Significance
Secretion of siderophores allows most microbes to assimilate ferric ions into their biological processes. Siderophores must be taken up into the cells, and chelated iron must be released. Here, we present the structure of an inner membrane siderophore reductase, FoxB, which is involved in the uptake of iron from ferrioxamine siderophores in Pseudomonas aeruginosa. Our structure reveals FoxB to be a di-heme membrane protein, which is able to reduce the iron in chelated ferric-siderophore complexes. In combination with in vivo uptake studies, these results offer insights into the function of this poorly characterized membrane protein family and its role in iron release from bacterial siderophores.
Keywords: integral membrane protein, reductase, heme, siderophore uptake, crystal structure
Abstract
Gram-negative bacteria take up the essential ion Fe3+ as ferric-siderophore complexes through their outer membrane using TonB-dependent transporters. However, the subsequent route through the inner membrane differs across many bacterial species and siderophore chemistries and is not understood in detail. Here, we report the crystal structure of the inner membrane protein FoxB (from Pseudomonas aeruginosa) that is involved in Fe-siderophore uptake. The structure revealed a fold with two tightly bound heme molecules. In combination with in vitro reduction assays and in vivo iron uptake studies, these results establish FoxB as an inner membrane reductase involved in the release of iron from ferrioxamine during Fe-siderophore uptake.
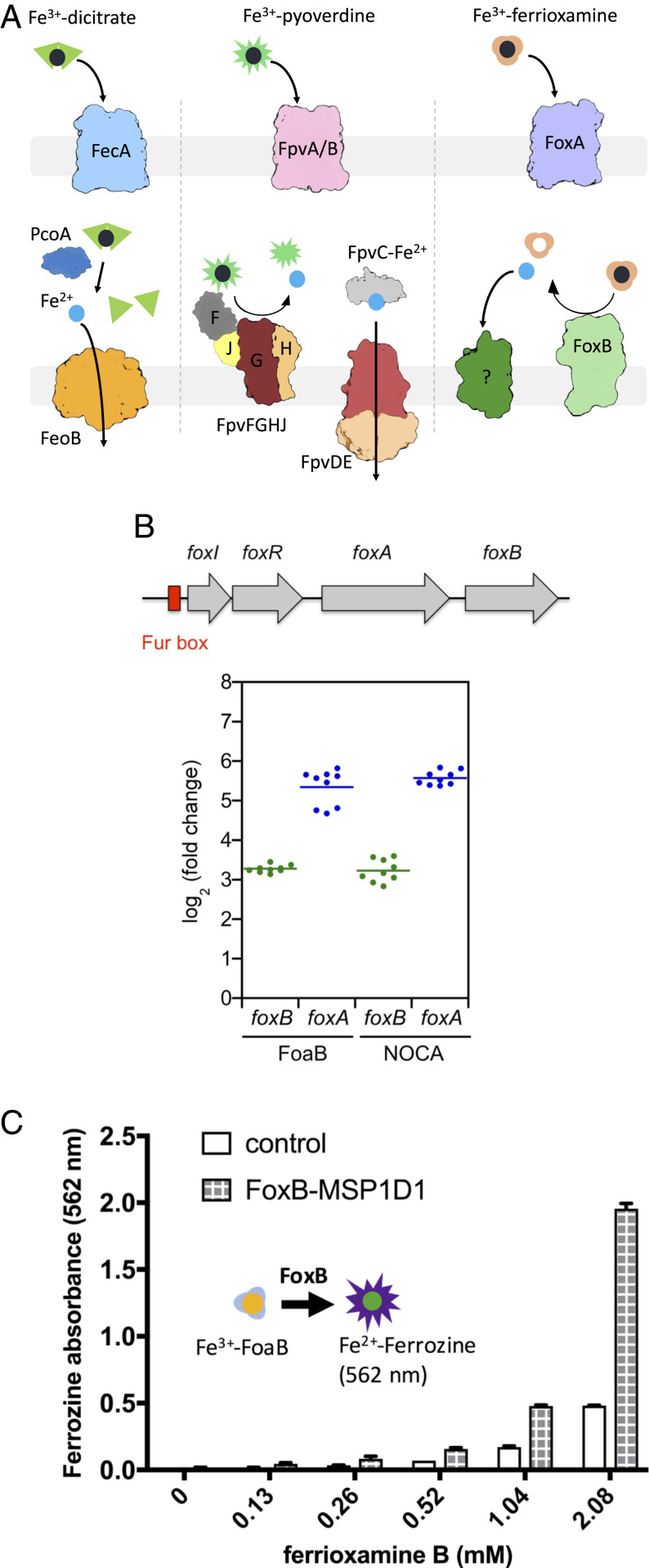
Most microbial communities rely on the bioavailability of iron for their survival. Due to its physicochemical properties, ferric iron exhibits low solubility at physiological pH, precipitating out as ferric oxides and hydroxides. The limited availability of this precious resource has led to severe competition among microbes to scavenge this extremely scarce micronutrient from the environment. Most bacterial species as well as several fungi secrete numerous small-molecule iron chelators, named siderophores, which bind ferric ion with extremely high affinity and selectivity while maintaining them in a soluble state. Once such a complex is formed, ferric-siderophores can be taken up into the cells (Fig. 1A). In gram-negative bacteria, siderophores are generally taken up by TonB-dependent transporters (TBDTs) present in the bacterial outer membrane (OM). Translocation across the OM relies on the coupling of ferri-siderophore–bound TBDTs to the energizing complex in the inner bacterial membrane composed of ExbB/ExbD/TonB proteins. Generally, a single TBDT recognizes a subset of siderophores within the same chemical family.
Fig. 1.
