Significance
Studies that experimentally examine evolution have provided critical insight in biology. Indeed, rigorous and replicated laboratory experiments have revealed parallel evolution and have proved essential to understanding evolutionary processes. Nonetheless, a typical limitation of such work is the lack of ecological realism. We report on a decade-long field experiment with a native plant (common evening primrose) that advances our understanding of how populations adapt to environmental change. In particular, we address whether the common strategy of seed dormancy significantly alters the evolutionary trajectory of populations. We find that population genetic diversity is restored following a period of sustained dormancy. Nonetheless, the impact of past natural selection persisted under some conditions.
Keywords: chemical ecology, common evening primrose Oenothera biennis, defense theory, experimental evolution, multigenerational experiment
Abstract
Dormancy has repeatedly evolved in plants, animals, and microbes and is hypothesized to facilitate persistence in the face of environmental change. Yet previous experiments have not tracked demography and trait evolution spanning a full successional cycle to ask whether early bouts of natural selection are later reinforced or erased during periods of population dormancy. In addition, it is unclear how well short-term measures of fitness predict long-term genotypic success for species with dormancy. Here, we address these issues using experimental field populations of the plant Oenothera biennis, which evolved over five generations in plots exposed to or protected from insect herbivory. While populations existed above ground, there was rapid evolution of defensive and life-history traits, but populations lost genetic diversity and crashed as succession proceeded. After >5 y of seed dormancy, we triggered germination from the seedbank and genotyped >3,000 colonizers. Resurrected populations showed restored genetic diversity that reduced earlier responses to selection and pushed population phenotypes toward the starting conditions of a decade earlier. Nonetheless, four defense and life-history traits remained differentiated in populations with insect suppression compared with controls. These findings capture key missing elements of evolution during ecological cycles and demonstrate the impact of dormancy on future evolutionary responses to environmental change.
Dormancy is a common life-history feature of many plants, animals, and microbes (1–5). The emergence of dormant propagules can influence population responses to environmental change in multiple ways, from rescuing apparently extirpated populations (6) to slowing the rate of evolution by reintroducing genotypes that were abundant prior to bouts of natural selection (1). Comparative phenotyping of current and resurrected dormant propagules can also reveal a signature of acute natural selection from the past or selection on particular traits during dormancy (7, 8). Complementing these population-level phenomena, dormancy is a critical but understudied life-history strategy in successional species, often putting individuals on pause for prolonged periods.
Although our understanding of dormancy has improved dramatically in recent decades (1, 2, 6, 9), the long-term consequences of evolution by natural selection on successional species with dormant periods remains largely unexplored (2, 10). Among genotypes within a population, fitness is the ultimate currency, but estimating long-term success is challenging, especially in species with such complex life-histories (4, 11–13). At the population level, does the evolutionary trajectory prior to dormancy persist and resume following resurrection from prolonged dormancy? Or, do changing selective factors and stochasticity during dormancy change the genetic structure that previously existed in populations?
A key impediment to answering these questions has been the difficulty in tracking genotype frequencies over time and through dormancy in field populations (14, 15). Retrospective studies (16, 17), landscape genetic approaches (18), and reciprocal transplant experiments have substantially improved our understanding of adaptation and its genetic basis in plants (19, 20), but no previous study to our knowledge has tracked adaptation in real time before and after a period of dormancy. In an experimental evolution study initiated in 2007, we demonstrated changes in genotype frequencies of an early successional plant (common evening primrose, Oenothera biennis Onagraceae) in response to insect herbivory in replicated field populations (Fig. 1; for study background, see Materials and Methods). Because this species has a genetic system that suppresses recombination and segregation of alleles (making most offspring clones of parents), we were able to track changes in genotype frequencies over several generations. We found that herbivory resulted in the evolution of later flowering and more defended genotypes that were less tolerant of interspecific competition (15). Although natural selection on these traits showed temporal variation over the first 5 y of the study (see table S1 in ref. 15), there was a strong response to selection. Due to the parallel changes that occurred in replicate populations and the observed relationship between fecundity and recruitment, evolution was attributable to natural selection and not genetic drift.
Fig. 1.
