Abstract

Epigenetics is nowadays a well-accepted area of research. In the last years, tremendous progress was made regarding molecules targeting EZH2, directly or indirectly. Recently tazemetostat hit the market after FDA-approval for the treatment of lymphoma. However, the impairment of EZH2 activity by orthosteric intervention has proven to be effective only in a limited subset of cancers. Considering the multiproteic nature of the PRC2 complex and the marked dependence of EZH2 functions on the other core subunits such as EED, in recent years, a new targeting approach ascended to prominence. The possibility to cripple the function of the PRC2 complex by interfering with its multimeric integrity fueled the interest in developing EZH2–EED protein–protein interaction and EED inhibitors as indirect modulators of PRC2-dependent methyltransferase activity. In this Perspective, we aim to summarize the latest findings regarding the development and the biological activity of these emerging classes of PRC2 modulators from a medicinal chemist’s viewpoint.
1. Introduction
The polycomb repressive complex 2 (PRC2) is part of the epigenetic machinery and belongs to the polycomb-group (PcG) of proteins that play a central role in how distinct expression patterns are positioned, maintained, and inherited in specialized cells of multicellular organisms.1 Due to its S-adenosyl-l-methionine (SAM)-dependent methyltransferase activity, the PRC2 was characterized as an epigenetic writer targeting the lysine 27 of histone H3 (H3K27) tail.2 However, the increasing understanding of how this post-translational modification (PTM) is directed or impeded on specific genomic sites by some of the PRC2 components has shed light on its additional reading functions.3,4 As a post-translational modifier, the PRC2 is able to catalyze with different propensity the mono- (me1), di- (me2), and trimethylation (me3) of H3K27;5,6 nonetheless, it can also target lysines from noncanonical substrates including JARID2, STAT3, and RARa.7−9 The H3K27 methylated states encode distinct epigenetic meanings, and whereas the H3K27me1 is ordinarily found in actively transcribed loci, the H3K27me3 mark is associated with a heterochromatin transition and silencing of developmentally important genes.10 In this regard, intriguing findings spotlighted that PRC2 not only installs de novo H3K27me3 tags but rather reinforces the established repressive state by fueling the so-called “spreading” of the H3K27me3 signal on already marked nucleosomes.11,12
Four interactors compose the human PRC2 complex: EZH2 or its homologue EZH1, which endows the complex with the histone methyltransferase (HMTase) capability, and three regulatory subunits (SUZ12, EED, and RbAp46/48).5,6,13 These noncatalytic members confer structural stability to the assembly and enhance its enzymatic activity and target recognition.6,13−15 However, the complex may also include facultative subunits, such as AEBP2, JARID2, the PCL1–3 proteins, PALI, and EPOP, which grant further regulating functions.16−20
EZH2 endows the PRC2 with catalytic competence through its structurally conserved C-terminal SET [Su(var)3–9, enhancer-of-zeste, and trithorax] domain.21 Although the SET domain is catalytically self-sufficient in other HMTase proteins, in the PRC2, it is unable to accomplish the methyl transfer by itself.14,22,23 When EZH2 is devoid of the core subunits EED and SUZ12, it is biologically unstable, and its SET domain switches to an inactive catalytic state that cripples the PRC2 methyltransferase capability and functions.24,25
EED carries out a further regulatory role by sensing the methylation status of already H3K27me3-tagged histones, exerting a positive allosteric control on EZH2 catalysis. Upon recognition of the H3K27me3 trimethylammonium motif in a shallow pocket of its β-propeller structure, EED prompts a structural reorganization of the SET domain, enabling a basal to stimulated catalysis shift and efficient H3K27me3 deposition by PRC2.3,26 Other trimethylated fragments were found to interact with EED in vitro, such as the stimulating JARID2-K116me3 (JARID2me3) and histone tail-derived peptides; albeit the latter do not establish any allosteric circuits.7,26,27 Furthermore, Muir and co-workers recently discovered that unmethylated H3K36 can increase the EZH2 activity through an ancillary sensing pocket, hence broadening the extent of this kind of feedback loop.4 Therefore, EZH2 may pass through distinct catalytic stages according to the PRC2 architecture and the chromatin context: an autoinhibited configuration when it lacks the EED and SUZ12 pair,23 a basal active state within the PRC2 ternary assembly,6 and an H3K27me3-dependent stimulated state.26,27
Dysregulation of the PRC2–H3K27me3 axis is linked to different diseases, including several cancers,28,29 viral infections,30,31 the Weaver syndrome,32,33 and inflammation processes.34 In cancer, EZH2 is generally considered oncogenic and, together with the other core subunits, was found to be overexpressed and correlated with poor prognosis in a multitude of solid tumors.35−37 Genome sequence analysis also revealed that gain-of-function mutations of EZH2 (Y641, A677,38 and A687) are abundant among varied forms of lymphomas.38−42 PRC2 components may also act as tumor suppressors and undergo loss-of-function or missense mutations in T-cell acute lymphoblastic leukemia and myelodysplastic syndromes.36,43,44
Taken together, these pieces of evidence endorsed the PRC2 as a top-ranked target for cancer treatment, and in this respect, extensive med-chem campaigns were launched to develop small molecular entities tackling its functions. As catalytic subunit, EZH2 was considered the ideal target to shut down abnormal PRC2 activities directly, and several SAM-competitive inhibitors, sharing a pharmacophoric 2-pyridone scaffold, have been developed with the time. Such inhibitors demonstrated HMTase-dependent activities both in vitro and in vivo, and some of them (CPI-1205, SHR2554, DS-3201b, CPI-0209) are presently undergoing phase I/II clinical investigations for solid or hematological tumors, as deeply discussed in recent reviews.45−47 Notably, 2020 saw the FDA-approval of the first SAM-competitive anti-EZH2 drug, EPZ-6438 or tazemetostat (Tazverik), which has been authorized for the care of locally advanced epithelioid sarcoma and follicular lymphoma.48−50 An overview of the EZH2 inhibitors currently in clinical trials is reported in Figure 1.
Figure 1.
Catalytic EZH2 inhibitors under phase I/II clinical studies.
More and more EZH2 inhibitor drugs are awaited in the near future; however, such agents are not free of drawbacks: they have proven efficacy in a limited subset of cancers, mainly in Y641 or A677 mutant lymphoma cells, and their prolonged administration aroused different adaptive cancerous response mechanisms, such as activation of the insulin-like growth factor 1 receptor (IGF-1R), MEK, and phosphoinositide-3-kinase (PI3K) pathways, that ultimately restricted the overall clinical outcome.51
Considering the multiproteic nature of the PRC2 complex and the strict dependence of EZH2 activity on the scaffolding and regulating roles of EED, in recent years, alternative inactivating strategies gained attention. On one side, the possibility to cripple the PRC2 complex functions by interfering with the intimate protein–protein interaction (PPI) between EZH2 and EED fueled the development of different chemotypes as EZH2–EED protein–protein interaction (PPI) inhibitors. These chemical agents exert a methyltransferase inhibitory activity on PRC2 by impeding the scaffolding role of EED on EZH2-SET domain correct folding, as discussed in the following related paragraph. On the other hand, as anticipated in a seminal viewpoint,52 a high throughput screening by Novartis demonstrated the “druggability” of the H3K27me3-recognizing cavity of EED as a means to allosterically inhibit EZH2 catalysis. The ensuing hit-optimization process has led to EED binders with improved physicochemical and biological properties in vivo, ultimately providing a clinical candidate (MAK683).53,54
In this Perspective, we aim to present these emerging classes of PRC2 modulators and to summarize the latest findings regarding their development and biological activity from a medicinal chemist’s viewpoint.
2. The PRC2 Core Complex Architecture and Its Transition between Catalytic States
EZH2, EED, and SUZ12 subunits interact through extended molecular surfaces, and different X-ray cocrystal structures appear in the literature showing the molecular contacts engaged in the core assembly, the target recognition, the nucleosomal recruitment, and the catalytic pocket organization.55−60 The human PRC2 core complex (EED, EZH2, and SUZ12-VEFS domain) was cocrystallized with the histone-derived oncogenic H3K27M21–33 false-substrate (H3K27M) lying in the substrate-binding furrow and the JARID2110–122-K116me3 peptide (JARID2me3) bound in the EED central pore (Figure 2A,B).59 This assembly has a three-fold structure and is functionally divided into a regulatory portion (corresponding to EED and the N-terminal segments of EZH2), a catalytic region (encompassing the C-terminal domains of EZH2 (CXC and SET)), and a middle section (bridging the previous ones and including the C-terminal α-helical bundle of SUZ12 (SUZ12-VEFS) and the EZH2-SANT2/MCSS domains).
Figure 2.
Overall architecture of the human PRC2 core complex and stabilization of its basal catalytic state. (A) Schematic representation of the PRC2 complex subunits. The colors of the EZH2 domains are in accordance with their representation in panels B and C. H3K27M and JARID2me3 peptide sequences are in one-letter code. Mutant methionine is underscored. (B) Structure of the catalytically active PRC2 core complex in cartoon and semitransparent surface representations (PDB 5HYN). PRC2 subunits and EZH2 domains are labeled and highlighted in varied colors. H3K27M and JARID2me3 peptides are in blue and yellow cartoon, respectively. SAH is in stick representation. (C) Rotated view (Y = 50°) of the complex with the EZH2 domains, forming the belt-like structure, labeled and highlighted in colors. (D) Superimposition of the human EZH2 SET domain in isolated inactive (PDB 4MI5, orange) and catalytically active conformation (PDB 5HYN, green). (E) Superimposition of the post-SET subdomain from the inactive (orange) and active conformation (green), showing its outward torsion that clears the H3K27M (blue) binding cleft.
In this assembly, the regulatory-adaptor protein EED occupies a central location and arranges multiple contacts with the different EZH2 segments. EED is a WD40-repeat-containing protein distinguished by a seven-bladed β-propeller architecture with one blade formed by a four-stranded antiparallel β-sheet (WD40 motif).61 The radial arrangement of the WD40 motif outlines two different-sized surfaces (top and bottom), which serve as a scaffold for the belt-like arrangement of the EZH2 N-terminal region (residues 10–246) on EED (Figure 2C). On one side, an extended α-helical stretch of EZH2 (EBD or EED binding domain) sits within a sharp gorge defined by the loops connecting the β-propeller blades and spans the length of the EED bottom surface.59−61 On the other side, EZH2 assumes a more unstructured conformation and interacts with EED through two flexible segments, SAL (SET activation loop) and SRM (stimulation-responsive motif). This “embracing” configuration is additionally secured by contacts given by the SBD (SANT1-binding domain) that intramolecularly docks into the distal SANT1 α-helix bundle and acts as a molecular clasp. EZH2 proceeds as a string of mainly α-helical domains (SANT1, MCSS, SANT2) with further stabilization roles and cementing SUZ12-VEFS domain to the core structure.59,60 Finally, the C-terminal CXC (or pre-SET) and SET domains compose the catalytic region of EZH2 and locate in close proximity to the SAL and SRM domains. Given the relevance as catalytic portion and the differential conformational plasticity, SET is additionally split into four subdomains, namely, SET-C, SET-I, SET-N, and post-SET, which delineate the substrate and cofactor binding sites.62
The described EED/SUZ12-dependent organization is essential to ensure the catalytic ability of the otherwise autoinhibited EZH2 subunit. Crystallographic alignments of the EZH2, alone or complexed with the core members, revealed consistent structural rearrangements at both the substrate and the cofactor binding sites, which become misfolded or self-seized by distinct SET portions.22,59,60,63 Indeed, in isolated EZH2, the SET-I and the post-SET subdomains undergo body rotations that bring them to fit within the histone binding cleft (orange structures in Figure 2D,E). This inactivating configuration demonstrates the critical role of EED as an adaptor protein since the organization around its β-propeller positions the SAL segment to stretch out at the border between EED and the buried surfaces of SET-I and SET-N and re-establishes a functional SET domain. As a result of these interactions, the SET-I α-helix rotates in a counterclockwise manner around two conserved “hinge” Gly residues (Gly643 and Gly681) and returns to its active orientation (green structure in Figure 2D).63 The substrate binding site is further released by an outward torsion of the post-SET region around its loop structure (residues 726–729) that simultaneously clears the lower part of the histone-recognizing channel and shapes the lid of the cofactor binding pocket (green structure in Figure 2E).23,63 These conformational changes together with the recognition of SAL-like structures in other SET-containing methyltransferases affirm the SAL domain as a key effector in mediating the SET transition toward an active catalytic state.63
Akin to the bottom side, the top surface of EED supplies additional sites of interaction with EZH2, and through the recognition of a quaternary ammonium moiety in its central cavity, it modulates the SET catalytic function in a context-specific manner. In this respect, Margueron et al. first reported a cocrystal structure of isolated EED with a H3K27me3 peptide embedded within the top gorge of the β-propeller, thus providing structural insights on its epigenetic “reading” function.26 This recognition is not limited to the H3K27me3 mark, and other trimethyllysine-containing ligands were discovered to bind to EED (Figure 3A).7,26,27 Nonetheless, not all of these histone-derived binders prompted an allosteric stimulation in vitro, and only H3K27me3 and JARID2me3 proved to increase, up to 3-fold, the nucleosomal methyltransferase activity of PRC2.7,27 Despite these differences, the EED recognition of trimethylated ligands relies on common binding features, revealed here by overlaying the poses of H3K27me3 and JARID2me3 within the shallow pocket of the β-propeller structure (Figure 3B).7,26 The ligands dock by their trimethylammonium moieties, which interact with a triad of aromatic amino acids (Phe97, Tyr148, and Tyr365) whose almost perpendicular arrangement defines the so-called “aromatic cage”. Besides these π–cation juxtapositions, the trimethyllysine side chains assume an extended conformation and position van der Waals interactions with the nearby Trp364 indole ring, which also interacts by polar contacts with the peptide backbones. Finally, the H3K27me3 and JARID2me3 are further stabilized on the EED surface by hydrogen bonds occurring between the rim of the cavity (Arg414) and the main-chain carbonyl groups flanking the Lys27me3 and Lys116me3. Since the aromatic cage organization is a structural trait common to other epigenetic “readers” (i.e., chromodomain proteins), the EED selectivity toward H3K27me3 and JARID2me3 can be explained considering two additional shallow hydrophobic ditches surrounding the central gorge of EED (highlighted in orange surface) where they orient van der Waals interactions through small aliphatic amino acids at i – 2 and i + 2 positions relative to the trimethylated lysine.12,64−66 Indeed, both of them contain an Ala residue at i + 2 position, whereas at the i – 2, the H3K27me3 peptide presents an additional Ala and JARID2 contains a Gln, which can recline to orient its aliphatic side chain to the local hydrophobic surface of EED (Figure 3B).7
Figure 3.
