variants with a range of EPZ015666 concentrations. For both of the KP1-R lines, the two variants (with or without continued selection pressure) had comparable proliferation rates in either the presence or absence of 10 µM EPZ015666 (SI Appendix, Fig. S2 J and K) and also showed indistinguishable resistance to EPZ015666 (Fig. 2I and SI Appendix, Fig. S2L). Thus, the resistance phenotypes of the R cell lines remain stable in the absence of EPZ015666.
We next considered the origin of the resistant cells. Given the apparent cellular heterogeneity in the parental cells, the rapid development of resistance, and the fact that preexisting populations are the typical source of resistance to targeted therapies, we hypothesized that our R lines arose from rare preexisting resistant cells. To address this, we independently infected three different KP1 starting populations (A, B, C) with the ClonTracr barcoding system (38) and expanded them for 12 divisions to ensure expansion of cells with unique barcodes. We harvested samples at t = 0 (n = 3/starting population) to allow for quantification of the initial barcodes and cultured parallel aliquots in 10 µM EPZ015666 (n = 3/starting population) or vehicle control (n = 3/starting population) following the same 12-d treatment and subsequent expansion protocol used to generate our R and C lines. We used sequencing to identify all of the barcodes that were conserved across all of the initial (n = 3) and vehicle-selected (n = 3) samples, yielding 3,514 (for the A population), 3,603 (B population), and 4,277 (C population) total unique barcodes. We then examined the barcode representation within the inhibitor-selected samples. If resistance was derived from rare, preexisting cells, we would enrich for a small number of barcodes, which would be shared between the replicates. In contrast, we found that the vast majority of barcodes were retained in each of the independent replicates for the A (80, 86, and 97%), B (89, 94, and 98%), and C (87, 88, and 88%) populations (SI Appendix, Fig. S2M). Moreover, there was no selective enrichment across the EPZ015666-treated replicates for any single barcode, and the relative abundances of the barcodes across the EPZ015666-selected samples was within the range seen in the vehicle control samples (Fig. 2J and SI Appendix, Fig. S2 N–P). Taken together, these data indicate that most, if not all, of the cells in the starting populations had an equal probability of giving rise to the drug-selected population. Thus, we conclude that stable resistance to PRMT5 inhibition is acquired in response to drug treatment via Lamarckian induction, as opposed to selection of a rare, preexisting drug-resistant population.
Cells Resistant to PRMT5 Inhibition Are Collaterally Sensitive to Paclitaxel.
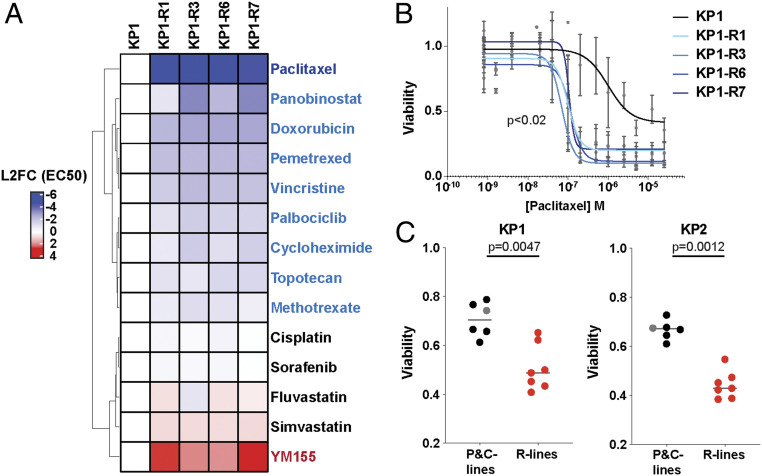
We wondered whether the induced PRMT5i-resistant (R) state altered the response to other chemotherapeutic agents, potentially creating clinically relevant collateral sensitivities or resistances. To address this question, the parental KP1 and four KP-R cell lines (KP1-R1, R3, R6, and R7) were treated for 5 d in triplicate with 13 doses of 14 standard-of-care drugs spanning different drug classes. We determined the EC50s and, for many of the drugs, observed markedly different responses between the parental and R lines that were remarkably consistent across all four R lines. Specifically, the four R cell lines all displayed collateral resistance to only one drug, YM155 (Fig. 3A). As YM155 is a transcriptional inhibitor of SURVIVIN/BIRC5, we compared the levels of SURVIVIN in cell extracts from KP1 and three R lines (SI Appendix, Fig. S3A). This revealed increased levels of SURVIVIN in the R lines (SI Appendix, Fig. S3A), offering a likely explanation for their YM155 resistance.
Fig. 3.
Cells resistant to PRMT5i display collateral sensitivity to paclitaxel. (A) The heatmap shows the L2FC in EC50 of KP1-R1, R3, R6, and R7 lines, relative to the KP1 control, for the indicated panel of 14 different drugs (n = 3 for each line and condition). Red is more resistant, and blue is more sensitive. (B) Graph shows the data generated by the screen of the response to paclitaxel of KP1 (black) versus KP1-R1, R3, R6, and R7 (blue) after a 5-d treatment. Viability is relative to DMSO control. Significance was determined using Welch’s t test. (C) Viability measured with resazurin after a 2-d treatment with 1 µM paclitaxel of KP1 (Left graph) or KP2 (Right graph) parental (gray), C (black; n = 5), and R cell lines (red, n = 7). For significance calculations (Mann–Whitney U test), the parental and C lines were collectively compared to the R lines.
Our drug screen also identified collateral sensitivities to several drugs (Fig. 3A). Of these, paclitaxel was the most striking, with the EC50s of all four R populations being 25-fold lower than the parental KP1 (Fig. 3B). To determine whether paclitaxel sensitivity was a general property of our resistant LUAD lines, we examined the paclitaxel response of all C and R lines. For both the KP1 and KP2 lines, the C lines and their relevant parental control showed similar moderate responses to paclitaxel after a 2-d treatment (Fig. 3C and SI Appendix, Fig. S3B). In stark contrast, five out of seven KP1-R lines, and all seven KP2-R lines, displayed heightened paclitaxel sensitivities (Fig. 3C and SI Appendix, Fig. S3B). Given this shared phenotype, we also tested the collateral resistance drug, YM155, and a second collateral sensitivity drug, vincristine, in a KP2 R line and showed that these altered responses were conserved (SI Appendix, Fig. S3 C and D). The highly consistent responses of various independently arising R populations suggested that acquired resistance to PRMT5 inhibition may occur via a shared mechanism.
Stmn2 Is Necessary for Resistance to PRMT5i and Subsequent Sensitivity to Paclitaxel.
