ABSTRACT
Membrane contact sites are critical junctures for organelle signaling and communication. Endoplasmic reticulum–plasma membrane (ER–PM) contact sites were the first membrane contact sites to be described; however, the protein composition and molecular function of these sites is still emerging. Here, we leverage yeast and Drosophila model systems to uncover a novel role for the Hobbit (Hob) proteins at ER–PM contact sites. We find that Hobbit localizes to ER–PM contact sites in both yeast cells and the Drosophila larval salivary glands, and this localization is mediated by an N-terminal ER membrane anchor and conserved C-terminal sequences. The C-terminus of Hobbit binds to plasma membrane phosphatidylinositols, and the distribution of these lipids is altered in hobbit mutant cells. Notably, the Hobbit protein is essential for viability in Drosophila, providing one of the first examples of a membrane contact site-localized lipid binding protein that is required for development.
KEY WORDS: Hobbit, ER–PM contact sites, Phosphatidylinositol, S. cerevisiae, Drosophila, Salivary gland, Regulated exocytosis
Summary: Membrane contact sites between organelles are critical regulators of cellular function. Here we identify the highly conserved Hob proteins as novel lipid-binding proteins at ER–PM contact sites.
INTRODUCTION
Eukaryotic cells are defined by the presence of distinct and specialized membrane-bound organelles, and this subcellular compartmentalization enables a wide range of complex cellular functions. In recent years, the importance of organelle membrane contact sites (MCS) for cellular physiology has become increasingly apparent and appreciated; MCS are critical sites for signaling, metabolic channeling, lipid trafficking, organelle fission or fusion, and Ca2+ homeostasis, among other functions (Prinz et al., 2020). MCS are ubiquitous; every type of eukaryotic cell contains them, and every organelle within a cell forms a functional contact site with at least one other organelle (Prinz et al., 2020; Shai et al., 2018; Valm et al., 2017). The endoplasmic reticulum (ER) in particular makes extensive contacts with other organelles, including the plasma membrane (PM), Golgi, mitochondria, peroxisomes, endosomes, lysosomes/vacuoles and lipid droplets (Wu et al., 2018).
ER–plasma membrane (ER-PM) contact sites were the first MCS to be described. These contact sites were identified in electron microscopy (EM) images of muscle cells in the 1950s (Porter and Palade, 1957). ER–PM contact sites are particularly ubiquitous and prevalent in the budding yeast Saccharomyces cerevisiae, with nearly 40% of the PM in contact with the ER (Manford et al., 2012; Pichler et al., 2001; West et al., 2011). Yeast ER–PM contact sites are maintained by six or seven conserved tethering proteins: the tricalbins (Tcb1, Tcb2 and Tcb3), two vesicle-associated membrane protein (VAMP)-associated proteins (VAPs; Scs2 and Scs22), the putative ion channel Ist2, and the more recently described Ice2 (Manford et al., 2012; Quon et al., 2018). Genetic deletion of six tethers (the tricalbins, VAPs and Ist2; Δtether strain) causes dramatic changes in ER morphology, with the collapse of cortical ER contacts and redistribution of the ER into the cytosol (Manford et al., 2012). In yeast, ER–PM contact sites play critical roles in lipid trafficking and homeostasis, including phosphatidylinositol trafficking and turnover (Manford et al., 2012; Stefan et al., 2011) and maintenance of proper lipid synthesis (Jorgensen et al., 2020; Omnus et al., 2016). However, surprisingly, ER–PM contact sites are not required for yeast viability under normal laboratory growth conditions (Manford et al., 2012), although these MCS are critical for survival under conditions of cellular stress (Collado et al., 2019; Omnus et al., 2016).
ER–PM contact sites are also present in metazoans, but the abundance and morphology of these MCS varies widely by cell type (Saheki and De Camilli, 2017). Like yeast, ER–PM contacts in metazoans are maintained by tethering proteins, including the extended synaptotagmins (E-Syts; orthologous to yeast tricalbins) (Giordano et al., 2013; Henne et al., 2015), VAPs (Murphy and Levine, 2016) and a number of other proteins that act as dynamic tethers in specific functional contexts or cell types (Eisenberg-Bord et al., 2016; Henne et al., 2015). The functions of ER–PM contact sites in metazoans are diverse but fall into two major categories: Ca2+ homeostasis and non-vesicular lipid transfer.
Store-operated Ca2+ entry (SOCE) is activated to replenish ER Ca2+ stores when they have been depleted (Hogan and Rao, 2015). SOCE is primarily mediated by ER-localized STIM proteins and PM-localized Orai Ca2+ channels, both of which were first discovered in RNAi screens conducted in Drosophila S2 cells (Feske et al., 2006; Liou et al., 2005; Roos et al., 2005; Vig et al., 2006; Zhang et al., 2006). SOCE is essential for development, as STIM and Orai knockouts are lethal in both mice and Drosophila (Baba et al., 2008; Cuttell et al., 2008; Gwack et al., 2008; Oh-hora et al., 2008; Pathak et al., 2017; Stiber et al., 2008). Several different lipid moieties undergo non-vesicular transfer at ER–PM contact sites, including sterols, glycerophospholipids and phosphatidylinositols (Henne et al., 2015; Saheki and De Camilli, 2017). However, knockout or mutation of most characterized lipid transfer proteins does not result in animal lethality, suggesting either that lipid transfer is not essential for development or that functionally redundant transfer proteins can compensate for one another.
The Hobbit (Hob) proteins are large (>2000 amino acid) proteins that are conserved throughout eukaryotes, but little is known about their molecular function. We identified hobbit in a genetic screen for Drosophila mutants that arrest development during metamorphosis with a small pupa phenotype, and found that hobbit function is required in professional secretory cells, including the insulin-producing cells (IPCs) and larval salivary glands, for regulated exocytosis (Neuman and Bashirullah, 2018). Phenotypes have also been observed upon mutation of the plant orthologs of hobbit. Mutation of Arabidopsis thaliana SABRE results in short roots with increased diameter and formation of ectopic root hairs; SABRE function is thought to be required for proper plant cell expansion and maintenance of planar cell polarity (Aeschbacher et al., 1995; Benfey et al., 1993; Pietra et al., 2013, 2015; Yu et al., 2012). Mutation of A. thaliana KIP, a putative paralog of SABRE, causes defects in pollen tube growth (Procissi et al., 2003), as does mutation of Zea mays APT1 (Xu and Dooner, 2006). Finally, mutation of the SABRE ortholog in the moss Physcomitrium patens causes stunted growth, defects in polarized growth, and failures in cell division (Cheng and Bezanilla, 2021). hobbit also has clear orthologs in vertebrates, and we have previously shown that expression of the human ortholog of hobbit (KIAA0100) fully rescues Drosophila hobbit mutant animals (Neuman and Bashirullah, 2018). However, although genetic screens in plants and insects have revealed phenotypic insights, the molecular function of hobbit and its orthologs has remained elusive.
Here, we use both yeast and Drosophila model systems to examine the function of hobbit. We find that Hobbit localizes to ER–PM contact sites in both yeast and Drosophila salivary gland cells, and this localization is mediated by an N-terminal membrane anchor and conserved C-terminal sequences. Our data also shows that ER–PM localization is required for hobbit function. Notably, the C-terminal Apt1 domain of Hobbit binds to phosphatidylinositols, and the distribution of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is altered in hobbit mutant cells. Together, these results demonstrate that Hobbit is a novel protein that localizes to ER–PM contact sites. We propose that Hobbit may be a lipid transfer protein whose function is required for animal development.
RESULTS
The S. cerevisiae orthologs of hobbit localize to ER–PM contact sites
The S. cerevisiae genome contains two predicted orthologs of hobbit, FMP27 and YPR117W. To begin to characterize these proteins in yeast, we generated GFP-tagged versions of both proteins under control of their endogenous promoters and examined where they localized within the cell. Both proteins localized in cortical patches or puncta (Fig. 1A; Fig. S1A), like other proteins that localize to ER–PM contact sites (Manford et al., 2012). We focused primarily on Fmp27–GFP because Ypr117w–GFP was expressed at barely detectable levels (Fig. S1A). We found that Fmp27–GFP colocalized with the pan-ER marker RFP–HDEL throughout the ER but was enriched in cortical patches that also colocalized with the ER–PM tether Tcb3–mCherry (Fig. 1A,B) (Manford et al., 2012), providing further evidence that Fmp27 is enriched at ER–PM contact sites. Some proteins that are enriched at ER–PM contact sites are anchored to the plasma membrane, while others are anchored to the ER (Hogan and Rao, 2015). To distinguish between these possibilities, we examined the localization of Fmp27–GFP in a strain that knocks out six ER–PM tether proteins (Δtether) (Manford et al., 2012). Fmp27–GFP was no longer present in cortical puncta in the Δtether background and instead localized to perinuclear and internal structures that colocalized with RFP–HDEL (Fig. 1C), indicating that Fmp27 localizes to collapsed ER in the Δtether mutant and confirming that Fmp27 is an ER-localized protein. FMP27 (for ‘found in mitochondrial proteome’) got its name from mitochondrial proteome analysis (Reinders et al., 2006), raising the possibility that this protein may also be present in mitochondria. We therefore examined the localization of FMP27–GFP with the mitochondrial marker Su9–DsRED but did not observe any colocalization (Fig. S1B), suggesting that Fmp27 may have been a contaminant in the mitochondrial proteome study. Overall, these results demonstrate that Fmp27 is an ER-localized protein enriched at ER–PM contact sites in yeast.
Fig. 1.
