Abstract
The inability of adult mammalian cardiomyocytes to proliferate underpins the development of heart failure following myocardial injury. Although the newborn mammalian heart can spontaneously regenerate for a short period of time after birth, this ability is lost within the first week after birth in mice, partly due to increased mitochondrial reactive oxygen species (ROS) production which results in oxidative DNA damage and activation of DNA damage response. This increase in ROS levels coincides with a postnatal switch from anaerobic glycolysis to fatty acid (FA) oxidation by cardiac mitochondria. However, to date, a direct link between mitochondrial substrate utilization and oxidative DNA damage is lacking. Here, we generated ROS-sensitive fluorescent sensors targeted to different subnuclear compartments (chromatin, heterochromatin, telomeres, and nuclear lamin) in neonatal rat ventricular cardiomyocytes, which allowed us to determine the spatial localization of ROS in cardiomyocyte nuclei upon manipulation of mitochondrial respiration. Our results demonstrate that FA utilization by the mitochondria induces a significant increase in ROS detection at the chromatin level compared to other nuclear compartments. These results indicate that mitochondrial metabolic perturbations directly alter the nuclear redox status and that the chromatin appears to be particularly sensitive to the prooxidant effect of FA utilization by the mitochondria.
Keywords: mitochondria, reactive oxygen species, metabolism
Heart failure, a leading cause of death worldwide, is precipitated by the inability of the adult mammalian heart to regenerate following injury. Although the adult mammalian heart is incapable of meaningful regeneration (1), neonatal mammalian cardiomyocytes retain a proliferative competency for the first few days of life (2), which underpins the transient regenerative potential of the newborn mammalian heart (3). The regulation of the cardiomyocyte cell cycle is, however, highly complex, as demonstrated by the multitude of pathways that have been implicated in this process (4, 5). We recently demonstrated that the postnatal switch from anaerobic glycolysis to fatty acid (FA)-dependent mitochondrial respiration induces DNA damage and activation of the DNA damage response pathway, which mediates cell-cycle arrest of postnatal cardiomyocytes (6, 7) Conversely, gradual exposure to severe systemic hypoxia or inhibition of mitochondrial FA oxidation decreases DNA damage and reactivates cardiomyocyte cell cycle in adult mice (7, 8). Although the role of mitochondria as a source of oxidative stress has been established since the 1960s, the mechanism by which mitochondrial reactive oxygen species (ROS) induce DNA damage is not well understood. Specifically, it is unclear whether mitochondrial ROS affect various nuclear compartments equally as part of a pattern of generalized oxidative stress, or whether specific subnuclear compartments are more susceptible to mitochondrial oxidative stress. Therefore, we set out to examine the spatial pattern of ROS in the nucleus as a function of inhibition of respiratory chain complexes and altering mitochondrial substrate utilization.
Results and Discussion
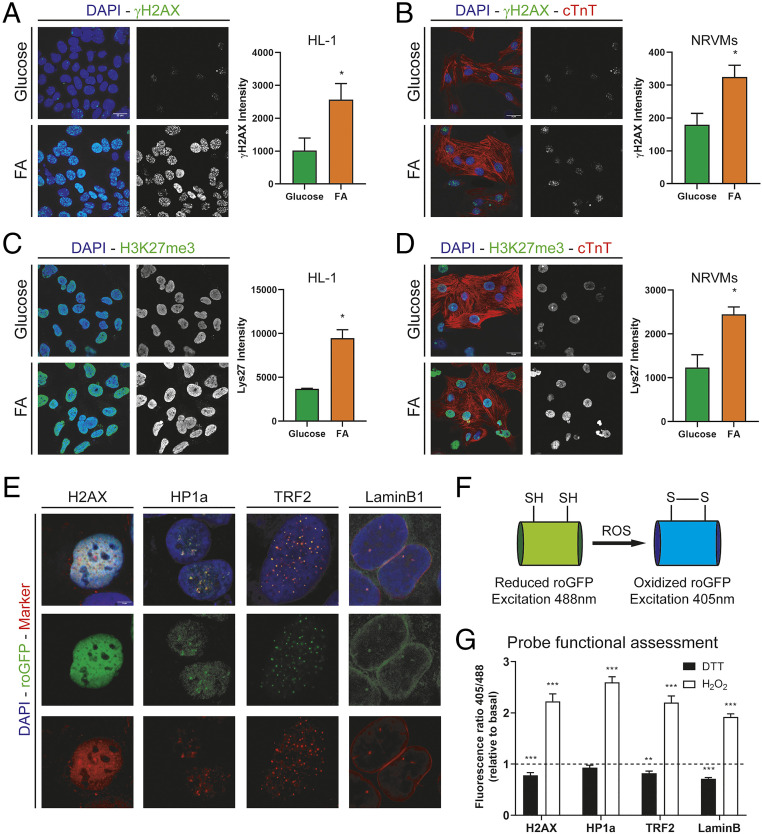