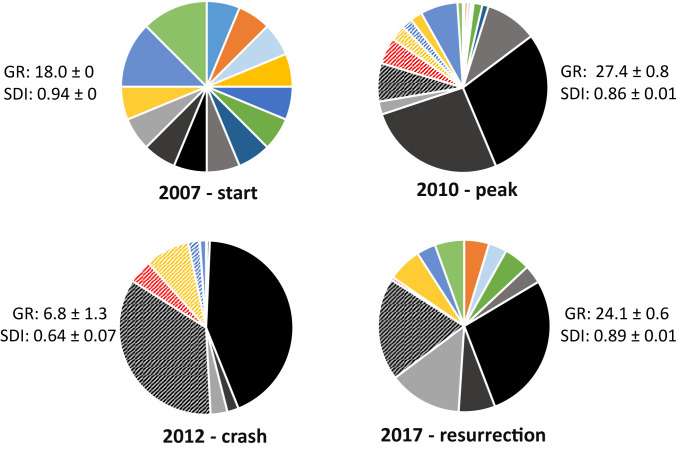
Temporal genotypic structure of common evening primrose (O. biennis) across all experimental evolution field populations (n = 16, treatments did not show differences in genotypic diversity). Pie charts show the global frequencies of genotypes >1% frequency summed across all 16 populations in each of the 4 y; hatched pattern indicates four novel outcrossed genotypes derived within the experiment. Gray–black colors represent the five genotypes with the strongest propensity to reproduce annually. Mean ± SE genotypic richness (GR) and Simpson’s diversity index (SDI) is shown on a per plot basis for all genotypes detected. SDI was calculated as 1 − the sum of proportional abundances for each genotype.
In our O. biennis populations, as in other early successional species, the original array of genotypes is expected to narrow over time, as an increasingly intense competitive environment imposes natural selection against noncompetitive genotypes (15). The shifting genetic composition of the population is dictated both by this selection and the ecological process of succession itself, reducing the overall population size. During this period, as the aboveground population builds and then declines, dormant seeds accumulate in the soil. When at some future point these seeds are resurrected by soil disturbance, and once again establish an aboveground population, the population is presumably composed of those genotypes that persisted as adult plants the longest as well as genotypes only present in early generations that were not strongly favored.
Here, we address how genotype-specific fecundity and predormancy evolutionary changes in response to a treatment (suppression of insect herbivory) influence O. biennis populations when they are resurrected after at least 5 y of dormancy. Our goal was to examine two key unanswered questions: the extent to which genetic diversity is gained or lost during a full successional cycle, and whether prior selection mediated by insects is maintained or erased during dormancy (21).
Results and Discussion
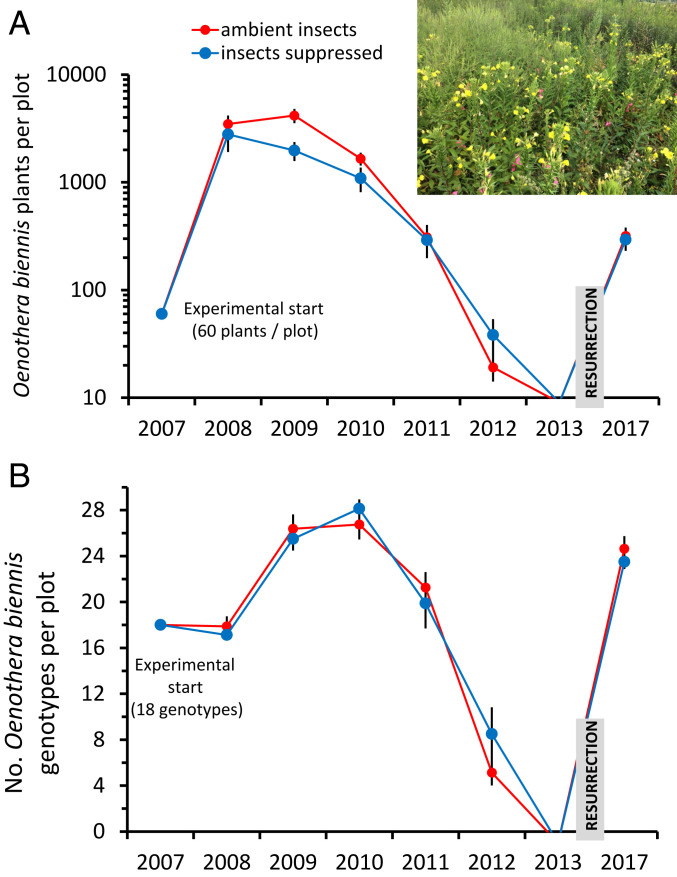
In our 16 experimental populations, plants were initiated with nearly identical abundance and frequency of 18 genotypes representing a diversity of antiherbivore defensive phenotypes and life-histories that ranged from annual to biennial (15) (Fig. 1). Per capita fecundity ranged ∼fourfold among genotypes in the first year of the experiment when plant productivity was high due to the absence of interspecific competition (SI Appendix, Fig. S1). In such early successional communities, competition, herbivory, and genotypic turnover typically increase until perennial vegetation suppresses colonizing species (15, 22–24). In our experiment, after five annual generations and densities upwards of 200 plants/m2, in 2012, experimental populations crashed to ∼2 plants/m2 and then quickly disappeared altogether (Fig. 2) (23). Cumulatively over the first 5 y of the experiment, an average of 225,000 fruits (∼25 million seeds) were deposited in each population. O. biennis seeds can stay dormant for 75 y in the soil, but about 10% of seeds lose viability per decade (25).