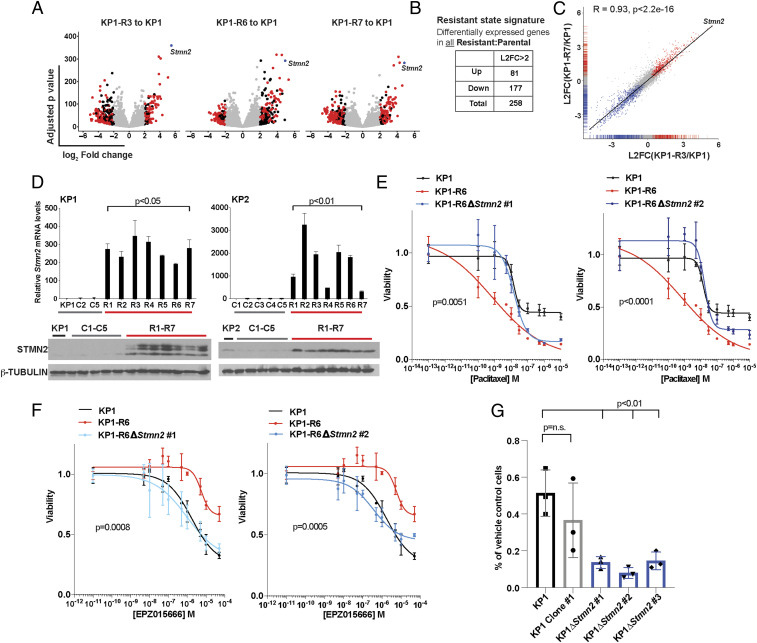
To probe the molecular mechanism of resistance to PRMT5 inhibition, we performed bulk RNA-sequencing on the parental KP1 and three of the KP1-R lines, KP1-R3, R6, and R7. We assessed differential expression using DESeq2 (39) between each R line and KP1 and compared the gene expression changes across the R lines. This revealed high concordance between all three R lines, including an overlap of 258 differentially expressed genes (log2 fold change [L2FC] >2, p < 0.05; Fig. 4 A–C and SI Appendix, Fig. S4A). Stmn2 stood out immediately from this list, as it was one of the top up-regulated genes in the R state signature across all three lines (Fig. 4 A and C and SI Appendix, Fig. S4A). Moreover, since STMN2 is known to regulate microtubule dynamics (40), it was an excellent candidate to account for the collateral sensitivity to paclitaxel that is a hallmark of the resistant state. To determine whether STMN2 expression was common to all R lines, we examined Stmn2 RNA and STMN2 protein levels across our full panel of parental, R, and C lines for both KP1 and KP2 and found that they were barely detected in the parental and C lines but highly up-regulated in each of the R lines (Fig. 4D). Thus, acquisition of STMN2 expression is a defining feature of the resistant state. To determine whether this was broadly applicable to PRMT5i resistance, we used our resistance selection protocol to develop KP1 resistant lines to the PRMT5i JNJ64619178 (JNJ R lines), which works through a distant mechanism from EPZ015666. These JNJ R lines also displayed STMN2 up-regulation and significantly increased sensitivity to paclitaxel (SI Appendix, Fig. S4B). Thus, these characteristics of the resistant cells are consequences of on-target inhibition of PRMT5.
Fig. 4.
Stmn2 is necessary for resistance to PRMT5i and subsequent sensitivity to paclitaxel. (A) Volcano plots showing the L2FC and adjusted p values of genes that are differentially expressed between KP1 and KP1-R3 (Left), KP1-R6 (Middle) or KP1-R7 (Right). Black indicates L2FC > 2, red indicates the 258 genes that showed significant differential expression in all three samples, and blue indicates Stmn2, which is one of the most up-regulated. Significance was determined using a Wald test. (B) Number of genes with significant (L2FC > 2, p < 0.05) up- or down-regulation in the R versus parental lines. (C) Correlation between the differentially expressed genes in KP1-R3 and -R7 indicating Stmn2 is one of the most correlated, highly differentially expressed genes. Significance determined by Pearson correlation, R = 0.93, p < 2.2e-16. (D) Comparison of Stmn2 mRNA levels relative to Gapdh (n = 3 technical replicates; significance established by Welch’s t test) and STMN2 protein levels (with β-TUBULIN loading control) in the C versus R lines for KP1 (Left) and KP2 (Right). (E) Graph shows the response after 4-d treatment to paclitaxel of KP1 (black), KP1-R6 (red), KP1-R6∆Stmn2 #1 (blue, Left), and #2 (blue, Right). Viability is relative to DMSO control. (F) Graph shows the response to EPZ015666 of KP1, KP1-R6, KP1-R6-∆Stmn2 #1, and #2 after a 5-d treatment. Viability is relative to DMSO control. (G) Quantification of the fraction of KP1 parental (black), KP1 Clone #1 (which retains wild-type Stmn2; gray), and KP1-∆Stmn2 clones (blue) that survived 12 d treatment with 10 µM EPZ015666 (compared to treatment with DMSO). For all panels, significance was determined using Student’s t test unless otherwise noted. n.s., not significant.
We then asked if Stmn2 is necessary for paclitaxel sensitivity. For this, we generated Stmn2 knockout clones in the KP1-R6 line using CRISPR-Cas9, confirming the deletion by both sequencing and Western blotting (SI Appendix, Fig. S4C). We treated these cells with a range of paclitaxel doses and found that the Stmn2 knockout clones completely lost their sensitivity to paclitaxel, such that they responded in a comparable manner to the parental KP1 line (p < 0.01, Fig. 4E). Thus, Stmn2 is required for the acquired sensitivity of the R lines to paclitaxel.
While STMN2 is an excellent candidate to explain the paclitaxel sensitivity of our R lines, a compelling biological rationale for its role in PRMT5i resistance was not apparent. We conducted bulk RNA-sequencing of parental KP1 cells with or without a 72-h treatment with EPZ015666 and found that Stmn2 was one of a panel of 173 genes that showed significant differential expression (L2FC >1, p < 0.05), indicating that its induction was an early response to PRMT5 inhibition. Indeed, Stmn2 was one of only 10 genes that was up-regulated in both the early PRMT5i response signature and the shared R state signature (SI Appendix, Fig. S4 D and E). Given Stmn2’s early up-regulation, we wondered whether it might actually contribute to acquisition of PRMT5i resistance, as opposed to just being a hallmark of the R state. Thus, we treated both KP1-R6 Stmn2 knockout clones with EPZ015666. Remarkably, Stmn2 deficiency completely reversed the resistance phenotype of the KP1-R6 cells (P < 0.001), such that the response of each knockout clone now resembled that of the parental KP1 line (Fig. 4F). We then asked whether Stmn2 was also necessary to establish resistance, using CRISPR-Cas9 to delete Stmn2 in the KP1 parental line. As confirmed by sequencing, we successfully isolated three Stmn2 knockout single-cell clones, as well as a single-cell clone that remained Stmn2 wild type (WT; SI Appendix, Fig. S4F), which we used as a control for single-cell cloning effects. These clones were each subjected to our resistance selection protocol. We initially quantified viable cells after 12 d of treatment with 10 µM EPZ015666, which is when resistant cells begin to grow out in the parental KP1 line. Gratifyingly, the three Stmn2 KO lines all had significantly fewer surviving cells than either the parental KP1 or WT Stmn2 clone (p < 0.01; Fig. 4G). Moreover, by 20 d, the Stmn2 WT clone had yielded a resistant population, but nothing grew out from any of the three Stmn2 KO lines (SI Appendix, Fig. S4G). Taken together, our data show that a single gene, Stmn2, is necessary for the establishment and maintenance of resistance to PRMT5 inhibition, as well as the collateral sensitivity to paclitaxel.
