Abstract
Background
Acute respiratory infections (ARI) are associated with a huge morbidity and mortality worldwide. Rhinoviruses (RVs) and Enteroviruses (EVs) are recognized as leading causes of ARI.
Objectives
The present study describes the molecular epidemiology of RVs and EVs in Cameroon over a 3‐year surveillance period.
Methods
From September 2011 to October 2014, nasopharyngeal swabs were collected from patients with influenza‐like illness (ILI) and severe acute respiratory infections (SARI). Two sub‐genomic regions of the EVs and RVs were targeted for molecular characterization. These included the most conserved 5′‐untranslated region (5′UTR) and the viral protein 4/viral protein 2 transition region (VP4/VP2).
Results
A total of 974 samples were collected. Children ≤5 years accounted for 85.7% (835/974) of all participants. Among them, 160 (16.4%) were positive for RVs and/or EVs. RVs and/or EVs were significantly more identified in ILI compared to SARI patients (P = .015). Both viruses co‐circulated all year long with a marked increase of occurrence during rainy and cold season. All RV species were found to circulate in Cameroon, with 6, 10 and 6 virus types belonging to the RV‐A, RV‐B and RV‐C, respectively. EV species identified comprised EV‐A (1 Coxsackie virus A5), EV‐B (1 Coxsackie virus A9 and 2 Coxsackie virus B1) and EV‐C (1 EV‐C117).
Conclusions
This study indicates a strong year‐round occurrence of EV and RV associated respiratory infections in Cameroon. Molecular characterization identified a wide variety of RVs and EVs in patients with ARI in Cameroon.
Keywords: Cameroon, Enterovirus, molecular characterization, respiratory infections, Rhinovirus
1. INTRODUCTION
Acute respiratory infections (ARI) constitute a major cause of morbidity and mortality worldwide. Although global mortality due to lower respiratory tract infection (LRTI) and upper respiratory tract infection (URTI) underwent a reduction of 21.1% and 42.1%, respectively, from 2007 to 2017, these infections still caused more than 2.5 million deaths in 2017.1 The burden of LRTI was worsened even more with the emergence of COVID‐19 in late 2019 which has already caused more than 2.4 million deaths worldwide in less than a year.2 Respiratory viruses are also responsible for huge economic losses due to the use of health care resources and absenteeism at work and school.3, 4, 5 Respiratory viruses, including Rhinoviruses (RVs) and Enteroviruses (EVs), are the most common cause of ARI. Rhinoviruses are the main cause of URTI worldwide with common cold being their main clinical presentation.6, 7 With the development of molecular detection assays, RVs have recently been shown to be also involved in a significant proportion of more severe LRTIs such as wheezing, bronchiolitis, asthma exacerbation and chronic obstructive pulmonary disease.8, 9, 10 Rhinoviruses have also been reported in apparently healthy individuals, possibly due to the prolonged viral shedding period.11, 12 Enteroviruses, on the other hand, have a broader tropism and can cause a wide range of human infections including acute respiratory infections, meningitis, encephalitis, gastroenteritis, acute flaccid paralysis or conjunctivitis.13, 14, 15 A new strain of EV‐D68 re‐emerged, circulated worldwide and caused particularly severe acute respiratory infections (SARI) and acute flaccid myelitis.16
Enteroviruses and RVs belong to the Picornaviridae family and the Enterovirus genus. According to the International Committee on Taxonomy of Viruses (ICTV), the Enterovirus genus comprises 15 species EV‐A to L and RV‐A to C.17 Based on the sequence diversity of the structural protein coding genes, individual viruses of the EV species are further assigned into virus types. Rhinoviruses consist of more than a hundred virus types. Virus type assignments based on the nucleotide sequence of the VP4/VP2 region correlate with the type assignment based on the VP1 region of EVs and RVs.18, 19
All RV and EV species have a global distribution and a year‐round circulation pattern with occasional peaks.20 In West Africa,21, 22, 23 Southern Africa,24, 25 Northern Africa,26, 27 Eastern Africa28, 29, 30 and Central Africa,31, 32, 33 multiple studies reported the circulation of EVs and RVs in patients with mild and severe respiratory infections. However, data on genetic diversity and circulating types remained limited. Most of the studies have been conducted outside of Central Africa and have focussed on strains of EV‐64.24, 34, 35, 36, 37, 38, 39, 40 In Cameroon, RVs and EVs have been shown to be among the most commonly viruses detected in patients with LRTI and URTI,31, 41 but the virus types involved remain unknown. In the present study, we report the occurrence and phylogenetic relatedness of EVs and RVs detected in samples from patients of all ages suffering from ARI in Cameroon based on the sequences of the VP4/VP2 genomic region.
2. METHODS
2.1. Study design
In this cross‐sectional study, we conducted a prospective recruitment of patients of all ages at Centre Hospitalier d'Essos in Yaounde, Cameroon. We included participants enrolled from September 2011 to October 2014. Ethical approval was obtained from the National Ethics Committee of Research for Human Health (N° 121/CNE/SE/2011) and an administrative authorization from the Ministry of Public Health of Cameroon. Patients were informed of the study objectives, constraints, risks and benefits of participating in the study and those who provided their informed consent were enrolled. We included outpatients and inpatients with acute respiratory infections. A case of ARI was considered as any patient with a fever <5 days in addition to a cough and/or sore throat. Participants who did not provide their consent were not included.
