Significance
Canonical, β-catenin–dependent Wnt signaling is well known to promote a variety of cancer types. However, the contribution of β-catenin’s cell adhesion function versus its transcriptional output to malignant tumor progression and metastasis has remained unexplored. We dissected β-catenin’s function in cell adhesion from its transcriptional output in Wnt signaling in the mouse mammary tumor virus (MMTV)–PyMT transgenic mouse model of metastatic breast cancer. We found that β-catenin’s dual functions are required for tumor cell survival. In contrast, its transcriptional activity is required for the regulation of epithelial–mesenchymal transition in vitro and metastasis formation in vivo. Altogether, the results delineate at a molecular and mechanistic level β-catenin’s contribution to malignant breast cancer progression.
Keywords: β-catenin, breast cancer, cell adhesion, metastasis, Wnt signaling
Abstract
During malignant progression, epithelial cancer cells dissolve their cell–cell adhesion and gain invasive features. By virtue of its dual function, β-catenin contributes to cadherin-mediated cell–cell adhesion, and it determines the transcriptional output of Wnt signaling: via its N terminus, it recruits the signaling coactivators Bcl9 and Pygopus, and via the C terminus, it interacts with the general transcriptional machinery. This duality confounds the simple loss-of-function analysis of Wnt signaling in cancer progression. In many cancer types including breast cancer, the functional contribution of β-catenin’s transcriptional activities, as compared to its adhesion functions, to tumor progression has remained elusive. Employing the mouse mammary tumor virus (MMTV)–PyMT mouse model of metastatic breast cancer, we compared the complete elimination of β-catenin with the specific ablation of its signaling outputs in mammary tumor cells. Notably, the complete lack of β-catenin resulted in massive apoptosis of mammary tumor cells. In contrast, the loss of β-catenin’s transcriptional activity resulted in a reduction of primary tumor growth, tumor invasion, and metastasis formation in vivo. These phenotypic changes were reflected by stalled cell cycle progression and diminished epithelial–mesenchymal transition (EMT) and cell migration of breast cancer cells in vitro. Transcriptome analysis revealed subsets of genes which were specifically regulated by β-catenin’s transcriptional activities upon stimulation with Wnt3a or during TGF-β–induced EMT. Our results uncouple the signaling from the adhesion function of β-catenin and underline the importance of Wnt/β-catenin–dependent transcription in malignant tumor progression of breast cancer.
The identification and functional characterization of the molecular pathways underlying the malignant progression of metastatic cancers remain prime goals for the development of innovative cancer therapies. Among many, the Wnt/β-catenin signaling pathway is critical for numerous processes during embryonic development and in adult tissue homeostasis, including stem cell maintenance and homeostatic self-renewal, cell differentiation, and regulation of tissue growth (1). While mutations of components of canonical Wnt signaling that lead to its activation are observed in many different cancer types, they are only rarely found in breast cancer. In contrast, in breast cancer, the Wnt pathway is found activated by either the increased expression of individual Wnt ligands or the loss of the expression of negative pathway regulators. In fact, Wnt1 has been first identified as an oncogene in murine mammary tumor virus-induced tumors in mice (2). In breast cancers, the genes encoding for negative pathway regulators, such as Wnt inhibitory factor 1, secreted frizzled-related protein, Dickkopf 1 as well as APC (adenomatous polyposis coli), AXIN1, and AXIN2, have been found hypermethylated and their expression levels reduced (3–7). As a consequence, increased nuclear β-catenin is detected in many breast tumors (8, 9). Consistent with these findings, the oncogenic potential of Wnt ligands and other canonical Wnt pathway components have been confirmed in the mammary gland (8). For example, the forced expression of stabilized β-catenin or of LRP5/6 (low density lipoprotein receptor-related protein 5/6) or the loss of APC in the mammary gland of transgenic mice leads to mammary gland hyperplasia or tumor formation (10–13).
β-catenin itself has a dual function: it is a crucial component of E-cadherin and other cadherin-mediated cell–cell adhesion complexes required for the structural integrity and functional polarization of epithelia and other tissue structures, yet it is also the key nuclear effector of the canonical Wnt signaling pathway. Upon Wnt pathway activation, β-catenin accumulates in the cytoplasm and translocates to the nucleus where it binds to TCF/LEF transcription factors (14, 15). Many different coactivators are known to bind to β-catenin and thereby to modulate its transcriptional activity. Some of them bind at the C terminus of β-catenin, among them CBP (CREB binding protein, CREBBP), p300 (E1A binding protein P300), TIP60 (lysin acetyltransferase 5, KAT5), SWI/SNF (SWItch/Sucrose Non-Fermentable), ISWI (imitation Switch), and the Mediator complex, and all together affect chromatin structure and nucleosome rearrangements and connect β-catenin to the general transcriptional machinery. Other coactivators bind the N terminus of β-catenin, including Bcl9 and Bcl9l, which in turn recruit the cofactors Pygopus 1 and 2 and thus indirectly connect β-catenin to histones (15–18).
The dual functions of β-catenin have made it difficult to specifically study its transcriptional activity independent from its adhesion function during tumor progression and metastasis formation. Recently, specific β-catenin mutant forms have been generated which ablate the transcriptional Wnt signaling activity of β-catenin without affecting its cell adhesion function (19). A D164A point mutation abrogates the binding of Bcl9 and Bcl9l to the N terminus of β-catenin, while a C-terminal truncation (ΔC) of β-catenin prevents the binding of C-terminal transcriptional cofactors required for chromatin remodeling and transcriptional activation. The combination of these two mutations in a double-mutant (dm) version of β-catenin completely abrogates Wnt/β-catenin transcriptional activity but preserves β-catenin’s functions in cell adhesion (19).
To study the contribution of β-catenin transcriptional activity to malignant breast cancer progression, epithelial–mesenchymal transition (EMT), and metastasis, we have compared the complete elimination of β-catenin to its replacement by the signaling-deficient dm version of β-catenin in tumor cells of the mouse mammary tumor virus (MMTV)–PyMT tumor mouse model of breast cancer. MMTV-PyMT transgenic mice exhibit a linear progression of mammary gland carcinogenesis from hyperplasia, further progressing via the adenoma stage to an early and then late carcinoma stage and the formation of lung metastasis (20). We found that the complete ablation of β-catenin resulted in massive tumor cell apoptosis. In contrast, the abrogation of the transcriptional output of β-catenin had more distinct effects: It increased tumor cell apoptosis and diminished malignant tumor progression and metastasis formation in vivo and repressed cell cycle progression and EMT in vitro. The results demonstrate that both of β-catenin’s functions are indispensable for mammary tumor cell survival, while its transcriptional output defines a critical node in the regulation of mammary gland tumor growth, malignant progression, and metastasis formation.
Results
Loss of β-catenin Leads to Tumor Cell Apoptosis.