Fig. 2.
Population and genotypic dynamics of common evening primrose (O. biennis) in replicate experimental evolution field populations. Populations fell dormant beginning in 2012, and the soil was disturbed in autumn of 2016. (A) Population sizes in the ambient insect (control) versus insect suppression treatments. Note that the y-axis is on a log scale. (Inset) O. biennis flowering in a resurrected plot, August 2017. (B) Genotypic richness over time. Shown are means ± SE.
Effects of Dormancy on Population Size and Genotypic Diversity.
After a minimum of 5 y of dormancy (i.e., of seeds produced between 2007 and 2012 that did not germinate during this time or thereafter), all 16 experimental populations were disturbed by shallow tilling of the soil in autumn 2016, resulting in germination during the following spring. Cumulative fruit input from 2007 to 2012 varied 40-fold per genotype per population (SI Appendix, Fig. S1). Genotype-specific per-capita reproduction in the first generation and cumulative genotypic seed input through 2012 were not correlated (n = 18, r = 0.18, P = 0.465; SI Appendix, Fig. S1), and the latter was much more variable among genotypes due to changes in relative abundance of plants over time (Fig. 2).
Despite differences in plant abundance between treatments prior to dormancy, population sizes were not different in the spring after soil disturbance (F1,14 = 0.072, P = 0.76, mean ± SE, 305 ± 43 O. biennis plants per population, Fig. 2). Thus, plant establishment after resurrection was microsite limited, as total fruit input per population did not predict the total abundance of plants in 2017 (F1,15 = 1.33, P = 0.27). In other words, a limited number of locations for germination and successful establishment constrained population abundance; nonetheless, microsite limitation may have favored differential genotypic success. Using microsatellite DNA markers to genotype >3,000 plants (∼190/population), we found that resurrected populations were missing three of the original genotypes but had several novel genotypes that were produced through outcrossing. Critically, populations had an average of 6.8 (±1.3) genotypes when the aboveground population crashed but had a genotypic richness of 24.1 ± 0.6 after resurrection (and the number of genotypes was not different between ambient and insect suppression treatments, F1,14 = 0.88, P = 0.36, Fig. 1). Thus, resurrection more than tripled genotypic richness (and diversity) compared with 2012 when O. biennis populations crashed and returned diversity to above the starting point of the experiment (Figs. 1 and 2B).
Trait Evolution Before and After Dormancy.