2.2. Laboratory analysis
We collected a nasopharyngeal swab from all participants in a tube containing 1 mL of viral transport medium. Sample was shipped to the laboratory in reverse cold chain and stored at minus 80°C until the time of analyses. Viral RNAs were extracted and purified using the QIAamp Viral RNA Mini kit (Qiagen) according to manufacturer's specifications. A preliminary detection of RVs and/or EVs was carried out by real‐time RT‐PCR using primers and probes from the commercial Respiratory Multi Well System Rhino&EV/Cc r‐gene (BioMérieux) according to the manufacturer's intructions. Most of these RV and/or EV positive samples had already been the subject of several other studies, and the remaining volumes were very limited.41, 42, 43, 44, 45 Overall, 43 EV and/or RV positive samples have swab volumes were sufficient to allow RV and/or EV amplification and sequencing and were therefore considered in this study. We amplified the VP4/VP2 region of 43 RV and/or EV positive samples using the RT‐PCR technique previously described by Linsuwanon et al,46 in 2009. This RT‐PCR technique generates an amplicon of 540 nucleotides encompassing a fragment of the 5′‐UTR gene, the VP4 gene and a fragment of the VP2 gene. RNAs were initially used as a template to synthesize complementary DNA using random primers and the SuperScript™ III First‐Strand Synthesis System kit (Invitrogen). Resulting complementary DNAs were subsequently amplified in a first round PCR using the AmpliTaq DNA polymerase kit (Invitrogen) using primers F484 (5′‐CGGCCCCTGAATGYGGCTAA‐3′) and R1126 (5′‐ATCHGGHARYTTCCAMCACCA‐3′). The products of this first PCR were finally amplified in a second round PCR using the FastStart™ Taq DNA Polymerase kit (Roche) using primers F587 (5′‐CTACTTTGGGTGTCCGTGTTTC‐3′) and R1126 (5′‐ATCHGGHARYTTCCAMCACCA‐3′). Amplicons from the second PCR by gel green stained 2% agarose gel electrophoresis. Second round PCR amplicons were sequenced using the BigDye Terminator 1.1® kit (Thermo Fisher Scientific) according to the manufacturer's recommendations.
2.3. Sequence analysis
Sequences were assembled and edited using EDITSEQ of the software Seqman™ II Lasergene (DNA). RV and EV reference sequences were retrieved from the GenBank database. Multiple sequence alignments were carried out using the CLUSTAL W method47 in the MEGA software version 6.48 Originally, we used the neighbour‐joining method with a 1000‐bootstrap replicates to generate an extended phylogenetic tree with all known EV types in one hand and all known RV types in the other hand (Figures S1 and S2). Then, the EV and RV sequence data sets were downsized by omitting most branches containing no studied sequences. Reduced data set was used for maximum likelihood phylogenetic analyses using the best fit evolution model which was General Time Reversible with a Gamma distribution and assuming that a certain fraction of sites was invariable on an evolutionary level (GTR + G + I). The reliability of the tree nodes was evaluated by 1000 bootstrap resampling. Nucleotide sequences obtained in the present study were submitted to GenBank under the registration numbers MN508757 to MN508783.
2.4. Statistical analysis
We collected clinical and sociodemographic data from participants using a standardized form. We analysed the data using R software version 3.6.0. We described the categorical variables in number and percentage and the continuous variables in mean and standard deviation. We used the chi‐square test to compare categorical variables and linear regression for continuous variables, and P values < .05 were considered statistically significant.
3. RESULTS
3.1. Sociodemographic and clinical characteristics of the patients enrolled
From September 2011 to October 2014, we recruited 974 participants in this study. The age of the participants ranged from 1 month to 59 years with a mean of 3.3 ± 5.9 years. The proportions of the age groups represented varied inversely depending on the age of the participants: <2 years, 597 (61.3%); 2‐5 years, 238 (24.4%); 5‐15 years, 108 (11.1%); and >15 years, 25 (2.6%). A total of 509 (52.3%) participants were male, and 464 (47.6%) were female. A total of 528 (54.2%) participants were outpatients, while 446 (45.8%) were inpatients.
3.2. Viral identification
Of the 974 swabs tested, 160 (16.4%) were positive for RVs and/or EVs (Table 1). The rate of RV/EV positivity in inpatients vs outpatient was significantly different (P = .015). The rate of RV/EV positivity with respect to the age (P = .023) and year of recruitment was also significantly different (P = .002). In contrast, no significant difference in RV/EV positivity was found with respect to age group (P = .291) or sex (P = .604). The most common symptoms recorded in participants were rhinorrhoea and cough. RVs and EVs were found to circulate throughout the year (Figure 1). Apart from fatigue (P = .030) and headaches (P = .004), all other symptoms recorded were not significantly associated with RV/EV infections.
TABLE 1.