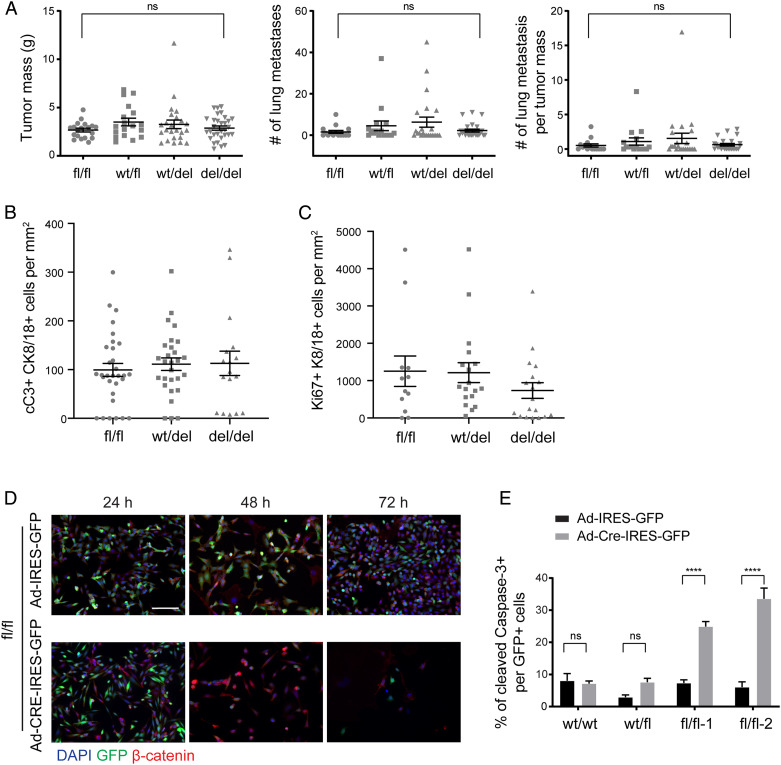
To assess the functional contribution of β-catenin to mammary gland tumor progression and metastasis formation, we employed the MMTV-PyMT mouse model of metastatic breast cancer (20). In MMTV-PyMT mice, β-catenin is highly expressed in all stages of mammary gland tumorigenesis, including hyperplasia, adenoma, and carcinoma, with reduced membrane localization and increased cytoplasmic and nuclear localization in invasive carcinomas (SI Appendix, Fig. S1A). To assess the functional contribution of β-catenin to mammary gland tumorigenesis, MMTV-PyMT mice were crossed with β-cateninfl/fl mice and with MMTV-Cre mice, expressing Cre recombinase exclusively in the mammary epithelial cells (21). β-cateninfl/fl;MMTV-PyMT control mice are designated as fl/fl genotype mice; β-cateninfl/fl;MMTV-PymT;MMTV-Cre mice are designated as del/del genotype mice. Unexpectedly, del/del mice did not show significant differences in tumor weights, in tumor progression from hyperplasia to adenoma to carcinoma, and in the number of lung metastases as compared to littermate fl/fl control mice (Fig. 1A and SI Appendix, Fig. S1 B–D) (22). However, genotype analysis revealed an incomplete deletion of the β-catenin (Ctnnb1) gene in tumors (SI Appendix, Fig. S2A). Consistent with this, immunofluorescence microscopy analysis of tumor sections revealed that β-catenin was still present in tumors of del/del mice (SI Appendix, Fig. S2B). To determine whether Cre recombinase was active in mammary tumor cells, we introduced an LSL-GFP reporter allele into the composite transgenic background. Quantifying the expression of GFP revealed a similar recombination efficiency between control fl/fl and experimental del/del mice at 5 wk of age. The Cre-mediated recombination efficiency of 20% by GFP expression seems low, specifically also in β-catenin wild-type (control) tumors, yet this may be due to the activity of the MMTV enhancer/promoter construct used here, which depends on hormonal induction in mammary epithelial cells. However, at 12 wk of age, the percentage of GFP-positive cells per tumor was found substantially reduced in del/del mice as compared to fl/fl control mice (SI Appendix, Fig. S2C).
Fig. 1.
Ablation of β-catenin expression leads to mammary tumor cell apoptosis. (A) Cre recombinase-mediated ablation of β-catenin in β-cateninfl/fl;MMTV-PyMT;MMTV-Cre triple-transgenic mice did not affect primary tumor growth and lung metastasis formation. Mouse numbers analyzed for primary tumor growth: β-cateninfl/fl (fl/fl), n = 19; β-cateninfl/wt (wt/fl), n = 18; β-cateninfl/wt;MMTV-Cre (wt/del), n = 24; β-cateninfl/fl;MMTV-Cre (del/del), n = 31. Mouse numbers analyzed for lung metastasis formation: β-cateninfl/fl (fl/fl), n = 15; β-cateninfl/wt (wt/fl), n = 16; β-cateninfl/wt;MMTV-Cre (wt/del), n = 23; β-cateninfl/fl;MMTV-Cre (del/del), n = 24. Data are displayed as mean ± SEM. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. ns, not significant. (B) Apoptotic mammary tumor cells were detected by immunofluorescence staining for cleaved Caspase-3 and CK8/18 for tumor cells in preneoplastic lesions of β-cateninfl/fl;MMTV-PyMT (fl/fl; wild-type control), β-cateninwt/fl;MMTV-PyMT;MMTV-Cre (wt/del; heterozygous knockout), and β-cateninfl/fl;MMTV-PyMT;MMTV-Cre (del/del; homozygous knockout) mice at 5 wk of age. The quantification of cleaved Caspase-3–positive/CK8/18-positive cells revealed only a moderate, nonsignificant increase in apoptotic tumor cells upon β-catenin deletion. Data are displayed as mean ± SEM, n = 3 to 4 mice. A representative image of the immunofluorescence staining is shown in SI Appendix, Fig. S2D. (C) Proliferating mammary tumor cells were detected by immunofluorescence staining for Ki67 and CK8/18 for tumor cells in preneoplastic lesions of β-cateninfl/fl;MMTV-PyMT (fl/fl; wild-type control), β-cateninwt/fl;MMTV-PyMT;MMTV-Cre (wt/del; heterozygous knockout), and β-cateninfl/fl;MMTV-PyMT;MMTV-Cre (del/del; homozygous knockout) mice at 5 wk of age. The quantification of Ki67-positive/CK8/18-positive cells revealed only a moderate, nonsignificant decrease in proliferating tumor cells upon β-catenin deletion. The dots in the graphs represent the individual imaging fields analyzed. Data are displayed as mean ± SEM, n = 3 to 4 mice. A representative image of the immunofluorescence staining is shown in SI Appendix, Fig. S2E. (D) Infection of primary tumor cell lines derived from β-cateninfl/fl;MMTV-PyMT tumors with Adenovirus-Cre-IRES-GFP resulted in rapid cell death of infected cells as compared to cells infected with an Adeno-IRES-GFP control virus. DAPI was used to visualize nuclei. (Scale bar, 100 μm.) (E) β-cateninwt/wt (wt/wt; Py2T), β-cateninwt/fl (wt/fl), and β-cateninfl/fl (fl/fl) cells were infected with either Adeno-Cre-IRES-GFP or Adeno-IRES-GFP. The quantification of the number of infected (GFP-positive) and apoptotic (cleaved Caspase-3–positive) cells revealed a significant increase in the numbers of apoptotic cells upon knockout of β-catenin. Data are displayed as mean ± SEM. The results represent four independent experiments. Statistical analysis was performed using one-way ANOVA multiple comparison test. ****P < 0.001; ns, not significant.
These results suggested that mammary tumor cells lacking β-catenin were being eliminated. We thus assessed the extent of apoptosis in tumors of fl/fl control as compared to tumors of wt/fl, wt/del, and del/del mice. Cleaved Caspase-3 staining was slightly but not significantly increased in tumor sections of 5-wk-old mammary gland–specific β-catenin knockout mice as compared to β-catenin wild-type mice (Fig. 1B and SI Appendix, Fig. S2D), while tumor cell proliferation was slightly but not significantly reduced (Fig. 1C and SI Appendix, Fig. S2E). The lack of significance might be due to the fact that an inefficient Cre-mediated recombination at early stages of tumorigenesis hampered the detection of recombined cells which were rapidly undergoing apoptosis, while nonrecombined cells started to overgrow.
In order to explore the possibility that mammary tumor cells might rapidly undergo apoptosis upon the loss of β-catenin expression, stable tumor cell lines were derived from tumors of β-cateninwt/wt;MMTV-PyMT (wt/wt) and of β-cateninfl/fl;MMTV-PyMT (fl/fl) mice. As expected, the infection of fl/fl cells with Adenovirus-Cre-IRES-GFP eliminated the β-catenin gene as assessed by genotyping. However, the loss of β-catenin expression also led to a loss of β-catenin–deficient cells and an increase in the amount of cleaved Caspase-3–positive cells as compared to fl/fl cells infected with a control Adenovirus-IRES-GFP or to wt/wt cells infected with Adenovirus-Cre-IRES-GFP (Fig. 1 D and E). Consistent with these results, we were not able to establish a stable mammary tumor cell line lacking β-catenin. Hence, β-catenin appears critically required for mammary tumor cell survival.
β-catenin Transcriptional Output Promotes Primary Tumor Growth.
