Abstract
The interferon regulatory factor 3 (IRF-3) gene encodes a 55-kDa protein which is expressed constitutively in all tissues. In unstimulated cells, IRF-3 is present in an inactive cytoplasmic form; following Sendai virus infection, IRF-3 is posttranslationally modified by protein phosphorylation at multiple serine and threonine residues located in the carboxy terminus. Virus-induced phosphorylation of IRF-3 leads to cytoplasmic to nuclear translocation of phosphorylated IRF-3, association with the transcriptional coactivator CBP/p300, and stimulation of DNA binding and transcriptional activities of virus-inducible genes. Using yeast and mammalian one-hybrid analysis, we now demonstrate that an extended, atypical transactivation domain is located in the C terminus of IRF-3 between amino acids (aa) 134 and 394. We also show that the C-terminal domain of IRF-3 located between aa 380 and 427 participates in the autoinhibition of IRF-3 activity via an intramolecular association with the N-terminal region between aa 98 and 240. After Sendai virus infection, an intermolecular association between IRF-3 proteins is detected, demonstrating a virus-dependent formation of IRF-3 homodimers; this interaction is also observed in the absence of virus infection with a constitutively activated form of IRF-3. Substitution of the C-terminal Ser-Thr phosphorylation sites with the phosphomimetic Asp in the region ISNSHPLSLTSDQ between amino acids 395 and 407 [IRF-3(5D)], but not the adjacent S385 and S386 residues, generates a constitutively activated DNA binding form of IRF-3. In contrast, substitution of S385 and S386 with either Ala or Asp inhibits both DNA binding and transactivation activities of the IRF-3(5D) protein. These studies thus define the transactivation domain of IRF-3, two domains that participate in the autoinhibition of IRF-3 activity, and the regulatory phosphorylation sites controlling IRF-3 dimer formation, DNA binding activity, and association with the CBP/p300 coactivator.
Interferons (IFNs) are a large family of multifunctional secreted proteins involved in antiviral defense, cell growth regulation, and immune activation (32). Virus infection induces the transcription and synthesis of multiple IFN genes (11, 23, 32); newly synthesized IFN interacts with neighboring cells through cell surface receptors and the Jak-STAT signalling pathway, resulting in the induction of over 30 new cellular proteins that mediate the diverse functions of the IFNs (6, 13, 15, 28). Among the many virus- and IFN-inducible proteins are the growing family of interferon regulatory transcription factors (IRFs), including IRF-1, IRF-2, IRF-3, IRF-4/Pip/ICSAT, IRF-5, IRF-6, IRF-7, ISGF3γ/p48, and ICSBP (21). All of the family members share a high degree of homology in the N-terminal DNA binding domain (DBD) with the five characteristic tryptophan repeats (21). Structurally, the Myb oncoproteins share homology with the IRF family, although their relationship to the IFN system is unclear (31). Recent evidence also demonstrates the presence of a virally encoded analogue of cellular IRFs in the genome of human herpesvirus 8 (25).
IRF-3 was originally identified as a member of IRF family on the basis of (i) homology with other IRF family members and (ii) binding to the IFN-stimulated regulatory element (ISRE) of the ISG15 promoter (1). This protein is distinct from cIRF-3, an avian protein which demonstrates homology to the IRF family members (10). IRF-3 is expressed constitutively in a variety of tissues, and the relative levels of IRF-3 mRNA do not change in virus-infected or IFN-treated cells. IRF-3 demonstrates a unique response to viral infection. Recent studies with IRF-3 demonstrate that virus- and double-stranded RNA (dsRNA)-inducible phosphorylation represents an important posttranslational modification, leading to cytoplasmic to nuclear translocation of phosphorylated IRF-3, association with the CBP/p300 coactivator, and stimulation of DNA binding and transcriptional activities (18, 20, 26, 33–35). Overexpression of IRF-3 significantly enhances virus-mediated expression of type I (alpha/beta) IFN and results in the induction of an antiviral state (14). Virus-induced phosphorylation of IRF-3 also represents a signal for proteasome-mediated degradation of IRF-3, since mutations altering serine and threonine residues at S396, S398, S402, T404, and S405 to alanines inhibit virus-induced IRF-3 phosphorylation and degradation, indicating that serine or threonine phosphorylation subsequent to viral infection signals degradation of this IRF protein (18). Treatment with proteasome inhibitors stabilizes IRF-3 protein levels, thus implicating the ubiquitin-proteasome pathway in virus-induced degradation of IRF-3. These biological features implicate IRF-3 as the immediate trigger of immediate-early IFN gene transcription which leads ultimately to the induction of the antiviral, growth regulatory, and immune modulatory functions of the IFN system (12). Recent studies indicate that virus-stimulated phosphorylation of IRF-3 results in the activation of the immediate-early IFN-α4 and IFN-β genes in murine cells. Once produced and secreted from the infected cell, IFN-α4 and IFN-β feed back on cells through the IFN receptor, stimulate the Jak-STAT pathway, and lead to the IFN-responsive activation of another member of the IRF family, IRF-7; up-regulation of IRF-7 production then mediates the induction of delayed type I, non-IFN-α4 gene expression (19).
Constitutively active and dominant-negative IRF-3 forms were generated by substitution of the phosphomimetic Asp for the Ser/Thr residues [IRF-3(5D)] and by deletion of the DBD [IRF-3(ΔN)]. Interestingly, endogenous human RANTES gene transcription was directly induced in tetracycline-inducible IRF-3(5D)-expressing cells or paramyxovirus-infected cells. Furthermore, the dominant-negative IRF-3(ΔN) inhibited virus-induced expression of the RANTES promoter. Specific mutagenesis of overlapping ISRE-like sites located between nucleotides −123 and −96 in the RANTES promoter reduced virus-induced and IRF-3-dependent activation, thereby demonstrating that at least one member of the chemokine superfamily is also targeted by IRF-3 activation (17).
Based on available data and by analogy with the properties of other IRF family members (21), it has been proposed that IRF-3 exists in closed conformation in the cytoplasm of uninfected cells, as observed with IRF-4 (4, 7). Virus-induced phosphorylation of IRF-3 may lead to a conformational change in IRF-3 that relieves autoinhibition and permits translocation to the nucleus, DNA binding, and interaction with the CBP/p300 coactivator. To validate this model, we now demonstrate that (i) a strong but atypical transactivation domain is located in the C terminus of IRF-3 between amino acids (aa) 134 and 394, corresponding in part to a proline-rich region and the IRF association domain (IAD); (ii) an intramolecular interaction occurs between the C-terminal 48 aa (aa 380 to 427) of IRF-3 and the N-terminal portion of IRF-3 (aa 98 to 240), an association that blocks IRF-3 DNA binding in unstimulated cells; (iii) an intermolecular association between IRF-3 molecules occurs after virus-mediated phosphorylation, leading to IRF-3 homodimer formation; and (iv) two classes of regulatory phosphorylation sites with differing effects on transactivation activity are located in the C-terminal region of IRF-3: Ser-Thr sites at positions 396 to 405 represent in vivo phosphoacceptor sites, while S385 and S386 play a regulatory role in controlling phosphorylation at the adjacent Ser-Thr sites and association with CBP/p300.
MATERIALS AND METHODS
Plasmid constructions and mutagenesis.
The wild-type and mutated forms of IRF-3 expression plasmids were described previously (18). For the construction of GAL4–IRF-3 chimeras (ΔN30, ΔN50, ΔN98, and ΔN197), the relevant IRF-3 cDNAs were created by PCR, and the resulting products were cloned into pSG424 in frame with the GAL4 DBD (1). Other GAL4–IRF-3 chimeras were subcloned from IRF-3/CMVBL plasmids (18) into pSG424. The (Gal4)5-TK/CAT reporter construct was described previously (1).
Yeast one-hybrid and two-hybrid analysis.