General data of the study participants
| Characteristics | Total | Rhinovirus/Enterovirus | P‐value | |
|---|---|---|---|---|
| Negative | Positive | |||
| No (974) | 814 (83.6) | 160 (16.4) | ||
| Gender | ||||
| Male | 509 (52.3) | 422 (51.8) | 87 (54.4) | .604 |
| Female | 464 (47.6) | 391 (48.0) | 73 (45.6) | |
| Age | ||||
| Median [IQR] | 1.7 [0.7‐3.0] | 2.0 [0.8‐4.0] | 1.4 [0.6‐3.0] | .023 |
| Age | ||||
| 0‐2 y | 597 (61.3) | 492 (60.4) | 105 (65.6) | .290 |
| 2‐5 y | 238 (24.4) | 200 (24.6) | 38 (23.8) | |
| 5‐15 y | 108 (11.1) | 93 (11.4) | 15 (9.4) | |
| >15 y | 25 (2.6) | 24 (2.9) | 1 (0.6) | |
| NA | 6 (0.6) | 5 (0.6) | 1 (0.6) | |
| Patient | ||||
| Inpatients | 446 (45.8) | 387 (47.5) | 59 (36.9) | .015 |
| Outpatients | 528 (54.2) | 427 (52.5) | 101 (63.1) | |
| Year | ||||
| 2011 | 73 (7.5) | 67 (8.2) | 6 (3.8) | .002 |
| 2012 | 429 (44.0) | 365 (44.8) | 64 (40.0) | |
| 2013 | 256 (26.3) | 219 (26.9) | 37 (23.1) | |
| 2014 | 216 (22.2) | 163 (20.0) | 53 (33.1) | |
| Symptoms | ||||
| Rhinorrhoea | 788 (80.9) | 650 (79.9) | 138 (86.3) | .199 |
| Cough | 819 (84.1) | 685 (84.2) | 134 (83.8) | .999 |
| Fatigue | 553 (56.8) | 474 (58.2) | 79 (49.4) | .030 |
| Wheezing | 369 (37.9) | 305 (37.5) | 64 (40.0) | 1.000 |
| Vomiting | 288 (29.6) | 244 (30.0) | 44 (27.5) | .318 |
| Breathlessness | 205 (21.0) | 170 (20.9) | 35 (21.9) | .935 |
| Diarrhoea | 191 (19.6) | 157 (19.3) | 34 (21.3) | 1.000 |
| Sore throat | 177 (18.2) | 156 (19.2) | 21 (13.1) | .100 |
| Cutaneous rash | 82 (8.4) | 63 (7.7) | 19 (11.9) | .200 |
| Headache | 138 (14.2) | 126 (15.5) | 12 (7.5) | .004 |
| Conjunctivitis | 102 (10.5) | 90 (11.1) | 12 (7.5) | .134 |
| Earache | 46 (4.7) | 37 (4.5) | 9 (5.6) | .501 |
| Arthralgia | 63 (6.5) | 57 (7.0) | 6 (3.8) | .184 |
| Myalgia | 39 (4.0) | 36 (4.4) | 3 (1.9) | .148 |
Data are number (%).
Abbreviation: NA, Not available.
FIGURE 1.
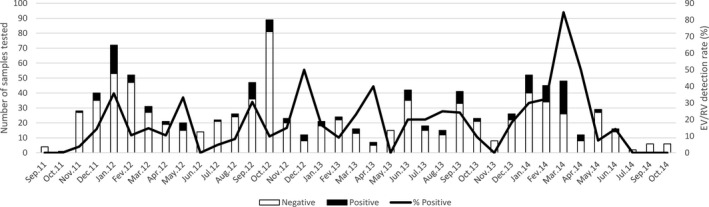
Seasonal distribution of Rhinoviruses and/or Enteroviruses detected in patients with acute respiratory infections during September 2011 till October 2014 in Cameroon. Primary y‐axis and bars describe the number of cases, while secondary y‐axis and lines describe the monthly detection rate
3.3. Genotyping
Overall, 43 RV/EV positive samples were available for molecular characterization in this study. The 5′UTR‐VP4‐VP2 region was amplified and sequenced successfully for 27 (62.8%) samples (22 RV and five EV) out of the 43 tested. The remaining 16 RV/EV samples from which these target viruses could not be sequenced exhibited a relatively high cycles threshold ranging from 31.5 to 37.5 in RV/EV real‐time PCR detection. Of these 16 samples that could not be successfully sequenced, three could not be amplified by conventional RT‐PCR and 13 produced inexploitable electrophoregrams with superimposed peaks.
Rhinovirus‐A, B and C species circulated in Cameroon during the 3 years of the study. This study identified six RV‐A, 10 RV‐B and six RV‐C (Table 2). The six RV types of the species RV‐A comprised A31, A76, A88, A81, along with two unassigned types. One of these unassigned RV‐A types was strikingly related to their closest matches from Tunisia, whereas the other one was featured closest phylogenetic relatedness with isolates from Ivory Coast (Figure 1). The 10 studied RVs of the RV‐B species comprised 3 B35, B42, B70, B83, B86, B93, B97 and one unassigned. The six virus types of the RV‐C species included RV‐C6, C8, C32, C40 and C42. All studied RV isolates segregated into diverse phylogenetic groups defined by their homotypic reference isolates (Figure 2). A nucleotide diversity of 2.9%‐19.1%, 0.3%‐35.0% and 0.0%‐41.0% was featured between the studied sequences and their homotypic reference sequences of the RV‐A, RV‐B and RV‐C species, respectively. Studied sequences and other reference sequences of the RV‐A, RV‐B and RV‐C displayed an amino acid diversity of 0.0%‐10.8%, 0.0%‐17.3% and 0.0%‐21.3%, respectively.
TABLE 2.
Characteristic of RV/EV successfully sequenced samples
| Characteristics | EV (5) | RV‐A (6) | RV‐B (10) | RV‐C (6) | Total |
|---|---|---|---|---|---|
| Types | Coxsackievirus‐B1 (2), Coxsackievirus‐A9 (1), Coxsackievirus‐A5 (1) and EV‐C117 (1) | A31, A76, A88, A81 and 2A non‐typed | 3 B35, B42, B70, B83, B86, B93 and B97 | C6, C8, C32, C40, and C42 | |
| Gender | |||||
| Male | 3 | 6 | 3 | 4 | 16 |
| Female | 2 | 0 | 7 | 2 | 11 |
| Age | |||||
| 0‐2 y | 2 | 4 | 6 | 5 | 17 |
| 2‐5 y | 2 | 1 | 2 | 1 | 6 |
| 5‐15 y | 1 | 1 | 2 | 0 | 4 |
| Patient | |||||
| Inpatients | 0 | 3 | 3 | 4 | 10 |
| Outpatients | 5 | 3 | 7 | 2 | 17 |
| Year | |||||
| 2011 | 0 | 1 | 0 | 0 | 1 |
| 2012 | 3 | 3 | 3 | 3 | 12 |
| 2013 | 0 | 2 | 4 | 2 | 8 |
| 2014 | 2 | 0 | 3 | 1 | 6 |
FIGURE 2.
