Significance
Protein homeostasis is critical for the proper growth and development of organisms. In ethylene biosynthesis, the protein abundance of ACC synthases (ACS), the rate-limiting enzymes in ethylene biosynthesis, is critical to control ethylene production and is regulated in part through crosstalk with other phytohormones. However, the underlying mechanism including regulatory components that integrate the crosstalk is poorly understood. This study identified a mechanism that employs the reciprocal degradation of two E3 ligase components to control the stability of ACS in a brassinosteroid and phosphoprotein-binding protein-dependent manner. The disruption of the reciprocal degradation between the E3 ligases compromises the survival of plants under carbon starvation. These findings have implications for understanding ethylene biosynthesis regulation, protein homeostasis, and its associated stress response.
Keywords: ethylene, ACC synthases, E3 ligases, reciprocal degradation, SINAT
Abstract
Ethylene influences plant growth, development, and stress responses via crosstalk with other phytohormones; however, the underlying molecular mechanisms are still unclear. Here, we describe a mechanistic link between the brassinosteroid (BR) and ethylene biosynthesis, which regulates cellular protein homeostasis and stress responses. We demonstrate that as a scaffold, 1-aminocyclopropane-1-carboxylic acid (ACC) synthases (ACS), a rate-limiting enzyme in ethylene biosynthesis, promote the interaction between Seven-in-Absentia of Arabidopsis (SINAT), a RING-domain containing E3 ligase involved in stress response, and ETHYLENE OVERPRODUCER 1 (ETO1) and ETO1-like (EOL) proteins, the E3 ligase adaptors that target a subset of ACS isoforms. Each E3 ligase promotes the degradation of the other, and this reciprocally antagonistic interaction affects the protein stability of ACS. Furthermore, 14–3-3, a phosphoprotein-binding protein, interacts with SINAT in a BR-dependent manner, thus activating reciprocal degradation. Disrupted reciprocal degradation between the E3 ligases compromises the survival of plants in carbon-deficient conditions. Our study reveals a mechanism by which plants respond to stress by modulating the homeostasis of ACS and its cognate E3 ligases.
Protein homeostasis is a core mechanism for maintaining cellular function, which enables organisms to rapidly respond to environmental stress in a specific manner. The ubiquitin–proteasome-mediated degradation pathway is one of the main pathways that govern protein homeostasis in cells and has been linked to diverse functions in plants, including hormone signaling, plant defense response, photomorphogenesis, and stress response (1–6).
The function of the gaseous hormone ethylene is largely regulated by the ubiquitin–proteasome system (7). In ethylene signaling, the abundance of positive regulators ETHYLENE-INSENSITIVE 2 (EIN2) and EIN3 is regulated by E3 ubiquitin ligases, EIN2-TARGETING PROTEIN 1 (ETP1) and ETP2 or EIN3-BINDING F-BOX PROTEIN 1 (EBF1) and EBF2, respectively (3, 8). Similar to the signaling pathway, in ethylene biosynthesis, the abundance of a subset of 1-aminocyclopropane-1-carboxylic acid (ACC) synthases (ACS), the rate-limiting enzymes in the pathway, is specifically regulated by ETHYLENE OVERPRODUCER 1 (ETO1) and its two paralogs, ETO1-like 1 (EOL1) and EOL2. ETO1/EOLs are components of a CULLIN3 E3 ligase, which specifically recognize type-2 ACS isoforms for rapid degradation via the 26S proteasome (9, 10). E3 ligase substrate-specificity subunits such as ETO1/EOLs determine the accessibility of E3 ligase complex to the substrate; thus, the abundance of ETO1/EOLs is an important regulatory factor for determining ACS stability in plants (11). The stability of EOL2 has been shown to be negatively regulated by 14–3-3, a family of phosphoprotein-binding proteins, though the underlying mechanism remains elusive (12).
Arabidopsis contains eight functional ACS isoforms that can be grouped into three types, namely type-1 (ACS2, 6), type-2 (ACS4, 5, 8, 9, and 11), and type-3 (ACS7), based on the presence or absence of phosphorylation sites in the C-terminal domains (9, 13, 14). The stability of different ACS is differentially regulated by diverse stimuli, including phytohormones (14–16). Most plant hormones regulate the protein stability of ACS with distinct effects on different ACS isoforms (15, 16). Brassinosteroid (BR) is one such hormone that regulates the protein stability of ACS. However, the underlying mechanisms and molecular components involved in the process are unknown (15). A component that likely plays a role in BR–ethylene crosstalk is the 14–3-3 proteins. The 14–3-3 proteins are evolutionally well-conserved dimeric proteins in all eukaryotic organisms and are involved in varied biological processes via phosphorylation-dependent protein–protein interactions (17–19). The Arabidopsis 14–3-3 family consists of 13 isoforms, and their roles have been suggested in a diverse range of physiological processes including BR signaling (20, 21), ethylene biosynthesis (12, 22), abiotic stress response (22–24), and light signaling (25). In the BR signaling pathway, 14–3-3 proteins interact with multiple BR signaling molecules, including BRASSINOID INSENSITIVE 1 (BRI1), BRI1 KINASE INHIBITOR 1 (BKI1), and BRI1-EMS SUPPRESSOR 1 (BES1)/BRASSINAZOLE-RESISTANT 1 (BZR1), resulting in the regulation of the BR signaling pathway (17). The role of 14–3-3 proteins has been implicated in ethylene biosynthesis via their interaction with ACS in rice, barley, and Arabidopsis (26–29). In Arabidopsis, 14–3-3 positively regulates the protein stability of ACS5 through increased turnover of EOL2 or through an ETO1/EOLs-independent mechanism (12); however, the detailed mechanism such as the stimuli triggering the 14–3-3–mediated regulation of ACS and ETO1/EOL stability or other regulatory components in the process remains unknown. Given the roles of 14–3-3 in ethylene biosynthesis and BR signaling, 14–3-3 could be a crosstalk point that integrates the interaction between ethylene biosynthesis and BR signaling in a phosphorylation-dependent manner.
SEVEN-IN-ABSENTIA (SINA) is a RING-type E3 ligase that has been linked to protein degradation and stress response in Drosophila, Mammalian, and plants (30–36). Arabidopsis contains a family of five SINA of Arabidopsis (SINAT) genes that encode two distinct clades of SINAT proteins (SINAT1 and 2; SINAT3, 4, and 5) based on sequence similarities (23). Several recent studies in different plant species have demonstrated that SINA family members play a role in response to abiotic and biotic stresses, including cold, drought, and pathogen invasion; some of these are linked to abscisic acid (ABA) or BR hormone signaling and autophagy, a highly conserved cellular degradation process linked to stress response (23, 33, 37). An autophagy receptor, DOMINANT SUPPRESSOR OF KAR 2 (DSK2), controls the degradation of BES1, a positive regulator for BR signaling, and SINAT2 participates in targeting BES1 for degradation via the DSK2 autophagy receptor under drought and fixed carbon starvation (33). SINAT E3 ligases also regulate the activity and/or stability of a subset of AUTOPHAGY-RELATED PROTEINS (ATGs) (30, 38). They also regulate the stability of FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1), the endosomal sorting complex required for the transport (ESCRT) component, thus controlling autophagy (30, 39–41). Intriguingly, SINAT2 has been identified as a putative 14–3-3–interacting protein along with EOL2 and several ACS isoforms through the proteomic profiling of purified complexes from Arabidopsis (27).
In this study, we investigated the regulatory mechanism for ACS5 protein stability through SINAT E3 ligases. Strikingly, ACS5 acts as a scaffold that tethers SINAT2 and EOL2 in a functional complex, increasing the stability of ACS5 via the reciprocal degradation of SINAT2 and EOL2. 14–3-3 activates the reciprocal degradation of SINAT2 and EOL2 through direct interaction with SINAT2 only in the presence of BR, thereby linking ethylene biosynthesis and BR signaling. Our results reveal a regulatory mechanism that allows the simultaneous fine-tuning of the protein abundance of ACS and its cognate E3 ligases, which is critical for stress response via autophagy.
Results
SINAT2 Positively Regulates BR-Induced Ethylene Biosynthesis.