The wild-type and mutated forms of IRF-3 cDNA were subcloned from IRF-3/CMVBL plasmids (18) into the TRP1 pAS2-1 vector (Clontech) in frame with the GAL4 DBD or into the LEU2 pACT2 vector (Clontech) in frame with the GAL4 transactivation domain. For yeast one-hybrid analysis, the GAL4–IRF-3 chimeras (in the pAS2-1 vector) were transformed into Saccharomyces cerevisiae Y190 (MATα ura3-52 his3-200 ade2-101 trp1-901 leu2-3,112 gal4Δ, met− gal80Δ URA3::GAL1UAS-GAL1TATA-lacZ) (Clontech) by the lithium acetate permeabilization method and selected for tryptophan prototrophy according to the Matchmaker protocol. Transactivation activity was detected by a colony lift filter assay as specified by the manufacturer.
Cell culture and transfections.
All transfections for chloramphenicol acetyltransferase (CAT) assay were carried out in human embryonic kidney 293 cells or COS cells grown in alpha modified Eagle medium (293) or Dulbecco’s modified Eagle medium (COS) (GIBCO-BRL) supplemented with 10% fetal bovine serum, glutamine, and antibiotics. Subconfluent cells were transfected with 2 μg of CsCl-purified CAT reporter and 4 μg of expression plasmids by the calcium phosphate coprecipitation method. The reporter plasmids were RANTES CAT, IFNβ CAT, PKR CAT, B4-TK CAT, and ISG15 CAT (27); the transfection procedures were previously described (16). At 48 h after transfections, total protein extracts were prepared, and 100 μg (IFNβ CAT, PKR CAT, and B4-TK CAT) or 5 μg (RANTES CAT and ISG15 CAT) of extract was assayed for CAT activity for 1 to 2 h at 37°C. CAT activity was normalized to β-galactosidase (β-Gal) activity. All transfections were performed three to six times. For mammalian one-hybrid analysis, 2 μg of (Gal4)5-TK/CAT or TK/CAT reporter plasmid was cotransfected with 4 μg of the GAL4–IRF-3 chimera-expressing plasmids, and CAT activity was assayed with 10 μg of total protein extract for 1 to 2 h at 37°C.
Western blot analysis of IRF-3.
To confirm expression of the transgenes, equivalent amounts of whole-cell extract (20 μg) were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in a 10% polyacrylamide gel. After electrophoresis, proteins were transferred to Hybond transfer membrane (Amersham) in a buffer containing 30 mM Tris, 200 mM glycine, and 20% methanol for 1 h. The membrane was blocked by incubation in phosphate-buffered saline (PBS) containing 5% dried milk for 1 h and then probed with polyclonal IRF-3 antibody (18), monoclonal Flag antibody M2 (Sigma), or Myc antibody 9E10 (8) in 5% milk–PBS at a dilution of 1:3,000. These incubations were done at 4°C overnight or at room temperature for 1 to 3 h. After four 10-min washes with PBS, membranes were reacted with a peroxidase-conjugated secondary goat anti-rabbit antibody (for IRF-3 antibody) or anti-mouse antibody (for Flag or Myc antibody) (Amersham) at a dilution of 1:2,500. The reaction was then visualized with the enhanced chemiluminescence (ECL) detection system as recommended by the manufacturer (Amersham Corp.).
Electrophoretic mobility shift assay (EMSA).
Whole-cell extracts were prepared 48 h after transfection with 5 μg of expression plasmids, as indicated for individual experiments. Cells were washed in PBS and lysed in a mixture containing 10 mM Tris-Cl (pH 8.0), 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 0.5% Nonidet P-40, 0.5 mM phenylmethylsulfonyl fluoride, leupeptin (10 μg/ml), pepstatin (10 μg/ml), aprotinin (10 μg/ml), chymostatin (0.5 ng/μl), and 0.25 μM microcystin. Equivalent amounts of whole-cell extract (20 μg) were assayed for IRF-3 or IRF-2 binding in gel shift analysis using a 32P-labeled double-stranded oligonucleotide corresponding to the ISRE of the IFN-α/β-inducible ISG15 gene (5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′). Complexes were formed by incubating the probe with 20 μg of each whole-cell extract. The binding mixture (20 μl) contained 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 50 mM NaCl, 2 mM dithiothreitol, 5% glycerol, 0.5% Nonidet P-40, and 10 μg of bovine serum albumin per μl; poly(dI-dC) (62.5 μg/ml) was added to reduce nonspecific binding. After 20 min of incubation with the probe, extracts were loaded on a 5% polyacrylamide gel (60:1 cross-link) prepared in 0.5× Tris-borate-EDTA. After running at 100 to 150 V for 3 h, the gel was dried and exposed to a Kodak film at −70°C overnight. To demonstrate the specificity of protein-DNA complex formation, the cell extract was preincubated with anti-CBP antibody A22 (Santa Cruz) and anti-IRF-3 antibody from Nancy Reich (34).
Immunoprecipitation and immunoblot analysis of interactions between different IRF-3 domains.
293 cells were cotransfected with expression plasmids encoding wild-type or mutated IRF-3 as indicated. Whole-cell extracts (200 to 300 μg) were prepared from cotransfected cells, and each extracts was incubated with 2 μl of anti-Myc antibody 9E10 or 0.5 μg anti-CBP antibody A22 cross-linked to 30 μl of protein A-Sepharose beads for 1 h at 4°C. Precipitates were washed five times with lysis buffer and eluted by boiling the beads for 3 min in 1× SDS sample buffer. Eluted proteins were separated by SDS-PAGE, transferred to Hybond transfer membranes, and incubated with anti-IRF-3, anti-Flag, anti-CBP, or anti-Myc antibody (1:1,000 to 1:3,000). Immunocomplexes were detected by using the ECL system.
Protein expression and purification.
Glutathione S-transferase (GST)–IRF-3C(381-427), GST–IRF-3C5D(381-427), and GST were expressed and isolated from Escherichia coli DH5α following a 3-h induction with 1 mM isopropyl-β-d-thiogalactopyranoside (Pharmacia) at 37°C. Bacterial extracts in PBS containing 1% Triton X-100 were incubated with glutathione-Sepharose beads (Pharmacia) for 20 min at room temperature. After three washes with PBS, the fusion proteins were stored on the beads with the addition of protease inhibitors.
RESULTS
IRF-3 contains a transactivation domain.
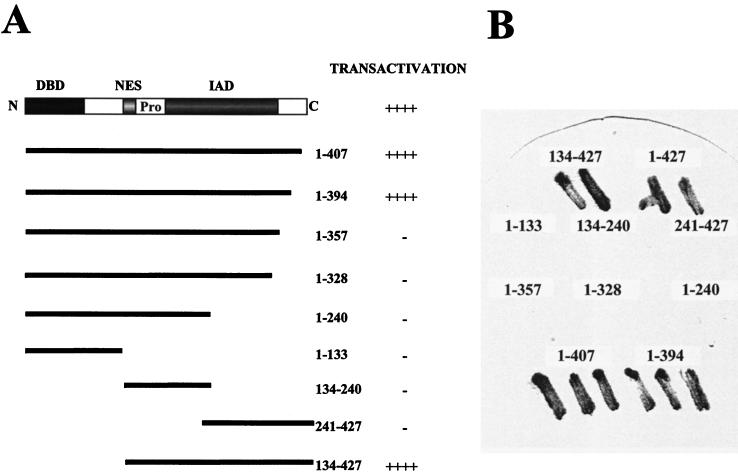
It has been suggested that IRF-3, like IRF-2 and ICSBP, does not contain a well-defined transactivation domain (1). However, in previous studies, deletion of the C-terminal 20 aa of IRF-3 stimulated transcription of IFN-β and ISG15 promoters about 10-fold (18), suggesting that C-terminal truncation may reveal an IRF-3 transactivation domain. During an unrelated two-hybrid genetic screen, it became clear that chimeric GAL4–IRF-3 (full-length IRF-3 fused in frame to the GAL4 DBD) strongly activated a GAL1-lacZ reporter gene in yeast one-hybrid analysis. Colonies from the yeast strain expressing the chimeric GAL4–IRF-3 protein turned blue within 30 min, as detected by the colony lift filter assay (Fig. 1), indicating that IRF-3 contained an intrinsic transactivation domain. To delineate the transactivation domain, we subcloned different segments of IRF-3 cDNA into the TRP1 pAS2-1 vector in frame with the GAL4 DBD and tested chimeric GAL4–IRF-3 proteins for the ability to activate transcription of GAL1-lacZ reporter in yeast strain Y190 (Clontech). Figure 1 demonstrates that the chimeric proteins containing IRF-3 aa 1 to 407, 1 to 394, and 134 to 427 stimulated transcription of GAL1-lacZ reporter, whereas chimeric proteins containing IRF-3 aa 1 to 133, 1 to 240, 1 to 328, 1 to 357, 134 to 240, and 241 to 427 did not activate transcription (colonies remained white or light blue after 24 h).