To determine whether changing mitochondrial substrate utilization can affect the degree of DNA damage we cultured HL1 cardiomyocyte cell line and neonatal rat ventricular myocytes (NRVMs) in presence of high levels of glucose (25 mM) or FA medium (100 µM sodium oleate, 100 µM sodium palmitate, and 3 mM l-carnitine) for 24 h and analyzed the levels of DNA damage by γH2AX staining (Fig. 1 A and B). Nuclear γH2AX intensity quantification showed higher values following FA utilization, indicating that mitochondrial FA oxidation results in increased DNA damage, which is supportive of previous reports (7). DNA repair is known to be associated with increased methylation of histone H3 at lysine 27 (9), and thus we analyzed H3K27me3 staining intensity upon FA treatment (Fig. 1 C and D). We found that trimethylation of H3K27 is increased after FA treatment compared to glucose. Next, we examined whether the increased amount of DNA damage upon FA treatment was due to increased ROS levels in the nucleus in real time using the ratiometric sensor roGFP. Since the nucleus is not a homogeneous structure, we directed the sensor to various nuclear subcompartments by using different fusion reporter tags including euchromatin (histone 2AX, H2AX), heterochromatin (heterochromatin protein 1α, HP1α), telomeres (telomeric repeat-binding factor 2, TRF2), and nuclear membrane (lamin B1). We used cells stably expressing these constructs to confirm proper localization of the sensor by costaining with specific markers and green fluorescent protein (GFP) fluorescence (Fig. 1E), confirming the proper localization of the probes within the nuclear subcompartments. roGFP is a modified GFP with two cysteine residues that are sensitive to oxidation. This results in alteration of its fluorescence in response to the redox environment where the reduced roGFP has a maximum fluorescence at 488 nm and the oxidized roGFP has a maximum fluorescence at 405 nm (Fig. 1F). By recording 405-nm and 488-nm fluorescence we are able to measure the ratio of oxidized to reduced sensor (405 nm/488 nm) independent of the total amount of sensor expressed (10). Fluorescence ratio quantification of NRVMs following dithiothreitol (DTT) (4 mM) or H2O2 (1 mM) for 10 min showed a decreased 405/488 ratio upon DTT treatment (probe reduction) and increased ratio upon H2O2 treatment (probe oxidation), confirming the functionality of the fusion sensors (Fig. 1G).
Fig. 1.
(A) γH2AX staining in HL1 cells treated with 25 mM glucose or 200 µM FA for 24 h. Graph shows γH2AX nuclear intensity quantification (n = 4). (B) γH2AX staining in NRVMs treated with 25 mM glucose or 200 μmM FA for 24 h. Graph shows γH2AX nuclear intensity quantification (n = 3). (C) Histone 3 trimethylated lysine 27 (H3K27me3) staining in HL1 cells treated with 25 mM glucose or 200 µM FA for 24 h. Graph shows γH2AX nuclear intensity quantification (n = 4). (D) Histone 3 trimethylated lysine 27 (H3K27me3) staining in NRVMs treated with 25 mM glucose or 200 µM FA for 24 h. Graph shows γH2AX nuclear intensity quantification (n = 3). (E) Fluorescence images of nuclear targeted probes. roGFP fluorescence is shown in green (Middle). Markers for subnuclear compartments (H2AX, euchromatin; HP1α, heterochromatin; TRF2, telomeres; and lamin B1, nuclear membrane) are shown in red (Bottom). (F) Schematic representation of roGFP in reduced and oxidized states. (G) Fluorescence ratio (405/488) per cell relative to basal levels (low glucose) in the presence of 4 mM DTT (black bars) or 1 mM H2O2 (white bars). For all graphs, bars represent mean ± SEM. ***P < 0.005, **P < 0.01, *P < 0.05; Student’s t test. (Scale bars, 20 µm.)