Allosteric binding site and activation of the PRC2 core complex. (A) JARID2me3 and H3K27me3 peptides binding to the EED central cavity. EED is represented in gray surface, and JARID2me3 (PDB 5HYN) and H3K27me3 (PDB 3IIW) are in yellow and magenta cartoons, respectively. (B) Enlarged view of H3K27me3 peptide superimposed to JARID2me3 cocrystallized with EED (PDB 5HYN). Peptide side chains are presented as sticks; EED is in semitransparent surface and cartoon. Amino acids delimiting the trimethyllysine aromatic cage are represented as sticks and labeled. Molecular hollows hosting the i + 2 and i – 2 amino acids from trimethylated lysine are highlighted in orange surface. Hydrogen bonds are in dashed black lines. (C) Allosteric stimulation of the SET catalytic function. EED is in gray surface, JARID2me3 peptide (yellow), SAL domain (magenta), SRM (violet), SET (green), and H3K27M peptide (blue) are in cartoon representation with key residues labeled and in sticks. Hydrogen bonds are shown in dashed black lines.
The mechanism of the allosteric activation of EZH2 catalysis takes advantage of the role of EED as mentioned earlier and has been elucidated at an atomic level in Chaetomium thermophilum and human trimeric PRC2, in complex with H3K27me3 and JARID2-K116me3 stimulating peptides, respectively.59,60 Both of them merged to a model centered on the role of the EZH2-SRM domain in communicating the allosteric inputs from the EED top surface to the SET catalytic region. In this dynamic, the EZH2 arrangement around EED brings the disordered SRM to a location near the β-propeller cavity, where it senses the presence of stimulating signals. In a PRC2 ternary complex at the basal state, the SRM domain is disordered and unresolved in the electron density map.60 However, when H3K27me3 or JARID2-K116me3 bind to EED, a set of side-chain contacts between these allosteric modulators (we show here JARID2-K116me3) and the SRM itself determine its conformational stabilization in an α-helical structure. This interaction is dominated by Arg115(JARID2) (or Arg26 in H3K27me3 peptide), which is an invariant feature of repressive trimethyllysine histone marks and establishes polar contacts both with the SRM helix (Asp140) and intramolecularly with Gln112 and Gln119 (Figure 3C). This ordered disposition causes the juxtaposing surfaces of the SRM and SET-I α-helices to tightly pack to each other and to converge to an overall sandwich-like arrangement, which is supposed to be responsible for the increased catalytic activity of the SET domain.
3. Inhibitors of EZH2–EED Protein–Protein Interaction
Considering the multiproteic nature of the PRC2 complex and the marked dependence of EZH2 functions on the other core subunits, in recent years, a new targeting approach ascended to prominence. The possibility to cripple the function of the PRC2 complex by interfering with its multimeric integrity fueled interest in developing protein–protein interaction (PPI) inhibitors as indirect anti-EZH2 agents. In this respect, a seminal study reported in 2013 the development of an all-hydrocarbon stapled peptide recapitulating the α-helical EED-binding domain (EBD) of EZH2 and disclosed the feasibility of hampering the interaction between EED and EZH2 to disable the PRC2 biological functions.67
The stabilized α-helix of EZH2 (SAH-EZH2) selectively inhibits H3K27 trimethylation by dose-responsively disrupting the EZH2–EED complex, evaluated through fluorescence polarization (FP) assay, and reducing EZH2 protein levels. Notably, the reduction of EZH2 protein level was not observed upon treatment with GSK126. The impairment of the EZH2–EED interaction led to proliferation arrest and myeloid differentiation in PRC2-dependent MLL-AF9 leukemia cells and remarkably affected the viability of cancer cells bearing EZH2 mutant variants. Given the two distinct mechanisms of action between a catalytic EZH2 inhibitor (GSK126) and the SAH-EZH2 peptide, the authors investigated whether the coadministration of the two compounds would enhance the antiproliferative effects. Indeed, upon the cotreatment of MLL-AF9 and KARPAS422 cells with SAH-EZH2 and GSK126, an apparent synergistic impairment of cell viability was observed.67 Although peptide-based chemotypes are often characterized by a poor pharmacokinetic (PK) profile, Orkin and co-workers demonstrated the therapeutic potential of targeting the binding interfaces between the PRC2 partners as a novel strategy to modulate their biological functions and paved the way for the development of small molecule PPI inhibitors.67
In a following study published in 2014, the crystal structure of EED in complex with an EZH2-EBD-resembling peptide (Figure 5A, PDB 2QXV) was used for docking-based virtual screening of an in-house database containing approximately 1000 known drugs by Luo’s research group.68
Figure 5.
X-ray cocrystal structures of EZH2 EBD domain and astemizole in complex with EED and structural optimization of astemizole-derived compounds (3–5). (A) X-ray crystal structure (PDB 2QXV) of EBD peptide in cartoon (teal) with EED in surface (white). The dashed square represents the EBD-binding site portion where astemizole is accommodated. (B) Binding mode of astemizole, in stick (magenta) along the bottom surface of EED, in cartoon (blue-white). The amino acids important for the binding are represented as sticks and labeled. (C) Structural optimization of astemizole-like compounds (3–5) and leading to DC-PRC2in-01 (5b).
Among them, astemizole (1, Figure 4), a second-generation FDA-approved (but later withdrawn from the market) antiallergy drug with histamine H1 receptor antagonist activity,69 showed the highest EZH2–EED interaction inhibitory activity, displacing the EZH2 peptide with a Ki of 23.01 μM in a competitive fluorescence polarization assay. To validate whether 1 directly bound to EED and competed with EZH2, additional methods, including fluorescence-based thermal shift and saturation transfer difference (STD) nuclear magnetic resonance (NMR) spectra, were used, and the results corroborated the first experiments. Considering the importance of the EZH2–EED interaction for the integrity of the PRC2 complex, 1 was also evaluated for assessing its capability to reduce PRC2 protein levels upon dissociation of the EZH2–EED complex. As expected, 1 treatment dose-dependently decreased the protein levels of EZH2, EED, and SUZ12 in both the DB (EZH2 mutant) and Toledo (wild-type EZH2) cell lines, which was in agreement with the reported effects of mutations able to disrupt these protein–protein interactions.24,70 Furthermore, low micromolar concentrations of 1 markedly impaired the proliferation of GCB-DLBCL cells carrying mutant or wild-type forms of EZH2. Of note, a synergistic effect was observed in both DB and Toledo cells upon the coadministration of 1 and the catalytic EZH2 inhibitor EPZ005687, corroborating the results observed for the stapled EZH2 peptide in the study above-mentioned.67
Figure 4.

Structures of astemizole (1) and wedelolactone (2).
In 2015, a different work described the discovery of the natural compound wedelolactone (2, Figure 4) being able to bind to EED with a micromolar affinity (KD = 2.82 μM), to block the EZH2–EED interaction in vitro, to induce the degradation of both EZH2 and EED proteins at 50 μM in HepG2, THP1, and K562 cells, and also to modulate the expression of detected PRC2 downstream targets and cancer-related genes.71
Very recently, Luo’s group published the cocrystal structure of EED in complex with 1 at 2.15 Å (residues 40–441) (PDB 7KXT).72 The structure (Figure 5B) elucidates the detailed binding mode of 1 to EED and provides insights for a structure-guided drug design that led to a novel EZH2–EED interaction inhibitor, DC-PRC2in-01 (5b). The compound 5b, bearing a pyrrolidine moiety instead of the piperidine as in 1, displayed a KD value versus EED protein of 5 μM, measured by surface plasmon resonance (SPR), and an IC50 of 4 μM, determined by fluorescence polarization competitive binding assay. As shown in Figure 5B, EED folds into a typical seven-bladed β-propeller structure with 1 binding to the bottom of the WD40-repeat domain, where EED recognizes the EBD domain of EZH2 (residues 32–70) (Figure 2B,C, see above),61 and differently from the EED binders that dock in the H3K27me3 binding pocket at the top of WD40-repeat domain of EED.
The binding site accommodating 1 extends along the EED cleft that harbors the N-terminal portion of the EZH2-EBD domain (dashed square in Figure 5A) and involves a series of mostly hydrophobic residues. Compound 1 binds to this site with the fluorobenzene group inserting deeply into a hydrophobic pocket formed by a cluster of 6 residues (Leu315, Cys330, Leu391, Val393, Pro396, and His397) (Figure 5B). Compared with the structure of the EZH2-EBD peptide bound to EED, this fluorobenzene group occupies the space of the hot-spot residue Phe42 of EZH2 (Figure 5A).61 Moreover, the benzimidazole moiety establishes interactions with hydrophobic residues Leu318, Met336, and Leu353, whereas the 4-methoxyphenethyl moiety engages in hydrophobic interactions with Leu246, Phe372, Trp373, and Gln374.72
The compound 1 scaffold was optimized according to two lines of intervention: (1) the N-benzyl and the terminal phenethyl moiety were modified with functional groups aiming to reinforce the hydrophobic interactions or to establish novel polar contacts within the connectivity of the original structure (3 and 4); (2) the distance between the nitrogen atoms of the 4-aminopiperidine linker was optimized through cyclic homology and ring-opening study of the piperidine moiety (5) (Figure 5C).
Among the N-benzyl modifications, only the para- and the meta,para-disubstitution with electron-withdrawing groups (EWGs) proved to be tolerated (3a–d), yielding a weak increase of potency (IC50 = 57–66 μM) with respect to 1 (IC50 = 74 μM), while on the phenethyl terminal only the meta-hydroxyl modification (4c) provided a similar improvement (IC50 = 62 μM). Manipulation of the 4-aminopiperidine moiety provided a significant improvement in the potency and highlighted the importance of maintaining a proper length and geometry in this portion (5a–d). In this regard, the piperidine ring replacement with pyrrolidine afforded compound 5b (DC-PRC2in-01), showing a 17-fold increase of potency (IC50 = 4 μM). This result was rationalized by molecular docking studies that hypothesized the formation of H-bonds between the 3-aminopyrrolidine nitrogen atoms and the surface of EED. Furthermore, while the cyclic homologation to azetidine (5a) still provided a more potent compound (IC50 = 22 μM) than 1, the piperidine ring-opening led to inactive compound (5c) whereas the pyrrolidine ring-opening (5d), bearing an ethylene spacer, was tolerated.
At a protein level, 5b was able to destabilize the PRC2 complex, thus leading to the dose-dependent depletion of mainly EED along with EZH2 and SUZ12 proteins up to 10 μM. Notably, 5b dose-dependently and specifically reduced the levels of H3K27me3 with respect to other H3 methyl marks, thus demonstrating the efficacy of the EZH2–EED interaction inhibitors in evoking epigenetic effect.72 Compound 5b dose-dependently impaired cell viability of Pfeiffer, KARPAS422, SU-DHL-4, and DB cell lines, harboring EZH2 heterozygous mutations, with IC50 values below 10 μM, by blocking the cell cycle in G0/G1 phase.
This study paves the way to (a) a new in-depth astemizole SAR investigation for increasing its potency and (b) the design of novel molecular models for disrupting the EZH2–EED interaction.
4. First Steps toward EED Binders
The evidence that trimethylated JARID2 protein binds to EED and allosterically stimulates the PRC2 methyltransferase activity prompted the initial attempts to address the EED top cavity for targeting the PRC2 functions.7 In this regard, Barnash et al. exploited a previously developed combinatorial peptide screening platform to readily generate a set of peptidomimetic EED binders inspired by JARID2-K116me3, eventually steering toward a 4-mer micromolar PRC2-allosteric inhibitor, UNC5114 (Figure 6A).73,74
Figure 6.
Chemical structure of JARID2-K116me3-inspired peptidomimetics and binding pose comparison between JARID-K116me3 and compound 8. (A) Evolution of SAR studies on 5-mer JARID2-K116me3 structure leading to 9 (UNC5114) and 10 (UNC5115). (B) Superimposition between structures of 13-mer JARID2-K116me3 (teal, PDB 4X3E) and 8 (violet, PDB 5TTW) in stick representation and complexed with EED (in gray surface and cartoon representations). EED interacting amino acids are labeled and represented as sticks. Polar contacts are represented as dashed black lines. Surfaces harboring chemical entities in i – 2 and i + 2 positions are highlighted in light orange.
The 13-mer JARID2-K116me3 peptide represented an advanced starting point in the optimization process since it exhibits a 10-fold higher affinity for EED compared with H3K27me3 in vitro (KD = 3 versus 40 μM, respectively) and it primarily interacts through five amino acids (QRKme3FA, 6) centered on the K116me3 with Gln114 and Ala118 providing auxiliary interactions (Figure 6A,B).7 With the aim to move toward a more druglike scaffold and revert adverse physicochemical properties, such as an overall +3 charge and the presence of disadvantageous polar functions, peptide 6 underwent two different rounds of combinatorial modifications. First, the i – 2 position was varied with apolar substituents able at the same time to mimic the Gln114 and to cap the cationic N-terminus. These chemical manipulations led to the identification of compound 7 that in isothermal titration calorimetry (ITC) measurements showed a 2-fold improvement (KD = 4.74 μM) with respect to 6. Second, a more robust round of optimization encompassed a larger portfolio of replacements and included the i + 2 position as variable element. A combination of the beneficial modifications into a common structure resulted in 8 exhibiting a further enhancement in potency (KD = 1.09 μM) and decreased +2 overall charge. Compound 8 was cocrystallized with EED and preserved the binding pose adopted by the 13-mer JARID2-K116me3 (Figure 6B), with the terminal Leu and Pro residues harbored in the same hydrophobic hollows hosting the Ala118 and the Gln116 from JARID2 peptide, respectively. Consistent with the negligible contribution to the β-propeller binding, the i – 1 position can hold a certain degree of variability, with the Ser side chain being unresolved and pointing toward the solvent.
The understanding of the structural determinants responsible for the 8–EED interaction assisted the last stage of the optimization toward a peptidomimetic structure. Indeed, the ensuing modifications revolved around the shortening of the amino acidic sequence and the replacement of the trimethyllysine moiety with a more cell-permeable function. In this respect, the substitution of the K116me3 cationic function with an N-methyl-N-isopropyl amine, together with the Pro-Ser dimer replacement with the bicyclic (2S,3aS,7aS)-octahydro-1H-indole afforded the tetrameric UNC5114 (9), featuring a sub-micromolar affinity (KD = 0.68 μM) and fewer amide bonds. Insertion of a piperidinyl nucleus in place of the C-terminal Leu brought about a cognate compound of 9, namely, UNC5115 (10) exhibiting a lower affinity (KD = 1.14 μM).