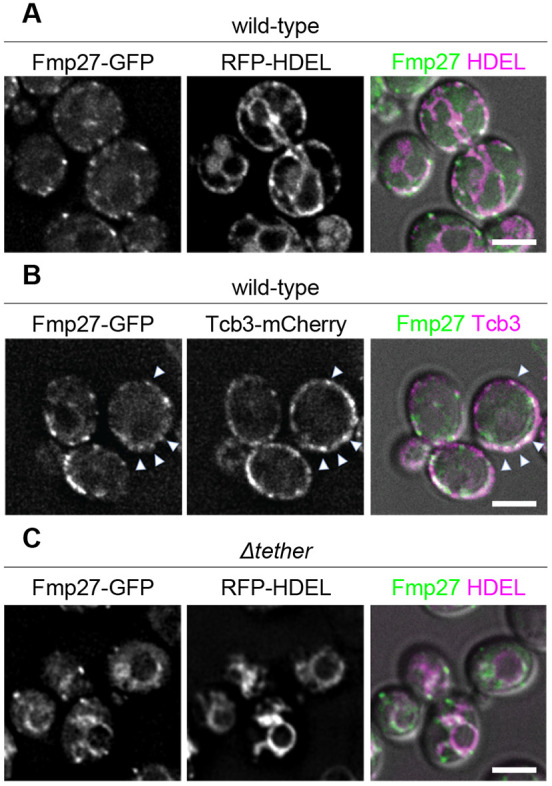
Fmp27 localizes to ER-PM contact sites. (A) Live-cell imaging of endogenously tagged Fmp27–GFP (green) shows that Fmp27 colocalizes with the pan-ER marker RFP–HDEL (magenta) and is enriched in puncta at the cell cortex. RFP–HDEL is expressed from a plasmid. (B) Live-cell imaging of Fmp27–GFP (green) with the ER–PM tether Tcb3–mCherry (magenta) confirms that Fmp27 is enriched at ER–PM contact sites (arrowheads highlight colocalization). (C) Live-cell imaging of endogenously tagged Fmp27–GFP (green) in the Δtether background with RFP-HDEL (magenta) shows that Fmp27 localizes to collapsed ER upon loss of ER-PM tethers. RFP–HDEL is expressed from a plasmid. Images shown are representative of three independent experiments with n≥3 images acquired per replicate. Scale bars: 2 µm.
Yeast hobbit localizes to the ER via an N-terminal membrane anchor
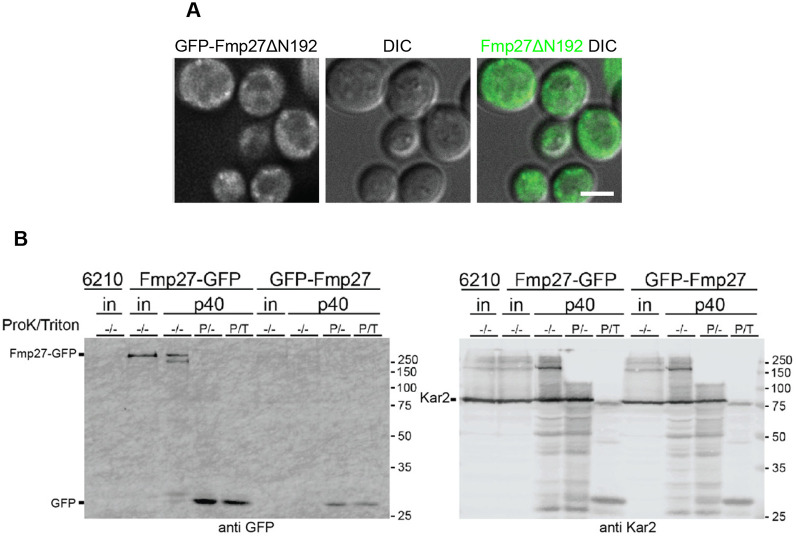
We next wanted to determine how Fmp27 localizes to the ER. Bioinformatic transmembrane prediction programs suggest that there may be a transmembrane domain at the N-terminus of the protein. Because Fmp27 has not been structurally characterized, we performed sequence alignments to identify conserved ‘blocks’ of primary sequence and used this analysis to identify break points for truncation studies (Fig. S1C). Consistent with a possible transmembrane domain at the N-terminus, deletion of the first 192 amino acids of Fmp27 (GFP–Fmp27ΔN192) abolished ER localization and caused the truncated protein to localize to the cytosol (Fig. 2A), indicating that Fmp27 is anchored to the ER by a transmembrane domain or hairpin at the N-terminus. We next analyzed the membrane topology of Fmp27. We isolated microsomes from cells expressing either Fmp27–GFP or GFP–Fmp27 and performed protease protection assays. In these assays, we would expect the luminal domain of Fmp27 to be protected from proteinase K, while GFP, which is a highly stable and tightly folded protein, should run as a free band on a gel if exposed to the cytosol. Note that GFP is resistant to degradation by proteinase K under these experimental conditions. GFP–Fmp27 was barely detectable in its full-length form (Fig. 2B), suggesting that N-terminal GFP tagging interferes with membrane insertion of the protein and may cause it to be degraded. In contrast, Fmp27–GFP was degraded after proteinase K treatment, as evidenced by the appearance of a free GFP band on the gel (Fig. 2B). The ER luminal protein Kar2 (Brodsky and Schekman, 1993; Rose et al., 1989) was used as a control to show that the ER lumen is protected from proteinase K under these conditions (Fig. 2B). These results demonstrate that Fmp27 is anchored to the ER via an N-terminal transmembrane domain or hairpin and the C-terminus of Fmp27 faces the cytosol.
Fig. 2.
Fmp27 localizes to the ER via an N-terminal membrane anchor with the C-terminus facing the cytosol. (A) Live-cell imaging of N-terminally truncated Fmp27 (GFP–Fmp27ΔN192; green) shows that the truncated protein localizes to the cytosol. N-terminal truncation was made at the endogenous FMP27 locus but expressed using the YPQ1 promoter. Images shown are representative of three independent experiments with n≥3 images acquired per replicate. Scale bar: 2 µm. (B) Left, anti-GFP western blot showing Fmp27–GFP and GFP–Fmp27 from cell lysate or the p40 fraction (microsomes). The p40 fraction was either mock-treated, treated with 0.12 µg/ml proteinase K, or 0.12 µg/ml proteinase K and 1% Triton X-100. The appearance of a free GFP band in the proteinase K-treated Fmp27–GFP p40 fraction shows that the C-terminus of Fmp27 faces the cytosol. Right, anti-Kar2 (ER lumen protein) western blot from the same samples in the left panel. ‘in’ represents input, ‘P’ represents that proteinase K was added, ‘T’ represents that Triton X-100 was added. ‘6210’ indicates a control strain lacking tagged Fmp27.
Fmp27 does not function as an ER–PM tether in yeast
Several ER–PM tethering proteins have been identified in yeast, and deletion of six or seven of them is required to significantly reduce the formation of these contact sites (Manford et al., 2012; Quon et al., 2018). However, even upon deletion of seven tethers, cortical ER is still occasionally observed, which could be due to incidental contact or to the function of unidentified tethers (Quon et al., 2018). The topology of Fmp27, with an N-terminal membrane anchor and cytosol-facing C-terminus, coupled with its enrichment at ER–PM contact sites, is consistent with a potential function as an ER–PM tether. To examine this possibility, we first generated cells with a knockout for both hobbit orthologs (fmp27Δ ypr117wΔ). fmp27Δ ypr117wΔ cells were viable with no obvious growth phenotypes on complete medium. Imaging of RFP–HDEL did not reveal any obvious ER morphology defects in fmp27Δ ypr117wΔ cells, in contrast to Δtether cells, where nearly all cortical ER is lost (Fig. 3A). We also used thin-section transmission electron microscopy (EM) to quantify ER-PM contacts in wild-type, fmp27Δ ypr117wΔ, and Δtether fmp27Δ ypr117wΔ (8 knockout) mutant cells (see a representative image in Fig. S1D). No significant differences were observed in the ratio of the amount of cortical ER to total PM between wild-type and fmp27Δ ypr117wΔ or between Δtether and Δtether fmp27Δ ypr117wΔ mutant cells (Fig. 3B). We also overexpressed FMP27 to look for increased ER-PM contacts; however, we observed a significant reduction in the ratio of the amount of cortical ER to PM in FMP27-overexpressing cells (Fig. 3B). Finally, we tested whether overexpression of FMP27 could rescue the loss of cortical ER in Δtether cells. We used fluorescence microscopy with RFP–HDEL to examine ER morphology and did not see a rescue of ER–PM contact site formation in Δtether cells overexpressing FMP27 (Fig. 3C). Taken together, these results demonstrate that Fmp27 localizes to ER–PM contact sites but does not appear to function as a tether.
Fig. 3.
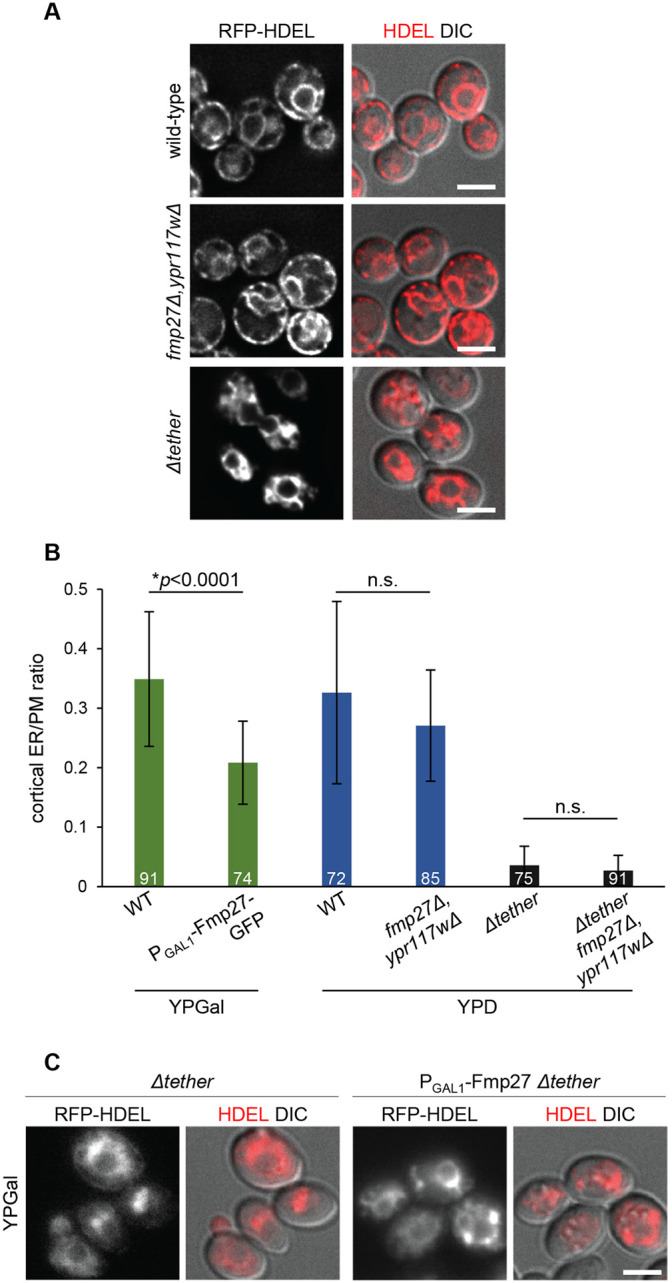
Fmp27 does not function as an ER-PM tether. (A) Live-cell imaging of RFP–HDEL (red) in control, fmp27Δ ypr117wΔ and Δtether cells showing that ER morphology is unaffected upon loss of both copies of the yeast ortholog of hobbit, in contrast to Δtether cells, where nearly all cortical ER is lost. (B) Quantification of the ratio of cortical ER to total PM in control (wild-type, WT), FMP27–GFP overexpression, fmp27Δ ypr117wΔ, Δtether, and Δtether fmp27Δ ypr117wΔ cells showing that overexpression of Fmp27 does not increase the number of cortical ER contacts (green bars); in fact, the number of contacts is significantly reduced. Loss of both Fmp27 and Ypr117w does not decrease the number of cortical ER contacts in a wild-type (blue bars) or Δtether background (gray bars). Cells were grown on YPGal medium to drive Fmp27 overexpression; otherwise, cells were grown on standard YPD medium. Graphs show mean±s.d.; statistics calculated by Welch's t-test; n.s., not significant. The number on each bar represents the number of cells for which the cortical-ER to PM ratio was measured. (C) Live-cell imaging of RFP–HDEL (red) in Δtether (left) and Δtether cells overexpressing Fmp27 (right) showing that overexpression of Fmp27 does not rescue cortical ER contacts in the Δtether background. Fmp27 is expressed under its endogenous promoter in the left panels and under the GAL1 promoter in right panels. Cells were collected from mid-log cultures grown in YPGal medium to drive Fmp27 overexpression. Images shown in A and C are representative of three independent experiments with n≥3 images acquired per replicate. Scale bars: 2 µm.