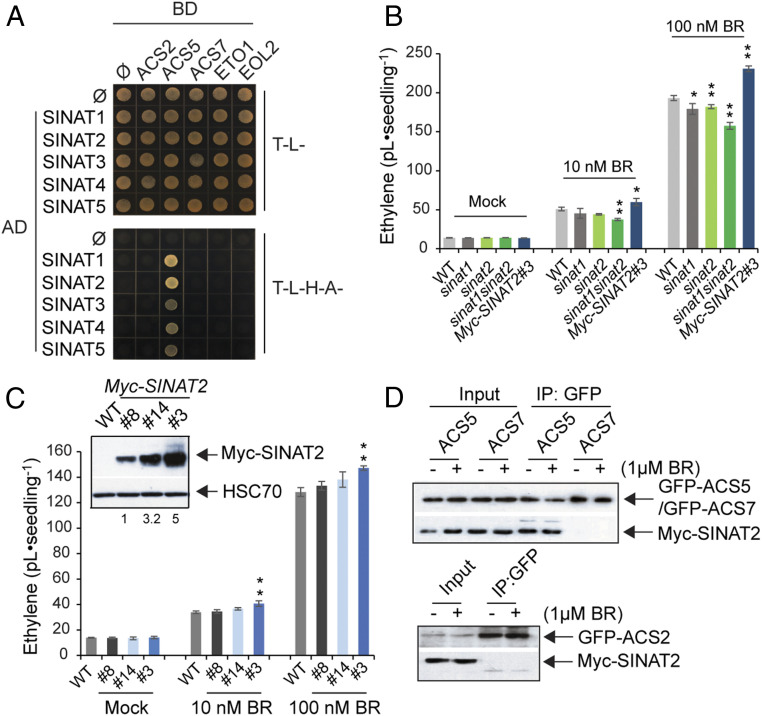
To further explore the mechanism that regulates ethylene biosynthesis, we performed yeast two-hybrid (Y2H) screening using ACS5 as bait and identified Seven-in-Absentia of Arabidopsis 2 (SINAT2) as one of the ACS5 interactors. SINAT2 specifically interacted with ACS5 and all other type-2 ACS but it did not interact with type-1 (ACS2 and ACS6) or type-3 ACS (ACS7) (Fig. 1A and SI Appendix, Fig. S1). ACS5 bound strongly with SINAT1 and 2 while showing a weak interaction with the other SINAT isoforms, suggesting that SINAT1 and 2 are the major SINAT isoforms that have a functional relationship with ACS5 and ethylene biosynthesis. SINAT1 and SINAT2 share over 90% amino acid sequence identity; therefore, we focused on SINAT2 in this study.
Fig. 1.
SINAT E3 ligases are positive regulators for BR-induced ethylene biosynthesis. (A) Y2H assays for the interaction between different ACS isoforms and SINAT E3 ligase isoforms. (B) BR-induced ethylene biosynthesis in dark-grown seedlings. Seedlings were grown on a medium with different concentrations of BR, and the accumulated ethylene was measured using gas chromatography. (C) BR-induced ethylene biosynthesis in seedlings expressing different levels of SINAT2 protein. (Inset) Expression levels of SINAT2 protein in independent transgenic lines expressing Myc-SINAT2 without BR treatment in the dark. The Myc-SINAT2 band intensity was normalized to HSC70 band, and the values were expressed relative to the ratio of Myc-SINAT2/HSC70 in no. 8, which was set to 1. (D) Co-IP of ACS5 and SINAT2 complex in N. benthamiana. The indicated combinations of tagged proteins were transiently expressed in N. benthamiana with or without BR treatment and used for co-IP analysis. GFP-ACS5 was immunoprecipitated with an anti-GFP antibody, and the resulting precipitates were further analyzed via immunoblotting using an anti-GFP or an anti-Myc antibody. Data represent the mean ± SD of three biological replicates (n = 3). *P < 0.05 and **P < 0.01 indicate a significant difference when samples were compared to the WT plants in each BR concentration using a Student’s t test.
SINAT E3 ligases have been shown to play a role in plant stress responses, including BR-mediated autophagy; however, the role of SINAT has not previously been shown to modulate ethylene biosynthesis (15, 16, 23, 30, 33). Since SINAT has a role in BR-mediated stress responses, we examined whether SINAT2 regulates ethylene biosynthesis in a BR-dependent manner. To test this, we measured the ethylene production in sinat1, sinat2, sinat1sinat2 double mutant, and seedlings that express Myc-tagged SINAT2 from CaMV 35S promoter (35S::Myc-SINAT2) in the presence or absence of BR. All dark-grown seedlings increased their ethylene production in response to BR (Fig. 1B). However, sinat1sinat2 showed significantly lower levels of BR-induced ethylene compared to the wild type (WT), suggesting that SINAT1/SINAT2 is a positive regulator for BR-induced ethylene biosynthesis. Unlike the sinat1sinat2, sinat3, sinat5, and sinat3sinat5, the double mutant produced comparable levels of BR-induced ethylene to the WT, which may be attributable to the weak interaction of SINAT3/SINAT5 with ACS5 (SI Appendix, Fig. S2). Consistent with their positive role in ethylene biosynthesis, the 35S::Myc-SINAT2 plants produced more ethylene with BR treatment (Fig. 1B), which was not associated with alterations in the transcript levels of ACS and ETO1/EOL2 (SI Appendix, Fig. S3). The moderate (but significant) reduction of ethylene levels in sinat1sinat2 may have been due to the functional redundancy among SINAT proteins that showed weak interaction with ACS5. However, the minor increase (∼20% of that in the WT) of BR-induced ethylene levels in 35S::Myc-SINAT2 was a conundrum, considering it was an overexpression line. Thus, we measured ethylene biosynthesis in three independent 35S::Myc-SINAT2 lines with significantly different expression levels of the SINAT2 protein (approximately three- to fivefold differences) (Fig. 1C). Interestingly, ethylene production in the independent lines was not significantly or only slightly different from each other (Fig. 1C), indicating the existence of feedback mechanisms that suppress SINAT2-mediated ethylene biosynthesis or the saturation of the system that controls SINAT2-mediated ethylene production. Since SINAT1 and SINAT2 played a positive role in BR-induced ethylene production, we examined whether ACS5-SINAT interaction was BR dependent. SINAT2 formed a complex with ACS5 regardless of BR but not with type-1 (ACS2) or type-3 (ACS7) ACS in plants (Fig. 1D). Together, these results suggest that SINAT2 is a positive regulator for BR-induced ethylene biosynthesis, but its interaction with ACS5 is not regulated by BR.
SINAT2 Forms a Complex with EOL2 Only in the Presence of ACS5 in Plants.
SINAT2 positively regulated BR-induced ethylene biosynthesis and only interacted with type-2 ACS isoforms, suggesting that it might negatively regulate ETO1/EOLs, the cognate E3 ligases for type-2 ACS isoforms. To determine this, we examined the interactions between SINAT and ETO1 and EOL2, one of the paralogs of ETO1, using a Y2H assay. ETO1 and EOL2 did not interact with any of the SINAT E3 ligases in the Y2H assay (Fig. 1A). Since ETO1 and EOLs have been shown to play a redundant role in ethylene biosynthesis regulation (40, 42), EOL2 was used as the ETO1/EOL representative in following results. Unlike the results of the Y2H assay, strikingly, SINAT2 and EOL2 were pulled down together when ACS5 was coexpressed in Nicotiana benthamiana, regardless of the BR treatment (Fig. 2A). Coexpression of a negative control type-3 ACS7 did not promote the in vivo interaction between SINAT2 and EOL2 (Fig. 2A). To further confirm the in vivo interaction of SINAT2 and ACS5 and subcellular localization of the interaction, BiFC analysis of SINAT2 and EOL2 was performed in the presence and absence of ACS5. Consistent with the results of the Y2H and co-immunoprecipitation (co-IP) analysis, SINAT2 and EOL2 interacted only in the presence of ACS5, and intriguingly, the interaction occurred at AUTOPHAGY-RELATED PROTEIN 8 (ATG8)-labeled autophagosomes, double-membraned vesicles containing damaged cellular materials to be degraded by autophagy (Fig. 2 B and C) (43). The colocalization analysis corroborated that the three proteins colocalized in cytoplasmic puncta, presumably autophagosomes (SI Appendix, Fig. S4). These results agreed with the subcellular localization of a subset of SINAT family at autophagosomes (40). In sum, the results demonstrate that ACS5 specifically mediates the interactions between SINAT2 and EOL2 E3 ligases in plants. Additionally, their interaction at autophagosomes suggests the autophagy-mediated protein stability regulation of SINAT/EOL2/ACS and the crosstalk between ethylene biosynthesis and autophagy.
Fig. 2.