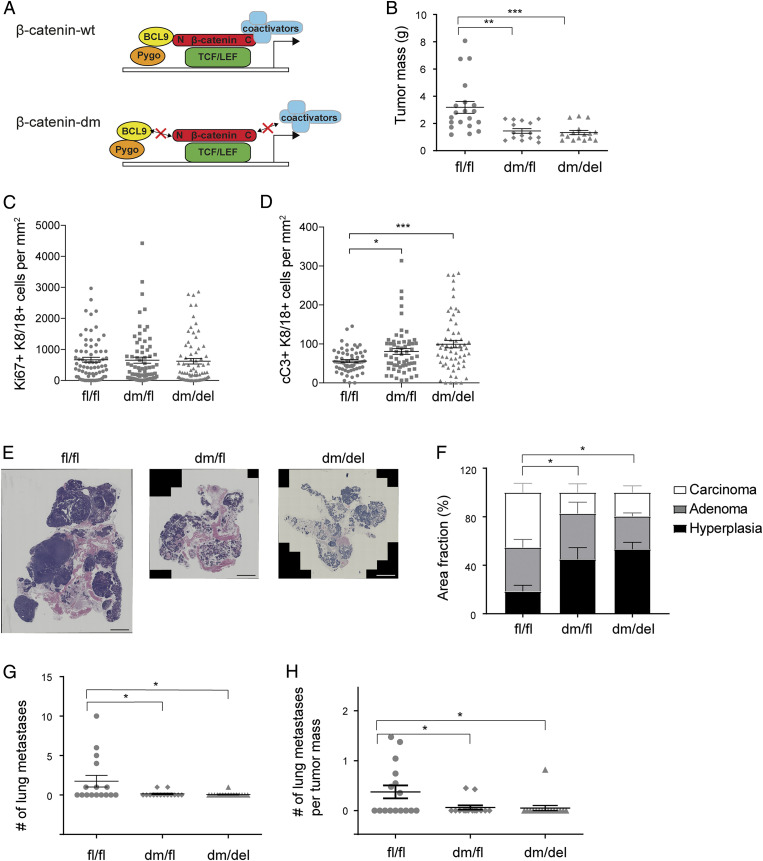
To specifically study the functional contribution of Wnt signaling-induced transcriptional activity of β-catenin to mammary gland tumor progression and metastasis formation, we employed a knock-in mouse line carrying a dm version of β-catenin (19). In this mutant version of β-catenin, the binding of the transcriptional cofactors Bcl9 and Bcl9l to its N terminus (D164A mutation) is abrogated and the C terminus of β-catenin (ΔC) is lacking, thus preventing the binding of cofactors required for chromatin remodeling and transcriptional activation (designated as β-catenindm/fl: dm/fl mice) (Fig. 2A). These mice were intercrossed with MMTV-PyMT mice and with MMTV-Cre mice. The resulting composite transgenic mice expressed either one allele of the dm form and one allele of a conditional (fl) but fully functional allele of β-catenin in mammary tumor cells (β-catenindm/fl;MMTV-PyMT = dm/fl mice) or exclusively the dm form of β-catenin in mammary tumor cells (β-catenindm/fl;MMTV-PyMT;MMTV-Cre: dm/del mice). At 12 wk of age, the composite transgenic mice expressing only the dm form of β-catenin showed a significantly lower tumor mass compared to transgenic mice expressing only wild-type β-catenin (fl/fl mice; Fig. 2B). Notably, composite transgenic mice harboring one allele of dm β-catenin and one fully functional allele of β-catenin (dm/fl mice) also showed reduced tumor growth, indicating a dominant-negative effect of the mutant form of β-catenin (see β-catenin Transcriptional Output Promotes Tumor Cell Proliferation). No such effect was apparent in heterozygous tumors carrying a wild-type and a floxed allele of β-catenin (wt/fl mice; Fig. 1A), thus excluding a gene dosage effect.
Fig. 2.
Loss of β-catenin cofactor binding impairs primary tumor growth, tumor progression, and metastasis formation. (A) Schematic representation of the mutant version of β-catenin used in this study. β-catenin can be divided into three distinct domains: an N-terminal region which binds Bcl9/9l, a central Armadillo (Arm) repeat region which binds TCF/LEF and E-cadherin, and a C-terminal region that binds several transcriptional coactivators. The dm version of β-catenin combines a D164A mutation in the first Arm repeat, which carries an amino acid change from aspartic acid to alanine, ablating the binding of Bcl9/9l to the N terminus, and a ΔC mutation, which carries a truncation of the C terminus of β-catenin, thereby preventing binding of C-terminal transcriptional coactivators. The red crosses on top of arrows highlight impaired interactions. It is important to note that the mutant form of β-catenin retains its cadherin-mediated cell adhesion function. (B) Primary tumor weights of mammary tumors of 12-wk-old MMTV-PyMT transgenic mice expressing in their mammary tumor cells wild-type (fl/fl) β-catenin, one wild-type and one mutant allele of β-catenin (dm/fl), or only a mutant allele of β-catenin (dm/del). Mouse numbers analyzed: fl/fl, n = 20; dm/fl, n = 15; dm/del, n = 16. The dots in the graphs represent the individual mice analyzed. Data are displayed as mean ± SEM. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. **P < 0.01; ***P < 0.005. (C) Quantification of the immunofluorescence staining for Ki67 positive and K8/18 positive on tumor sections from β-cateninfl/fl;MMTV-PyMT (fl/fl), β-catenindm/fl;MMTV-PyMT (dm/fl), and β-catenindm/fl;MMTV-PyMT;MMTV-Cre (dm/del) mice at 12 wk of age. n = 5 mice, 12 imaging fields per mouse. Dots in the graphs represent the individual imaging fields analyzed. Data are displayed as mean ± SEM. (D) Quantification of the immunofluorescence staining for cC3 positive and K8/18 positive on tumor sections from β-cateninfl/fl;MMTV-PyMT (fl/fl), β-catenindm/fl;MMTV-PyMT (dm/fl), and β-catenindm/fl;MMTV-PyMT;MMTV-Cre (dm/del) mice at 12 wk of age. n = 5 mice, 12 imaging fields per mouse. The dots in the graphs represent the individual imaging fields analyzed. Data are displayed as mean ± SEM. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. *P < 0.05; ***P < 0.005. (E) Representative images of histological tumor sections from β-cateninfl/fl;MMTV-PyMT (fl/fl), β-catenindm/fl;MMTV-PyMT (dm/fl), and β-catenindm/fl;MMTV-PyMT;MMTV-Cre (dm/del) mice at 12 wk of age stained with hematoxylin and eosin and visualized by wide-field stitching microscopy. (Scale bar, 2 mm.) (F) Quantification of tumor stages in mice expressing wild-type (fl/fl) β-catenin, one wild-type and one mutant allele of β-catenin (dm/fl), or only the mutant allele of β-catenin (dm/del). Mouse numbers analyzed: fl/fl, n = 14; dm/fl, n = 12; dm/del, n = 12. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. *P < 0.05. (G) Quantification of the number of lung metastases in mice expressing in their mammary tumors in wild-type (fl/fl) β-catenin, one wild-type and one mutant allele (dm/fl), or only a mutant allele (dm/del). Numbers of mice analyzed: fl/fl, n = 16; dm/fl, n = 14; dm/del, n = 16. Data are displayed as mean ± SEM. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. *P < 0.05. (H) Metastatic index (number of lung metastases per primary tumor mass) of 12-wk-old MMTV-PyMT transgenic mice expressing in their mammary tumor cells in wild-type (fl/fl) β-catenin, one wild-type and one mutant allele (dm/fl), or only the mutant allele (dm/del). Mouse numbers analyzed: fl/fl, n = 16; dm/fl, n = 14; dm/del, n = 16. Data are displayed as mean ± SEM. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. *P < 0.05.
Immunofluorescence microscopy analysis of tumor sections for Ki67 and keratin (K) 8/18 as indicators of tumor cell–specific proliferation revealed that the reduced tumor growth in β-catenin mutant-expressing mice was not due to reduced tumor cell proliferation (Fig. 2C and SI Appendix, Fig. S3A). Staining for cleaved Caspase-3 and K8/18 as markers for tumor cell–specific apoptosis revealed a significant increase in the amount of apoptotic tumor cells in the mice expressing dm β-catenin (Fig. 2D and SI Appendix, Fig. S3A). Hence, β-catenin–mediated transcriptional output appeared to moderately contribute to tumor cell survival.
β-catenin Transcriptional Output Is Required for Metastasis Formation.
We next investigated whether the impairment of β-catenin transcriptional output in dm mutant tumors had any effect on malignant tumor progression and metastasis formation. Histopathological quantification of the various stages of tumor progression identifiable in the PyMT tumor model (hyperplasia, adenoma, and carcinoma) (22) revealed that mice expressing the dm form of β-catenin (dm/fl and dm/del) showed a significantly lower carcinoma burden than control mice (fl/fl), suggesting a delay in tumor progression (Fig. 2 E and F).