To shed light on the effect on PRC2 methyltransferase activity, the UNC5114 and UNC5115 were functionally evaluated in a scintillation proximity assay where they proved to decrease the catalytic activity up to 20% of the saturating concentration of ligand. Additional biochemical evaluations validated the allosteric mechanism of action for 9 and 10, and their strict dependence on the aromatic cage recognition for binding was substantiated by the loss of PRC2 inhibition in F97A, W364A, and Y365A EED mutants.
5. Discovery of Hit Compounds as EED Binders
First reports on hit identification of small molecular entities as EED binders were released by Novartis researchers in 2017. The development of a homogeneous time resolved fluorescence (HTRF) assay for a five-component PRC2 complex enabled the high-throughput screening (HTS) of approximately 1.4 million compounds.75 The assay was based on the detection of H3K27me0 to H3K27me2 conversion of a substrate peptide (histone H3, residues 21–44) and used a europium cryptate labeled antibody as a FRET donor. Given the inability of this preliminary test to discriminate between SAM competitive and noncompetitive PRC2 inhibitors, different biochemical investigations were carried out to define the mechanism of action of the identified hits. Compounds with IC50 < 50 μM were first evaluated in the same HTRF assay using different SAM concentrations to ascertain the activity cofactor independence. Then, the allosteric nature of the PRC2 inhibition was confirmed, measuring the binding disruption between a resin-bound EED protein (residues 1–441) and a H3K27me3 peptide (residues 19–33) in an AlphaScreen format. By means of this biochemical crossroads, different low-micromolar range H3K27me3-competitive PRC2 inhibitors were identified, belonging to different scaffolds, and a representative set of these (11–16) was disclosed (Figure 7A).76 Alternatively, in 2019, researchers from AstraZeneca developed a fluorescence polarization-based assay to identify EED binder hits with a single run of experiments readily. Their assay employed a previously reported EED binder as a displacement probe with the advantage of leading to hits with the same mechanism of action and less unspecific binding events.77,78
Figure 7.
Chemical structures of identified EED binders (11–16). (A) Chemical structures of compounds 11–16 showing the moieties exploring the “deep pocket” highlighted in orange and the scaffolds interacting with the aromatic cage highlighted in green; (B) Overall structure of EED–EBD–(11–16) ternary complex; EED is represented in semitransparent surface and cartoon (gray) (PDB 5H19), EBD peptide is represented in cartoon (violet), and hit compounds are represented in differently colored sticks. (C) Binding modes of 11–16 within the H3K27me3 pocket of EED (gray). Structures are shown as stick representations within X-ray cocrystal structure of 11 with EED (PDB 5H17). The residues delimiting the sides and the bottom of the binding gorge are labeled and represented as thin sticks. (D) Binding mode of H3K27me3 peptide (violet cartoon) within the upper cavity of EED (in gray cartoon and surface) (PDB 3IIW). The H3K27me3 side chains are represented as sticks. PDB codes for the other hit compounds: 12 (PDB 5H19), 13 (PDB 5H15), 14 (PDB 5H14), 15 (PDB 5H13), and 16 (PDB 5H25).
Compounds 11–16 were crystallized in complex with EED and an α-helical strand resembling the EZH2-EBD domain (residue 40–68). These cocrystal structures revealed that the compounds dock in the H3K27me3 binding pocket of EED (Figure 7B), albeit relying on an “induced fit” in the aromatic cage compared to the H3K27me3-bound configuration (Figure 7C,D). Some of the key residues governing the width of the cavity (Tyr364 and Trp365) were engaged in relevant side chain rotations to accommodate the diverse structures of 11–16, while Phe97 and Tyr148 were unperturbed. Similar molecular movements were also observed for Arg367, which in H3K27me3-bound EED is shielded by Tyr365 but with EED binder interaction lies at the bottom of the cavity and swings between two different conformations. The combination of movements of Tyr365 and Arg367 determines the loss of their reciprocal π–cation stacking interaction, which is a feature of the apo-structure of EED. Arg367 does not shape the wall of the aromatic cage; however, it fulfils the role of “gatekeeper” residue for the access to the interior of the EED β-propeller hole, thus defining an additional site of interaction, referred to as “deep pocket” or “induced pocket”. The observed conformational changes between the inhibitor-bound and the apo-structures of EED evidenced a certain adaptability of the β-propeller cavity in modeling its width and depth toward different scaffolds and furnished an explanation for the chemical diversity of the binders identified by Li et al.75
Compounds 11–16, although displaying different structural cores, shared several points of interaction and the overall orientation within the binding site. With the sole exception of 16, all the compounds are endowed with electron-rich aromatic systems (highlighted in orange in Figure 7A) to interact with the Arg367 guanidinium group by cation−π stacking in a face-to-face (11–14) or edge-to-face orientation (15). The central scaffolds of 11–16 (highlighted in green in Figure 7A), despite not being rationalizable in a common pharmacophore, locate within the space housing the trimethylated lysine of H3K27 and establish π–π stacking (12 and 14–16) or cation−π and van der Waals interactions (11 and 13) with the amino acids outlining the sides of the aromatic cage (Phe97, Tyr365, and Tyr148). The adaptability of the allosteric binding site also reflects the possibility of granting distinct interactions between different functional groups of 11–15 and ancillary amino acids projecting on the inner surface of the H3K27me3 binding cavity (see ref (91) for further details). The optimization of these contacts was exploited to improve the binding affinity and to develop more efficient EED binders starting from the hits 11–13.
5.1. Structural Optimization of EED210
In the wake of the hit discovery campaign reported by Li et al.,75 the H3K27me3-competitive PRC2 inhibitor EED210 (11, Figure 7A) was selected for further optimization.79 EED210 exhibited a modest micromolar inhibition of the PRC2 complex in the enzymatic assay (IC50 = 2.5 μM), and in the cocrystal structure with EED, it docked in the H3K27me3-binding cavity with an interaction mode similar to that observed for the other identified hits (Figure 7C).75 With the aim to optimize the binding efficiency and to converge to a synthetically more accessible scaffold, EED210 underwent structural manipulations according to a deconstruction–reconstruction approach (Figure 8). Preservation of the pharmacophoric elements along with simplification of the tricyclic core of EED210 led to the piperidine-based compound 17. Despite its essential structure, 17 retained the ability to inhibit PRC2 (IC50 = 95 μM) and afforded an increase in ligand efficiency (LE) and lipophilic efficiency (LipE) with respect to the parent compound 11, thus indicating an improved balance between fragment affinity and lipophilicity.80,81
Figure 8.
Deconstruction and reconstruction of 11 (EED210) and its derivatives (17–21). LE and LipE indexes were calculated according to the following formula: LE = (1.36 × pIC50)/HAC and LipE = pIC50 – cLogP. HAC = heavy atom count.
These results found confirmation in structural analysis (Figure 9A) where 17 was shown to retain the ability to embed within the remodelled H3K27me3 binding site and its 3-methoxyphenyl portion leveraged deep-pocket interactions with the Arg367. Interestingly, the protonated piperidine ring perfectly docked within the aromatic cage where it mimicked the presence of the H3K27me3 quaternary amine and contacted the Tyr365 and Glu238 by water-mediated H-bonds. From this minimized fragment, the authors started a stepwise reconstruction process to elicit improved chemical–physical properties, which involved the methoxyphenyl moiety and the core interacting with the aromatic cage (Figure 8). In this concern, given the relevance of π–cation interactions with the Arg367 guanidinium group for efficient binding, the deep pocket was probed with electron-rich aromatic systems characterized by the methoxy group shifted in different positions or where the oxygen atom was included in a bicyclic core (18a–d). Among these, the methoxy group in meta (17) or in ortho (18a) position represented the sole tolerated options, whereas the replacements with benzofuran regioisomers (18b,c) or benzodioxole ring (18d) afforded up to a 2-fold increase or retention of enzymatic potency (IC50 = 35–95 μM) together with improved LE and LipE.
Figure 9.
X-ray cocrystal structures of 17, 19d, and 20b within EED structure. (A) Binding mode of 17 (teal) within the H3K27me3 pocket of EED (gray); interacting residues are labeled and represented as sticks (PDB 5U5K). (B) Binding mode of 19d (green) within the H3K27me3 pocket of EED (PDB 5U5T). (C) Binding mode of 20b (violet) within H3K27me3 pocket of EED (PDB 5U62). In all three cases, water molecules are labeled and reported as cyan balls; inter- and intramolecular H-bonds are represented as dashed black lines.
Subsequently, attention was paid to the possibility of replacing the point charge-mediated π–cation stacking with interactions arising from a charge delocalized on π-aliphatic or π-aromatic systems. Maintaining the previous beneficial deep-pocket moieties, the piperidine secondary amine was converted into a guanidine group or fused within a tetrahydroimidazo[1,5-a]pyridine (Figure 8). Combining the π-aliphatic system with selected deep-pocket moieties resulted in a general increase of the PRC2 inhibition, especially for the benzofuran- or benzodioxole-bearing compounds (19b–d), which were characterized by single-digit micromolar range potency (IC50 = 2.1–3.9 μM). Importantly, these compounds showed a significant improvement in terms of lipophilic efficiency (LipE = 3.05–3.51) that reflects a better exploitation of direct contacts rather than unspecific lipophilic interactions to increase ligand binding. The cocrystal structure of 19d with EED (Figure 9B) explained the observed improvements and revealed that the guanidinium group, while juxtaposed with Tyr148 and Tyr365, facilitates H-bonds with Tyr148, Asn194, and Glu238, either directly or via a water molecule. These polar contacts teamed up with the deep-pocket interactions featured by the parent fragment 17 and were responsible for the remarkable increase of the binding potency. In ensuing assessments, 19a–d proved to inhibit the H3K27 methylation in kidney cancer (G-401) cells. However, due to permeability issues, they did not proceed in further development. Cellular permeation is negatively affected by strong positive charge; therefore, it was speculated that selecting moieties with appropriate pKa values might provide a guideline to properly mimic the H3K27me3 cationic nature without dampening the permeation capability. Looking for a protonable source with π aromatic delocalization potential, the authors reported a following series of compounds bearing a 3-amino- (20a,b) or 1-isopropyl-3-amino-tetrahydroimidazo[1,5-a]pyridine (21a,b) (Figure 8). This choice allowed a reduction of the number of H-bond donors and mitigated the high pKa values disclosed by the previous series of compounds (guanidine pKa > 11 versus aminoimidazole pKa = 9.2). When tested in an enzymatic setup, these compounds preserved their inhibitory activities in single-digit (20a,b and 21a) to sub-micromolar range (21b) and importantly demonstrated a marked enhancement of cell permeation in a cellular Caco-2 model. Inhibitory effects on H3K27 methylation status in G-401 cells were similarly maintained, and notably, the sub-micromolar inhibitor 21b showed a dose-dependent antiproliferative effect in Pfeiffer cells (IC50 = 3.4 μM), thus proving that allosteric inhibition of EED might impair cellular growth in lymphoma cells. EED–20b binary complex (Figure 9C) provided structural insights to the presumable binding pose of 21b and, in general, for this series of compounds. Compound 20b engages the same deep-pocket interactions of its parent compound 19d employing the benzodioxole ring and fills the aromatic cage triad through its 5,6,7,8-tetrahydroimidazo[1,5-a]pyridine core that establishes face-to-face π–cation stackings with Tyr148 and Tyr365. Contrarily, the amino group of 20b could not protrude to directly contact Asn194 and Glu238 polar groups and interacted with these moieties via a water molecule network. Finally, in the EED–21a cocrystal structure (reported by the authors but not included in the present review), the 4-isopropyl pendant bulged outside the EED cavity toward the solvent and was accounted as a plausible position for further future modifications.79
5.2. Structural Optimization of EED162
In a further investigation, Huang et al. focused on compound 12 (EED162) and started a structure-based hit optimization campaign on its core structure.82 The high-resolution cocrystal structure (PDB 5H19) of EED162 revealed that the binding into the H3K27me3 pocket relies on the establishment of a set of π–π interactions and hydrogen bonds with the EED counterpart (Figure 10A). An aromatic face-to-face π–π juxtaposition between the electron-deficient [1,2,4]triazolo[4,3-a]pyridine and the electron-rich Tyr148 and Tyr365 of the aromatic cage held in place the central scaffold of 12 in the middle of the β-propeller pocket. The so-obtained binding geometry enables the furan-2-yl in C5 of the core to dock into the “deep pocket” where the furan ring positions cation−π and edge-to-face π–π interactions with the guanidinium group of Arg367 and Tyr365, respectively. This molecular portion penetrates into a predominantly hydrophobic region of EED, outlined by Leu240, Asp310, and Arg367, where the 5-aminomethylene linker faces the entrance of the “induced pocket” and undergoes polar contacts with the Asn194 side chain. Other structural elements additionally contribute to the interaction: the endocyclic N2 nitrogen affords a hydrogen bond with Lys211 on the rim of the H3K27me3 pocket, whereas the cyano group in C6 interacts with the backbone of Tyr365 and Arg414 via water-mediated H-bonds. Remarkably, the C7/C8-fused N-benzylpiperidine is not essential for the binding and confers a detrimental lipophilicity to a solvent-exposed molecular portion, thus representing a suitable position for modification in a deconstruction–reconstruction approach.82 Initial fragmentation of the EED162 structure led to the compounds 22 and 23 (Figure 11), lacking the furan-2-yl-methylenamine in C5 and N-benzylpiperidine in C7–C8, respectively, and demonstrated the importance of leveraging “deep pocket” interactions. Indeed, while 23 (IC50 = 1 μM) retained the same activity range of 12 (IC50 = 4.03 μM) in biochemical assay, compound 22, which was unable to penetrate deeply into the inner H3K27me3 pocket, was inactive (IC50 > 100 μM).
Figure 10.
Binding interactions and X-ray crystal structure of EED–EBD–12 (EED162) and EED–EBD–16 ternary complexes. (A) Binding mode of 12 is presented in orange sticks within the H3K27me3-binding pocket of EED in white surface (on the left) and polar contacts within EED aromatic cage (on the right) (PDB 5H19). Interacting amino acids are labeled and shown in gray stick representation, whereas water molecules are depicted as cyan balls, and inter- and intramolecular H-bonds are represented as dashed black lines. (B) Superimposition of binding modes of 12 and 16 within the cavity of EED. Compound 16 is represented in pink; the corresponding EED cocrystal structure is in a pink cartoon (PDB 5H25), and the amino acids outlining the rim of the pocket are labeled and represented as sticks.