SINAT2 E3 ligases regulate the protein stability of ACS5 and EOL2 in a BR-dependent manner. (A) Co-IP of SINAT2 and EOL2. The indicated combinations of tagged proteins were transiently expressed in N. benthamiana with and without BR treatment. Protein extracts from the infiltrated leaves were used for co-IP analysis. GFP-EOL2 was immunoprecipitated with an anti-GFP antibody, and the resulting precipitates were further analyzed via immunoblotting using an anti-Myc antibody. (B) BiFC of SINAT2 and EOL2 in the presence and absence of ACS in transiently transformed N. benthamiana. Instead of mCherry-ACS5, mCherry-bZIP was transformed as a transfection control. (Scale bar, 50 µm.) (C) Colocalization of ATG8 and BiFC fluorescence of SINAT2 and EOL2 in the presence of ACS5. (Scale bar, 50 µm.) (D and E) SINAT2 positively regulates the protein stability of ACS5 in a BR-dependent manner. Protein extracts from Arabidopsis mesophyll protoplasts transformed with plasmid-expressing YFP-ACS5, and increasing amounts of plasmid-expressing cYFP-SINAT2 were analyzed via immunoblotting with an anti-GFP antibody. Relative quantities (Rel. quantities) represent the ratio of the intensity of YFP-ACS5 to GFP band relative to the ratio of YFP-ACS5/GFP bands from a control sample expressing YFP-ACS5 without c-YFP-SINAT2 with or without BR (D). The half-life analysis of ACS5 in protoplasts from WT, sinat1sinat2, and 35S::Myc-SINAT2 plants (E). Protoplasts were transformed with plasmid-expressing Myc-ACS5 with or without BR. At Time 0, CHX was added, and the total proteins were extracted at the indicated times, and then total protein extract from the protoplasts was analyzed by immunoblotting using the indicated antibodies. GFP was coexpressed as a transformation control. The experiments were repeated at least three times with similar results. Relative quantities (Rel. quantities) represent the ratio of the intensity of Myc-ACS5 to the GFP band relative to the ratio of Myc-ACS5/GFP bands from a control sample, which was set to 1. (F) SINAT2 destabilizes EOL2 in the presence of BR. Protein extracts from Arabidopsis protoplasts transformed with plasmid-expressing GFP-EOL2, and increasing amounts of plasmid-expressing cYFP-SINAT2 were analyzed via immunoblotting with an anti-GFP antibody. GFP was coexpressed as a transformation control. Rel. quantities represent the ratio of the intensity of GFP-EOL2 to GFP band relative to the ratio of GFP-EOL2/GFP bands from a control sample expressing GFP-EOL2 without cYFP-SINAT2 with or without BR. (G) Auto-ubiquitination assay for SINAT2. Bacteria expressed and purified MBP-tagged WT SINAT, SINAT2C63S, and free MBP proteins were incubated with E1, E2, and ubiquitin, and the ubiquitination of SINAT2 was analyzed via immunoblotting with an anti-MBP (Upper) or anti-ubiquitin (Bottom) antibody. The smeared and laddered bands indicate the ubiquitinated WT MBP-SINAT2. (H and I) SINAT2 ubiquitinates ACS5 and EOL2 E3 ligases. Immunoprecipitated YFP-ACS5 (H) and GFP-EOL2 (I) from N. benthamiana were incubated with purified SINAT2, E1, E2, and ubiquitin, and YFP-ACS5 and GFP-EOL2 were immunoprecipitated with an anti-GFP antibody and further analyzed via immunoblotting using an anti-Ub antibody.
SINAT2 Regulates the Stability of EOL2 and ACS5 in a BR-Dependent Manner.
To elucidate the consequence of SINAT2 interaction with ACS5 and EOL2 in plants, we investigated whether SINAT2 regulates the protein stability of both proteins. We transiently expressed an increasing amount of cYFP-fused SINAT2 (cYFP-SINAT2) with either YFP-ACS5 or GFP-EOL2 in Arabidopsis mesophyll protoplasts and subsequently treated the protoplasts with or without exogenous BR. The steady-state levels of ACS5 decreased as the expression of SINAT2 increased in the protoplasts under no BR treatment, indicating that SINAT2 negatively regulates ACS5 protein stability when overexpressed (Fig. 2D). However, in the presence of BR, the negative effects of SINAT2 on ACS5 stability were significantly alleviated (Fig. 2D), suggesting that BR suppresses the SINAT2-mediated degradation of ACS5. Corroborating these results, ACS5 protein stability was differentially regulated by BR in protoplasts isolated from the WT, sinat1sinat2, or SINAT2 overexpression line (35S::Myc-SINAT2) (Fig. 2E). BR treatment reduced the turnover of ACS5 in protoplasts from WT and 35S::Myc-SINAT2 plants but not in sinat1sinat2 protoplasts. Similar to ACS5, SINAT2 reduced the steady-state levels of ACS2 but not of the ACS7 protein (SI Appendix, Fig. S5). This may be attributed to the heterodimerization among the ACS isoform and the long half-life of ACS7 (15). In contrast to ACS2 and ACS5, SINAT2 did not affect the stability of GFP-EOL2 in the absence of BR; however, upon the BR treatment, the steady-state levels of GFP-EOL2 declined proportionally to the increased expression of SINAT2 (Fig. 2F). These results demonstrate that SINAT2 increases and decreases the protein stability of ACS5 and EOL2, respectively, in a BR-dependent manner, which may play a role in a BR-induced increase in ethylene biosynthesis in the dark. Our results also showed that SINAT2 could negatively regulate ACS stability when plants express an excess amount of SINAT2.
The SINAT2-mediated regulation of ACS5 and EOL2 protein stability indicated that ACS5 and EOL2 may be the in vivo substrates of SINAT2. To determine this, we initially examined the E3 ligase activity of WT SINAT2 and a mutant SINAT2 with a C63S mutation (SINAT2C63S). Cys63 in the RING motif is highly conserved among SINAT isoforms, and the substitution of the Cys63 to Ser has previously shown to abolish the E3 ligase activity of SINAT5 (SI Appendix, Fig. S6A) (44). Consistent with a previous study, MBP-tagged WT SINAT2 (MBP-SINAT2), but not MBP-SINAT2C63S, exhibited self-ubiquitination, demonstrating its biochemical function as an E3 ligase (Fig. 2G). Next, we conducted an in vitro ubiquitination assay using immunoprecipitated YFP-ACS5 or GFP-EOL2, which was transiently expressed in N. benthamiana, and in vitro purified SINAT proteins. The immunoblotting analysis showed that SINAT2 ubiquitinated both ACS5 and EOL2, whereas SINAT2C63S was unable to ubiquitinate either protein (Fig. 2 H and I). In support of this, SINAT2C63S did not influence the steady-state levels of ACS2, ACS5, or EOL2, regardless of BR, indicating that SINAT2-mediated ubiquitination is required for regulating the stability of ACS5 and EOL2 (SI Appendix, Fig. S6 B–D). These results suggest that ACS5 and EOL2 are in vivo substrates of SINAT2 E3 ligase.
SINAT2 and EOL2 Undergo Reciprocal Antagonistic Degradation, and This Process Requires Type-2 ACS5/ACS9 and BR.
The ACS5-mediated interaction between SINAT2 and EOL2 and BR-induced ethylene biosynthesis suggested that both E3 ligases may undergo reciprocal degradation in a BR-dependent manner. To assess this possibility, we first determined the effect of BR on SINAT2 and EOL2 protein stability. We treated dark-grown 35S::Myc-SINAT2/WT seedlings with BR for 2 h followed by cycloheximide (CHX) treatment and measured the loss of SINAT2 protein over time. BR significantly increased the turnover of SINAT2 (Fig. 3A and SI Appendix, Fig. S7A). We observed that the remaining fraction of Myc-SINAT2 was greatly reduced at the later time point after CHX treatment, presumably due to a low ratio of free SINAT2 to bound SINAT2 in the SINAT2/ACS/EOL2 complex. By contrast (and consistent with a previous study (45)), BR neither stimulated SINAT2 turnover nor affected SINAT2 transcript levels in light-grown seedlings (SI Appendix, Fig. S8). Similar to SINAT2, BR promoted the degradation of EOL2 protein in the dark (Fig. 3B and SI Appendix, Fig. S7B) but not in the light (SI Appendix, Fig. S8), suggesting a different regulatory mechanism for the protein stability of SINAT2 and EOL2 under the light. Contrary to SINAT2 and EOL2, but corroborating previous studies, BR increased the protein abundance of ACS5 in the dark (15, 16). BR also stabilized ACS5 in the light (SI Appendix, Fig. S8), which is similar to the results of a prior study demonstrating the light-induced stabilization of ACS5 during a dark-to-light transition (46).
Fig. 3.