An analysis of the incidence of lung metastasis, the predominant metastatic spread in MMTV-PyMT mice, revealed that the complete abrogation of Wnt signaling activity in the dm form of β-catenin significantly reduced metastasis formation (Fig. 2 G and H and SI Appendix, Fig. S3B). Transgenic mice harboring one allele of the dm form and one allele of a conditional but fully functional allele of β-catenin (dm/fl mice) also showed reduced tumor progression and metastasis, again indicating a dominant-negative effect of the mutant form of β-catenin (see β-catenin Transcriptional Output Promotes Tumor Cell Proliferation). These results suggest a critical requirement of β-catenin’s transcriptional output for primary mammary tumor growth and its malignant tumor progression and metastasis formation.
β-catenin Transcriptional Output Promotes Tumor Cell Proliferation.
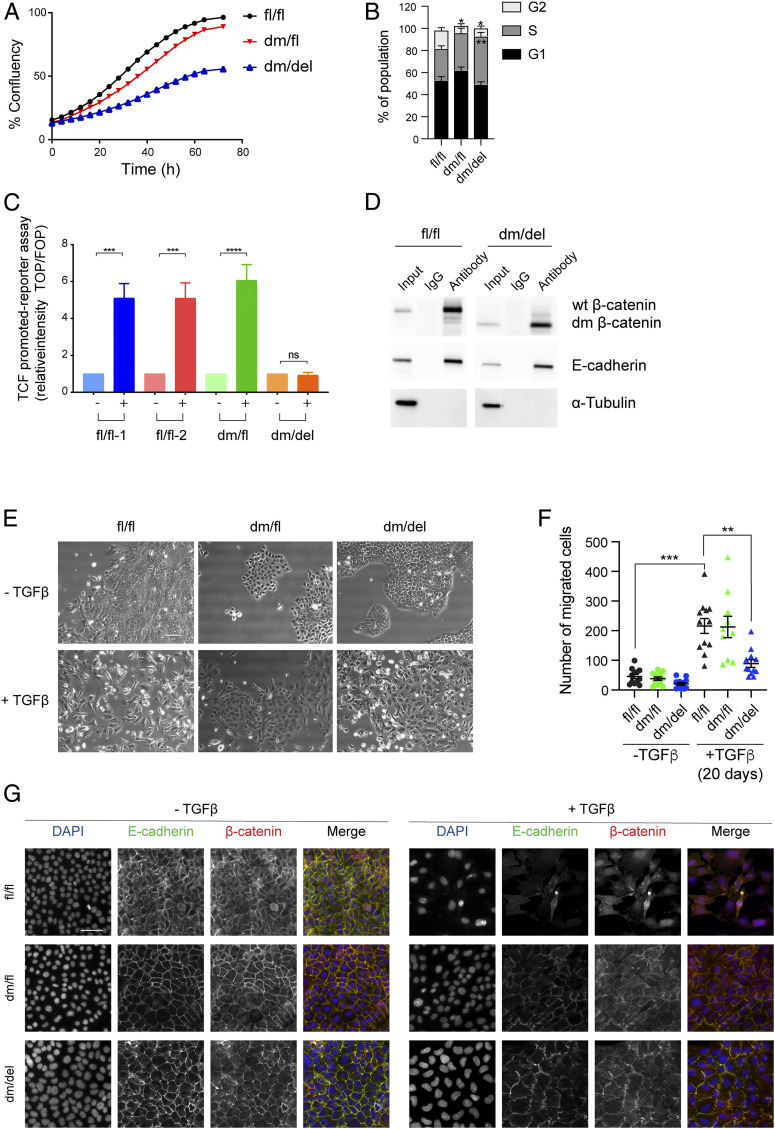
To examine the effects of dm β-catenin on canonical Wnt activity and to validate the results observed with tumor cell apoptosis in the transgenic mouse model, we established epithelial cell lines from dm β-catenin heterozygous (dm/fl) tumors. The floxed allele of β-catenin was then removed by infection with Adeno-CRE-IRES-GFP, and cells expressing only the mutant version of β-catenin were enriched by flow cytometry sorting for GFP-positive cells and validated by genotyping (SI Appendix, Fig. S4A). To assess whether the alterations in cell death observed in β-catenin mutant-expressing tumors in vivo can also be observed in cultured cells in vitro, we determined the growth curves for wild-type β-catenin fl/fl and dm mutant β-catenin (dm/del) cell lines. Consistent with the results on primary tumor growth, the cells expressing the dm form of β-catenin exhibited reduced cell numbers than a cell line expressing wild-type β-catenin (Fig. 3A and SI Appendix, Fig. S4B). Cell cycle analysis by flow cytometry revealed that the cells expressing dm β-catenin were stalled in the S phase of the cell cycle (Fig. 3B and SI Appendix, Fig. S4C). We therefore explored whether cell cycle arrest may be due to senescence induction, yet the activity of acidic β-galactosidase, a marker for cell senescence, was rather reduced in dm β-catenin–expressing cells as compared to wild-type β-catenin–expressing cells (SI Appendix, Fig. S4 D and E). These results suggest that the lack of transcriptional output stalls cell cycle progression in mammary tumor cells.
Fig. 3.
Loss of β-catenin cofactor binding impairs Wnt signaling activity, tumor cell proliferation, and EMT. (A) Comparison of the growth curves of mammary tumor cell lines expressing wild-type (fl/fl) β-catenin, one wild-type and one mutant allele of β-catenin (dm/fl), or only the mutant allele of β-catenin (dm/del). (B) Mammary tumor cells expressing the dm version of β-catenin (dm/del or dm/fl) are stalled in the S phase of the cell cycle. Cell cycle analysis by flow cytometry quantified from the data shown in SI Appendix, Fig. S4C. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. *P < 0.05; **P < 0.01. (C) Wnt-induced β-catenin–mediated transcriptional activity as determined by superTOPflash/ superFOPflash promoter reporter assay. Cell lines derived from tumors of the various genotype MMT-PyMT transgenic mice were treated with Wnt3a, and Wnt-dependent luciferase activity was determined. fl/fl-1 and fl/fl-2 = two independent cell lines expressing wild-type β-catenin; dm/fl = cell line expressing wild-type β-catenin and the dm β-catenin; dm/del = cell line expressing only the dm version of β-catenin. The results represent four independent experiments. Data are displayed as mean ± SEM. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. ***P < 0.005; ****P < 0.001; ns, not significant. (D) The mutant form of β-catenin still binds E-cadherin at the adhesion junctions. Coimmunoprecipitation using anti–β-catenin or rabbit IgG control in cell lines expressing wild-type (fl/fl) or the dm form of β-catenin (dm/del) and subsequent immunoblotting against β-catenin, E-cadherin, and α-tubulin as a loading control for the input. (E) Mammary tumor cell lines expressing mutant β-catenin are partially impaired in undergoing a TGF-β–induced EMT. Wild-type β-catenin–expressing cell lines (fl/fl) readily undergo TGF-β–induced morphological changes to a mesenchymal cell phenotype, while dm β-catenin–expressing cells (dm/fl or dm/del) only form filopodia without showing any specific mesenchymal cell morphology. (Scale bar, 100 μm.) (F) Migration of mammary tumor cells expressing wild-type β-catenin (fl/fl), cells expressing one allele of β-catenin and one allele of dm β-catenin (dm/fl), and cells expressing exclusively dm β-catenin (dm/del) in the absence of TGF-β or upon treatment with TGF-β for 20 d was determined in a Transwell migration assay. Data are displayed as mean ± SEM. Statistical analysis was performed using ordinary one-way ANOVA multiple comparison test. **P < 0.01; ***P < 0.005. (G) Immunofluorescence staining for β-catenin (red) and the EMT-related marker E-cadherin (green) reveals a reduced expression of the epithelial markers E-cadherin at the cell membrane in wild-type β-catenin–expressing cells (fl/fl), while in dm β-catenin–expressing cells (dm/fl and dm/del), E-cadherin and β-catenin are maintained at the cell junctions. DAPI was used to visualize nuclei. (Scale bar, 50 μm.)
We next assessed the levels of canonical Wnt signaling output in wild-type β-catenin–expressing and dm β-catenin–expressing cell lines by transfecting the cells with superTOPflash or superFOPflash promoter reporter constructs and by treatment with or without Wnt3a. Indeed, the cells expressing exclusively the dm form of β-catenin (dm/del) completely lacked Wnt3a-induced β-catenin/TCF-dependent transcriptional output, while the cells expressing only wild-type β-catenin (fl/fl) or both wild-type and dm β-catenin (dm/fl) showed high Wnt3a-induced transcriptional output (Fig. 3C). Notably, the dm form of β-catenin could still be observed in the nucleus of cells expressing it, suggesting that the lack of transcriptional output was not due to a lack of nuclear localization but rather to a failure to interact with its transcriptional cofactors (SI Appendix, Fig. S5A).