Membrane topology of hobbit is conserved in Drosophila
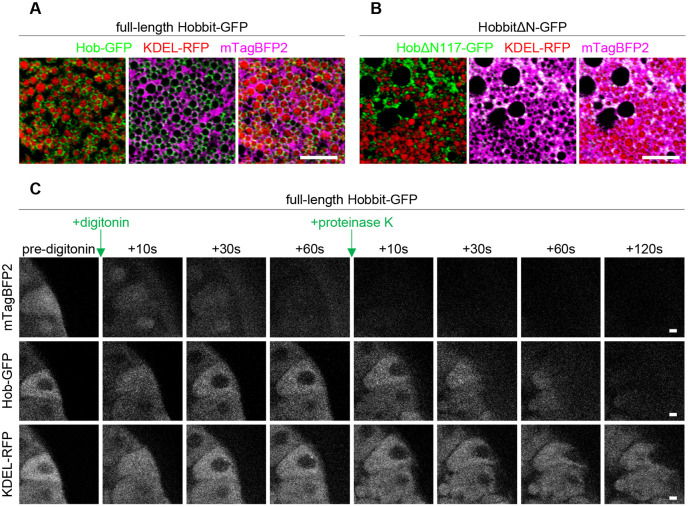
Since we did not observe any obvious phenotypes in yeast that provide clues to the function of hobbit at ER–PM contact sites, we turned to Drosophila, where mutation of hobbit results in a dramatic reduction in animal body size, lethality during metamorphosis and defects in regulated exocytosis in multiple cell types, including the insulin-producing cells and the larval salivary glands (Neuman and Bashirullah, 2018). We first wanted to determine whether the ER membrane localization of Hobbit was conserved in flies. Our previous work demonstrated that Hobbit localizes to the ER (Neuman and Bashirullah, 2018). High-resolution confocal microscopy enabled us to determine that Hobbit is present on the ER membrane in the larval salivary glands; we observed Hobbit–GFP-enriched around the ER luminal marker KDEL–RFP but non-overlapping with cytoplasmic mTagBFP2 (Fig. 4A). Additionally, Kyte–Doolittle hydrophobicity analysis (Kyte and Doolittle, 1982) shows a short, highly hydrophobic region at the N-terminus of the Hobbit protein (Fig. S2A), consistent with a possible transmembrane anchor. Like yeast, deletion of a conserved block of sequence at the N-terminus resulted in the loss of ER localization, as HobΔN117–GFP colocalized with cytoplasmic mTagBFP2 (Fig. 4B; Fig. S2B). Importantly, ubiquitous overexpression of hobΔN117-GFP did not rescue the small body size or lethality of hobbit mutant animals (see Fig. 6C), suggesting that ER localization is critical for hobbit function. In contrast, we have previously reported that ubiquitous overexpression of full-length hobbit-GFP inserted at the same genomic locus as hobΔN117-GFP fully rescues both body size and lethality in hobbit mutant animals (Neuman and Bashirullah, 2018). Our next goal was to determine whether fly Hobbit had the same membrane topology as yeast Fmp27, and we conducted an imaging-based protease protection assay to analyze the topology. Addition of the selective detergent digitonin, which permeabilizes the plasma membrane but not intracellular membranes (Lorenz et al., 2006), resulted in the release of cytosolic mTagBFP2, while Hobbit–GFP and KDEL–RFP were unaffected (Fig. 4C; Movie 1). This further confirms that Hobbit is anchored to the ER membrane. Subsequent addition of proteinase K resulted in the progressive loss of Hobbit–GFP signal, while KDEL–RFP was unaffected (Fig. 4C), confirming that the C-terminus of fly Hobbit faces the cytosol.
Fig. 4.
ER membrane localization and topology of Hobbit are conserved in Drosophila. (A) Live-cell imaging of full-length Hobbit–GFP (green), the ER lumen marker KDEL–RFP (red) and cytosolic mTagBFP2 (magenta) in the Drosophila larval salivary glands at the onset of metamorphosis (0 h after puparium formation, PF) shows that Hobbit–GFP localizes to the ER membrane. Full genotype: UAS-KDEL-RFP/+; Sgs3>hob-GFP/UAS-mTagBFP2. (B) Live-cell imaging of N-terminally truncated HobbitΔN117–GFP (green), the ER lumen marker KDEL–RFP (red) and cytosolic mTagBFP2 (magenta) at 0 h PF shows that HobbitΔN117–GFP localizes to the cytosol. Full genotype: UAS-KDEL-RFP/+; Sgs3>hobΔN117-GFP/UAS-mTagBFP2. Images in A and B show a single slice from a z-stack comprising three optical sections at a 0.28 µm step size. (C) Imaging-based protease protection assay shows that the C-terminus of Drosophila Hobbit faces the cytosol. Cytosolic mTagBFP2 (top) rapidly diffuses out of the cells after permeabilization with digitonin, while Hobbit–GFP (middle) and KDEL–RFP (bottom) are unaffected. Hobbit–GFP (tagged at the C-terminus) is degraded after subsequent addition of proteinase K, while KDEL–RFP is unaffected. Note that the cells flatten after addition of proteinase K. Experiment was conducted using 0 h PF glands. Full genotype: UAS-KDEL-RFP/+; Sgs3>hob-GFP/UAS-mTagBFP2. Images shown are representative of three independent experiments with n≥10 salivary glands from independent animals analyzed per genotype. Scale bars: 5 µm (A,B); 10 µm (C).
Fig. 6.
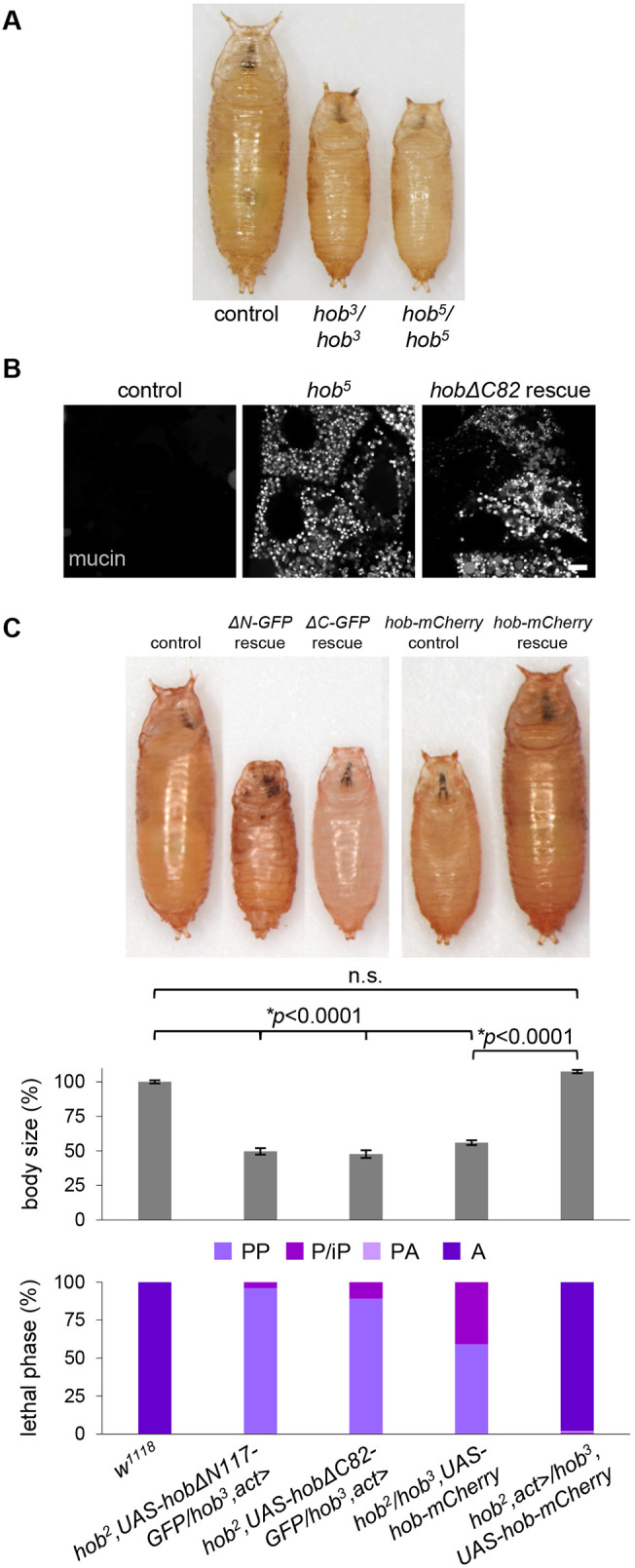
ER–PM localization is required for hobbit function. (A) Images of control (w1118), homozygous hob3, and homozygous hob5 pupae showing that hob5 mutant animals display the same small body size as hob3 mutant animals. Pupae are representative of three independent experiments with n≥50 animals examined per replicate. (B) Live-cell imaging of mucins in control, homozygous hob5 and hobΔC82 rescue salivary glands dissected from prepupal animals showing that mucins are secreted in controls but not in hob5 mutant or hobΔC82 rescue animals. Full genotypes: control, Sgs3-GFP/+. hob5: Sgs3-GFP/+; hob5/hob5. hobΔC82 rescue, Sgs3-GFP/+; hob2, UAS-hobΔC82-mCherry; hob3, act>. Images shown are representative of three independent experiments with n≥10 salivary glands from independent animals analyzed per genotype. Scale bar: 10 µm. (C) Body size quantification and lethal phase analysis for control (w1118), hobΔN117-GFP rescue (hob2, UAS-hobΔN117-GFP/hob3, act>), hobΔC82-GFP rescue (hob2, UAS-hobΔC82-GFP/hob3, act>), hob-mCherry rescue control (hob2/hob3, UAS-hob-mCherry) and hob-mCherry rescue (hob2, act>/hob3, UAS-hob-mCherry) showing that ubiquitous overexpression of neither the N- nor C-terminal truncation of Hobbit rescues the small body size or lethality of hobbit mutant animals. In contrast, ubiquitous overexpression of full-length Hobbit–mCherry fully rescues the small body size and lethality of hobbit mutant animals. Body size quantified by pupa volume expressed as a percentage relative to control (100%). Data shown as mean±s.e.m., n=100 animals per genotype. Statistics calculated by an unpaired, two-tailed Student's t-test. Note that the three pupae on the left were captured in a different image from the two on the right; both were imaged at the same magnification. PP, prepupa; P/iP, pupa/incomplete pupa; PA, pharate adult; A, adult.
