Significance
Wnt signaling plays essential roles in embryonic patterning, stem cell renewal, and cell cycle progression from G1 to S phase via the regulation of β-catenin target genes. Here, we show that Wnt signaling also promotes timely execution of mitosis. We demonstrate that the Wnt signaling transducer Dishevelled recruits the mitotic kinesin KIF2A and mediates its binding to the mitotic spindle. KIF2A is a microtubule depolymerase that controls chromosome alignment and congression during mitosis. Consequently, we found that inhibition of Wnt signaling leads to KIF2A-dependent chromosome congression and alignment delay in somatic and pluripotent stem cells.
Keywords: Wnt signaling, mitosis, Dishevelled, chromosome alignment, Kinesin-13
Abstract
Canonical Wnt signaling plays critical roles in development and tissue renewal by regulating β-catenin target genes. Recent evidence showed that β-catenin–independent Wnt signaling is also required for faithful execution of mitosis. However, the targets and specific functions of mitotic Wnt signaling still remain uncharacterized. Using phosphoproteomics, we identified that Wnt signaling regulates the microtubule depolymerase KIF2A during mitosis. We found that Dishevelled recruits KIF2A via its N-terminal and motor domains, which is further promoted upon LRP6 signalosome formation during cell division. We show that Wnt signaling modulates KIF2A interaction with PLK1, which is critical for KIF2A localization at the spindle. Accordingly, inhibition of basal Wnt signaling leads to chromosome misalignment in somatic cells and pluripotent stem cells. We propose that Wnt signaling monitors KIF2A activity at the spindle poles during mitosis to ensure timely chromosome alignment. Our findings highlight a function of Wnt signaling during cell division, which could have important implications for genome maintenance, notably in stem cells.
The canonical Wnt signaling pathway plays essential roles in embryonic development and tissue homeostasis (1, 2). In particular, Wnt signaling governs stem cell maintenance and proliferation in many tissues, and its misregulation is a common cause of tumor initiation (3, 4).
Wnt ligands bind Frizzled (FZD) receptors and the coreceptors low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6) (5). The activated receptor complexes cluster on Dishevelled (DVL) platforms and are internalized via caveolin into endosomes termed LRP6 signalosomes, which triggers sequential phosphorylation of LRP6 by GSK3β and CK1γ (6–10). LRP6 signalosomes recruit the β-catenin destruction complex, which contains the scaffold proteins AXIN1 and adenomatous polyposis coli, the kinases CK1α and GSK3β, and the E3 ubiquitin ligase β-TrCP (11). This recruitment inhibits GSK3β and releases β-TrCP, which leads to β-catenin stabilization and nuclear translocation in a IFT-A/KIF3A–dependent manner (12–16). LRP6 signalosomes mature into multivesicular bodies, sequestering the Wnt receptors together with GSK3β, thereby maintaining long-term activation of the Wnt pathway and promoting macropinocytosis (14, 17–21). In contrast to Wnt ligands, the Wnt inhibitor Dickkopf-related protein 1 (DKK1) induces the clathrin-dependent internalization and turnover of LRP5/6 and thereby abrogates canonical Wnt signaling (22).
LRP6 signalosome formation peaks in mitosis (23, 24). On the one hand, the LRP6 competence to respond to Wnt ligands is promoted during G2/M by a priming phosphorylation at its intracellular domain by CDK14/16 and CCNY/CCNYL1 (24, 25). On the other hand, CDK1 phosphorylates and recruits B-cell CLL/lymphoma 9 (BCL9) to the mitotic LRP6 signalosomes (23). BCL9 protects the signalosome from clathrin-dependent turnover, thereby sustaining basal Wnt activity on the onset of mitosis.
Mitotic Wnt signaling not only modulates β-catenin (24) but increasing evidence suggests that it promotes a complex posttranslational program during mitosis (26). For instance, we have shown that mitotic Wnt signaling promotes stabilization of proteins (Wnt/STOP), which is required for cell growth and ensures chromosome segregation in somatic and embryonic cells (23, 26–31). Particularly, basal Wnt/STOP activity maintains proper microtubule plus-end polymerization rates during mitosis, and its misregulation leads to whole chromosome missegregation (31, 32). Furthermore, mitotic Wnt signaling controls the orientation of the spindle (33) and promotes asymmetric division in stem cells through components of the LRP6 signalosome (34). Accordingly, several Wnt components functionally associate with centrosomes, kinetochores, and the spindle during mitosis (25, 33, 35, 36). Consequently, both aberrant up-regulation or down-regulation of Wnt signaling have been associated with chromosome instability (CIN) (31, 32, 35, 37), which is a hallmark of cancer (38). Despite the importance of these processes for tissue renewal and genome maintenance, the targets and specific functions of mitotic Wnt signaling remain largely uncharacterized.
Kinesin family member 2A (KIF2A) is a member of the kinesin-13 group (KIF2A,B,C) of minus-end microtubule depolymerases (39–41). KIF2A is essential for the scaling of the spindle during early development (42) and plays critical roles in neurogenesis by modulating both cilium disassembly and neuronal wiring (43–47). In dividing cells, KIF2A was thought to be required for the assembly of a bipolar spindle due to a small interfering RNA (siRNA) off-target effect (48, 49). Current evidence supports a role of KIF2A in microtubule depolymerization at the spindle poles, which can generate pulling forces on attached kinetochores, thereby ensuring the congression, alignment, and segregation of chromosomes (50–56). Genetic depletion of KIF2A in mouse leads to neonatal lethality and to severe brain malformations, including microcephaly (43, 44, 57). KIF2A recruitment to microtubules is tightly coordinated by several protein kinases (45, 47, 50–52, 58–60). For instance, phosphorylation of KIF2A at several sites by Polo-like kinase 1 (PLK1) stimulates its recruitment to and activity at the spindle (45, 58, 61). On the other hand, Aurora kinase A and B inhibit KIF2A activity and restrict its subcellular localization during mitosis (50, 58, 60).
Here, we show that mitotic Wnt signaling promotes chromosome congression and alignment in prometaphase by recruiting KIF2A to the spindle in both somatic cells and pluripotent stem cells. We found that KIF2A is recruited by the LRP6 signalosome during mitosis. Mechanistically, we identified that KIF2A clusters with DVL via the N-terminal and motor domains of the depolymerase. We show that Wnt signaling controls KIF2A interaction with PLK1, which is critical for KIF2A localization at the spindle poles. We propose that basal Wnt signaling ensures timely chromosome congression and alignment prior cell division by modulating the spindle minus-end depolymerization dynamics through KIF2A.
Results
Mitotic Wnt Signaling Recruits KIF2A to the Spindle.
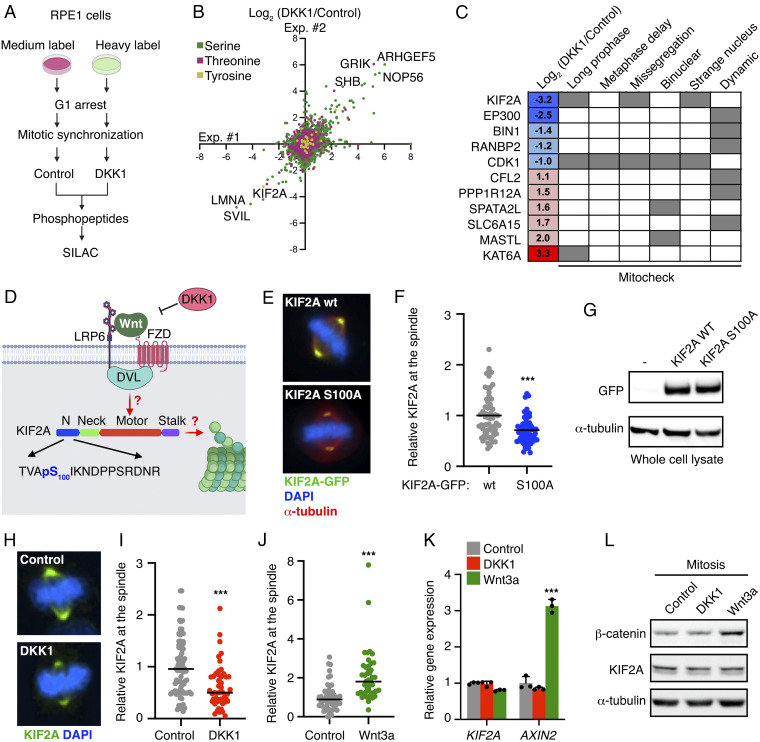
Mitosis is driven by posttranslational modifications of proteins, most notably phosphorylation (62). To identify direct targets and functions of Wnt signaling in mitosis, we performed a phosphoproteome-wide analysis using stable isotope labeling with amino acids in cell culture–based mass spectrometry (SILAC-MS) (Fig. 1A). In detail, we synchronized SILAC-labeled immortalized RPE1-hTert (RPE1) cells at G1/S phase, released the cells into the cell cycle and harvested cells synchronized in mitosis with the kinesin-5 inhibitor dimethylenastron (DME). To inhibit basal canonical Wnt signaling, synchronized cells were treated with DKK1 ligands for 1.5 h before harvesting (Fig. 1 A and B and SI Appendix, Fig. S1 A–C). Phosphorylated peptides were enriched from whole cell lysates, followed by their identification using ultra-high-performance liquid chromatography-tandem mass spectrometry (MS).