The Combination of PRMT5i and Paclitaxel Is a Potent and Synergistic Therapy.
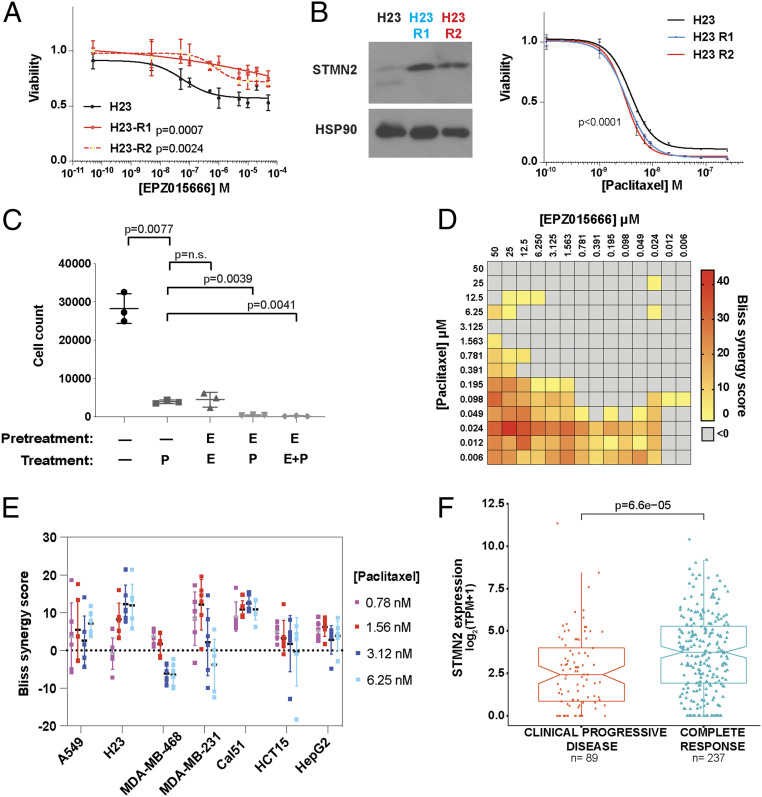
We wanted to determine whether our findings on PRMT5i-resistant cells are relevant to human cell lines. Using our resistance protocol, we developed two EPZ015666-resistant lines from the human LUAD cancer cell line, H23, which harbors KRASG12C and tp53M246I mutations (Fig. 5A). Importantly, these resistant lines (H23-R1, R2) also have both an increase in STMN2 levels and a significant increase in sensitivity to paclitaxel (Fig. 5B). Thus, PRMT5i resistance by STMN2 up-regulation, and the consequent collateral sensitivity to paclitaxel, also occurs in human lung cancer lines.
Fig. 5.
The combination of PRMT5i and paclitaxel is a potent and synergistic therapy. (A) Graph shows the response of H23, H23-R1, and H23-R2 to a 10-point curve of EPZ015666 after a 5-d treatment. Viability relative to DMSO control. (B, Left) Western blot for STMN2 of H23, H23-R1, and H23-R2. (Right) Graph shows the response of H23, H23-R1, and H23-R2 to a 10-point curve of paclitaxel. (A and B) Significance was determined using Student’s t test. (C) Number of KP1 cells surviving after being pretreated with 10 µM EPZ015666 (E) or with vehicle control (—) for 4 d, then treated for 2 d with vehicle control, 0.025 µM paclitaxel (P) alone, 10 µM EPZ015666 (E) alone, or E + P, sequentially or simultaneously, and then allowed to recover in the absence of drug for 4 d before counting (n = 3 replicates/condition). Significance was determined using Welch’s t test. (D) The heatmap shows the drug synergy between EPZ015666 and paclitaxel, which was determined after a 3-d treatment over a range of 14 drug concentrations and assessed by Bliss synergy scores. Synergy is defined as a score greater than zero (colored). Data shown is one of three representative experiments. (E) Graph shows Bliss synergy score of human cancer cell lines (lung: A549, H23; breast: MDA-MB-468, MDA-MB-231, CAL51; liver: HepG2; colon: HCT15) after pretreatment with vehicle or 10 µM of EPZ015666 for 3 d and then treatment with 0.78, 1.56, 3.12, or 6.25 nM of paclitaxel for 3 d (n = 6 replicates per condition). (F) Pancancer TCGA analysis showing the correlation between STMN2 levels (log2TPM+1) and clinical response to taxane treatment. Significance was determined using the Wilcoxon rank-sum test.
Given that paclitaxel sensitivity is coupled to PRMT5i resistance, via STMN2, we hypothesized that combining paclitaxel with EPZ015666 treatment should prevent resistant cells from emerging, creating an effective up-front combination therapy. We first tested this on our KP1 line. Since the phenotypic response to EPZ015666 has slower kinetics than to paclitaxel, we included a 4-d pretreatment phase with or without 10 µM EPZ015666 prior to treatment with DMSO, 0.025 µM paclitaxel, 10 µM EPZ015666, or a combination of EPZ015666 and paclitaxel for an additional 2 d. The cells were then allowed to recover for 4 d in the absence of drug, before the surviving populations were quantified (Fig. 5C). We saw a significant reduction in the number of surviving cells in response to paclitaxel alone (7-fold) or EPZ015666 alone (7.5-fold), and combining these treatments, either sequentially or together, yielded a further marked reduction (>10-fold; Fig. 5C). To determine whether the response to EPZ015666 and paclitaxel was additive or synergistic, we performed 14-point dose–response curves of both drugs. Calculation of the Bliss synergy score showed synergy, particularly at low doses of paclitaxel and EPZ015666 (Fig. 5D). To see whether we observed drug synergy broadly across different contexts, we extended this analysis to a panel of human tumor lines, including lung, breast, liver, and colon cancer. For this, we pretreated for 3 d with 10 µM EPZ015666 followed by a 3-d treatment with 10 µM EPZ015666 plus a range of paclitaxel concentrations. Remarkably, we saw synergy for all of these cell lines (Fig. 5E). The maximum occurred at different concentrations of paclitaxel (Fig. 5E and SI Appendix, Fig. S5A), as might be expected considering the epigenetic and transcriptomic differences across these diverse tumor lines. Together, PRMT5i and paclitaxel are a potent and synergistic drug combination.