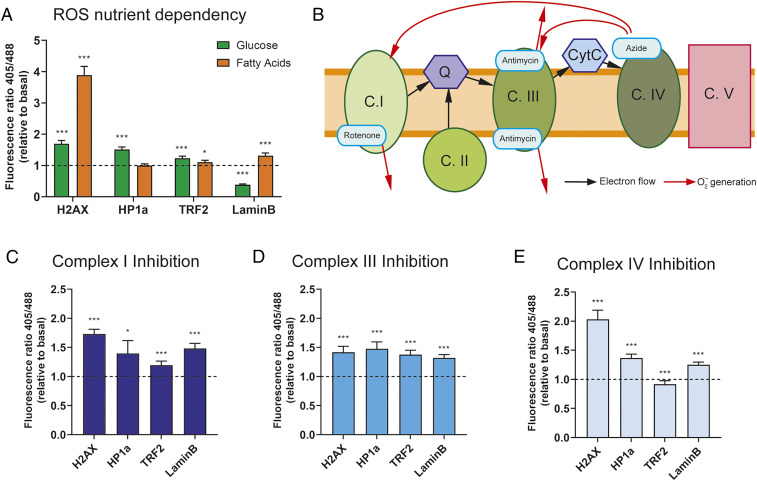
To assess the role of FA oxidation in subnuclear ROS levels, NRVMs expressing the sensors were first imaged under low-glucose medium (1 mM) as basal condition. After 5 min of acquisition time, cells were incubated with high-glucose-containing medium (25 mM) or FA medium (100 µM sodium oleate, 100 µM sodium palmitate, and 3 mM l-carnitine) for 10 min. Ratio quantification showed increased probe oxidation upon glucose treatment for all compartments except nuclear membrane compared to baseline. Intriguingly, FA utilization resulted in a marked increase in oxidation of the chromatin-targeted probe compared to other subcompartments (Fig. 2A). These results suggest that the observed increased DNA damage upon FA utilization is in part secondary to increased ROS levels in the euchromatin.
Fig. 2.
(A) Fluorescence ratio (405 nm/488 nm) per cell relative to basal levels (low glucose) in presence of 25 mM glucose (green bars) or 200 µM FA (orange bars). (B) Schematic of mitochondrial electron transport chain with inhibitor binding sites and ROS generation flow (red arrows). (C–E) Fluorescence ratio (405 nm/488 nm) per cell relative to basal levels (low glucose) in presence of 10 µM rotenone (C), 30 µM antimycin A (D) or 20 mM sodium azide (E). For all graphs, bars represent mean ± SEM. ***P < 0.005, *P < 0.05; Student’s t test.