FIG. 1.
Transactivation of IRF-3 in yeast one-hybrid analysis. (A) Schematic illustration of the IRF-3 protein showing the DBD, NES, proline-rich domain (Pro), and IAD. The wild-type and mutated forms of IRF-3 in the chimeric proteins and their transactivation activities are indicated. ++++, blue colony formation within 30 min; −, white or light blue colonies after 24 h. (B) Representative plate illustrating β-Gal activity. The plasmids encoding chimeric proteins were transformed into yeast strain Y190 carrying GAL1-lacZ reporter; transactivation activity was detected by colony lift filter assay according to the Matchmaker (Clontech) protocol.
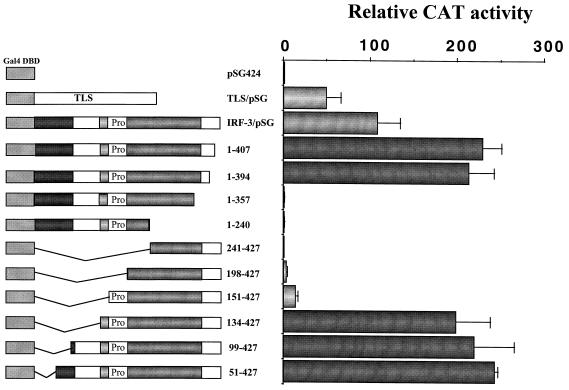
To investigate whether this activation domain also functions in mammalian cells, segments of IRF-3 were inserted into the pSG424 vector in frame with the GAL4 DBD (1). The transactivation activity of the chimeric GAL4–IRF-3 proteins was tested in 293 and COS cells, using a reporter gene that contains five GAL4 binding sites upstream of the minimal thymidine kinase (TK) promoter. In 293 cells, the chimeric protein containing full-length IRF-3 stimulated transcription over 100-fold compared with the pSG424 vector alone (Fig. 2). The positive control TLS/pSG424, which encodes the GAL4-TLS-1-267 chimeric protein (36), stimulated transcription 40- to 60-fold. Chimeric proteins containing the entire region of IRF-3 between aa 134 and 394, including aa 1 to 407, 1 to 394, 134 to 427, 99 to 427, and 51 to 427, activated reporter gene activity up to 200-fold (Fig. 2), whereas chimeric proteins containing IRF-3 aa 1 to 240, 1 to 357, and 241 to 427 did not activate transcription. The chimeric protein containing IRF-3 aa 198 to 427 and 151 to 427 activated transcription only weakly (Fig. 2). Immunoblot analysis of cell extracts revealed that all of the transfected plasmids tested were expressed (data not shown). Reporter gene activation by GAL4–IRF-3 chimeric proteins was dependent on the presence of GAL4 DNA binding sites, since the chimeric proteins did not activate the minimal TK promoter alone (data not shown). These studies delineate a transactivation domain in IRF-3, localized to a continuous stretch of amino acids between positions 134 and 394; the N-terminal limit appears to be localized between aa 134 and 151, while the C-terminal boundary is between aa 357 and 394.
FIG. 2.
Analysis of intrinsic transactivation activities of various IRF-3 regions fused to the yeast GAL4 DBD. 293 cells were transfected with the (Gal4)5-TK/CAT reporter plasmid and various GAL4–IRF-3 chimeric expression plasmids as indicated. CAT activity was analyzed at 48 h posttransfection with 10 μg of total protein extract for 1 to 2 h at 37°C. Relative CAT activity was measured as fold activation (relative to the basal level of the reporter gene in the presence of pSG424 vector after normalization to cotransfected β-Gal activity); the values represent the average of three experiments with variability shown by the error bars.
Characterization of two autoinhibitory domains.
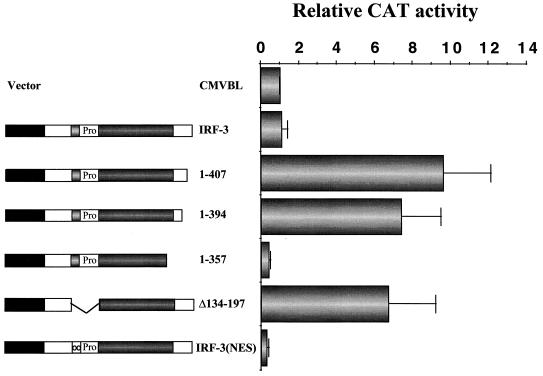
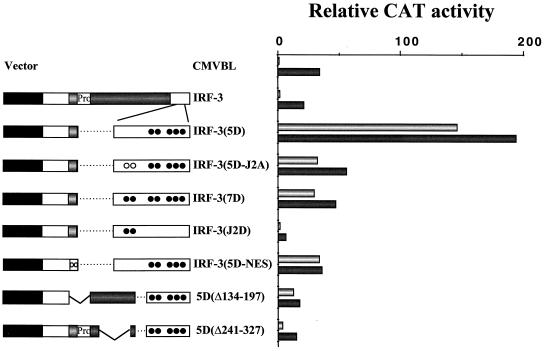
We next tested a series of IRF-3 mutants for transcriptional activity by reporter gene assay in 293 (Fig. 3) and COS (data not shown) cells. The human RANTES promoter linked to CAT was used as a reporter gene, since the RANTES promoter was shown recently to be regulated by IRF-3 (17). Wild-type IRF-3 (wt IRF-3) did not activate the RANTES promoter, whereas deletion of the C-terminal 20 aa of IRF-3 (aa 408 to 427) stimulated expression 10-fold, and IRF-3(394) also activated transcription 7-fold. Further deletion to aa 357 did not stimulate gene expression but rather slightly repressed RANTES promoter activity (Fig. 3). Furthermore, IRF-3 with an internal deletion of aa 134 to 197, which comprises the proline-rich region and the nuclear export signal (NES) element, also stimulated transcription from the RANTES promoter sevenfold (Fig. 3). Similarly, IRF-3 alone weakly activated the ISG15 promoter, and deletion of the C-terminal by 20 or 33 aa generated an IRF-3 that stimulated the ISG15 promoter 12- and 6-fold, respectively. IRF-3(1-357) did not stimulate expression but rather slightly repressed ISG15 (18) (data not shown). These results indicate that two regions of IRF-3, the C-terminal 48 aa (aa 380 to 427) and the internal region from aa 134 to 197, both contributed to inhibition of IRF-3 transactivation potential.
FIG. 3.