ACS-mediated reciprocal degradation of SINAT2 and ETO1/EOLs. (A and B) BR promotes the turnover of SINAT2 or EOL2. Dark-grown seedlings expressing Myc-SINAT2 or GFP-EOL2 were treated with or without 1 μM BR and subjected to protein degradation assays in the presence of CHX (250 μM). The seedlings were collected at the indicated time after the CHX treatment and further analyzed. Rel. quantities represent the ratio of the intensity of Myc-SINAT2 or GFP-EOL2 band to HSC70 band signals, and these values are expressed relative to the time 0 value, which was set to 1. The graphs show the quantification of the remaining fraction of SINAT2 or EOL2 after the treatment of CHX over time from three replicates. (C) BR regulates the protein stability of ACS5, SINAT2, and EOL2 via the 26S proteasome pathway. Dark-grown seedlings that were 3 d old expressing Myc-ACS5, GFP-EOL2, and Myc-SINAT2 were treated with or without 1 μM BR for 2 h in the presence or absence of MG132 (100 μM), and the steady-state levels of the proteins were analyzed via immunoblotting using α-Myc, α-GFP, or α-HSC70 (loading control) antibody. (D) Wortmannin treatment alters the protein abundance of SINAT2 and EOL2 but not that of ACS5. Dark-grown seedlings that were 3 d old were treated with or without wortmannin (10 μM) for 3 h, and the total protein extracts were analyzed by immunoblotting using anti-Myc, -GFP, or HSC70 antibody. (E) The protein stability of SINAT2 is negatively regulated by ACS5 and ACS9 in the presence of BR. Seedlings expressing Myc-SINAT2 in different acs mutant backgrounds were treated with or without 1 μM BR for 2 h, and the steady-state levels of Myc-SINAT2 were analyzed via immunoblotting using an anti-Myc antibody. Rel. quantities represent the ratio of the intensity of Myc-SINAT2 to the HSC70 band, and these values were expressed relative to the ratio of the intensity of the cognate Myc-SINAT2/HSC70 bands in the same background with no BR treatment, which was set to 1. (F) Quantitative gene expression analysis for SINAT2 in seedlings in E. Data represent the mean ± SD of three biological replicates (n = 3), each with three technical replicates. (G and H) SINAT2 and ETO1/EOLs reciprocally regulate each other’s protein stability. Dark-grown seedlings expressing Myc-SINAT2 in eto1eol1eol2 or GFP-EOL2 in sinat1sinat2 mutants were treated with different concentrations of BR for 2 h, and the steady-state levels of Myc-SINAT2 or GFP-EOL2 were analyzed via immunoblotting using an anti-Myc or anti-GFP antibody. HSC70 was used as a loading control. All immunoblotting analyses were performed at least three times with similar results. (I) In vivo ubiquitination of EOL2 and SINAT2. Arabidopsis protoplasts were cotransfected with the indicated combinations of plasmids expressing HA-UBQ3, YFP-EOL2, Myc-SINAT2, and V5-ACS5 and incubated for 16 h followed by treatment with 20 µM MG132 and with or without 1 µM BR. The ubiquitinated EOL2 and SINAT2 were detected by an anti-GFP and -Myc immunoblotting after immunoprecipitation with anti-HA magnetic beads (top two panels). The total ubiquitinated proteins and myc-SINAT2, YFP-EOL2, and V5-ACS5 in input samples were analyzed by immunoblotting using anti-HA, -Myc, -GFP, and -V5 tag antibodies, respectively (Bottom two panels).
Furthermore, we examined if the protein stability of SINAT2 is regulated by the ubiquitin–proteasome pathway. MG132 treatment completely blocked BR-induced SINAT2 degradation in dark-grown seedlings (Fig. 3C), demonstrating that BR negatively regulates SINAT2 protein stability through the 26S proteasome in an identical manner to the regulation of ACS5 and EOL2 stability. Given that SINAT2, ACS5, and EOL2 colocalized at autophagosomes, we also determined whether autophagy controls the protein stability of the three proteins. The treatment with wortmannin, an autophagy inhibitor, significantly increased the steady-state levels of SINAT2 and EOL2 proteins but not that of ACS5 at the concentration used in this study (Fig. 3D). This implies that the protein stability of SINAT2 and EOL2 is also directly or indirectly regulated by the autophagy pathway.
Regardless of BR treatment, SINAT2 and EOL2 only formed a complex in the presence of ACS5, and both proteins specifically interacted with type-2 ACS proteins (Figs. 1A and 2A and SI Appendix, Fig. S1) (47, 48). This led to a hypothesis that ACS5, and perhaps other type-2 ACS isoforms, promote the reciprocal degradation between these two E3 ligases by bringing them into close proximity with each other. To determine this, we generated transgenic plants expressing Myc-SINAT2 in different ACS loss-of-function (LOF) mutant backgrounds, including acs5, acs5acs9, and acs2acs6, and we examined the effect of ACS LOF on the BR-induced degradation of SINAT2. BR treatment markedly reduced the steady-state levels of Myc-SINAT2 in the 35S::Myc-SINAT2/WT plants (Fig. 3E). However, BR-induced reduction in Myc-SINAT2 protein levels was slightly mitigated in the acs5 and acs2acs6 mutants compared to that in the WT background (Fig. 3E). Interestingly, the removal of ACS5 and ACS9, the closest homolog of ACS5, completely blocked BR-induced SINAT2 degradation without altering the SINAT2 transcripts (Fig. 3F). These results suggest that ACS5 and ACS9 are involved in BR-induced SINAT2 degradation by tethering SINAT2 and EOL2 in a complex.
To further validate the reciprocal degradation of SINAT2 and EOL2 and the role of BR, we examined the effect of a lack of SINAT1/SINAT2 or ETO1/EOL1/EOL2 on the protein stability of EOL2 and SINAT2, respectively. Unlike its effect in WT plants, BR did not influence the protein stability of the EOL2 in the sinat1sinat2 background (Fig. 3G). Similarly, the protein stability of SINAT2 in the eto1eol1eol2 mutant was not influenced by BR (Fig. 3H).
In support of these results, in vivo ubiquitination assays demonstrated that the polyubiquitination of SINAT2 and EOL2 depends on each other and is significantly enhanced in the presence of ACS5 and BR treatment (Fig. 3I and SI Appendix, Fig. S9). Together, these results showed that each E3 ligase is required for the BR-induced degradation of the other, and ACS5 plays a role in promoting the reciprocal degradation between the two E3 ligases. Therefore, we concluded that the BR-induced degradation of SINAT2 and EOL2 occurs through the reciprocal antagonistic degradation between SINAT2 and EOL2.
T147 in the 14–3-3 Binding Motif in SINAT2 Is Required for the Activation of the BR-Induced Reciprocal Degradation of SINAT2 and EOL2.
The data thus far indicated that the interaction between ACS5, EOL2, and SINAT2 is BR independent. However, the reciprocal degradation between SINAT2 and EOL2 and its associated change in ethylene biosynthesis were BR dependent. These results suggested the existence of a missing component that conveys BR signals into the SINAT2-ETO1/EOLs regulatory circuit to activate the reciprocal degradation between the two E3 ligases. A likely candidate for the missing component is a 14–3-3 protein based on the known function of 14–3-3 in BR signaling (20, 21) and ethylene biosynthesis (12, 22) and a potential 14–3-3 binding motif in SINAT E3 ligases (Fig. 4A). Computational analyses predicted that SINAT2 contains a 14–3-3-binding motif (CRFRPY[T]147CPYA), including a potential phosphorylation site in the substrate binding and dimerization domain (49, 50). This motif is highly conserved among all SINAT isoforms (Fig. 4A). To determine whether 14–3-3 activates the reciprocal degradation of SINAT2 and EOL2, thus increasing the stability of ACS5, we first examined the involvement of 14–3-3 in BR-induced ethylene biosynthesis. To this end, we treated dark-grown seedlings expressing Myc-ACS5 (35S::Myc-ACS5) with R18 peptide, a competitive inhibitor for 14–3-3 protein–protein interaction. R18 treatment reduced a BR-induced increase in ACS5 protein levels (Fig. 4B). R18 treatment also significantly reduced BR-induced ethylene biosynthesis in etiolated WT and 35S::Myc-ACS5 seedlings (Fig. 4 C and D). Furthermore, we found that seedlings overexpressing Myc-14–3-3ω (35S::Myc-14–3-3ω), a 14–3-3 isoform that was previously shown to control ACS and EOL2 stability (12), produced significantly higher ethylene than the WT in response to BR (Fig. 4E). Similar to the 35S::Myc-ACS5 seedlings, R18 significantly attenuated BR-induced ethylene biosynthesis in 35S::Myc-14–3-3ω (Fig. 4F). These results suggest that among the 13 Arabidopsis 14–3-3 isoforms, at least 14–3-3ω is involved in BR-induced ACS stability regulation and ethylene biosynthesis. Of note, the abundance of 14–3-3ω was down-regulated by BR and the light-to-dark transition (Fig. 4 G, H, and I). Next, we analyzed the interaction between 14–3-3ω and all five SINAT isoforms using a Y2H assay (12). None of the SINAT E3 ligases bound to 14–3-3ω in the yeast system (Fig. 4J). SINAT2, however, was pulled down with 14–3-3ω in the presence of BR in plants (Fig. 4K), showing that BR promotes the binding of 14–3-3ω to SINAT2. To assess the role of T147 residue in the 14–3-3-binding motif in SINAT2 and its connection to BR signaling, we mutagenized T147 to Ala (T147A) and examined the interaction of SINAT2T147A with 14–3-3ω using a co-IP assay. Unlike WT SINAT2, SINAT2T147A did not form a stable complex with 14–3-3ω in the presence of BR in plants (Fig. 4K), demonstrating that T147 of SINAT2 is required for its interaction with 14–3-3ω.