To examine the multigenerational fitness of genotypes with differing traits, we regressed genotype-specific mean values of plant abundance after resurrection in 2017 against four traits previously demonstrated to exhibit rapid evolution in these populations: fruit chemical defense, the likelihood of annual reproduction, an index of flowering phenology, and an index of competitive ability (15, 26). For both fruit chemical defense and annual life-history, strong evidence of past differential selection between treatments remained after resurrection (SI Appendix, Fig. S3 and Table S1). The two other traits that showed initial rapid evolution in response to herbivory (flowering phenology and competitive ability) were not predictive of postdormancy plant abundance (SI Appendix, Fig. S2 and Table S1), indicating a possible relaxation of selection on these traits during dormancy (or genetically correlated traits disfavored during the dormant phase). Overall, the four traits were largely genetically uncorrelated, with the exception of a modest relationship (Pearson r = ∼0.5) between fruit chemical defense and the two life-history traits (annual reproduction and flowering phenology) (SI Appendix, Table S2).
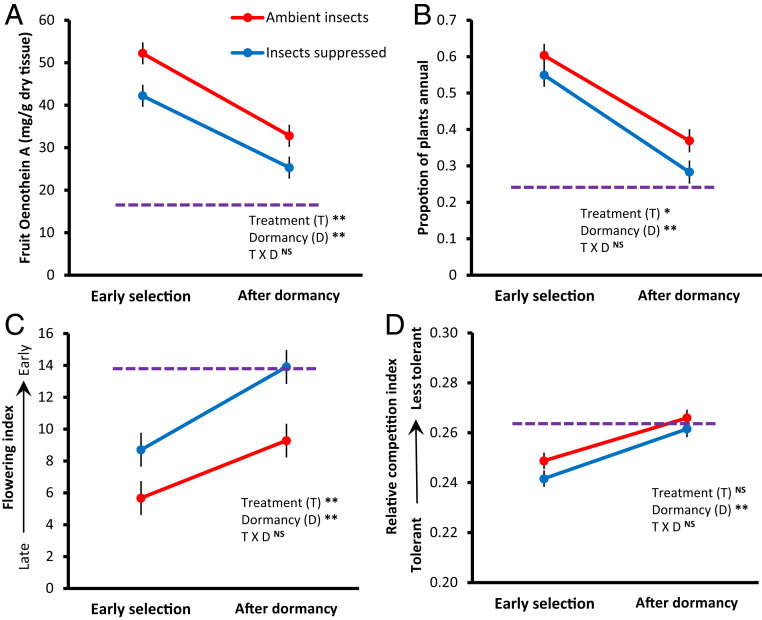
To test for evolutionary shifts between the early generations (predormancy) and the resurrected populations (postdormancy), we compared the estimated population phenotypes for these same four traits. We used genotype-specific trait values measured in common gardens (15) multiplied by genotype frequencies for each replicate population. Population phenotypes were estimated before and after dormancy to estimate the shift through dormancy. Due to changes in genotype frequencies, predicted phenotypes of the four traits shifted an average of 40% from before compared with after dormancy (Fig. 3). In all cases, this shift pushed the resurrected populations toward the starting population phenotypes when genotype frequencies were equal. This regression toward initial population phenotypes was not likely caused by the first generation of highly equal reproduction, as only ∼5% of the cumulative fruits were produced in the first generation, and the genetic structure of populations dramatically differed at the end of the experiment (Fig. 1).
Fig. 3.
Phenotypic evolution of four traits in O. biennis field populations before (2011, early selection) and after (2017) successional dormancy. (A) fruit chemical defense by a toxic ellagitannin, (B) likelihood of annual reproduction, (C) an index of flowering phenology, and (D) an index of plant competitive ability. All traits except annual reproduction were significantly differentiated prior to dormancy. Mean population phenotypes were derived by genotype-specific phenotypic means multiplied by genotype frequencies in each of eight replicate control (ambient insects) and eight insect-suppression populations. All populations were initiated 10 y prior to soil disturbance with nearly equal genotype frequencies. Estimated population phenotypes at the start of the experiment in 2007 are shown by the purple dashed line. Panels show means ± SE; NS = not significant, *P < 0.05, **P < 0.01
Despite the striking phenotypic changes before compared with after dormancy (Fig. 3), a strong signature of past evolution in response to herbivory was maintained (for fruit chemical defense and flowering phenology, but not competitive ability). Additionally, annual life-history, which was not statistically different between treatments previously (15), showed a significant difference between treatments after dormancy based on estimated population phenotypes. Furthermore, field sampling of annual reproduction in 2017 (after resurrection) matched this estimate for the evolution of life-history (the proportion of annual versus biennial plants observed in 2017 was highly skewed by past insect suppression treatment, mean ± SE proportion annuals: ambient insect populations = 0.52 ± 0.04, insect suppression populations = 0.32 ± 0.04, F1,14 = 9.787, P = 0.007). Thus, although evolutionary divergence due to insect herbivory was not erased by plant dormancy, the increased abundance of some genotypes that had been previously selected against, as well as the addition of dominant outcrossed genotypes (Fig. 1), changed the mean phenotype of resurrected populations. This subtly weakened the selective effects of herbivores on some traits (phenology and competitive ability) and maintained or enhanced it for others (defense chemistry and annual life-history).
