Significance
The experiments here advance understanding of the function of the SAMD9L gene and protein in innate immune mechanisms in resisting virus infection and in the pathogenesis of inflammatory, hematological, and neurological disorders. The clinical syndrome defined in two children with de novo truncating SAMD9L mutations expands the phenotypes in this newly recognized autoinflammatory disorder. Analysis of cells expressing normal or mutant SAMD9L reveals the protein represses protein translation, with the truncating mutations greatly exaggerating this activity. The experiments find equally potent gain of function caused by the truncating mutations or a recurrent missense mutation associated with clinically milder ataxia and pancytopenia syndromes, demonstrating that diverse clinical manifestations can arise from mutations that appear cell-biologically equivalent.
Keywords: translational repression, virus sensing, pathogen-associated molecular pattern receptor, autoinflammatory disease, nucleotide-binding and oligomerization domain
Abstract
Sterile α motif domain-containing protein 9-like (SAMD9L) is encoded by a hallmark interferon-induced gene with a role in controlling virus replication that is not well understood. Here, we analyze SAMD9L function from the perspective of human mutations causing neonatal-onset severe autoinflammatory disease. Whole-genome sequencing of two children with leukocytoclastic panniculitis, basal ganglia calcifications, raised blood inflammatory markers, neutrophilia, anemia, thrombocytopaenia, and almost no B cells revealed heterozygous de novo SAMD9L mutations, p.Asn885Thrfs*6 and p.Lys878Serfs*13. These frameshift mutations truncate the SAMD9L protein within a domain a region of homology to the nucleotide-binding and oligomerization domain (NOD) of APAF1, ∼80 amino acids C-terminal to the Walker B motif. Single-cell analysis of human cells expressing green fluorescent protein (GFP)-SAMD9L fusion proteins revealed that enforced expression of wild-type SAMD9L repressed translation of red fluorescent protein messenger RNA and globally repressed endogenous protein translation, cell autonomously and in proportion to the level of GFP-SAMD9L in each cell. The children’s truncating mutations dramatically exaggerated translational repression even at low levels of GFP-SAMD9L per cell, as did a missense Arg986Cys mutation reported recurrently as causing ataxia pancytopenia syndrome. Autoinflammatory disease associated with SAMD9L truncating mutations appears to result from an interferon-induced translational repressor whose activity goes unchecked by the loss of C-terminal domains that may normally sense virus infection.
Sterile α motif domain-containing protein 9 (SAMD9) and SAMD9-like (SAMD9L) (1) are paralogous type-I interferon-responsive genes (2, 3) with significant mutual functional redundancy and incompletely understood roles in the restriction of virus replication (2–6). SAMD9 has been shown to be important to restricting myxoma, vaccinia, and West Nile viruses (4–8), while SAMD9L is important to the pathogenesis of vaccinia (2) and Japanese encephalitis viruses (9) and in the control of hepatitis B–associated hepatocellular carcinoma (10). The genes are located contiguously on chromosome 7q21.3 in humans, arose by duplication after the divergence of Marsupialia from Placentalia (1), have ancestors in basal metazoan invertebrate species, and encode proteins of similar length (1,584 and 1,589 amino acids, respectively) and 58% sequence identity (11).
Computer-based predictions of SAMD9 proteins indicate they belong to the STAND protein family (11), which includes the apoptosome-forming protein APAF1 and the nuclear factor κB–activating noddosome protein NOD2. APAF1 and NOD2 proteins contain a C-terminal sensor domain that binds inducer ligands cytochrome c or muramyl dipeptide, respectively, which trigger polymerization mediated by a central nucleotide-binding and oligomerization domain (NOD) to activate N-terminal effector domains (12). In SAMD9/SAMD9L the central NOD is flanked by C-terminal tandem TPR repeats and OB-fold that might serve as RNA sensors and by N-terminal domains that may enhance oligomerization and mediate effector functions: SAM motif, putative RNA/DNA-binding AlbA_2 domain, and a domain related to SIR2 family histone deacetylases (Fig. 1A) (11). However, the nature of SAMD9L inducers or effectors is obscure.
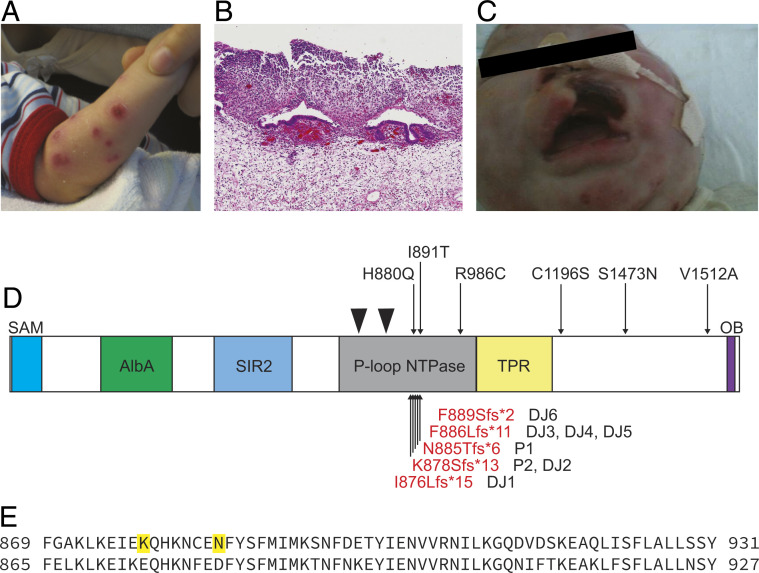
Fig. 1.
Autoinflammatory disease caused by de novo truncating SAMD9L mutations. (A) Deep erosive, scarring skin lesions in P1. (B) Neutrophilic infiltrates in section of perforated bowel from P1. (C) Tissue necrosis of the upper lip in P2. (D) SAMD9L protein domains and location of de novo mutations in P1 and P2, compared with location of published ATXPC missense mutations [black, Tesi et al. (16), Gorcenco et al. (25), Pastor et al. (24), Cheah et al. (36), Wong et al. (37)] and SAAD truncating mutations [red, de Jesus et al. (17)]. Walker A and B motifs at Leu739-Thr749 and Val798-Asp804 are denoted by black triangles. (E) Amino acid sequence of homologous region in SAMD9L (Top, NP_689916.2) and SAMD9 (Bottom, NP_060124.2) lacking truncating mutations in gnomAD. Locations of truncating mutations in P2 and P1 are shown in bold.
SAMD9 and SAMD9L are components of the hallmark antiviral type-1 interferon response of many cells (13), with SAMD9L messenger RNA (mRNA) undergoing a manyfold increase following stimulation with interferon beta (2). SAMD9 forms polymeric rods (14) consistent with other SAM domain-containing proteins which promote self-oligomerization (11), and this property may be what allows SAMD9 to participate in the formation of antiviral stress granules (5), cytoplasmic storage sites containing translationally silenced messenger ribonucleoproteins that can be released to resume translation after stress subsides. SAMD9-mediated granule formation is antagonized by poxvirus-encoded proteins that directly bind SAMD9 (5). In the absence of these viral antagonists, poxvirus infection triggers SAMD9-dependent formation of cytoplasmic granules containing translationally repressed viral mRNAs and a collapse in polyribosomes containing viral and host-cell mRNA (4–6). Translational repression by SAMD9/SAMD9L may inhibit growth factor signaling (15) and cell-cycle progression (10), suppressing cell proliferation (16) and stimulating inflammation. (17).
