Abstract
The Smads are a family of nine related proteins which function as signaling intermediates for the transforming growth factor β (TGF-β) superfamily of ligands. To discern the in vivo functions of one of these Smads, Smad3, we generated mice harboring a targeted disruption of this gene. Smad3 null mice, although smaller than wild-type littermates, are viable, survive to adulthood, and exhibit an early phenotype of forelimb malformation. To study the cellular functions of Smad3, we generated Smad3 null mouse embryonic fibroblasts (MEFs) and dermal fibroblasts. We demonstrate that null MEFs have lost the ability to form Smad-containing DNA binding complexes and are unable to induce transcription from the TGF-β-responsive promoter construct, p3TP-lux. Using the primary dermal fibroblasts, we also demonstrate that Smad3 is integral for induction of endogenous plasminogen activator inhibitor 1. We subsequently demonstrate that Smad3 null MEFs are partially resistant to TGF-β’s antiproliferative effect, thus firmly establishing a role for Smad3 in TGF-β-mediated growth inhibition. We next examined cells in which Smad3 is most highly expressed, specifically cells of immune origin. Although no specific developmental defect was detected in the immune system of the Smad3 null mice, a functional defect was observed in the ability of TGF-β to inhibit the proliferation of splenocytes activated by specific stimuli. In addition, primary splenocytes display defects in TGF-β-mediated repression of cytokine production. These data, taken together, establish a role for Smad3 in mediating the antiproliferative effects of TGF-β and implicate Smad3 as a potential effector for TGF-β in modulating immune system function.
Transforming growth factor β (TGF-β) is a multifunctional polypeptide hormone which has diverse effects on a variety of cell types to regulate many complex multicellular systems (46). The complexity and diversity of TGF-β’s function is demonstrated through its multiple roles in immune system suppression, wound healing, fibrosis, development, and oncogenesis. Many of these global effects of TGF-β stem from its ability to regulate cellular proliferation, differentiation, and gene expression (38). One of the most studied aspects of TGF-β function is its ability to inhibit the proliferation of many different cell types, including cells of epithelial, endothelial, neuronal, hematopoietic, and lymphoid origins (46, 31).
These effects of TGF-β are mediated through its interaction with cell surface receptors. By binding to its type I and type II serine/threonine kinase receptors, TGF-β induces the phosphorylation and activation of the type I receptor by the type II receptor (57, 58). The type I receptor kinase can then phosphorylate cytoplasmic substrates, including members of the Smad family of proteins, which function as intermediates in the signaling pathways for the TGF-β superfamily of ligands (3, 4, 12, 18, 39). Originally identified in genetic screens for TGF-β effectors in Drosophila (49) and Caenorhabditis elegans (47), the mammalian Smad family now consists of nine structurally related proteins, Smad1 to Smad9. The identification and characterization of these proteins has provided valuable insights into the early events involved in TGF-β-mediated signal transduction.
The highly related Smad2 and Smad3 serve as substrates for the type I TGF-β receptor kinase (14, 30, 36, 42, 53, 62, 65). Upon phosphorylation, these two Smads bind to their common partner, Smad4, to form Smad2-Smad4 and Smad3-Smad4 complexes. These complexes then translocate to the nucleus (1, 29, 33, 41, 66). Clues to the nuclear function of these Smad complexes came from studies describing an intrinsic transcriptional activity of the C-terminal domain of the Smads (29, 59). Subsequently, overexpression of particular combinations of Smads was shown to activate transcription from a number of TGF-β-responsive promoters, including the plasminogen activator inhibitor 1 (PAI-1) promoter and the reporter construct 3TP-lux (11, 29, 65).
The role of the Smads as putative transcription factors was strengthened by the finding that Smad3-Smad4 complexes and the Drosophila Mad are sequence-specific DNA binding proteins which on binding DNA can activate transcription (11, 25, 63, 64). In addition to a direct DNA binding activity, the Smads can be targeted to specific promoter sequences through their interaction with other transcription factors, as demonstrated by the finding that Smad2-Smad4 complexes bind to the transcription factor FAST-1 in response to activin and TGF-β (7, 34). In addition, recent studies have implicated a functional interaction between the Smad3-Smad4 complex and the AP1 family of transcription factors (32, 63, 67).