Fig. 1.
Mitotic Wnt signaling recruits KIF2A to the spindle. (A) Scheme of SILAC-MS analysis to identify changes in the phosphoproteome upon Wnt inhibition by DKK1 in mitotic cells. (B) Scatter plot comparing the differential counts of phosphopeptides of two biological replicates. Axes show the log2-fold change in phosphopeptide counts between control and DKK1 treatment in mitotic RPE1 cells. Phosphopeptides are color-coded for their modified residue. (C) Table containing the log2-fold change in the phosphopeptide counts of the indicated proteins from our performed MS screen, and the mitotic phenotypes identified for their siRNAs in the Mitocheck database. (D) Scheme showing the Wnt receptor complex and KIF2A. The phosphopeptide sequence identified in A and B is shown below with KIF2A phospho-S100, which is down-regulated by DKK1, highlighted in blue. (E) Representative immunofluorescence microscopy images showing overexpressed EGFP-KIF2A protein levels (green) in HeLa cells during metaphase. (F) Quantification of overexpressed KIF2A wt and S100A at the spindle of mitotic HeLa cells from E. Means of n > 61 cells per condition of a representative experiment of n = 3 independent experiments are shown. (G) Representative Western blots showing lysates from HeLa cells transfected as indicated. (H) Representative immunofluorescence microscopy images showing endogenous KIF2A protein levels (green) in RPE1 cells during metaphase treated as indicated 1.5 h before mitosis. (I and J) Quantification of spindle associated KIF2A in mitotic RPE1 cells upon 1.5 h treatments with control or DKK1 conditioned media (I) and with control or Wnt3a conditioned media (J). In I, mean of n > 43 cells per condition of a representative experiment of n = 3 independent experiments is shown. In J, mean of n > 44 cells per condition from n = 2 independent experiments that were pooled are shown. (K) qPCR analysis of KIF2A and AXIN2 expression levels in G2/M arrested RPE1 cells upon 1.5 h treatment with the indicated conditioned media. Data are displayed as mean ± SD of 3 replicates. (L) Representative Western blots from n = 3 independent experiments showing cytoplasmic lysates from RPE1 cells synchronized in mitosis and treated for 1.5 h with the indicated conditioned media.
We quantified 12,208 phospho-sites in two independent replicate experiments (Fig. 1B and Supplemental Dataset 1A). We observed a high quantitative reproducibility between the replicate experiments (Fig. 1B) and identified 21 up-regulated and 73 down-regulated phosphopeptides upon treatment with DKK1 (Supplemental Dataset 1B). Among the DKK1-regulated proteins, we found a moderate enrichment of proteins involved in cytoskeleton organization (P = 1.3 × 10−4) and regulation of cell cycle arrest (P = 6.7 × 10−4), suggesting an active role of Wnt signaling in mitotic progression (Fig. S1D and Supplemental Dataset 1C). To select Wnt target candidates with potential functions in mitosis, we examined the Mitocheck database (63), which provides a comprehensive dataset of mitotic phenotypes obtained upon a genome-wide siRNA screen. Five of our candidates (KIF2A, KAT6A, MASTL, CDK1, and SPATA2L) displayed specific mitotic phenotypes (Fig. 1C). For this study, we focused on KIF2A, which is required for chromosome congression, alignment and segregation of chromosomes (42, 50–55).
Our MS analysis revealed that DKK1 treatment during mitosis reduced the phosphorylation of KIF2A serine 100 by >9-fold (Fig. 1 B–D and Supplemental Dataset 1D). This uncharacterized phospho-site has been shown in previous proteomics analyses to be reduced upon PLK1 inhibition (61), and it is located in the N-terminal region, which modulates KIF2A recruitment to the spindle (50, 52, 61) (Fig. 1D). Notably, we found that mutation of serine 100 to alanine reduced KIF2A localization at the spindle, without affecting its total protein levels (Fig. 1 E and G). Hence, we examined by immunofluorescence whether Wnt signaling controls endogenous KIF2A localization during mitosis (Fig. 1D). Importantly, inhibition of Wnt signaling by DKK1 for 1.5 h during G2 to M-phase markedly reduced spindle-bound KIF2A in metaphase in RPE1 cells (Fig. 1 H and I), thereby phenocopying the KIF2A S100A phospho-inactive mutant. Conversely, exogenous stimulation with Wnt3a during G2/M boosted the localization of endogenous KIF2A at the spindle (Fig. 1J).
Mitotic Wnt signaling has been shown to regulate its targets by different mechanisms including via β-catenin–dependent transcription, Wnt-dependent stabilization of proteins (Wnt/STOP), and direct interactions with components of the LRP6 signalosome (25, 26). Neither DKK1 nor Wnt3a treatment impacted KIF2A transcriptional or protein levels (Fig. 1 K and L). Furthermore, we have previously reported that Wnt/STOP regulates microtubule plus-end dynamics in mitosis (31, 32). However, knockdown of KIF2A did not impact this function (SI Appendix, Fig. S1E). Taken together, these results exclude a role of Wnt/STOP and β-catenin in KIF2A regulation and suggest that Wnt signaling directly promotes KIF2A recruitment to the mitotic spindle.
KIF2A Is Recruited by LRP6 Signalosomes in Mitosis.
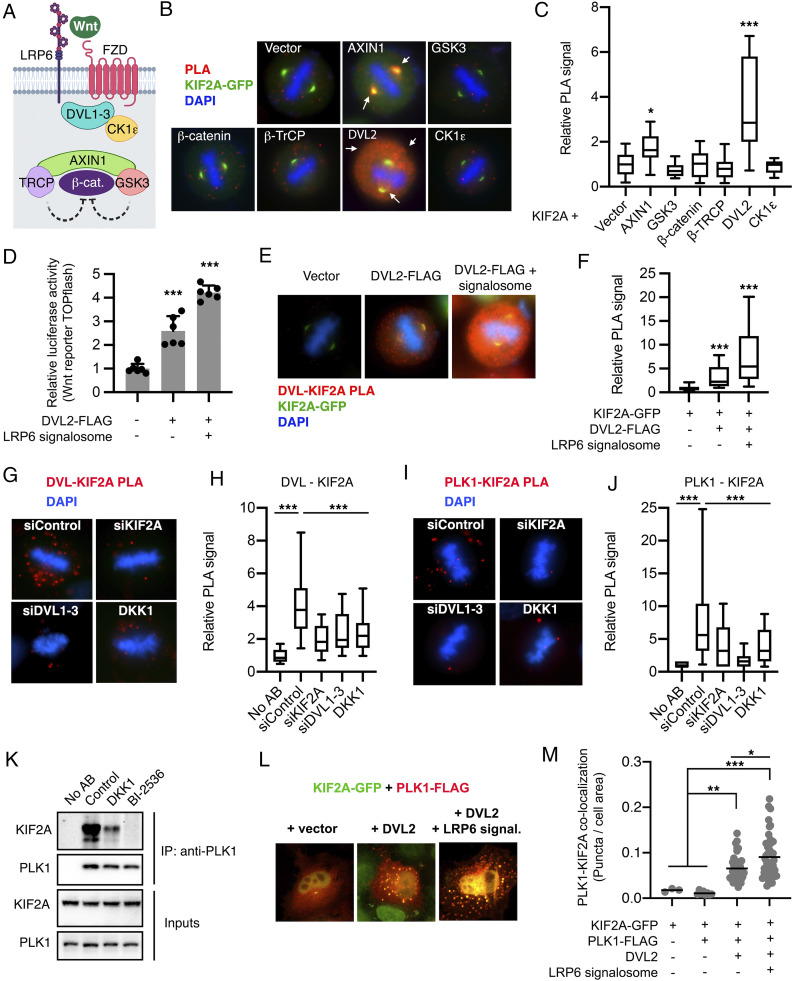
We next examined whether any Wnt signaling transduction components interact with KIF2A by proximity ligation assays (PLA) using validated antibodies in HeLa cells (Fig. 2A). We identified in situ KIF2A complexes with the signalosome scaffolding proteins AXIN1 at the spindle, and specially with DVL2 in the cytoplasm of mitotic cells (Fig. 2 B and C, complexes are shown in red). On the other hand, GSK3β, CK1ε, β-TRCP, and β-catenin only displayed background signal (Fig. 2 B and C). Importantly, coexpression of Wnt3a, FZD8, AXIN1, and LRP6, which induce LRP6 signalosomes (Fig. 2D) (10), boosted KIF2A recruitment to DVL2 during mitosis (Fig. 2 E and F). Furthermore, additional PLAs revealed in situ interaction between endogenous DVL (DVL1-3) and KIF2A in the cytoplasm of mitotic cells, which was further validated by knockdown of DVL1-3 and KIF2A. Critically, endogenous DVL–KIF2A in situ interaction in mitotic cells was significantly impaired also upon inhibition of Wnt signaling by DKK1 (Fig. 2 G and H).