Predicting Long-Term Fitness.
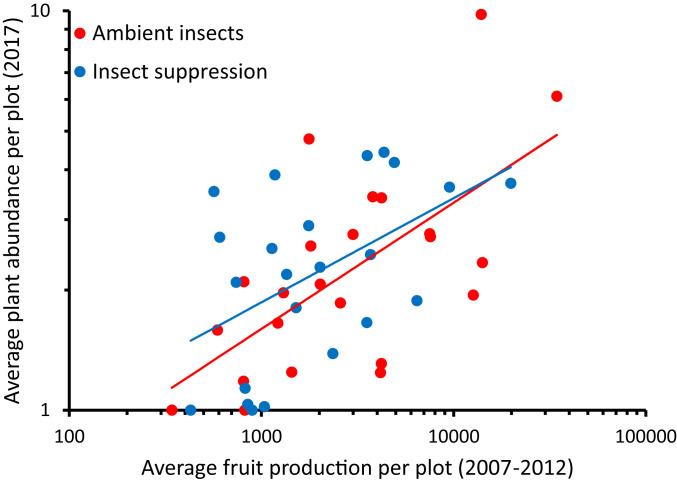
In early successional species, the long-term fitness of a given genotype may be shaped by initial per-capita fecundity that occurs after populations establish and face relatively little competition, by cumulative fecundity over several generations when genotypes may have variable abundance, or by their proportional recruitment out of the seedbank when disturbance allows re-establishment of populations. To understand the drivers of long-term fitness, we assessed the ability of genotype-specific per-capita fecundity (estimated in the first year of the experiment and cumulatively in the years prior to populations crashing; SI Appendix, Fig. S1) to predict postdormancy genotypic abundance. Across the eight replicate populations of each treatment, only past cumulative fecundity predicted the genotype-specific abundance of plants after resurrection (Fig. 4); this effect was consistent in both ambient and insect-suppression populations.
Fig. 4.
Predicting long-term fitness of common evening primrose (O. biennis) genotypes in field populations. Plant abundance in 2017 (after soil disturbance) is the average of eight control (ambient insects) and eight insect-suppression plots. Shown are the original genotypes and five novel outcrossed genotypes originating in the experiment (note the log scales, statistical model includes genotype as a random effect; average cumulative fruit production: r2 = 0.88, F1,36.44 = 10.853, P < 0.01). The relationship between cumulative fecundity and plant abundance was not different between treatments (F1,24.36 = 0.539NS, interaction F1,21.77 = 0.346NS).
Although it is known that recruitment out of the seedbank can be genetically variable (12), the impact of variation in dormancy on recruitment dynamics has largely been studied in interspecific comparisons (27, 28). We thus also tested the effect of two indices of recruitment for predicting plant abundance in 2017: seed dormancy measured from a previous 1-y field experiment (26) and the proportion of previously produced seeds that germinated and established in the current experiment (SI Appendix, Fig. S3). Variation in proportional recruitment among genotypes could occur because of differences in mortality in the seedbank or variation in dormancy among genotypes, which we cannot distinguish. Proportional recruitment was not genetically correlated with our previous measure of dormancy (n = 18, r = −0.196, P = 0.435) (26). Neither dormancy nor proportional recruitment of genotypes was predictive of plant abundance in 2017; thus, genotypic differences in recruitment were not enough to overcome effects of cumulative seed production. Thus, the long-term fitness of genotypes was primarily influenced by the combined effects of their per-capita fecundity and their abundances across years.