Fig. 4.
14–3-3 activates the reciprocal degradation of SINAT2 and EOL2 by interacting with SINAT2 in the presence of BR. (A) A partial amino acid sequence comparison among Arabidopsis SINAT E3 ligases. The underlined amino acid sequences are a potential 14–3-3–binding motif predicted by the 14–3-3–Pred program (50). An asterisk indicates a potential phosphorylation site. (B) R18 decreases the BR-induced increase in ACS5 protein abundance. Dark-grown seedlings that were 3 d old were treated with or without BR (1 µM) or R18 peptide (200 µg/mL) for 2 h, and the steady-state levels of ACS5 were analyzed by immunoblotting. Rel. quantities indicate the ratio of intensity of Myc-ACS5 to the HSC70 band relative to the ratio of Myc-ACS5/HCS70 bands with no BR and R18 treatment, which was set to 1. (C and D) BR-induced ethylene biosynthesis is inhibited by R18. Seedlings that were 3 d old were treated with or without BR (0.1 µM) or R18 (200 µg/mL) for 1 d in the dark, and then the accumulated ethylene was measured using gas chromatography. (E) Ethylene production from WT and a transgenic line expressing Myc-14–3-3ω from a dexamethasone-inducible promoter. (F) R18 attenuates BR-induced ethylene biosynthesis in 35S::Myc-14–3-3 seedlings. (G and H) BR increases the turnover of 14–3-3ω. The steady-state levels of 14–3-3ω at different concentrations of BR were analyzed by immunoblotting (G). 35S::Myc-14–3-3 seedlings were grown on growth medium with or without BR (1 µM) for 3 d and treated with CHX at time 0. The total protein extracts were prepared at the indicated times and then analyzed by immunoblotting using the indicated antibodies (H). (I) A light-to-dark transition decreases 14–3-3 protein abundance. Light-grown seedlings that were 7 d old expressing Myc-14–3-3ω were transferred to darkness for 1 or 2 d, and the total proteins were analyzed by immunoblotting. L, light; D1, dark 1 d; D2, dark 2 d. (J) A Y2H assay for the interaction between 14–3-3 and SINAT2 isoforms. (K) Co-IP of 14–3-3 and SINAT2. N. benthamiana leaves were infiltrated with agrobacterium, coexpressing the indicated proteins with or without BR. Total protein extracts were immunoprecipitated with anti-GFP, and the resulting precipitates were analyzed via immunoblotting using an anti-GFP (Upper) anti-Myc antibody (Bottom). (L and M) Mutant SINAT2 with T147A does not respond to BR to control the stability of ACS or EOL2. Protein extracts from protoplasts expressing a fixed amount of YFP-ACS5 or GFP-EOL2 with an increasing amount of YFP-SINAT2T147A in the presence or absence of BR were analyzed via immunoblotting using an anti-GFP antibody. The GFP signal was used as a loading control. Rel. quantities represent the ratio of the intensity of YFP-ACS5 (L) or GFP-EOL2 (M) to GFP band relative to the ratio of YFP-ACS5/GFP (L) or GFP-EOL2/GFP (M) bands from a control sample expressing YFP-ACS5 (L) or GFP-EOL2 (M) without c-YFP-SINAT2 with or without BR. (N) The T147A mutation in SINAT2 reduced BR-induced ethylene biosynthesis. Seedlings were grown on an Murashige and Skoog medium with or without BR for 3 d, and the accumulated ethylene was measured using gas chromatography. Data represent the mean ± SD of three biological replicates (n = 3). Letters indicate differences at a P < 0.05 significance level using ANOVA analysis with a Tukey post hoc correction. *** indicates a significant difference (P < 0.0001) when samples were compared to the WT control with BR treatment using a Student’s t test.
To further elucidate the in vivo function of 14–3-3 in the reciprocal degradation of SINAT2 and ETO1/EOLs, we investigated whether SINAT2T147A still possesses the ability to control the stability of ACS5 and EOL2 in a BR-dependent manner. When overexpressed in protoplasts, SINAT2T147A negatively regulated ACS5 stability (Fig. 4L), indicating that the T147A mutation did not affect the E3 ligase activity of SINAT2T147A. Interestingly, unlike WT SINAT2, BR did not block SINAT2T147A-stimulated ACS5 turnover (Fig. 4L). Conversely, SINAT2T147A failed to destabilize EOL2 in the presence of BR (Fig. 4M). Together, these results showed that the BR sensitivity of turnover of ACS and EOL2 requires T147 of SINAT2, and BR-induced phosphorylation on T147 likely plays a role in this process. Furthermore, 35S::Myc-SINAT2T147A/sinat1sinat2 seedlings produced only 50% of the ethylene produced in 35S::Myc-SINAT2/sinat1sinat2 and the WT seedlings in the presence of BR (Fig. 4N). This result showed that T147 in SINAT2 is required for BR-induced ethylene biosynthesis.
Plants with a Mutated T147 of SINAT2 Are Hypersensitive to Fixed Carbon Starvation.
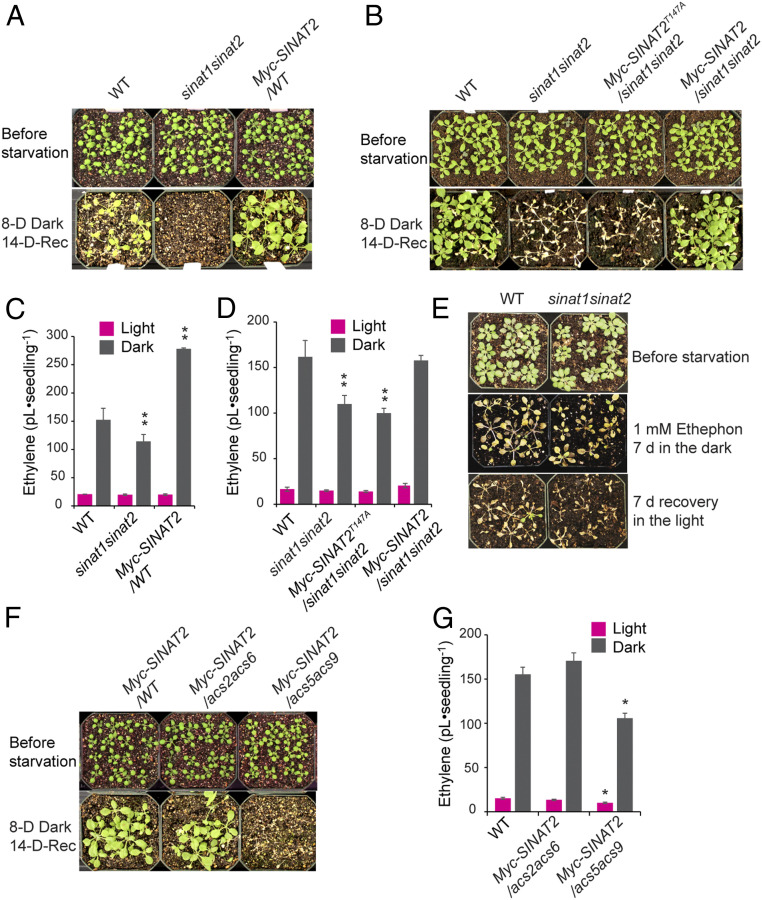
SINAT2 is involved in responses to drought and fixed carbon starvation by promoting the degradation of the BR-responsive transcription factor BES1, which is mediated by the autophagy receptor DSK2 (33). Many autophagy mutants are hypersensitive to or have reduced survival under conditions of nitrogen and/or carbon starvation (51). Therefore, we examined the function of SINAT1/SINAT2 on carbon starvation using the sinat1sinat2 mutant and transgenic lines expressing WT SINAT2 in a WT background (35S::Myc-SINAT2/WT). After 8 d of dark treatment followed by 2 wk of recovery in light, most of the WT and 35S::Myc-SINAT2 plants had recovered from the stress, whereas the sinat1sinat2 mutant completely died (Fig. 5A and SI Appendix, Fig. S10). These results showed that SINAT2 is required for the response to carbon starvation, which is consistent with the previous study (33). Furthermore, we determined the effect of the T147A mutation in SINAT2 on carbon starvation using transgenic lines expressing Myc-SINAT2T174A or Myc-SINAT2 in the sinat1sinat2 background. Unlike 35S::Myc-SINAT2/sinat1sinat2 and the WT plants, 35S::Myc-SINAT2T147A/sinat1sinat2 did not recover from the carbon starvation, showing results identical to that of sinat1sinat2 (Fig. 5B). These results demonstrated that T147 residue in SINAT2 plays a key role in the response to carbon starvation.