Human SAMD9L gene-associated disease is perplexing because it is not readily aligned with the evidence of SAMD9/SAMD9L as an innate antiviral defense mechanism. One or the other gene may be missing from individual species (e.g., lack of SAMD9 in mice) (1), while in humans homozygous obligate loss-of-function mutations are found in databases of normal variation (18), suggesting that lack of one gene may be tolerated in humans as well, although there are reports of biallelic loss of SAMD9 potentially causing familial tumoral calcinosis (19, 20). However, while biallelic loss of one gene may be tolerated, in cis heterozygous loss of both SAMD9 and SAMD9L genes from the same chromosome through deletion or copy number variants results in myelodysplastic syndrome in humans (21), while loss of the SAMD9L locus alone has the same effect in mice that do not possess SAMD9 (15). The inference is that having some activity of SAMD9 or SAMD9L function on each chromosome 7 may be more important than the total quantum of these genes or gene products. On the other hand, the two genes are unlikely to subserve exactly identical functions, given that the diseases caused by similar defects in SAMD9 and SAMD9L manifest with overlapping but different clinical phenotypes (22, 23).
Until recently heterozygous mutations in SAMD9L had been known to underlie two syndromes: the familial ataxia pancytopaenia (ATXPC) syndrome, which manifests with features of B-cell immunodeficiency and cerebral calcifications, and familial myelodysplastic syndrome/monosomy 7, where there is no neurological phenotype (22, 24). All of the reported disease-associated variants in SAMD9L associated with these entities have been gain-of-function (GOF) missense mutations (16, 22, 25), clustering in the NOD domain (16, 22, 24) (Fig. 1A). Recently a third SAMD9L-associated clinical entity has been identified, caused by truncating de novo mutations within a very narrow region of the SAMD9L NOD domain. These variants are associated with an acute, early childhood syndrome of neutrophil inflammation of the skin (panniculitis), interstitial lung inflammation, greatly elevated blood C-reactive protein (a laboratory marker of systemic inflammation), and progressive decline in circulating B lymphocytes and natural killer (NK) cells. However, unlike a similar syndrome caused by mutations in the PSMB8 proteasome subunit, increased expression of interferon-induced mRNAs was seen in the blood of only two of five severely ill patients with SAMD9L truncating mutations (17). This new disease entity has been called SAMD9L-associated autoinflammatory disease (SAAD) (17).
Key unresolved questions are how SAMD9L mutations cause disease, whether truncating SAMD9L mutations causing SAAD differ functionally from SAMD9L missense mutations causing myelodysplastic syndrome or ATXPC syndrome, and why SAAD resembles autoinflammatory diseases with exaggerated interferon production yet does not display consistent signs of elevated interferon signaling. Here we studied two de novo SAMD9L mutations causing SAAD in two new pediatric cases. In transfected human cells we show that increasing expression of wild-type (WT) SAMD9L protein causes a progressive inhibition of protein translation, in a cell-autonomous manner that does not interfere with translation in cocultured cells not expressing elevated SAMD9L. SAAD-causing truncating mutations lower the threshold for this translation-inhibiting activity so that it is profound even in cells expressing low levels of SAMD9L protein. A missense mutation found recurrently in ATXPC syndrome had comparably exaggerated translation-inhibition activity to the truncating mutations. The results are consistent with other STAND proteins and other sensors of microbial infection, where activation of effector function depends on a threshold amount of protein for polymerization, with polymerization autoinhibited by the C-terminal sensor domain until released by binding an inducer.
Results
Clinical Cases.
P1 is a 10-y-old boy, the child of nonconsanguineous Chinese-Australian parents, who was previously reported as part of a case series of neonatal Sweet’s syndrome (26). Briefly, he presented shortly after birth with fever and raised inflammatory markers, hepatosplenomegaly, and widespread erythematous plaques on his skin which developed into deep ulcerative, scarring lesions (Fig. 1A). Skin biopsy showed extensive neutrophilic leukocytoclasis with panniculitis, consistent with Sweet’s syndrome. He went on to have noninfective epiglottitis and perforated bowel associated with neutrophilic infiltrates (Fig. 1B), calcification of the basal ganglia, thrombocytopaenia, and anemia requiring regular blood transfusions, neutrophilia, and trilineage dysplasia with few blasts but no clonal cytogenetic abnormality (Table 1 and SI Appendix, Fig. S1). He had B-cell immunodeficiency with low immunoglobulin (Ig)G and absent IgA and IgM. Flow cytometry on blood and bone marrow at 1 mo failed to identify B-lineage cells (SI Appendix, Fig. S2). Unexpectedly, after the first year of life he began to produce platelets and red cells and no longer needs transfusions. Peripheral blood testing at aged 4 y demonstrated substantial recovery of mature B cells with a normal phenotype and normal IgG, IgA, and IgM. A more extensive history is available in SI Appendix, Fig. S1. Since age 4 y he has had limited hematological, immune, or inflammatory involvement, with the exception of mildly low platelets, but has been largely lost to formal follow-up for a number of years.
Table 1.
SAMD9L-SAAD clinical features
| Patient | P1 | P2 | DJ1 (17) | DJ2 | DJ3 | DJ4 | DJ5 | DJ6 |
| SAMD9L variant | c.2654delA; p.N885Tfs*6 | c.2633delA; p.K878fs*13 | c.2626delA; p.I876Lfs*15 | c.2633delA; p.K878fs*13 | c.2658_2659delTT; p.F886Lfs*11 | c.2658_2659delTT/ F886Lfs*11 | c.2658_2659delTT/ F886Lfs*11 | c.2666delT; p.F889Sfs*2 |
| Ethnicity | Chinese | Korea | NS | NS | NS | NS | NS | NS |
| Cons | No | No | NS | NS | NS | NS | NS | NS |
| Sex | M | F | M | NS | NS | NS | NS | NS |
| Age at presentation | <1wk | <1 wk | NS | NS | NS | NS | NS | NS |
| Rash macroscopic | +++ scarring | +++ scarring | Panniculitis/lipoatrophy | Panniculitis | Panniculitis | Panniculitis | Panniculitis | Panniculitis |
| Biopsy of rash | Neutrophil dermatosis | Leukocytoclast vasculitis | Neutrophilic panniculitis | Neutrophilic panniculitis | Neutrophilic panniculitis | Neutrophilic panniculitis | Neutrophilic panniculitis | Neutrophilic panniculitis |
| Lip erosion | + | + | NS | NS | NS | NS | NS | NS |
| ILD | No | No | No | No | Yes | Yes | Yes | Yes |
| HSM | + | + | + | + | + | + (with portal hypertension) | NS | Splenomegaly |
| MRI brain change | Basal ganglia calcification | Basal ganglia calcification | Subcortical white matter change | NS | Basal ganglia calcifications – bilateral white matter disease | Perinatal bleed/basal ganglia calcifications | NS | CT NAD |
| Epiglottitis | + | + | NS | NS | NS | NS | NS | NS |
| Hemoglobin | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | NS |
| Platelets | ↓ | ↓ | ↑ then ↓ | ↓ | ↓ | ↓ | ↓ | ↓ |
| Neutrophils | Normal | ↑ | NS | NS | NS | NS | NS | NS |
| Inflammation markers | ↑ | ↑ | NS | NS | NS | NS | NS | NS |
| Ferritin (maximum) | 4,305 µg/L | 10,619 ng/mL | NS | NS | NS | NS | NS | NS |
| % B cells | <1% | Absent | ↑ then ↓ | ↓ | ↓ | ↓ | ↓ | ↓ |
| Bone marrow B cells | Low | Low | Low | Markedly low | NS | ↓ (<5%) | NS | NS |
| NK cells | ↓ | ↓ | NS | NS | NS | ↓ | ↓ | ↓ |
| IgG | ↓ | ↓* 663 mg/dL | N | ↑ | N | NR | NR | ↓ |
| IgA | ↓ | ↓ | ↑ | ↑ | N | NR | NR | NR |
| IgM | ↓ | ↓ | N | N | ↓ | NR | NR | NR |
| T cell no. and function | N | Abnormal PHA | NS | NS | NS | NS | Normal numbers decreased prolif | Normal numbers |
| Viral infections | CMV viraemia | Coronavirus HKU1, RSV, parainflu, adenovirus, rhinovirus | Rhinovirus, RSV | RSV, parainfluenza | NS | NS | NS | |
| Treatment | Prednisolone MMF (? PR) | BMT | Prednisolone (PR) Tocilizumab (PR) Baracitinib (PR) | Etanercept (PR) IVIG (PR) | Cyclosporin (MR) Etanercept (MR) Adalimumab (MR) Anakinra (MR) BMT (Successful) | Anakinra (MR) IVIG | IVIG | IVIG BMT (successful) |
| Outcome | Alive 11 y; immune Hematology/inflammatory features self-resolved | Died in BMT | Alive at reporting | Alive at reporting | Alive post-BMT | Died 6 y | Died 2 mo | Alive after -BMT |
NS = not stated; MMF = mycophenolate mofetil; PR = partial response; MR = minimal response; HSM, hepatosplenomegaly; ILD, interstitial lung disease; BMT, bone marrow transplant; Cons, consanguinity; PHA, phytohemagglutinin; IVIG, intravenous immunoglobulin; Prolif, proliferation.