Functional characterization of foxB. (A) Overview of relevant Fe3+-siderophore transport pathways in P. aeruginosa. (B) Expression of foxB and foxA in the presence of FoaB or NOCA in minimal medium. (Top) Organization of fox genes in P. aeruginosa genome. foxA encodes for TBDT transporter localized at the OM, foxI encodes for sigma factor, foxR encodes for anti-sigma factor localized at the IM, and foxB encodes for protein localized at the IM. A fur box is present upstream of the foxI gene. (Bottom) PAO1 wild-type was grown in CAA medium overnight and then resuspended in fresh CAA medium at OD600nm = 0.1 and treated or not with 10 μM FoaB or 10 μM NOCA for 8 h. Total RNA was extracted and retrotranscribed in complementary DNA. Expression of genes was normalized to the uvrD reference gene in each condition. The expression of foxA and foxB was measured in FoaB or NOCA condition and related to the untreated condition. Results were represented in log2 fold change expression. Graphical representation of the result of three biological experiments each performed in triplicate. The bars represent the means. (C) FoaB reduction assay using ferrozine absorption to monitor Fe2+ release from the siderophore. Absorbance was measured at 562 nm for a range of ferrioxamine concentrations and FoxB in MSP1D1 (3 μM) and 15 mM DTT. Plots represent an average of three independent assays with SE shown.
The route of ferric-siderophores across the inner membrane (IM) is less straightforward and differs across many bacterial species and siderophore chemistries. Generally, once inside the periplasmic space, ferric-siderophore complexes are recognized by the dedicated periplasmic-binding proteins for delivery to IM transporters for uptake into the cytoplasm. The most common family of IM transporters involved in the uptake of ferric-siderophores are ATP-binding cassette (ABC) importers. Several ABC transporters implicated in siderophore transport have been identified and characterized to date (1, 2).
In order to assimilate the captured iron into their biological processes, the ferric-siderophore complex must be dissociated. Several mechanisms for iron release from the chelated siderophores have been postulated, which finally all lead to iron reduction, associated either with a siderophore hydrolysis (3), chemical modification of the siderophore such as acetylation (4), or proton-mediated iron release (3, 5, 6). Hydrolysis of siderophores is carried out by dedicated esterases, which fragment the siderophore, lowering the stability of the interaction with iron and facilitating subsequent dissociation steps. To date, several siderophore hydrolases and esterases have been identified, including Fes, IroD, and IroE found in Escherichia coli and Salmonella (7, 8), PfeE from Pseudomonas aeruginosa (9), and Cee from Campylobacter jejuni (10). Some of these proteins are cytoplasmic, whereas others are localized in the periplasm.
The bacterial cell compartment of iron reduction and release from the siderophore complex also differs based on the microbe and siderophore. Several siderophore reductases have been identified in various bacteria and fungi. For example, E. coli have FhuF, an Fe-S cluster–containing protein that targets ferric complexes with coprogen, ferrichrome, and ferrioxamine B (FoaB) (11). Additionally, YqjH, another siderophore reductase found in E. coli, as well as its homolog ViuB from Vibrio cholera use NADPH-FAD cofactors to reduce iron in ferric-triscatecholates complexes such as enterobactin, aerobactin, vibriobactin, and ferric dicitrate (12). In gram-positive mycobacteria, IrtAB, a fusion of an ABC transporter and a reductase, is responsible for both the uptake and subsequent reduction of several mycobacterial siderophores (13). All these siderophore reductases function in the cytoplasm after the translocation of the ferric-chelate across the bacterial IM.
In stark contrast, certain siderophores rely on iron release within the periplasmic space. For example, in P. aeruginosa pyoverdine and citrate siderophores dissociate their iron in the periplasm for subsequent uptake via a dedicated Fe transporter, implying the existence of siderophore-specific periplasmic reductases (Fig. 1A) (5, 14, 15). Recent studies have identified the four proteins FpvG, FpvH, FpvF, and FpvJ, which co-occur in the pyoverdine uptake operon, that are responsible for the reduction of iron in the ferric-pyoverdine complex (15). The IM FpvG is the reductase, but it needs the two other IM proteins FpvH and FpvK and the periplasmic FpvJ protein for its full activity (Fig. 1A).
Pseudomonas aeruginosa is a ubiquitous gram-negative bacterium that has emerged as an opportunistic human pathogen with significant clinical implications. It is one of the leading causes in high mortality rates arising from nosocomial infections. It uses two major siderophores, namely, pyoverdine (PVD) and pyochelin (PCH), to obtain iron from the environment. At the same time, it can also utilize foreign siderophores (exosiderophores) as part of its highly adaptable lifestyle. One such exosiderophore family consists of hydroxamate polydentates named ferrioxamines produced by members of Streptomyces family and several fungi. P. aeruginosa, like numerous gram-negative bacteria, is able to take up ferrioxamines via a dedicated TBDT FoxA in an act of siderophore piracy (16, 17). Interestingly, our recent studies have suggested that FoaB but not ferrioxamine E (nocardamine [NOCA]) uses an additional unidentified TBDT for its uptake into P. aeruginosa (18). Generally, gram-negative bacteria use ABC transporters fhuDBC or hmuUVT in a species-specific manner to transport hydroxamate siderophores across the IM (19, 20). However, neither of these uptake systems have been described or characterized in P. aeruginosa.
Here, we report on the crystal structure of FoxB, which belongs to a large, poorly understood family of IM oxidoreductases associated with siderophore uptake and processing. The protein possesses a novel fold with the transmembrane (TM) domain harboring di-hemes, indicating a role as IM reductase involved in Fe-siderophore uptake.
Results and Discussion
Characterization of FoxB as Reductase.