Fig. 5.
The T147A mutation in SINAT2 and overexpression of SINAT2 in the acs5acs9 mutant lead to a hypersensitive response to carbon starvation. (A, B, and F) A fixed carbon starvation assay. Seedlings that were 2 wk old were transferred to total darkness for 8 d and returned to the light for 2 wk for recovery. (C, D, and G) Ethylene biosynthesis in seedlings in response to carbon starvation. Light-grown seedlings that were 7 d old grown on a growth medium without sucrose were capped for 7 d or transferred to total darkness for 7 d, and the accumulated ethylene was measured via gas chromatography. Data represent the mean ± SD of three biological replicates (n = 3). *P < 0.05 and **P < 0.001 indicate the significant difference when samples were compared to the WT in dark or light conditions using a Student’s t test. (E) Ethephon treatment does not rescue the hypersensitive carbon starvation phenotypes of sinat1sinat2. Light-grown seedlings that were 3 wk old were treated with or without 1 mM ethephon and further kept in the dark for 7 d followed by 1-wk recovery in the light.
Carbon Starvation–Induced Ethylene Production Is Significantly Reduced in Plants Expressing SINAT2T147A.
The T147A mutation in SINAT2 inhibited the BR-mediated stabilization and destabilization of ACS and EOL2, respectively, which led to reduced BR-induced ethylene biosynthesis. To determine whether the compromised survival of sinat1sinat2 and 35S::Myc- SINAT2T147A/sinat1sinat2 in response to dark treatment is associated with a reduction in ethylene production, we measured ethylene biosynthesis in the seedlings before and after the dark treatment. All light-grown sinat1sinat2 and 35S::Myc-SINAT2/WT seedlings produced comparable levels of ethylene production to the WT seedlings (Fig. 5C). However, dark-induced ethylene production was significantly decreased and increased in the sinat1sinat2 and 35S::Myc-SINAT2/WT plants, respectively, compared to the WT seedlings (Fig. 5C). Like the sinat1sinat2 mutant, dark treatment induced significantly less ethylene in the 35S::Myc-SINAT2T147A/sinat1sinat2 seedlings compared to the WT seedlings (Fig. 5D), despite the similar levels of ethylene in the light-grown seedlings. The dark treatment–induced changes in ethylene production in these plants were similar to the alteration in ethylene biosynthesis mediated by BR in the dark (Figs. 1B and 4N). In support of the role of 14–3-3 interaction with SINAT2 in ethylene biosynthesis, the dark treatment of 35::Myc-14–3-3ω plants produced a higher amount of ethylene than the WT plants (SI Appendix, Fig. S11). These results showed that the response of a plant to carbon starvation involves increased ethylene production, which requires SINAT1/SINAT2 with functional T147 residue.
The Disrupted Reciprocal Degradation of SINAT2 and EOL2 Leads to the Reduced Ethylene Levels in Plants with Reduced SINAT Function.
To further examine the correlation between ethylene biosynthesis, carbon starvation response, and the role of SINAT2, we examined the carbon starvation response and its associated ethylene production in eto1eol1eol2, 35S::Myc-SINAT2/eto1eol1eol2, 35S::Myc-SINAT2/acs2cs6, and 35S::Myc-SINAT2/acs5acs9. Both eto1eol1eol2 and 35S::Myc-SINAT2/eto1eol1eol2 showed enhanced recovery from carbon starvation compared to the WT, which was correlated to increased ethylene production in light and during the dark treatment (SI Appendix, Fig. S12). We speculate that the reduced ethylene biosynthesis in 35S::Myc-SINAT2/eto1eol1eol2 compared to eto1eol1eol2 may have been due to the negative role of SINAT2 on ACS stability when it is overexpressed in plants due to a lack of ETO1/EOLs.
If the higher levels of ethylene attribute to the enhanced recovery of plants from carbon starvation, ethylene treatment would rescue the hypersensitive response of plants with low ethylene levels. To test this, we treated 3-wk-old sinat1sinat2 seedlings, which produce a low amount of ethylene during carbon starvation (Fig. 5 C and D), with ethephon followed by a 7-d dark treatment (Fig. 5E). Ethephon treatment inhibited the recovery of WT from dark treatment and did not rescue the hypersensitive response of sinat1sinat2, suggesting that the lower levels of ethylene are not the main cause of the hypersensitivity of the sinat1sinat2 to carbon starvation. To further confirm the role of ethylene in the process, we examined the carbon starvation response of the constitutive ethylene response ctr1-2 and ethylene-insensitive ein2-5 mutants (SI Appendix, Fig. S13). The ctr1-2 failed to recover from dark treatment; however, the ein2-5 showed a similar recovery as the WT. These results suggest that ethylene levels and ethylene signaling are unlikely responsible for the hypersensitive carbon starvation phenotypes of plants with the reduced SINAT function.
In the absence of ACS5 and ACS9, the negative role of BR on the destabilization of SINAT2 protein was greatly diminished (Fig. 3E). Thus, we examined the effect of increased protein abundance of SINAT2 in the acs5acs9 mutant background during carbon starvation. To this end, we first examined the carbon starvation phenotypes and ethylene production of acs2acs6 and acs5acs9. Both the acs2acs6 and acs5acs9 mutant produced a significantly lower amount of ethylene than the WT with a more notable decrease in the acs5acs9 during carbon starvation (SI Appendix, Fig. S14). The acs2acs6 and acs5acs9 mutants exhibited similar levels of growth recovery from starvation compared to the WT (SI Appendix, Fig. S14), although a smaller number of both the mutants recuperated more slowly than the WT plants. The similar recovery of the acs5acs9 and acs2acs6 despite different levels of ethylene biosynthesis during starvation may have resulted from the redundant role of other type-2 ACS proteins that interact with SINAT2 in the acs5acs9 mutant (SI Appendix, Figs. S1 and S14). Interestingly, unlike 35S::Myc-SINAT2/acs2cs6, 35S::Myc-SINAT2/acs5acs9 plants did not recover from carbon starvation and produced significantly lower levels of ethylene than the WT and 35S::Myc-SINAT2/acs2acs6 during starvation (Fig. 5 F and G and SI Appendix, Fig. S10). We also found that the higher levels of Myc-SINAT2 were expressed in the acs5acs9 background than in the WT and acs2acs6 after 5 d of dark treatment (SI Appendix, Fig. S15), which is similar to the response of these plants to BR (Fig. 3E). Considering the negative roles of SINAT2, which promotes the degradation of a subset of ATG proteins in carbon starvation (30, 38), the increased protein abundance of SINAT2 in acs5acs9 may have negatively impacted carbon starvation response. In sum, these results suggest that a plant responds to carbon starvation by regulating SINAT2/EOL2/ACS5 protein homeostasis, which results in the fine-tuning of ethylene biosynthesis.