P2, a girl, was the first child of nonconsanguineous parents from South Korea who developed extensive scarring skin lesions in the neonatal period characterized by leukocytoclastic panniculitis, raised inflammatory markers, hepatosplenomegaly, persistent fever, basal ganglia calcification, neutrophilia, anemia, thrombocytopaenia, and almost no B cells (Table 1). She developed tissue necrosis around the upper lip (Fig. 1C) and upper airway obstruction thought secondary to noninfective epiglottitis. She developed a significant pulmonary disease and, sadly, died during bone marrow transplant.
Whole-Genome Sequencing of Family Trios.
Whole-genome sequencing (WGS) was performed as part of a study of children with early-onset autoimmune or autoinflammatory disease. Both families underwent trio WGS, which identified two different de novo truncating frameshift mutations within the same short coding region of SAMD9L (Fig. 1 D and E). P1 variant is chr7-92762631-T-; NM_152703.4:c.2654delA; (p.Asn885Thrfs*6), which does not appear on the gnomAD database of normal variation. The P2 variant is chr7-92762652-T-; NM_152703.4:c.2633delA; (p.Lys878Serfs*13), which is also absent from gnomAD. These variants are located within a domain spanning amino acids 712 to 998 identified as homologous to the NOD or P-loop ATPase domain of APAF1 (11) (Fig. 1A), and p.Lys878Serfs*13 is identical to a variant reported in association with SAAD (17). The p.Asn885Thrfs*6 is novel but resides close to other SAMD9L-SAAD truncating variants, Ile876Leufs*15, Phe886Leufs*11, and Phe889Serfs*2 (Fig. 1D). While each of the frameshift mutations yields different C-terminal peptide sequences, they all truncate the normal SAMD9L protein 71 to 85 amino acids C-terminal to the Walker B motif. In APAF1, this would correspond to truncating the NOD domain after the nucleotide binding domain (NBD) and helical domain 1 (HD1), but before the winged helical domain (27, 28).
Extended analysis of whole-genome data from P1 failed to demonstrate evidence of a second somatic mutation, copy number variant, or region of uniparental disomy, which might explain the improvements in myeloid and lymphoid lineage function from the end of the first year of life. Other than DNA there was no patient tissue available to explore other potential methods of reversion.
SAMD9L-SAAD Frameshift Mutations Exert a GOF Effect on Cell Proliferation.
Missense variants in SAMD9L causing ATXPC have previously been shown to reduce proliferation above that seen by WT SAMD9L in transient transfection systems, indicating a GOF effect (16). To determine if frameshift truncating variants from patients P1 an P2 might also cause a GOF, 293FT cells were transiently transfected with vectors encoding green fluorescent protein (GFP) fused to the N terminus of SAMD9L, either WT, the P1 and P2 truncating variants Asn885fs and Lys878fs, or an Arg986Cys missense variant located at the C-terminal end of the predicted NOD/P-type ATPase domain. Arg986Cys is a recurring ATXPC mutation described in 1) a multigeneration ATXPC family where some affected individuals had profound B cell and NK cell deficiency from infancy while others had later onset ataxia and myelodysplastic syndrome (16), 2) a two-generation family with pancytopenia and no neurological abnormalities (24), 3) an isolated case of longstanding cerebellar ataxia with transient cytopenia (29), and 4) as a de novo mutation in an adult diagnosed with ataxia (30). Dilution of cell trace violet (CTV) fluorescent dye was used to measure cell division 72 h posttransfection by flow cytometry.
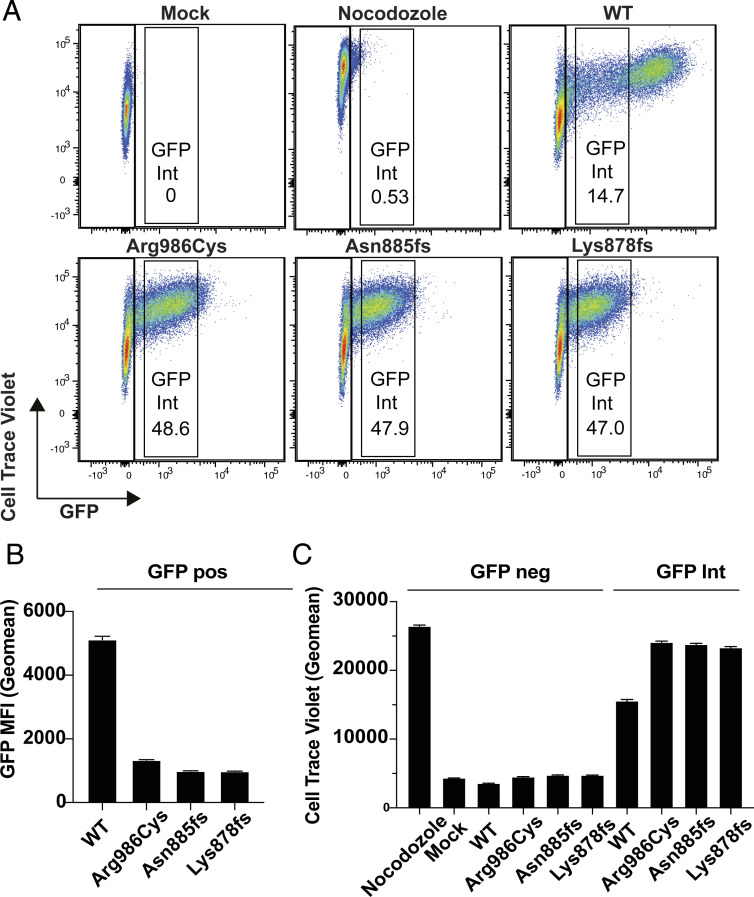
Accumulation of the truncating or missense SAMD9L variant proteins in single cells was greatly reduced compared with WT as evidenced by a fivefold drop in mean GFP fluorescence per cell (Fig. 2 A and B). Gates were applied on cells with these intermediate levels of GFP (GFPint) to compare single cells expressing comparable levels of WT or mutant SAMD9L protein and dilution of CTV analyzed in these matched cells. The GFP-negative cells in each culture acted as an internal control cell population, dividing and diluting CTV down to the mean fluorescence intensity exhibited by mock-transfected cells, regardless of the SAMD9L vector expressed in the GFP+ cells in that culture (Fig. 2 A and C). CTV fluorescence was not diluted in GFP− cells from cultures treated with the antimitotic microtubule-blocking drug nocodazole. Comparable inhibition of CTV dilution occurred in GFPhi cells expressing WT SAMD9L, whereas GFPint cells with WT protein had intermediate inhibition of CTV dilution (Fig. 2 A and C). GFPint cells expressing the missense variant and both truncated variants had significantly higher CTV mean fluorescence than GFPint cells expressing WT SAMD9L, consistent with a GOF effect (Fig. 2C).