Speculation and Conclusion.
Ultimately, the persistence of genotypes in populations with dormancy is likely to be driven by natural selection (resulting from a combination of per-capita fecundity and abundance) during the early successional phase or by being well-adapted to conditions in the seedbank. In our experiment with O. biennis, some genotypes were certainly highly favored by selection (e.g., annual and outcrossed genotypes, Fig. 1, refs. 26, 29) and others made a strong comeback after dormancy despite their absence during most of succession (e.g., colorfully coded genotypes in 2017 pie of Fig. 1.). Given the large aboveground populations, millions of seeds in the seedbank, and the parallel changes among replicate populations, genetic drift was unlikely to play a strong role in affecting our results. Thus, even though there may have been relaxed selection for some genotypic traits like competitive ability during dormancy, competitive genotypes likely had other unfavorable traits in terms of persistence during dormancy (as genetic drift would have likely amplified the presence of previously favored competitive genotypes). Our earlier results indicating variable natural selection on defense and life-history traits among years during the early successional phase (table S1 in ref. 15), along with results from this study, indicate that selection both before and during dormancy contributes to the genetic structure of populations after resurrection.
Recruitment from a dormant propagule bank is a common and critical aspect of the biology of many organisms, especially in desert annuals and early successional species (1, 2, 6). Genotypes in such environments are typically subject to strong natural selection due to changing biotic and abiotic conditions. In our study, long-term genotypic fitness was largely determined by cumulative fecundity across generations, which is the product of per-capita fecundity and annual relative flowering abundance. Per-capita reproduction or the proportional recruitment of seeds out of the seedbank by themselves did not predict our results. Thus, snapshot measures of fecundity or germination may be poor reflections of long-term fitness (9, 12). At the same time, we demonstrated that although selection and succession eroded genotypic diversity, dormancy refreshed diversity as predicted by theory (21) and resurrection returned populations toward mean phenotypes at the colonization stage. A longer period of dormancy may have revealed selection on particular genotypes, further introduced changes in population genetic structure, or altered the connection between past and future evolution. Nonetheless, we have shown that natural selection during cycles of ecological succession leaves an evolutionary legacy, and that the cycle itself, and dormancy in particular, renews a population’s evolutionary potential after resurrection.
Materials and Methods
O. biennis flowers open for a single day, and seeds produced through self-pollination (which typically occurs in O. biennis before flowers open) are genetic clones of their parent (30). Cross-pollination (i.e., outcrossing) occasionally occurs in O. biennis, which results in progeny with an entirely linked set of paternal haploid chromosomes associated with a new maternal haploid set of chromosomes (again, with no recombination), creating seeds that represent a single novel outcrossed genotype (29). In 2007, we established the experimental populations of O. biennis in 16 replicate 13.5-m2 plots (spaced a minimum of 10 m apart) that were plowed and sprayed twice with the herbicide glyphosate (Roundup, Monsanto) prior to planting. Although pollinators could easily move between replicate plots, seed dispersal was local, as O. biennis seeds simply drop from mature fruits. Plots were protected from deer herbivory by 2-m-tall mesh fencing.
There was no evidence of dormant seeds at our site prior to the experiment, as no entirely novel genotypes were detected by sequencing. Plots were each initiated with 60 seedlings composed of 18 genotypes (details in ref. 15). Then, 8 of the 16 experimental populations were randomly assigned to an insect suppression treatment and were sprayed biweekly every year (2007 to 2015) during the growing season (April through October) with the insecticide esfenvalerate (Asana XL, Dupont). The ambient insect populations were sprayed on the same schedule with water as a control for the water in the insecticide spray. Populations were not weeded or otherwise manipulated after experimental populations were established in 2007.