Discussion
In this study, we uncovered a mechanism in which ACS abundance was regulated by the reciprocal antagonistic degradation of SINAT2 and EOL2 E3 ligases in a BR-dependent manner. We found that a subset of ACS acts as a scaffold that tethers SINAT2 and EOL2, which promotes the degradation of SINAT2 and EOL2, opening an opportunity to investigate the role of ACS beyond its canonical function in ethylene biosynthesis. It would be interesting to discover more biological processes that utilize ACS as a scaffold and to determine whether the enzyme activity of ACS is required for bridging SINAT2 and EOL2. Our findings unraveled the unique protein stability regulation mechanism that consists of multiple layers of negative feedback loops that enable the simultaneous and stringent control of the protein stability of multiple proteins. The substrate-dependent reciprocal degradation of E3 ligases also introduces a new aspect of protein stability regulation and demonstrates how homeostasis between E3 ligases and their substrates are maintained in cells.
Our findings also revealed a mechanism that regulates the crosstalk between ethylene and BR. Here, we showed that 14–3-3 likely plays a role in the reciprocal degradation between SINAT2 and EOL2 by conveying BR signal into the SINAT-ETO1/EOL regulatory circuit. All five SINAT isoforms possessed a strong 14–3-3–binding motif (CRFRPY[T]147CPYA) with a potential phosphorylation site (T147 on SINAT2) (Fig. 4A). The T147A mutation in SINAT2, which did not affect the E3 ligase activity of SINAT2, inhibited the reciprocal degradation-mediated ACS stability regulation in the presence of BR. These results suggest that the BR-induced phosphorylation of T147 of SINAT2 promotes 14–3-3 recruiting to SINAT2, thereby bridging SINAT2 and EOL2 for reciprocal degradation. It is also likely that EOL2 could undergo BR-mediated phosphorylation because 14–3-3 also directly binds to EOL2 and regulates the stability of EOL2 (12). The critical role of T147 residue in SINAT2 on ACS and EOL2 turnover regulation in the presence of BR suggests that BR-activated kinases, such as BRI1 and BAK1, could be involved in this regulatory process. Further studies on identifying kinases that are activated by BR and phosphorylate SINAT and/or ETO1/EOLs would allow more insights into BR-induced 14–3-3–mediated ACS protein stability regulation and ethylene–BR crosstalk. Intriguingly, we found that BR decreases 14–3-3 protein abundance in the dark and during the light-to-dark transition (Fig. 4 G, H, and I). These results were unexpected considering that 14–3-3 stimulates the reciprocal degradation of SINAT2 and EOL2 in the presence of BR. However, the dark- or BR-induced down-regulation of 14–3-3 stability could be another layer of the negative feedback loop that maintains the cellular protein homeostasis of ACS and its cognate E3 ligases during carbon starvation by limiting the availability of 14–3-3 proteins. Moreover, multiple 14–3-3 isoforms in Arabidopsis may also increase the complexity of the reciprocal antagonistic regulation of SINAT2 and EOL2. Arabidopsis contains 13 different 14–3-3 isoforms of which we examined the role of 14–3-3ω in this study. Given that 35::Myc-SINAT2T147A seedlings showed a hypersensitive response to carbon starvation, other 14–3-3 isoforms could have a similar role in this process. However, several studies have demonstrated that different 14–3-3 isoforms often play a distinctive role in plants, which may be related to their substrate specificity (25, 52–54). In addition to 14–3-3ω, SINAT2, EOL2, and ACS5 exist as a multigene family; thus, further studies on whether the role of SINAT2/EOL2/ACS5/14–3-3ω can be extrapolated to the other family members of each protein will provide more insight into the regulation of ethylene biosynthesis in response to stress.
Fixed carbon starvation assays demonstrated a link between the BR-activated SINAT/EOL2 regulatory circuit and plant stress responses. Plants without SINAT1 and SINAT2 and those expressing SINAT2 with the T147A mutation showed hypersensitive responses to carbon starvation. The measurement of the ethylene production in these plants revealed that both plants produced significantly lower levels of ethylene than the WT plants after dark treatment. Consistent with these results, ethylene-overproducing mutants showed enhanced recovery from carbon starvation (SI Appendix, Fig. S12). However, ethephon treatment did not rescue the hypersensitive carbon starvation phenotypes of the sinat1sinat2 (Fig. 5E) and inhibited the recovery of WT seedlings from starvation. Corroborating these results, ctr1-2 displayed a hypersensitive response to carbon starvation (SI Appendix, Fig. S13). Together, these results suggest that neither ethylene levels nor ethylene signaling plays a role in the hypersensitivity of the sinat1sinat2 to carbon starvation. Based on our biochemical analysis of the SINAT2-mediated regulation of ACS stability, the lower ethylene levels and hypersensitive carbon starvation response of sinat1sinat2 and 35S::Myc-SINAT2T147A/sinat1sinat2 are likely due to the disrupted the cellular protein homeostasis of ACS5/SINAT2/EOL. The disrupted cellular protein homeostasis of ACS/SINAT2/EOL2 may further lead to the disturbance of the homeostasis of other proteins that play a role in carbon starvation such as BES1 (33). A recent study has shown that SINAT2 promotes BES1 degradation during carbon starvation, and SINAT RNA interference lines displayed a hypersensitivity to carbon starvation, which may result from the overaccumulation of BES1 protein (33).
In addition, a subset of SINAT proteins has been shown to be involved in regulating the stability and activity of ATG proteins that are essential for autophagosome formation and stress responses (30, 38). SINATs also promote the ubiquitination of the ESCRT component FREE1, which regulates vacuolar protein transport and the vacuolar degradation of autophagosomes and is involved in the plant response to iron deficiency (39–41). Our results showed that the abundance of SINAT2 is also regulated by the reciprocal degradation of SINAT2 and ETO1/EOLs (Fig. 3). Thus, the hypersensitive response of 35S::Myc-SINATT147A and 35S::Myc-SINAT2/acs5acs9 to carbon starvation may have been, in part, a result of an enhanced SINAT2-mediated down-regulation of ATG proteins or FREE1; both the T147 mutation in SINAT2 and the lack of ACS5 and ACS9 disrupt the SINAT2-ETO1/EOL reciprocal degradation, likely leading to an increase SINAT2 stability. Intriguingly, 35S::Myc-SINAT2/eto1eol1eol2 and eto1eol1eol2 showed an opposite carbon starvation response to 35S::Myc-SINAT2/acs5acs9 despite the possibility that both plants would produce an increased amount of SINAT2 (Fig. 5F and SI Appendix, Fig. S12). One possible explanation for this discrepancy is that stabilized type-2 ACS proteins in the absence of ETO1/EOL1/EOL2 negatively regulate SINAT2 stability in a manner independent of ETO1EOL1EOL2, presumably via other unknown E3 ligases or degradation mechanism(s).
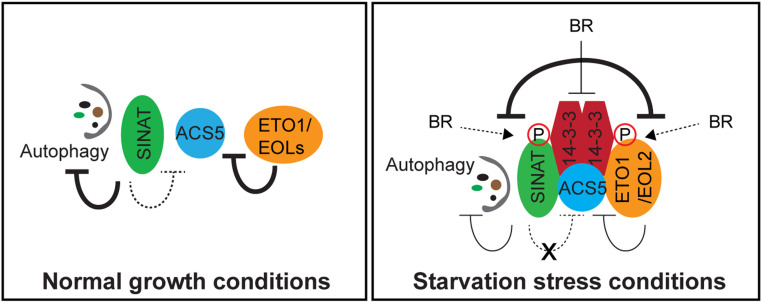
We propose a model in which SINAT2 and ETO1/EOLs control the stability of ACS and themselves differently during normal growth and stress conditions (Fig. 6). During normal growth, a plant maintains low levels of ethylene to promote vegetative growth. This can be mainly achieved via the down-regulation of type-2 ACS by ETO1/EOLs. SINAT E3 ligases may negatively act on ACS stability when it is stabilized by other stimuli such as light; otherwise, they mainly suppress autophagy likely by inhibiting a subset of ATG proteins, thus maintaining the basal level of autophagy. When under stress, the stress-activated BR signaling pathway stimulates the reciprocal degradation of SINAT2 and ETO1/EOLs; 14–3-3 interacts with the BR-induced phosphorylated SINAT2, and perhaps the phosphorylated form of ETO1/EOL as well, bridging the BR signaling pathway and ethylene biosynthesis. The activation of the reciprocal degradation between the E3 ligases leads to an increase in type-2 ACS protein abundance. The reciprocal degradation between the E3 ligases also enhances the activation of autophagy via increased protein stability or the activity of ATG proteins that result from reduced SINAT2 protein levels. The reciprocal antagonistic regulation of SINAT2 and EOL2 may be negatively regulated by the BR-mediated destabilization of 14–3-3, presenting the dual function of 14–3-3 in the process. It is of interest to determine whether SINAT2 or EOL2 is a cognate E3 ligase for 14–3-3.
Fig. 6.