Removal of the IRF-3 C terminus stimulates intrinsic IRF-3 transactivation potential. 293 cells were transfected with RANTES CAT reporter plasmid and various IRF-3 plasmids expressing truncated forms of IRF-3 as indicated. CAT activity was analyzed at 48 h posttransfection with 5 μg of total protein extract for 1 to 2 h at 37°C. Relative CAT activity was measured as fold activation (relative to the basal level of reporter gene in the presence of CMVBL vector after normalization to cotransfected β-Gal activity); the values represent the average of three experiments with standard deviation shown by the error bars.
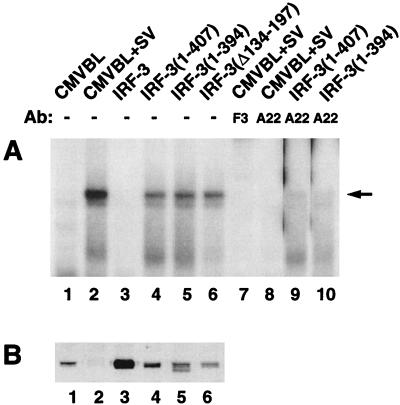
One possibility is that the C-terminal region of IRF-3 inhibits DNA binding capacity through a physical interaction with the N terminus, as demonstrated for IRF-4/Pip (4). To correlate gene activation by IRF-3 and DNA binding to the ISRE site, we transiently transfected wild-type and mutated forms of IRF-3 into 293 cells and determined their DNA binding capacity by EMSA using whole-cell extracts and an ISRE oligonucleotide from the ISG15 gene. In cells transfected with vector alone, IRF-3 DNA binding activity was detected only after virus infection (Fig. 4A, lanes 1 and 2); as expected, extracts from cells transfected with full-length IRF-3 did not bind DNA even though the protein was expressed at a high level (Fig. 4A and B, lanes 3). Deletion of the carboxy-terminal 20 or 33 aa of IRF-3 or, surprisingly, deletion of the region from aa 134 to 197 enabled binding of IRF-3 mutants to the ISRE site in the absence of viral induction (Fig. 4A and B, lanes 4 to 6). These protein-DNA complexes, which were previously characterized in detail (18, 34, 35), contained the CBP coactivator, as confirmed by supershift analysis using the anti-CBP antibody A22 (Fig. 4A, lane 8). Supershift analysis with an anti-IRF-3 antibody (34) demonstrated that this protein-DNA complex also contained IRF-3 (Fig. 4A, lane 7). C-terminal deletion of IRF-3 also resulted in the formation of an IRF-3–CBP DNA binding complex (Fig. 4A, lanes 5 and 6) without virus infection; this complex was also shifted by anti-CBP antibody A22 (Fig. 4A, lanes 9 and 10) or by the anti-IRF-3 antibody (data not shown). Transfected and endogenous IRF-3 levels in cell extracts were detected by immunoblot analysis (Fig. 4B, lanes 1 to 5) except for IRF-3(Δ134-197), which was not recognized by the IRF-3 antibody (Fig. 4B, lane 6). Thus 20 aa in the C-terminal region of IRF-3 (aa 407 to 427) and the region from aa 134 to 197 containing the proline-rich domain and the NES element were required to inhibit IRF-3 DNA binding activity.
FIG. 4.
C-terminal IRF-3 truncation induces constitutive DNA binding. (A) EMSA was performed on whole-cell extracts (20 μg) derived from 293 cells transfected with various IRF-3 expression plasmids (5 μg of each) as indicated above the lanes. At 24 h posttransfection, cells were infected with Sendai virus (SV) for 12 h or left uninfected as indicated. The 32P-labeled probe corresponds to the ISRE of the ISG15 gene (5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′). The position of the IRF-3–CBP complex is indicated by the arrow. Anti-CBP antibody (Ab) A22 (lanes 8 to 10) and anti-IRF-3 antibody F3 (lane 7) were added as indicated to demonstrate the presence of CBP and IRF-3 in the high-molecular-weight protein-DNA complex. (B) Whole-cell extracts (20 μg) from panel A were analyzed by immunoblotting with anti-IRF-3 antibody.
Intramolecular interactions between the N- and C-terminal autoinhibitory domains.
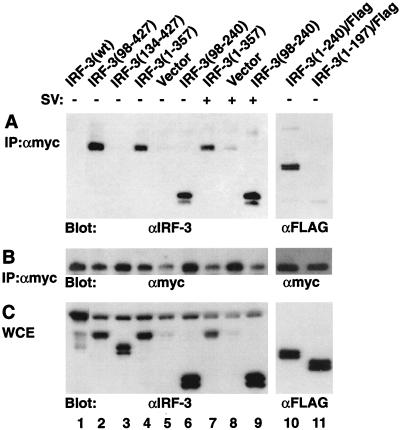
Based on the above observations, we reasoned that an intramolecular contact between the N- and C-terminal domains of IRF-3 may result in autoinhibition of DNA binding. Interactions between distinct IRF-3 domains were investigated by coimmunoprecipitation using 293 cells cotransfected with truncated Myc-tagged IRF-3(328-427) and N- or C-terminally deleted IRF-3 expression plasmids (Fig. 5). After immunoprecipitation of Myc-tagged IRF-3 from cell extracts with anti-Myc antibody (18), immunoblot analysis revealed that IRF-3(98-427) or IRF-3(1-357), lacking the N-terminal 97 aa or the C-terminal 70 aa, coprecipitated with Myc-tagged IRF-3(328-427) (Fig. 5A, lanes 2 and 4) but not with preimmune serum (data not shown). Interaction required part of the N-terminal DBD since IRF-3(134-427), which contains the full C-terminal region but lacks the DBD, did not immunoprecipitate with Myc tag antibody (Fig. 5A, lane 3); also, no endogenous or transfected full-length IRF-3 coprecipitated with the Myc-tagged C-terminal IRF-3 (Fig. 5A, lanes 1 to 6).
FIG. 5.
Intramolecular association between the N- and C-terminal domains of IRF-3. 293 cells were transfected with Myc-tagged IRF-3(328-427) and expression plasmids encoding IRF-3 deletions (5 μg of each plasmid) as indicated above the lanes. At 24 h posttransfection, cells were infected with Sendai virus (SV) (+) for 12 h or left uninfected (−). (A) Whole-cell extracts (200 μg) were immunoprecipitated (IP) with anti-Myc antibody 9E10 (αmyc); immunoprecipitated complexes were run on an SDS–10% polyacrylamide gel and subsequently probed with anti-IRF-3 antibody (lanes 1 to 9) or anti-Flag antibody M2 (lanes 10 and 11). (B) The same membrane was also reprobed with anti-Myc antibody 9E10. (C) Whole-cell extracts (WCE; 20 μg) were run on an SDS–10% polyacrylamide gel and probed with anti-IRF-3 antibody (lanes 1 to 9) or anti-Flag antibody (lanes 10 and 11) to assess the level of transgene expression.
To localize further the region within the N-terminus of IRF-3 that interacted with the C terminus, different IRF-3 deletions were tested for interaction with Myc–IRF-3(328-427). Since the IRF-3 antibody failed to recognize IRF-3 forms lacking the region between aa 150 and 240, some Flag-tagged IRF-3 deletion mutants were used. IRF-3(1-240) and IRF-3(98-240) both associated with Myc–IRF-3(328-427) (Fig. 5A, lanes 6 and 10). Interaction required the entire proline-rich domain, the NES region, and part of the IAD, since the Flag-tagged IRF-3(1-197) did not coprecipitate with Myc-tagged IRF-3(328-427) (Fig. 5A, lane 11). Immunoblot analysis of cell extracts revealed that all of the transfected plasmids were expressed in transfected cells (Fig. 5C). Virus infection had no effect on the interaction between these truncated proteins (Fig. 5A, lanes 7 to 9), perhaps due to phosphorylation of only a portion of the Myc-tagged IRF-3(328-427), since an immunoblot with anti-Myc antibody indicated that most of the protein in immunocomplexes was unphosphorylated (Fig. 5B). In vitro GST pull-down experiments also demonstrated that Myc-tagged IRF-3(1-357) bound to a GST–IRF-3(380-427) fusion protein but not to GST alone (data not shown). Thus the C-terminal autoinhibitory domain is capable of physically interacting with the N-terminal region of IRF-3 located between aa 98 and <240; intramolecular association maintains cytoplasmic IRF-3 in a closed, non-DNA binding conformation.