Fig. 2.
Inhibition of cell division in single cells expressing different levels of GFP-SAMD9L fusion protein. Dilution of CTV was used to measure cell division by 293FT cells labeled with CTV 3 h prior to transfection or treatment with the microtubule inhibitor nocodazole as a positive control. Cells were either mock-transfected or transfected with vectors encoding N-terminal GFP-tagged SAMD9L, either with WT sequence, the missense variant Arg986Cys, or the Asn885fs and Lys878fs truncating variants from P1 or P2, and analyzed 72 h later by flow cytometry. (A) Representative flow cytometric plots showing CTV and GFP fluorescence in individual cells. (B) GFP geometric mean fluorescence intensity (MFI, error bars 95% CI) gated on all GFP-positive cells. (C) CTV geometric MFI in cells gated as shown in A, either GFP-negative cells or cells with comparable, intermediate levels of GFP-SAMD9L fusion protein (GFP Int). Results are representative of three independent experiments.
Transfected SAMD9L Proteins Decrease the Expression of Cotransfected Reporter Red Fluorescent Protein (RFP).
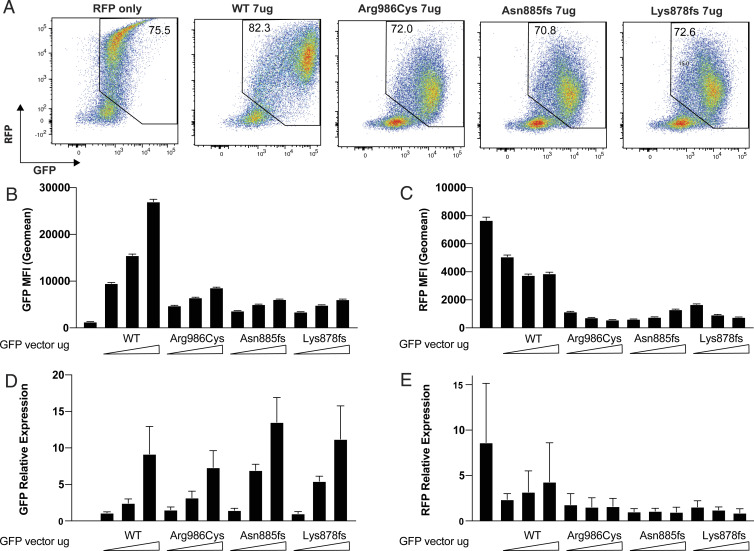
We next asked if the diminished accumulation of the missense and frameshift SAMD9L proteins in transfected single cells compared with WT was limited to the GFP-fusion protein, perhaps reflecting protein instability, or might reflect a more global effect on transcription or translation of other genes and proteins. To distinguish these alternatives, a plasmid expressing an RFP transcribed from the retroviral mouse stem cell virus (MSCV) promoter was cotransfected with varying amounts of the DNA vector encoding GFP-tagged SAMD9L. GFP and RFP protein and mRNA expression were measured 48 h posttransfection by flow cytometry and qPCR. RFP protein expression was reduced from a geometric mean fluorescence per cell of 7,678 when transfected alone to 3,883 when an equivalent amount of WT SAMD9L vector DNA was cotransfected (Fig. 3 A and C). Mean RFP expression was further reduced to 578, 1,312, and 766 in cells expressing the missense and two truncating variants, respectively. As seen previously, accumulation of the GFP-tagged truncating variants is lower than that of the missense variant and this is reflected in slightly smaller reductions in RFP (Fig. 3 A and B). Transfection of higher amounts of the truncating plasmids is required to achieve similar GFP expression levels and reduction in RFP (Fig. 3 B and C).
Fig. 3.
Mutant GFP-SAMD9L inhibits RFP expression from a cotransfected vector. 293FT cells were transfected with 7 μg RFP retroviral vector (MSCV promoter) alone or in combination with a 2, 4.5, or 7 μg vector encoding GFP-tagged SAMD9L with the indicated mutations or WT. After 48 h, relative amounts of RFP and GFP protein were measured in single cells by flow cytometry, and RFP and GFP mRNA was measured by qPCR in pooled cells from the same cultures. (A) Representative flow cytometric plots showing RFP and GFP fluorescence in single cells transfected with equal vector amounts. Numbers show the percent of cells within the RFP+/GFP+ gate. (B) GFP geometric MFI (error bars 95% CI) in GFP/RFP-positive cells. (C) RFP geometric MFI in GFP/RFP positive cells. (D) GFP mRNA relative expression (mean ± SD) measured by qPCR. (E) RFP mRNA relative expression (mean ± SD) measured by qPCR. Results are representative of three independent experiments.
Accumulation of GFP and RFP mRNA was measured by qPCR in pooled cells from each transfected culture. GFP mRNA amount increased with increasing dose of transfected plasmid (Fig. 3D), whereas GFP protein accumulation showed a more modest increase in the same cultures (Fig. 3B). RFP mRNA was lower in cultures transfected with GFP-SAMD9L, especially missense or truncating mutant SAMD9L, although the decrease in RFP mRNA was less than the decrease in mean RFP protein per cell (Fig. 3 C and E). Collectively these results establish that mutant SAMD9L inhibits accumulation of an independent mRNA and protein expressed in the same cell but do not resolve if inhibition is at the level of transcription or translation.
Translational Repression by SAMD9L against Cytomegalovirus (CMV)-Based RFP Expression.