Fig. 2.
KIF2A is recruited by the LRP6 signalosome in mitosis. (A) Scheme of the Wnt signaling transduction components analyzed in this figure. (B) Representative microscopy images of in situ PLAs in mitotic HeLa cells transfected with EGFP-KIF2A and the indicated FLAG-tagged constructs. (C) Box plot showing the quantification of the PLA signal from B normalized to the empty vector control. Data are displayed as median PLA signal flanked by upper and lower quartile, followed by the 10% to 90% extremes of n > 18 mitotic cells. (D) TOPflash reporter assays in HeLa cells upon cotransfection with empty vector, DVL2, or DVL2 together with the rest of the components of the LRP6 signalosome (Wnt3a/LRP6/FZD8/AXIN1). Data are displayed as mean ± SD of 6 biological replicates. (E and F) Representative microscopy images of in situ PLAs in mitotic HeLa cells transfected as indicated, and its corresponding box plot showing the quantification of the PLA signal. Data are displayed as indicated in C for n > 65 cells. LRP6 signalosome: Wnt3a/LRP6/FZD8/AXIN1. (G) Representative microscopy images of in situ PLAs of endogenous proteins in mitotic HeLa cells. (H) Box plot showing the quantification of the endogenous PLA signal from G. Data are displayed as median PLA signal flanked by upper and lower quartile, followed by the 10% to 90% extremes of n > 26 mitotic cells representing n = 2 independent experiments. (I and J) Representative microscopy images of in situ PLAs of endogenous proteins in mitotic HeLa cells quantified as indicated in H. (K) Representative Western blots from n = 3 independent experiments showing coimmunoprecipitations of endogenous proteins from HeLa cells synchronized in prometaphase and treated with control, DKK1 or the PLK1 inhibitor BI-2536, as indicated in the methods section. (L) Representative immunofluorescence microscopy images from n = 2 independent experiments showing HeLa cells transfected with EGFP-KIF2A (green), PLK1-FLAG (stained red) and the indicated constructs. (M) Dot plots showing all single cells of n = 60 cells/condition from L, that displayed colocalization of EGFP-KIF2A and PLK1-FLAG in interphase puncta, and the number of puncta per cell area including their mean values.
To further validate the association between DVL and KIF2A, we performed immunofluorescence experiments. As shown previously, ectopic expression of DVL leads to the formation of puncta in interphase representing high molecular complexes through the DIX domains of DVL, which mimic LRP6 signalosomes (10, 64) (SI Appendix, Fig. S2A). Notably, DVL1-3 recruited KIF2A to these puncta (SI Appendix, Fig. S2A), further supporting a role of the signalosomes recruiting KIF2A through DVL (Fig. 2F). Next, we examined which domains of KIF2A are involved in its recruitment to the signalosomes (SI Appendix, Fig. S2B). Deletion of the KIF2A Neck or Stalk domains, which modulate the motor activity and dimerization respectively (59), did not impair KIF2A localization in DVL2 puncta. On the other hand, deletion of the N-terminal (KIF2AΔN) or the motor domain (KIF2AΔmotor and KIF2A N-term) abolished the colocalization of KIF2A in DVL2 puncta (SI Appendix, Fig. S2 C and D). Interestingly, both domains integrate signals that modulate KIF2A activity and localization, including by PLK1 (45, 47, 50, 52, 58, 61). Strikingly, S100A mutation largely abolished KIF2A recruitment to DVL2 puncta (SI Appendix, Fig. S2 E and F), phenocopying the deletion of the whole N-terminal domain of KIF2A (SI Appendix, Fig. S2C). Thus, we conclude that KIF2A requires its motor domain and the serine 100 at the N-terminal domain to functionally interact with DVL.
DVL is known to associate with PLK1 during mitosis (33). Importantly, PLK1 inhibition was shown to reduce both KIF2A activity and S100 phosphorylation (45, 51, 58, 61). Hence, we investigated whether Wnt mediates KIF2A interaction with PLK1. First, we identified the association of endogenous KIF2A and PLK1 in the cytoplasm of mitotic cells by PLA experiments, which was strongly reduced upon knockdown of DVL1-3 or DKK1 treatment (Fig. 2 I and J). Second, we performed coimmunoprecipitation experiments with endogenous proteins from cells synchronized in prometaphase. As recently reported (51), PLK1 coprecipitated KIF2A (Fig. 2K). Importantly, treatment of synchronized mitotic cells with DKK1 or the PLK1 inhibitor BI-2536 strongly reduced PLK1–KIF2A interaction (Fig. 2K). In agreement with these results, we found that ectopic expression of LRP6 signalosome components was sufficient to recruit PLK1 to DVL (SI Appendix, Fig. S3 A and B), and to promote PLK1-KIF2A colocalization in DVL puncta in interphase cells (Fig. 2 L and M).
Serine 100 does not match a PLK1 consensus phospho-site (SI Appendix, Fig. S3C) (65). However, PLK1 directly phosphorylates several sites in KIF2A, including the N-terminal and the motor domains (45, 47, 61), which we found to be required for KIF2A interaction with DVL (SI Appendix, Fig. S2 C and D). Given that PLK1 is required for KIF2A recruitment to microtubules (51, 58, 61) and that PLK1 inhibition reduces serine 100 phosphorylation, we suggest that PLK1 functions upstream of other kinase(s) regulating KIF2A activity and localization, including through serine 100.
Taken together, these results indicate that LRP6 signalosomes recruit KIF2A and promote its interaction with PLK1.
Wnt Signaling Promotes Chromosome Congression and Alignment through KIF2A.
Both the N-terminal and motor domains of KIF2A are required for its localization and activity (50, 52, 59), and are essential for the functional interaction with DVL (SI Appendix, Fig. S2 C and D). Besides, our data indicate that inhibition of mitotic Wnt signaling reduces KIF2A association with the spindle and PLK1 (Fig. 1 H and I and 2 I and J). Hence, we next investigated whether Wnt signaling controls chromosome congression and alignment through KIF2A.
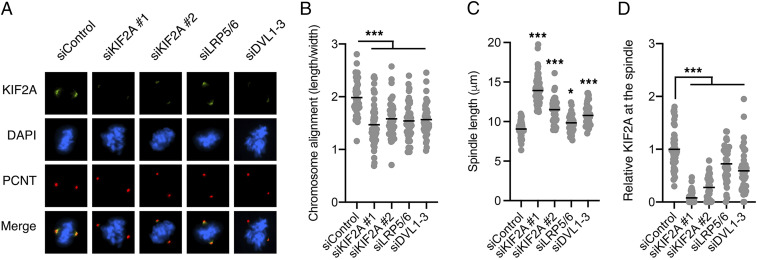
Knockdown of KIF2A by two independent siRNAs resulted in elongated spindles and misaligned chromosomes or incomplete chromosome congression in mitosis (Fig. 3 A–D and SI Appendix, Fig. S4A), similarly as reported previously (51, 52, 54, 58). Knockdown of the signalosome components LRP5/6 and DVL1-3 reduced the recruitment of KIF2A to the mitotic spindle in RPE1 cells (Fig. 3 A and D and SI Appendix, Fig. S4A), as we showed for DKK1 (Fig. 1 H and I). Knockdown of LRP5/6 and DVL also led to elongated spindles and misaligned chromosomes (Fig. 3 A–C), which suggests a role of Wnt signaling in promoting chromosome congression and alignment. On the other hand, knockdown of KIF2A, LRP5/6, or DVL neither promoted centrosome duplication nor impacted bipolar spindle assembly (SI Appendix, Fig. S4 B and C). These results are consistent with the function of KIF2A in spindle depolymerization (42, 43, 49, 51, 52, 55, 58, 59), which generates pulling forces on attached kinetochores (54, 56). Accordingly, knockdown of KIF2A, LRP5/6, or DVL resulted in recruitment of the spindle checkpoint protein BUBR1 to the kinetochores of misaligned chromosomes (SI Appendix, Fig. S4D), which suggests a loss of tension at the sister kinetochores (66).