Apace with the rapid development of the understanding of the Smads on a biochemical level, the role of the Smads in development and diseases is beginning to be understood. Recently, mouse models for both Smad2 and Smad4 function have been described (43, 52, 55, 61). Mice with homozygous targeted disruptions of these genes are embryonic lethal at day 9.5 and days 6.5 to 8.5, respectively. Thus, these Smads play critical, nonredundant roles in early embryonic development. The early embryonic lethality of these mice, however, renders the functional analysis of these molecules in the adult animals impossible in this system and makes their study on a cellular level difficult.
In humans, the role of Smad2 and Smad4 as tumor suppressor genes is now well established (6, 14–16, 40, 44, 45, 48). Concurrent with the identification of the Smads through genetic screens, Smad4 was identified as a tumor suppressor gene, which is deleted in about 50% of pancreatic carcinomas. In addition to pancreatic cancers, Smad4 mutations have also been discovered in breast, ovary, head and neck, esophagus, colon, and lung cancers. Not only are Smad4 mutations found in spontaneous cancers, but recent reports show that inherited juvenile colon cancer can derive from the inheritance of a single mutant Smad4 allele (19). In addition, Smad2 is mutated in several types of cancers, including colon cancers and head and neck cancers (14, 44). To date, Smad3 has not been reported to be mutated in human cancers (2, 45). These data, together with their role as intermediates in the TGF-β signaling pathway, clearly implicate Smad2 and Smad4 as playing an important function in cell growth regulation.
The cellular functions of the Smads have largely been inferred from the occurrence of mutations in human diseases and from cellular studies employing the use of Smad dominant negatives and Smad overexpression in Smad-deficient cell lines which likely harbor additional genetic lesions. Thus, the physiological functions of Smad3, particularly its potential involvement in mediating the TGF-β antiproliferative effect, remain speculative. To address the biological functions of Smad3, we generated mice harboring a targeted disruption of the Smad3 gene. Unlike the Smad2 and Smad4 null mice, Smad3 null mice are viable and survive to adulthood, demonstrating distinct roles for the three Smad proteins during mouse development. In addition, Smad3 null mice are smaller than wild-type littermates and have an incompletely penetrant joint formation abnormality. At the cellular level, we focused our study initially on defining the role of Smad3 in TGF-β signal transduction in the mouse embryonic fibroblast (MEF) and dermal fibroblast model systems. Here we show that Smad3 is required for activation of a TGF-β-responsive promoter, 3TP-lux, and the endogenous PAI-1 gene and, more importantly, acts as an integral effector of TGF-β-mediated inhibition of cellular proliferation. We next focused on the cell types with highest Smad3 expression, specifically cells of lymphoid origin, and found that under specific conditions, the antiproliferative effects of TGF-β on isolated Smad3 null splenocytes are lost. In addition, we found that the inhibition of anti-CD3 (αCD3)-stimulated cytokine production by TGF-β in primary splenocytes is markedly blunted due to the absence of Smad3. Taken together, these findings implicate Smad3 as a critical effector in TGF-β-mediated inhibition of cellular proliferation and a potential effector for TGF-β regulation of immune system function.
MATERIALS AND METHODS
Smad3 gene disruption.
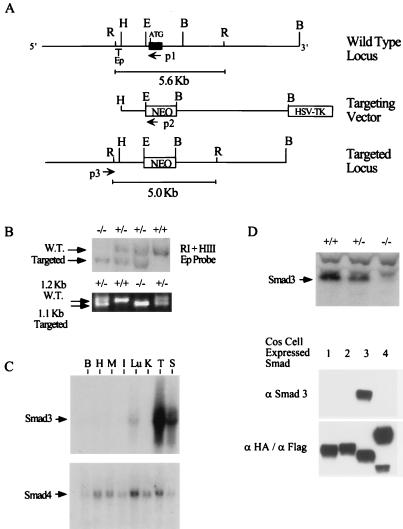
The Smad3 gene was isolated from a 129/sv mouse genomic library by using the 5′ end of the human Smad3 cDNA as a probe. An isolated 15-kb genomic clone was used for the creation of a Smad3 targeting vector. Briefly, a 1.0-kb EheI-HindIII fragment was cloned into the XhoI site of the vector pPNT (54). A 6.0-kb BamHI fragment was next cloned into the resulting construct. This produced a targeting vector which, when inserted into the genome, replaces the sequence between EheI and BamHI with a neomycin expression cassette, as diagrammed in Fig. 1A. The resulting targeting vector was linearized with HindIII and electroporated into 129/sv embryonic stem (ES) cells. Screening of neomycin-resistant clones was performed by PCR with the following primers: the common primer (P3; GTC TTT GAG GCC CGT TTT CTG C), a primer from the targeted sequence (P1; CTG GGG TGG TAA TGC ACT TGG), and a primer in the PGK promoter (P2; CAT GCT CCA GAC TGC CTT GGG). PCR of the wild-type allele results in a 1.2-kb product. PCR of the targeted allele results in a 1.1-kb product. Positive clones were confirmed by Southern blotting of EcoRI-digested genomic DNA probed with an EcoRI-HindIII fragment immediately adjacent to the sequences used in the targeting vector. The wild-type allele of Smad3 produces a 5.6-kb fragment. The targeted allele produces a 5.0-kb fragment.