Our previous biochemical and structural studies have implicated the TBDT FoxA in the uptake of ferrioxamines B and E across the OM in P. aeruginosa. The foxA gene shares its operon with genes encoding for sigma and antisigma factors foxI and foxR, respectively, which regulate the operon expression through siderophore-dependent signaling cascades involving the signaling domain of FoxA (Fig. 1B). The operon possesses an additional gene coding for an uncharacterized IM protein, FoxB, belonging to the conserved family of iron-regulated membrane proteins, termed the PepSY_TM family (COG3182). FoxB and its distant orthologs have been implicated in siderophore uptake but with little functional understanding of their activity (21). Recent studies with a paralogous protein FpvG, found in the pyoverdine operon, have concluded FpvG to be an integral membrane siderophore reductase of ferric pyoverdine (Fig. 1A) (15). Therefore, we reasoned that FoxB could also act as a ferrioxamine reductase.
Upon overexpression and purification of FoxB from E. coli membranes, it became evident that the protein possesses heme groups with a typical ultraviolet–visible (UV-Vis) spectrum having a Soret band and a broad Q-band, characteristic of heme-bound proteins. Reducing treatment of FoxB with sodium dithionite resulted in the shift of the Soret peak from 414 nm to 429 nm, along with a sharp appearance of distinct peaks in the Q-band region (SI Appendix, Fig. S1A). Oxidation of FoxB with potassium ferricyanide reverted the spectrum to its original state, indicating that FoxB purifies in an oxidized state. Reducing the activity of FoxB and subsequent iron dissociation from FoaB was followed using the colorimetric ferrozine assay, which detects free Fe2+ ions in solution. Purified FoxB had no detectable reductive capacity in any detergents. However, after reconstitution into MSP1D1 nanodiscs, the reductive activity of FoxB could be measured, using a range of Fe3+-FoaB concentrations (Fig. 1C). Ferrozine forms a complex with Fe2+, resulting in absorbance at 562 nm and a color change of solution. The increase in absorbance at 562 nm could be followed over time in the presence of FoxB, dithiothreitol (DTT), and Fe3+-FoaB, indicating that Fe2+ ions were formed by the reduction of the ferric sideophore.
Overall Structure of FoxB.
To gain further structural insights into the reductase function of FoxB, we have determined the crystal structure of FoxB using single-wavelength anomalous dispersion (SAD) in combination with molecular replacement (MR). Initial SAD phases based on the calculated selenium and iron sites (SI Appendix, Fig. S2) allowed the building of ∼70% of the backbone, revealing the overall fold of FoxB and placement of heme groups. Some sequence assignment could be made based on the positions of Se atoms; however, due to the low number of methionine residues (5 total out of 382 residues) and low resolution of the datasets, further side-chain tracing could not be completed. We then submitted our sequence to the Critical Assessment of Structure Prediction (CASP14) competition (target T1058) and used the model predicted by the best-performing group (later identified as AlphaFold2/DeepMind) (22) for MR. With this model, we obtained a clear MR solution, which could then be used for MR-SAD, resulting in a good electron density map for the entire protein (SI Appendix, Fig. S3).
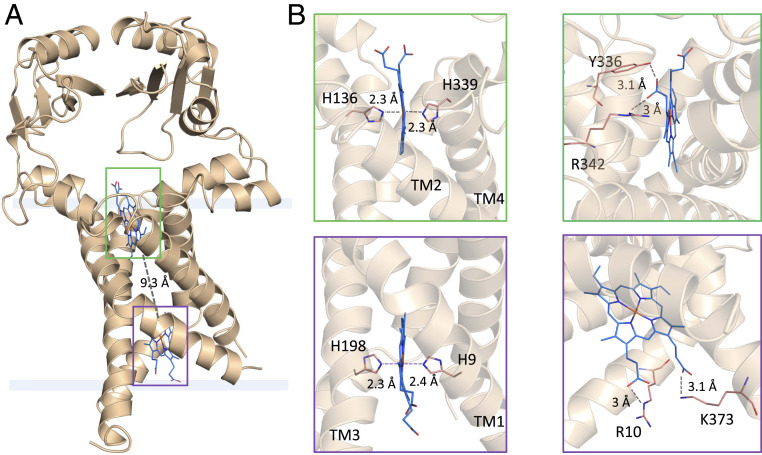
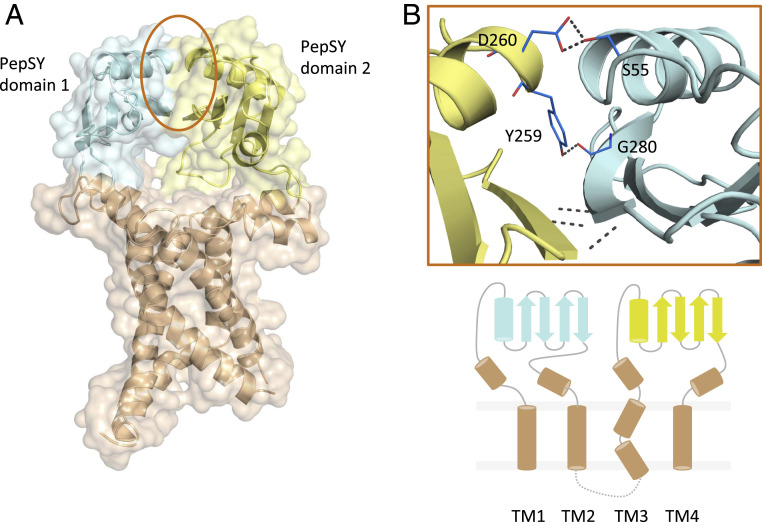
The structure consists of a four TM helical bundle capped at the periplasm by two PepSY (peptidase propeptide and YpeB domain/Pfam:PF03413) domains (Fig. 2). The cytoplasmic residues forming a loop between TM2 and TM3 (residues 172 to 188) were absent from the electron density and omitted from the final model. The TM bundle wraps around two b-type hemes coordinated noncovalently by highly conserved His residues protruding from the TM domains (Fig. 2 and SI Appendix, Fig. S4). The iron in both heme groups is in the octahedral coordination state. Both hemes are found at the edges of the lipid bilayers with propionate groups protruding toward the solvent environment. The two heme groups are separated by a 9.3 Å edge-to-edge distance (Fig. 2A), with two highly conserved Trp residues potentially mediating the electron transfer within the TM core of FoxB. Single-point alanine mutations of W206, adjacent to the cytoplasmic heme, and W157, located in the center of the TM bundle (SI Appendix, Fig. S3B), lower the reducing activity of FoxB by 48% and 26% respectively, relative to wild-type FoxB (SI Appendix, Fig. S5A). The double alanine mutant exhibited ∼17% activity, confirming a crucial role of the Trp residues in the electron transfer reaction.