Fig. 3.
Basal Wnt signaling promotes chromosome congression and alignment. (A) Representative immunofluorescence microscopy images from n = 4 independent experiments showing mitotic RPE1 cells transfected with the indicated siRNAs and stained with DAPI and antibodies against KIF2A and pericentrin (PCNT). (B–D) Quantification of chromosome alignment (median), distance between spindle poles (mean), and KIF2A immunofluorescence (mean) of all single cells from A. In B, the length occupied by the chromosomes at the metaphase plate was divided by the width toward the spindle poles. In C, spindle length was measured between pericentrin-stained centrosomes. n > 25 mitotic cells per condition.
Defects in chromosome congression or alignment can lead to CIN (56). However, recent evidence on loss of KIF18A shows that chromosome misalignment does not necessarily lead to aneuploidy, unless kinetochore attachments are affected (67). We found that most cells (∼70%) lost one or more chromosomes upon prolonged loss of KIF2A or LRP5/6 over 30 d (SI Appendix, Fig. S4 E and F). Our data are consistent with previous evidence on the roles of Wnt signaling in promoting chromosomal stability (31). However, given the complex functions of both KIF2A and WNT during mitosis, it is likely that different mechanisms converge into their roles in chromosome stability.
To confirm a role of Wnt signaling in chromosome congression and alignment, we performed live cell imaging experiments (SI Appendix, Fig. S5). Knockdown of KIF2A or LRP5/6 in RPE1 cells stably transfected with EGFP-H2B resulted in 1) a mild delay in the congression of all chromosomes to the spindle equator after nuclear envelope breakdown (NEB) (SI Appendix, Fig. S5 A–C, referred to as congression delay), as well as 2) one or more chromosomes being clearly detached from the central forming metaphase plate (SI Appendix, Fig. S5 A–C, referred to as alignment/metaphase delay). In both instances, RPE1 cells eventually completed congression and progressed to anaphase.
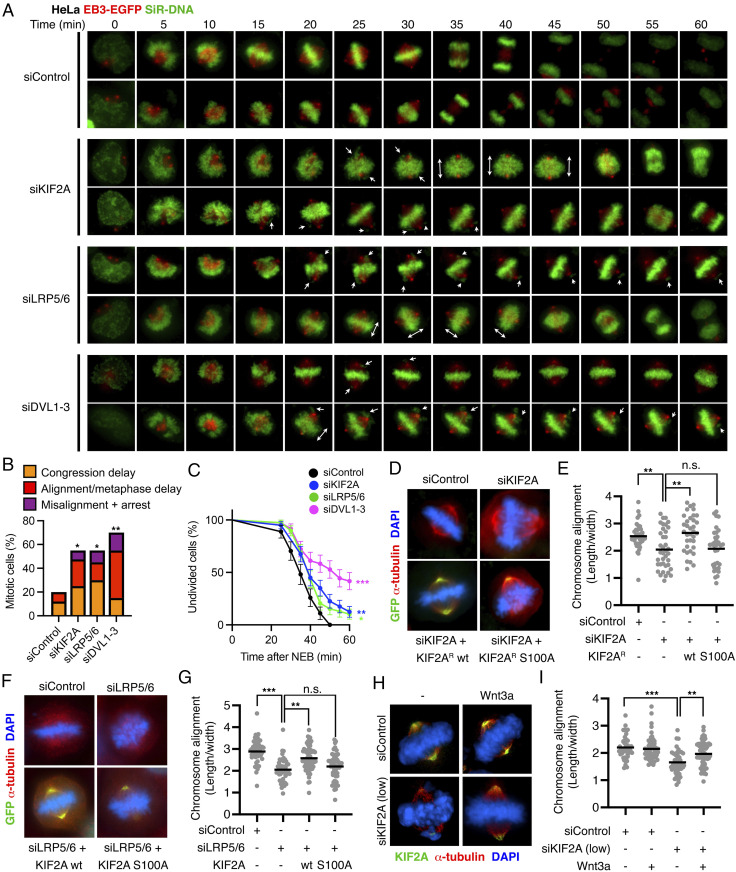
We also investigated HeLa cells, stably transfected with EB3-EGFP and stained with SiR-DNA, which allowed us to monitor centrosome and spindle orientation during live cell imaging. In these cells, knockdown of KIF2A led to congression or alignment delay in over 50% of divisions (Fig. 4 A and B and SI Appendix, Fig. S6A), resulting in longer mitoses, or even mitotic arrest in 10% of the cases (Fig. 4 B and C). We obtained similar results upon knockdown of LRP5/6 and DVL (Fig. 4 A–C), as well as DKK1 treatment (SI Appendix, Fig. S6 B and C). The more severe phenotypes in HeLa compared to RPE1 cells could be due to additional stress derived from aneuploidy in the cancer cell line, which is also reflected in higher basal errors.
Fig. 4.
Wnt signaling promotes chromosome congression and alignment through KIF2A. (A) Representative time lapses of mitotic HeLa cells stably expressing EB3-EGFP, which marks the microtubule plus ends and centrosomes. Cells were transfected with the indicated siRNAs and stained with SiR-DNA before imaging to label the chromosomes. (B) Analysis of congression and alignment delay in n = 20 HeLa cells from A. (C) Analysis of mitotic progression from NEB to anaphase onset in HeLa cells from A. (D–I) Quantification of chromosome alignment in mitotic HeLa cells transfected with siRNA, cotransfected with empty or EGFP-KIF2A plasmid or treated as indicated for 1.5 h. In D, F, and H, representative immunofluorescence microscopy images of n ≥ 2 independent experiments are shown. In E, G, and I, the length occupied by the chromosomes at the metaphase plate was divided by the width toward the spindle poles in n > 35 mitotic cells per condition. All single cells and the median values are shown.
Analyses of metaphases in fixed HeLa cells further confirmed that knockdown of KIF2A in HeLa cells leads to chromosome misalignment (Fig. 4 D and E). We generated siRNA insensitive KIF2A constructs and found that wild-type (wt) KIF2A, but not KIF2A S100A, rescued siKIF2A induced defects (Fig. 4 D and E), confirming the specificity of the siRNA, as well as the importance of the S100 phospho-site for KIF2A activity. Inhibition of Wnt signaling by DKK1 or knockdown of LRP5/6 phenocopied siKIF2A effects leading to chromosome misalignment in mitosis (Fig. 4 F and G and SI Appendix, Fig. S6 D and E). Importantly, ectopic expression of wt KIF2A, but not KIF2A S100A, rescued both DKK1 and siLRP5/6 effects (Fig. 4 F and G and SI Appendix, Fig. S6 D and E). Next, we partially knocked down KIF2A, which still resulted in chromosome misalignment (Fig. 4 H and I and SI Appendix, Fig. S6F). In agreement with a role of mitotic Wnt signaling controlling KIF2A recruitment to the spindle, acute Wnt3a treatment in mitosis restored KIF2A levels at the spindle under low siKIF2A conditions and rescued the misalignment (Fig. 4 H and I and SI Appendix, Fig. S6F).