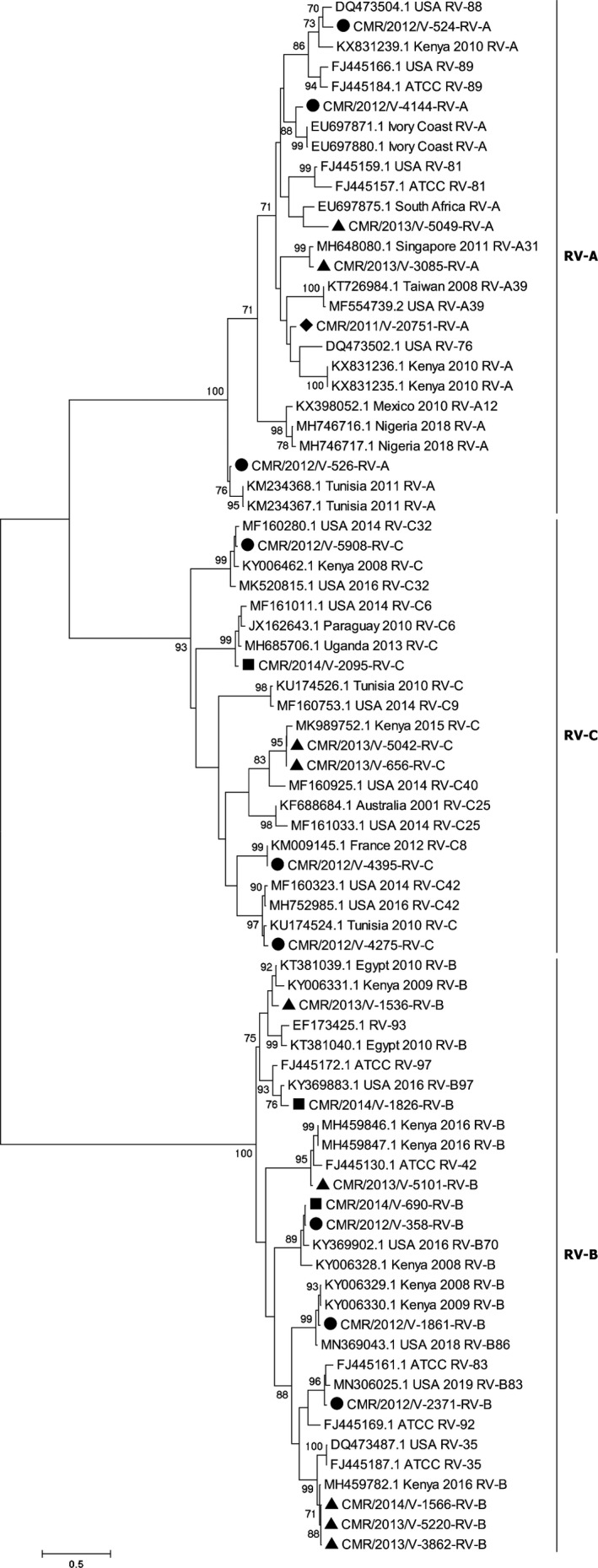
Phylogenetic tree of Rhinoviruses strains detected in Yaoundé, Cameroon, from 2011 to 2014. Multiple sequence alignment was performed with Clustal W. The rooted tree was generated based on VP4/VP2 gene nucleotide sequences using the maximum likelihood method and the best model evolution (General Time Reversible) with a Gamma distribution and assuming that a certain fraction of sites was invariable on an evolutionary level in Mega 6. The scale bars represent the frequency of nucleotide substitutions, and the numbers on the nodes of the branches are determined values for resampling bootstrap after 1000 iterations. Only values ≥70% are presented. The reference sequences of different continents, obtained on GenBank, are identified from the left to the right with the access number, the origin country and the species attributed by the author. The strains of the current study are designated from left to right by CMR (Cameroon), the year of detection and the laboratory number. Cameroon sequences are shown with (♦), (●), (▲) and (■) for the 2011, 2012, 2013 and 2014 sequences, respectively
The five successfully sequenced EV isolates were from outpatients. The phylogenetic tree showed that the five EVs belonged to the Coxsackievirus‐B1 (2), Coxsackievirus‐A9 (1), Coxsackievirus‐A5 (1) and EV‐C117 (1) types (Figure 2). A nucleotide sequence similarity of 1.4%‐65.7% was recorded between the studied and reference EV sequences. As expected, all studied EV isolates segregated into diverse phylogenetic groups defined by their homotypic reference isolates (Figure 3).
FIGURE 3.