Intermolecular interactions between IRF-3 after virus infection.
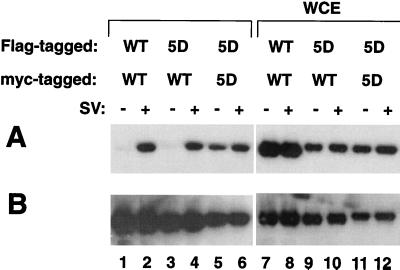
Given the presence of an IAD in the C-terminal region of IRF-3, we examined whether IRF-3 can form homodimers in uninfected or virus-infected cells (Fig. 6). Following transient cotransfection of 293 cells with two tagged forms of IRF-3 (Myc–IRF-3 and Flag–IRF-3), immunoprecipitation of Myc–IRF-3 with anti-Myc antibody 9E10 was performed, followed by immunoblot analysis of the immunoprecipitate with anti-FLAG antibody M2. Interestingly, Flag-tagged IRF-3 coimmunoprecipitated with Myc-tagged IRF-3 in Sendai virus-infected cell extracts (Fig. 6A, lane 2) but not in uninfected cell extracts (Fig. 6A, lane 1), indicating a requirement for virus-induced IRF-3 phosphorylation prior to dimerization. Interestingly, Flag-tagged IRF-3(5D), a constitutively active form of IRF-3 with substitution of the Ser/Thr cluster at aa 396 to 405 with the phosphomimetic Asp (18), associated with Myc-tagged IRF-3(5D) in the absence of virus infection (Fig. 6A, lane 5). However, the interaction between IRF-3(5D) and wtIRF-3 was detected only in the virus-infected cells (Fig. 6A, lanes 3 and 4), suggesting that both IRF-3 molecules must be phosphorylated prior to homodimer formation. The expression of transfected Flag-tagged IRF-3 proteins is shown in Fig. 6A, lanes 7 to 12; the membrane shown in Fig. 6A was also blotted with anti-Myc antibody to confirm expression of the second tagged IRF-3 form (Fig. 6B).
FIG. 6.
IRF-3 homodimer formation after virus infection. 293 cells were transfected with Flag-tagged and Myc-tagged IRF-3 expression plasmids (5 μg) as indicated above the lanes. At 24 h posttransfection, cells were infected with Sendai virus (SV) for 12 h (+) or left uninfected (−). (A) Whole-cell extracts (WCE; 200 μg) were immunoprecipitated with anti-Myc antibody 9E10; immunoprecipitated complexes were run on an SDS–10% polyacrylamide gel and subsequently probed with anti-Flag antibody M2. (B) The membrane in panel A, reprobed with anti-Myc antibody 9E10 to assess the level of expression of transfected protein.
Chimeric GAL4–IRF-3 transactivation activity is increased upon virus infection.
In a previous study, the inducible phosphorylation sites of IRF-3 were located to the region ISNSHPLSLTSDQ, between aa 395 and 407 (18), although another study identified S385 and S386 as critical residues for triggering virus-induced phosphorylation (35). To determine the relationship between these residues, phosphorylation, and transactivation activity, chimeric proteins containing the GAL-4 DBD and IRF-3 point mutations were examined in coexpression assays. As shown in Table 1, point mutants S402/404/405A (3A), S396/398/402/404/407A (5A), and S396/398/402/404/407D (5D) activated transcription up to 200-fold, about 2-fold higher than wtIRF-3, while mutants S396/398A (2A), S385/386A (J2A), and S385/386D (J2D) had lower transactivation activities. The NES mutant was a very weak activator (less than 10-fold induction), whereas the phosphomimetic IRF-3(5D) restored the transactivation activity of the NES mutant. Although previous studies indicated that chimeric GAL4–IRF-3 activated a promoter containing UASG or five GAL4 DNA binding sites in a virus infection-dependent manner (33, 35), in 293 cells, Sendai virus infection increased transactivation of only some GAL4–IRF-3 chimeric proteins: 1.5-fold for full-length IRF-3; 2- to 3-fold for IRF-3(151-427), IRF-3(2A), and IRF-3(J2D); and 4- to 5-fold for IRF-3(NES) (Table 1). Similar results were obtained for COS cells (data not shown).
TABLE 1.
Stimulation of transactivation activity of the GAL4–IRF-3 chimeras by Sendai virusa
| Cotransfected plasmid | Relative CAT activity
|
|
|---|---|---|
| Control | Sendai virus | |
| pSG424 | 1 | 1.3 ± 0.4 |
| IRF-3 | 108 ± 27 | 157 ± 36 |
| IRF-3(407) | 228 ± 23 | 237 ± 26 |
| IRF-3(394) | 213 ± 29 | 243 ± 33 |
| IRF-3(151-427) | 14.2 ± 3.3 | 35 ± 7.1 |
| IRF-3(134-427) | 198 ± 40 | 217 ± 57 |
| IRF-3(99-427) | 219 ± 56 | 227 ± 46 |
| IRF-3(2A) | 69 ± 20 | 148 ± 40 |
| IRF-3(3A) | 190 ± 39 | 206 ± 45 |
| IRF-3(5A) | 193 ± 42 | 206 ± 46 |
| IRF-3(5D) | 229 ± 44 | 264 ± 24 |
| IRF-3(J2A) | 77 ± 14 | 85 ± 24 |
| IRF-3(J2D) | 69 ± 18 | 172 ± 36 |
| IRF-3(NES) | 4.9 ± 2.5 | 22 ± 8.1 |
| IRF-3(5D-NES) | 160 ± 17 | 139 ± 33 |
293 cells were transfected with 2.5 μg of (Gal4)5-TK-CAT reporter plasmid and 5 μg of the various GAL4–IRF-3 chimeric expression plasmids as indicated. At 24 h posttransfection, cells were infected with Sendai virus for 16 h or left uninfected. CAT activity was analyzed at 48 h posttransfection with 10 μg of total protein extract for 1 h at 37°C. Relative CAT activity was measured as fold activation (relative to the basal level of reporter gene in the presence of pSG424 vector after normalization with cotransfected β-Gal activity); the values represent the average of three experiments ± standard deviation.
C-terminal IRF-3 phosphorylation stimulates transactivation potential.
Next, the effects of specific mutations in the C-terminal phosphorylation sites with respect to transactivation potential were examined. As shown previously, substitution of the Ser/Thr cluster at aa 396 to 405 with the phosphomimetic Asp generated a strong, constitutive transactivator IRF-3(5D) that stimulated the RANTES, IFN-β, and other promoters more than a 100-fold (17, 18), whereas substitution of S385 and S386 with Asp did not activate transcription compared to wtIRF-3 (Fig. 7). However, using IRF-3(5D) as the starting construct, the S385/386A substitution [IRF-3(5D-J2A)] decreased IRF-3(5D) transactivation of the RANTES promoter from 160- to 25-fold (Fig. 7). Strikingly, the S385/386D substitution to generate IRF-3(7D) suppressed IRF-3(5D) transactivation activity to a similar level (Fig. 7). As in the case of chimeric GAL4–IRF-3, the L145/146A substitution [IRF-3(5D-NES)] also reduced the transactivation activity of IRF-3(5D). Furthermore, IRF-3(5DΔ134-197) or IRF-3(5DΔ241-327), in which either the NES and proline-rich domain or part of the IAD was deleted, also lost transactivation activity (Fig. 7). Virus infection strongly activated reporter gene expression in cells cotransfected with wtIRF-3 or vector alone but only slightly enhanced the IRF-3(5D)-mediated activation (Fig. 7).