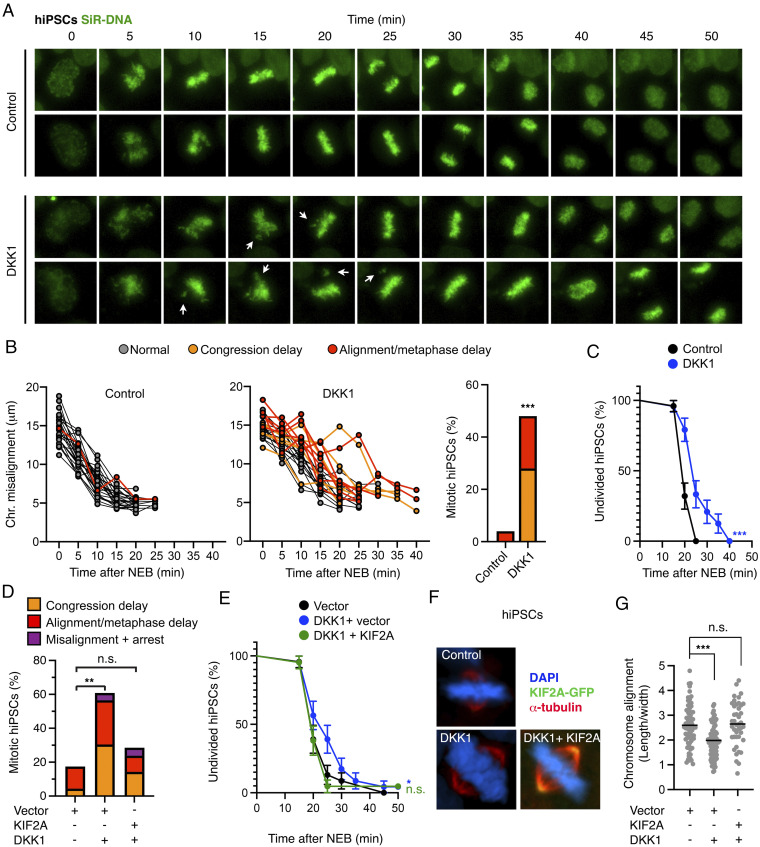
Finally, given the importance of Wnt signaling in stem cell proliferation and maintenance (1, 68), we investigated whether Wnt also promotes faithful execution of mitosis in human induced pluripotent stem cells (hiPSCs) through the regulation of chromosome congression and alignment. Inhibition of basal Wnt signaling in hiPSCs by DKK1 resulted in 1) chromosome congression delay in 28% of divisions compared to 0% of controls and 2) the misalignment of single chromosomes in 20% of divisions compared to 4% of controls (Fig. 5 A and B). Consequently, DKK1 treatment delayed progression from NEB to anaphase (Fig. 5C) but did not promote mitotic arrest. Notably, ectopic expression of KIF2A was sufficient to rescue chromosome congression and alignment delay induced by DKK1 (Fig. 5 D and E and SI Appendix, S7 A and B). As in the case of RPE1 and HeLa cells, analyses of fixed hiPSCs confirmed the induction of chromosome misalignment upon inhibition of autocrine Wnt signaling by DKK1 as well as by the Wnt secretion inhibitor LGK-974 (SI Appendix, Fig. S7 C and D). Ectopic expression of KIF2A also rescued the chromosome misalignment induced by DKK1 in fixed hiPSCs in immunofluorescence experiments (Fig. 5 F and G).
Fig. 5.
Wnt signaling promotes chromosome congression and alignment via KIF2A in hiPSCs. (A) Representative time lapses of mitotic hiPSCs stained with SiR-DNA and treated with control or DKK1 conditioned medium. (B) Analysis of congression and alignment delay in n = 25 hiPSCs from A. (C) Analysis of mitotic progression from NEB until anaphase in hiPSCs from A. (D) Analysis of congression and alignment delay in n > 20 hiPSCs transfected with empty vector or EGFP-KIF2A and treated with or without DKK1. See images and detailed analyses in SI Appendix, Fig. S7 A and B. (E) Analysis of mitotic progression from NEB until anaphase in hiPSCs from D and SI Appendix, Fig. S7 A and B. (F) Representative immunofluorescence microscopy images of mitotic hiPSCs transfected with empty vector or EGFP-KIF2A and treated with the Wnt inhibitor DKK1 for 1.5 h. (G) Quantification of the chromosome alignment from F. The length occupied by the chromosomes at the metaphase plate was divided by the width toward the spindle poles in n > 42 mitotic cells per condition. The median value is indicated. Representative images and analyses of n = 3 independent experiments.
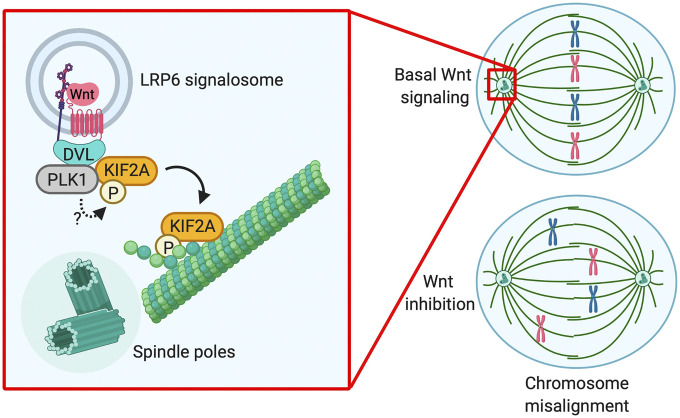
Taken together, these results indicate that Wnt signaling promotes chromosome congression and alignment by recruiting KIF2A to the spindle in somatic cells and pluripotent stem cells (Fig. 6).
Fig. 6.
Wnt signaling induces KIF2A interaction with and activation by PLK1 to promote congression and alignment of chromosomes during mitosis. Scheme of the suggested model for Wnt modulating chromosome congression in mitosis. Basal Wnt signaling during G2/M induces DVL clustering in LRP6 signalosomes, which serves as platforms for PLK1 and KIF2A interaction. Downstream of PLK1, phosphorylation of KIF2A at S100 by an unknown kinase further stabilizes the interaction, leading to KIF2A activation and transfer to the spindle poles. Finally, by monitoring KIF2A at the mitotic spindle, Wnt signaling ensures timely chromosome congression and alignment in mitosis.
Discussion
The roles of Wnt signaling in G1/S progression are well established (25), but its functions during cell division are poorly understood. The main discovery of our study is that Wnt signaling promotes chromosome congression and alignment during mitosis by controlling KIF2A localization to the spindle poles. Our results support the following model: Wnt activity during G2/M promotes DVL clustering in LRP6 signalosomes. DVL recruits KIF2A through its motor and N-terminal domains, thereby facilitating its interaction with and activation by PLK1. Downstream of PLK1, phosphorylation of KIF2A at S100 by an unknown kinase further promotes the interaction with DVL and subsequent binding to the spindle. Finally, by monitoring KIF2A levels at the spindle poles, Wnt ensures timely chromosome alignment (Fig. 6).
Our results further highlight a critical role of canonical Wnt signaling in modulating spindle dynamics during mitosis by different mechanisms. First, our previous research identified that Wnt/STOP signaling increases microtubule plus-end dynamics, which leads to whole chromosome missegregation and aneuploidy (31, 32). Second, this study shows a role of Wnt regulating KIF2A, a depolymerase functioning at the microtubule minus ends, i.e., at the spindle poles. Notably, both mechanisms converge to ensure proper chromosome alignment and segregation (39, 52, 59, 69). Interestingly, recent evidence on KIF18A shows that chromosome misalignment does not necessarily lead to aneuploidy (67). Hence, it would be important to further characterize the different processes controlled by KIF2A during mitosis and assess their relationship with chromosome missegregation and CIN. A standing question is whether the Wnt-mediated control of KIF2A plays a role in spindle scaling during development (42), especially considering that Wnt/STOP signaling is also required during the first embryonic cleavages (30). Furthermore, it is possible that these mechanisms may contribute to the Wnt conserved roles in spindle orientation and asymmetric cell division (33, 34, 70–72).
Another important conclusion from our study is that Wnt signaling not only regulates proliferation and self-renewal (68, 73), but also controls faithful execution of mitosis in pluripotent stem cells. Our research is consistent with—and suggests a mechanism for—previous evidence showing that Wnt secretion controls genome stability in mouse embryonic stem cells (74). Interestingly, it has been shown recently that self-renewal of pluripotent cells is tightly linked to their genome integrity (75). Hence, mitotic Wnt signaling may contribute to the known TCF/β-catenin functions in stem cell renewal by promoting faithful execution of mitosis.
Finally, our data indicates that KIF2A can be recruited by DVL also in interphase. Given the critical roles of KIF2A in neurogenesis by modulating both cilium disassembly and neuronal wiring (43–47), which are also key biological processes monitored by Wnt signaling (71, 76, 77), it would be important to explore whether Wnt modulates KIF2A’s functions beyond mitosis.
Methods
Cell Culture.
HeLa, HEK293T, and L cells (American Type Culture Collection [ATCC]) were cultured in DMEM (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin. RPE1-hTert cells (ATCC) were cultured in DMEM-F12 medium (Gibco) supplemented with 10% FBS, 1% penicillin/streptomycin and 4% Sodium Bicarbonate. All cells were grown at 37 °C with 5% CO2. HeLa Kyoto cells, stably expressing EB3-EGFP, were purchased from CLS (depositor: J. Ellenberg). The RPE1-hTert were stably transfected using H2B-EGFP (a kind gift from S. Gaubatz, University of Würzburg, Germany) and single cell clones were selected with medium containing Geneticin (G418). The hiPSCs were a gift from Kyung-Min Noh and were cultured in Essential E8 Medium (Thermo Scientific) supplemented with Penicilin/Streptomycin (78). Revitacell Supplement (Thermo Scientific) was added for the first 24 h after plating. Medium was changed every day and cells were split every 4 to 5 d using versene solution. All the experiments were carried out with hiPSCs below passage 13.
Control and Wnt3a conditioned media were obtained from stably transfected L cells (ATCC) as described in the manufacturer’s protocol. DKK1 conditioned medium was produced by transiently transfecting HEK293T cells with pCS-FLAG-DKK1 plasmid (a gift from C. Niehrs) using the calcium-phosphate method (15 µg plasmid per 10 cm dish). Medium was changed 7 h post transfection and harvested 48 h after transfection.