FIG. 1.
Targeted disruption of Smad3. (A) Smad3 genomic structure and targeting strategy. The Smad3 genomic clone used for the creation of the targeting vector is diagrammed. The black box denotes the first exon of Smad3. ATG denotes the initiating methionine. The first exon coding sequence was replaced by a neomycin expression cassette (NEO), creating an EcoRI digest size difference between the wild-type and targeted loci. Ep denotes the EcoRI-HindIII DNA fragment used as a probe for Southern blotting. P1, P2, and P3 denote the locations of primers used for PCR screening. Restriction sites are abbreviated as follows: R, EcoRI; H, HindIII; E, EheI; B, BamHI. HSV-TK, herpes simplex virus thymidine kinase. (B) Southern and PCR detection of the targeted allele. The targeted allele can be distinguished from the wild type (W.T.) both by Southern blotting of EcoRI-digested genomic DNA with the Ep probe and by PCR using the primers indicated in panel A. (C) Northern blot analysis of Smad3 and Smad4. Northern analyses were performed on RNA derived from multiple tissues of an adult (2-month-old) C57BL/6 mouse and a 3′ untranslated sequence probe for Smad3 and a coding-sequence probe for Smad4. Organs are abbreviated as follows: B, brain; H, heart; M, skeletal muscle; I, small intestine; Lu, lung; K, kidney; T, thymus; S, spleen. (D) Western blot analysis of Smad3 expression. The top panel shows Western analysis using an antibody created against a peptide in the central linker domain of Smad3 on thymic protein extract from wild-type, heterozygous, and knockout mice. The bottom two panels demonstrate the specificity of this antibody among overexpressed Smad family members. HA-tagged Smad1, Smad2, and Smad4 and Flag-tagged Smad3 were overexpressed in COS cells from which protein extract were isolated and used for Western analysis. Identical blots were probed with the Smad3-specific antibody (middle panel) and a mixture of αHA and αFlag (bottom panel). In all panels of all figures, +/+ denotes Smad3 wild type, +/− denotes Smad3 heterozygous, and −/− denotes Smad3 null.
Primary fibroblast and immune cell culture.
Primary fibroblasts were cultured from day 14 embryos. Embryos were mechanically disrupted by passage through an 18-gauge needle and plated on gelatin-coated 10-cm-diameter plates in Dulbecco modified Eagle medium (DMEM) with 20% heat-inactivated fetal bovine serum (FBS) and penicillin-streptomycin (P-S) (Gibco BRL, Gaithersburg, Md.). Confluent cells were trypsinized and further carried in medium containing 10% FBS. In all experiments, compared wild-type and null cells represent littermate embryos at the same passage number.
Primary dermal fibroblasts were isolated from 2-day-old mice. Trunk skin was removed, washed three times in phosphate-buffered saline (PBS) containing kanamycin, amphotericin, penicillin, and streptomycin (KAPS), and incubated overnight at 4°C in 0.25% trypsin (Worthington Biochemical, Freehold, N.J.) in PBS-KAPS. The skins were then incubated at 37°C for 20 min. The trypsin was next neutralized with 20% FBS, and the skins were then washed in DMEM–10% FBS. The trypsinized skins were next placed in individual 10-cm-diameter dishes, and the epidermis was peeled off and discarded. The resulting dermis layers were mechanically dissociated, and 10 ml of DMEM–10% FBS–P-S was added to each dish. Cells were then incubated at 37°C in 5% CO2 until dermal fibroblasts became confluent (3 to 4 days). The dermal fibroblasts were carried in DMEM–10% FBS–P-S and genotyped, and passage 5 cells were used in the PAI-1 assay.