FIG. 7.
Analysis of IRF-3 point mutations for transactivation potential. 293 cells were transfected with RANTES CAT reporter plasmid and various IRF-3 plasmids expressing C-terminal point mutations as indicated in on the left. At 24 h posttransfection, cells were infected with Sendai virus for 16 h (dark bars) or left uninfected (light bars). The point mutations are indicated by open circles (mutated to Ala) and closed circles (mutated to Asp): 2A, S396A/S398A; 3A, S402A/T404A/S405A; 5A, S396A/S398A/S402A/T404A/S405A; 5D, S396D/S398D/S402D/T404D/S405D; J2A, S385A/S386A; J2D, S385D/S386D; and NES, L145A/L146A. CAT activity was analyzed at 48 h posttransfection with 5 μg of total protein extract for 1 to 2 h at 37°C. Relative CAT activity was measured as fold activation (relative to the basal level of reporter gene in the presence of CMVBL vector after normalization to cotransfected β-Gal activity); the values represent the average of three experiments with variability shown in the error bars.
The transactivation potential of IRF-3 point mutations was also analyzed in assays using the IRF-3-responsive IFN-β, ISG15, PKR, and B4-TK promoters. As shown previously (18), IRF-3 alone weakly activated the IFN-β promoter, but conversion of the C-terminal Ser/Thr residues at aa 396 to 405 to Asp generated a constitutively active IRF-3: with the IFN-β promoter, 80- to 100-fold induction was observed, whereas IRF-3(J2D) did not stimulate IFN-β gene expression. Again remarkably, IRF-3(5D-J2A) produced only 10- to 15-fold induction, while IRF-3(7D) activated the IFN-β promoter about 5-fold. With the PKR, ISG15, and B4-TK promoters, quantitatively different results were obtained, although the different IRF-3 point mutations generated qualitatively similar results with the various IRF-3-inducible promoters (data not shown).
C-terminal IRF-3 phosphorylation stimulates DNA binding.
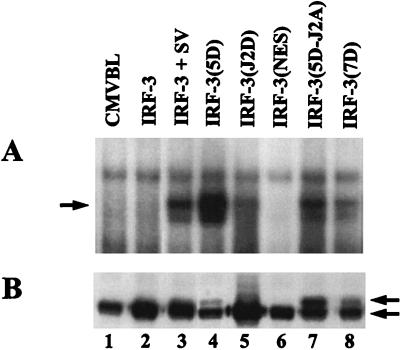
IRF-3 mutant proteins were also analyzed by EMSA to correlate transactivation with DNA binding activity. As expected, IRF-3(5D) associated with DNA, whereas wtIRF-3 did not (Fig. 8A, lanes 2 and 4). Interestingly, the NES mutant did not bind DNA (Fig. 8A, lane 6); IRF-3(J2D), with the S385/386D mutation had poor DNA binding activity despite being expressed at a high level (Fig. 8A and B, lanes 5). Substitution of S385 and S386 with either Ala or Asp to create IRF-3(5D-J2A) and IRF-3(7D) respectively, reduced IRF-3(5D) DNA binding activity more than 10-fold (Fig. 8A; compare lanes 7 and 8 with lane 4), although both proteins were expressed at higher levels than IRF-3(5D) (Fig. 8B; compare lanes 7 and 8 with lane 4). Despite a lower steady-state level of IRF-3(5D) due to constitutive turnover (18), IRF-3(5D) nonetheless possessed strong DNA binding activity compared to other IRF-3 forms (Fig. 8A; compare lane 4 with lanes 7 and 8). These experiments illuminate an interesting relationship between the Ser/Thr phosphoacceptor cluster (aa 396 to 405) and the adjacent S385 and S386, which are not apparent phosphorylation targets but rather play a regulatory role in the phosphorylation events at the C-terminal end of IRF-3.
FIG. 8.
Effects of IRF-3 point mutations on DNA binding activity. (A) EMSA was performed on whole-cell extracts (20 μg) derived from 293 cells transfected with various IRF-3 expression plasmids as indicated. At 24 h posttransfection, cells were infected with Sendai virus for 12 h or left uninfected. The 32P-labeled probe corresponds to the ISRE of the ISG15 gene: 5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′. (B) Whole-cell extracts (20 μg) from panel A were analyzed by immunoblotting with anti-IRF-3 antibody. IRF-3(5D), IRF-3(5D-J2A), and IRF-3(7D) proteins migrated more slowly than wtIRF-3, at a position consistent with phosphorylated IRF-3; arrows indicate positions of the different forms of IRF-3.
IRF-3 association with CBP/p300.
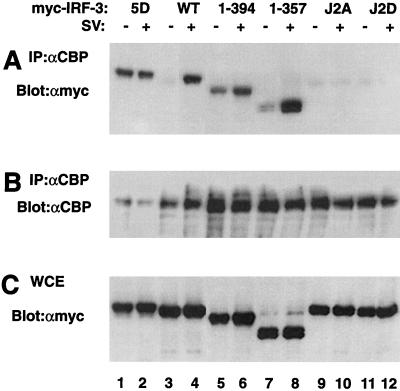
Finally, the effects of specific IRF-3 mutations on IRF-3–CBP interactions were investigated by coimmunoprecipitation using 293 cells transfected with Myc-tagged IRF-3 forms. After immunoprecipitation of CBP from cell extracts with anti-CBP antibody A22 (Fig. 9B), immunoblot analysis with anti-Myc antibody 9E10 revealed that Myc-tagged IRF-3 associated with CBP in virus-infected cells (Fig. 9A, lane 4) but not in unstimulated cells (Fig. 9A, lane 3) as demonstrated previously (18, 34, 35). As expected, IRF-3(5D) constitutively associated with the CBP coactivator, regardless of virus infection (Fig. 9A, lanes 1 and 2). Substitution of S385 and S386 with either Ala or Asp [IRF-3(J2A) or IRF-3(J2D), respectively] also interfered virus mediated IRF-3–CBP interactions (Fig. 9A, lanes 9 to 12). Surprisingly, deletion of the C-terminal domain of IRF-3 in the constructs IRF-3(394) and IRF-3(357) resulted in the association of IRF-3 with CBP/p300 in the absence of virus infection (Fig. 9A, lanes 5 and 7); interaction with CBP/p300 was further stimulated three- to fivefold by Sendai virus infection (Fig. 9A, lanes 6 and 8). These experiments demonstrate that conversion of the IRF-3 Ser/Thr cluster at aa 396 to 405 to Asp results in constitutive interaction with CBP/p300, whereas the mutation of S385 and S386 to either Ala or Asp blocks IRF-3 association with CBP. Removal of the C-terminal autoinhibitory domain of IRF-3 partially overcomes the inhibition to IRF-3–CBP interaction and permits association of IRF-3 with CBP/p300.
FIG. 9.
Effects of specific mutations in the C-terminal inducible phosphorylation sites on IRF-3–CBP complex formation. 293 cells were transfected with Myc-tagged IRF-3 expression plasmids (5 μg) as indicated above the lanes. At 24 h posttransfection, cells were infected with Sendai virus (SV) for 12 h (+) or left uninfected (−). Whole-cell extracts (300 μg) were immunoprecipitated with anti-CBP antibody A22 (αCBP). Immunoprecipitated complexes (A and B) or 20 μg of whole-cell extracts (WCE) (C) were run on an SDS–8% polyacrylamide gel and subsequently probed with anti-Myc antibody (A and C). The membrane in panel A was reprobed with anti-CBP antibody 9E10 (B) to verify expression of the transfected genes.