Of note, the simultaneous N- and C-terminal mutations in the dm of β-catenin did not affect its interaction with E-cadherin: immunoprecipitation experiments (Fig. 3D) and colocalization at the cell membranes (SI Appendix, Fig. S5B) demonstrated that the interaction with E-cadherin was retained by dm β-catenin. Since we were not able to obtain cell lines without any β-catenin expression, we conclude that it is β-catenin’s adhesion function which is critical for general mammary tumor cell survival.
Moreover, as already suspected in the experiments described above, the mutant version of β-catenin exerted a dominant-negative effect on cell mitosis as determined by phospho-histone 3 (pH3) staining when transiently transfected in a β-catenin wild-type cell line (wt/wt; Py2T) (SI Appendix, Fig. S5 C and D). These data, together with the diminished growth and metastasis formation of tumors expressing one mutant allele and one wild-type allele of β-catenin (Fig. 2B), indicate that the dm form of β-catenin exerts a dominant-negative effect on tumor cell proliferation. Interestingly however, this dominant-negative effect was not observed when determining Wnt3a-induced β-catenin/TCF-dependent transcriptional output in the superTOPflash/superFOPflash promoter reporter assay (Fig. 3C).
β-catenin Transcriptional Output Is Required for EMT.
Since EMT has been regarded as a potential mechanism underlying metastatic spread, we assessed whether the loss of β-catenin coactivator binding and, thus, the loss of canonical Wnt signaling in the dm form of β-catenin might affect EMT. Mammary tumor cells expressing wild-type β-catenin (fl/fl), expressing wild-type and dm β-catenin (dm/fl), or exclusively expressing dm β-catenin (dm/del) were treated for 4 d with TGF-β, and their ability to undergo EMT was analyzed. As expected, fl/fl cells converted from an epithelial to a more mesenchymal cell morphology upon TGF-β treatment, whereas dm/fl and in particluar dm/del cells retained their epithelial morphology and cluster formation yet exhibited substantial filopodia formation (Fig. 3E). Also, long-term (20 d) TGF-β treatment did not induce a full mesenchymal morphology of dm/del cells (SI Appendix, Fig. S6A). Moreover, small interfering RNA–mediated depletion of E-cadherin induced an EMT-like change in cell morphology in fl/fl cells but not in dm/del cells (SI Appendix, Fig. S6A). Only in the combination of TGF-β and the knockdown of E-cadherin, a slight increase in mesenchymal morphology was observed in dm/del cells. Finally, while cell migration of fl/fl cells was markedly induced by TGF-β, this was moderately observed with dm/fl cells but not at all with dm/del cells (Fig. 3F). As expected, immunofluorescence microscopy analysis for different EMT-related markers (E-cadherin, β-catenin, ZO-1, and vimentin) upon 4 d of TGFβ treatment of fl/fl cells revealed an increased expression of the mesenchymal marker vimentin and the reduced expression of the epithelial markers E-cadherin, β-catenin, and ZO-1 at the cell membranes (Fig. 3G and SI Appendix, Fig. S6B). In contrast, dm/del cells and to a lesser extent dm/fl cells, failed to undergo EMT upon TGF-β treatment: they retained E-cadherin, β-catenin, and ZO-1 at the cell membranes and did not increase the expression of vimentin (Fig. 3G and SI Appendix, Fig. S6B).
Since EMT has been implicated in the gain of stem cell–like traits and cell plasticity, we next assessed the functional contribution of β-catenin transcriptional activity to mammosphere formation, a well-established assay to characterize mammary stem cell–like cells. Surprisingly, dm/del cells formed more mammospheres than fl/fl cells, in particular after stimulation with TGF-β (SI Appendix, Fig. S7 A and B). Flow cytometry for the expression of CD44 and CD24 as markers of cancer stem cells supported this observation in that after TGF-β stimulation, 36.1% of dm/del cells exhibited a CD44highCD24low murine stem cell–like marker expression, whereas only 2.68% of fl/fl cells did so (SI Appendix, Fig. S7 C and D).
Since EMT has also been shown to contribute to therapy resistance, we next tested the role of β-catenin transcriptional activity in tumor cells’ response to conventional chemotherapies before and after TGF-β treatment. Treatment with paclitaxel and doxorubicine and less so with cyclophosphamide showed more resistance to chemotherapy by fl/fl cells regardless of TGF-β treatment, while dm/del cells were readily killed by the chemotherapeutics (SI Appendix, Fig. S8).
The impaired ability of dm/del cells to undergo a full EMT in vitro are consistent with the reduction in metastasis-observed dm β-catenin–expressing composite transgenic mice in vivo (Fig. 2 G and H). Thus, the reduced capability of dm/del cells to undergo EMT may account for the reduced metastatic burden observed. However, the observations that, after TGF-β treatment, dm/del cells exhibit more stem cell–like traits than fl/fl cells but are more rapidly killed by chemotherapy are unexpected and raise the possibility that cancer cell stemness and chemoresistance may be mechanistically uncoupled by the loss of β-catenin–mediated transcriptional output (Discussion).
β-catenin Mediates a Dominant Wnt-Induced Transcriptional Signature.
While the ablation of all cofactor binding in dm β-catenin resulted in a complete loss of Wnt signaling activity (Fig. 3C), it was not clear whether all endogenous target genes were similarly affected. Hence, to assess the overall deficiency of β-catenin–mediated Wnt signaling and to identify the target genes affected by dm β-catenin, breast tumor cell lines expressing wild-type (fl/fl) or dm β-catenin (dm/del) were subjected to next generation RNA sequencing. The cells were treated for 3 or 4 d with Wnt3a or TGF-β, respectively. At these late time points, EMT-related phenotypic changes were first sufficiently apparent for analysis (23), and we were motivated in assessing the functional contribution of β-catenin’s transcriptional activity to the cellular process of EMT and not to the early events of TGF-β signaling. Wnt3a has been chosen because it is a widely used recombinant Wnt ligand to trigger canonical Wnt signaling.
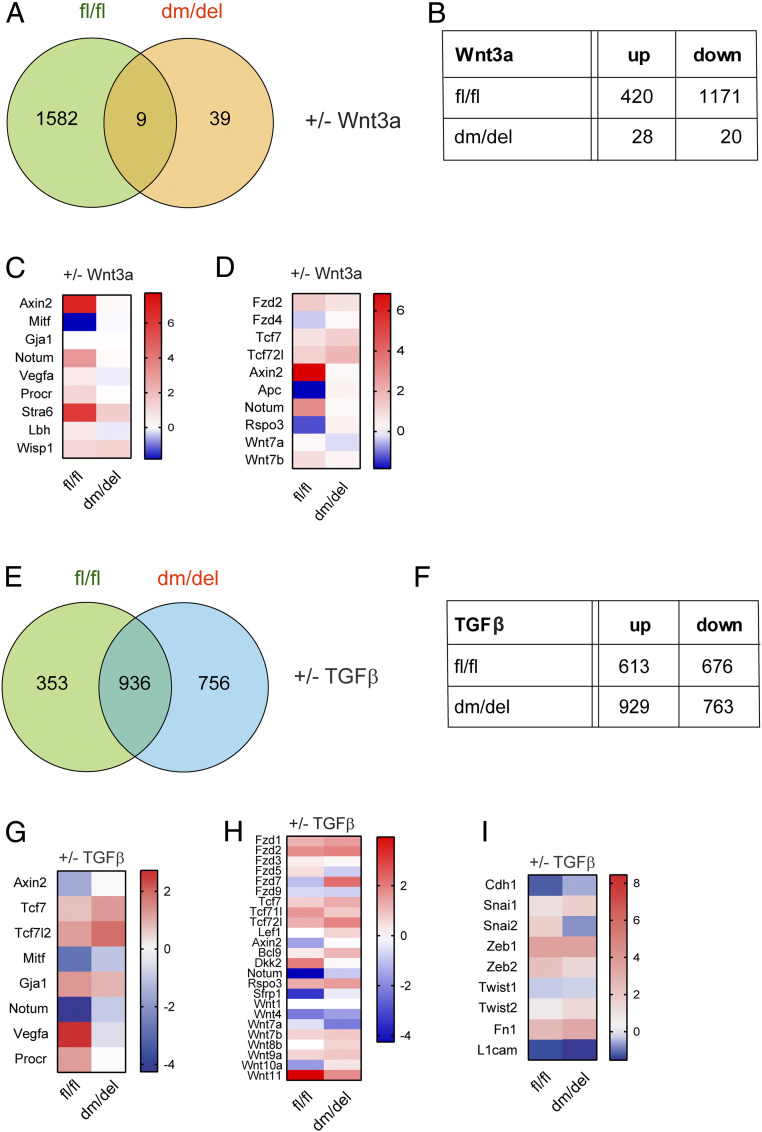
In wild-type cells, treatment with Wnt3a lead to the up-regulation of 420 and the down-regulation of 1,171 genes (Fig. 4 A and B). In the dm β-catenin mutant cells, substantially less genes were differentially expressed upon Wnt3a treatment. Consistent with the TOPFLASH assay, in dm/del cells, the Wnt response was essentially abolished: only 28 genes were up-regulated, and 20 genes were down-regulated (Fig. 4 A and B), and merely nine genes were found to be concomitantly regulated by wild-type and dm β-catenin (Fig. 4A and SI Appendix, Dataset S1).