Primary splenocytes, thymocytes, and purified splenic B and T cells were generated from the spleens of 2- to 4-month-old mice. Lymphocytes were isolated by mechanical dissociation in the culture medium described below. Large debris was removed, and erythrocytes were lysed by complete resuspension of pelleted cells in 145 mM ammonium chloride–17 mM Tris (pH 7.5). Cells were subsequently washed in PBS and resuspended in their final culture medium for all experiments described: RPMI with the addition of 10% heat-inactivated FBS, P-S, and 0.1 mM β-mercaptoethanol.
Northern and Western blotting.
Northern blotting for Smad3 was performed on RNA prepared from adult C57BL/6 mouse organs by homogenizing the indicated tissues in Trizol reagent (Gibco BRL, Gaithersburg, Md.) as specified by the manufacturer. Ten micrograms of total RNA was resolved on a formaldehyde-agarose gel, which was subsequently transferred by capillary action to a nylon membrane (Hybond, Amersham Life Science) and visualized by methylene blue staining to confirm RNA loading and quality. Blots were probed with a mouse Smad3 cDNA probe created by random priming (Prime-It II, Stratagene, La Jolla, Calif.) of a HindIII-EcoRI fragment containing sequences entirely in the Smad3 3′ untranslated region. The membrane was subsequently reprobed with a human Smad4 cDNA probe containing the entire coding sequence for human Smad4.
For Western blotting, all cell and organ lysates were prepared in a Nonidet P-40-based lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 50 mM NaF, 0.5% Nonidet P-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 10 mM β-glycerophosphate, 0.2 mM sodium molybdate, protease inhibitors). For Western blotting of thymus tissue, whole thymus was homogenized in 1 ml of lysis buffer and equal protein amounts from animals of each genotype were analyzed. Western blotting of fibroblasts was performed by lysis of 106 fibroblasts from 10-cm-diameter tissue culture plates in 200 μl of lysis buffer and analyzing equal protein amounts for each genotype. Western analyses of equal amounts of protein extracts from splenic B cells purified by using Dynabeads- Mouse pan T (Dynal, Lake Success, N.Y.) and of splenic T cells purified by using mouse T-cell enrichment columns (R&D Systems, Minneapolis, Minn.) were performed with a Smad3-specific antibody. Western analyses of αCD3-stimulated splenocytes were performed on splenocytes isolated as described above and cultured at a density of 107 cells in 2 ml of medium in six-well tissue culture plates in the presence 5 μg of αCD3 (01081D; PharMingen, San Diego, Calif.) with and without 100 pM TGF-β for 24 (cyclin E cyclin-dependent kinase [Cdk2], and p27 western blots) or 48 h (retinoblastoma protein [Rb] Western blots). Cells were lysed in 200 μl of lysis buffer, and equal protein amounts were analyzed. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western analyses were performed with the following antibodies; αcyclin E (M-2), αCdk2 (M-2), αp27 (C-19), αp21 (C-19), αp15 (C-20), and αCDC25A (144), all from Santa Cruz Biotechnology, Inc., and αRb (14001A; PharMingen). Smad3 Western analyses were performed with a rabbit polyclonal antibody created against the Smad3-specific peptide DAGSPNLSPNPMSPAHNNLD. Crude rabbit serum was further purified on an antigen affinity column. The specificity of this antibody was assessed by Western analysis of equal protein amounts of COS cell extract in which the following tagged human Smads were individually overexpressed: Flag-tagged Smad1, Flag-tagged Smad2, hemagglutin epitope (HA)-tagged Smad3, and Flag-tagged Smad4. Cytomegalovirus promoter-driven constructs encoding the various Smads were transiently transfected into COS cells by using a standard DEAE-dextran protocol as previously described (10). Expression of the various Smads was confirmed by Western analysis of equal amounts of appropriately transfected COS extract, using a mixture of αHA (Boehringer Mannheim, Indianapolis, Ind.) and αFlag (Eastman Kodak, New Haven, Conn.) antibodies.
Cdk2 kinase assays.
Splenocytes were isolated and cultured as described above for Western blotting. Whole-cell lysates were generated after 24 h of TGF-β treatment in the same manner as for Western analysis. Cdk2 kinase activity immunoprecipitated with a polyclonal αCdk2 antibody (M-2; Santa Cruz Biotechnology) from equal protein amounts for each condition described was measured on the substrate histone H1 as previously described (10).