Finally, since STMN2 is necessary for paclitaxel sensitivity, we wondered whether STMN2 levels could have clinical relevance beyond PRMT5i treatment as a predictive indicator of the efficacy of paclitaxel treatment in patients. Considering all tumor types, we analyzed The Cancer Genome Atlas (TCGA) data of patients who had been treated with taxanes and had reported outcomes (SI Appendix, Fig. S5B). Irrespective of PRMT5 levels, we found a strong correlation between STMN2 levels and patient response, with STMN2 levels being significantly higher (p = 6.6 × 10−5) in patients showing a complete response (CR) versus no response (clinical progressive disease; CPD) to taxane treatment (Fig. 5F and SI Appendix, Fig. S5 C and D). We also examined STMN1 levels, as high STMN1 has been reported to confer resistance to paclitaxel (41, 42), and found that these were not significantly different between these patient cohorts (SI Appendix, Fig. S5E). The STMN2:STMN1 ratio had similar, but less predictive, power than STMN2 levels alone, indicating that STMN2, not STMN1, is the key feature (SI Appendix, Fig. S5E). Thus, STMN2 levels correlate with patient response to taxane therapy.
Discussion
This study describes the generation of PRMT5i-resistant cell lines. Our data clearly establish that these cells are not preexisting but instead are specifically induced by exposure to PRMT5i. Thus, we conclude that they arise via Lamarckian induction, a rarity for cancer resistance mechanisms. Notably, we observed one consistent mechanism of resistance for KP mutant LUAD, as judged by the adoption of a shared and stable transcriptional program. KP mutant LUAD cells have been shown to exist in various transcriptional states, which reflect reactivation of embryonic transcriptional programs along the normal lung developmental trajectory and correlate with tumor progression (43). Our resistant state signature does not resemble any of these known signatures, reinforcing the notion that it is drug induced. Importantly, our data show that this state creates vulnerabilities to standard-of-care chemotherapies, especially the taxane, paclitaxel. Remarkably, we were able to identify a single gene, Stmn2, as being required for both the observed taxane sensitivity and the establishment and maintenance of the PRMT5i-resistant state. Finally, we show that human cancer cell lines can also develop PRMT5 resistance that is characterized by STMN2 expression and paclitaxel sensitivity and show a synergistic response to PRMT5i and paclitaxel treatment.
We were surprised to find that the degree of PRMT5 inhibition is comparable between the parental KP line and its resistant variants, at least at the gross level. This raises the question of how the resistant cells are able to cope with the loss of SDMA modifications on PRMT5 targets, especially when the parental cells are so profoundly affected. Our data show that the resistant state ameliorates the PRMT5i-induced proliferation defects that occur in the parental KP LUAD lines, even though there is no detectable difference in the basal proliferation rate of the two populations. However, the cause of this bypass remains to be uncovered. We cannot rule out that one or more key PRMT5 targets have subtle differences in the level of methylation in resistant versus parental cells, such that sufficient function remains in the former but not the latter setting. However, it seems more likely that the acquired cell state and consequent gene expression profile of the R lines alters the regulatory landscape in a way that somehow reduces dependence on SDMA-modified PRMT5 targets. Understanding the nature of this compensation will provide key insight into how and why PRMT5 is important for tumorigenesis.
Our understanding of the resistance phenotypes was greatly informed by the drug screen, and we believe that there is strong rationale for employing this as a general strategy in probing mechanisms of resistance. First, it revealed that the resistance state induced a collateral sensitivity to various drugs, particularly the taxane, paclitaxel. This uncovers collateral effects between a nongenetic resistant state and standard-of-care chemotherapies. Second, combining our drug-screen findings with gene expression analyses led us to prioritize Stmn2. Notably, there was no logical reason to hypothesize that STMN2 would be involved in PRMT5i resistance, but it was an obvious candidate to explain the collateral sensitivity to taxanes because both play roles in microtubule biology. However, quite remarkably, our data showed that STMN2 mediates both paclitaxel sensitivity and PRMT5i resistance. The mechanism by which STMN2 contributes to PRMT5i resistance remains to be established, including whether this reflects its role in microtubule biology or another, unappreciated function. We note that Stmn2 is one of a handful of genes that is highly expressed in the resistant cells and also up-regulated in parental cells as an early response to PRMT5i treatment. Since Stmn2 is required for acquisition of the PRMT5i-resistant state, it could serve to enable and maintain the epigenetic shift that creates the R state and/or be more directly involved in conferring resistance, such as by compensating for loss of PRMT5 activity.
Our study of STMN2 also has significant potential clinical relevance. Our data show that PRMT5i and taxanes can be an effective and synergistic up-front combination therapy in murine and human cancer cell lines. Interestingly, one of the ongoing clinical trials is testing the PRMT5i PF-06939999 in non-small cell lung cancer (NSCLC) patients together with the taxane docetaxel (31). The rationale for this choice is not disclosed, but it is likely because docetaxel is the standard-of-care second-line therapy for advanced NSCLC. Our discoveries provide a clear biological rationale for this combination and predict a successful outcome. Equally important are our insights regarding STMN2. The literature contains extensive evidence in human cells that the STMN2 paralog, STMN1, influences tumorigenesis in many tumor types (44), including showing that it promotes resistance to taxanes (41, 42). In contrast, published roles of STMN2 in cancer were limited to implications of STMN2 phosphorylation in metastatic potential (45) and the inclusion of STMN2 in a panel of biomarkers associated with prognosis of ovarian and lung cancer (46, 47). Our analyses of TCGA patient data revealed a significant correlation between high STMN2 levels and CR to taxane treatment, independent of the levels of PRMT5. Importantly, the TCGA data were not limited to LUAD but instead included all tumor types that received taxane treatment and reported outcomes, such that breast cancer was the majority tumor type. Taxanes are primarily used in the treatment of breast cancer, NSCLC, and prostate cancer, which together make up an estimated 30 to 40% of new cancer cases in the United States (48). Our findings raise the possibility that STMN2 levels might serve as a predictive biomarker for either taxane response or cotreatment with PRMT5i and taxanes, upon Food and Drug Administration approval, and certainly provide clear rationale for investigating STMN2’s role in cancer.