Fig. 4.
Ablating the binding of N- and/or C- terminal coactivators to β-catenin affects the expression of specific Wnt target genes. Cell lines derived from tumors of MMTV-PyMT transgenic mice either expressing wild-type β-catenin (fl/fl) or exclusively the dm version of β-catenin (dm/del) were treated with Wnt3a or TGF-β for 3 and 4 d, respectively, and then subjected to next generation RNA sequencing. (A) Venn diagram showing the overlap of differentially expressed genes among the two β-catenin genotype cell lines upon treatment with Wnt3a. (B) Number of differentially up- and down-regulated genes upon Wnt3a treatment in the fl/fl and the dm/del genotype cell lines. (C and D) Heat maps showing the log2 fold changes of selected canonical Wnt target genes (C) and Wnt ligands and canonical Wnt pathway components (D) being differentially expressed between fl/fl and dm/del cells upon Wnt3a treatment. Red represents up-regulated expression, and blue represents down-regulated expression. RNA changes with P ≤ 0.05 and a fold change ≥ 1.5 were considered differentially expressed. (E) Venn diagram showing the overlap of differentially expressed genes among the two β-catenin genotype cell lines upon treatment with TGF-β. (F) Number of differentially up- and down-regulated genes upon TGF-β treatment in the fl/fl and the dm/del genotype cell lines. (G–I) Heat maps showing the log2 fold changes of selected Wnt target genes (G), of Wnt signaling components (H), and of genes encoding for EMT regulators (I) being differentially expressed between fl/fl and dm/del cells upon TGF-β treatment. Red represents up-regulated expression, and blue represents down-regulated expression. RNA changes with P ≤ 0.05 and a fold change ≥ 1.5 were considered differentially expressed.
To identify putative direct Wnt target genes, genes with TCF-binding sites in their promoter regions (taken from https://web.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes) were selected from the list of differentially expressed genes and in addition compared to known Wnt target genes previously identified in the mammary gland (24) (Fig. 4 C and D). Most canonical Wnt target genes appeared to be regulated only in wild-type cells. However, some direct Wnt target genes, such as Tcf7, Tcf7l2, Stra6, and Wisp1, were still up-regulated in dm/del cells upon Wnt treatment but to a much lesser extent as in wild-type cells (Fig. 4 C and D).
β-catenin Affects Mainly Wnt Target Genes during TGF-β–Induced EMT.
Next, we sought to identify genes that are transcriptionally regulated by β-catenin during TGF-β–induced EMT. The majority of changes in gene expression induced by TGF-β were not affected by the dm mutation in β-catenin, suggesting that most of TGF-β–induced pathways acted independently of β-catenin signaling (Fig. 4 E and F). However, several Wnt target genes and genes encoding for Wnt signaling effectors showed differential expression upon TGF-β–mediated EMT induction between wild-type and dm β-catenin–expressing cells (Fig. 4 G and H). An analysis of the expression of EMT-related genes revealed that Cdh1 was only significantly down-regulated in the wild-type cells and not in dm β-catenin cells, whereas mesenchymal marker genes, such as Fn1, Snai1, Zeb1, Zeb2, and Twist2, were up-regulated in all cell lines. In contrast, Twist1 expression was down-regulated by TGF-β in all cells, and Snai2 expression was up-regulated in wild-type cells, whereas it was rather down-regulated in the β-catenin mutant cells (Fig. 4I and SI Appendix, Dataset S2).
These results show that TGF-β affects the expression of direct Wnt target genes and also of Wnt pathway components during the induction of EMT. However, some of the genes show an opposite regulation of expression by TGF-β as compared to Wnt3a treatment, and, hence, their regulation appears context specific. This complexity was also found when analyzing the expression of some of the classical Wnt target genes in primary tumors of fl/fl or dm/del mice. Here, the expression of Wnt targets seemed to be reduced in the dm tumors yet only with limited significance (SI Appendix, Fig. S9A), most likely due to the high stromal contribution in MMTV-PyMT mammary tumors. As expected, Wnt target genes widely failed to be regulated by TGF-β in the dm/del cell lines (SI Appendix, Fig. S9B).
The results suggest that the binding of N-terminal and C-terminal coactivators of β-catenin affect the expression of direct Wnt target genes and other genes during TGF-β–induced EMT, likely explaining the failure of dm β-catenin–expressing cells to undergo an TGF-β–induced EMT.
Discussion
We have dissected the structural and transcriptional functions of β-catenin during tumor progression and metastasis formation in the MMTV-PyMT mouse model of metastatic breast cancer. First, we report that the complete absence of any β-catenin protein results in apoptosis of mammary gland tumor cells in vivo and in established mammary tumor cell lines in vitro. Consistent with our observations, the loss of β-catenin in mouse embryonic stem cells has been reported to lead to increased cell death (25). Moreover, ablation of β-catenin expression in head and neck squamous cell carcinomas and lung cancer cell lines also provokes cell death (26, 27).
To specifically investigate the functional contribution of β-catenin’s transcriptional functions to malignant breast mammary tumor progression and metastasis, we have employed a knock-in mouse line carrying a dm allele that lacks the capability of β-catenin to bind Bcl9/Bcl9l at its N terminus (D164A) and to the general transcription machinery at its C terminus (ΔC) yet retains its functions in cadherin-mediated cell adhesion (19). In contrast to the complete loss of β-catenin, the cells expressing the dm β-catenin mutant form survive, despite an almost complete loss of Wnt-induced transcriptional activity. This result indicates that β-catenin–mediated cell adhesion is more relevant for cell survival than β-catenin–mediated signaling activity and that the complete ablation of β-catenin may thus give a false picture when analyzing β-catenin–mediated transcriptional activities. Still, abolishing the binding of the N-terminal and C-terminal coactivators significantly repressed primary tumor growth in the MMTV-PyMT mouse model of metastatic breast cancer because of an increase in tumor cell apoptosis. In contrast, reduced tumor cell proliferation is found in established cell lines expressing dm β-catenin, in part based on an S phase arrest of tumor cells. Canonical Wnt signaling is known to promote cell proliferation at different levels (28, 29). The levels of β-catenin itself vary during the cell cycle; β-catenin cytoplasmic and nuclear fractions increase in S phase and peak at G2/M phase, while they decrease as the cells reenter G1 (30). In addition, β-catenin is also involved in centrosome regulation and the formation of bipolar mitotic spindles (30, 31).
The absence of β-catenin–mediated transcriptional activity in dm β-catenin also delayed malignant tumor progression and diminished lung metastasis in the MMTV-PyMT model of breast cancer, also in a dominant-negative manner. These suppressive effects of dm β-catenin on malignant tumor progression in vivo were also found with TGF-β–induced EMT in cell lines in vitro. While wild-type cell lines readily underwent EMT upon TGF-β treatment, dm/del cells exhibited a major impairment in undergoing EMT. These findings are in line with previous studies in which Bcl9/9l deficiency, also in combination with Pygopus deficiency, is reported to repress Wnt-driven tumorigenesis and to reduce the tumor load in mouse models of myeloma and colorectal and intestinal cancers and also to induce differentiation of tumor cells, thereby repressing tumor malignancy (7, 12, 32, 33). Most of these recent results have been found in colorectal and intestinal cancers in which mutations of Wnt signaling components frequently drive oncogenesis. These mutations are rather rare in breast cancer, yet activated canonical Wnt signaling has been demonstrated to promote breast cancer development (3–7). Here, we have shown that β-catenin–mediated transcriptional output is required for the growth, malignant progression, and metastasis formation in the MMTV-PyMT mouse model of breast cancer, highlighting the functional importance of Wnt signaling in yet another malignant cancer type.