PAI-1 assay.
Dermal fibroblasts were plated at a density of 106 cells per 10-cm-diameter plate in the fibroblast culture medium described above and incubated overnight at 37°C. Cells were then incubated in methionine-free DMEM–0.5% FBS–P-S for 4 h and then treated with 100 pM TGF-β for 6 h. During the last 2 h of TGF-β treatment, the fibroblasts were labeled with [35S]methionine (100 μCi/ml). The cells were then washed with PBS and removed by lysis in three washes of 10 mM Tris-HCl (pH 8.0)–0.5% sodium deoxycholate–1 mM phenylmethylsulfonyl fluoride, and the resulting plate-bound extracellular matrix washed a final time with PBS. The amount of matrix associated PAI-1 was assessed by scraping the plates in SDS-PAGE loading buffer containing dithiothreitol and resolving the protein on an 10% polyacrylamide gel. Gels were subsequently dried, and autoradiography was performed.
Thymidine incorporation assays.
Fibroblasts of the indicated genotypes were plated at a density of 20,000 cells/well in six-well tissue culture plates in DMEM–10% FBS and incubated in the presence or absence of 100 pM TGF-β for 24 or 48 h as indicated. For the last 4 h of culture, 5 μCi of [3H]thymidine was added to the culture, and thymidine incorporation was assayed as previously described (10). For mixed wild-type and Smad3 null experiments, the indicated percentage of each cell type was plated in six-well plates to a total cell number of 20,000/ml. Thymidine incorporation was assayed after 48 h as described above.
Thymidine incorporation of splenocytes was performed on cells isolated as described above. Isolated splenocytes were plated at a density of 5 × 105 cells in 200 μl of medium in 24-well plates and stimulated with lipopolysaccharide (LPS; 10 μg/ml; Sigma, St. Louis, Mo.), anti-immunoglobulin M (αIgM; 5 μg/ml; Cappel, Durham, N.C.) and interleukin-4 (IL-4; 12.5 U/well; PharMingen) or αCD3 (2.5 μg/ml; PharMingen) in the presence or absence of 100 pM TGF-β and cultured for 48 h; 5 μCi of [3H]thymidine was added to the culture for the last 4 h. Thymidine incorporation was assayed by harvesting cells with a PHD cell harvester (Cambridge Technologies, Inc.).
Luciferase assays.
Fibroblasts (200,000/well) from each genotype were seeded into six-well tissue culture plates. Cells were transfected by using a standard DEAE-dextran transfection protocol as previously described (10) with the indicated amounts of DNAs (Fig. 4B). Cells were cotransfected with 0.25 μg of a cytomegalovirus-driven β-galactosidase reporter vector to normalize for transfection efficiency. Transfections with the expression and reporter plasmids used here have been previously described (63).
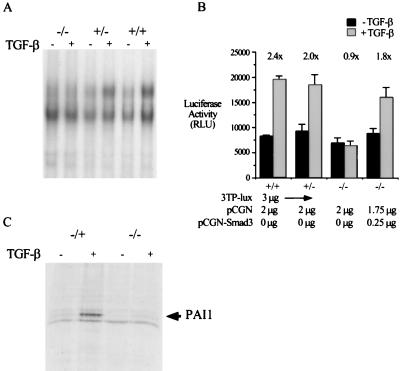
FIG. 4.
Smad3 is required for TGF-β-mediated Smad-containing DNA binding complex formation and activation of 3TP-Lux in primary MEFs and for TGF-β-mediated induction of the PAI-1 gene in primary dermal fibroblasts. (A) Loss of a Smad-containing DNA binding complex in the Smad3 null MEFS. EMSAs were performed with nuclear extract from MEFs of the indicated genotype, either treated with TGF-β for 30 min or untreated, and a probe derived from the TGF-β-responsive region of the promoter-reporter construct, p3TP-lux. The arrow indicates the TGF-β-inducible DNA binding complex. (B) Smad3 is required for induction of the p3TP-lux reporter construct. The indicated DNAs were transfected into MEFs of the indicated genotype. Twelve hours after transfection, the cells were treated with 100 pM TGF-β for an additional 24 h, and TGF-β-induced luciferase activity (relative luciferase units [RLU]) from this reporter construct was assayed. Bars represent the average luciferase activity of duplicate transfections in a single experiment; error bars represent the standard deviation. Fold induction by TGF-β is indicated over each set of bars. (C) Smad3 is an integral component of the TGF-β-mediated induction of the endogenous PAI-1 gene. Smad3 heterozygote and null primary dermal fibroblasts were treated with TGF-β for 8 h. The arrow represents [35S]methionine-labeled, extracellular matrix-associated PAI-1, assayed as described in Materials and Methods.