Finally, we believe that our study offers paradigms for thinking about the consequences of targeting epigenetic regulators in cancer treatment. In concert with the existing data on BET inhibitor resistance, our data strongly suggest that resistance will occur primarily through nongenetic mechanisms. Specifically, we hypothesize that disrupting regulation of the epigenome will increase the plasticity of tumor cells, heightening their ability to shift to alternate states that can enable drug resistance. One prediction of this model is that epigenetic inhibitors may be predisposed to yield poor clinic responses and/or a high rate of relapse. Indeed, this has been observed for BET inhibitors. However, extrapolating from our current study, we predict that acquisition of resistance will enable induced collateral sensitivities as a result of these state shifts. In this manner, resistance could actually be advantageous, from a treatment standpoint, if the new state is more treatable than the initial tumor. These predictions suggest that drugs against epigenetic regulators, such as BET and PRMT5, may be relatively ineffective when used alone but rendered highly effective when combined with therapies that target the resistant state. This provides strong rationale for probing the resistance phenotypes of other classes of epigenetic inhibitors and for utilizing drug screens to determine whether the acquisition of collateral sensitivities is a common hallmark of these states.
Materials and Methods
Cell Culture and Chemicals.
Murine KP LUAD cell lines were previously reported (49, 50): KP393T5 (KP1), KP1233 (KP2), KP373T1, KP1234T4, KP393T3, KP1234T3, KP393T4, and KP2677; or generated for this study: KP3414B, KP4823F, KP4825A, and KP4825D. Human cells used included the following: H23, A549, CAL51, MDA-MB-453, MDA-MB-231, HepG2, and HCT15. Cells were cultured in Dulbecco's modified Eagle's media (DMEM) or Roswell Park Memorial Institute (RPMI) media (H23, HCT15) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. For figures with a specified cell count, this was conducted using CountBright Absolute Counting Beads (ThermoFisher Scientific, C36950). Small-molecule inhibitors were obtained from the following: L. Garraway (EPZ015666; high pressure liquid chromatography [HPLC] trace shown in SI Appendix, Supplementary Methods), Selleck Chemicals (GSK3326595 and JNJ64619178), MilliporeSigma (paclitaxel, vincristine, fluvastatin, DMSO), LC Laboratories (doxorubicin, palbociclib, panobinostat, pemetrexed, sorafenib), and Tocris Biosciences (cisplatin, cycloheximide, methotrexate, simvastatin, topotecan, YM155).
Generation of Resistant Cell Lines.
For both the KP1 and KP2 lines, 12 10-cm plates were each plated with 4 × 104 cells. The next day, seven plates were treated with 10 µM EPZ015666 and five plates were treated with 0.01% DMSO (vehicle control). On day 4 of treatment, and again on day 8, the cells were trypsinized, and 4 × 104 cells were replated in the same treatment conditions. On day 12, the surviving cells were allowed to grow out in the continued presence of DMSO or EPZ015666 treatment, splitting when they approached confluence, to yield the resistant (EPZ-treated) and control (DMSO-treated) cell lines. The H23 R lines and KP1 JNJ R lines were plated and split at the same densities and frequencies. The H23 R lines were treated with 10 µM EPZ015666, the JNJ R lines were treated with the following concentrations of JNJ64619178: 0.01 µM (J1) and 0.5 µM (J2).
Dose–Response Curves.
Viability was measured using RealTime-Glo MT Cell Viability Assay (Promega, G9712) or, when indicated, resazurin sodium salt (Sigma-Aldrich). For RealTime-Glo dose–response curves of PRMT5i, 150 cells per well were plated in 150 µL media in a white-clear bottom 96-well plate. At 1 d after plating, 50 µL of a 4× solution of the final drug concentration in media was mixed into each well. RealTime-Glo was added after 2.5 d and luminescence measured every 12 h between days 3 and 6. For paclitaxel, vincristine, and YM155, 1,000 cells per well were plated, and the respective drug was added 1 d later. RealTime-Glo was added after 1.5 d of treatment and readings taken between days 2 and 4. For resazurin (used at 0.008 mg/mL), fluorescence was measured 6 h after addition at an excitation of 550 nm and emission of 600 nm.
For the H23 cell lines, 2.5 × 103 cells per well were plated for EPZ015666 dose–response curves, and 7.5 × 103 cells per well were plated for paclitaxel dose–response curves. After 5 (EPZ) or 3 (paclitaxel) d of treatment, CellTiter-Glo (Promega G7570) was used according to manufacturer’s instructions.
All luminescence and fluorescence was read using a Tecan M200 Pro.
Viability normalization.
Background luminescence (RealTime-Glo) or fluorescence (resazurin) was determined by measurement of media-only wells and subtracted from other viability measurements. Viability readings were normalized by dividing the background corrected median readings of the drug-treated cells by those of the relevant DMSO control.
Nonlinear fit model.
Normalized viability data from the dose–response curves was fit to the following 4-parameter hill curve to find EC50s:
where Ainf and A0 are the viabilities at the highest and lowest doses, respectively. This calculation was done using Prism, except for the drug screen, which used MATLAB.
Figures.
Dose–response curves show the fit to the model with mean and SD of the raw data. For input into statistical analyses, area under the curve was calculated with Prism and used to determine differences between dose–response curves. Percent growth inhibition reported is the percent change between the growth (over days 3 to 5) of cells treated with x concentration compared to the growth of the cells treated with 0 µM drug.
Flow Cytometry.
Flow cytometry was conducted using BD Fortessa or BD Celesta and analyzed using FloJo 10.5.3.
Cell Proliferation Assay.
Cell proliferation was assessed using the CellTrace Violet Cell Proliferation Kit (Thermo Fisher C34571) and triplicate samples for each condition: t = 0 (no drug) control versus the 0.01% DMSO or 10 µM EPZ015666 treated time points. In each case, cells were harvested, and the fluorescent population signal was analyzed via flow cytometry (channel = BV421). The mean fluorescent signal for the DMSO and EPZ015666-treated samples were normalized to the initial control. The relative proliferation graphed is the reciprocal of the normalized mean fluorescent signal.
Annexin-V Assay.
Annexin-V positivity was assayed using RealTime-Glo Annexin-V Apoptosis Assay (Promega JA1000). Cleaved caspase 3 and 7 (CC3/CC7) activity was measured using the Caspase-Glo 3/7 Assay (Promega G8091). Luminescence readings for both were taken using a Tecan M200 Pro plate reader.
Crystal Violet.
Cells were plated as described for dose–response curves and resazurin used to determine the viability after treatment. After reading fluorescence, the resazurin was aspirated, and the cells were washed first with phosphate buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min and then with water and stained with 0.1% crystal violet (MilliporeSigma) for 20 min. Plates were washed in water, dried, and imaged.
Competition Assay.