Figure 11.
Overview of structural optimizations of compound 12 (EED162). Evolution of SAR investigations on compound 12 leading to scaffold I (8-aryl-[1,2,4]triazolo[4,3-a]pyridine (24)), and scaffold II (8-aryl-[1,2,4]triazolo[4,3-c]pyrimidine (25)), followed by further optimizations and definition of scaffold III (31) and IV (32). Ligand efficiency (LE) and lipophilic efficiency (LipE) indexes were calculated according to the following formula: LE = (1.36 × pIC50)/HAC and LipE = pIC50 – cLogP. HAC = heavy atom count.
Structural analysis of the HTS hit 16 (Figure 10B) suggested that an edge-to-face π–π stacking between its C5 phenyl ring and Phe97 might provide a beneficial interaction for the affinity. This compound, even though devoid of the pivotal structural features seen in 12, such as an electron-rich aryl group in C5 position and an H-bond acceptor in N2, elicited a single-digit micromolar inhibition of the EED–H3K27me3 interaction (IC50 = 6.04 μM) comparable to that of 12 (Figure 7A). Superimposition of the crystal structures of 12 and 16 revealed that an aryl substitution in the C8 position of 12 might correspond to that shown in C5 by compound 16 (Figure 10B). As a result, the combination of structural features from compounds 16 and 23 led to the structure 24, characterized by a 5,8-disubstituted bicyclic aromatic module (scaffold I, Figure 11) and displaying a 60-fold improvement of potency (IC50 = 0.062 μM).
Interestingly, the replacement of the cyano group in C6 of structure 24 with an endocyclic nitrogen atom led to compound 25, endowed with similar biochemical potency (IC50 = 0.052 μM) and better ligand efficiency (LE = 0.45), thus yielding an alternative core for SAR studies (scaffold II, Figure 11). Scaffold hopping investigations on rings A and B of 25 revealed that a certain positioning of nitrogen atoms on the core structure also played a relevant role. Whereas the removal of N1 (26) caused a 5-fold decrease of activity, when the nitrogen atom was removed from position 2 or 6 (27–29), the potency consistently decreased in the micromolar range as a result of a loss of hydrogen bond acceptors (see compound 25 in Figure 11 for the core numbering). Reduction of the number of heteroatoms as well as their mutual positions within the ring was also detrimental (28 and 30), but this can be ascribed to the decreased electron deficiency of the [1,2,4]triazolo[4,3-a]pyridine core, which is necessary to establish effective π–π contacts with Tyr148 and Tyr365 residues.45
Huang et al. also reported that functionalization on the 3′- or 4′-position of the C8 aryl group on structure 24 with a dimethylamino group (31a) or other substituted amines (data not published) provided a further increase of the biochemical inhibition potency (31a, IC50 = 0.013 μM).82 The inclusion of the dimethylaminomethylene moiety on the previously identified [1,2,4]triazolo[4,3-a]pyridine and [1,2,4]triazolo[4,3-c]pyrimidine cores defined the scaffolds III and IV, which were used to carry out SAR investigations focusing on the “deep pocket” moiety (ring C, Figure 11). In this regard, different aromatic modules were evaluated, such as electron-rich (31a/32a, 31b/32b, 31c/32c) or electron-deficient five-membered rings (31d/32d, 31e/32e) and structures with different steric hindrance including 3′-methyl- or 3′-chlorothiophenyl (31f/32f, 31g/32g) and 6-membered phenyl or 2′-methoxyphenyl rings (31h/32h, 31i/32i). Despite the good tolerance exhibited by electron-rich aromatic substitutions, these SAR studies identified the furan-2-yl ring as a critical element for potent PRC2 methyltransferase inhibition. Given the nanomolar activity of 31a and 32a, both showing an IC50 = 0.013 μM in a biochemical setting, the ensuing investigations focused on the solvent-exposed ring D functions to enhance the biological and PK profiles. The considered compounds (33–35, Figure 12A) showed nanomolar range activities, and among them, the 4′-methansulfonyl compound 35 (EED226) stood out as the best candidate for late in vivo studies (biochemical IC50 = 0.022 μM, KD = 82 and 114 nM for EED and PRC2, respectively, in ITC evaluation). Notably, 35 exhibited complete oral bioavailability and a reasonable half-life (t1/2 = 2.2 h) due to its very low in vitro and in vivo hepatic clearance as predicted by the MLM (mouse liver microsome) assay.82
Figure 12.
Main compounds obtained during SAR investigations on ring D and binding mode representation of EED–EBD–EED226 (35) ternary complex. (A) Structures of compounds 33–35 and their pharmacokinetic properties. (B) Binding mode of 35 (green) within the H3K27me3 pocket of EED (gray); interacting residues are labeled and represented as sticks, whereas water molecules are shown as cyan balls, and inter and intramolecular H-bonds are shown as dashed black lines (PDB 5GSA). (C) Binding mode of 35, in green sticks, embedded in EED cavity shown in transparent white surface representation. Residues surrounding the binding pocket are represented in sticks, water molecules are represented as cyan balls, and inter- and intramolecular hydrogen bonds are represented as dashed black lines.
Qi et al. solved the crystal structure of EED in complex with 35 and the EBD peptide (PDB 5GSA) (Figure 12B), revealing that the compound retains all the fundamental interactions of the parent compound 12 and establishes “deep-pocket” interactions with the EED cavity by the furan-2-yl moiety.83 As for the cyano group in C6 of structure 12, the endocyclic N6 nitrogen atom interacts with various residues (Phe97, Arg367, Arg414) via a water-mediated H-bond network. These water molecules fill the remaining volume in the deep pocket formerly occupied by the C6 cyano group and are essential to anchor 35 within the induced binding pocket (Figure 12C). The C8 (4′-methansulfonyl)phenyl moiety is mostly solvent exposed, but along its trajectory, its phenyl ring engages in an edge-to-face π–π interaction with the Phe97 side chain.83 In further biochemical evaluations, 35 was confirmed to compete with H3K27me3 for EED in AlphaScreen-based assessment and proved to be selective for the PRC2 complex over 67 different proteins including methyltransferases, kinases, GPCRs, ion channels, nuclear receptors, and transporters.83
After showing that 35 was able to induce a dose-dependent global decrease of both H3K27me3 and H3K27me2 levels in G401 tumor cells, the authors compared its cellular effects with those of EI1, a SAM-competitive EZH2 inhibitor. In particular, gene expression microarray studies were performed to evaluate the induction of expression pattern changes at a transcriptome level. In KARPAS422 cells, EI1 and 35 raised mRNA levels of the same genes in a dose-dependent manner, and these alterations were linked to a decrease of H3K27me3 deposition on promoter regions of corresponding genes.
The robust anticancer activity of 35 resulted in a dose- and time-dependent effect on the proliferation of KARPAS422 cells, a DLBCL cell line holding the Y641N EZH2 gain-of-function mutation. Subcutaneous administration of 35 in the mouse xenograft model of KARPAS422 demonstrated good tolerability with no apparent side effects. Moreover, 35 induced slower growth and reduction of the tumor volume after dosing for 21 days at different concentrations twice a day and complete tumor regression after dosage for 32-days at 40 mg/kg twice a day. Interestingly, in a SAM-dependent drug-resistant WSU-DLCL2 cell pool (W-R10) and clones (W-R10-#2, #5 and #22) containing Y111N and F120L EZH2 mutations, 35 inhibited the proliferation with a similar potency to that with the wild-type PRC2 complex; whereas in the same experiments, EI1 and the drug tazemetostat exhibited no cell growth inhibition. Finally, the combination of 35 and EI1 demonstrated synergistic antiproliferative effects, thus suggesting that coadministration of an EED binder and a SAM-competitive EZH2 inhibitor might provide more effective anticancer activity.83
Notably, 35 was also successfully converted into a bivalent chemical degrader able to efficiently and selectively degrade EED as much as EZH2 and SUZ12 proteins in a ubiquitin proteasome-dependent fashion.84 To achieve this goal, the authors developed PROTACs endowed with linkers of different lengths to ensure a functional proximity and orientation between the EED- and the VHL-binding modules. Among the synthesized PROTACs, the one able to simultaneously bind to EED and induce its degradation (UNC6852) was further biologically assessed and proven to lessen H3K27me3 levels and to hamper cell growth when tested in Pfeiffer and DB cell lines.84
5.3. Development of EEDi-5285
A multidisciplinary team from the University of Michigan and Ascentage Pharma began a structure-guided discovery program of EED binders arising from a precursor of 35, which in the end resulted in exceptionally potent, efficacious, and orally active compounds EEDi-5285 (39d) and EEDi-1056 (39f).85 Possessing a good antitumor activity in EZH2 mutant lymphoma models as well as in clones resistant to SAM-dependent EZH2 inhibitors, 35 represented a compelling core structure for further optimization. The structure-based campaign started from the previously reported inhibitor 26 (see Figure 11), which exhibited moderate competitive activity against EED–H3K27me3 binding in AlphaScreen assay (IC50 = 0.115 μM) and reasonable anticancer activity in KARPAS422 lymphoma cells (IC50 = 2.6 μM).85 Since the binding affinity to EED strongly depends on π–π stacking interactions with the electron-rich Tyr148/Tyr365 pair in the aromatic cage, the authors initially introduced small electron-withdrawing groups (EWGs; 36a,b) on the imidazo[1,5-c]pyrimidine core of 26. The methylamide-bearing 36b was determined to have biochemical potency comparable to 26, whereas the ethyl ester derivative 36a was twice as potent (IC50 = 0.059 μM). Given the metabolic instability of the furan ring, further modifications concerned the moiety exploring the EED “deep pocket”. To reinforce the π–cation stacking interaction with the Arg367 guanidinium function at the bottom of the β-propeller cavity, an electron-rich 2-methoxy group was added to the phenyl ring (37b, Figure 13), and this compound displayed an IC50 value of 0.086 μM toward EED binding and IC50 of 1.9 μM against KARPAS422 cells, thus proving to be slightly more potent than the corresponding unsubstituted phenyl ring (37a). As a chemical probe, a fluorine atom was introduced at the C6 position of ortho-, meta-, and para-methoxy phenyl rings (37c–e). Although these modifications did not generally translate into a very strong improvement of activity in biochemical and cellular settings (KARPAS422), when a 2-fluoro-6-methoxyphenyl moiety was introduced (37c), a 1.7-fold increase of biochemical potency (IC50 = 0.050 μM) and an improvement of over 6-fold in cellular activity (KARPAS422 IC50 = 0.3 μM) was observed.
Figure 13.
Panel of structural optimizations of compounds 36–39 and leading to EEDi-5285 (39d) and EEDi-1056 (39f).
Further, the authors investigated the effects of phenyl ring replacement with electron-rich bicyclic systems (37f,g) by cycling the methoxy group with the phenyl ring on the deep binding pocket. Interestingly, the position of the oxygen in the dihydrobenzofuran nucleus had a significant impact on EED binding, with 37g being 7-fold more potent than the corresponding regioisomer 37f (IC50 = 0.029 μM and 0.207 μM, respectively). Moreover, the introduction of a fluorine atom at the C5 position of the 2,3-dihydrobenzofuran group (37h), described already in a previous Novartis patent,78 resulted in excellent IC50 values against EED (IC50 = 0.018 μM) and in cellular testing (IC50 = 0.012 μM), hence improving the antiproliferative activity in KARPAS422 more than 200-fold compared to 26.
The encouraging results obtained with the substitution pattern of 37h prompted the team to exhaustively probe the effect of EWGs in C1 of the imidazo[1,5-c]pyrimidine scaffold (38a–d). In this regard, N-methylcarboxamide, trifluoromethyl, methansulfonyl, and dimethylphosphine oxide were considered fruitful groups to strengthen the π–π interaction with the Tyr148/Tyr365 pair, and 38c stood out as the most potent of the series in a biochemical setting (IC50 = 0.0007 μM).
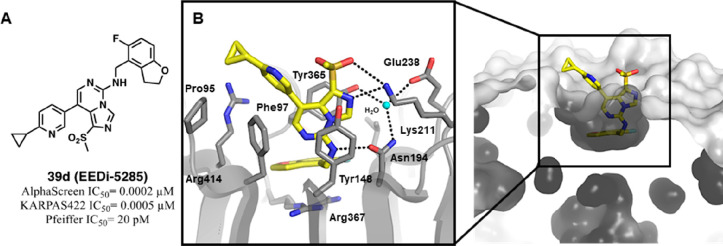
Considering the predominant aromatic nature of 38c, the next chemical manipulations were made around the C8 phenyl ring projecting outward from the EED cavity to improve the chemical–physical properties of the series. The effect of the aromatic ring in C8 was largely investigated by inserting suitable functional groups aiming either to increase the overall solubility of the compounds or to allow additional interactions over the external surface of EED (39a–f). In this regard, initial attempts led to compounds bearing a methansulfonyl group (39a), as in 35, although this modification yielded limited binding improvement. Second, the phenyl extension was replaced with a variably decorated pyridine-3-yl ring to improve the solubility and simultaneously maintain favorable edge-to-face contacts with Phe97 (39b–f). Because the previously developed binder 35 established hydrophobic interactions with the external surface of EED, the pyridine-3-yl ring and the C8 position of the central scaffold (structures not shown, for further details see ref (85)) were decorated with mostly aliphatic moieties. Among these, the derivative endowed with a 4′-cyclopropyl group, 39d, showed sub-nanomolar activity both on isolated EED (IC50 = 0.0002 μM) and in cell growth inhibition studies on KARPAS422 and Pfeiffer cells (IC50 = 0.0005 μM and 20 pM, respectively), thus being endorsed as the best candidate for further biological studies. Compound 39f was also selected to be characterized by comparable biophysical affinity (IC50 = 0.0005 μM) and cellular antiproliferative activities (KARPAS422 IC50 = 0.0028 μM and Pfeiffer IC50 = 70 pM).
The binding mode of 39d was determined via high-resolution cocrystal structure with EED (Figure 14), and comparison with the parent compound 35 rationalizes its extraordinarily potent interaction. Compound 39d embeds within the H3K27me3 binding site through its 5-fluoro-2,3-dihydrobenzofuran group, which entirely fills the “deep pocket,” excluding the transient water molecules previously stabilizing the 35 structure (Figure 12B,C). Moreover, the 2,3-dihydrobenzofuran moiety establishes a stronger interaction with the Arg367 guanidinium group due to a better π–cation juxtaposition. At the entrance of the EED cavity, the methansulfonyl pendant in C1 provides a hot spot for H-bonding to the Lys211 side chain, thus strengthening the docking within the aromatic cage. Finally, the solvent exposed 3-pyridinyl engages in edge-to-face π–π interactions with Phe97 and orients its cyclopropyl moiety toward Pro95 where van-der-Waals interactions are positioned (Figure 14).