Notably, the mutant version of β-catenin exerts a dominant-negative effect over wild-type β-catenin in the parameters of tumor growth, EMT, cell migration, and metastasis formation measured in vivo and in vitro. It is even more surprising to find that this dominant-negative effect is not observed in the TCF promoter reporter assay. We speculate that the plasmid-borne reporter assay may respond differently to the stoichiometry of the equal expression of mutant and wild-type β-catenin and thus does not detect the dominant-negative impact of dm β-catenin.
Since the loss of cofactor interaction in dm β-catenin effectively abrogated Wnt-induced transcriptional output, our results confirm that N- and C-terminal coactivators conjointly contribute to the expression of specific Wnt targets genes. However, the dependency of the expression of individual Wnt target genes on specific cofactors and combinations thereof may vary in a cell context-dependent manner. Consistent with these results, in mouse intestinal tumors, only a subset of Wnt target genes has been found down-regulated in Bcl9/9l-deficient tumors (7). In conclusion, the results show that β-catenin regulates its target genes by interacting with N- and C-terminal coactivators, and the loss of all coactivator binding leads to an almost complete loss of β-catenin transcriptional activity.
We found that β-catenin’s transcriptional activity is required for malignant breast cancer progression, and EMT induced upon TGF-β treatment. Not unexpectedly, in contrast to Wnt3a-induced gene expression, TGF-β–induced gene expression was much less affected by the loss of cofactor binding in dm β-catenin cells. The expression of some EMT-related transcription factors and mesenchymal markers, known to be regulated by canonical Wnt signaling (18, 34), was induced upon TGF-β treatment regardless of β-catenin mutation. However, the expression of the EMT transcription factor Snai2 was only induced in β-catenin wild-type cells and not in dm β-catenin cells. Accordingly, the expression of Cdh1 was down-regulated only in wild-type cells and not in mutant cells. Hence, β-catenin’s interaction with its N- and C-terminal coactivators affected the induction of target genes and thus EMT and metastasis. In the context of EMT, TGF-β also induced changes in the expression of Wnt pathways components, such as Wnt ligands, Wnt receptors, and Wnt pathway transcription factors (Tcf7, Tcf7l1, Tcf7l2, and Lef1). In β-catenin wild-type cells, the expression of the canonical Wnt target genes Axin2 and Notum was down-regulated upon TGF-β treatment, whereas it was up-regulated by Wnt3a. Notably, the expression of these canonical Wnt target genes did not specifically correlate with the presence of dm β-catenin in primary tumors (SI Appendix, Fig. S9A), and Axin2 expression has been reported to be inversely correlated with breast carcinogenesis (35). Hence, the regulation of target genes via β-catenin’s N- or C-terminal coactivators seem highly context dependent or also regulated by other transcriptional mechanisms, for example, not only by Wnt ligands but also by TGF-β and other cytokines of the tumor microenvironment. A functional interaction of Wnt and TGF-β signaling during EMT and other pathophysiological processes has been demonstrated before (36–38), and our results with cultured cell lines underscore the potential importance of this crosstalk.
Another curious finding of our work has been that dm β-catenin rather increased tumor cell stemness as assessed by the analysis of mammosphere formation and stem cell marker expression, while it decreased EMT, metastasis, and chemotherapy responses. Apparently, the dm version uncouples hallmarks which thus far have been all implicated in the overall process of EMT. However, the recent findings that EMT occurs in multiple steps and hybrid stages which represent different phenotypic capabilities (the EMT continuum) may explain the uncoupling of EMT-related features (39). In other words, wild-type β-catenin mammary tumor cells can undergo a full EMT, and cells may mainly reside in more mesenchymal state, while dm β-catenin cells may be arrested in a hybrid stage, which exhibits more traits of stemness and less of metastasis and chemoresistance.
In this study, we show that specifically abrogating the binding of N- and C-terminal coactivators of β-catenin results in a complete loss of transcriptional output and affects mammary tumor cell proliferation, tumor growth, EMT, malignant progression, and metastasis formation. In contrast, the complete loss of β-catenin’s signaling and its adhesion function leads to apoptosis of mammary tumor cells, suggesting a key role of β-catenin–mediated adhesion in tumor cell survival. The results suggest the possibility of therapeutically targeting breast cancer by interfering with the interaction of β-catenin with its transcriptional cofactors, avenues which are already being explored (40, 41).
Materials and Methods
Also refer to SI Appendix, Supplementary Methods for more details.
Antibodies and Reagents.
The following antibodies were used: E-cadherin (13-1900, Thermo Fisher Scientific, used for immunofluorescence staining), E-cadherin (610182, Transduction Laboratories, used for immunoblotting), α-tubulin (T-9026, Sigma-Aldrich), fibronectin (F3648 Sigma-Aldrich), vimentin (NB300-223, Novus Biologicals), Zonula occludens-1 (617300, Thermo Fisher Scientific), β-catenin (06-734, Cell Signaling Technology, used for immunofluorescence staining), β-catenin (NBP-32239, Novus Biologicals, used for immunoprecipitation), Caspase-3 (Asp-175) (5A1E) (9664, Cell Signaling Technology), phospho-Histone 3 (Ser10) (06-570, Merck), cytokeratin 8 + 18 (Fitzgerald Industries, 20R-CP004), Ki67 (Invitrogen, 14–5698-82), Myc-Tag (9B11) (2276, Cell Signaling Technology), Alexa Fluor 568 Phalloidin (A12380; Invitrogen), and DAPI (D9542, Sigma-Aldrich).
Recombinant murine Wnt3a (315-20, PeproTech) and recombinant human TGF-β1 (240-B, R&D Systems) were also used.
Mouse Experiments.
Mouse colonies were kept at the animal facility of the Department of Biomedicine (DBM), University of Basel, Basel, Switzerland. All animal experiments were carried out in accordance with the guidelines of the Swiss Federal Veterinary Office and the Cantonal Veterinary Office of Basel-Stadt. To examine the conditional ablation of β-catenin in breast cancer cells, β-cateninfl/wt mice were crossed with MMTV-PyMT (a kind gift of N. Hynes, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) (20, 22) and MMTV-Cre (a kind gift of Lothar Hennighausen, NIH, Bethesda, MD) mice (21). β-cateninfl/fl; MMTV-PyMT and MMTV-Cre mice obtained were then crossed with the β-catenin knock-in mutant mouse strain β-catenindm/wt. All mouse lines were maintained in a Friend Virus B NIH Jackson (FVB/N) genetic background, and experimental mice were always compared to littermate controls. To monitor MMTV-Cre–mediated recombination, β-cateninfl/fl; MPY, MMTV-Cre mice were crossed with GFP reporter mice (R26-LSL-GFP, kindly provided by V. Taylor, DBM, University of Basel). For the quantification of recombination, whole tumor sections were imaged with a Zeiss Axio Imager Scanning Microscope (10× magnification). GFP-positive areas and total tumor areas (DAPI-positive area) were quantified using ImageJ. All experiments were performed with female mice. For analysis of the mammary gland, mice were euthanized at 5 wk and before a tumor volume of 1,500 mm3 was reached, usually at 12 to 13 wk of age.
Cell and Tumor Genotyping.
To extract genomic DNA, cells from a confluent 10-cm Petri dish were trypsinized, washed in phosphate-buffered saline, and pelleted by centrifugation. DNA was extracted using GenElute Mammalian Genomic DNA Miniprep Kits (G1N70, Sigma-Aldrich) according to the manufacturer’s protocol. Standard PCR was performed using the following primers: for the β-catenin floxed and mutant allele, sense primer RM41 (5′-AAG GTA GAG TGA TGA AAG TTG TT-3′) and antisense primer RM42 (5′-CAC CAT GTC CTC TGT CTA TTC-3′) were used, generating 324–base pair (bp) and 221-bp products from the floxed and mutant alleles, respectively. To detect the floxed allele, sense primer RM68 (5′-AAT CAC AGG GAC TTC CAT ACC AG-3′) and antisense primer RM69 (5′-GCC CAG CCT TAG CCC AAC T-3′) were used generating a 631-bp product from the deleted allele (42).
RNA Sequencing and Analysis.