Resistant or control cells were infected with lentivirus containing empty vector pULTRA-Hot, which expresses mCherry. High-expressing mCherry R or C cells were sorted using a BD FACSAria and mixed with (unlabeled) parental cells at a 10:90 ratio. A total of 3.5 × 104 cells were then plated in 6-cm dishes in triplicate and grown in 0.01% DMSO control, 10 µM EPZ015666, or 10 µM GSK3326595. Every 5 d, each plate was trypsinized. A total of 3.5 × 104 cells were replated into 6-cm dishes, and the remainder were subjected to flow cytometry (channel = PE-Cy5) to determine the representation of mCherry-positive cells.
qRT-PCR.
Total RNA was isolated and purified using RNeasy Mini (Qiagen). RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Thermo Fisher). Real-time PCR reactions were performed using FAST-SYBR Green on a StepOnePlus Real-Time PCR System (Applied Biosystems). Data were analyzed using the ∆∆CT method, and relative messenger RNA (mRNA) levels were normalized to Gapdh levels. The following primers were used: Gapdh F: 5′-ATGGTGAAGGTCGGTGTGA; Gapdh R: 5′- AATCTCCACTTTGCCACTGC; Stmn2 F: 5′-CTGATCTGCTCCTGCTTCTAC; Stmn2 R: 5′- CTGAGCCTCTTGAGACTTTCTT.
Western Blotting.
For the Western blot of pan-SDMA over different concentrations of EPZ015666, 5 × 103 KP1 cells were plated in 6-cm culture dishes and 1 d later treated with 0.01% DMSO over the following concentrations of EPZ015666: 0.1, 1, 5, and 10 µM. Cells were harvested after 5 d of treatment. For the Western blot of pan-SDMA over time, 5 × 103 KP1 and KP1-R6 cells were plated in 6-cm culture dishes and 1 d later harvested or treated with 0.01% DMSO or 10 μM EPZ015666 and then harvested 1 to 5 d later. For harvesting and lysis, cells were washed twice with PBS, harvested via scraping, pelleted at 4 °C, resuspended in 200 μL radioimmunoprecipitation assay (RIPA) lysis buffer [50 mM Tris [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS)] supplemented with protease inhibitor mixture (MilliporeSigma 11697498001), and lysed by three rounds of sonication. The cell debris was pelleted at 4 °C and the protein concentration of the lysate determined using the Pierce BCA Protein Assay Kit (Thermo Fisher 23225). A total of 50 μg protein was mixed with standard 2× Laemmli buffer and run on 12% SDS-polyacrylamide gels before transfer onto nitrocellulose membranes.
To assess pan-SDMA and PRMT5, immunoblotting was conducted using anti-SDMA (Cell Signaling Technology, 13222), anti-PRMT5 (Cell Signaling Technology, 79998), and anti-GAPDH (Thermo Fisher AM4300) primary antibodies, anti-rabbit 800 nm (LI-COR, NE 925-32212) secondary antibodies, and imaged on the LI-COR Odyssey. Fluorescent signal was quantified using Image Studio Version 5.2.5.
To assess STMN2 and SURVIVIN, immunoblotting was conducted with anti-STMN2 (Thermo Fisher 720178), anti-SURVIVIN (Thermo Fisher MA1-16838), anti–β-TUBULIN (MilliporeSigma T7816), and anti-HSP90 (BD Biosciences, 610418) primary antibodies and anti-rabbit (MilliporeSigma GENA934) and anti-mouse (MilliporeSigma GENA931) secondary antibodies and the signals visualized with enhanced chemiluminescence (WesternLightning Plus-ECL, PerkinElmer) using film or imaged with a Biorad ChemiDoc MP.
Drug Screen.
Cells were plated in triplicate 384-well plates with 250 cells per well. The next day they were treated with 13-point dose–response curves (SI Appendix, Table S1). After 5 d of treatment with the respective drugs, cell viability was measured using resazurin and EC50s calculated as described above. Log2 fold changes of EC50s were calculated for each R line relative to the KP1 parental line. These analyses were performed using MATLAB.
Synergy Experiment.
For KP1 cells, 2 × 103 KP1 or KP1-R6 cells were plated in triplicate 384-well plates. The next day, cells were treated for 3 d with simultaneous 14-point dose–response curves of EPZ015666 and paclitaxel (as indicated in Fig. 5D) alone and in all possible combinations, along with no drug controls.
For human cell lines, 1 × 103 cells per well were plated in 96-well plates. The next day, cells were treated with 10 µM EPZ015666 or vehicle control (0.01% DMSO) for 3 d. After 3 d, the cells were further treated with 0, 0.78, 1.56, 3.12, or 6.25 nM paclitaxel in replicates of six for an additional 3 d.
For both, viability was measured using CellTiter-Glo (Promega). Bliss synergy was calculated using the following formula: , where EA and EB are the killings observed for drugs A and B individually at each dose, and EAB is the killing of the combination. EA, EB, and EAB are defined as 1 minus viability at the respective dose. The difference between the observed EAB and the expected EAB (times 100) is shown as the Bliss synergy score in Fig. 5D, where any score above zero marks a synergistic pair of drug concentrations.
Barcoding Experiment.
Cells were barcoded using the ClonTracr library (Addgene 67267) described previously (38). KP1 cells were plated into three plates and the library transduced into the cells using spinoculation to yield <10% infection. A total of 5 × 105 RFP-positive cells were sorted and expanded for 12 more doublings to yield three separate barcoded populations (A, B, C). For each, we plated nine aliquots of 8 × 105 cells. Three aliquots were collected 2 d later to sample the initial barcode representation. The remainder were treated with 0.01% DMSO (n = 3) or 10 µM EPZ015666 (n = 3). These cells were split and expanded every 4 d for 12 d and then harvested. Pooled genomic DNA was extracted (MilliporeSigma G1N70), amplified for Illumina sequencing using primers containing adapters as previously described (38), and sequenced using an Illumina NextSeq. Reads were filtered for high-quality sequence and adapter trimmed and barcodes identified and quantified using the method described here: https://github.com/SGarg-Lab/lineage-entropy/tree/master/Fastq_Decomplexation. Barcodes with a Hamming distance <5 were collapsed to allow for PCR and sequencing errors and then counts were normalized by dividing the raw counts by the number of counts per sample and multiplying by the total number of counts across all samples. For each population, we identified barcodes that were present above a threshold of 50 counts (threshold determination in SI Appendix, Supplementary Methods) in all three initial samples and all three final DMSO-treated samples. We then determined which of these barcodes were present in the EPZ-treated samples, also above a threshold of 50 counts/sample. Barcode abundance was calculated as the normalized count for that barcode divided by the sum of the normalized counts for that sample.
siRNA Knockdown.