Figure 14.
Chemical structure of EEDi-5285 (39d) and its binding mode representation within EED. (A) Chemical structure of EEDi-5285 (39d). (B) Cocrystal structure of compound EEDi-5285 (yellow) with the H3K27me3 pocket of EED (gray); interacting residues are labeled and represented as sticks, whereas water molecule is shown as a cyan ball, and intermolecular H-bonds are represented as dashed black lines (PDB 6W7F).
To evaluate the in vivo efficacy as antitumor agents, EEDi-5285 (39d) and the cognate compound EEDi-1056 (39f) were selected for explorative PK investigations. Both of them achieved acceptable plasma concentrations with oral bioavailability of 75% and 69%, respectively, and terminal T1/2 of approximately 2 h in a mouse model. In ensuing in vivo pharmacodynamic (PD) studies, 39d and 39f were evaluated in a KARPAS422 xenograft mouse model at doses of 50 and 100 mg/kg via daily oral administration. Notably, in a 28 day regimen, both of them induced complete tumor regression with minimal weight loss and were capable of a long-lasting activity since no tumor relapse was observed after 72 days by the end of the treatment.
5.4. Structural Optimization of Compound EED709
When Novartis began the hit-discovery campaigns described above, AbbVie Inc. researchers similarly embarked on a high-throughput screening campaign to identify PRC2 allosteric inhibitors.86 Interestingly, employing a thermal shift assay (TSA)-based screening platform, they independently identified the same hit compound previously reported by Li et al.,75 EED709 (13, Figure 7A), as a sub-micromolar EED binder. This pyrrolidine-based hit was confirmed as an EED ligand by a time-resolved (TR)-FRET counter-screen, exhibiting a Ki of 0.6 μM, and its allosteric mechanism was ascertained on trimeric PRC2 in vitro. To shed light on its binding mode and guide chemical manipulations in a structure-based drug design approach, EED709 was crystallized with EED (Figure 15B). As seen for the other binders, the central cavity of EED undergoes significant remodelling upon ligand recognition, especially around Tyr365 and Arg367, and the newly formed space allows compound 13 to dock in the β-propeller structure. In this arrangement, the pyrrolidine core fits within the aromatic cage, directing the 2-methoxybenzyl moiety toward the Arg367, where it juxtaposes with the cationic guanidino group. The two pyrrolidine substituents in C3 and C4 protrude away from each other, the N,N-dimethylamino function contacts the Tyr148, Asn194, and Glu238 by a water network, and the N-methylindole flanks the rim of the EED pore where it engages in mostly hydrophobic interactions.
Figure 15.
Chemical structure of 13 (EED709) and its binding mode representation within EED. (A) Chemical structure of compound 13 (EED709). (B) Cocrystal structure of compound 13 (light green) with the H3K27me3 pocket of EED (gray); interacting residues are labeled and represented as sticks, water molecules are shown as cyan balls, and intermolecular H-bonds are represented as dashed black lines (PDB 5U69).
Starting from these insights, Curtin et al. began SAR studies along two lines of intervention (Figure 16): first, they probed the EED deep pocket with different mono- and bicyclic aromatic pendants (40 and 41); second, they investigated the solvent-exposed portion by replacing the N-methylindole with polar functionalities (42 and 43).86 The reported chemotypes presented two stereocenters at the C3 and C4 of the pyrrolidine (see Figure 15A for the core numbering) and, due to synthetic reasons, were obtained as a trans-racemic mixture. In addition to appraising the EED binding by TR-FRET assay, the most potent compounds were also tested in cancer cell lines responsive to EZH2 inhibition (i.e., G-401 and OCILY cells) to assess the impairment of the H3K27 trimethylation. Effects on proliferation in Pfeiffer cells, harboring EZH2 activating-mutations, were also evaluated. Initial efforts to enhance binding affinity revealed that replacing the 2-methoxyphenyl pendant with 2,6-disubstituted aromatic rings had a relevant impact on potency, leading to EED binders in the double digit nanomolar range (40a–d). The affinity improvement was related not only to electronic effects provided by the presence of halogens on the benzyl ring but more likely to the 2,6-substitution pattern, as observed comparing the 2-fluoro-5-methyl (40a) and 2-fluoro-6-methyl (40d) derivatives with the 2,6-dihalogenated 40b,c. One step forward in this regard was represented by 41a where the annulation between the ortho position and benzylic methylene led to a conformationally locked 7-fluoroindane analog with retained affinity in TR-FRET assay and increased inhibition of H3K27 trimethylation in kidney cancer cells (G-401 IC50 = 0.29 μM) and B cell lymphomas (OCILY IC50 = 0.66 μM). The indane moiety insertion induced an additional stereocenter; however, only the S-indane enantiomer showed low-nanomolar potency (41a being over 10-fold more potent than 41b). In the attempt to reduce the lipophilicity of the series, SAR studies around the indane core and other bicyclic rings were also undertaken (41c–f). Nevertheless, despite the cLogP demonstrating better values (3.2 for 41f versus a mean value of 4–5 for 40a–d and 41a,b), these modifications elicited a significant loss of affinity, up to10-fold, suggesting scarce space for improvement of the pharmacokinetic profile in this moiety.
Figure 16.
Structural modifications of compound 13 (EED709) leading to compounds 40–43. All compounds are trans-racemic mixtures in C3–C4 unless otherwise noted with an asterisk. In the case of 43a and 43c, the trans racemic mixture was resolved, but the compound is an epimer at the indane stereocenter. #IC50 values of H3K27 trimethylation inhibition over 6 days in tumor cells.
Med-chem efforts on the solvent-exposed area led to better results in terms of balance between EED binding and lipophilicity. Indeed, projecting out of the H3K27me3 binding site, this moiety was more amenable to structural manipulations. So, placing polar extensions on the 40d core structure (by replacing the N-methyindole scaffold) and using different para-substituted bicyclic linkers yielded single-digit to sub-nanomolar range ligands (42a–c) with reduced cLogP values (from 3.5 to −0.6). Finally, when some of the prior functionalities were combined with the 7-fluoroindan-1-yl pendant, the so-obtained EED binders exhibited sub-nanomolar potency in a TR-FRET assay and nanomolar cellular activities (43a–c). This series of compounds were obtained as trans-racemic mixtures, and only 43a and 43c were resolved as 3R,4S diastereomers, epimeric at the C1′ indane stereocenter.86 In particular, 43c (A395) resulted in the most promising one and exhibited double digit nanomolar inhibitory activities either on H3K27 methylation in cancer G-401 and OCILY-19 cells or on Pfeiffer cell proliferation.
Determination of its cocrystal structure with EED demonstrated that 43c preserves the key interactions of EED709 and engages in enhanced π–cation interactions with Arg367 through its electron-rich 7-fluoro-indane cycle. Its trans configuration is determinant since it allows the dimethylamino moiety to take part in a water-mediated network of H-bonds with Tyr148, Asn194, and Glu238 while projecting the polar 4-methansulfonyl-piperazine extension out of the EED central pore toward a solvent-exposed area (Figure 17).87
Figure 17.
Chemical structure of 43c (A395) and its binding mode representation within EED. (A) Chemical structure of compound 43c (A395). (B) Binding mode of compound 43c (A395) (magenta) within the H3K27me3 pocket of EED (gray); interacting residues are labeled and represented as sticks, water molecules are shown as cyan balls, and intermolecular H-bonds are represented as dashed black lines (PDB 5K0M).
High-throughput thermal shift assay against EED protein proved that 43c markedly stabilized EED (ΔTm = 13 °C at 50 μM), thus confirming its binding. In additional biophysical SPR experiments, 43c bound to EED in 1:1 stoichiometry displaying a KD value of 0.0015 μM. Moreover, evaluation of its inhibitory activity on the three-membered EZH2–EED–SUZ12 complex in a radioactivity-based assay revealed an IC50 of 0.0018 μM. When profiled against other epigenetic targets, 43c proved to be selective for PRC2 up to 50 μM concentration over a panel of 32 different methyltransferases, including lysine, arginine, and DNA methyltransferases, and other epigenetic readers with trimethyl-lysine-recognizing capability. In cellular settings, 43c blocked the H3K27 di- and trimethylation in rhabdoid tumor cells with IC50 values of 0.390 and 0.090 μM, respectively, and this inhibition was selective over other histone target methylation. Regarding in vivo studies, He et al. tested 43c in a DLBCL Pfeiffer xenograft model by comparing its antitumor efficacy with the orthosteric EZH2 inhibitor GSK126, which due to PK issues, was administered subcutaneously at a dose of 300 mg/kg twice per week. In this experimental model, tumor growth inhibition (TGI) was reduced by 84%, while GSK126, administered at its established dosage protocol, elicited a TGI of 62%. Importantly, 43c was less prone to induce resistance in Pfeiffer and KARPAS422 DLBCL cell lines and in drug-resistant clones retained antiproliferative activity,87 thus underscoring the potential of PRC2 allosteric inhibitors as an alternative therapeutic option to SAM-competitive EZH2 inhibitors.
6. Recent Advancements in EED Binder Development
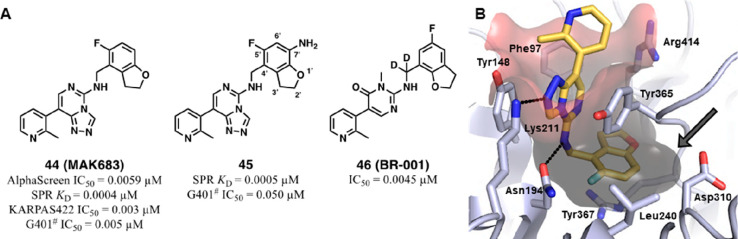
The above-discussed allosteric PRC2 inhibitors are excellent chemical tools proving the efficacy of tackling the EED H3K27me3-recognizing capability to cripple the PRC2 functions in vitro and in vivo. Following these efforts, Novartis developed another EED binder, MAK683 (44, Figure 18A), which is currently in phase I/II clinical trials for the treatment of DLBCL, nasopharyngeal carcinoma (NPC), or other advanced solid tumors (NCT02900651). MAK683 structure was reported in recent literature in Novartis’ patents together with other potent EED binders78,88 and by Huang et al.89 Similarly to EED226, it is characterized by a triazolo[4,3-c]pyrimidine core, but it features a 5-fluoro-2,3-dihydrobenzofuran ring, replacing the metabolically labile furan, which interestingly was shared in 39d (EEDi-5285) as a fruitful “deep pocket” moiety. Compound 44 exhibited potent inhibition of EED–H3K27me3 peptide interaction in AlphaScreen experiments (IC50 = 0.0059 μM) and induced nanomolar antiproliferative effects in B cell lymphoma KARPAS422 cells (IC50 = 0.003 μM).78 Given its promising potential, very recently, 44 was employed to develop an EED-targeted PROTAC and drive the core PRC2 complex components to protein degradation, although with different kinetics.90 The solvent-exposed portion of 44 was exploited to functionalize the EED binder with the VHL warhead, and this structural modification proved to marginally alter the EED affinity and PRC2 inhibition in vitro. TR-FRET in vitro experiments revealed that EED–PROTAC–VHL ternary complex formation occurred only in the presence of PROTAC with the correct stereochemistry. Notably, the authors hypothesized that the diverse rate of protein degradation observed for EED, EZH2, and SUZ12 might imply different underlying mechanisms. On one side, EED is primarily targeted by the ubiquitin proteasome system; on the other, EZH2 and SUZ12 protein depletion might occur as a result of their instability outside the PRC2 complex or be induced by secondary ubiquitylation of a target lysine on EZH2 and SUZ12 upon EED–PROTAC–VHL ternary complex formation.90
Figure 18.
Chemical structures of 44–46 and binding mode of MAK683 within EED. (A) Chemical structures of 44 (MAK683), 45, and 46 (BR-001). (B) Binding mode of compound 44 (MAK683) (gold) within the H3K27me3 pocket (in deep-salmon surface) of EED (blue-white cartoon); interacting residues are labeled and represented as sticks, intermolecular H-bonds are represented as dashed black lines (PDB 6YVJ). The dark arrow points to the EED surface facing the C7′ position of the dihydrobenzofuran ring of MAK683. #IC50 values of H3K27 trimethylation inhibition over 6 days in tumor cells.
Moreover, very recently, AstraZeneca researchers have launched a structure-guided optimization of 44 to enhance its unexpected poor solubility (5 μM at pH 7.4) and physicochemical properties.91 To achieve indications for structural optimizations, 44 was cocrystallized with EED (Figure 18B), and the 3D model was used as a platform for free energy perturbation (FEP) studies directed toward the C7′ position of the dihydrobenzofuran ring. The interaction surface facing C7′ presented an inward hollow (highlighted by a black arrow in Figure 18B) and was assumed to be amenable to insertion of small moieties (-F, -NH2, -OH, -CN) able to increase the compound solubility without dampening the binding affinity. A significant improvement of solubility (69 μM at pH 7.4) was obtained with 45 (Figure 18A), which retained excellent affinity in SPR experiments (KD = 0.0005 μM) with respect to 44 (KD = 0.0004 μM) but was 10-fold less efficient in inhibiting H3K27 trimethylation in G401 cells (45 IC50 = 0.05 μM versus 44 IC50 = 0.005 μM).91
Starting from EED226, another potent and selective EED binder was recently developed by Zou and collaborators from Shanghai Blueray Biopharma.54 Compound 46 (BR-001) (Figure 18A) arose from a scaffold-hopping approach and featured a pyrimidone core structure bearing a 5-fluoro-2,3-dihydrobenzofuran-7-yl moiety at the C2 position (Figure 18A). Structural analysis on isolated EED uncovered a typical binding pose for 46, with the 3-methylpyrimid-4-one scaffold lodging within the Phe97–Tyr148–Tyr365 aromatic cage where it engages in π–π stacking interactions. Protruding from this position, its benzofuran-7-yl fits within the induced-pocket of the β-propeller pore, whereas the 2-methylpyrimidin-3-yl projects outward to the solvent. In competition binding assays on EED, BR-001 displaced H3K27me3 with a low-nanomolar potency (IC50 = 0.0045 μM), and this activity translated well in a cellular setting (KARPAS422) where the compound dose-dependently inhibited H3K27 trimethylation and cell viability. To assess antiproliferative effects in a panel of cancer cell lines, 46 was tested in DLBCL cell lines, harboring EZH2-activating mutations, along with other cancer cell types characterized by EZH2 overexpression.