Fig. 2.
Overall structure of FoxB. (A) FoxB structure reveals a four-helix TM-fold topped with periplasmic PepSY domains. The membrane-spanning region is indicated, and both heme molecules are boxed. (B) Di-heme coordination by the TM domain of FoxB. All four TM helices contribute to heme binding. Both hemes are found at the edges of the lipid bilayers with propionate groups protruding toward the solvent environment. Coordinating residues toward the Fe atoms or propionate groups are indicated for both hemes in two orientations.
The positioning of the TM bundle in the lipid bilayer was analyzed by the Orientations of Proteins in Membranes (OPM) web server (23) and suggests a tilt angle of 20° with a hydrophobic thickness of 30.4 Å (SI Appendix, Fig. S6). TM3 is kinked in the middle by a nonconserved stretch of G201-G202 residues, splitting it into two TM helices, TM3a and TM3b. The kink causes TM3a to bend by 66° perpendicular to the lipid bilayer plane and positions H198 to act as an apical ligand coordinating Fe within the cytoplasmic heme group.
Both propionate groups of the cytoplasmic heme coordinate the conserved residues R10 and K373 through electrostatic or H-bond forces. At the periplasmic side, a single heme propionate group is within an H-bonding distance of a conserved R342 and a nonconserved Y336 (Fig. 2).
The periplasmic PepSY domains form extensive interdomain contacts through β-augmentation between the β-sheets of individual domains as well as additional side-chain H-bond contacts from the adjacent α-helices (D260 and S55) (Fig. 3). The buried surface area for the interdomain complex is 479.3 Å2. Both PepSY domains enclose a large, solvent-accessible cavity located above the TM domain (SI Appendix, Fig. S7). This cavity is rich in aromatic amino acids along with a cluster of charged residues in PepSY domain 1. The cluster of negatively charged residues is located on the periplasmic surface of domain 1 (residues 33 to 117) and a loop from the second PepSY domain (residues 297 to 300) (SI Appendix, Fig. S8A). Using in-silico docking through AutoDock Vina, we were able to place both FoaB and NOCA inside this periplasmic cavity, clustered by the aromatic and polar residues of PepSY domain 2 (SI Appendix, Fig. S7B). The docked ligands are located above the propionate groups of the periplasmic heme. Siderophores and one of the propionate groups are bridged by Y336, which is located at the base of the binding pocket.
Fig. 3.
Periplasmic PepSY domains within FoxB. (A) Two PepSY domains (residues 46 to 118 and 239 to 325, respectively) form the periplasmic part of the protein and interact via several hydrogen bonds (shown in B). Overall secondary structure of FoxB is also shown.
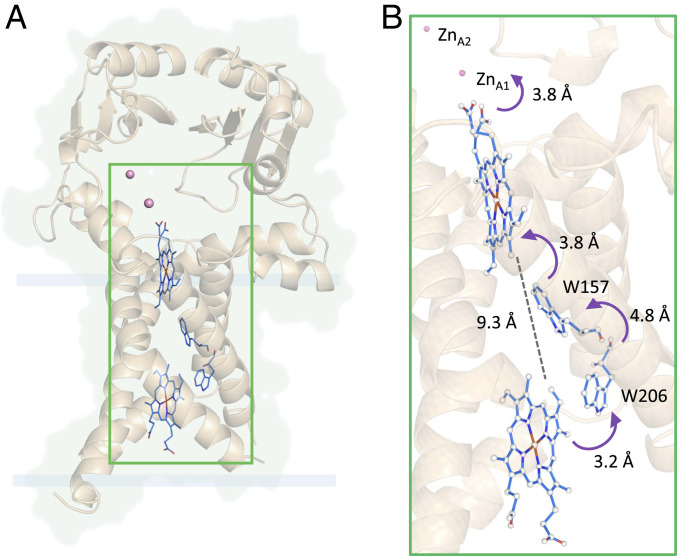
We could identify two Zn ions in one of the molecules of FoxB (ZnA1 and ZnA2) as well a single Zn atom within the second FoxB molecule (ZnB). The crystallization conditions contain 100 mM ZnCl2, which we believe is the source for bound Zn ions. No crystals of FoxB could be obtained without Zn supplementation. ZnA1 and ZnA2 are coordinated by a tetrad of His residues, which form a small tunnel out of the periplasmic cavity. The position of ZnA1 and ZnB are equivalent but not identical; both ions are in the vicinity of the propionate groups of the periplasmic heme group (SI Appendix, Fig. S8B).
A Dali search suggests a very distant structural similarity of the FoxB TM domains to the E. coli superoxide:ubiquinone oxidase (pdb:5oc0) and fumarate reductase (pdb:1e7p) with an rmsd of 5.6 Å and 5.5 Å, respectively (SI Appendix, Fig. S9A). Both of these are heme-containing membrane proteins involved in redox reactions. An analysis of the PepSY domains from FoxB against the available structures of PepSY-containing proteins suggests a highly conserved fold (SI Appendix, Fig. S9B). The function of these proteins is currently poorly understood, with experimental evidence pointing toward the inhibition of peptidase activity through protein–protein interactions. However, these proteins are believed to not possess any TM domains and regulate specific peptidases as stand-alone entities (24).
FoaB and NOCA Induce foxB and foxA Expression.