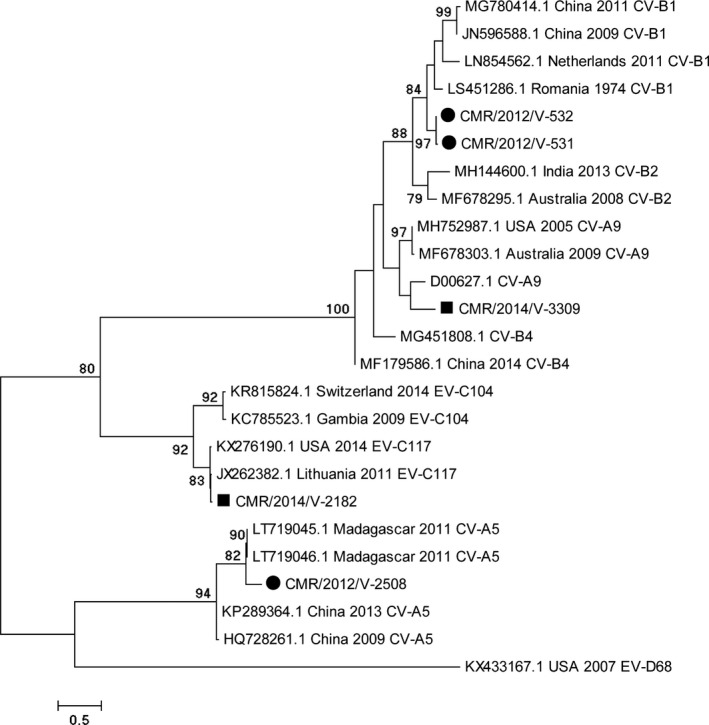
Phylogenetic tree of Enteroviruses strains detected in Yaoundé, Cameroon, from 2011 to 2014. Multiple sequence alignment was performed with Clustal W. The rooted tree was generated based on VP4/VP2 gene nucleotide sequences using the maximum likelihood method and the best model evolution (General Time Reversible) with a Gamma distribution and assuming that a certain fraction of sites was invariable on an evolutionary level in Mega 6. The scale bars represent the frequency of nucleotide substitutions, and the numbers on the nodes of the branches are determined values for resampling bootstrap after 1000 iterations. Only values ≥70% are presented. The reference sequences of different continents, obtained on GenBank, are identified from the left to the right with the access number, the origin country and the species attributed by the author. The strains of the current study are designated from left to right by CMR (Cameroon), the year of detection and the laboratory number. Cameroon sequences are shown with (●) and (■) for the 2012 and 2014 sequences, respectively
4. DISCUSSION
This study reports for the first time in Cameroon the molecular characterization of RV and EV strains detected in hospitalized and ambulatory patients of all ages with acute respiratory infections. A total of 16.4% of patients tested in this study presented with RV and/or EV infections. Data analysis showed a high prevalence of RV/EV in outpatients and a year‐round circulation of RV/EV strains in Cameroon. The strains of RV and EV successfully sequenced suggested a high diversity of RV and EV types circulating in Cameroon.
The prevalence of these two viruses varied with respect to multiple factors including, patient age, clinical presentation, type of sampling, climate, time of year, type of detection assay, species/types/genotypes of the studied viruses, the type of sample tested and many other factors.27, 49, 50 As expected from previous studies, RV and EV detection rates were lower in adults compared to children likely because adult have experienced multiple past exposure to these widespread viruses and are thus less susceptible to infections. Accordingly, Aston et al49 in a study including adults >18 years hospitalized with pneumonia in Malawi reported a detection frequency as low as 3.7% (17/455) for RV and 1.1% (5/455) for EV by PCR using nasopharyngeal swabs. As in this study a high RV/EV detection rate at 25.3% (41/162) was also found for EV in bronchial, throat and/or nasal specimens from children <5 years with acute respiratory infections in Senegal.50 Like in most regions of the world including Africa, RV/EV circulated throughout the year during this study.51, 52 However, EVs in temperate countries show peaks in summer and fall, while in tropical countries peaks are observed in the rainy season.52 RVs also exhibit the same annual circulation pattern but with peaks recorded in autumn and spring.53, 54 The symptoms (fatigue and headaches) found associated with RV/EV positivity in this study are general signs corresponding to several other diseases. This suggests that it is difficult to derive a fair interpretation. However, none of the respiratory infections specific symptoms were associated with RV/EV positivity. This is because all of the patients had acute respiratory infections, and the RV/EV negatives are thus positive for other respiratory pathogens that cause similar signs. Successful sequencing rates of RV and/or EV positive samples ranging from 23.6% to 100% can be obtained in studies.51, 52, 55, 56, 57 Brini Khalifa et al27 in a study including children <1 year with acute respiratory infections in Tunisia reported a RV detection rate in nasopharyngeal swabs as high as 51.8% (267/515) by PCR, thus indicating that your age is a key factor associated to RV and EV infection. This large variability in the RV and/or EV detection and sequencing rates in studies depends on several factors including mainly the detection assay used, viral load, type of samples tested and target genes. Altogether, this and previous studies indicate that the occurrence of RV and EV is more frequent high in children compared to adults.
In this study, only 62.8% (27/43) of the RV and EV detected by real‐time PCR were successfully sequenced. Samples that could not amplify in conventional PCR were those with low viral loads indicated by high cycles thresholds ranging from 31.5 to 37.5. This is consistent with the fact that real‐time RT‐PCR is more sensitive than conventional RT‐PCR.