Hobbit localizes to ER–PM contact sites in Drosophila cells
Fmp27 is highly enriched at ER–PM contact sites; therefore, we wanted to determine whether Hobbit was present at these sites in Drosophila cells. The protein composition of ER–PM contact sites is poorly understood in Drosophila; therefore, we developed a new marker for these sites based on the structure of the Stim protein. Drosophila Stim and its human ortholog STIM1 are well-characterized ER membrane proteins that localize to ER–PM contact sites when ER Ca2+ stores are depleted (Hogan and Rao, 2015). To generate a Stim reagent that constitutively labels ER–PM junctions, we changed D155 and D157 of Drosophila Stim (orthologous to D76 and D78 in human STIM1) to alanine (A) residues (StimDDAA-GFP). These amino acids are required for Ca2+ binding within the EF hand domain of Stim, and mutation of these sites locks Stim in an extended conformation that enables it to interact with Orai Ca2+ influx channels specifically at ER–PM contact sites (Liou et al., 2005; Lunz et al., 2019; Zhang et al., 2005). Accordingly, we find that StimDDAA–GFP localized in puncta that are present at the plasma membrane in larval salivary gland cells (Fig. 5A), consistent with the expected pattern and localization of ER-PM contact sites. Importantly, full-length Hobbit–mCherry strongly colocalized with StimDDAA–GFP puncta, indicating that fly Hobbit is enriched at ER–PM contact sites (Fig. 5B,D). Genetic rescue experiments confirmed that the hobbit-mCherry transgene produces functional protein (Fig. 6C). Additionally, we examined the localization of StimDDAA–GFP in the salivary glands of hobbit mutant animals, using alleles we have previously reported (Neuman and Bashirullah, 2018). StimDDAA–GFP appeared to localize normally in hobbit mutant cells (Fig. 5A), indicating that ER–PM contact sites still form in the absence of hobbit. Taken together, these results demonstrate that Hobbit localization to ER–PM contact sites is conserved between yeast and flies.
Fig. 5.
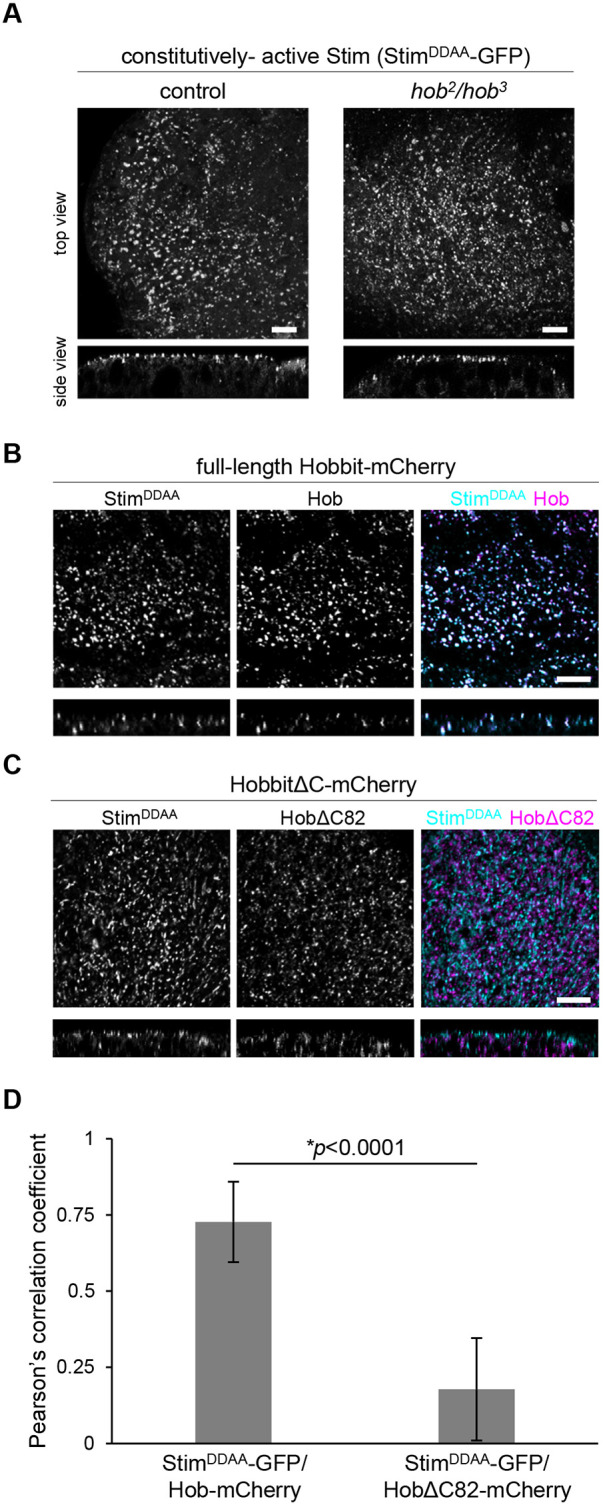
The C-terminus of Hobbit is required for localization to ER–PM contact sites. (A) Live-cell imaging of constitutively active Stim (StimDDAA-GFP) in control and hobbit mutant salivary gland cells at the onset of metamorphosis (0 h after puparium formation, PF) showing that this protein localizes to puncta at the plasma membrane. Top images show a maximum intensity projection of 20 optical slices from a z-stack comprising 71 total slices (control) or 57 total slices (hobbit mutant) at a 0.36 µm step size. Bottom images show a transverse section (xz slice) from the z-stacks shown above. Full genotypes: control, UAS-StimDDAA-GFP/+; Sgs3>/+ and hobbit mutant, UAS-StimDDAA-GFP/+; hob2, Sgs3>/hob3. (B) Live-cell imaging of constitutively-active Stim (StimDDAA–GFP, cyan) and full-length Hobbit (Hobbit–mCherry, magenta) in wandering L3 (wL3) salivary glands showing that Hobbit and constitutively active Stim colocalize at ER–PM contact sites. Top images show a maximum intensity projection of 10 optical slices from a z-stack comprising 31 total slices at a 0.36 µm step size. Bottom images show a transverse section (xz slice) from the z-stacks shown above. Full genotype: UAS-StimDDAA-GFP/+; Sgs3>hob-mCherry/+. (C) Live-cell imaging of constitutively-active Stim (StimDDAA-GFP, cyan) and C-terminally truncated Hobbit (HobbitΔC82–mCherry, magenta) in wandering L3 (wL3) salivary glands showing that HobbitΔC82 no longer localizes to ER–PM contact sites. Top images show a maximum intensity projection of 10 optical slices from a z-stack comprising 31 total slices at a 0.35 µm step size. Bottom images show a transverse section (xz slice) from the z-stacks shown above. Full genotype: UAS-StimDDAA-GFP/+; Sgs3>hobΔC82-mCherry/+. Images in A–C were acquired as z-stacks beginning outside of the salivary gland cell and extending inward through the plasma membrane. Images shown are representative of three independent experiments with n≥10 salivary glands from independent animals analyzed per genotype. Scale bars: 5 µm. (D) Quantification of colocalization of StimDDAA–GFP with full-length Hob–mCherry and HobΔC82–mCherry. y-axis shows Pearson's correlation coefficient; x-axis shows the proteins analyzed. Graphs show mean±s.d. from n=10 salivary glands isolated from independent animals. Statistics calculated by unpaired, two-tailed Student's t-test.