To enrich cells in prometaphase, cells were first synchronized in G1/S for 24 h with 2 mM thymidine. After washing and releasing for 5 h in culture medium, cells were synchronized in mitosis using 2 µM DME for 4 h. Treatments were applied in the presence of DME 1.5 h prior to harvesting.
siRNA Transfection.
HeLa cells were transfected with 30 nM and RPE1 cells with 50 nM siRNA (Sigma Aldrich or Dharmacon) using Lipofectamine RNAiMAX transfection reagent (Thermo Scientific). 48 h after transfection, cells were further analyzed. The following siRNAs were used: siControl (MISSION siRNA Universal Negative Control #1 (SIC001) for imaging experiments and siGENOME Non-Targeting siRNA #5 (D-001210-05-20) for Western blots), siKIF2A #1 (SASI_Hs02_00319177, GAAGCUAUUCUUGAGCAAA), siKIF2A #2 (GGAAUGGCAUCCUGUGAAA) (58), siKIF2C (SASI_Hs01_00111628), siLRP5/6 (1:1 mix of SASI_Hs01_00086821 and SASI_Hs01_00039493), and siDVL1-3 (mix of 1:1:1 SASI_Hs01_00142403, SASI_Hs01_00142404 and SASI_Hs01_0004203).
DNA Transfection and Expression Constructs.
Cells were transfected with 250 to 800 ng DNA per well of a 12-well plate or 500 ng per well of a 6-well plate using X-tremeGENE 9 DNA transfection reagent (Roche), and harvested after 24 h.
FLAG-GSK3, FLAG-β-catenin, FLAG-DVL1, FLAG-mDvl2, FLAG-DVL3, FLAG-CK1ε, LRP6, mFzd8, AXIN1, Myc-DVL2, pCS2+, pCS-FLAG-DKK1, TOPflash and Renilla plasmids were kindly provided by C. Niehrs (DKFZ Heidelberg, Germany). pEGFP-KIF2A (#52401), 3xFLAG-DVL2 (#24802), FLAG-AXIN1 (#109370) and FLAG-β-TrCP (#10865) plasmids were obtained from Addgene. The KIF2A-phospho-mutant (KIF2A S100A), truncation mutants and siKIF2A resistant KIF2A constructs were generated by PCR and blunt-end cloning. More details and the primers that were used are included in the SI Appendix, Supplemental Methods.
MS Sample Preparation and Analysis.
RPE1 cells were seeded in 15 cm dishes, synchronized in prometaphase, and treated with in-house produced control (Medium) or DKK1 (Heavy) SILAC media (79) for 1.5 h before harvesting. The experiment was performed in duplicates. Phosphoproteome analysis was performed as described previously (80). In brief, proteins were extracted by full lysis of cells, digested by protease, enriched for phosphopeptides with TiO2 beads, and analyzed by MS. For the downstream analysis, only peptides with fold changes > 2 in both replicate experiments were considered significant. Further details on the MS procedures are provided in the SI Appendix, Supplemental Methods.
Immunofluorescence.
RPE1 or HeLa cells were seeded on coverslips in 12-well plates. Cells were transfected with siRNA for 48 h, transfected with DNA for 24 h and treated with conditioned medium for 1.5 h, as indicated. Cells were fixed with 2% paraformaldehyde in PBS and stained with antibodies against the indicated proteins and DAPI.
The following primary antibodies were used: rabbit anti–α-tubulin (Sigma Aldrich, SAB4500087), mouse anti-KIF2A (Santa Cruz, sc-271471), rabbit anti-Pericentrin (Abcam, ab4448), rabbit anti-FLAG (Invitrogen, PA1-984B), rabbit anti-DVL 2 [N1N3] (GeneTex, GTX111156), mouse anti-BUBR1 (BD Bioscience, 612502), guinea pig anti-CENPC (MBL, PD03). The following secondary antibodies were employed: anti-mouse AF488 (Thermo Scientific, A21202), anti-rabbit AF488 (Thermo Scientific, A21206), anti-mouse AF594 (Thermo Scientific, A21203), anti-rabbit AF594 (Thermo Scientific, A21207), and anti–guinea pig Cy3 (Millipore, AP193C).
Coverslips were imaged with a Nikon Eclipse Ti using a 60× objective with oil immersion and the NIS Elements software. Data were analyzed using ImageJ 2.0.0. To access KIF2A levels at the spindle, a threshold for the KIF2A signal was set to only detect KIF2A at the poles. To determine chromosome congression and alignment in mitosis, the length of the forming metaphase plate was divided by its width toward the spindle poles. Cells for which not both poles were distinctively visible, including tilted cells, were excluded from the analyses.
To assess KIF2A recruitment to DVL interphase puncta (SI Appendix, Fig. S2), the ratio of cells showing KIF2A in puncta was quantified relative to the total number of cells expressing DVL. To detect colocalization of KIF2A and PLK1 in interphase puncta in relation to signalosome overexpression (Fig. 2 L and M and SI Appendix, S3 A and B) the number of puncta per cell was counted and divided by its total cell area.
Western Blotting.
For Western blotting, cells were lysed in full lysis buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.05% SDS, 1 mM β-mercaptoethanol, 2 mM EDTA, 1× protease phosphatase inhibitor mixture [Thermo Scientific]) or cytoplasmic lysis buffer (PBS supplemented with 0.05% saponin, 10 mM β-mercaptoethanol, 2 mM EDTA, 1× protease phosphatase inhibitor mixture). The cleared lysates were mixed with 4× NuPAGE LDS sample buffer (Thermo Scientific) containing 50 mM DTT, resolved on 8% NuPAGE gels and transferred to nitrocellulose membranes. For Western blot experiments the following antibodies were used: mouse anti–α-tubulin (Sigma Aldrich, T9026), mouse anti–β-catenin (BD Bioscience, 610153), mouse anti-KIF2A (Santa Cruz, sc-271471), rabbit anti-LRP6 (Cell Signaling, 2560S), rabbit anti-DVL (Merck Millipore, ABD122), chicken anti-GFP (Abcam, ab13970), and mouse anti-FLAG M2 (Sigma Aldrich, F1804).
Real-Time PCR.
Cells were synchronized as described above, treated with control, DKK1 or Wnt3a conditioned medium for 1.5 h and harvested. Total cellular RNA was extracted by using the RNeasy Plus Mini Kit (QIAGEN) and cDNA was synthesized from 500 ng of total RNA using the Bioline SensiFAST cDNA Synthesis Kit. Quantitative PCR was performed with the SensiFAST SYBR Hi-ROX Kit (Bioline) using a StepOnePlus 96-well plate reader (Applied Biosystems). Gene expression was normalized to GAPDH. The primers used in this study are described in the SI Appendix, Supplemental Methods.
Proximity Ligation Assay (PLA).
HeLa cells were seeded on coverslips in 12-well plates. Cells were transfected with siRNA for 48 h, transfected with DNA for 24 h and treated with conditioned medium for 1.5 h, as indicated. Cells were fixed with 2% paraformaldehyde in PBS and PLAs were performed with the Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma Aldrich) according to the manufacturer’s protocol.
For the overexpression PLAs, EGFP-KIF2A and a FLAG-tagged component were overexpressed and the following antibodies were used: rabbit anti-GFP (Abcam, ab290), mouse anti-FLAG M2 (Sigma Aldrich, F1804). For the endogenous PLAs, mouse anti-KIF2A (Santa Cruz, sc-271471), rabbit anti-DVL (Merck Millipore, ABD122), and rabbit anti-PLK1 (Abcam, ab14209) were used.
Coverslips were imaged with a Nikon Eclipse Ti using a 60× objective with oil immersion and the NIS Elements software. Data were analyzed using ImageJ 2.0.0. The PLA signal was quantified by the number of PLA complexes per cell (Fig. 2 C, H, and J) or the total fluorescence signal per cell (Fig. 2F). Values are displayed relative to the control.
Wnt Reporter Assay (TOPflash).
RPE1 or HeLa cells were seeded into 96-well plates. RPE1 cells were transfected with 50 ng DNA per well, including 5 ng TOPflash luciferase and 1 ng Renilla luciferase. HeLa cells were transfected with 100 ng of DNA containing 30 ng TOPflash luciferase, 20 ng Renilla luciferase and, where indicated, 25 ng Myc-DVL2 or 25 ng Myc-DVL2, 22 ng LRP6, 2 ng mouse Fzd8 and 1.25 ng AXIN1. An empty vector (pCS2+) was added to fill up the reactions to the final DNA mass. The transfection was performed using X-tremeGENE 9 DNA transfection reagent (Roche) according to manufacturer instructions. After 24 h, cells were treated overnight as indicated, and lysed with 1× Passive Lysis Buffer (Promega) on a shaker for 15 min at 4 °C. Lysates were analyzed using a Dual-Luciferase Reporter Assay System (Promega) with a Tecan Microplate Reader (Tecan Infinite M1000). TOPflash signals were normalized to the Renilla reporter and Wnt activity was calculated relative to the control condition.