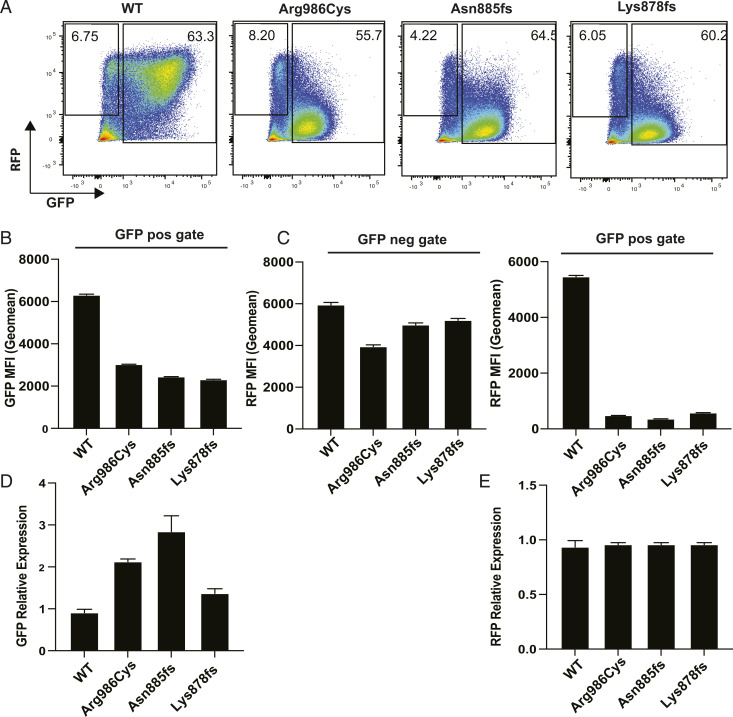
RFP expression in the previous experiments employed a retroviral vector. To rule out the potential influence of the retroviral sequences on transfected protein transcription or translation, transfections were repeated with RFP expressed from the same vector backbone as the SAMD9L constructs employing a CMV promoter. pcRFP (CMV-based RFP plasmid) was transfected in combination with equal quantities of the SAMD9L constructs into 293FT cells. GFP and RFP protein and mRNA expression were measured 48 h posttransfection by flow cytometry and qPCR. GFP+ cells expressing the missense or truncating SAMD9L variants exhibited greatly decreased RFP per cell (Fig. 4A), with the geometric mean fluorescence decreased to 480, 359, and 578, respectively, contrasting with mean RFP fluorescence of 5,465 in GFP+ cells expressing WT SAMD9L (Fig. 4 C, Right). A distinct subset of cells in each culture were GFP− yet RFP+, consistent with these cells having been transfected with RFP vector alone (Fig. 4A). These GFP− cells showed little decrease in RFP, despite coculture with GFP+ cells expressing mutant SAMD9 (Fig. 4 C, Left). Thus, the inhibitory effect of mutant SAMD9L on accumulation of RFP is a cell-autonomous process that does not interfere with RFP accumulation in cocultured cells not expressing the mutant protein.
Fig. 4.
Mutant GFP-SAMD9L inhibits RFP protein but not mRNA expressed from a cotransfected CMV-based RFP vector. 293FT cells were transfected with an RFP vector employing a CMV promoter alone or in combination with an equivalent amount of vector encoding GFP-tagged SAMD9L with the indicated mutations or WT. After 48 h, relative amounts of RFP and GFP protein were measured in single cells by flow cytometry and RFP and GFP mRNA measured by qPCR in pooled cells from the same cultures. (A) Representative flow cytometric plots showing RFP and GFP fluorescence in single cells transfected with equal vector amounts. Numbers show the percent of cells within the RFP+ GFP− gate and the GFP+ gate. (B) GFP geometric MFI (error bars 95% CI) in GFP+ cells. (C) RFP geometric MFI in RFP+GFP- cells (Left) and in GFP+ cells (Right). (D) GFP mRNA relative expression (mean ± SD) measured by qPCR. (E) RFP mRNA relative expression (mean ± SD) measured by qPCR. Results are representative of three independent experiments.
Accumulation of GFP mRNA (Fig. 4D) and RFP mRNA (Fig. 4E), measured by qPCR in the same transfected cell cultures, showed no decrease in RFP mRNA in cultures transfected with mutant SAMD9L. Thus, when RFP is expressed from a CMV promoter vector, mutant SAMD9L does not diminish RFP mRNA but profoundly inhibits its translation into protein.
SAMD9L Exerts a Protein Concentration–Dependent Global Translational Block Exaggerated by Truncation.
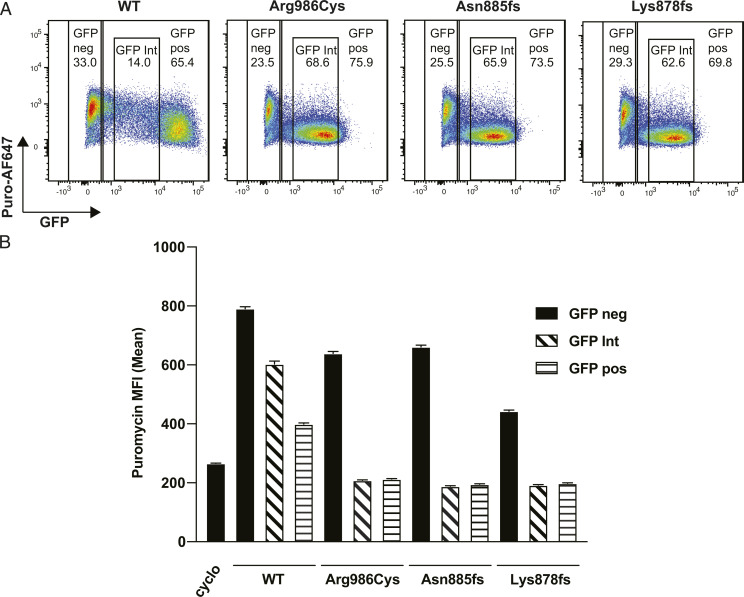
We next addressed whether the SAMD9L-induced translational block was limited to GFP and RFP proteins transcribed from transfected plasmid DNA or reflected global interference with endogenous protein translation. To measure global protein translation in single cells by flow cytometry we used the SUnSET method where nascent translated type-2 transmembrane proteins are pulse-labeled for 10 min with the nonextendible aminoacyl transfer RNA analog puromycin and detected on the cell surface after a chase period by cell surface staining with fluorescent antibody to puromycin (31). 293FT cells were labeled with puromycin for 10 min 48 h posttransfection, followed by a 50-min incubation with fresh media before the cells were harvested and stained with antipuromycin antibody. Since this timepoint precedes the peak of cell surface puromycin staining, which peaks from 1 to 3 h (31), decreased fluorescence is likely to reflect decreased translation and not decreased protein stability. As a positive control for translation inhibition, 293FT cells were treated with 100 µM cycloheximide for 4 h prior to the addition of puromycin. In cultures transfected with GFP-SAMD9L WT, puromycin mean fluorescence was 791 in GFP− cells in the culture but decreased to 400 and 603 in GFP+ and GFP intermediate gates, respectively, which was still higher than the mean fluorescence of 250 for cycloheximide-treated cells (Fig. 5 A and B). This effect was exaggerated in GFPint cells expressing the missense and truncating variants, with puromycin mean fluorescence decreasing to levels comparable to cycloheximide treatment (Fig. 5 A and B). Strikingly, there was little inhibition of translation in GFP− cells in the same cultures, indicating that missense and truncating SAMD9L mutant proteins inhibit translation cell-autonomously and not through induction of a cell-to-cell mediator.
Fig. 5.