A Fur box is present upstream of foxI gene (sigma factor) and represses the transcription of fox genes in the presence of iron. In iron-deficient conditions, foxA expression is induced (16); this expression is enhanced when the exosiderophores FoaB and NOCA are present in the P. aeruginosa environment (16, 18). The up-regulation of foxA transcription and expression is dependent on the presence of FoxI (16). It has been shown that FoaB induces the self-cleavage of the anti-sigma factor FoxR that certainly leads to the release of the sigma factor FoxI (25).
To check whether foxB transcription is induced in the presence of FoaB and NOCA (as is the case for foxA), P. aeruginosa PAO1 strain was grown in casamino acid (CAA) iron-deficient medium with and without 10 μM FoaB and NOCA. After 8 h, total RNA was extracted and foxA and foxB transcription were measured by qRT-PCR. As previously described (18), an increase in foxA transcription in the presence of both FoaB and NOCA was observed (log2 fold change increase of 5.34 ± 0.45 for ferrioxamine and 5.57 ± 0.18 for NOCA) (Fig. 1B). In parallel, foxB transcription was also increased in the presence of FoaB (log2 fold change increased 3.28 ± 0.09) and NOCA (log2 fold change increased 3.23 ± 0.28), demonstrating that transcription of both foxA and foxB genes are regulated by the presence of FoaB and NOCA.
foxB Deletion Affects Iron Acquisition By NOCA.
We have previously shown that iron acquisition by NOCA was completely dependent on FoxA OM transporter expression, but iron uptake by FoaB was only partially dependent on this transporter (18). To investigate the role of FoxB in these two iron acquisition pathways, 55Fe uptake was monitored in the presence of these two exosiderophores in a PVD- and PCH-deficient P. aeruginosa strain (ΔpvdFΔpchA) and its corresponding foxA and foxB deletion mutants (ΔpvdFΔpchA∆foxA and ΔpvdFΔpchAΔfoxB) (SI Appendix, Fig. S10). Strains unable to produce pyoverdine and pyochelin were used to avoid any 55Fe uptake by these two siderophores. These strains were grown in iron restricted CAA medium with 10 μM FoaB and NOCA to induce the expression of foxA and foxB, and the uptake assays were initiated by the addition of 500 nM 55Fe-FoaB or 55Fe-NOCA.
Similar rates as previously described for 55Fe uptake in the presence of FoaB and NOCA in ΔpvdFΔpchA cells were observed (18). In the foxA deletion mutant (ΔpvdFΔpchAΔfoxA), transport of 55Fe-NOCA was totally abolished and partially affected for 55Fe-FoaB as previously described: ferri-NOCA is transported across the OM only by FoxA, while ferri-FoaB is transported by FoxA and at least another TBDT (18). foxB deletion had no effect on 55Fe assimilation in the presence of FoaB but affected iron uptake by NOCA, resulting in ∼50% inhibition (SI Appendix, Fig. S10). This finding indicates that FoxB plays a role in iron assimilation by NOCA but is not essential for the FoaB iron uptake pathways. FoxB can probably be replaced by another protein that has a redundant function in ferri-NOCA as well as in ferri-FoaB uptake pathways. Redundancy among TBDTs toward siderophore uptake is common and could also exist among homologous FoxB reductases. An alternative explanation is that iron-loaded FoaB could be taken up directly into the cytoplasm by an uncharacterized IM transporter. In E. coli, iron-loaded hydroxamate siderophores such as FoaB are taken up via the FhuBCD ABC transporter, which is not present in P. aeruginosa. However, an operon consisting of genes PA2912-2914 encodes for a putative ABC transporter with distant homology to HmuUVT in P. aeruginosa. HmuUVT has been implicated in heme and hydroxamate siderophore uptake in Sinorhizobium meliloti (19).
Members of the COG3182 Family Are Widely Distributed across the Bacterial Kingdom.
A search for FoxB paralogues in the P. aeruginosa PAO1 genome revealed the existence of up to five additional operons associated with siderophore or metal ion uptake bearing COG3182 gene members. Each operon also has a dedicated TBDT adjacent to the COG3182 gene. The operons and their associated gene members are outlined in SI Appendix, Fig. S11. They include the pyochelin operon, putative mycobactin operon, and three additional “orphan” operons with no known siderophore substrates (ferrioxamine and pyoverdine operons are excluded).
Members of the COG3182 family are widespread across the bacterial kingdom, including proteobacteria, firmicutes, bacteroides, and cyanobacteria. In gram-negative bacteria, COG3182 members are often in genomic proximity of a TBDT associated with the uptake of siderophores and metal ions as judged by the COmparative Gene Neighborhoods Analysis Tool analysis. In several instances, we find gene fusions between COG3182 members and sulfite reductases. Therefore, we suspect that ferric-siderophore reduction or dissociation in the periplasm is much more common than previously assumed. Unlike FoxB, FpvG, which reduces iron chelated by pyoverdine and belongs to the COG3182 family, requires FpvH for an optimal activity. The released ferrous iron is then picked up by FpvC and delivered to the ABC transporter FpvDE (15). Similar to FoxB, fpvG mutants reduce 55Fe uptake by pyoverdine in P. aeruginosa by ∼30 to 50%. Interestingly, deletion of FpvDE also reduces iron uptake by 40 to 50%, suggesting that some iron finds its way into the cells via a different IM transporter.
Conclusion
Siderophores act to deliver iron, an essential micronutrient, to bacteria and fungi. Uptake and translocation of siderophores has received significant scientific attention, yet the precise mechanisms of iron release from these chelated complexes still remain poorly understood. In this study, we report on the structure of FoxB, which belongs to a large, uncharacterized family of integral membrane oxidoreductases that assist in the assimilation of iron from the captured ferric-siderophore complexes in the periplasm through the reduction and release of iron from siderophores.