Key results of this experiment through 2012, when populations crashed, have been previously reported (15, 26, 29). In October 2016, each of the plots was tilled to a depth of ∼20 cm using a Mashio disk harrow. Germination, establishment, and flowering of O. biennis was assessed May to September 2017, and leaf tissue from all established plants was taken for genotyping in July and August. A total of 3,027 plants were genotyped (mean of 189 per plot, range of 95 to 268). We followed previous methods for genotyping using four microsatellite DNA markers that diagnostically identified the 18 genotypes (details in refs. 15, 26). We identified the parents of novel outcrossed genotypes by determining all possible microsatellite haplotypes that comprised the Renner complexes of the original asexual genotypes. This analysis was facilitated by the high level of heterozygosity associated with our genotypes of O. biennis (29).
Genotype frequencies were calculated as the proportion of total plants sequenced in each plot in each year. As an estimate of per-capita fitness in the first generation, we counted all fruits for the original 60 plants in each population. However, beginning in 2009, populations were too large to sample every plant, so we subsampled populations by counting fruits on at least 50 randomly chosen individuals from each plot (or all bolters if less than 50 were present), identifying their genotypes as well. We then multiplied genotype frequency by the total number of individuals estimated within each population to estimate the number of plants of each genotype in each plot. We estimated the total per-plot fruit production for each genotype in each year by multiplying genotype-specific estimates of fruit production by our estimate of the number of reproductive individuals of that genotype in each population. Although seed counts were impractical in this experiment, fruit number is a good proxy for total fecundity (26).
Mean population phenotypes were calculated by multiplying genotype-specific means (for defense chemistry, likelihood of annual reproduction, flowering phenology, and competitive ability, each reported from common garden studies in ref. 15) by frequencies for each genotype within each plot; these values were then summed. Thus, for each plot (n = 16, half treated with insect suppression), a single value was calculated and used in ANOVAs. In these models, we include insect suppression treatment, dormancy, and their interaction as fixed effects; the dormancy factor compared 2011 population phenotype values with those from 2017. We included “population” as a random effect to control for the repeated measure of the population phenotype before and after dormancy.
In 2017, annual reproduction was directly recorded in the field, and we confirmed that differences between treatments represented an evolutionary response by multiplying genotype-specific means by genotype frequencies. To evaluate the drivers of trait evolution, we conducted genetic correlations (based on genotype means and Pearson correlations) between the four phenotypic traits. We also calculated total genotypic selection (plant abundance in 2017 regressed against genotype-determined trait values) for the four traits using analysis of covariance. In particular, an interaction between traits and experimental insect suppression indicates divergent outcomes of selection caused by insect herbivory.
All analyses were based on linear models and were conducted in JMP Pro-14. As indicated in figure legends and SI Appendix, plant genotype was included, where appropriate, in models as a random effect. Data were checked for normality of residuals, and data transformations are reported in the text.
Supplementary Material
Acknowledgments
We are grateful to M. T. J. Johnson and J.-P. Salminen, who collaborated on the initial phase of this project; Gunnar Glover for help with field work; and T. C. Coverdale, M. A. Geber, N. G. Hairston, A. R. Johnson, X. López-Goldar, J. S. Thaler, N. Whiteman, and S. X. Zhang for comments on the manuscript. Three anonymous reviewers and the editor also suggested substantial improvements. Molecular work for the project was conducted in the Evolutionary Genetics Core Facility within Ecology and Evolutionary Biology at Cornell University. This study was supported by US NSF Grants DEB-0950231 and IOS-1907491.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026212118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Hairston N. G. Jr, De Stasio B. T., Rate of evolution slowed by a dormant propagule pool. Nature 336, 239–242 (1988). [Google Scholar]
- 2.Orsini L., et al., The evolutionary time machine: Using dormant propagules to forecast how populations can adapt to changing environments. Trends Ecol. Evol. 28, 274–282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rengefors K., Karlsson I., Hansson L. A., Algal cyst dormancy: A temporal escape from herbivory. Proc. R. Soc. Lond. Ser. B Biol. Sci. 265, 1353–1358 (1998). [Google Scholar]