Interestingly, leukemia, gastric tumor, and brain tumor cells with altered levels of EZH2 did not respond to EZH2 inhibition by BR-001. Resistance to EZH2 inhibition was formerly linked to increased MLL1 expression level and H3K27ac upregulation. However, this effect was correlated to the induction of H3K27 acetylation rather than to MLL1 overexpression alone. In vivo KARPAS422 and Pfeiffer xenograft model studies demonstrated the ability of BR-001 to achieve 85% and 96% tumor growth inhibition, respectively, along with dose-dependent H3K27me3 level decrease and increased expression of PRC2-pertinent genes. Finally, the authors unveiled an unprecedented immunomodulatory mechanism of action for BR-001 and most likely for EZH2 allosteric inhibitors in general.54 Indeed, BR-001 resulted in robust upregulation of the CXCL10 chemokine levels in colorectal carcinoma cells that eventually led to CD8+ T cell recruitment to the tumor in murine models, which might expand the biological application of this antitumor agent.
7. Conclusions
In the last years, tremendous progress has been made in the development of small molecules directly or indirectly targeting EZH2. Recently, the EZH2 catalytic inhibitor tazemetostat hit the market after FDA approval for locally advanced epithelioid sarcoma and follicular lymphoma.48−50 However, the work for medicinal chemists is not finished yet as orthosteric EZH2 inhibitors induce resistance despite being already selective toward many other methyltransferases.
In 2014, after a structure-based virtual screening, the Luo group discovered astemizole (1) as the first-in-class compound able to disrupt the EZH2–EED interaction.72 Very recently, Luo’s group has provided the cocrystal structure of the EED–1 complex clarifying its binding mode: 1 is able to bind to the bottom of the WD40-repeat domain of EED, thus hampering its binding to EZH2, but differently from EED inhibitors that bind to the H3K27me3 binding pocket at the top of WD40-repeat domain of EED.72 In the same work, a first SAR investigation on 1 revealed that a cyclic homology study led to a significant improvement in potency with compound 5b that was the most potent (IC50 = 4.21 μM) among the novel analogues. Importantly, this new compound proved the ability to destabilize the PRC2 complex, leading to the degradation of EED, EZH2, and SUZ12 proteins,72 differently from the EED inhibitors, that contrarily stabilize the PRC2.83,87
The EED–1 crystal structure highlighted that the EZH2–EED interaction interface could be targeted by small molecules and built the basis for future structure-based optimization and development of novel chemical entities able to display such a mechanism of action. In this regard, recent literature has already started to reveal novel structures able to disrupt the EZH2–EED PPI, and in the near future, we will observe the development of these early hits into optimized chemical tools.92,93 Given the extent of the EZH2–EED contact, we cannot exclude that among these chemical entities, there might be some able to dock outside or partially overlapping with the 1 binding site, thus broadening the molecular surface amenable for protein–protein disruption.
As seen above, Novartis published an interesting whole medicinal chemistry approach starting from a HTS campaign and identifying two hit compounds, 11 (EED210, IC50 = 2.5 μM) and 12 (EED162, IC50 = 4.03 μM). Both compounds underwent structural optimizations: particularly, the optimization campaign on 12 resulted in a very potent, selective compound, 35 (EED226, IC50 = 0.022 μM), with excellent in vitro and in vivo properties. EED226 impaired proliferation in EZH2 orthosteric inhibitor-resistant cancer cells and also achieved complete tumor regression in a mouse model. Of note, when 35 and the orthosteric EZH2 inhibitor EI1 were combined, synergistic antiproliferative effects were observed hence providing a more efficient anticancer strategy by combining two PRC2 modulators with two different mechanisms of action.
Very recently, from the University of Michigan and Ascentage Pharma, an optimization study on compound 35 led to an extremely potent and orally active compound, EEDi-5285 (39d, IC50 = 0.0002 μM). In in vivo studies, 39d was evaluated through daily oral administration in a KARPAS422 xenograft mouse model, where it demonstrated complete tumor regression with minimal weight loss and also showed long-lasting activity since no tumor relapse was monitored after 72 days by the end of the treatment.
Concerning compound 11, the optimization is still in its infancy, especially compared to the first series, as more med-chem optimization needs to be carried out before an evaluation in vivo.
Interestingly, AbbVie discovered almost in parallel via their high throughput screening the same hit 13 (EED709) that was identified by Novartis. The following structural optimization campaign led to compound 43c (A395) with a Ki value of 0.0004 μM. When tested in a DLBCL Pfeiffer xenograft model, tumor growth was reduced by 84%. Notably, less resistance was observed during 43c treatment of Pfeiffer and KARPAS422 DLBCL cell lines, therefore confirming that the PRC2 allosteric inhibitors (EED binders) could help to overcome the resistance induced by the EZH2 catalytic inhibitors in cancer therapy.
A point to raise is the following: not only the optimization of already discovered hit compounds (EZH2–EED interaction inhibitors and EED binders), but also the discovery of additional targetable structures of this complex will be a feasible way to improve PRC2 modulation. In these regards, a surface dip (Glu579 pocket) that senses the methylation status of H3K36 has been discovered as an additional allosteric mechanism not yet fully understood. The methylation status of H3K36 does not affect the binding affinity of PRC2 for H3K27 but the catalytic activity. In this regard, unmethylated H3K36 increases PRC2 catalytic turnover; contrarily, when H3K36 is methylated, it results in diminished turnover. This explains why H3K27 and H3K36 are methylated in a mutually exclusive manner. The existence of this Glu579 pocket could pave the way for further innovative allosteric EZH2 modulators.4 Moreover, it would be fascinating to evaluate whether targeting the third essential protein within the PRC2, SUZ12, could also offer a chance to modulate the methyltransferase activity of PRC2. In the years to come, after successful clinical validation and the approval of the first EZH2 catalytic inhibitor tazemetostat and the growing body of literature around the PRC2 complex, we can expect further advancements in the understanding of its complex biological functions as well as its direct or indirect modulation through small molecules.
Noteworthy, the emerging “targeted degradation” approach was also applied to PRC2. Jin and co-workers, who described the first EZH2 degraders, claimed that solely inhibiting EZH2 without decreasing protein level was insufficient to reduce breast cancer cell proliferation. Contrarily, the EZH2 degraders were able to completely suppress tumor growth in a triple-negative MDA-MB-468 breast cancer mouse model.94 The same approach was applied by AstraZeneca researchers who reported the first proteolysis targeting chimeras (PROTACs) directed to EED that were capable of leading its degradation.90
Interestingly, their EED–PROTAC model, in addition to degrading EED, was proven to likewise induce the degradation of EZH2 and SUZ12 proteins, similarly to the EZH2–EED interaction inhibitor 1 and compound 5b, and to markedly arrest cell proliferation of KARPAS422 cells containing the gain of function Y641N EZH2 mutation.
These additional and very recent findings confirm that PRC2 can be nowadays considered a “hot target” in cancer chemotherapy research. Both EZH2 and EED degradation by EZH2–EED interaction impairment (5b) or by EZH2–EED–PROTAC offer a significant additional opportunity to block the PRC2 oncogenic activity, above all in the treatment of cancer types resistant to the EZH2 orthosteric inhibitors.
Moreover, it is worth mentioning that although PRC2 is considered a major complex in tumor therapy, its biological relevance in cancers different from lymphomas still remains to be elucidated. Furthermore, the synergistic effect observed in cellular settings upon the combination between the EZH2–EED interaction inhibitor 1 and the EZH2 inhibitor EPZ005687 but also between the EED binder 35 and EZH2 inhibitor EI1 prompt further in vitro and in vivo studies for in-depth antitumor property validation. Finally, it could be interesting to investigate the combination or to develop hybrid molecules acting as PRC2 orthosteric and allosteric inhibitors or degraders and on other epigenetic targets involved in the same or converging biological pathways of PRC2 such as HDAC or LSD1.95
Acknowledgments
This work was supported by AIRC2016 IG-19162, MIUR, PON R&I 2014-2020-AIM1873131-2 funds, Sapienza Ateneo Project 2020, RG120172B8E53D03. We thank Dr. Roberta Tesch (Goethe University, Frankfurt) for the technical assistance in the manuscript preparation.
Glossary
Abbreviations Used
- AEBP2
adipocyte enhancer-binding protein
- DLBCL
diffuse large B-cell lymphoma
- EBD
EED binding domain
- EED
embryonic ectoderm development
- EPOP
elongin BC and polycomb repressive complex 2 associated protein
- EZH2
enhancer of zeste homologue 2
- FEP
free energy perturbation
- FRET
fluorescence resonance energy transfer
- GPCR
G protein-coupled receptor
- H3K27
histone 3 lysine 37
- HAC
heavy atom count
- HDAC
histone deacetylase
- HMTase
histone methyltransferase
- HTRF
homogeneous time-resolved fluorescence
- IGF-1R
insulin-like growth factor 1 receptor
- ITC
isothermal titration calorimetry
- JARID2
jumonji and AT-rich interaction domain containing 2
- LE
ligand efficiency
- LipE
lipophilic efficiency
- LSD1
lysine-specific histone demethylase 1
- MEK
mitogen-activated protein kinase kinase
- MLM
mouse liver microsome
- NPC
nasopharyngeal carcinoma
- PALI
phylogeny and alignment of homologous protein structures
- PcG
polycomp group
- PCL1–3
polycomb-like proteins 1–3
- PI3K
phosphoinositide-3- kinase
- PPI
protein–protein interaction
- PRC1/2
polycomb repressive complex 1/2
- PROTAC
proteolysis targeting chimera
- PTM
post-translational modification
- RbAp46/48
retinoblastoma associated protein 46/48
- RARa
RAR-related orphan receptor alpha
- SAH-EZH2
stabilized α-helix of EZH2
- SAL
SET activation loop
- SAM
S-adenosyl-l-methionine
- SBD
SANT1-binding domain
- SET
Su(var.)3–9, enhancer-of-zeste and trithorax
- SRM
stimulation-responsive motif
- STD
saturation transfer difference
- STAT3
signal transducer and activator of transcription 3
- SPR
surface plasmon resonance
- SUZ12
suppressor of zeste 12
- TGI
tumor growth inhibition
- TR
time-resolved
- TSA
thermal shift assay
- VHL
Von Hippel–Lindau
Biographies
Stefano Tomassi graduated in Medicinal Chemistry at the University of Rome “La Sapienza” in 2009 and received his Ph.D. in Pharmaceutical Sciences at the University of Naples Federico II in 2014. In 2014/2015, he was a Postdoctoral Fellow at the Department of Chemistry and Chemical Biology of the TU Dortmund under the supervision of Prof. D. Rauh. After holding Postdoctoral positions at the University of Campania Luigi Vanvitelli, he was appointed as Assistant Professor at the Department of Pharmacy of the University of Naples Federico II (2019). His research activity has been focusing on the design and synthesis of epigenetic and kinase modulators and integrin and chemokine receptor ligands and development of innovative synthetic methodologies in peptide chemistry.
Annalisa Romanelli graduated in Pharmaceutical Chemistry and Technologies with a master thesis in medicinal chemistry and spent the last three years in the research group of Prof. Antonello Mai at Sapienza University of Rome as a Ph.D. student in Life Sciences. Recently she has concluded her Ph.D. working on the design and synthesis of small molecules modulating the activity of the principal epigenetic enzymes. In particular, she focused her attention on multitarget drug discovery, developing rationally designed compounds able to simultaneously inhibit two targets both involved in the onset and progression of cancer pathology.
Clemens Zwergel is currently a Postdoctoral Fellow at the Department of Drug Chemistry and Technologies, Sapienza University of Rome. In the last years, he proved to be a true European citizen: he moved after his license to practice as Pharmacist from his country of origin (Germany) to Exter (U.K.) for a Diploma in Pharmaceutical Sciences. Before moving to Italy, he was then a MarieCurie fellow in France (Metz), where he obtained his EuroPhD within the RedCat network. Since 2010, his main research interest lies mainly in the design and synthesis of analogs of natural compounds as well as modulators of epigenetic enzymes with potential applications in cancer, neurodegenerative, metabolic, and infectious diseases.
Sergio Valente is currently associate professor at Department of Drug Chemistry and Technologies, Sapienza University of Rome. He received a degree in Pharmacy and obtained his Ph.D. in Pasteur Science at the same University. Then, he completed postdoctoral research in medicinal chemistry within a REDCAT Marie Curie project at the University of Lorraine-Metz, France. Back in Italy, from 2011 to 2020, he has been assistant professor in medicinal chemistry at Sapienza University. His main research activity focuses on the design and synthesis of small molecules as epigenetic target modulators for cancer, neurodegenerative and metabolic pathologies, and bacterial, viral, and parasitic infection treatment. Recently, his activity has also covered the design and synthesis of chemical probes for chemoproteomic studies addressed to targets and interactors discovery.
Antonello Mai graduated in Pharmacy at Sapienza University of Rome, Italy, in 1984. He received his Ph.D. in 1992 in Pharmaceutical Sciences, with a thesis entitled “Researches on New Polycyclic Benzodiazepines Active on Central Nervous System”, advisor Prof. M. Artico. In 1998, he was appointed Associate Professor of Medicinal Chemistry at the same University. In 2011, Prof. Mai was promoted to Full Professor of Medicinal Chemistry at the Faculty of Pharmacy and Medicine, Sapienza University of Rome. His research interests include the synthesis and biological evaluation of new bioactive compounds, in particular small molecule modulators of epigenetic targets, chemical probes, and dual- and multitargeting compounds. Additionally, he is working in the fields of antibacterial/antimycobacterial, antiviral, and CNS agents.
Author Contributions
# S.T. and A.R. are co-first-authors. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Special Issue
Published as part of the Journal of Medicinal Chemistry special issue “Epigenetics 2022”.