Based on the structure of FoxB, we propose a through-space electron pathway involving the heme molecule on the cytoplasmic side of the TM region, two conserved Trp residues (W157/W206) located in TM2/3a, the periplasmic heme, and two Zn2+ ions that may occupy/substitute the Fe3+-siderophore binding site. All residues involved are aligned within a 5 Å distance (Fig. 4). Our mutagenesis studies confirm the roles of W157 and W206 amino acids in electron transfer, with the removal of both residues resulting in almost a complete loss of activity. This through-space electron tunneling pathway acts to deliver electrons via the heme groups to the periplasmic cavity where the siderophore would be located. The in-silico docking of FoaB and NOCA into this cavity places both siderophores above the periplasmic heme (SI Appendix, Fig. 7B) into a pocket that is rich in hydrophobic and aromatic residues similar to hydroxamate binding pockets of TBDTs (17, 26). A reduction of the Fe(III) to Fe(II) in the siderophore lowers the thermodynamic stability of the complex, which would facilitate iron dissociation by proton-mediated exchange or competitive chelation of iron from the Fe(II)-siderophore complex (27).
Fig. 4.
Potential electron pathway in FoxB reductase. The proposed through-space electron pathway comprises the heme molecule on the cytoplasmic side of the TM region, two highly conserved Trp residues (W157/W206) located in TM2/3a, the periplasmic heme, and two Zn2+ ions that may occupy/substitute the Fe3+-siderophore binding site. All residues involved are aligned within a distance of 5 Å.
The transporter responsible for Fe(II) uptake from FoxB-processed ferrioxamines is presently not known. There are numerous transporters in the IM responsible for Fe(II) uptake in gram-negative bacteria. For example, FeoB is involved in the acquisition of iron from ferric citrate (5), with no role in the transport of iron derived from FoaB or pyoverdine. Aside from FeoB, other transporter families involved in ferrous iron uptake in gram-negative bacteria include the EfeU family (28), MntH family of divalent metal ion/H+ symporters (part of the larger Nramp family of transporters) (29), and broad specificity Zip family divalent transporters. Their association with siderophore-dependent iron acquisition has not been shown to date.
Materials and Methods
Chemicals.
FoaB and the protonophore carbonyl cyanide m-chlorophenylhydrazone were purchased from Sigma-Aldrich, and 55FeCl3 was purchased from Perkin-Elmer. NOCA was purified as previously described (30).
Bacterial Strains and Growth Conditions.
The P. aeruginosa PAO1 strains used in this study are listed in SI Appendix, Table S2. Bacteria were initially grown in Luria-Bertani (LB) medium overnight at 30 °C. Next, bacteria were washed and resuspended in iron-deficient CAA medium containing 5 g ⋅ L−1 low-iron CAA (Difco), 1.46 g ⋅ L−1 K2HPO4 3H2O, and 0.25 g ⋅ L−1 MgSO4 7H2O and were grown overnight at 30 °C.
Plasmid and Strain Constructions.
For pEXG2foxB construction, a 1,449-bp insert containing flanking sequences of foxB gene (736 bp upstream and 715 bp downstream foxB gene) was amplified by PCR from genomic DNA of P. aeruginosa PAO1 using the specific primers listed in SI Appendix, Table S4 with High-Fidelity DNA polymerase (Thermo-Fisher Scientific). Primers were designed to generate XhoI and HindIII restriction sites at the 5′ end and 3′ end of the PCR product. The PCR product was cloned into pEXG2 vector using XhoI and HindIII restrictions sites. Ligation was performed using T4 DNA ligase (Thermo-Fisher Scientific). pEXG2foxB plasmid was transformed in E. coli strain TOP10 (Invitrogen). Mutation in the chromosomal genome of P. aeruginosa ΔpvdFΔpchA was generated as previously described (31) by triparental mating. Mutants were selected and verified by PCR and sequencing.
RNA Extractions.
Bacteria were grown overnight in CAA medium. Afterward, bacteria were pelleted, washed, and resuspended at OD600 nm = 0.1 in fresh CAA medium with or without 10 μM NOCA or FoaB. Cells were cultivated at 30 °C for 8 h. Then, 2.5 × 108 cells were mixed with two volumes of RNAprotect Bacteria Reagent (Qiagen). Samples were lysed in Tris-EDTA (pH 8.0) containing 15 mg/mL lysozyme (Sigma-Aldrich) for 15 min at 25 °C. Lysates were homogenized with QIAshredder kit (Qiagen), and total RNAs were extracted with RNeasy mini kit (Qiagen). After treatment with DNase (RNase-Free DNase Set, Qiagen), RNAs were purified with an RNeasy Mini Elute cleanup kit (Qiagen).
qRT-PCR Analysis.
RNA (1 μg) was reversed transcribed with High-Capacity RNA-to-cDNA Kit (Applied Biosystems) in accordance with the manufacturer’s instructions. The expression of genes was measured in a SpetOne Plus Instrument (Applied Biosystems), with Power Sybr Green PCR Master Mix (Applied Biosystems) and the appropriate primers (SI Appendix, Table S3). The uvrD expression was used as an internal control. For a given gene in each strain, the transcript levels were normalized with respect to those for uvrD and were expressed as a base two logarithms of the ratio (fold change) relative to the reference conditions.
Iron Uptake.
Siderophore-55Fe complexes were prepared as previously described (32) with a ratio 20:1 for siderophore:iron in 50 mM Tris⋅HCl (pH 8.0). After overnight growing in CAA medium, cells were washed and resuspended in fresh CAA medium at OD600 nm of 0.1 in the absence of 10 μM NOCA or FoaB to induce foxA and foxB expression and grown overnight at 30 °C. Afterward, bacteria were washed with 50 mM Tris⋅HCl (pH 8.0) and diluted to an OD600 nm of 1.0. Finally, bacteria were incubated with 500 nM siderophore-55Fe at 30 °C. At 0, 15, 30, 45, 60, and 120 min, aliquots were removed, cells were harvested by centrifugation, and the radioactivity was monitored in the bacteria pellet as previously described for iron uptake by pyochelin (32). The assay was carried out as well with bacteria incubated during 15 min with 200 µM carbonyl cyanide m-chlorophenylhydrazone in 50 mM Tris⋅HCl (pH 8.0) (OD600 nm of 1.0) before the addition of 500 nM siderophore-55Fe.