The C-terminus of Hobbit is required for ER–PM localization
We have previously reported five mutant alleles of hobbit; all are nonsense mutations scattered throughout the 2300-amino-acid protein (Neuman and Bashirullah, 2018) (Fig. S3A). Notably, one of these mutations, hob5, is only 82 amino acids away from the C-terminus of the protein, and animals with this mutant allele are phenotypically indistinguishable from animals with other nonsense mutations located much closer to the N-terminus, with a small body size, lethality during metamorphosis, and cell-autonomous defects in regulated exocytosis in the larval salivary glands (Fig. 6A,B). These results suggest that critical functional elements are likely encoded in the final 82 amino acids of the Hobbit protein. C-terminal protein sequences are required for enrichment of other proteins at ER–PM contact sites (Giordano et al., 2013); therefore, we hypothesized that the C-terminal 82 amino acids may be required for Hobbit enrichment at ER–PM contact sites. To test this idea, we generated both GFP- and mCherry-tagged overexpression constructs that delete the final 82 amino acids of the Hobbit protein (hobΔC82-GFP and hobΔC82-mCherry). These truncated proteins still localized to the ER membrane and topology was unaffected (Fig. S3B,C, Movie 2). However, strikingly, HobΔC82–mCherry did not significantly colocalize with StimDDAA–GFP (Fig. 5C,D), indicating that ER–PM contact site enrichment was lost upon deletion of the C-terminus of Hobbit. Additionally, ubiquitous overexpression of HobΔC82 did not rescue the small body size or lethality of hobbit mutant animals (Fig. 6C), nor did it rescue regulated exocytosis defects in the larval salivary glands (Fig. 6B). We also examined the effect of Fmp27 truncation in yeast cells. As we observed with fly hobbit, deletion of a conserved block of sequence at the C-terminus of Fmp27 strongly reduced cortical ER–PM localization (Fig. S4). Overall, these results demonstrate that ER–PM localization is required for hobbit function.
The C-terminal Apt1 domain of Hobbit binds to PM phosphatidylinositols
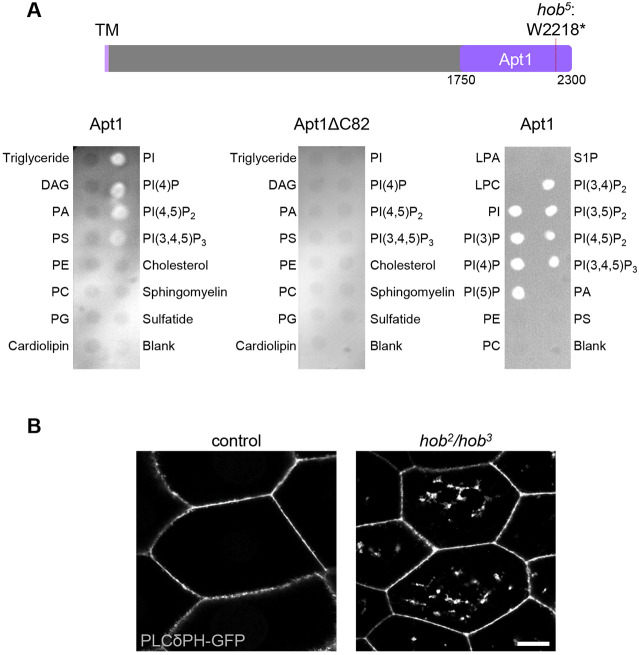
Sequence analysis using the protein domain database Pfam (Mistry et al., 2021) predicts that the C-terminus of Hobbit contains an Apt1 domain, a protein domain that was first identified in the APT1 protein, which is the Z. mays ortholog of hobbit. This domain appears to only be present in Hobbit and its orthologs (see Discussion section for additional details). Recent analysis of yeast Atg2, Vps13 and human VPS13A using the HHPred program indicates that these proteins also contain an Apt1 domain (Kaminska et al., 2016; Kolakowski et al., 2020; Rzepnikowska et al., 2017); however, the presence of this domain is not predicted in these proteins by Pfam, perhaps because the length and primary sequence diverge significantly from the Hobbit Apt1 domain (Fig. S5). The Apt1 domains of Atg2, Vps13 and VPS13A bind to phosphatidylinositols (Kaminska et al., 2016; Kolakowski et al., 2020; Rzepnikowska et al., 2017), raising the possibility that Hobbit may also bind to these lipids. To test this hypothesis, we expressed recombinant Hobbit Apt1 domain in E. coli, purified it, and tested it for binding on membrane lipid strips. Strikingly, full-length Hobbit Apt1 bound to phosphatidylinositol (PI), PI(4)P, PI(4,5)P2, and PI(3,4,5)P3, whereas Apt1 lacking the C-terminal 82 amino acids (Apt1ΔC82) did not bind to any of these lipids (Fig. 7A). These lipid moieties are known to be enriched at the plasma membrane (Balla, 2013), suggesting that binding to plasma membrane phosphatidylinositols may play a role in promoting the ER–PM localization of Hobbit. We also examined the binding of full-length Apt1 to all phosphatidylinositol moieties using PIP strips and found that Apt1 bound to all PIP moieties (Fig. 7A). Finally, we examined the subcellular distribution of one of these lipids, PI(4,5)P2, in control and hobbit mutant cells using the fluorescently tagged binding reporter PLCδPH–GFP (Verstreken et al., 2009). As expected, PLCδPH–GFP was strongly enriched at the plasma membrane in both control and hobbit mutant cells; however, PLCδPH–GFP also accumulated in large intracellular compartments in hobbit mutant cells (Fig. 7B), suggesting that hobbit plays a functional role in regulating the steady-state subcellular distribution of PI(4,5)P2.
Fig. 7.
Hobbit binds to phosphatidylinositols and affects PI(4,5)P2 distribution. (A) Top, schematic depicting the Apt1 domain of Hobbit and the location of the hob5 nonsense mutation. Note that Pfam annotates the Apt1 domain as amino acids 1774–2239, but we included extra upstream and downstream sequences to ensure proper folding of the domain. TM, transmembrane domain. Bottom left, lipid blot binding analysis on membrane lipid strips showing that full-length Apt1 binds to phosphatidylinositol (PI), PI(4)P, PI(4,5)P2, and PI(3,4,5)P3. Bottom middle, lipid blot binding analysis on membrane lipid strips showing that Apt1ΔC82 does not bind to any of these lipids. Both the left and middle blots were imaged at the same time with identical acquisition parameters. Bottom right, lipid blot binding analysis on PIP strips shows that full-length Apt1 binds to all phosphatidylinositol (PI) moieties but does not bind to other membrane lipids. All blots shown are representative of three experimental replicates with independent protein preparations used for each replicate. DAG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; LPA, lysophosphatidic acid; LPC, lysophosphocholine; S1P, sphingosine-1-phosphate. (B) Live-cell imaging of the PI(4,5)P2 marker PLCδPH–GFP in control and hobbit mutant wandering L3 (wL3) salivary gland cells showing that this lipid accumulates in large intracellular compartments in hobbit mutant cells. Full genotypes: control, Sgs3>/UAS-PLCδPH-GFP and hob mutant, hob2, Sgs3>/hob3, UAS-PLCδPH-GFP. Images shown are representative of three independent experiments with n≥10 salivary glands from independent animals analyzed per genotype. Scale bar: 20 µm.
DISCUSSION
ER–PM contact sites are critical junctures for Ca2+ and lipid homeostasis in all eukaryotic cells. Here, we have identified Hobbit as a novel protein that is enriched at ER–PM contact sites in both yeast and Drosophila. The topology of the Hobbit protein and the requirement for specific C-terminal sequences in ER–PM localization are also evolutionarily conserved. Our data demonstrate that the C-terminal Apt1 domain of Hobbit binds directly to phosphatidylinositols, and hobbit appears to play a role in regulating the subcellular distribution of these lipids. Loss of hobbit function results in lethality in Drosophila; therefore, hobbit represents a novel example of a lipid-binding protein that is essential for animal development.
The Pfam database (Mistry et al., 2021) annotates the Apt1 domain as a Golgi body localization domain. This annotation comes from analysis of the subcellular localization of a short C-terminal fragment of Z. mays APT1, which colocalized with a Golgi marker in vivo (Xu and Dooner, 2006). However, our data, coupled with analysis of yeast Atg2 and Vps13 and human VPS13A (Kaminska et al., 2016; Kolakowski et al., 2020; Rzepnikowska et al., 2017), suggests that the Apt1 domain represents a novel phosphatidylinositol-binding domain. PI(4)P, one of the lipids that Hobbit Apt1 bound to in our study, is most highly enriched in the plasma membrane and the Golgi (Vermeer et al., 2009), suggesting that the Z. mays protein fragment may have been enriched at the Golgi due to PI(4)P binding. Interestingly, the specificity for phosphatidylinositols appears to change among divergent Apt1 domains. The fly Hobbit Apt1 domain bound strongly to all phosphatidylinositol moieties, while the Apt1 domain of yeast Atg2 bound strongly to PI(3)P and more weakly to PI(3,5)P2 and PI(4,5)P2 (Kaminska et al., 2016), the yeast Vps13 Apt1 domain bound specifically to PI(3)P (Rzepnikowska et al., 2017), and the human VPS13A Apt1 domain bound strongly to PI(3)P and PI(5)P and more weakly to PI(4)P (Kolakowski et al., 2020). Thus, Apt1 domains may represent a novel family of phosphatidylinositol-binding domains with varying specificity.
Given that Hobbit binds to phosphatidylinositols and that PI(4,5)P2 distribution appears to be altered in hobbit mutant cells, we propose that hobbit may be a novel lipid transfer protein at ER–PM contact sites. Hobbit also has homology to Atg2 and Vps13 in the Apt1 domain; both of these proteins mediate lipid transfer at MCS (Kumar et al., 2018; Li et al., 2020; Maeda et al., 2019; Osawa et al., 2019; Valverde et al., 2019). Additionally, our data show that Hobbit binds to all phosphatidylinositol moieties, some of which are not enriched at the PM (Balla, 2013), raising the possibility that Hobbit may localize to multiple types of ER MCS. Indeed, in addition to the puncta at the plasma membrane, we also observe Hobbit in puncta at other subcellular locations in the larval salivary glands, a finding potentially consistent with localization to other ER MCS. A number of known lipid transfer proteins localize to multiple types of MCS; for example, VPS13A localizes to both ER–mitochondria and ER–lipid droplet contact sites (Ugur et al., 2020). Future work will be required to examine Hobbit localization to other types of ER MCS and to directly test whether Hobbit functions as a lipid transfer protein and, if so, which lipids are transferred by Hobbit. If hobbit does function as a lipid transfer protein, it would be the first such example of a lipid transfer protein that is essential for animal development.