The pattern of RV/EV circulation throughout the year observed in this study is very common in most studies.51, 52, 57 Regardless of the type of sample considered and the sequencing rate of RV strains detected in most studies in Africa, RV‐A and RV‐C strains are generally predominant compared to RV‐B species.30, 51, 52, 55, 56, 57 However, RV‐B strains were predominant in this study. However, this result should be considered with caution since there is a potential bias associated to the lack of sufficient sample volume for most RV and EV detected and the low viral loads preventing successful sequencing of some samples. The EV types identified in this study have already been described in other studies recruiting patients with acute respiratory infections.39, 58, 59, 60, 61, 62 In particular, the EV‐C117 which was first described in 2012 in a child with pneumonia60 was found identified in a ILI case in this study. Apart from CV‐B1, other EV type identified in ARI patients in this study has not been reported in previous studies among healthy and acute flaccid paralysis patients in Cameroon.63, 64, 65 It is noteworthy that we identify no EV‐D68 isolate despite the epidemiological context of the emergence EV‐68 in patients with acute respiratory infections in 2014.16
The small number of typed RV/EV positive samples and the potential bias in the samples available for molecular typing represent the main limitations of this study. Although apparently unrealistic, systematic typing of RV and EV strains in Cameroon would have likely revealed a more comprehensive genetic landscape of the RV and EV diversity in Cameroon. Nevertheless, the findings of this study suggest that RV and EV are common in Cameroon and they feature a high genetic variability in term of co‐circulating RV and EV types and species. Larger studies are still needed to further explore the molecular epidemiology of RV and EV in ARI patients in Cameroon using extended sample collections and more powerful sequencing approaches such as next generation sequencing.
In conclusion, the findings of the present study suggest an important level of annual circulation of respiratory RV/EV in Cameroon. Molecular characterizations have revealed a large diversity of RV/EV strains.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTION
Sebastien Kenmoe: Data curation (lead); Formal analysis (lead); Investigation (lead); Methodology (lead); Project administration (lead); Validation (lead); Writing‐original draft (lead); Writing‐review & editing (lead). Serge Alain Sadeuh‐Mba: Methodology (supporting); Validation (supporting); Writing‐review & editing (supporting). Marie‐Astrid Vernet: Methodology (lead); Validation (supporting); Writing‐review & editing (supporting). Véronique Penlap Beng: Methodology (supporting); Validation (supporting); Writing‐review & editing (supporting). Astrid Vabret: Methodology (lead); Project administration (lead); Supervision (lead); Validation (supporting); Writing‐review & editing (supporting). Richard Njouom: Conceptualization (lead); Methodology (supporting); Project administration (lead); Supervision (lead); Writing‐review & editing (supporting).
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/irv.12851.
Supporting information
Figure S1
Figure S2
ACKNOWLEDGEMENTS
We thank all laboratory and hospital staff involved data and respiratory samples collection used for this study.
Kenmoe S, Sadeuh‐Mba SA, Vernet M‐A, Penlap Beng V, Vabret A, Njouom R. Molecular epidemiology of Enteroviruses and Rhinoviruses in patients with acute respiratory infections in Yaounde, Cameroon. Influenza Other Respi Viruses. 2021;15:641–650. 10.1111/irv.12851
Funding information
This work was funded by the US Department of Health and Human Services (DHHS) grant number 6 DESP060001‐01‐01 via the International Network of Pasteur Institutes and the “Institut de Microbiologie et de Maladies Infectieuses” (IMMI) in France.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- 1.GBD 2017 Causes of Death Collaborators . Global, regional, and national age‐sex‐specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1736‐1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20:533‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non‐influenza‐related viral respiratory tract infection in the United States. Arch Intern Med. 2003;163:487‐494. [DOI] [PubMed] [Google Scholar]
- 4.Nichol KL, Heilly SD, Ehlinger E. Colds and influenza‐like illnesses in university students: impact on health, academic and work performance, and health care use. Clin Infect Dis. 2005;40:1263‐1270. [DOI] [PubMed] [Google Scholar]
- 5.Lambert SB, Allen KM, Carter RC, Nolan TM. The cost of community‐managed viral respiratory illnesses in a cohort of healthy preschool‐aged children. Respir Res. 2008;9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heikkinen T, Järvinen A. The common cold. Lancet. 2003;361:51‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitkäranta A, Hayden FG. Rhinoviruses: important respiratory pathogens. Ann Med. 1998;30:529‐537. [DOI] [PubMed] [Google Scholar]
- 8.Calvo C, García‐García ML, Blanco C, Pozo F, Flecha IC, Pérez‐Breña P. Role of rhinovirus in hospitalized infants with respiratory tract infections in Spain. Pediatr Infect Dis J. 2007;26:904‐908. [DOI] [PubMed] [Google Scholar]
- 9.Papadopoulos NG, Bates PJ, Bardin PG, et al. Rhinoviruses infect the lower airways. J Infect Dis. 2000;181:1875‐1884. [DOI] [PubMed] [Google Scholar]
- 10.Kenmoe S, Kengne‐Nde C, Ebogo‐Belobo JT, Mbaga DS, Modiyinji AF, Njouom R. Systematic review and meta‐analysis of the prevalence of common respiratory viruses in children < 2 years with bronchiolitis in the pre‐COVID‐19 pandemic era. PLoS One. 2020;15:e0242302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peltola V, Waris M, Kainulainen L, Kero J, Ruuskanen O. Virus shedding after human rhinovirus infection in children, adults and patients with hypogammaglobulinaemia. Clin Microbiol Infect. 2013;19:E322‐E327. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs SE, Lamson DM, St. George K, Walsh TJ. Human Rhinoviruses. Clin Microbiol Rev. 2013;26:135‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tapparel C, Siegrist F, Petty TJ, Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol. 2013;14:282‐293. [DOI] [PubMed] [Google Scholar]