It is well established that inhibition of respiratory chain complexes is associated with increased ROS production (11). Here we utilized this phenomenon to confirm whether the location of ROS generation at the level of respiratory chain complexes plays a role in the spatial distribution of ROS within the nucleus. To achieve this, we used specific inhibitors of respiratory chain complexes I, III, or IV and determined the effect on nuclear ROS detection in NRVMs. After basal conditions imaging (low glucose), cells were treated with complex I, III, or IV inhibitors for 10 min, as schematized in Fig. 2B. NRVMs treated with rotenone (10 µM, complex I inhibitor) showed increased oxidation in all compartments, with significantly higher oxidation detected in the chromatin-targeted sensor compared to heterochromatin or telomeres, while the increase compared to nuclear membrane probe was not significant (Fig. 2C). Complex III inhibition with antimycin A (30 µM) showed an increased oxidation pattern that was homogeneous among the different locations (Fig. 2D). Finally, azide-induced complex IV inhibition (20 mM) resulted in increased oxidation in all compartments except telomere, with the highest oxidation detected in the chromatin compartment (Fig. 2E). It is important to note here that it is expected that complex IV inhibition would result in increased ROS production at both complex I and complex III levels, but not directly at complex IV. These results indicate that all mitochondrial respiratory complexes can increase nuclear ROS levels. Overall, these inhibition studies support our previous results (Fig. 2A) indicating that mitochondrial respiration is a major source of nuclear oxidation. In addition, our observation that FAO markedly increases chromatin ROS and chromatin oxidation could at least partially explain why inhibition of FAO in the adult heart reduces oxidative DNA damage (7).
In summary, our results demonstrate that FA utilization in cardiomyocytes increases oxidative DNA damage in the nucleus, which is at least in part secondary to increased nuclear ROS levels, particularly at the level of the chromatin. Overall, this brief report demonstrates that mitochondrial-mediated ROS generation is sensed by the nucleus, which could play important roles both as a signaling mechanism, as well as a source of oxidative stress.
Materials and Methods
Cell Culture.
The HL1 cell line was maintained and FA treated as previously described (12). NRVMs were isolated and cultured as described previously (13).
Adeno-Associated Virus Production and Infection.
The plasmids pTR-GNP for generating adeno-associated virus (AAV) expression cassette and pDP6rs for packaging AAV6 were gifts from Roger Hajjar (Icahn School of Medicine at Mount Sinai, New York) (14).
Statistical Analysis.
Normalized ratio values (relative to basal conditions) were compared using Student’s t test. Significance was considered at P < 0.05.
Supplementary Material
Acknowledgments
I.M.-M. is supported by an Alfonso Martin Escudero Foundation Fellowship. This work was supported by NIH grants R01HL138983-02 (to H.A.S., L.I.S., and A.A.) and R01AG053341 (to A.A.) and Cancer Prevention and Research Institute of Texas grant RP190435 (to H.A.S. and A.A.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101674118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Senyo S. E., et al., Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soonpaa M. H., Kim K. K., Pajak L., Franklin M., Field L. J., Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 271, H2183–H2189 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Porrello E. R., et al., Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbas N., Perbellini F., Thum T., Non-coding RNAs: Emerging players in cardiomyocyte proliferation and cardiac regeneration. Basic Res. Cardiol. 115, 52 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yester J. W., Kühn B., Mechanisms of cardiomyocyte proliferation and differentiation in development and regeneration. Curr. Cardiol. Rep. 19, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puente B. N., et al., The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 157, 565–579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardoso A. C., et al., mitochondrial substrate utilization regulates cardiomyocyte cell cycle progression. Nat. Metab. 2, 167–178 (2020). [PMC free article] [PubMed] [Google Scholar]
- 8.Nakada Y., et al., Hypoxia induces heart regeneration in adult mice. Nature 541, 222–227 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., et al., Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J. Cell Sci. 131, jcs215525 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Dooley C. T., et al., Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 279, 22284–22293 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Loschen G., Flohé L., Chance B., Respiratory chain linked H(2)O(2) production in pigeon heart mitochondria. FEBS Lett. 18, 261–264 (1971). [DOI] [PubMed] [Google Scholar]
- 12.Crewe C., Schafer C., Lee I., Kinter M., Szweda L. I., Regulation of pyruvate dehydrogenase kinase 4 in the heart through degradation by the lon protease in response to mitochondrial substrate availability. J. Biol. Chem. 292, 305–312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Y. G., et al., Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation 114, 1159–1168 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammoudi N., Ishikawa K., Hajjar R. J., Adeno-associated virus-mediated gene therapy in cardiovascular disease. Curr. Opin. Cardiol. 30, 228–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.