Immunoprecipitation.
HeLa cells were synchronized in prometaphase as described above, treated with control medium, DKK1 medium or 1 µM BI-2536 (PLK1 inhibitor, Selleckchem) for 1.5 h, and endogenous PLK1–KIF2A interaction was analyzed by immunoprecipitation (IP). One day before the IP, protein G agarose (Roche) was washed three times with IP lysis buffer (25 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, 5% glycerol, 1× protease/phosphatase inhibitor mixture [Thermo Scientific], 4 units/mL DNase I [NEB]) and mixed 1:1 with IP lysis buffer containing 5% BSA. For each IP with roughly 1 mg protein lysate, 5 µg anti-PLK1 antibody (Santa Cruz, sc-17783) was mixed with 20 μL of washed beads in BSA-lysis buffer and incubated in rotation at 4 °C overnight. The next day, cells were lysed with IP lysis buffer for 15 min on ice. The lysates were cleaned with 20 μL of washed agarose for 1 h at 4 °C in rotation. One-thirteenth of each lysate was saved as the “input” and prepared with NuPAGE sample buffer including DTT for gel electrophoresis by 10 min heating at 95 °C. The antibody-agarose-complex from the day before was added to the lysates and IP performed for 4 h at 4 °C in rotation. Beads were washed three times with lysis buffer containing 10 µM MG132 and proteins were eluted in 1× NuPAGE sample buffer with 12.5 mM DTT for 10 min at 95 °C. Samples were analyzed by Western blotting as described above using rabbit anti-PLK1 (Bethyl Laboratories, A300-251A) and rabbit anti-KIF2A (Novus Biologicals, NB500-180).
Bipolar Spindle Maintenance and Centrosome Amplification.
The analysis of bipolar spindle maintenance was done as previously described (49). In detail, HeLa cells were seeded in 12-well plates with coverslips and transfected with 30 nM siRNA against KIF2A, LRP5/6, DVL1-3, as well as the positive control KIF2C (MCAK). One day after transfection, cells were synchronized in G1/S by a 2.5 mM thymidine treatment for 24 h, released for 7.5 h, and synchronized in metaphase with 5 µM MG132 for 2.5 h. Lastly, the Eg5 inhibitor DME (2 µM) was applied for 1.5 h. Cells were fixed with 2% paraformaldehyde and stained with mouse anti–α-tubulin (Sigma Aldrich, T9026), rabbit anti-Pericentrin (Abcam, ab4448), and DAPI. Coverslips were imaged with a Nikon Eclipse Ti using a 60× objective with oil immersion and the NIS Elements software. Data were analyzed using ImageJ 2.0.0. The presence of monopolar spindles as well as number of centrosomes per cell was quantified and displayed relative to the total number of cells analyzed. Note that Eg5 inhibition induces monopolar spindles in prometaphase; however, when cells are arrested in metaphase before its application, formation of monopolar spindles requires a concomitant loss of a component required for spindle maintenance.
Karyotype Analysis.
RPE1 cells were transfected every 5 d with 10 nM siRNA against KIF2A, LRP5/6 and scrambled (Control) over 30 d and karyotype analyses were performed as described previously (31). In brief, cells were treated with 2 μM DME for 4 h to enrich for mitotic cells. Cells were pelleted, washed with PBS, and incubated in hypotonic medium (40% DMEM-F12, 60% H2O) at RT for 15 min. Cells were fixed in Carnoy’s solution (methanol:acetic acid = 3:1). Chromosomes were spread onto glass slides and stained with Giemsa solution. Chromosome number variability was determined by chromosome counting.
Live Cell Imaging.
RPE1 cells stably expressing H2B-EGFP, HeLa cells stably expressing EB3-EGFP or wt hiPSCs were seeded in 10-well CELLview slides (Greiner Bio-One). Where indicated, cells were transfected with 30 nM siRNA for 48 h. For imaging, medium was replaced by DMEM-F12 (RPE1) or DMEM (HeLa) medium containing Hepes and no phenol red (Gibco), supplemented with 10% FBS, 1% penicillin/streptomycin and 1% sodium pyruvate. HiPS cells were cultured in Essential E8 Medium (Thermo Scientific), supplemented with penicillin/streptomycin, during image acquisition. Cells were incubated with 500 nM SiR-DNA (Spirochrome AG, SC007) 1 h prior to and during the experiment. In preliminary analyses, we validated that this concentration of SiR-DNA did not induce mitotic delay or phenotypes in hiPSCs and HeLa cells. Treatments with control or DKK1 conditioned medium were applied just before starting the imaging for HeLa cells and overnight for hiPSCs. Live cell imaging was performed using an automated Nikon Eclipse Ti2 inverted microscope equipped with a 40× water immersion objective (Nikon Apo LWD, NA 1.15) and a NEO sCMOS camera (Andor). Multipoint acquisition was controlled by NIS Elements 5.1 software. 5 z-stacks with 2-µm interval were recorded every 5 min for up to 5 h in a preheated chamber (STXG-WSKM, Tokai Hit) at 37 °C and 5% CO2. Images were analyzed using ImageJ 2.0.0 software. Mitotic timing (time from NEB until the onset of anaphase), chromosome misalignment (chromosomal width), as well as phenotypic aberrations in metaphase were analyzed. Phenotypes were scored as “chromosome congression delay” for cells failing to recruit all chromosomes at the spindle equator in a timely manner, “alignment/metaphase delay” characterized as one or more chromosomes being clearly detached from the forming metaphase plate, or “misalignment + arrest” for cells with congression or alignment defects leading to mitotic exit or arrest beyond 75 min.
FACS.
RPE1 cells were synchronized in mitosis as described above and treated for 1.5 h with control SILAC Medium or DKK1 SILAC Heavy media. Cells were harvested and fixed for FACS analysis measuring the DNA content (propidium iodide) and mitotic phospho-epitopes (MPM-2) as described previously (69).
Determination of Microtubule Plus-End Assembly Rates.
Microtubule plus-end growth rates were determined by tracking EB3-EGFP in living cells as described previously (69). Briefly, cells were transfected 48 h prior to the measurement with siRNA as indicated and pEGFP-EB3 (kindly provided by L. Wordeman, University of Washington), seeded onto glass bottom dishes (Ibidi) and treated with 2 μM DME for 2 h before measurement. Live imaging was performed using a Deltavision ELITE microscope (GE Healthcare) with a 60× objective. Images were recorded every 2 seconds while cells were incubated at 37 °C and 5% CO2. Images were deconvolved using SoftWorx 5.0/6.0 software and average assembly rates were calculated from 200 individual microtubules (20 individual microtubules per cell, n = 10 cells).
Image Data Processing.
Raw images were imported to Fiji (ImageJ 2.0.0) prior to their export to Photoshop 2020 for panel arrangement. Linear changes in contrast or brightness were equally applied to all controls and across the entire images. The models and schemes were created with BioRender.com.
Statistical Analyses.
Data are shown as mean ± SD or median ± upper and lower quartile with 10% to 90% extremes, as indicated in the figure legends. Where indicated, Student’s t tests (two groups) or one-way ANOVA analyses with Tukey correction (three or more groups) were calculated using Prism v8. Live cell imaging phenotypes were analyzed using Fisher Exact tests. The curves showing the time from NEB until chromosome separation were analyzed with Mantel-Cox log-rank tests using Prism v8. Significance is indicated as *P < 0.05, **P < 0.01, ***P < 0.001, or n.s., not significant.