- 14.Messacar K, Asturias EJ, Hixon AM, et al. Enterovirus D68 and acute flaccid myelitis‐evaluating the evidence for causality. Lancet Infect Dis. 2018;18:e239‐e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyda A, Stelzer‐Braid S, Adam D, Chughtai AA, MacIntyre CR. The association between acute flaccid myelitis (AFM) and Enterovirus D68 (EV‐D68) ‐ What is the evidence for causation? Euro Surveill. 2018;23:17‐00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm‐Hansen CC, Midgley SE, Fischer TK. Global emergence of enterovirus D68: A systematic review. Lancet Infect Dis. 2016;16:e64‐e75. [DOI] [PubMed] [Google Scholar]
- 17.Lukashev AN, Vakulenko YA, Turbabina NA, Deviatkin AA, Drexler JF. Molecular epidemiology and phylogenetics of human enteroviruses: Is there a forest behind the trees? Rev Med Virol. 2018;28:e2002. [DOI] [PubMed] [Google Scholar]
- 18.Wisdom A, Leitch ECM, Gaunt E, Harvala H, Simmonds P. Screening respiratory samples for detection of human Rhinoviruses (HRVs) and Enteroviruses: comprehensive VP4‐VP2 typing reveals high incidence and genetic diversity of HRV species C. J Clin Microbiol. 2009;47:3958‐3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piralla A, Baldanti F, Gerna G. Phylogenetic patterns of human respiratory Picornavirus species, including the newly identified group C Rhinoviruses, during a 1‐year surveillance of a hospitalized patient population in Italy. J Clin Microbiol. 2011;49:373‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noor A, Krilov LR. Enterovirus infections. Pediatr Rev. 2016;37:505‐515. [DOI] [PubMed] [Google Scholar]
- 21.Niang MN, Diop NS, Fall A, et al. Respiratory viruses in patients with influenza‐like illness in Senegal: focus on human respiratory adenoviruses. PLoS One. 2017;12:e0174287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolawole O, Oguntoye M, Dam T, Chunara R. Etiology of respiratory tract infections in the community and clinic in Ilorin, Nigeria. BMC Res Notes. 2017;10:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagare A, Maïnassara HB, Issaka B, Sidiki A, Tempia S. Viral and bacterial etiology of severe acute respiratory illness among children < 5 years of age without influenza in Niger. BMC Infect Dis. 2015;15:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellferscee O, Tempia S, Walaza S, et al. Enterovirus genotypes among patients with severe acute respiratory illness, influenza‐like illness and asymptomatic individuals in South Africa, 2012‐2014. J Med Virol. 2017;89:1759‐1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taipale A, Pelkonen T, Roivainen M, et al. Human rhino‐ and enteroviruses in children with respiratory symptoms in Luanda, Angola. Paediatrics and International Child Health. 2014;34:128‐132. [DOI] [PubMed] [Google Scholar]
- 26.Meligy B, Sayed A, Ismail DK, Kamal D, Abdel‐Latif W, Erfan DM. Detection of viral acute lower respiratory tract infection in hospitalized infants using real‐time PCR. Gaz Egypt Paediatr Assoc. 2016;64:13‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brini Khalifa I, Hannachi N, Guerrero A, et al. Demographic and seasonal characteristics of respiratory pathogens in neonates and infants aged 0 to 12 months in the Central‐East region of Tunisia. J Med Virol. 2019;91:570‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann J, Rabezanahary H, Randriamarotia M, et al. Viral and atypical bacterial etiology of acute respiratory infections in children under 5 years old living in a rural tropical area of Madagascar. PLoS One. 2012;7:e43666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho A, Aston SJ, Jary H, et al. Impact of HIV on the burden and severity of influenza illness in Malawian adults: a prospective cohort and parallel case‐control study. Clin Infect Dis. 2017;66:865‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Annamalay A, Lanaspa M, Khoo S, et al. Rhinovirus species and clinical features in children hospitalised with pneumonia from Mozambique. Tropical Med Int Health. 2016;21:1171‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njouom R, Yekwa EL, Cappy P, Vabret A, Boisier P, Rousset D. Viral etiology of influenza‐like illnesses in Cameroon, January–December 2009. J Infect Dis. 2012;206:S29‐S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lekana‐Douki SE, Nkoghe D, Drosten C, Ngoungou EB, Drexler JF, Leroy EM. Viral etiology and seasonality of influenza‐like illness in Gabon, March 2010 to June 2011. BMC Infect Dis. 2014;14:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobossi Serengbe G, Gody J‐C, Fioboy R, Nakoune E. [Viral etiology of acute childhood respiratory infections in Bangui, Central African Republic]. Arch Pediatr. 2015;22:324‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fall A, Jallow MM, Kebe O, et al. Low circulation of subclade A1 Enterovirus D68 strains in senegal during 2014 North America outbreak. Emerg Infect Dis. 2019;25:1404‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fall A, Ndiaye N, Jallow MM, et al. Enterovirus D68 subclade B3 circulation in Senegal, 2016: detection from influenza‐like illness and acute flaccid paralysis surveillance. Sci Rep. 2019;9:13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellferscee O, Treurnicht FK, Tempia S, et al. Enterovirus D68 and other enterovirus serotypes identified in South African patients with severe acute respiratory illness, 2009‐2011. Influenza Other Respir Viruses. 2017;11:211‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubi JA, Mutocheluh M, Bonney JHK, Ampofo WK, Odoom JK. Molecular detection of enterovirus D68 among children with acute respiratory tract infection in Ghana. Afr J Lab Med. 2019;8:732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.L'Huillier AG, Kaiser L, Petty TJ, et al. Molecular epidemiology of human Rhinoviruses and Enteroviruses highlights their diversity in Sub‐Saharan Africa. Viruses. 