Mutant GFP-SAMD9L inhibits global protein translation cell-autonomously. Incorporation of puromycin into nascent proteins in single cells expressing varying levels of GFP-SAMD9L was quantified by flow cytometry. 293FT cells were transfected with vector encoding GFP-tagged SAMD9L with the indicated mutations or WT. Puromycin was added to cultures 48 h posttransfection for 10 min and puromycin-containing media was replaced with fresh media and cultures incubated for 50 min prior to staining with monoclonal antibody to puromycin coupled to AF647 (Puro-AF647). (A) Representative flow cytometry plots showing puromycin fluorescence intensity in single cells expressing different levels of GFP-SAMD9L protein. Numbers show percent cells within the gates for GFP−, GFP+, or GFP-intermediate cells. (B) Puromycin-AF647 MFI (error bars 95% CI) in GFP−ve cells contrasted with GFP+ve and cells with an intermediate level of GFP fluorescence, expressing the indicated SAMD9L proteins. Results are representative of three independent experiments.
Discussion
The findings above provide insights into the function of SAMD9L in innate immunity and the pathogenesis of autoinflammatory diseases associated with SAMD9L mutations. The experiments show that enforced expression of WT SAMD9L protein represses protein translation in uninfected human cells, cell-autonomously, with greater repression observed in cells with higher SAMD9L protein. These results extend the findings of translational repression by SAMD9 in poxvirus-infected cells (6). Since SAMD9L is a hallmark interferon-induced mRNA (mSigDB M5911) (13), increased SAMD9L protein in cells exposed to interferon may mediate some of the translational repression effects of interferon signaling and explain how it serves to restrict viral replication. The single-cell flow cytometric analyses here nevertheless reveal that truncating and missense variants found in children with autoinflammatory disease induce much greater translational repression at lower SAMD9L protein amounts per cell. While this article was in preparation, a complementary study of different SAMD9L truncating mutations reached similar conclusions (32). As discussed below, these results point to allosteric regulation of SAMD9L by its C terminus. In other STAND domain proteins, the central NOD is activated to polymerize into effector disks or fibers by binding an inducer at their C-terminal sensor domain. It seems likely that poxvirus infection is sensed by the putative RNA/DNA binding OB-fold at the SAMD9L C terminus and/or the TPR domains. We hypothesize that, for innate immunity, infection of other cells is communicated cell-to-cell via interferon secretion to induce SAMD9L mRNA in uninfected cells, and the resulting increase in SAMD9L protein normally remains poised to act cell-autonomously, repressing mRNA translation upon sensing infection within the cell.
The conclusion that normal SAMD9L protein is poised but inhibited from potent translational repression in uninfected cells is supported by the potent increase in this activity caused by the truncating mutations studied here. It is striking that the SAMD9L truncating mutations found in children with SAAD cluster in a small region that appears not to be permissive to truncating mutations in the gnomAD database of normal human variation (18). There are 43 heterozygous, obligate truncating mutations in SAMD9L (e.g., frameshift, stop-gained) affecting 383 of >240,000 alleles, with the longest sequence containing no obligate loss-of-function variants being p.869–931 (Fig. 1E) that includes all of the SAMD9L-SAAD truncating variants and several of the missense mutations in ATXPC such as p.H880Q20, p.S888Y36, and p.I891T (16, 25). The paralogue SAMD9 also has many heterozygous loss-of-function alleles in gnomAD (>1,300), with the longest stretch without an obligate truncating allele (p.874–936) overlapping extensively with the invariant region in SAMD9L (Fig. 1E). This region is 70% homologous, greater than between the two proteins overall, and shows higher mean conservation across 100 species on the UCSC genome browser (33). In the recently published study by Allenspach et al., a range of experimentally created stop-gain mutations in the same region of SAMD9L also dramatically exaggerated translational repression when expressed from a self-cleaving GFP-T2A-SAMD9L bicistronic vector (32). The approach taken here extends and complements these findings by expressing SAMD9L proteins directly fused to GFP, making it possible to compare the degree of translational repression with the relative amount of SAMD9L protein in single cells.
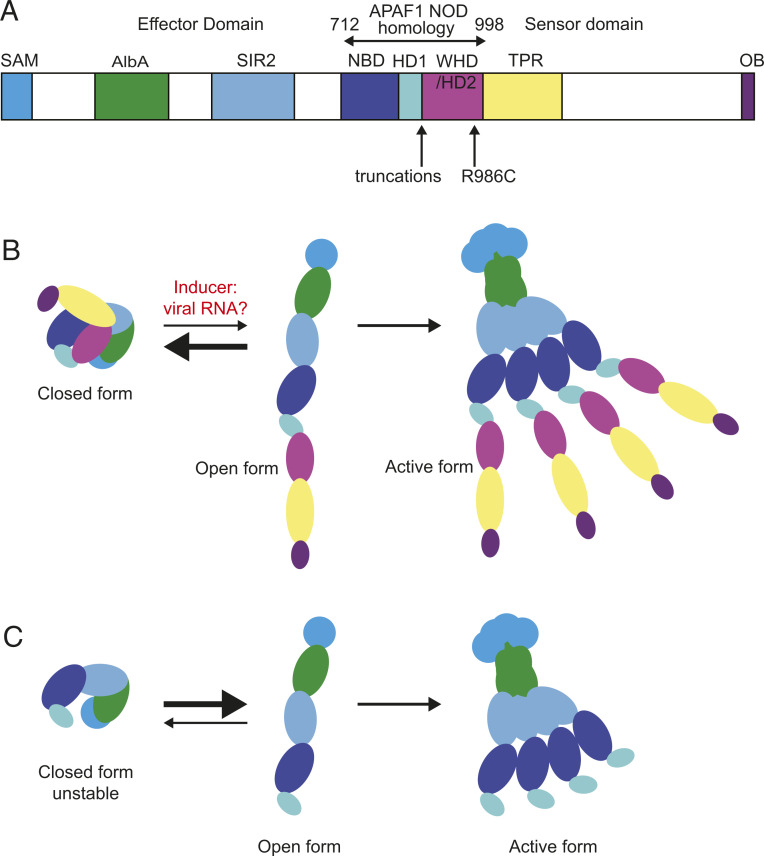
The region where the SAMD9L disease-causing truncations occur is 70 to 85 amino acids C-terminal to the Walker B motif, in an APAF1 NOD-homology region corresponding to the boundary in APAF1 between a module comprising the NBD and HD1 and a module comprising the winged helix domain (WHD) and helical domain 2 (HD2; Fig. 6A). Polymerization of APAF-1 is centrally mediated by extensive contacts between the NBDs of individual monomers. Upon cytochrome c binding to the C-terminal sensor domain of APAF1, the NBD–HD1 module rotates relative to the WHD/HD2 module, to convert the monomer from a closed to open state that exposes the NBD oligomerization interfaces. This shift allows polymerization, with the NBD–HD1 module forming the core of the apoptosome disk and the N-terminal effector CARD domain polymerized in a ring above (27, 28). By homology, we hypothesize that binding of viral RNA to the TPR domain triggers a similar rotation and opening of SAMD9L followed by polymerization into active fibers that repress mRNA translation (Fig. 6B). Given the homology of the NOD domains of SAMD9L and APAF1 and the location of the truncating mutations at the region corresponding to the rotation joint in APAF1, it is likely the truncating mutations preserve polymerizing activity of the core NBD–HD module while eliminating allosteric inhibition normally imposed by the WHD/HD2 and TPR domains (Fig. 6C).
Fig. 6.