Protein Expression and Crystallization.
A full-length foxB gene from P. aeruginosa PAO1 strain was cloned into a pnEK vector with a tobacco etch virus (TEV) cleavable N-terminal His6-tag. The protein was expressed in E. coli C43 (DE3) cells grown in terrific broth medium. Cells were grown at 37 °C to an OD600 of 1 and induced with 0.1 mM isopropyl β- d-1-thiogalactopyranoside (IPTG) followed by overnight growth at 20 °C.
Total membranes were prepared after cell lysis and solubilized in 1% (wt/vol) dodecyl maltopyranoside (DDM) in TBS buffer (20 mM Tris [pH 7.5], 350 mM NaCl, and 10% glycerol) for 1 h at 4 °C. Solubilized membranes were applied onto Ni2+-NTA resin and washed with TBS buffer + 0.03% DDM and 40 mM imidazole; the protein was eluted with 300 mM imidazole, and TEV was added overnight (1:5 wt/wt TEV:protein). The next day, FoxB was reverse purified to remove TEV, the cleaved His6-tag, and any noncleaved FoxB protein. The protein was then concentrated using a 50-kDa cutoff concentrator and injected onto a Superdex S200 10/300 increase column equilibrated with TBS buffer + 0.2% decyl maltopyranoside without glycerol. Purified FoxB protein in TBS buffer + 0.2% decyl maltopyranoside was concentrated to 15 mg/mL and crystallized using sitting-drop vapor diffusion.
Ferrozine Reduction Assay.
To check the reducing activity of FoxB, the protein was purified as described above using DDM and then incorporated into MSP1D1 nanodiscs in a 1:2:40 ratio (FoxB:MSP1D1:POPC) using previously established protocols. Briefly, proteins and lipids were mixed in the stated ratio, and biobeads (0.5 mg/mL) were added for 4 h at 4 °C. Subsequently, the assembled protein was removed from biobeads and purified further using a Superdex S200 10/300 column equilibrated in Hepes pH 7, 350 mM NaCl buffer (buffer B).
For the ferrozine assays, FoxB in nanodiscs (final concentration 3 µM) was mixed with varying concentrations of Fe3+-FoaB in buffer B, supplemented with 0.5 mM ferrozine and 30 mM DTT. The mixture was incubated at 37 °C for 45 min, and absorbance at 562 nm was then measured. Ferrozine forms a stable magenta-colored complex with an absorption peak at 562 nm with ferrous iron (33). As a negative control, FoxB was omitted from the reaction mixture.
Mutant Activity Measurement.
Single-point mutants of proposed electron transport residues W157A, W206A, and a double W157A/W206A were cloned, overexpressed, and purified in the same manner as the wild-type protein. All mutants were then incorporated into MSP1D1 and their activity assessed with respect to wild-type FoxB in MSP1D1 nanodiscs. Absorption spectra were also measured to ensure that mutations did not perturb the heme environment and overall protein fold. We noticed that some mutants tended to precipitate at 37 °C during the ferrozine-based ferrioxamine reduction; therefore, the activity assays were carried out at 22 °C with 50 mM TCEP for 4 to 5 h.
Structure Determination.
The structure of FoxB was determined using X-ray crystallography. Diffraction data were collected at 100 K at the PETRA III/DESY P11, P13, and P14 beamlines. Datasets were processed with XDS (34) and integrated using AIMLESS (35). The final data were derived from two merged datasets, collected with 0.1° and 0.2° oscillation, respectively, from different crystal regions. Due to diffraction anisotropy, StarANISO webserver (36) was used to perform anisotropy correction. Heavy atom sites were calculated with SHELX C/D (37). Further density modification was carried out using RESOLVE as part of the PHENIX package (38). A model generated by the AlphaFold2/DeepMind group during the CASP14 competition (target T1058) (22) was used for MR and subsequent MR-SAD. Anomalous difference maps were also used to validate the model. The final model was refined using COOT (39) and REFMAC (40) with thermal libation and screw-rotation and jelly body parameters. Model refinement showed high R-factors at low resolution, which is likely the underlying cause for slightly high overall R-factors (Rwork/Rfree = 0.26/0.298 for 3.35 Å resolution). In addition, some ice/lune effects were observed around the 3.8 Å resolution range. All data collection and refinement statistics are summarized in SI Appendix, Table S1.
Siderophore Docking Analysis.
In-silico docking of siderophores into FoxB was carried out using AutoDock Vina (41). Box size (48 Å, 28 Å, 26 Å) encompassing the entirety of the periplasmic PepSY domains and the cavity they form above the TM domain was selected as the search space for putative siderophore-binding sites. Siderophores FoaB and NOCA were used as ligands. The exhaustivity parameter was set to nine. Ligand and protein coordinates were prepared using AutoDockTools (http://mgltools.scripps.edu/), polar hydrogens were added, and any nonprotein molecules with the exception of heme groups were removed from the FoxB structure. The top docking solutions with were chosen for visual inspection, and the highest scored docking mode was selected. Both of the docked siderophores exhibited very similar binding modes within FoxB.
Supplementary Material
Acknowledgments
We are grateful to the staff at beamlines P11 (DESY), P13, and P14 (EMBL, Hamburg) and thank members of the H.T. laboratory for helpful discussions. We acknowledge access to the Sample Preparation and Characterization Facility of EMBL. We thank the AlphaFold2 team (DeepMind/Google) for their excellent model (CASP14 target ID: T1058) and Andriy Kryshtafovych (University of California Davis) for making CASP14 results available. This research was funded by the excellence cluster “The Hamburg Centre for Ultrafast Imaging–Structure, Dynamics and Control of Matter at the Atomic Scale” of the Deutsche Forschungsgemeinschaft (DFG EXC 1074).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101952118/-/DCSupplemental.