2015;7:6412‐6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opanda SM, Wamunyokoli F, Khamadi S, Coldren R, Bulimo WD. Genotyping of enteroviruses isolated in Kenya from pediatric patients using partial VP1 region. Springerplus. 2016;5:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokarz R, Firth C, Madhi SA, et al. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;93:1952‐1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenmoe S, Tchendjou P, Vernet M, et al. Viral etiology of severe acute respiratory infections in hospitalized children in Cameroon, 2011–2013. Influenza Other Respir Viruses. 2016;10:386‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenmoe S, Vernet M‐A, Le Goff J, Penlap VB, Vabret A, Njouom R. Molecular characterization of human adenovirus associated with acute respiratory infections in Cameroon from 2011 to 2014. Virol J. 2018;15:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kenmoe S, Vernet M‐A, Miszczak F, et al. Genetic diversity of human respiratory syncytial virus isolated among children with acute respiratory infections in Southern Cameroon during three consecutive epidemic seasons, 2011–2013. Trop Med Health. 2018;46:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenmoe S, Vernet M‐A, Njankouo‐Ripa M, Penlap VB, Vabret A, Njouom R. Phylogenic analysis of human bocavirus detected in children with acute respiratory infection in Yaounde, Cameroon. BMC Res Notes. 2017;10:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenmoe S, Vernet M‐A, Penlap Beng V, Vabret A, Njouom R. Phylogenetic variability of Human Metapneumovirus in patients with acute respiratory infections in Cameroon, 2011–2014. J Infect Public Health. 2019;13:606‐612. [DOI] [PubMed] [Google Scholar]
- 46.Linsuwanon P, Payungporn S, Samransamruajkit R, et al. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J Infect. 2009;59:115‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673‐4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725‐2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aston SJ, Ho A, Jary H, et al. Etiology and risk factors for mortality in an adult community‐acquired pneumonia cohort in Malawi. Am J Respir Crit Care Med. 2019;200:359‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Assane D, Makhtar C, Abdoulaye D, et al. Viral and bacterial etiologies of acute respiratory infections among children under 5 years in Senegal. Microbiol Insights. 2018;11:1178636118758651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esposito S, Daleno C, Baggi E, et al. Circulation of different rhinovirus groups among children with lower respiratory tract infection in Kiremba, Burundi. Eur J Clin Microbiol Infect Dis. 2012;31:3251‐3256. [DOI] [PubMed] [Google Scholar]
- 52.Fall A, Dia N, Kébé O, et al. Enteroviruses and Rhinoviruses: molecular epidemiology of the most influenza‐like illness associated viruses in Senegal. Am J Trop Med Hyg. 2016;95:339‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monto AS. Epidemiology of viral respiratory infections. Am J Med. 2002;112:4‐12. [DOI] [PubMed] [Google Scholar]
- 54.Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT‐PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006;78:644‐650. [DOI] [PubMed] [Google Scholar]
- 55.Annamalay AA, Abbott S, Sikazwe C, et al. Respiratory viruses in young South African children with acute lower respiratory infections and interactions with HIV. J Clin Virol. 2016;81:58‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milanoi S, Ongus JR, Gachara G, Coldren R, Bulimo W. Serotype and genetic diversity of human rhinovirus strains that circulated in Kenya in 2008. Influenza Other Respir Viruses. 2016;10:185‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Onyango CO, Welch SR, Munywoki PK, et al. Molecular epidemiology of human rhinovirus infections in Kilifi, Coastal Kenya. J Med Virol. 2012;84:823‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puenpa J, Mauleekoonphairoj J, Linsuwanon P, et al. Prevalence and characterization of Enterovirus infections among pediatric patients with hand foot mouth disease, herpangina and influenza like illness in Thailand, 2012. PLoS One. 2014;9:e98888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdelkhalek I, Seghier M, Yahia AB, et al. Molecular epidemiology of coxsackievirus type B1. Arch Virol. 2015;160:2815‐2821. [DOI] [PubMed] [Google Scholar]
- 60.Daleno C, Piralla A, Scala A, et al. Complete genome sequence of a novel human Enterovirus C (HEV‐C117) identified in a child with community‐acquired pneumonia. J Virol. 2012;86:10888‐10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du J, Zhu T, Zhuang L, et al. Identification and complete genome characterization of human enterovirus 117 from a child with pneumonia in China. Arch Virol. 2019;164:1489‐1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang Z, Tsatsral S, Liu C, et al. Respiratory infection with enterovirus genotype C117, China and Mongolia. Emerg Infect Dis. 2014;20:1075‐1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ayukekbong JA, Fobisong C, Lindh M, Nkuo‐Akenji T, Bergström T, Norder H. Molecular analysis of enterovirus in Cameroon by partial 5′UTR‐VP4 gene sequencing reveals a high genetic diversity and frequency of infections. J Med Virol. 2014;86:2092‐2101. [DOI] [PubMed] [Google Scholar]
- 64.Ayukekbong J, Kabayiza J‐C, Lindh M, et al. Shift of Enterovirus species among children in Cameroon–identification of a new enterovirus, EV‐A119. J Clin Virol. 2013;58:227‐232. [DOI] [PubMed] [Google Scholar]
- 65.Sadeuh‐Mba SA, Bessaud M, Massenet D, et al. High frequency and diversity of species C Enteroviruses in cameroon and neighboring countries. J Clin Microbiol. 2013;51:759‐770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