Model for SAMD9L regulation based on mechanism of APAF1 regulation. (A) Predicted domains of SAMD9L and region of homology with APAF1 NOD from ref. 11. In APAF1, the NOD domain comprises the NBD, HD1, and WHD/HD2. By analogy with other STAND domain proteins, the TPR domain is likely to be the sensor and the AlbA/SIR domains the effectors. (B) Model for physiological activation based on APAF1 (12), where cytochrome c binding to the sensor domain causes the NBD/HD1 module to rotate relative to the WHD/HD2/sensor domain, exposing the oligomerization interface of the NBD. In SAMD9L, the physiological trigger for this rotation may be viral RNA binding to the TPR domain. (C) Model for pathological activation of SAMD9L by truncating mutations. In the absence of the WHD/HD2 and TPR domains to obscure the NBD oligomerization interface, the truncated protein adopts the open conformation and multimerizes into the active form inhibiting mRNA translation in the absence of viral RNA triggering.
A key question is whether or not SAMD9L missense and truncating mutations are functionally distinct. A number of different syndromes have been related to heterozygous GOF SAMD9L mutations. Missense mutations in SAMD9L like Arg986Cys studied here underlie the familial ATXPC syndrome, which includes features of cytopenias, B cell immunodeficiency, cerebral calcifications, and rash (16, 25, 34). ATXPC mutations cluster toward the C-terminal end of the protein, with the most frequently reported mutations substituting residues 896 and 880 in the C-terminal part of the NOD domain (16, 22, 24). Truncating mutations are found in SAAD (SAMD9L-SAAD), which has many of the features of ATXPC including cytopenias, immunodeficiency, and cerebral calcification. However, the clinical syndrome of ATXPC appears milder and many patients remain relatively well into adult life or display only ataxia or subclinical cytopenia and transmit the mutation through several generations as autosomal dominant defects (16, 22, 25), and in one series 30% of mutation carriers were asymptomatic (24). SAMD9L-SAAD by comparison appears not to be inherited, being caused by de novo mutations, manifesting an early onset of disease with an aggressive and inflammatory presentation including reports of severe interstitial lung disease (17).
Set against this apparent clinical difference in severity, the experiments here demonstrate that the most frequently described missense mutation in ATXPC syndrome and also in isolated ataxia or cytopenia families, Arg986Cys, caused a comparably large exaggeration of translational repression activity to two different SAAD-syndrome truncating mutations. This conclusion is reinforced by a recent finding that another familial ATXPC missense mutation, His880Gln, exaggerated translational repression comparable to a Phe886Leufs11 mutation responsible for neonatal-onset autoinflammatory disease (32). The two children studied here displayed the classic ATXPC features of myeloid suppression, B cell deficiency, and cerebral calcification but also severe erosive dermatitis with lip erosions, sterile epiglottitis, intestinal neutrophilic infiltrates, interstitial lung disease, and inflammation. Differentiating between symptoms of SAMD9L-SAAD that resemble ATXPC and those that do not may be a useful lens with which to view this disease. The non-ATXPC symptoms most closely resemble a disease associated with OAS1 GOF mutations, which cause hyperactivation of a molecular pathway involved in innate recognition of double-stranded RNA and activation of type-I interferons (35) and which may present with erosive dermatitis and interstitial lung disease. Given the evidence that SAMD9L-SAAD resembles an interferonopathy (17), it is possible that the truncating variants may impact molecular pathways involved in ATXPC manifesting as suppression of cell proliferation and also inflammatory pathways involved in diseases resembling OAS1 GOF. Because the missense mutations retain the C-terminal domains, they may disrupt allosteric regulation similarly to truncation but retain compensatory mechanisms to inhibit SAMD9L auto-activation and polymerization that are not apparent in transiently transfected cells. However, it is also conceivable that the missense and truncating mutations within the SAMD9L NOD domain have identical biochemical consequences, and the clinical differences in severity and organs affected may reflect other genetic and environmental differences such as exposure to particular viruses.
In conclusion, this study extends the clinical phenotype of SAMD9L-associated autoinflammatory disease and demonstrates that SAMD9L blocks its own and other protein’s translation in uninfected cells when expressed at sufficient levels per cell and that this cell-autonomous function is greatly exaggerated by both missense ATXPC and truncating SAAD mutations within the NOD putative polymerization domain. These autoinflammatory diseases appear to result from an interferon-induced translational repressor whose activity goes unchecked by the loss of putative virus-sensing domains.
Materials and Methods
Patient Samples.
Approval for this study was obtained from the human research ethics committees of South Eastern Sydney Local Health Network HREC Ref 11/107; informed consent was obtained from all participants for human experiments described in this study.
Genome Sequencing.
Parent-proband trio genomes were sequenced on the Illumina HiSeq X platform using DNA isolated from whole blood using a QIAsymphony DSP DNA Midi kit (Qiagen). Libraries were generated using the KAPA Hyper PCR-Free kit (Roche). Raw reads were aligned to the hs37d5 reference using BWA-MEM v0.7.10-r789 and sorted and duplicate-marked with Novosort v1.03.01 (Novocraft Technologies). The GATK suite v3.3-0-g37228af was used for local indel realignment and base quality score recalibration. gVCFs generated with GATK HaplotypeCaller were joint-called as trios using GATK GenotypeGVCFs, and variants recalibrated using GATK Variant Quality Score Recalibrator (VQSR). VCF files were annotated with Variant Effect Predictor (VEP) v76 using the LoFTEE and dbNSFP plugins (including CADD v1.3) and assembled into GEMINI databases (v0.18.3).
Cell Culture and Transfection.
293FT cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 200 mM GlutMAX, 10 mM Gibco MEM Non-Essential Amino Acids, and 100 mM pyruvate in 37% CO2. Stock cultures were maintained in 500 µg/mL Geneticin (Life Technologies). 293FT cells were transiently transfected by either forward or reverse transfection (Lipofectamine 2000; Thermo Fisher Scientific). WT SAMDL9 along with the missense variant Arg986Cys (16) and the two truncating variants from patients 1 and 2 were synthesized and cloned as N-terminal fusions into NotI/XhoI sites of pcDNA3.1+N-GFP (Genscript). RFP was expressed from pMSCV-CY3 (TagRFP) (a kind gift from Robert Brink, Garvan Institute of Medical Research, Darlinghurst NSW, Australia) or pcDNA3.1(+) (Genscript)
Proliferation Assays.
Cells were labeled for 20 min with 1 mM of CellTraceViolet (Thermo Fisher Scientific) in prewarmed phosphate-buffered saline (PBS), washed with PBS, spun, and replated in fresh prewarmed media and transfected 3 h later. Cells were either treated with nocodazole at 20 µg/mL for 24 h or transfected with GFP-tagged SAMD9L constructs. Fourteen micrograms of plasmid DNA was transfected in 10-cm dishes with 50 µL lipofectamine. The cells were harvested 72 h after transfection, stained with eBioscience Fixable Viability Dye eFluor 780 (Life Technologies) for 30 min on ice, washed twice in PBS, and analyzed by flow cytometry on an LSR II analyzer (BD Pharmingen). Cytometer files were analyzed with FlowJo software (FlowJo LLC).
Cotransfection Assays.
Seven micrograms of RFP DNA was mixed with up to 7 µg SAMD9L constructs ± pcDNA3.1(−) to a total of 14 µg. Cells were harvested and resuspended at 6.4 × 105 cells per mL and combined with transfection complexes in 10-cm dishes. Media was replaced 5 h posttransfection. The cells were harvested 72 h after transfection, a small fraction was used to prepare RNA for qPCR, and the remaining cells were stained with eBioscience Fixable Viability Dye eFluor 780 (Life Technologies) for 30 min on ice and washed twice in PBS and analyzed by flow cytometry on a LSR II analyzer (BD Pharmingen). Cytometer files were analyzed with FlowJo software (FlowJo LLC).