KP1 cells were plated at 150 cells per well in a 96-well dish. On the next day, cells were treated with a dose–response curve of EPZ015666. On the following 2 d, cells were transfected with siRNA (25 nM) and MISSION siRNA Transfection reagent (Sigma S1452) according to manufacturer’s instructions. After 5 d of treatment with EPZ015666, viability was measured using CellTiter-Glo. Control siRNA used was MISSION siRNA Universal Negative Control #1 (Sigma SIC001). siPRMT5#1: (Sigma NM_013768; SASI_Mm01_00193421). siPRMT5#2: (Sigma NM_013768; SASI_Mm01_00193422)
Generation of Stmn2 Knockout Cell Lines.
KP1 and KP1-R6 cells were infected with a lentiviral vector (pLV[2CRISPR]-hCas9:T2A:Hygro-U6gRNAStmn2 #1-U6gRNAStmn2 #2) that contained hCas9 and two single guide RNAs (sgRNAs) targeting murine Stmn2 (Vector Builder VB191016-1257adw). Infected pools were single-cell cloned. Stmn2 was sequenced (for KP1 and KP1-R6 ∆-Stmn2 clones), and STMN2 protein expression was analyzed (for KP1-R6 ∆-Stmn2 clones). sgRNA sequences were as follows: Stmn2 #1: TGCAGGGGACTCACCGTCGT; Stmn2 #2: TGTTGATGTTGCGCGGCTCC.
Patient Data (TCGA) Analysis.
The manually curated TCGA drug-response data (51) was filtered for patients treated with taxane therapy (paclitaxel n = 184 or docetaxel n = 140) and for complete responders and nonresponders (i.e., samples annotated as CPD); samples annotated as partial responders and stable disease were excluded for this analysis. The correlation between STMN1, STMN2, PRMT5 expression, or STMN2:STMN1 ratio and the measure of response to taxanes was calculated by the Wilcoxon rank-sum test.
RNA-Sequencing and Differential Gene Expression Analysis.
Cells were grown in 0.01% DMSO (KP1 and KP1-R6) or 10 µM EPZ015666 (KP1) for 72 h. Total RNA was isolated and purified using the RNeasy Mini kit (Qiagen) and quality controlled using the Fragment Analyzer (Agilent). RNA sequencing (RNA-Seq) libraries were prepared using the Kapa mRNA HyperPrep kit starting with 100 ng total RNA using one-third reaction volumes and 14 cycles of PCR. Completed libraries were quantified using the Fragment Analyzer and qPCR before multiplexing and sequencing using 40 nucleotide single-end reads on an Illumina HiSeq2000.
Single-end RNA-Seq reads were aligned to a transcriptome derived from the mm10 primary assembly with an ensembl v.88 annotation using STAR version 2.5.3a (52). Gene expression was summarized using RNA-Seq by expectation-maximization (RSEM) version 1.3.0 (53) and samtools version 1.3 (54). Differential expression analysis was done with R version 3.4.4 using DESeq2 1.18.1 (55) and normal log fold change shrinkage (39). The resulting data were parsed and assembled using Tibco Spotfire Analyst version 7.11.1. Gene expression differences at the total gene level were considered significant at an adjusted p value <0.05. DESeq2 analysis results are shown in Dataset S1. To determine the degree of similarity between the expression profiles of the resistant lines (KP1-R3, R6, and R7) compared to the parental (KP1), the Pearson correlation coefficient, and respective p value, were calculated for each pairwise combination of resistant lines (R3 to R6, R3 to R7, and R6 to R7) using the log2 fold change for each gene.
Quantification, Statistical Analysis, and Graphics Plotting Software.
R 3.4.4, 3.6.0, and 3.6.3, GraphPad PRISM 8.2.1 and MATLAB 2018a were used to perform statistical analyses. GraphPad PRISM 8.2.1, MATLAB 2018a, R 3.6.0, and Adobe Illustrator 24.2.1 and Photoshop 2014.2.2 were used for generating and plotting figures.
Statistical Analysis.
Data show the mean with SD (error bars). All statistical analyses were performed using Prism software as Student’s t test unless the variance was significantly different between the two samples (calculated with the F-test) in which case it is then specified as Welch’s t test. For comparative analyses, Mann–Whitney U test was used when data points were from different cell lines (not replicates), for example comparing R and C/P lines. Full compilation of statistical tests and results is in Dataset S2. For statistical significance, p < 0.05 was considered statistically significant, p = n.s. refers to not significant (p > 0.05).
Supplementary Material
Acknowledgments
We thank the Robert A. Swanson (1969) Biotechnology Center and particularly the High-Throughput (HTP) Sciences (J. Cheah), Genomics and Bioinformatics (C. Whittaker), and Flow Cytometry Cores for technical assistance; D. Karl, V. Gocheva, and S. Naranjo for providing cell lines; and J.A.L.’s laboratory members, especially A. Amsterdam, T. Parisi, S. Hu, and M. Wilson for ideas and comments. Funding support was from the National Cancer Institute for the Koch Institute Support (core) Grants P30-CA14051, PO1 CA42063 (J.A.L.), T32 CA009216 (S.G.), and R01 CA233477-01 (M.T.H.); the Koch Institute Frontier Research Program and the Koch Institute Director’s Anonymous Discretionary Research Funds (J.A.L. and M.T.H.); the Ludwig Center at the Massachusetts Institute of Technology (MIT) (J.A.L. and M.T.H.); the MIT Center for Precision Cancer Medicine (M.T.H.); the Charles W. and Jennifer C. Johnson Clinical Investigator Award (S.G.); the NIH Pre-Doctoral Training Grant GM007287 (H.S.M., C.E.F., and S.D.); NSF Graduate Research Fellowship (H.S.M.); and David H. Koch Fellowship in Cancer Research (S.D.). J.A.L. is the Virginia and D.K. Ludwig Professor for Cancer Research at the Koch Institute at MIT.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024055118/-/DCSupplemental.
Data Availability
KP1 (KP393T5) and KP1-R3,6,7 RNA-Seq data are available from the Gene Expression Omnibus under accession no. GSE157715 (57).