Data Availability
Structural coordinates and structural factors have been deposited in the RCSB Protein Data Bank under accession number 7ABW (SI Appendix, Table S1).
References
- 1.Schalk I. J., Guillon L., Fate of ferrisiderophores after import across bacterial outer membranes: Different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 44, 1267–1277 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Rohrbach M. R., Braun V., Köster W., Ferrichrome transport in Escherichia coli K-12: Altered substrate specificity of mutated periplasmic FhuD and interaction of FhuD with the integral membrane protein FhuB. J. Bacteriol. 177, 7186–7193 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miethke M., Marahiel M. A., Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannauer M., Barda Y., Mislin G. L., Shanzer A., Schalk I. J., The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome. J. Bacteriol. 192, 1212–1220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall B., Stintzi A., Gilmour C., Meyer J. M., Poole K., Citrate-mediated iron uptake in Pseudomonas aeruginosa: Involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology (Reading) 155, 305–315 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Schröder I., Johnson E., de Vries S., Microbial ferric iron reductases. FEMS Microbiol. Rev. 27, 427–447 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Zhu M., Valdebenito M., Winkelmann G., Hantke K., Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology (Reading) 151, 2363–2372 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lin H., Fischbach M. A., Liu D. R., Walsh C. T., In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127, 11075–11084 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perraud Q., et al., A key role for the periplasmic PfeE Esterase in iron acquisition via the siderophore enterobactin in Pseudomonas aeruginosa. ACS Chem. Biol. 13, 2603–2614 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Zeng X., Mo Y., Xu F., Lin J., Identification and characterization of a periplasmic trilactone esterase, Cee, revealed unique features of ferric enterobactin acquisition in Campylobacter. Mol. Microbiol. 87, 594–608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matzanke B. F., Anemüller S., Schünemann V., Trautwein A. X., Hantke K., FhuF, part of a siderophore-reductase system. Biochemistry 43, 1386–1392 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Miethke M., Hou J., Marahiel M. A., The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 50, 10951–10964 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Arnold F. M., et al., The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 580, 413–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenwald J., et al., Real time fluorescent resonance energy transfer visualization of ferric pyoverdine uptake in Pseudomonas aeruginosa. A role for ferrous iron. J. Biol. Chem. 282, 2987–2995 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Ganne G., et al., Iron release from the siderophore pyoverdine in Pseudomonas aeruginosa involves three new actors: FpvC, FpvG, and FpvH. ACS Chem. Biol. 12, 1056–1065 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Llamas M. A., et al., The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188, 1882–1891 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josts I., Veith K., Tidow H., Ternary structure of the outer membrane transporter FoxA with resolved signalling domain provides insights into TonB-mediated siderophore uptake. eLife 8, e48528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Normant V., et al., Nocardamine-dependent iron uptake in Pseudomonas aeruginosa: Exclusive involvement of the FoxA outer membrane transporter. ACS Chem. Biol. 15, 2741–2751 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Cuív P. O., Keogh D., Clarke P., O’Connell M., The hmuUV genes of Sinorhizobium meliloti 2011 encode the permease and ATPase components of an ABC transport system for the utilization of both haem and the hydroxamate siderophores, ferrichrome and ferrioxamine B. Mol. Microbiol. 70, 1261–1273 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Kingsley R. A., et al., Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl. Environ. Microbiol. 65, 1610–1618 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuív P. O., Keogh D., Clarke P., O’Connell M., FoxB of Pseudomonas aeruginosa functions in the utilization of the xenosiderophores ferrichrome, ferrioxamine B, and schizokinen: Evidence for transport redundancy at the inner membrane. J. Bacteriol. 189, 284–287 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callaway E., ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature 588, 203–204 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeats C., Rawlings N. D., Bateman A., The PepSY domain: A regulator of peptidase activity in the microbial environment? Trends Biochem. Sci. 29, 169–172 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Bastiaansen K. C., Otero-Asman J. R., Luirink J., Bitter W., Llamas M. A., Processing of cell-surface signalling anti-sigma factors prior to signal recognition is a conserved autoproteolytic mechanism that produces two functional domains. Environ. Microbiol. 17, 3263–3277 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Grinter R., Lithgow T., Determination of the molecular basis for coprogen import by Gram-negative bacteria. IUCrJ 6, 401–411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrington J. M., Crumbliss A. L., The redox hypothesis in siderophore-mediated iron uptake. Biometals 22, 679–689 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Grosse C., et al., A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol. Microbiol. 62, 120–131 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Makui H., et al., Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol. Microbiol. 35, 1065–1078 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Meyer J.-M., Abdallah M. A., The siderochromes of non-fluorescent Pseudomonads: Production of nocardamine by Pseudomonas stutzeri. Microbiology 118, 125–129 (1980). [Google Scholar]
- 31.Perraud Q., et al., Phenotypic adaption of Pseudomonas aeruginosa by hacking siderophores produced by other microorganisms. Mol. Cell. Proteomics 19, 589–607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoegy F., Schalk I. J., Monitoring iron uptake by siderophores. Methods Mol. Biol. 1149, 337–346 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Stookey L. L., Ferrozine—A new spectrophotometric reagent for iron. Anal. Chem. 42, 779–781 (1970). [Google Scholar]
- 34.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans P. R., An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tickle I. J., et al., The STARANISO Server (Global Phasing Ltd., Cambridge, UK, 2018). [Google Scholar]
- 37.Sheldrick G. M., A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov G. N., et al., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trott O., Olson A. J., AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural coordinates and structural factors have been deposited in the RCSB Protein Data Bank under accession number 7ABW (SI Appendix, Table S1).