Supplementary Material
Acknowledgments
We thank C. Niehrs, K. M. Noh, J. Ellenberg, and T. W. Holstein for sharing reagents and cells. We thank the Nikon Imaging Center at the University of Heidelberg for access to microscopes and for technical help. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - SFB 1324 - Project number 331351713 (Project B03 to S.P.A. and H.B.). A.d.J.-S. holds a Humboldt Research Fellowship for Postdoctoral Researchers. A.B. was recipient of a junior award by the Schmeil-Stiftung Heidelberg.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108145118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Clevers H., Loh K. M., Nusse R., Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Niehrs C., On growth and form: A Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137, 845–857 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Nusse R., Clevers H., Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Bugter J. M., Fenderico N., Maurice M. M., Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat. Rev. Cancer 21, 5–21 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Tamai K., et al., LDL-receptor-related proteins in Wnt signal transduction. Nature 407, 530–535 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Zeng X., et al., A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438, 873–877 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson G., et al., Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature 438, 867–872 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Cruciat C. M., et al., Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science 327, 459–463 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto H., Komekado H., Kikuchi A., Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of beta-catenin. Dev. Cell 11, 213–223 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Bilic J., et al., Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316, 1619–1622 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Aberle H., Bauer A., Stappert J., Kispert A., Kemler R., beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16, 3797–3804 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S. E., et al., Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science 340, 867–870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li V. S., et al., Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 149, 1245–1256 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Taelman V. F., et al., Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143, 1136–1148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metcalfe C., Mendoza-Topaz C., Mieszczanek J., Bienz M., Stability elements in the LRP6 cytoplasmic tail confer efficient signalling upon DIX-dependent polymerization. J. Cell Sci. 123, 1588–1599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong L. T., et al., Kinesin-2 and IFT-A act as a complex promoting nuclear localization of β-catenin during Wnt signalling. Nat. Commun. 9, 5304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tejeda-Muñoz N., Albrecht L. V., Bui M. H., De Robertis E. M., Wnt canonical pathway activates macropinocytosis and lysosomal degradation of extracellular proteins. Proc. Natl. Acad. Sci. U.S.A. 116, 10402–10411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrecht L. V., et al., GSK3 inhibits macropinocytosis and lysosomal activity through the Wnt destruction complex machinery. Cell Rep. 32, 107973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H., Vick P., Hedtke J., Ploper D., De Robertis E. M., Wnt signaling translocates Lys48-linked polyubiquitinated proteins to the lysosomal pathway. Cell Rep. 11, 1151–1159 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Vinyoles M., et al., Multivesicular GSK3 sequestration upon Wnt signaling is controlled by p120-catenin/cadherin interaction with LRP5/6. Mol. Cell 53, 444–457 (2014). [DOI] [PubMed] [Google Scholar]
- 21.García de Herreros A., Duñach M., Intracellular signals activated by canonical Wnt ligands independent of GSK3 inhibition and β-catenin stabilization. Cells 8, E1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao B., et al., LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 411, 321–325 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Chen J., et al., CDK1-mediated BCL9 phosphorylation inhibits clathrin to promote mitotic Wnt signalling. EMBO J. 37, e99395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson G., et al., Cell cycle control of Wnt receptor activation. Dev. Cell 17, 788–799 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Niehrs C., Acebron S. P., Mitotic and mitogenic Wnt signalling. EMBO J. 31, 2705–2713 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acebron S. P., Niehrs C., β-catenin-independent roles of Wnt/LRP6 signaling. Trends Cell Biol. 26, 956–967 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Madan B., et al., Temporal dynamics of Wnt-dependent transcriptome reveal an oncogenic Wnt/MYC/ribosome axis. J. Clin. Invest. 128, 5620–5633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinze L., et al., Synthetic lethality of Wnt pathway activation and asparaginase in drug-resistant acute leukemias. Cancer Cell 35, 664–676.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acebron S. P., Karaulanov E., Berger B. S., Huang Y. L., Niehrs C., Mitotic wnt signaling promotes protein stabilization and regulates cell size. Mol. Cell 54, 663–674 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Huang Y. L., Anvarian Z., Döderlein G., Acebron S. P., Niehrs C., Maternal Wnt/STOP signaling promotes cell division during early Xenopus embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, 5732–5737 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolz A., Neufeld K., Ertych N., Bastians H., Wnt-mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation. EMBO Rep. 16, 490–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y. C., et al., Wnt10b-GSK3β-dependent Wnt/STOP signaling prevents aneuploidy in human somatic cells. Life Sci. Alliance 4, e202000855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kikuchi K., Niikura Y., Kitagawa K., Kikuchi A., Dishevelled, a Wnt signalling component, is involved in mitotic progression in cooperation with Plk1. EMBO J. 29, 3470–3483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habib S. J., et al., A localized Wnt signal orients asymmetric stem cell division in vitro. Science 339, 1445–1448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadjihannas M. V., et al., Aberrant Wnt/beta-catenin signaling can induce chromosomal instability in colon cancer. Proc. Natl. Acad. Sci. U.S.A. 103, 10747–10752 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fumoto K., Hoogenraad C. C., Kikuchi A., GSK-3beta-regulated interaction of BICD with dynein is involved in microtubule anchorage at centrosome. EMBO J. 25, 5670–5682 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodde R., et al., Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 3, 433–438 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Kops G. J., Weaver B. A., Cleveland D. W., On the road to cancer: Aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5, 773–785 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Manning A. L., et al., The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell 18, 2970–2979 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakhoum S. F., et al., Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walczak C. E., Gayek S., Ohi R., Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 29, 417–441 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Wilbur J. D., Heald R., Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife 2, e00290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Homma N., et al., Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell 114, 229–239 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Homma N., et al., KIF2A regulates the development of dentate granule cells and postnatal hippocampal wiring. eLife 7, e30935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyamoto T., et al., The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. 10, 664–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W., et al., Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 10, 2612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa T., Hirokawa N., Microtubule destabilizer KIF2A undergoes distinct site-specific phosphorylation cascades that differentially affect neuronal morphogenesis. Cell Rep. 12, 1774–1788 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Ganem N. J., Compton D. A., The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 166, 473–478 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanenbaum M. E., et al., Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. 19, 1703–1711 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Uehara R., et al., Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J. Cell Biol. 202, 623–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu R., et al., Mitosis-specific MRN complex promotes a mitotic signaling cascade to regulate spindle dynamics and chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 115, E10079–E10088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ali A., Veeranki S. N., Chinchole A., Tyagi S., MLL/WDR5 complex regulates Kif2A localization to ensure chromosome congression and proper spindle assembly during mitosis. Dev. Cell 41, 605–622.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Yi Z. Y., et al., Kif2a regulates spindle organization and cell cycle progression in meiotic oocytes. Sci. Rep. 6, 38574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon H. J., Park J. E., Song H., Jang C. Y., DDA3 and Mdp3 modulate Kif2a recruitment onto the mitotic spindle to control minus-end spindle dynamics. J. Cell Sci. 129, 2719–2725 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Eagleson G., et al., Kif2a depletion generates chromosome segregation and pole coalescence defects in animal caps and inhibits gastrulation of the Xenopus embryo. Mol. Biol. Cell 26, 924–937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maiato H., Gomes A. M., Sousa F., Barisic M., Mechanisms of chromosome congression during mitosis. Biology (Basel) 6, E13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poirier K., et al., Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 45, 639–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang C. Y., Coppinger J. A., Seki A., Yates J. R. III, Fang G., Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J. Cell Sci. 122, 1334–1341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trofimova D., et al., Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate. Nat. Commun. 9, 2628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knowlton A. L., Vorozhko V. V., Lan W., Gorbsky G. J., Stukenberg P. T., ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr. Biol. 19, 758–763 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santamaria A., et al., The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell Proteomics 10, M110.004457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen J. V., et al., Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Neumann B., et al., Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwarz-Romond T., et al., The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat. Struct. Mol. Biol. 14, 484–492 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Nakajima H., Toyoshima-Morimoto F., Taniguchi E., Nishida E., Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 278, 25277–25280 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Musacchio A., Hardwick K. G., The spindle checkpoint: Structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3, 731–741 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Fonseca C. L., et al., Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase. J. Cell Biol. 218, 1148–1163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blauwkamp T. A., Nigam S., Ardehali R., Weissman I. L., Nusse R., Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat. Commun. 3, 1070 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ertych N., et al., Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat. Cell Biol. 16, 779–791 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walston T., et al., Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev. Cell 7, 831–841 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Sugioka K., Mizumoto K., Sawa H., Wnt regulates spindle asymmetry to generate asymmetric nuclear β-catenin in C. elegans. Cell 146, 942–954 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Ciruna B., Jenny A., Lee D., Mlodzik M., Schier A. F., Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature 439, 220–224 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.ten Berge D., et al., Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13, 1070–1075 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Augustin I., et al., Autocrine Wnt regulates the survival and genomic stability of embryonic stem cells. Sci. Signal. 10, eaah6829 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Su J., et al., Genomic integrity safeguards self-renewal in embryonic stem cells. Cell Rep. 28, 1400–1409.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hall A. C., Lucas F. R., Salinas P. C., Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell 100, 525–535 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Weiner A. T., et al., Endosomal Wnt signaling proteins control microtubule nucleation in dendrites. PLoS Biol. 18, e3000647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen G., et al., Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ong S. E., et al., Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Borisova M. E., Wagner S. A., Beli P., Mass spectrometry-based proteomics for quantifying DNA damage-induced phosphorylation. Methods Mol. Biol. 1599, 215–227 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.