References
- Schuettengruber B.; Bourbon H. M.; Di Croce L.; Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 2017, 171, 34–57. 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Margueron R.; Reinberg D. The polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poepsel S.; Kasinath V.; Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 2018, 25, 154–162. 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani K. S.; Jain S. U.; Ge E. J.; Diehl K. L.; Lundgren S. M.; Muller M. M.; Lewis P. W.; Muir T. W. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 8295–8300. 10.1073/pnas.1819029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R.; Wang L.; Wang H.; Xia L.; Erdjument-Bromage H.; Tempst P.; Jones R. S.; Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 2002, 298, 1039–1043. 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A.; Nishioka K.; Erdjument-Bromage H.; Tempst P.; Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev. 2002, 16, 2893–2905. 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanulli S.; Justin N.; Teissandier A.; Ancelin K.; Portoso M.; Caron M.; Michaud A.; Lombard B.; da Rocha S. T.; Offer J.; Loew D.; Servant N.; Wassef M.; Burlina F.; Gamblin S. J.; Heard E.; Margueron R. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell 2015, 57, 769–783. 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A.; Shen X.; Ma Q.; Cao J.; von Gise A.; Zhou P.; Wang G.; Marquez V. E.; Orkin S. H.; Pu W. T. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012, 26, 37–42. 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M.; Lee J. S.; Kim H.; Kim K.; Park H.; Kim J. Y.; Lee S. H.; Kim I. S.; Kim J.; Lee M.; Chung C. H.; Seo S. B.; Yoon J. B.; Ko E.; Noh D. Y.; Kim K. I.; Kim K. K.; Baek S. H. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell 2012, 48, 572–586. 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Ferrari K. J.; Scelfo A.; Jammula S.; Cuomo A.; Barozzi I.; Stutzer A.; Fischle W.; Bonaldi T.; Pasini D. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 2014, 53, 49–62. 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- Oksuz O.; Narendra V.; Lee C. H.; Descostes N.; LeRoy G.; Raviram R.; Blumenberg L.; Karch K.; Rocha P. P.; Garcia B. A.; Skok J. A.; Reinberg D. Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell 2018, 70, 1149–1162. 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. H.; Bracken A. P.; Pasini D.; Dietrich N.; Gehani S. S.; Monrad A.; Rappsilber J.; Lerdrup M.; Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008, 10, 1291–1300. 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Margueron R.; Li G.; Sarma K.; Blais A.; Zavadil J.; Woodcock C. L.; Dynlacht B. D.; Reinberg D. Ezh1 and ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518. 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R.; Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004, 15, 57–67. 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Holoch D.; Margueron R. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 2017, 42, 531–542. 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Kim H.; Kang K.; Kim J. AEBP2 as a potential targeting protein for polycomb repression complex PRC2. Nucleic Acids Res. 2009, 37, 2940–2950. 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; Margueron R.; Ku M.; Chambon P.; Bernstein B. E.; Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010, 24, 368–380. 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K.; Margueron R.; Ivanov A.; Pirrotta V.; Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol. Cell. Biol. 2008, 28, 2718–2731. 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E.; Jerman E.; Healy E.; Ito S.; Holoch D.; Oliviero G.; Deevy O.; Glancy E.; Fitzpatrick D. J.; Mucha M.; Watson A.; Rice A. M.; Chammas P.; Huang C.; Pratt-Kelly I.; Koseki Y.; Nakayama M.; Ishikura T.; Streubel G.; Wynne K.; Hokamp K.; McLysaght A.; Ciferri C.; Di Croce L.; Cagney G.; Margueron R.; Koseki H.; Bracken A. P. A Family of vertebrate-specific polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol. Cell 2018, 70, 408–421. 10.1016/j.molcel.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Liefke R.; Karwacki-Neisius V.; Shi Y. EPOP interacts with elongin BC and USP7 to modulate the chromatin landscape. Mol. Cell 2016, 64, 659–672. 10.1016/j.molcel.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S.; Eisenhaber F.; O’Carroll D.; Strahl B. D.; Sun Z. W.; Schmid M.; Opravil S.; Mechtler K.; Ponting C. P.; Allis C. D.; Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Antonysamy S.; Condon B.; Druzina Z.; Bonanno J. B.; Gheyi T.; Zhang F.; MacEwan I.; Zhang A.; Ashok S.; Rodgers L.; Russell M.; Gately Luz J. Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-ray crystallographic analysis of the EZH2-SET domain. PLoS One 2013, 8, e84147 10.1371/journal.pone.0084147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Zeng H.; Dong A.; Li F.; He H.; Senisterra G.; Seitova A.; Duan S.; Brown P. J.; Vedadi M.; Arrowsmith C. H.; Schapira M. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One 2013, 8, e83737 10.1371/journal.pone.0083737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery N. D.; Yee D.; Chen A.; Kalantry S.; Chamberlain S. J.; Otte A. P.; Magnuson T. The murine polycomb group protein eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 2005, 15, 942–947. 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Pasini D.; Bracken A. P.; Jensen M. R.; Denchi E. L.; Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R.; Justin N.; Ohno K.; Sharpe M. L.; Son J.; Drury W. J. 3rd.; Voigt P.; Martin S. R.; Taylor W. R.; De Marco V.; Pirrotta V.; Reinberg D.; Gamblin S. J. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009, 461, 762–767. 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.; Bian C.; Yang W.; Galka M.; Ouyang H.; Chen C.; Qiu W.; Liu H.; Jones A. E.; MacKenzie F.; Pan P.; Li S. S.; Wang H.; Min J. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 19266–19271. 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H.; Roberts C. W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Chng W. J. EZH2 abnormalities in lymphoid malignancies: underlying mechanisms and therapeutic implications. J. Hematol. Oncol. 2019, 12, 118. 10.1186/s13045-019-0814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K.; Das B.; Dobrowolski C.; Karn J. Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. mBio 2017, 8, e00133. 10.1128/mBio.00133-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A. W.; Dronamraju R.; Potjewyd F.; James K. S.; Winecoff D. K.; Kirchherr J. L.; Archin N. M.; Browne E. P.; Strahl B. D.; Margolis D. M.; James L. I. Evaluation of EED inhibitors as a class of PRC2-targeted small molecules for HIV latency reversal. ACS Infect. Dis. 2020, 6, 1719–1733. 10.1021/acsinfecdis.9b00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K.; Hanks S.; Ruark E.; Zachariou A.; Duarte S. D. V.; Ramsay E.; Snape K.; Murray A.; Perdeaux E. R.; Seal S.; Loveday C.; Banka S.; Clericuzio C.; Flinter F.; Magee A.; McConnell V.; Patton M.; Raith W.; Rankin J.; Splitt M.; Strenger V.; Taylor C.; Wheeler P.; Temple K. I.; Cole T.; Douglas J.; Rahman N. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2011, 2, 1127–1133. 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa E.; Higashimoto K.; Sakai Y.; Numakura C.; Okamoto N.; Matsunaga S.; Ryo A.; Sato Y.; Sanefuji M.; Ihara K.; Takada Y.; Nishimura G.; Saitsu H.; Mizuguchi T.; Miyatake S.; Nakashima M.; Miyake N.; Soejima H.; Matsumoto N. Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum. Mutat. 2017, 38, 637–648. 10.1002/humu.23200. [DOI] [PubMed] [Google Scholar]
- Stazi G.; Taglieri L.; Nicolai A.; Romanelli A.; Fioravanti R.; Morrone S.; Sabatino M.; Ragno R.; Taurone S.; Nebbioso M.; Carletti R.; Artico M.; Valente S.; Scarpa S.; Mai A. Dissecting the role of novel EZH2 inhibitors in primary glioblastoma cell cultures: effects on proliferation, epithelial-mesenchymal transition, migration, and on the pro-inflammatory phenotype. Clin. Epigenet. 2019, 11, 173. 10.1186/s13148-019-0763-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wassef M.; Margueron R. The multiple facets of PRC2 alterations in cancers. J. Mol. Biol. 2017, 429, 1978–1993. 10.1016/j.jmb.2016.10.012. [DOI] [PubMed] [Google Scholar]
- Comet I.; Riising E. M.; Leblanc B.; Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 2016, 16, 803–810. 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Qi J.; Reyes J. M.; Li L.; Rao P. K.; Li F.; Lin C. Y.; Perry J. A.; Lawlor M. A.; Federation A.; De Raedt T.; Li Y. Y.; Liu Y.; Duarte M. A.; Zhang Y.; Herter-Sprie G. S.; Kikuchi E.; Carretero J.; Perou C. M.; Reibel J. B.; Paulk J.; Bronson R. T.; Watanabe H.; Brainson C. F.; Kim C. F.; Hammerman P. S.; Brown M.; Cichowski K.; Long H.; Bradner J. E.; Wong K. K. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discovery 2016, 6, 1006–1021. 10.1158/2159-8290.CD-16-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe M. T.; Graves A. P.; Ganji G.; Diaz E.; Halsey W. S.; Jiang Y.; Smitheman K. N.; Ott H. M.; Pappalardi M. B.; Allen K. E.; Chen S. B.; Della Pietra A. 3rd.; Dul E.; Hughes A. M.; Gilbert S. A.; Thrall S. H.; Tummino P. J.; Kruger R. G.; Brandt M.; Schwartz B.; Creasy C. L. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 2989–2994. 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap D. B.; Chu J.; Berg T.; Schapira M.; Cheng S. W.; Moradian A.; Morin R. D.; Mungall A. J.; Meissner B.; Boyle M.; Marquez V. E.; Marra M. A.; Gascoyne R. D.; Humphries R. K.; Arrowsmith C. H.; Morin G. B.; Aparicio S. A. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 2011, 117, 2451–2459. 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle T. J.; Knutson S. K.; Jin L.; Kuntz K. W.; Pollock R. M.; Richon V. M.; Copeland R. A.; Scott M. P. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett. 2011, 585, 3011–3014. 10.1016/j.febslet.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Majer C. R.; Jin L.; Scott M. P.; Knutson S. K.; Kuntz K. W.; Keilhack H.; Smith J. J.; Moyer M. P.; Richon V. M.; Copeland R. A.; Wigle T. J. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012, 586, 3448–3451. 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- Morin R. D.; Johnson N. A.; Severson T. M.; Mungall A. J.; An J.; Goya R.; Paul J. E.; Boyle M.; Woolcock B. W.; Kuchenbauer F.; Yap D.; Humphries R. K.; Griffith O. L.; Shah S.; Zhu H.; Kimbara M.; Shashkin P.; Charlot J. F.; Tcherpakov M.; Corbett R.; Tam A.; Varhol R.; Smailus D.; Moksa M.; Zhao Y.; Delaney A.; Qian H.; Birol I.; Schein J.; Moore R.; Holt R.; Horsman D. E.; Connors J. M.; Jones S.; Aparicio S.; Hirst M.; Gascoyne R. D.; Marra M. A. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C.; Chagraoui J.; Krosl J.; Gendron P.; Wilhelm B.; Lemieux S.; Boucher G.; Chagnon P.; Drouin S.; Lambert R.; Rondeau C.; Bilodeau A.; Lavallee S.; Sauvageau M.; Hebert J.; Sauvageau G. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012, 26, 651–656. 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T.; Chase A. J.; Score J.; Hidalgo-Curtis C. E.; Bryant C.; Jones A. V.; Waghorn K.; Zoi K.; Ross F. M.; Reiter A.; Hochhaus A.; Drexler H. G.; Duncombe A.; Cervantes F.; Oscier D.; Boultwood J.; Grand F. H.; Cross N. C. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010, 42, 722–726. 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Martin M. C.; Zeng G.; Yu J.; Schiltz G. E. Small molecule approaches for targeting the polycomb repressive complex 2 (PRC2) in Cancer. J. Med. Chem. 2020, 63, 15344–15370. 10.1021/acs.jmedchem.0c01344. [DOI] [PubMed] [Google Scholar]
- Dockerill M.; Gregson C.; O'Donovan D. H. Targeting PRC2 for the treatment of cancer: an updated patent review (2016 - 2020). Expert Opin. Ther. Pat. 2021, 31, 119–135. 10.1080/13543776.2021.1841167. [DOI] [PubMed] [Google Scholar]
- Fioravanti R.; Stazi G.; Zwergel C.; Valente S.; Mai A. Six years (2012–2018) of researches on catalytic EZH2 inhibitors: the boom of the 2-pyridone compounds. Chem. Rec. 2018, 18, 1818–1832. 10.1002/tcr.201800091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson S. K.; Warholic N. M.; Wigle T. J.; Klaus C. R.; Allain C. J.; Raimondi A.; Porter Scott M.; Chesworth R.; Moyer M. P.; Copeland R. A.; Richon V. M.; Pollock R. M.; Kuntz K. W.; Keilhack H. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 7922–7927. 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson S. K.; Kawano S.; Minoshima Y.; Warholic N. M.; Huang K. C.; Xiao Y.; Kadowaki T.; Uesugi M.; Kuznetsov G.; Kumar N.; Wigle T. J.; Klaus C. R.; Allain C. J.; Raimondi A.; Waters N. J.; Smith J. J.; Porter-Scott M.; Chesworth R.; Moyer M. P.; Copeland R. A.; Richon V. M.; Uenaka T.; Pollock R. M.; Kuntz K. W.; Yokoi A.; Keilhack H. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 2014, 13, 842–854. 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- Hoy S. M. Tazemetostat: first approval. Drugs 2020, 80, 513–521. 10.1007/s40265-020-01288-x. [DOI] [PubMed] [Google Scholar]
- Bisserier M.; Wajapeyee N. Mechanisms of resistance to EZH2 inhibitors in diffuse large B-cell lymphomas. Blood 2018, 131, 2125–2137. 10.1182/blood-2017-08-804344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. Y.; Wang S. Allosteric inactivation of polycomb repressive complex 2 (PRC2) by inhibiting its adapter protein: embryonic ectodomain development (EED). J. Med. Chem. 2017, 60, 2212–2214. 10.1021/acs.jmedchem.7b00287. [DOI] [PubMed] [Google Scholar]
- ClinicalTrial NCT02900651. https://www.clinicaltrials.gov/ct2/show/NCT02900651 (16/06/2021).