The most conspicuous phenotype in Drosophila hobbit mutant animals is a dramatic reduction in body size caused by failure to secrete insulin from the IPCs; hobbit mutant animals also exhibit defects in regulated exocytosis of mucin-like ‘glue’ proteins from the larval salivary glands (Neuman and Bashirullah, 2018). Interestingly, mutation of the Z. mays and A. thaliana orthologs of hobbit causes defects in root hair tip growth and pollen tube growth, and both of these processes are highly dependent upon secretion (Guan et al., 2013; Procissi et al., 2003; Xu and Dooner, 2006). Thus, there appears to be an evolutionarily shared requirement for hobbit function in cells with high secretory loads. However, our data presented here suggests that hobbit plays a role in the regulation of phosphatidylinositol distribution, indicating that the effects on secretion may be indirect. Phosphatidylinositols play essential roles in regulating intracellular trafficking, both in defining membrane identity and in ensuring the recruitment of the proper subset of proteins to the appropriate organelle membrane (Balla, 2013); therefore, disruption of phosphatidylinositol subcellular distribution would be expected to interfere with membrane trafficking. We do not observe widespread membrane-trafficking defects in hobbit mutant cells; instead, regulated exocytosis appears to be specifically affected. The impact of phosphatidylinositol dynamics on regulated exocytosis in hobbit mutant cells remains an important open question.
MATERIALS AND METHODS
Yeast media and genetics
Standard recipes were used for YPD (complete medium) and synthetic drop out medium (Sherman, 2002). YPGal medium was prepared like YPD but contained 2% galactose instead of 2% dextrose. Drop out medium was used for selection and retention of plasmids. All imaging experiments were performed using synthetic complete or drop out medium. Gene deletions and GFP fusions were made following standard genetic techniques (Longtine et al., 1998).
Yeast imaging
1 ml of mid-log cultures grown in synthetic medium were spun down (12,000 g for 1 min), and all but ∼50 µl of medium were aspirated. The pellet was then re-suspended in the remaining medium, resulting in a concentrated suspension of cells. Cells were then imaged on a DeltaVision RT system, using a DV Elite CMOS camera and a 100× objective. Image deconvolution was performed using DeltaVision software (softWoRx 6.5.2).
Yeast protease protection assay
Cells (40 ODs) of cells were collected from mid-log cultures grown in YPD. Cell pellets were re-suspended in pre-spheroplast medium (100 mM Tris-HCl pH 8, 10 mM DTT) and incubated for 10 min at room temperature. Cells were spun down and re-suspended in 4 ml spheroplast buffer (0.1 M phosphocitrate, 1.0 M sorbitol and 0.5 mg/ml zymolyase) and incubated at 30°C for 30 min. Cells were then spun down (2000 g for 5 min) and washed with 4 ml 0.1 M phosphocitrate, 1 M sorbitol. To lyse cells, cells were spun down (2000 g for 5 min) and re-suspended in 4.3 ml lysis buffer (50 mM Tris-HCl pH 7.5, 0.6 M sorbitol), then the suspension was dounced 15 times. The lysis product was spun at 500 g for 5 min to remove intact cells, and 200 μl of the supernatant was taken as the input sample. The remaining supernatant was centrifuged at 40,000 g for 10 min. The supernatant was discarded, and the pellet was resuspended in 100 µl lysis buffer. The resuspended pellet was then split into three tubes of 33 µl each. Tube one was used as a mock experiment. The second tube was the protease protection experiment, and contained 0.12 µg/ml proteinase K (Invitrogen, 25530049). The third tube was the protease positive control experiment and contained 0.12 µg/ml proteinase K and 1% Triton X-100 to disrupt membranes. The tubes were incubated for 25 min at room temperature. To destroy proteinase K function, 3 µl 0.2 M PMSF was added, followed by 33 µl 2× sample buffer (150 mM Tris-HCl, pH 6.8, 7 M urea, 10% SDS, 24% glycerol, Bromophenol Blue). The input fraction was precipitated in 10% trichloroacetic acid (TCA) and re-suspended in 100 µl 2× sample buffer. Samples were heated at 42°C for 7 min then loaded on an 11% SDS page gel for western blotting using standard procedures. Mouse anti-GFP (B2) antibody (Santa Cruz Biotechnology, sc-9996) was diluted to 1:500 and rabbit anti-Kar2 antibody (a gift from R. Schekman, University of California at Berkeley, CA, USA) was diluted to 1:20,000.
Electron microscopy and analysis of ER–PM contact sites in yeast
Yeast cells were grown overnight in YPD or YPGal and harvested during mid-log growth phase. Cell fixing and staining were performed as previously described (Jorgensen et al., 2020). For quantification of ER–PM contacts, the researcher was blind to the strain and growth conditions of cells in the EM images. ImageJ was used to measure the length of the PM from each cell by tracing the PM using the freehand line tool, then measuring the length of each ER–PM contact site using the same tool. The sum of the ER–PM contact measurements was then divided by the total length of the PM of each image. These values were used to calculate the average cER to PM ratio for each strain.
Sequence alignments and conservation analysis
To identify highly conserved protein sequences in Fmp27, the S. cerevisiae Fmp27 protein sequence was aligned with the orthologous protein sequence from Kazachstania naganishii, Tetrapisispora blattae, Kuraishia capsulata, Candida albicans, Cercospora beticola, Neurospora crassa, Ustilago maydis, Aspergillus flavus and Schizosaccharomyces pombe. The multiple sequence alignment was performed using MAFFT version 7 (Katoh et al., 2018; Kuraku et al., 2013). For Drosophila Hobbit, an alignment was performed between Drosophila melanogaster hobbit and its orthologs in Drosophila willistoni, Drosophila ananassae, Drosophila erecta, Drosophila simulans, Drosophila sechellia, Drosophila grimshawi, Drosophila virilis, Drosophila mojavensis and Drosophila busckii using ClustalOmega (Sievers et al., 2011). ClustalOmega was also used to align the Apt1 domains. For both yeast and Drosophila, the alignments were visualized in Jalview (Waterhouse et al., 2009) and conservation scores were extracted at each position to generate the identity plots.
Drosophila stocks, husbandry, and body size and lethal phase analysis
The following fly stocks were obtained from the Bloomington Drosophila Stock Center: UAS-KDEL-RFP, UAS-mTagBFP2, Sgs3-GAL4, Sgs3-GFP, act-GAL4, UAS-PLCδPH-GFP. UAS-hobbit-GFP and the hobbit mutant alleles were previously described (Neuman and Bashirullah, 2018). We have used the ‘>’ symbol in genotypes as shorthand for ‘GAL4’, which acts as a driver. All experimental crosses were grown on standard cornmeal-molasses medium in uncrowded bottles or vials kept in an incubator set to 25°C. Body size quantification was performed as previously described (Neuman and Bashirullah, 2018). Pupa images were captured on an Olympus SXZ16 stereomicroscope coupled to an Olympus DP72 digital camera with DP2-BSW software. For lethal phase analysis, pupae were allowed to age on grape agar plates at 25°C for 1 week; then, Bainbridge and Bownes staging criteria (Bainbridge and Bownes, 1981) were used to determine lethal phase.
Drosophila transgene generation
To generate UAS-hobbitΔC82-GFP, the pENTR 1A-hobbit plasmid (Neuman and Bashirullah, 2018) was digested with SpeI and NotI. The following primers were annealed, phorphorylated, and ligated into pENTR 1A-hobbit: 5′-CTAGTCATACCTACGCTGGAGTATCACAATGTGACGAAGC-3′ and 5′-GGCCGCTTCGTCACATTGTGATACTCCAGCGTAGGTATGA-3′. The resulting plasmid (pENTR 1A-hobbitΔC82) was sequence-verified and recombined into the Gateway cloning destination vector pBID-UASC-G-GFP (Neuman and Bashirullah, 2018; Wang et al., 2012) using LR Clonase (Invitrogen). Successful recombination was confirmed by sequencing. To generate UAS-hobbitΔC82-mCherry, mCherry was first amplified from pLV-mCherry (Addgene) using the following primers: 5′-ACAGGTACCAGTGAGCAAGGGCGAGGAGGAT-3′ and 5′-ACATCTAGATACTTGTACAGCTCGTCCATG-3′. The resulting PCR product was digested with KpnI and XbaI and ligated into pBID-UASC-G-GFP (thereby replacing GFP with mCherry). The pENTR 1A-hobbitΔC82 entry clone was recombined into pBID-UASC-G-mCherry as described above and sequence verified. Similarly, the pENTR 1A-hobbit entry vector was recombined into pBID-UASC-G-mCherry to generate UAS-hobbit-mCherry and was sequence-verified. To generate UAS-hobbitΔN117-GFP, pENTR 1A-hobbit was digested with KpnI and AgeI. The resulting ∼0.8 kb fragment was used as a template for PCR with the following primers: 5′-AAAGGTACCCAACATGGCCTCGGAGGCGAAGGGTGT-3′ and 5′-AAAACCGGTCGATGTCCTCCGCTTGGCCT-3′. The resulting 0.37 kb fragment was digested with KpnI and AgeI and ligated into pENTR 1A-hobbit; the pENTR 1A-hobbitΔN117 entry vector was recombined into pBID-UASC-G-GFP as described above and sequence-verified. All final plasmids were injected into VK00027 flies for phiC31-mediated site-directed integration using standard techniques (Rainbow Transgenic Flies, Inc.). To generate UAS-StimDDAA-GFP transgenic flies, plasmid LD45776 containing the cDNA sequence of Drosophila Stim Isoform A was obtained from the Drosophila Genomics Resource Center (DGRC). The Stim coding sequence was cloned by PCR using primers 5′-CACCATGCGAAAGAATACCATTTGGAAC-3′ and 5′-TTCCGTGGCAAGCAGCGAAAAGTTC-3′ and ligated into pENTR/D-TOPO (Invitrogen). Site-directed mutagenesis (Stratagene QuikChange XL) was then used to convert the codons encoding D155 and D157 into codons for alanine using the primer 5′-GCTTGCATCGTCAGCTAGCTGATGCCGATAATGGAAACATCG-3′ and its reverse complement, followed by sequence confirmation. The StimDDAA coding sequence was then recombined into the pPWG destination vector (Carnegie Drosophila Gateway Vector Collection), which introduces a UASp promoter and C-terminal EGFP tag, using LR Clonase. The final plasmid was injected into w1118 embryos for generation of transgenic animals by random transposable element insertion.