- 4.Shoemaker W. R., Lennon J. T., Evolution with a seed bank: The population genetic consequences of microbial dormancy. Evol. Appl. 11, 60–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pake C. E., Venable D. L., Seed banks in desert annuals: Implications for persistence and coexistence in variable environments. Ecology 77, 1427–1435 (1996). [Google Scholar]
- 6.Evans M. E., Dennehy J. J., Germ banking: Bet-hedging and variable release from egg and seed dormancy. Q. Rev. Biol. 80, 431–451 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Hairston N. G., et al., Rapid evolution revealed by dormant eggs. Nature 401, 446 (1999). [Google Scholar]
- 8.Hendry A. P., Schoen D. J., Wolak M. E., Reid J. M., The contemporary evolution of fitness. Annu. Rev. Ecol. Evol. Syst. 49, 457–476 (2018). [Google Scholar]
- 9.Metcalf C. J., Pavard S., Why evolutionary biologists should be demographers. Trends Ecol. Evol. 22, 205–212 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Yamamichi M., Hairston N. G., Rees M., Ellner S. P., Rapid evolution with generation overlap: The double-edged effect of dormancy. Theor. Ecol. 12, 179–195 (2019). [Google Scholar]
- 11.Ehrlén J., Fitness components versus total demographic effects: Evaluating herbivore impacts on a perennial herb. Am. Nat. 162, 796–810 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Taylor M. A., et al., Interacting effects of genetic variation for seed dormancy and flowering time on phenology, life history, and fitness of experimental Arabidopsis thaliana populations over multiple generations in the field. New Phytol. 216, 291–302 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Fournier-Level A., et al., Predicting the evolutionary dynamics of seasonal adaptation to novel climates in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 113, E2812–E2821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Primack R. B., Kang H., Measuring fitness and natural selection in wild plant populations. Annu. Rev. Ecol. Syst. 20, 367–396 (1989). [Google Scholar]
- 15.Agrawal A. A., Hastings A. P., Johnson M. T. J., Maron J. L., Salminen J. P., Insect herbivores drive real-time ecological and evolutionary change in plant populations. Science 338, 113–116 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Franks S. J., Sim S., Weis A. E., Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. U.S.A. 104, 1278–1282 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geerts A., et al., Rapid evolution of thermal tolerance in the water flea Daphnia. Nat. Clim. Chang. 5, 665–668 (2015). [Google Scholar]
- 18.Wang J., et al., A major locus controls local adaptation and adaptive life history variation in a perennial plant. Genome Biol. 19, 72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price N., et al., Combining population genomics and fitness QTLs to identify the genetics of local adaptation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 115, 5028–5033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ågren J., Oakley C. G., McKay J. K., Lovell J. T., Schemske D. W., Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 110, 21077–21082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellner S., Hairston N. G. Jr, Role of overlapping generations in maintaining genetic variation in a fluctuating environment. Am. Nat. 143, 403–417 (1994). [Google Scholar]
- 22.Kalske A., Kessler A., Population-wide shifts in herbivore resistance strategies over succession. Ecology 101, e03157 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Agrawal A. A., Hastings A. P., Fines D. M., Bogdanowicz S., Huber M., Insect herbivory and plant adaptation in an early successional community. Evolution 72, 1020–1033 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Carson W. P., Root R. B., Top-down effects of insect herbivores during early succession: Influence on biomass and plant dominance. Oecologia 121, 260–272 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Darlington H. T., The 70-year period for beal seed viability experiment. Am. J. Bot. 38, 379–381 (1951). [Google Scholar]
- 26.Agrawal A. A., Johnson M. T. J., Hastings A. P., Maron J. L., Experimental evolution of plant life-history traits and its eco-evolutionary feedback to seed predator populations. Am. Nat. 181, S35–S45 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Ooi M. K., Auld T. D., Denham A. J., Climate change and bet‐hedging: Interactions between increased soil temperatures and seed bank persistence. Glob. Chang. Biol. 15, 2375–2386 (2009). [Google Scholar]
- 28.Edwards G., Crawley M., Herbivores, seed banks and seedling recruitment in mesic grassland. J. Ecol. 87, 423–435 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Maron J. L., Johnson M. T. J., Hastings A. P., Agrawal A. A., Fitness consequences of occasional outcrossing in a functionally asexual plant (Oenothera biennis). Ecology 99, 464–473 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Johnson M. T., The contribution of evening primrose (Oenothera biennis) to a modern synthesis of evolutionary ecology. Popul. Ecol. 53, 9–21 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.