Translation Assays.
Cells were labeled 48 h posttransfection and assessed for global protein translation using the SUnSET (surface sensing of translation) method (31). Briefly, cells were treated with 10 µg/mL puromycin (P8833; Sigma-Aldrich) for 10 min, and cells were washed and incubated with fresh media for 50 min. As a positive control for translation inhibition, untransfected cells were treated with cycloheximide at 100 µg/mL for 4 h prior to treatment with puromycin. Treated cells were harvested and stained with eBioscience Fixable Viability Dye and AF-647-labeled anti-puromycin antibody (MABE343-AF647; Merck) on ice for 30 min, washed twice in PBS, and analyzed by flow cytometry on an LSR II analyzer (BD Pharmingen). Cytometer files were analyzed with FlowJo software (FlowJo LLC).
qPCR.
RNA was prepared using the Qiagen RNeasy mini kit which included a DNaseI digestion step. Prior to the complementary DNA (cDNA) preparation the RNA was spiked with MCF7 cell line RNA in order to use ESR1 as an endogenous control; this was done to control for potential transcriptional effects of the transfected SAMD9L. MCF7 expresses high levels of ESR1, which was confirmed to be absent in 293FT cells. RNA was reverse-transcribed using the Bio-Rad iScript cDNA Synthesis Kit. Primers were designed to GFP (forward 5-ACGTAAACGGCCACAAGTTC-3′, reverse 5′-AAGTCGTGCTGCTTCATGTG-3′), RFP (forward 5-AACACCGAGATGCTGTACCC-3′, reverse 5′-GCTGCTCGACGTAGGTCTCT-3′) and ESR1 (forward 5-TGGAGATCTTCGACATGCTG-3′, reverse 5′-GCTGGACAGAAATGTGTACACTC-3′). Real-time PCR was performed on an Applied Biosystems Quantstudio7 instrument using KAPA SYBR FAST qPCR Master Mix. The expression of GFP and RFP was calculated using the ∆∆Ct method relative to ESR1.
Supplementary Material
Footnotes
Competing interest statement: P.E.G., J.B.Z., and A.F. are coauthors of a 2021 J. Clin. Invest. article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110190118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Lemos de Matos A., Liu J., McFadden G., Esteves P. J., Evolution and divergence of the mammalian SAMD9/SAMD9L gene family. BMC Evol. Biol. 13, 121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng X., et al., A paralogous pair of mammalian host restriction factors form a critical host barrier against poxvirus infection. PLoS Pathog. 14, e1006884 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka M., Shimbo T., Kikuchi Y., Matsuda M., Kaneda Y., Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int. J. Cancer 126, 1982–1991 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Sivan G., Ormanoglu P., Buehler E. C., Martin S. E., Moss B., Identification of restriction factors by human genome-wide RNA interference screening of viral host range mutants exemplified by discovery of SAMD9 and WDR6 as inhibitors of the vaccinia virus K1L-C7L-mutant. MBio 6, e01122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., McFadden G., SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J. Virol. 89, 1925–1931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivan G., Glushakow-Smith S. G., Katsafanas G. C., Americo J. L., Moss B., Human host range restriction of the vaccinia virus C7/K1 double deletion mutant is mediated by an atypical mode of translation inhibition. J. Virol. 92, e01329-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., et al., A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio 4, e00385-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., Wennier S., Zhang L., McFadden G., M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J. Virol. 85, 3270–3282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L. K., Chai F., Li H. Y., Xiao G., Guo L., Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J. Proteome Res. 12, 2666–2678 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Wang Q., et al., SAMD9L inactivation promotes cell proliferation via facilitating G1-S transition in hepatitis B virus-associated hepatocellular carcinoma. Int. J. Biol. Sci. 10, 807–816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mekhedov S. L., Makarova K. S., Koonin E. V., The complex domain architecture of SAMD9 family proteins, predicted STAND-like NTPases, suggests new links to inflammation and apoptosis. Biol. Direct 12, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danot O., Marquenet E., Vidal-Ingigliardi D., Richet E., Wheel of life, wheel of death: A mechanistic insight into signaling by STAND proteins. Structure 17, 172–182 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Liberzon A., et al., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight M. J., Leettola C., Gingery M., Li H., Bowie J. U., A human sterile alpha motif domain polymerizome. Protein Sci. 20, 1697–1706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagamachi A., et al., Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell 24, 305–317 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Tesi B., et al., Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood 129, 2266–2279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jesus A. A., et al., Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J. Clin. Invest. 130, 1669–1682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch L., Exploring human genomic diversity with gnomAD. Nat. Rev. Genet. 21, 448 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Chefetz I., et al., Normophosphatemic familial tumoral calcinosis is caused by deleterious mutations in SAMD9, encoding a TNF-alpha responsive protein. J. Invest. Dermatol. 128, 1423–1429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Topaz O., et al., A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am. J. Hum. Genet. 79, 759–764 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asou H., et al., Identification of a common microdeletion cluster in 7q21.3 subband among patients with myeloid leukemia and myelodysplastic syndrome. Biochem. Biophys. Res. Commun. 383, 245–251 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Chen D. H., et al., Ataxia-pancytopenia syndrome is caused by missense mutations in SAMD9L. Am. J. Hum. Genet. 98, 1146–1158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narumi S., et al., SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat. Genet. 48, 792–797 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Pastor V. B., et al., Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica 103, 427–437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorcenco S., et al., Ataxia-pancytopenia syndrome with SAMD9L mutations. Neurol. Genet. 3, e183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray P. E., Bock V., Ziegler D. S., Wargon O., Neonatal Sweet syndrome: A potential marker of serious systemic illness. Pediatrics 129, e1353–e1359 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Riedl S. J., Li W., Chao Y., Schwarzenbacher R., Shi Y., Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434, 926–933 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Cheng T. C., Hong C., Akey I. V., Yuan S., Akey C. W., A near atomic structure of the active human apoptosome. eLife 5, e17755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaughan D., et al., Ataxia pancytopenia syndrome due to SAMD9L mutation presenting as demyelinating neuropathy. J. Peripher. Nerv. Syst. 25, 433–437 (2020). [DOI] [PubMed] [Google Scholar]
- 30.King-Robson J., et al., Ataxia-pancytopenia syndrome due to a de novo SAMD9L mutation. Neurol. Genet. 7, e580 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt E. K., Clavarino G., Ceppi M., Pierre P., SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Allenspach E. J., et al., Germline SAMD9L truncation variants trigger global translational repression. J. Exp. Med. 218, e20201195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong CK, et al. The UCSC Interaction Browser: Multidimensional data views in pathway context. Nucleic Acids Res. 41, W218–W224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thunström S., Axelsson M., Leukoencephalopathia, demyelinating peripheral neuropathy and dural ectasia explained by a not formerly described de novo mutation in the SAMD9L gene, ends 27 years of investigations - A case report. BMC Neurol. 19, 89 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho K., et al., Heterozygous mutations in OAS1 cause infantile-onset pulmonary alveolar proteinosis with hypogammaglobulinemia. Am. J. Hum. Genet. 102, 480–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheah J. J. C., et al., A novel germline SAMD9L mutation in a family with ataxia-pancytopenia syndrome and pediatric acute lymphoblastic leukemia. Haematologica 104, e318–e321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong J. C., et al., Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight 3, 121086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.