Protein expression, purification and lipid blots
The Apt1 fragment of the Hobbit protein (amino acids 1750–2300) and Apt1ΔC82 (amino acids 1750–2218) were amplified from the pENTR 1A-hobbit plasmid (described above) using the following primers: Apt1 F 5′-ATCCGGTACCCAACATGGTAGTCTCAGAGACTGTTGGAGCTTTCTTGAGCGAC-3′ and Apt1 R 5′-GCATAAGCTTCTAATGATGATGATGATGATGGCTGCCGCCGCCGCCCAGCTCCTCCTTGCTCTCCCGG-3′; Apt1ΔC82 F 5′-ATCCGGTACCCAACATGGTAGTCTCAGAGACTGTTGGAGCTTTCTTGAGCGAC-3′ and Apt1ΔC82 R 5′-GCATAAGCTTCTAATGATGATGATGATGATGGCTGCCGCCGCCGCCCCACGTCACATTGTGATACTCCAGCG-3′. The reverse primers include a C-terminal five amino acid spacer and a 6×-His tag. The resulting PCR products were digested with KpnI and HindIII, ligated into pTRC99a (Amann et al., 1988) and sequence verified. Plasmids were transformed into E. coli EXPRESS BL21(DE3) chemically competent cells (Lucigen). A dense overnight culture was diluted 1:100 into 15 ml LB (Miller) with 100 µg/ml ampicillin. Cultures were grown at 37°C to A600 ∼0.4, then 200 μM IPTG (final concentration) was added and cultures continued shaking for 3 h at 37°C. Cultures were then centrifuged for 5 min at 5000 g, and pellets were frozen at −80°C. For protein purification, pellets were thawed on ice for 15 min, resuspended in 3 ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8) with 1 mg/ml lysozyme (final concentration), 1.5 µl Benozoase nuclease (Sigma) and 1 µl protease inhibitor cocktail (Sigma P8849), incubated on ice for 30 min, and centrifuged for 30 min at 10,000 g at 4°C. 1.5 ml of Ni-NTA agarose (Qiagen) was washed three times with lysis buffer. Cleared lysate and washed Ni-NTA agarose were combined and incubated for 1 h at 4°C while rotating on a nutator. The slurry was loaded onto a gravity column, washed two times with 1 ml wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8), and eluted four times with 0.5 ml elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8). Proteins were visualized on 4–20% Mini-PROTEAN TGX precast gels (BioRad) and concentration was determined using the Bradford Protein Assay (BioRad). For lipid blotting experiments, Membrane Lipid Strips (Echelon Biosciences) or PIP Strips (Echelon Biosciences) were blocked in phosphate-buffered saline (PBS) with 0.1% Tween-20 (PBST) and 3% BSA overnight while rotating at 4°C. The strips were then incubated for 1 h at room temperature with 1.5 mg of purified protein diluted in PBST with 3% BSA to a 15 ml final volume. A 1:10,000 dilution of mouse anti-(6×)His (Invitrogen) primary antibody in PBST with 3% BSA was applied for 1 h at room temperature, followed by a 1:20,000 dilution of anti-mouse-IgG HRP-conjugated secondary antibody (Promega) for 1 h at room temperature. Washes with PBST were performed between each step. Lipid strips were then incubated for 5 min with SuperSignal West Pico PLUS Chemiluminescent Reagent (Thermo Fisher Scientific) and visualized using a UVP ChemiDoc-It2. Image brightness and contrast were optimized post acquisition using Adobe Photoshop CS6.
Confocal microscopy in Drosophila salivary glands
All images were obtained from live, unfixed tissues. Salivary glands were dissected from animals of the appropriate developmental stage and genotype in PBS and mounted in 1% low-melt agarose (Apex Chemicals) made in PBS. Tissues were imaged for no more than 15 min after mounting, and at least 10 salivary glands were imaged per experiment. Imaging was carried out at room temperature. Images were acquired using an Olympus FV3000 laser scanning confocal microscope (20× objective, NA 0.75 or 100× oil immersion objective, NA 1.49) with FV31S-SW software. The pinhole was closed down to 0.8 Airy disc for images acquired at high resolution (Fig. 4A,B, Fig. S3B). Images obtained as z-stacks, as indicated in the figure legends, and were deconvolved using three iterations of the Olympus CellSens Deconvolution for Laser Scanning Confocal Advanced Maximum Likelihood algorithm. Brightness and contrast were optimized post acquisition using Olympus FV31S-SW software. Pearson's correlation coefficients were calculated using Olympus CellSens software. For imaging-based protease protection assays, salivary glands were adhered to a plastic coverslip in a minimal quantity of PBS; after at least 30 s of time-lapse image acquisition using the Olympus FV3000 resonant scan head, 50 µl of 0.05% digitonin (Invitrogen) was added, followed by an additional at least 1 min of imaging. Then we added 75 µl of 50 µg/ml proteinase K (Thermo Fisher Scientific), followed by at least 2 min of imaging.
Supplementary Material
Acknowledgements
Reagents obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and Drosophila Genomics Resource Center (NIH 2P40OD010949) were used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.D.N., J.R.J., S.D.E., A.B.; Methodology: S.D.N., J.R.J., S.D.E., A.B.; Validation: S.D.N., J.R.J., S.D.E., A.B.; Investigation: S.D.N., J.R.J., A.T.C., J.T.S., J.E.S.; Data curation: S.D.N., J.R.J.; Writing - original draft: S.D.N.; Writing - review & editing: S.D.N., J.R.J., J.T.S., S.D.E., A.B.; Visualization: J.R.J., S.D.E., A.B.; Supervision: S.D.E., A.B.; Project administration: S.D.E., A.B.; Funding acquisition: J.T.S., S.D.E., A.B.
Funding
This work was supported in part by the National Institutes of Health (GM123204 to A.B.) and by a Cornell University Research Grant to S.D.E. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.259086
References
- Aeschbacher, R. A., Hauser, M. T., Feldmann, K. A. and Benfey, P. N. (1995). The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev. 9, 330-340. 10.1101/gad.9.3.330 [DOI] [PubMed] [Google Scholar]
- Amann, E., Ochs, B. and Abel, K.-J. (1988). Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69, 301-315. 10.1016/0378-1119(88)90440-4 [DOI] [PubMed] [Google Scholar]
- Baba, Y., Nishida, K., Fujii, Y., Hirano, T., Hikida, M. and Kurosaki, T. (2008). Essential function for the calcium sensor STIM1 in mast cell activation and anaphylactic responses. Nat. Immunol. 9, 81-88. 10.1038/ni1546 [DOI] [PubMed] [Google Scholar]
- Bainbridge, B. S. P. and Bownes, M. (1981). Staging the metamorphosis of Drosophila melanogaster. Development 66, 57-80. 10.1242/dev.66.1.57 [DOI] [PubMed] [Google Scholar]
- Balla, T. (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019-1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey, P. N., Linstead, P. J., Roberts, K., Schiefelbein, J. W., Hauser, M. T. and Aeschbacher, R. A. (1993). Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119, 57-70. 10.1242/dev.119.1.57 [DOI] [PubMed] [Google Scholar]
- Brodsky, J. L. and Schekman, R. (1993). A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123, 1355-1363. 10.1083/jcb.123.6.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. and Bezanilla, M. (2021). SABRE populates ER domains essential for cell plate maturation and cell expansion influencing cell and tissue patterning. eLife 10, e65166. 10.7554/eLife.65166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado, J., Kalemanov, M., Campelo, F., Bourgoint, C., Thomas, F., Loewith, R., Martínez-Sánchez, A., Baumeister, W., Stefan, C. J. and Fernández-Busnadiego, R. (2019). Tricalbin-mediated contact sites control ER curvature to maintain plasma membrane integrity. Dev. Cell 51, 476-487.e7. 10.1016/j.devcel.2019.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttell, L., Vaughan, A., Silva, E., Escaron, C. J., Lavine, M., Van Goethem, E., Eid, J.-P., Quirin, M. and Franc, N. C. (2008). Undertaker, a Drosophila junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell 135, 524-534. 10.1016/j.cell.2008.08.033 [DOI] [PubMed] [Google Scholar]
- Eisenberg-Bord, M., Shai, N., Schuldiner, M. and Bohnert, M. (2016). A tether is a tether is a tether: tethering at membrane contact sites. Dev. Cell 39, 395-409. 10.1016/j.devcel.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Feske, S., Gwack, Y., Prakriya, M., Srikanth, S., Puppel, S.-H., Tanasa, B., Hogan, P. G., Lewis, R. S., Daly, M. and Rao, A. (2006). A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441, 179-185. 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Giordano, F., Saheki, Y., Idevall-Hagren, O., Colombo, S. F., Pirruccello, M., Milosevic, I., Gracheva, E. O., Bagriantsev, S. N., Borgese, N. and De Camilli, P. (2013). PI(4,5)P2-Dependent and Ca2+-Regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494-1509. 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y., Guo, J., Li, H. and Yang, Z. (2013). Signaling in pollen tube growth: crosstalk, feedback, and missing links. Mol. Plant 6, 1053-1064. 10.1093/mp/sst070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwack, Y., Srikanth, S., Oh-hora, M., Hogan, P. G., Lamperti, E. D., Yamashita, M., Gelinas, C., Neems, D. S., Sasaki, Y., Feske, S.et al. (2008). Hair loss and defective T- and B-cell function in mice lacking ORAI1. Mol. Cell. Biol. 28, 5209-5222. 10.1128/MCB.00360-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne, W. M., Liou, J. and Emr, S. D. (2015). Molecular mechanisms of inter-organelle ER-PM contact sites. Curr. Opin. Cell Biol. 35, 123-130. 10.1016/j.ceb.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Hogan, P. G. and Rao, A. (2015). Store-operated calcium entry: Mechanisms and modulation. Biochem. Biophys. Res. Commun. 460, 40-49. 10.1016/j.bbrc.2015.02.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, J. R., Tei, R., Baskin, J. M., Michel, A. H., Kornmann, B. and Emr, S. D. (2020). ESCRT-III and ER–PM contacts maintain lipid homeostasis. Mol. Biol. Cell 31, 1302-1313. 10.1091/mbc.E20-01-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska, J., Rzepnikowska, W., Polak, A., Flis, K., Soczewka, P., Bala, K., Sienko, M., Grynberg, M., Kaliszewski, P., Urbanek, A.et al. (2016). Phosphatidylinositol-3-phosphate regulates response of cells to proteotoxic stress. Int. J. Biochem. Cell Biol. 79, 494-504. 10.1016/j.biocel.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Katoh, K., Rozewicki, J. and Yamada, K. D. (2018). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160-1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakowski, D., Kaminska, J. and Zoladek, T. (2020). The binding of the APT1 domains to phosphoinositides is regulated by metal ions in vitro. Biochim. Biophys. Acta Biomembr. 1862, 183349. 10.1016/j.bbamem.2020.183349 [DOI] [PubMed] [Google Scholar]