References
- 1.Holohan C., Van Schaeybroeck S., Longley D. B., Johnston P. G., Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Angus S. P., Zawistowski J. S., Johnson G. L., Epigenetic mechanisms regulating adaptive responses to targeted kinase inhibitors in cancer. Annu. Rev. Pharmacol. Toxicol. 58, 209–229 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Shaffer S. M., et al., Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell C. C., Gilan O., Principles and mechanisms of non-genetic resistance in cancer. Br. J. Cancer 122, 465–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fong C. Y., et al., BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell C. C., et al., Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat. Commun. 10, 2723–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisco A. O., et al., Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat. Commun. 4, 2467–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim C., et al., Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173, 879–893.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugo W., et al., Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell 162, 1271–1285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinohara K., et al., KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 34, 939–953.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zawistowski J. S., et al., Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the P-TEFb complex. Cancer Discov. 7, 302–321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A., et al., Longitudinal single-cell RNA sequencing of patient-derived primary cells reveals drug-induced infidelity in stem cell hierarchy. Nat. Commun. 9, 4931–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu S., et al., Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robey R. W., et al., Histone deacetylase inhibitors: Emerging mechanisms of resistance. Mol. Pharm. 8, 2021–2031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisserier M., Wajapeyee N., Mechanisms of resistance to EZH2 inhibitors in diffuse large B-cell lymphomas. Blood 131, 2125–2137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pál C., Papp B., Lázár V., Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 23, 401–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stopa N., Krebs J. E., Shechter D., The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell. Mol. Life Sci. 72, 2041–2059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tee W.-W., et al., Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 24, 2772–2777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan-Penebre E., et al., A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 11, 432–437 (2015). [DOI] [PubMed] [Google Scholar]
- 20.US National Library of Medicine , ClinicalTrials.gov (2020). https://clinicaltrials.gov/ct2/show/NCT02783300. Accessed 15 August 2020.
- 21.US National Library of Medicine , ClinicalTrials.gov (2020). https://clinicaltrials.gov/ct2/show/NCT03614728.
- 22.Lin H., Luengo J. I., Nucleoside protein arginine methyltransferase 5 (PRMT5) inhibitors. Bioorg. Med. Chem. Lett. 29, 1264–1269 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Lin H., et al., Discovery of potent and selective covalent protein arginine methyltransferase 5 (PRMT5) inhibitors. ACS Med. Chem. Lett. 10, 1033–1038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu K., et al., Rational design, synthesis and biological evaluation of novel triazole derivatives as potent and selective PRMT5 inhibitors with antitumor activity. J. Comput. Aided Mol. Des. 33, 775–785 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Zhu K., et al., Identification of 5-benzylidene-2-phenylthiazolones as potent PRMT5 inhibitors by virtual screening, structural optimization and biological evaluations. Bioorg. Chem. 81, 289–298 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Bonday Z. Q., et al., LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med. Chem. Lett. 9, 612–617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q., et al., Identification of a novel protein arginine methyltransferase 5 inhibitor in non-small cell lung cancer by structure-based virtual screening. Front. Pharmacol. 9, 173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong G.-M., et al., Selective small-chemical inhibitors of protein arginine methyltransferase 5 with anti-lung cancer activity. PLoS One 12, e0181601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao R., et al., Potent, selective, and cell active protein arginine methyltransferase 5 (PRMT5) inhibitor developed by structure-based virtual screening and hit optimization. J. Med. Chem. 60, 6289–6304 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Prabhu L., et al., Adapting AlphaLISA high throughput screen to discover a novel small-molecule inhibitor targeting protein arginine methyltransferase 5 in pancreatic and colorectal cancers. Oncotarget 8, 39963–39977 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Hu W., Yuan Y., Protein arginine methyltransferase 5 (PRMT5) as an anticancer target and its inhibitor discovery. J. Med. Chem. 61, 9429–9441 (2018). [DOI] [PubMed] [Google Scholar]
- 32.US National Library of Medicine , ClinicalTrials.gov (2020). https://clinicaltrials.gov/ct2/show/NCT03573310. Accessed 15 August 2020.
- 33.US National Library of Medicine , ClinicalTrials.gov (2020). https://clinicaltrials.gov/ct2/show/NCT03854227. Accessed 20 December 2020.
- 34.US National Library of Medicine , ClinicalTrials.gov (2020). https://clinicaltrials.gov/ct2/show/NCT03886831. Accessed 20 December 2020.
- 35.Yang W., et al., Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41(Database issue) D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun C. J., et al., Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell 32, 411–426.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibrahim R., et al., Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum. Pathol. 45, 1397–1405 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Gu Z., et al., Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem. J. 446, 235–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhang H.-E. C., et al., Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 21, 440–448 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manna T., Grenningloh G., Miller H. P., Wilson L., Stathmin family protein SCG10 differentially regulates the plus and minus end dynamics of microtubules at steady state in vitro: Implications for its role in neurite outgrowth. Biochemistry 46, 3543–3552 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Alli E., Bash-Babula J., Yang J.-M., Hait W. N., Effect of stathmin on the sensitivity to antimicrotubule drugs in human breast cancer. Cancer Res. 62, 6864–6869 (2002). [PubMed] [Google Scholar]
- 43.Bao P., et al., High STMN1 expression is associated with cancer progression and chemo-resistance in lung squamous cell carcinoma. Ann. Surg. Oncol. 24, 4017–4024 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Snyder E. L., et al., Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol. Cell 50, 185–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biaoxue R., Xiguang C., Hua L., Shuanying Y., Stathmin-dependent molecular targeting therapy for malignant tumor: The latest 5 years’ discoveries and developments. J. Transl. Med. 14, 279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Q., et al., PAK4 kinase-mediated SCG10 phosphorylation involved in gastric cancer metastasis. Oncogene 33, 3277–3287 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Wang Y., Lei L., Chi Y.-G., Liu L.-B., Yang B.-P., A comprehensive understanding of ovarian carcinoma survival prognosis by novel biomarkers. Eur. Rev. Med. Pharmacol. Sci. 23, 8257–8264 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Grieve S., et al., Immunohistochemical validation study of 15-gene biomarker panel predictive of benefit from adjuvant chemotherapy in resected non-small-cell lung cancer: Analysis of JBR.10. ESMO Open 5, e000679-e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.American Cancer Society , Cancer Facts & Figures 2020 (American Cancer Society, Atlanta, GA, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winslow M. M., et al., Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gocheva V., et al., Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc. Natl. Acad. Sci. U.S.A. 114, E5625–E5634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moiso E., Manual curation of TCGA treatment data and identification of potential markers of therapy response. medRxiv [Preprint] (2021). 10.1101/2021.04.30.21251941 (Accessed 9 May 2021). [DOI]
- 53.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106–R112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mueller H. S., et al., Acquired resistance to PRMT5 inhibition and the consequent collateral sensitivity to paclitaxel are both Stmn2 dependent.Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157715. Deposited 9 September 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Mueller H. S., et al., Acquired resistance to PRMT5 inhibition and the consequent collateral sensitivity to paclitaxel are both Stmn2 dependent.Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157715. Deposited 9 September 2020.
Supplementary Materials
Data Availability Statement
KP1 (KP393T5) and KP1-R3,6,7 RNA-Seq data are available from the Gene Expression Omnibus under accession no. GSE157715 (57).