- Dong H.; Liu S.; Zhang X.; Chen S.; Kang L.; Chen Y.; Ma S.; Fu X.; Liu Y.; Zhang H.; Zou B. An allosteric PRC2 inhibitor targeting EED suppresses tumor progression by modulating the immune response. Cancer Res. 2019, 79, 5587–5596. 10.1158/0008-5472.CAN-19-0428. [DOI] [PubMed] [Google Scholar]
- Ciferri C.; Lander G. C.; Maiolica A.; Herzog F.; Aebersold R.; Nogales E. Molecular architecture of human polycomb repressive complex 2. eLife 2012, 1, e00005 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinath V.; Faini M.; Poepsel S.; Reif D.; Feng X. A.; Stjepanovic G.; Aebersold R.; Nogales E. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 2018, 359, 940–944. 10.1126/science.aar5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N. V.; Pei X. Y.; Zhang W.; Sparkes M.; Vicente-Garcia J.; Pratap J. V.; McLaughlin S. H.; Ben-Shahar T. R.; Verreault A.; Luisi B. F.; Laue E. D. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 2008, 16, 1077–1085. 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges F. W.; Prusty A. B.; Faty M.; Stutzer A.; Lingaraju G. M.; Aiwazian J.; Sack R.; Hess D.; Li L.; Zhou S.; Bunker R. D.; Wirth U.; Bouwmeester T.; Bauer A.; Ly-Hartig N.; Zhao K.; Chan H.; Gu J.; Gut H.; Fischle W.; Muller J.; Thoma N. H. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 2011, 42, 330–341. 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Justin N.; Zhang Y.; Tarricone C.; Martin S. R.; Chen S.; Underwood E.; De Marco V.; Haire L. F.; Walker P. A.; Reinberg D.; Wilson J. R.; Gamblin S. J. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016, 7, 11316. 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L.; Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 2015, 350, aac4383 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.; Xing X.; Hu M.; Zhang Y.; Liu P.; Chai J. Structural basis of EZH2 recognition by EED. Structure 2007, 15, 1306–1315. 10.1016/j.str.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Xiao B.; Wilson J. R.; Gamblin S. J. SET domains and histone methylation. Curr. Opin. Struct. Biol. 2003, 13, 699–705. 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Brooun A.; Gajiwala K. S.; Deng Y. L.; Liu W.; Bolanos B.; Bingham P.; He Y. A.; Diehl W.; Grable N.; Kung P. P.; Sutton S.; Maegley K. A.; Yu X.; Stewart A. E. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun. 2016, 7, 11384. 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.; Zhang Y.; Xu R. M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003, 17, 1823–1828. 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W.; Wang Y.; Jacobs S. A.; Kim Y.; Allis C. D.; Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Cioaba M. A.; Min J. Structure and function of histone methylation binding proteins. Biochem. Cell Biol. 2009, 87, 93–105. 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- Kim W.; Bird G. H.; Neff T.; Guo G.; Kerenyi M. A.; Walensky L. D.; Orkin S. H. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat. Chem. Biol. 2013, 9, 643–650. 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X.; Chen L.; Jiao L.; Jiang X.; Lian F.; Lu J.; Zhu K.; Du D.; Liu J.; Ding H.; Zhang N.; Shen J.; Zheng M.; Chen K.; Liu X.; Jiang H.; Luo C. Astemizole arrests the proliferation of cancer cells by disrupting the EZH2-EED interaction of polycomb repressive complex 2. J. Med. Chem. 2014, 57, 9512–9521. 10.1021/jm501230c. [DOI] [PubMed] [Google Scholar]
- Janssens M. M. Astemizole. A nonsedating antihistamine with fast and sustained activity. Clin Rev. Allergy 1993, 11, 35–63. 10.1007/BF02802293. [DOI] [PubMed] [Google Scholar]
- Montgomery N. D.; Yee D.; Montgomery S. A.; Magnuson T. Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J. Mol. Biol. 2007, 374, 1145–1157. 10.1016/j.jmb.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Gao S.; Li J.; Liu D.; Sheng C.; Yao C.; Jiang W.; Wu J.; Chen S.; Huang W. Wedelolactone disrupts the interaction of EZH2-EED complex and inhibits PRC2-dependent cancer. Oncotarget 2015, 6, 13049–13059. 10.18632/oncotarget.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D.; Xu D.; Zhu L.; Stazi G.; Zwergel C.; Liu Y.; Luo Z.; Li Y.; Zhang Y.; Zhu K.; Ding Y.; Liu J.; Fan S.; Zhao K.; Zhang N.; Kong X.; Jiang H.; Chen K.; Zhao K.; Valente S.; Min J.; Duan W.; Luo C. Structure-guided development of small-molecule PRC2 inhibitors targeting EZH2-EED interaction. J. Med. Chem. 2021, 64, 8194–8207. 10.1021/acs.jmedchem.0c02261. [DOI] [PubMed] [Google Scholar]
- Barnash K. D.; Lamb K. N.; Stuckey J. I.; Norris J. L.; Cholensky S. H.; Kireev D. B.; Frye S. V.; James L. I. Chromodomain ligand optimization via target-class directed combinatorial repurposing. ACS Chem. Biol. 2016, 11, 2475–2483. 10.1021/acschembio.6b00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnash K. D.; The J.; Norris-Drouin J. L.; Cholensky S. H.; Worley B. M.; Li F.; Stuckey J. I.; Brown P. J.; Vedadi M.; Arrowsmith C. H.; Frye S. V.; James L. I. Discovery of peptidomimetic ligands of EED as allosteric inhibitors of PRC2. ACS Comb. Sci. 2017, 19, 161–172. 10.1021/acscombsci.6b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Zhang H.; Zhang M.; Zhao M.; Feng L.; Luo X.; Gao Z.; Huang Y.; Ardayfio O.; Zhang J. H.; Lin Y.; Fan H.; Mi Y.; Li G.; Liu L.; Feng L.; Luo F.; Teng L.; Qi W.; Ottl J.; Lingel A.; Bussiere D. E.; Yu Z.; Atadja P.; Lu C.; Li E.; Gu J.; Zhao K. Discovery and molecular basis of a diverse set of polycomb repressive complex 2 inhibitors recognition by EED. PLoS One 2017, 12, e0169855 10.1371/journal.pone.0169855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Gu X. J.; Hao Q.; Fan H.; Li L.; Zhou S.; Zhao K.; Chan H. M.; Wang Y. K. A liquid chromatography/mass spectrometry-based generic detection method for biochemical assay and hit discovery of histone methyltransferases. Anal. Biochem. 2013, 443, 214–221. 10.1016/j.ab.2013.08.029. [DOI] [PubMed] [Google Scholar]
- Read J. A.; Tart J.; Rawlins P. B.; Gregson C.; Jones K.; Gao N.; Zhu X.; Tomlinson R.; Code E.; Cheung T.; Chen H.; Kawatkar S. P.; Bloecher A.; Bagal S.; O’Donovan D. H.; Robinson J. Rapid identification of novel allosteric PRC2 inhibitors. ACS Chem. Biol. 2019, 14, 2134–2140. 10.1021/acschembio.9b00468. [DOI] [PubMed] [Google Scholar]
- Chan H. M.; Xingnian F. U.; Gu X.-j. J.; Huang Y.; Li L.; Mi Y.; Qi W.; Sendzik M.; Sun Y.; Wang L.. Triazolopyridine compounds and uses thereof. WO 2017221092A1, 2017.
- Lingel A.; Sendzik M.; Huang Y.; Shultz M. D.; Cantwell J.; Dillon M. P.; Fu X.; Fuller J.; Gabriel T.; Gu J.; Jiang X.; Li L.; Liang F.; McKenna M.; Qi W.; Rao W.; Sheng X.; Shu W.; Sutton J.; Taft B.; Wang L.; Zeng J.; Zhang H.; Zhang M.; Zhao K.; Lindvall M.; Bussiere D. E. Structure-guided design of EED binders allosterically inhibiting the epigenetic polycomb repressive complex 2 (PRC2) methyltransferase. J. Med. Chem. 2017, 60, 415–427. 10.1021/acs.jmedchem.6b01473. [DOI] [PubMed] [Google Scholar]
- Schultes S.; et al. Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discovery Today: Technol. 2010, 7, e157 10.1016/j.ddtec.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Leeson P. D.; Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discovery 2007, 6, 881–890. 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Zhang J.; Yu Z.; Zhang H.; Wang Y.; Lingel A.; Qi W.; Gu J.; Zhao K.; Shultz M. D.; Wang L.; Fu X.; Sun Y.; Zhang Q.; Jiang X.; Zhang J.; Zhang C.; Li L.; Zeng J.; Feng L.; Zhang C.; Liu Y.; Zhang M.; Zhang L.; Zhao M.; Gao Z.; Liu X.; Fang D.; Guo H.; Mi Y.; Gabriel T.; Dillon M. P.; Atadja P.; Oyang C. Discovery of first-in-class, potent, and orally bioavailable embryonic ectoderm development (EED) inhibitor with robust anticancer efficacy. J. Med. Chem. 2017, 60, 2215–2226. 10.1021/acs.jmedchem.6b01576. [DOI] [PubMed] [Google Scholar]
- Qi W.; Zhao K.; Gu J.; Huang Y.; Wang Y.; Zhang H.; Zhang M.; Zhang J.; Yu Z.; Li L.; Teng L.; Chuai S.; Zhang C.; Zhao M.; Chan H.; Chen Z.; Fang D.; Fei Q.; Feng L.; Feng L.; Gao Y.; Ge H.; Ge X.; Li G.; Lingel A.; Lin Y.; Liu Y.; Luo F.; Shi M.; Wang L.; Wang Z.; Yu Y.; Zeng J.; Zeng C.; Zhang L.; Zhang Q.; Zhou S.; Oyang C.; Atadja P.; Li E. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 2017, 13, 381–388. 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- Potjewyd F.; Turner A. W.; Beri J.; Rectenwald J. M.; Norris-Drouin J. L.; Cholensky S. H.; Margolis D. M.; Pearce K. H.; Herring L. E.; James L. I. Degradation of Polycomb Repressive Complex 2 with an EED-Targeted Bivalent Chemical Degrader. Cell Chem. Biol. 2020, 27, 47–56. 10.1016/j.chembiol.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rej R. K.; Wang C.; Lu J.; Wang M.; Petrunak E.; Zawacki K. P.; McEachern D.; Fernandez-Salas E.; Yang C. Y.; Wang L.; Li R.; Chinnaswamy K.; Wen B.; Sun D.; Stuckey J.; Zhou Y.; Chen J.; Tang G.; Wang S. EEDi-5285: An exceptionally potent, efficacious, and orally active small-molecule inhibitor of embryonic ectoderm development. J. Med. Chem. 2020, 63, 7252–7267. 10.1021/acs.jmedchem.0c00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin M. L.; Pliushchev M. A.; Li H. Q.; Torrent M.; Dietrich J. D.; Jakob C. G.; Zhu H.; Zhao H.; Wang Y.; Ji Z.; Clark R. F.; Sarris K. A.; Selvaraju S.; Shaw B.; Algire M. A.; He Y.; Richardson P. L.; Sweis R. F.; Sun C.; Chiang G. G.; Michaelides M. R. SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding. Bioorg. Med. Chem. Lett. 2017, 27, 1576–1583. 10.1016/j.bmcl.2017.02.030. [DOI] [PubMed] [Google Scholar]
- He Y.; Selvaraju S.; Curtin M. L.; Jakob C. G.; Zhu H.; Comess K. M.; Shaw B.; The J.; Lima-Fernandes E.; Szewczyk M. M.; Cheng D.; Klinge K. L.; Li H. Q.; Pliushchev M.; Algire M. A.; Maag D.; Guo J.; Dietrich J.; Panchal S. C.; Petros A. M.; Sweis R. F.; Torrent M.; Bigelow L. J.; Senisterra G.; Li F.; Kennedy S.; Wu Q.; Osterling D. J.; Lindley D. J.; Gao W.; Galasinski S.; Barsyte-Lovejoy D.; Vedadi M.; Buchanan F. G.; Arrowsmith C. H.; Chiang G. G.; Sun C.; Pappano W. N. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 2017, 13, 389–395. 10.1038/nchembio.2306. [DOI] [PubMed] [Google Scholar]
- Liu B.; Huang Y.; Mao L.; Wang L.; Waykole L. M.; Zhang L.. Crystalline forms of a triazolopyrimidine compound. WO 2017219948A1, 2017.
- Huang D.; Tian S.; Qi Y.; Zhang J. Z. H. Binding modes of small-molecule inhibitors to the EED pocket of PRC2. ChemPhysChem 2020, 21, 263–271. 10.1002/cphc.201900903. [DOI] [PubMed] [Google Scholar]
- Hsu J. H.; Rasmusson T.; Robinson J.; Pachl F.; Read J.; Kawatkar S.; O'Donovan D. H.; Bagal S.; Code E.; Rawlins P.; Argyrou A.; Tomlinson R.; Gao N.; Zhu X.; Chiarparin E.; Jacques K.; Shen M.; Woods H.; Bednarski E.; Wilson D. M.; Drew L.; Castaldi M. P.; Fawell S.; Bloecher A. EED-targeted PROTACs degrade EED, EZH2, and SUZ12 in the PRC2 complex. Cell Chem. Biol. 2020, 27, 41–46. 10.1016/j.chembiol.2019.11.004. [DOI] [PubMed] [Google Scholar]
- O'Donovan D. H.; Gregson C.; Packer M. J.; Greenwood R.; Pike K. G.; Kawatkar S.; Bloecher A.; Robinson J.; Read J.; Code E.; Hsu J. H.; Shen M.; Woods H.; Barton P.; Fillery S.; Williamson B.; Rawlins P. B.; Bagal S. K. Free energy perturbation in the design of EED ligands as inhibitors of polycomb repressive complex 2 (PRC2) methyltransferase. Bioorg. Med. Chem. Lett. 2021, 39, 127904. 10.1016/j.bmcl.2021.127904. [DOI] [PubMed] [Google Scholar]
- Misawa K.; Yamaotsu N.; Hirono S. Identification of novel EED-EZH2 PPI inhibitors using an in silico fragment mapping method. J. Comput.-Aided Mol. Des. 2021, 35, 601–611. 10.1007/s10822-021-00378-6. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Du D. H.; Wang J.; Cai X. Q.; Deng A. X.; Nosjean O.; Boutin J. A.; Renard P.; Yang D. H.; Luo C.; Wang M. W. Identification of catalytic and non-catalytic activity inhibitors against PRC2-EZH2 complex through multiple high-throughput screening campaigns. Chem. Biol. Drug Des. 2020, 96, 1024–1051. 10.1111/cbdd.13702. [DOI] [PubMed] [Google Scholar]
- Ma A.; Stratikopoulos E.; Park K. S.; Wei J.; Martin T. C.; Yang X.; Schwarz M.; Leshchenko V.; Rialdi A.; Dale B.; Lagana A.; Guccione E.; Parekh S.; Parsons R.; Jin J. Discovery of a first-in-class EZH2 selective degrader. Nat. Chem. Biol. 2020, 16, 214–222. 10.1038/s41589-019-0421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanelli A.; Stazi G.; Fioravanti R.; Zwergel C.; Di Bello E.; Pomella S.; Perrone C.; Battistelli C.; Strippoli R.; Tripodi M.; Del Bufalo D.; Rota R.; Trisciuoglio D.; Mai A.; Valente S. Design of first-in-class dual EZH2/HDAC inhibitor: biochemical activity and biological evaluation in cancer cells. ACS Med. Chem. Lett. 2020, 11, 977–983. 10.1021/acsmedchemlett.0c00014. [DOI] [PMC free article] [PubMed] [Google Scholar]