DISCUSSION
The DNA binding and transactivation properties of transcription factor IRF-3 are induced by virus infection or dsRNA treatment (18, 33–35), although the molecular mechanisms underlying IRF-3 activation remain to be elucidated. Several novel aspects of IRF-3 structure and function are highlighted in the present study: (i) an IRF-3 transactivation domain which contains a NES sequence, a proline-rich domain, and an IAD is located between aa 134 and 394; (ii) two autoinhibitory domains, one located at the carboxy-terminal end of IRF-3 (aa 380 to 427) and a second region between aa 98 and 240, interact to create in uninfected cells a closed conformation of IRF-3 that inhibits IRF-3 DNA binding; (iii) virus infection results in C-terminal phosphorylation of IRF-3, which relieves the conformational constraints imposed by autoinhibitory domain interactions and results in the formation of IRF-3 homodimers; and (iv) substitution of the Ser/Thr residues at aa 396 to 405 with the phosphomimetic amino acid Asp, but not the adjacent S385 and S386, generates a protein with constitutive DNA binding and transactivation activities, as well as the potential for homodimer formation. Together with previous studies, these results indicate that the DNA binding and transcriptional activation properties of IRF-3 are tightly regulated by the combination of intramolecular and intermolecular interactions, virus-mediated phosphorylation, and subcellular localization.
The observation that deletion of the C-terminal 20 aa of IRF-3 to create IRF-3(407) stimulated transcription of IFN-β and ISG15 promoters suggested the possibility that IRF-3 contained an activation domain (18). When the full-length IRF-3 protein was fused in frame to the GAL4 DBD, the chimeric GAL4–IRF-3 protein activated a GAL1-lacZ reporter construct in a yeast one-hybrid analysis. Furthermore, the chimeric GAL4–IRF-3 protein also activated a reporter gene containing five GAL4 binding sites inserted upstream of the minimal TK promoter in mammalian cells. Similarly, Ronco et al. reported that chimeric GAL4–IRF-3 protein activated a reporter gene containing five GAL4 binding sites in unstimulated cells (24), although other groups using similar approaches either failed to identify a transactivation domain or found that virus infection was required to activate the chimeric protein (1, 33, 35). Deletion studies then localized the transactivation domain of IRF-3 to a region between aa 134 and 394, which contains a NES sequence (aa 134 to 150), a proline-rich segment (aa 151 to 197), and an IAD homology domain (199 to 375). In this regard, the activation domain of IRF-3 is similar to that of IRF-4, which also contains a proline-rich segment (aa 151 to 237) and an IAD (aa 237 to 412) (4). In both cases, the activation domain is unusually long, comprising 250 to 300 aa.
Full-length IRF-3, prepared from unstimulated cells, was unable to bind DNA (33, 35), whereas IRF-3(5D), a constitutively activated form of IRF-3, associated with DNA and stimulated transcription. Furthermore, deletion the C-terminal 20 aa or the sequence between aa 134 and 197 also permitted protein-DNA interaction and a low level of transcriptional activation (5- 10-fold, versus the 100- 200-fold stimulation observed following virus infection). Coimmunoprecipitation studies demonstrated that the C-terminal region of IRF-3 between aa 328 and 427 was able to interact with N-terminal region between aa 98 and 240, thus supporting the idea that an intramolecular association between the two autoinhibitory domains controls cytoplasmic latency of IRF-3 in uninfected cells.
IRF-3 is also regulated by subcellular localization in response to virus infection (18, 33–35); however, deletion of the C-terminal 20 aa [IRF-3(407)] did not alter the subcellular localization, as determined by green fluorescent protein fluorescence (data not shown). This result was somewhat surprising since it would be expected that once autoinhibition is relieved, IRF-3 may transit to the nucleus, via an as yet unidentified nuclear localization sequence. Nuclear import may be counterbalanced by active export from the nucleus, mediated by the NES element in IRF-3 (18, 35). Interestingly, although mutation of the NES element results in the nuclear accumulation of IRF-3, the nuclear localization of IRF-3 NES mutant is not sufficient to stimulate transcription, indicating that an event in addition to nuclear localization, presumably phosphorylation and/or dimerization, is required for full IRF-3 transcriptional activity.
In previous studies, the inducible phosphorylation sites of IRF-3 were localized to the region ISNSHPLSLTSDQ, between aa 395 and 407 (18), whereas another study identified S385 and S386 as the critical residues for triggering virus-induced phosphorylation (35). To resolve this apparent discrepancy, we demonstrated that phosphorylation of Ser/Thr cluster between aa 395 and 407 but not the S385 and S386 residues plays an important role in IRF-3 DNA binding and transactivation activities, based on two observations: (i) substitution of the Ser/Thr residues at aa 396 to 405 with the phosphomimetic Asp generated a protein with constitutive DNA binding and transactivation activities, while the S385/386D mutant neither bound to DNA nor activated transcription; and (ii) on SDS-PAGE, IRF-3(5D) behaved as a phosphoprotein, migrating more slowly than unphosphorylated wild-type protein, whereas IRF-3 S385/386D comigrated with endogenous IRF-3. However, the S385 and S386 residues do play a critical regulatory role with regard to virus-induced phosphorylation of the adjacent Ser/Thr cluster located at aa 396 to 405, since substitution of these two serines with alanine blocked virus-induced phosphorylation and IRF-3–CBP interaction (Fig. 9 and reference 35). Interestingly, substitution of these two serines with aspartic acid also blocked virus-induced IRF-3–CBP complex formation. These two serine residues are also important for transactivation activity, since substitution with either Ala or Asp dramatically reduced the IRF-3(5D) transactivation activity. One possibility consistent with our observations is that S385 and S386 represent a part of the docking site for the interaction with CBP/p300 coactivator and/or may be involved in the interaction with the kinase(s) that ultimately phosphorylates IRF-3 at the downstream Ser/Thr sites.
S385 and S386 lie within the consensus sites for phosphorylation of casein kinase 2 (S/TxxE/D/Yp/Sp) and Golgi apparatus casein kinase (SxE/Sp), respectively (22). Substitution of the E388 residue with A388 eliminated the consensus phosphorylation sites for these two constitutive protein kinases; however, the E388A mutation had essentially no effect on the activity of IRF-3(5D) (data not shown), indicating that these two kinases played no role in S385/S386 phosphorylation or in IRF-3 activity.
Following its C-terminal Ser/Thr phosphorylation in virus-infected cells, IRF-3 formed homodimers in vivo as detected by coimmunoprecipitation. Although the precise regions of IRF-3 involved in dimerization have yet to be delineated, preliminary studies indicate that a truncated protein of 1 to 240 aa can still form dimers, suggesting that only a portion of the IAD may be required for formation of IRF-3 homodimers in virus-infected cells. As shown previously for other IRF family members, the IAD region in ICSBP is responsible for its interaction with IRF-1 and IRF-2 (2, 29), and the equivalent region in ISGF3γ also mediates ISGF3α-γ interaction (30). In the case of IRF-4, the IAD region of IRF-4 mediates the interaction with the Ets factor PU.1 to generate a unique B-cell-specific heterodimer (4, 7) IRF-3 was also shown previously to associate with IRF-7, and this heteromeric association may cooperatively activate the transcription from the IFN-β promoter (33).
Recent studies indicate that type I IFN genes may be subdivided into two groups: immediate-early genes such as IFN-α4 and IFN-β, activated in response to virus by a protein synthesis-independent pathway, and delayed-type genes, including other IFN-α subtypes whose expression is protein synthesis dependent. Posttranslational activation of IRF-3, NF-κB, and other factors results in IFN-α4 and IFN-β gene expression. Once produced and secreted from the infected cell, the immediate-early IFNs act through the IFN receptor to stimulate the Jak-STAT pathway, leading to the IFN-responsive activation of the delayed-type IFN genes. This secondary response is mediated through the ISGF3-dependent induction of another IRF family member, IRF-7. Like IRF-3 activation, IRF-7 activation requires virus-mediated C-terminal phosphorylation for induction of the delayed-type IFN genes (19). These studies raise the interesting possibility that differential type I IFN gene regulation may be controlled in part by distinct IRF-3 and IRF-7 homo- and heterodimer combinations with different affinities for ISRE- and IRF-like sites in responsive promoters.