Established cell lines (β-cateninfl/fl, β-catenindm/fl, and β-catenindm/del) were treated either with 100 ng/mL Wnt3a for 3 d or with 2 ng/mL TGF-β for 4 d. Untreated cells served as control. Biological duplicates were prepared, and total RNA was isolated using the miRNeasy Mini Kit (Qiagen, 217004) with on-column deoxyribonuclease digestion according to the manufacturer’s instructions. RNA quality control was performed using RNA ScreenTape on an Agilent 4200 TapeStation, and RNA concentration was measured using Quanti-iT RiboGreen RNA Assay Kit (Life Technologies). RNA sequencing libraries were prepared from total RNA using poly(A) enrichment using 200 ng input RNA with TruSeq stranded mRNA Sample prep (Illumina). RNA quality control was performed on a fragment analyzer using the DNF-473–33-SS NGS Fragment 1- to 6,000-bp kit. RNA sequence libraries were sequenced on a NextSeq 500 using the 75-Cycle High Output Kit (Illumina). Single-end RNA sequencing reads were mapped to the mouse genome assembly, version mm10, using RNA-STAR (PubMed identification [PMID]: 23104886) with default parameters except for allowing only unique hits to genome (outFilterMultimapNmax = 1) and filtering reads without evidence in spliced junction table (outFilterType=”BySJout”). Using RefSeq mRNA coordinates from University of California, Santa Cruz (http://genome.ucsc.edu, downloaded in December 2015) and the qCount function from QuasR package (version 3.12.1) (PMID: 25417205), we quantified gene expression as the number of reads that started within any annotated exon of a gene. The differentially expressed genes were identified using the edgeR package (version 1.10.1) (PMID: 19910308). Genes with false discovery rate ≤ 0.05 and minimum log2 fold change of ± 1.0 were considered statistically significant and included in further analysis.
Statistical Analysis.
Statistical analysis and graphs were generated using GraphPad Prism 7.02 software. All data are presented as mean ± SEM.
Supplementary Material
Acknowledgments
Parts of the paper were adapted/excerpted from the first author’s PhD dissertation. We thank E. Panoussis for technical support, C. Beisel and the Department of Biosystems Science and Engineering sequencing facility for RNA sequencing, and P. Lorenz and the DBM Mattenstrasse microscopy facility for technical support. This work was supported by the Swiss NSF Grants 310030B_163471 (G.C.) and 310030B_173331 (K.B.), the Swiss NSF Sinergia Grant (G.C. and K.B.), the Swiss Cancer League Grant KFS-3479-08-2014 (G.C.), and the Krebsliga Beider Basel Grant 03-2013 (G.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020227118/-/DCSupplemental.
Data Availability
The RNA sequencing data are deposited on the Gene Expression Omnibus database under GSE148843.
References
- 1.Nusse R., Clevers H., Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Cohen J. C., Shank P. R., Morris V. L., Cardiff R., Varmus H. E., Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell 16, 333–345 (1979). [DOI] [PubMed] [Google Scholar]
- 3.Szeto W., et al., Overexpression of the retinoic acid-responsive gene Stra6 in human cancers and its synergistic induction by Wnt-1 and retinoic acid. Cancer Res. 61, 4197–4205 (2001). [PubMed] [Google Scholar]
- 4.Rieger M. E., Sims A. H., Coats E. R., Clarke R. B., Briegel K. J., The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol. Cell. Biol. 30, 4267–4279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Q., et al., Prevalence of protein C receptor (PROCR) is associated with inferior clinical outcome in breast invasive ductal carcinoma. Pathol. Res. Pract. 213, 1173–1179 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Wang D., et al., Protein C receptor stimulates multiple signaling pathways in breast cancer cells. J. Biol. Chem. 293, 1413–1424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deka J., et al., Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res. 70, 6619–6628 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Gordon M. D., Nusse R., Wnt signaling: Multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 281, 22429–22433 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Grumolato L., et al., Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 24, 2517–2530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhmetshina A., et al., Activation of canonical Wnt signalling is required for TGF-β-mediated fibrosis. Nat. Commun. 3, 735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., Tian X. J., Xing J., Signal transduction pathways of EMT induced by TGF-β, SHH, and WNT and their crosstalks. J. Clin. Med. 5, 41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani M., et al., BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 69, 7577–7586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry D. C., Levi L., Noy N., Holo-retinol-binding protein and its receptor STRA6 drive oncogenic transformation. Cancer Res. 74, 6341–6351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein E. A., Assoian R. K., Transcriptional regulation of the cyclin D1 gene at a glance. J. Cell Sci. 121, 3853–3857 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J., Chen Y., Olopade O. I., MYC and breast cancer. Genes Cancer 1, 629–640 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busby M., Hallett M. T., Plante I., The complex subtype-dependent role of Connexin 43 (GJA1) in breast cancer. Int. J. Mol. Sci. 19, 693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Heyden M. A., et al., Identification of connexin43 as a functional target for Wnt signalling. J. Cell Sci. 111, 1741–1749 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Heuberger J., Birchmeier W., Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb. Perspect. Biol. 2, a002915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valenta T., et al., Probing transcription-specific outputs of β-catenin in vivo. Genes Dev. 25, 2631–2643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy C. T., Cardiff R. D., Muller W. J., Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner K. U., et al., Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25, 4323–4330 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin E. Y., et al., Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer-Schaller N., et al., A hierarchical regulatory landscape during the multiple stages of EMT. Dev. Cell 48, 539–553.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Yu Q. C., Verheyen E. M., Zeng Y. A., Mammary development and breast cancer: A Wnt perspective. Cancers (Basel) 8, 65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raggioli A., Junghans D., Rudloff S., Kemler R., Beta-catenin is vital for the integrity of mouse embryonic stem cells. PLoS One 9, e86691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang H. W., et al., Knockdown of β-catenin controls both apoptotic and autophagic cell death through LKB1/AMPK signaling in head and neck squamous cell carcinoma cell lines. Cell. Signal. 25, 839–847 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Guo L., Ou S., Ma X., Zhang S., Lai Y., MACC1 silencing inhibits cell proliferation and induces cell apoptosis of lung adenocarcinoma cells through the β-catenin pathway. Neoplasma 65, 552–560 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Wang X., Jia Y., Fei C., Song X., Li L., Activation/proliferation-associated protein 2 (caprin-2) positively regulates CDK14/cyclin Y-mediated lipoprotein receptor-related protein 5 and 6 (LRP5/6) constitutive phosphorylation. J. Biol. Chem. 291, 26427–26434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi K., Niikura Y., Kitagawa K., Kikuchi A., Dishevelled, a Wnt signalling component, is involved in mitotic progression in cooperation with Plk1. EMBO J. 29, 3470–3483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olmeda D., Castel S., Vilaró S., Cano A., Beta-catenin regulation during the cell cycle: Implications in G2/M and apoptosis. Mol. Biol. Cell 14, 2844–2860 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bahmanyar S., Guiney E. L., Hatch E. M., Nelson W. J., Barth A. I., Formation of extra centrosomal structures is dependent on beta-catenin. J. Cell Sci. 123, 3125–3135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mieszczanek J., van Tienen L. M., Ibrahim A. E. K., Winton D. J., Bienz M., Bcl9 and Pygo synergise downstream of Apc to effect intestinal neoplasia in FAP mouse models. Nat. Commun. 10, 724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gay D. M., et al., Loss of BCL9/9l suppresses Wnt driven tumourigenesis in models that recapitulate human cancer. Nat. Commun. 10, 723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valenta T., Hausmann G., Basler K., The many faces and functions of β-catenin. EMBO J. 31, 2714–2736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H., et al., SOX7 co-regulates Wnt/β-catenin signaling with Axin-2: Both expressed at low levels in breast cancer. Sci. Rep. 6, 26136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Micalizzi D. S., Farabaugh S. M., Ford H. L., Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia 15, 117–134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalluri R., Weinberg R. A., The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X., Unwinding a path to nuclear beta-catenin. Cell 127, 40–42 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Yang J.et al.; EMT International Association (TEMTIA) , Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng M., et al., Pharmacological inhibition of β-catenin/BCL9 interaction overcomes resistance to immune checkpoint blockades by modulating Treg cells. Sci. Adv. 5, eaau5240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang G., et al., Targeted disruption of the BCL9/β-catenin interaction by endosomal-escapable nanoparticles functionalized with an E-cadherin-derived peptide. Nanotechnology 31, 115102 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Brault V., et al., Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data are deposited on the Gene Expression Omnibus database under GSE148843.