A model for the SINAT2-ETO1/EOL–mediated regulation of ACS stability in response to starvation stress. Under normal growth conditions, the protein stability of ACS5 and other type-2 ACS isoforms is negatively regulated by ETO1/EOLs, resulting in low levels of ethylene in Arabidopsis plants. SINAT E3 ligases suppress autophagy by downregulating the activity and stability of a subset of AUTOPHAGY-RELATED PROTEINs, thus maintaining the basal levels of autophagy. The SINAT E3 ligase family also may negatively regulate the stability of ACS5 when their expression is induced by stimuli. Under stress conditions such as carbon starvation, BR-induced phosphorylation of SINAT2 and/or ETO1/EOLs recruits 14–3-3 to the complex, which activates the reciprocal degradation of the E3 ligases, thus increasing the ACS5 protein abundance and autophagy activity by reducing SINAT stability. BR negatively regulates the reciprocal antagonistic regulation of SINAT2/EOL2/ACS complex via a negative feedback loop in which BR decreases 14–3-3 protein abundance. The thickness of the lines indicates the strength of the inhibition. Solid or dotted lines represent experimentally confirmed or hypothetical results, respectively. Arrows and blunt lines indicate activation or inhibition, respectively.
In conclusion, our findings revealed that the regulation of ACS protein stability is a key mechanism that governs the crosstalk between ethylene biosynthesis, BR, and autophagy. ACS protein stability is regulated by diverse inputs, including plant hormones and environmental signals. Given that, ACS could be a central molecular component that can transduce environmental signals into the cells via crosstalk with phytohormone pathways, thereby changing cellular energy levels via autophagy (55).
Materials and Methods
Detailed information, including plant materials and growth conditions, plasmid construction, steady-state protein level analysis using the Arabidopsis protoplast system, protein degradation assays, co-IP, fixed carbon starvation assays, Y2H, bimolecular fluorescence complementation and colocalization analysis, fluorescence microscope analysis, immunoblot analysis, ubiquitination assays, coimmunoprecipitation, qRT-PCR, protein expression and purification, R18 treatment, and ethylene measurement is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Alison Delong for the critical reading of the manuscript and Yuan-Chi Chen for supporting experiments. This work was supported by grants from Purdue Startup and NSF (MCB-1817286, to G.M.Y.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.M.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011900118/-/DCSupplemental.
Data Availability
All study data are included in the paper and SI Appendix.
References
- 1.Moon J., Parry G., Estelle M., The ubiquitin-proteasome pathway and plant development. Plant Cell 16, 3181–3195 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smalle J., Vierstra R. D., The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Gagne J. M., et al., Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 6803–6808 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren H., et al., Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 4722–4731 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C. W., et al., The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18, 1084–1098 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu K., Yang W., E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 58, 1461–1476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClellan C. A., Chang C., The role of protein turnover in ethylene biosynthesis and response. Plant Sci. 175, 24–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiao H., Chang K. N., Yazaki J., Ecker J. R., Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23, 512–521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon G. M., New insights into the protein turnover regulation in ethylene biosynthesis. Mol. Cells 38, 597–603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booker M. A., DeLong A., Producing the ethylene signal: Regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 169, 42–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connor H. F., Huibregtse J. M., Enzyme-substrate relationships in the ubiquitin system: Approaches for identifying substrates of ubiquitin ligases. Cell. Mol. Life Sci. 74, 3363–3375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon G. M., Kieber J. J., 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel J. P., Woeste K. E., Theologis A., Kieber J. J., Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. U.S.A. 95, 4766–4771 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chae H. S., Faure F., Kieber J. J., The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15, 545–559 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H. Y., Chen Y. C., Kieber J. J., Yoon G. M., Regulation of the turnover of ACC synthases by phytohormones and heterodimerization in Arabidopsis. Plant J. 91, 491–504 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Hansen M., Chae H. S., Kieber J. J., Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 57, 606–614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camoni L., Visconti S., Aducci P., Marra M., 14-3-3 proteins in plant hormone signaling: Doing several things at once. Front Plant Sci 9, 297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaspert N., Throm C., Oecking C., Arabidopsis 14-3-3 proteins: Fascinating and less fascinating aspects. Front Plant Sci 2, 96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oecking C., Jaspert N., Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12, 760–765 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Gampala S. S., et al., An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu H., et al., Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catalá R., et al., The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu G., et al., FERONIA phosphorylates E3 ubiquitin ligase ATL6 to modulate the stability of 14-3-3 proteins in response to the carbon/nitrogen ratio. J. Exp. Bot. 70, 6375–6388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y. S., et al., Sugar starvation-regulated MYBS2 and 14-3-3 protein interactions enhance plant growth, stress tolerance, and grain weight in rice. Proc. Natl. Acad. Sci. U.S.A. 116, 21925–21935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayfield J. D., Folta K. M., Paul A. L., Ferl R. J., The 14-3-3 proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiol. 145, 1692–1702 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao Y., Du Y., Jiang L., Liu J. Y., Interaction between ACC synthase 1 and 14-3-3 proteins in rice: A new insight. Biochemistry (Mosc.) 72, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Chang I. F., et al., Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9, 2967–2985 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S. J., et al., A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J. Exp. Bot. 64, 4343–4360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander R. D., Morris P. C., A proteomic analysis of 14-3-3 binding proteins from developing barley grains. Proteomics 6, 1886–1896 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Qi H., et al., Arabidopsis SINAT proteins control autophagy by mediating ubiquitylation and degradation of ATG13. Plant Cell 32, 263–284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peralta D. A., Araya A., Gomez-Casati D. F., Busi M. V., Over-expression of SINAL7 increases biomass and drought tolerance, and also delays senescence in Arabidopsis. J. Biotechnol. 283, 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Den Herder G., et al., Seven in absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiol. 148, 369–382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nolan T. M., et al., Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, 33–46.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagano Y., et al., Siah1/SIP regulates p27(kip1) stability and cell migration under metabolic stress. Cell Cycle 10, 2592–2602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang J., Liu N., Tanaka N., Abe K., Expressions of hypoxic stress sensor proteins after transient cerebral ischemia in mice. J. Neurosci. Res. 90, 648–655 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Matsuzawa S., Takayama S., Froesch B. A., Zapata J. M., Reed J. C., p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: Suppression by BAG-1. EMBO J. 17, 2736–2747 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao Y., et al., The tumor necrosis factor receptor-associated factor (TRAF)-like family protein SEVEN IN ABSENTIA 2 (SINA2) promotes drought tolerance in an ABA-dependent manner in Arabidopsis. New Phytol. 202, 174–187 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Qi H., et al., TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 29, 890–911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao C., et al., Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 112, 1886–1891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia F. N., et al., SINAT E3 ubiquitin ligases mediate FREE1 and VPS23A degradation to modulate abscisic acid signaling. Plant Cell 32, 3290–3310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Z., et al., SINAT E3 ligases regulate the stability of the ESCRT component FREE1 in response to iron deficiency in plants. J. Integr. Plant Biol. 62, 1399–1417 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Christians M. J., et al., The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57, 332–345 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall R. S., Vierstra R. D., Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Xie Q., et al., SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Yang M., et al., SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 41, 47–58.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo D. H., Yoon G. M., Light-induced stabilization of ACS contributes to hypocotyl elongation during the dark-to-light transition in Arabidopsis seedlings. Plant J. 98, 898–911 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Yoshida H., Nagata M., Saito K., Wang K. L., Ecker J. R., Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 5, 14 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida H., et al., The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol. Biol. 62, 427–437 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Den Herder G., Yoshida S., Antolín-Llovera M., Ried M. K., Parniske M., Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 24, 1691–1707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madeira F., et al., 14-3-3-Pred: Improved methods to predict 14-3-3-binding phosphopeptides. Bioinformatics 31, 2276–2283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung T., Phillips A. R., Vierstra R. D., ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62, 483–493 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Wang P. H., et al., The glutamate receptor-like protein GLR3.7 interacts with 14-3-3ω and participates in salt stress response in Arabidopsis thaliana. Front Plant Sci 10, 1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tseng T. S., Whippo C., Hangarter R. P., Briggs W. R., The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell 24, 1114–1126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambeck I., et al., Kinetic analysis of 14-3-3-inhibited Arabidopsis thaliana nitrate reductase. Biochemistry 49, 8177–8186 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Park C., Lee H. Y., Yoon G. M., The regulation of ACC synthase protein turnover: A rapid route for modulating plant development and stress responses. Curr. Opin. Plant Biol. 63, 102046 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the paper and SI Appendix.