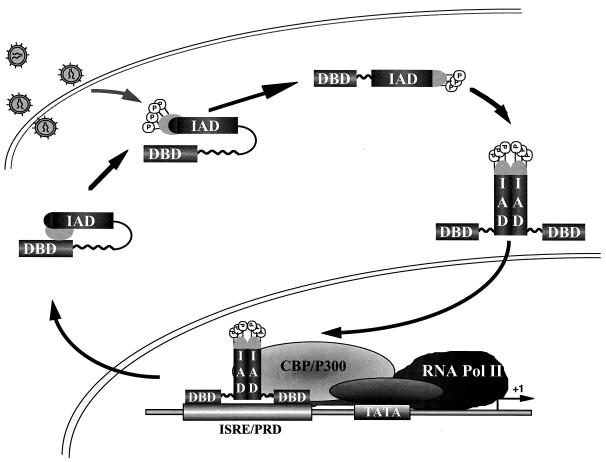
We propose a model in which virus-inducible C-terminal phosphorylation alters protein conformation and permits IRF-3 dimerization, nuclear translocation, and primary activation of IFN- and IFN-responsive genes (Fig. 10). In uninfected cells, ubiquitously expressed IRF-3 exists in the cytoplasm; intramolecular association between the two autoinhibitory regions maintains IRF-3 in a latent state by masking both DBD and IAD regions of the protein, as observed with IRF-4 (4, 7). Virus-induced phosphorylation at the Ser/Thr residues at the aa 396 to 405 cluster leads to a conformational change in IRF-3 that relieves C-terminal autoinhibition and exposes both DBD and IAD regions. Phosphorylated IRF-3 then forms homodimers, mediated by a portion of the IAD region. IRF-3 dimers translocate to the nucleus via an unidentified nuclear localization sequence and bind to DNA at ISRE- and PRDI/PRDIII-containing promoters. Phosphorylation is also necessary for IRF-3 association with the chromatin remodeling activity of CBP/p300. The presence of a NES element ultimately shuttles IRF-3 from the nucleus and terminates the initial activation of virus-responsive promoters. The phosphorylated form of IRF-3 exported from the nucleus may now be susceptible to proteasome-mediated degradation. This scenario shares several features with the protein synthesis-independent activation of NF-κB, complements the finding that IRF-3 and CBP/p300 are components of the dsRNA-inducible complex DRAF (5, 34), and indicates that IRF-3 may be a previously identified virus-inducible complex (3, 9). In conclusion, these studies provide new structural and functional insights regarding a member of the IRF transcription factor family that represents a direct inducer of type I IFNs, the chemokine RANTES, and potentially other cytokines required for the orchestration of an effective immune response against virus infection.
FIG. 10.
Schematic representation of IRF-3 activation and dimerization by virus-induced phosphorylation. IRF-3 exists in the cytoplasm of uninfected cells. Intramolecular association between the C terminus and the DBD maintains IRF-3 in a latent state by masking both DBD and IAD regions of the protein. Virus-induced phosphorylation (P) of the Ser/Thr residues in the aa 396 to 405 cluster leads to a conformational change in IRF-3 that relieves C-terminal autoinhibition and exposes both DBD and IAD regions. The opened conformation of IRF-3 is now able to homodimerize; translocation to the nucleus leads to DNA binding at ISRE- and PRDI/PRDIII-containing promoters. The presence of a NES element ultimately shuttles IRF-3 from the nucleus and terminates the initial activation of virus-responsive promoters. Pol, polymerase.
ACKNOWLEDGMENTS
We thank Paula Pitha, Dimitrios Thanos, Nancy Reich, and Illka Julkunen for reagents used in this study and members of the Molecular Oncology Group, Lady Davis Institute, for helpful discussions.
This research was supported by grants from the Cancer Research Society and Medical Research Council of Canada. R.L. was supported in part by a Fraser Monat McPherson fellowship from McGill University, Y.M. was supported by an FRSQ studentship, and J.H. was supported by an MRC Senior Scientist award.
REFERENCES
- 1.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bovolenta C, Driggers P H, Marks M S, Medin J A, Politis A D, Vogel S N, Levy D E, Sakaguchi K, Appella E, Coligan J E, Ozato K. Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc Natl Acad Sci USA. 1994;91:5046–5050. doi: 10.1073/pnas.91.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bragança J, Génin P, Bandu M-T, Darracq N, Vignal M, Cassé C, Doly J, Civas A. Synergism between multiple virus-induced-factor-binding elements involved in the differential expression of IFN-A genes. J Biol Chem. 1997;272:22154–22162. doi: 10.1074/jbc.272.35.22154. [DOI] [PubMed] [Google Scholar]
- 4.Brass A, Kehrli E, Eisenbeis C, Storb U, Singh H. Pip, a lymphoid restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 5.Daly C, Reich N C. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol Cell Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 7.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 8.Evan G I, Bishop J M. Isolation of monoclonal antibodies specific for the human c-myc proto-oncogene product. Mol Cell Biol. 1985;4:2843–2850. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Génin P, Bragança J, Darracq N, Doly J, Civas A. A novel PRDI and TG binding activity involved in virus-induced transcription of IFN-A genes. Nucleic Acids Res. 1995;23:5055–5063. doi: 10.1093/nar/23.24.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant C E, Vasa M Z, Deeley R G. cIRF-3, a new member of the interferon regulatory factor (IRF) family that is rapidly and transiently induced by dsRNA. Nucleic Acids Res. 1995;23:2137–2146. doi: 10.1093/nar/23.12.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiscott J, Nguyen H, Lin R. Molecular mechanisms of interferon beta gene induction. Semin Virol. 1995;6:161–173. [Google Scholar]
- 12.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. A role for IRF-3 transcription factor in triggering the activation of interferon gene expression. J. Interferon Cytokine Res., in press. [DOI] [PubMed]
- 13.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 14.Juang Y T, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factory-3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy D E. Interferon induction of gene expression through the Jak-Stat pathway. Semin Virol. 1995;6:181–190. [Google Scholar]
- 16.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin R, Heylbroeck C, Genin P, Pitha P, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marie I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen H, Hiscott J, Pitha P M. The growing family of IRF transcription factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 22.Pinna L A, Meggio F. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog Cell Cycle Res. 1997;3:77–97. doi: 10.1007/978-1-4615-5371-7_7. [DOI] [PubMed] [Google Scholar]
- 23.Pitha P M, Au W-C. Induction of interferon alpha gene expression. Semin Virol. 1995;6:151–159. [Google Scholar]
- 24.Ronco L, Karpova A, Vidal M, Howley P. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P. Nucleotide sequence of the kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of IRF family transcription factor IRF-3 in virus-induced activation of IFN-β gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 27.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type 1 interferon gene expression by interferon regulatory factor 3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 28.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 29.Sharf R, Meraro D, Azriel A, Thornton A M, Ozato K, Petricoin E F, Larner A C, Schaper F, Hauser H, Levi B-Z. Phosphorylation events modulate the ability of interferon consensus sequence binding protein to interact with interferon regulatory factors and to bind DNA. J Biol Chem. 1997;272:9785–9792. doi: 10.1074/jbc.272.15.9785. [DOI] [PubMed] [Google Scholar]
- 30.Veals S A, Santa Maria T, Levy D E. Two domains of ISGF3γ that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol Cell Biol. 1993;13:196–206. doi: 10.1128/mcb.13.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veals S A, Schindler C, Leonard D, Fu X-Y, Aebersold R, Darnell J E, Jr, Levy D E. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 33.Wathelet M G, Lin C H, Parakh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 34.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoneyama M, Suhara W, Fukuhara Y, Fukada M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]