- Kumar, N., Leonzino, M., Hancock-Cerutti, W., Horenkamp, F. A., Li, P. Q., Lees, J. A., Wheeler, H., Reinisch, K. M. and De Camilli, P. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625-3639. 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku, S., Zmasek, C. M., Nishimura, O. and Katoh, K. (2013). aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41, W22-W28. 10.1093/nar/gkt389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J. and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105-132. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- Li, P. Q., Lees, J. A., Patrick Lusk, C. and Reinisch, K. M. (2020). Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J. Cell Biol. 219, e202001161. 10.1083/jcb.202001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, J., Kim, M. L., Won, D. H., Jones, J. T., Myers, J. W., Ferrell, J. E. and Meyer, T. (2005). STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235-1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Lorenz, H., Hailey, D. W. and Lippincott-Schwartz, J. (2006). Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Methods 3, 205-210. 10.1038/nmeth857 [DOI] [PubMed] [Google Scholar]
- Lunz, V., Romanin, C. and Frischauf, I. (2019). STIM1 activation of Orai1. Cell Calcium 77, 29-38. 10.1016/j.ceca.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Maeda, S., Otomo, C. and Otomo, T. (2019). The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 8, e45777. 10.7554/eLife.45777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford, A. G., Stefan, C. J., Yuan, H. L., MacGurn, J. A. and Emr, S. D. (2012). ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129-1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., Tosatto, S. C. E., Paladin, L., Raj, S., Richardson, L. J.et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412-D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S. E. and Levine, T. P. (2016). VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1861, 952-961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Neuman, S. D. and Bashirullah, A. (2018). Hobbit regulates intracellular trafficking to drive insulin-dependent growth during Drosophila development. Development 145, dev161356. 10.1242/dev.161356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-hora, M., Yamashita, M., Hogan, P. G., Sharma, S., Lamperti, E., Chung, W., Prakriya, M., Feske, S. and Rao, A. (2008). Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432-443. 10.1038/ni1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omnus, D. J., Manford, A. G., Bader, J. M., Emr, S. D. and Stefan, C. J. (2016). Phosphoinositide kinase signaling controls ER-PM cross-talk. Mol. Biol. Cell 27, 1170-1180. 10.1091/mbc.E16-01-0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, T., Kotani, T., Kawaoka, T., Hirata, E., Suzuki, K., Nakatogawa, H., Ohsumi, Y. and Noda, N. N. (2019). Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26, 281-288. 10.1038/s41594-019-0203-4 [DOI] [PubMed] [Google Scholar]
- Pathak, T., Trivedi, D. and Hasan, G. (2017). CRISPR-Cas-induced mutants identify a requirement for dSTIM in larval dopaminergic cells of Drosophila melanogaster. G3 Genes, Genomes, Genet. 7, 923-933. 10.1534/g3.116.038539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler, H., Gaigg, B., Hrastnik, C., Achleitner, G., Kohlwein, S. D., Zellnig, G., Perktold, A. and Daum, G. (2001). A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268, 2351-2361. 10.1046/j.1432-1327.2001.02116.x [DOI] [PubMed] [Google Scholar]
- Pietra, S., Gustavsson, A., Kiefer, C., Kalmbach, L., Hörstedt, P., Ikeda, Y., Stepanova, A. N., Alonso, J. M. and Grebe, M. (2013). Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat. Commun. 4, 2779. 10.1038/ncomms3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra, S., Lang, P. and Grebe, M. (2015). SABRE is required for stabilization of root hair patterning in Arabidopsis thaliana. Physiol. Plant. 153, 440-453. 10.1111/ppl.12257 [DOI] [PubMed] [Google Scholar]
- Porter, K. R. and Palade, G. E. (1957). Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J. Biophys. Biochem. Cytol. 3, 269-300. 10.1083/jcb.3.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, W. A., Toulmay, A. and Balla, T. (2020). The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21, 7-24. 10.1038/s41580-019-0180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procissi, A., Guyon, A., Pierson, E. S., Giritch, A., Knuiman, B., Grandjean, O., Tonelli, C., Derksen, J., Pelletier, G. and Bonhomme, S. (2003). Kinky Pollen encodes a Sabre-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. Plant J. 36, 894-904. 10.1046/j.1365-313X.2003.01933.x [DOI] [PubMed] [Google Scholar]
- Quon, E., Sere, Y. Y., Chauhan, N., Johansen, J., Sullivan, D. P., Dittman, J. S., Rice, W. J., Chan, R. B., Di Paolo, G., Beh, C. T.et al. (2018). Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS Biol. 16, e2003864. 10.1371/journal.pbio.2003864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders, J., Zahedi, R. P., Pfanner, N., Meisinger, C. and Sickmann, A. (2006). Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5, 1543-1554. 10.1021/pr050477f [DOI] [PubMed] [Google Scholar]
- Roos, J., DiGregorio, P. J., Yeromin, A. V., Ohlsen, K., Lioudyno, M., Zhang, S., Safrina, O., Kozak, J. A., Wagner, S. L., Cahalan, M. D.et al. (2005). STIM1, an essential and conserved component of store-operated Ca 2+ channel function. J. Cell Biol. 169, 435-445. 10.1083/jcb.200502019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., Misra, L. M. and Vogel, J. P. (1989). KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57, 1211-1221. 10.1016/0092-8674(89)90058-5 [DOI] [PubMed] [Google Scholar]
- Rzepnikowska, W., Flis, K., Kaminska, J., Grynberg, M., Urbanek, A., Ayscough, K. R. and Zoladek, T. (2017). Amino acid substitution equivalent to human choreaacanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate. Hum. Mol. Genet. 26, 1497-1510. 10.1093/hmg/ddx054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki, Y. and De Camilli, P. (2017). Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 86, 659-684. 10.1146/annurev-biochem-061516-044932 [DOI] [PubMed] [Google Scholar]
- Shai, N., Yifrach, E., Van Roermund, C. W. T., Cohen, N., Bibi, C., Ijlst, L., Cavellini, L., Meurisse, J., Schuster, R., Zada, L.et al. (2018). Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat. Commun. 9, 1761. 10.1038/s41467-018-03957-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (2002). Getting started with yeast. Methods Enzymol. 350, 3-41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J.et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan, C. J., Manford, A. G., Baird, D., Yamada-Hanff, J., Mao, Y. and Emr, S. D. (2011). Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144, 389-401. 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Stiber, J., Hawkins, A., Zhang, Z.-S., Wang, S., Burch, J., Graham, V., Ward, C. C., Seth, M., Finch, E., Malouf, N.et al. (2008). STIM1 signalling controls store-operated calcium entry required for development and contractile function in skeletal muscle. Nat. Cell Biol. 10, 688-697. 10.1038/ncb1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur, B., Hancock-Cerutti, W., Leonzino, M. and De Camilli, P. (2020). Role of VPS13, a protein with similarity to ATG2, in physiology and disease. Curr. Opin. Genet. Dev. 65, 61-68. 10.1016/j.gde.2020.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm, A. M., Cohen, S., Legant, W. R., Melunis, J., Hershberg, U., Wait, E., Cohen, A. R., Davidson, M. W., Betzig, E. and Lippincott-Schwartz, J. (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162-167. 10.1038/nature22369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, D. P., Yu, S., Boggavarapu, V., Kumar, N., Lees, J. A., Walz, T., Reinisch, K. M. and Melia, T. J. (2019). ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218, 1787-1798. 10.1083/jcb.201811139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer, J. E. M., Thole, J. M., Goedhart, J., Nielsen, E., Munnik, T. and Gadella, T. W. J.Jr. (2009). Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 57, 356-372. 10.1111/j.1365-313X.2008.03679.x [DOI] [PubMed] [Google Scholar]
- Verstreken, P., Ohyama, T., Haueter, C., Habets, R. L. P., Lin, Y. Q., Swan, L. E., Ly, C. V., Venken, K. J. T., De Camilli, P. and Bellen, H. J. (2009). Tweek, an evolutionarily conserved protein, is required for synaptic vesicle recycling. Neuron 63, 203-215. 10.1016/j.neuron.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig, M., Peinelt, C., Beck, A., Koomoa, D. L., Rabah, D., Koblan-Huberson, M., Kraft, S., Turner, H., Fleig, A., Penner, R.et al. (2006). CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science (80-.) 312, 1220-1223. 10.1126/science.1127883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.-W., Beck, E. S. and McCabe, B. D. (2012). A modular toolset for recombination transgenesis and neurogenetic analysis of Drosophila. PLoS ONE 7, e42102. 10.1371/journal.pone.0042102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. and Barton, G. J. (2009). Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189-1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M., Zurek, N., Hoenger, A. and Voeltz, G. K. (2011). A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193, 333-346. 10.1083/jcb.201011039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Carvalho, P. and Voeltz, G. K. (2018). Here, there, and everywhere: the importance of ER membrane contact sites. Science (80- ) 361, eaan5835. 10.1126/science.aan5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. and Dooner, H. K. (2006). The Maize aberrant pollen transmission 1 Gene Is a SABRE/KIP Homolog required for pollen tube growth. Genetics 172, 1251-1261. 10.1534/genetics.105.050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Luo, N., Sun, L. and Liu, D. (2012). HPS4/SABRE regulates plant responses to phosphate starvation through antagonistic interaction with ethylene signalling. J. Exp. Bot. 63, 4527-4538. 10.1093/jxb/ers131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. L., Yu, Y., Roos, J., Kozak, J. A., Deerinck, T. J., Ellisman, M. H., Stauderman, K. A. and Cahalan, M. D. (2005). STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437, 902-905. 10.1038/nature04147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. L., Yeromin, A. V., Zhang, X. H.-F., Yu, Y., Safrina, O., Penna, A., Roos, J., Stauderman, K. A. and Cahalan, M. D. (2006). Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. USA 103, 9357-9362. 10.1073/pnas.0603161103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.