EMSAs.
For electrophoretic mobility shift assays (EMSAs), nuclear extracts prepared from 106 fibroblasts of the indicated genotype either treated or untreated with 100 pM TGF-β for 1 h were incubated with a probe derived from an NdeI-Sph1 fragment of 3TP-lux (63). Nuclear extract preparation and gel shift conditions were exactly as previously described (62).
RNase protection analysis of cytokine expression.
Primary splenocytes were isolated as described above. Splenocytes (7.5 × 106) were plated in 2 ml of medium and stimulated with 5 μg of αCD3 in the presence or absence of 100 pM TGF-β for 48 h in individual wells of a six-well plate. The cells were harvested, and total RNA was isolated (RNeasy; Qiagen, Santa Clarita, Calif.). Cytokine RNA levels were assessed by RNase protection assays using a RiboQuant multiprobe kit (45024K/mCK-1; PharMingen) on equal amounts of RNA (7.5 μg) for each culture condition as specified by the manufacturer. Equal amounts and quality of RNA were confirmed through the quantification of the protection fragments of two housekeeping genes provided in the multiprobe template set, L32 and GAPDH.
FACS analysis.
Fluorescence-activated cell sorting (FACS) analysis was performed on live splenocytes and thymocytes isolated as described above. Approximately 105 pelleted cells were resuspended in 100 μl of PBS with 5% heat-inactivated FBS with the inclusion of the antibodies indicated in each figure: phycoerythrin (PE)-αCD4 (01065B), fluorescein isothiocyanate (FITC)-αCD8 (01044D), and FITC-αB220 (01124D) (all from PharMingen), plus PE-αIgM and 7-amino actinomycin D (7AAD; Molecular Probes, Eugene, Oreg.). After 30 min, these cells were washed once in PBS–5% FBS and analyzed. The purity of the isolated splenic B and T cells used in the thymidine incorporation assays was assessed by FACS analysis with the use of PE-αB220 and FITC-labeled anti-T-cell receptor beta chain (01304D and 01125B, respectively; PharMingen). Viable cells were identified by exclusion of 7AAD (Molecular Probes) staining.
RESULTS
Smad3 null mice are viable.
To generate a targeted disruption of Smad3, we first screened a murine 129/sv genomic library with sequences in the amino terminus of Smad3 to obtain a 14-kb genomic clone. This clone contains the first exon of Smad3, including the initiating methionine and the first 69 amino acids. A targeting vector was created by replacing the first exon and part of the first intron with a PGK-neomycin expression cassette. Proper insertion of this targeting vector into the mouse genome removes the initiating ATG, making the production of full-length Smad3 impossible. In addition, this insertion does not disrupt any sequences 5′ to the RNA transcriptional start site (Fig. 1A).
Using standard ES cell technology, Smad3 mutant heterozygous 129 ES lines were generated by transfection of the described targeting vector. Initial screening for proper insertion in neomycin-resistant clones was determined by Southern blotting and PCR as indicated in Fig. 1B. Three percent of neomycin-resistant ES cell clones had a properly targeted Smad3 allele. These ES cells were then used to create 129-C57BL/6 chimeric founder mice. When bred to C57BL/6 females, mice generated from one of these lines transmitted the mutant Smad3 allele at a frequency of 50%, with 100% of offspring being derived from the 129 stem cells. Heterozygous mice from these matings were subsequently mated to produce Smad3 null mice. Smad3 null mice are born to F1 heterozygotes at a frequency of 20.7%, the same frequency as for wild-type mice (297 Heterozygote, 106 wild-type, and 103 KO knockout mice). The near Mendelian inheritance of wild-type and targeted Smad3 alleles suggests no embryonic lethality of the Smad3 null mice. Thus, in sharp contrast to the Smad2 and Smad4 deficiencies, Smad3 is not essential for embryonic development. This F2 generation of mice was used for the experiments described below.
To identify organs with highest Smad3 expression, we first performed multiple-tissue Northern analysis. Unlike Smad2 and Smad4, Smad3 has an expression pattern which varies with tissue types, with highest levels of expression in the spleen and thymus (Fig. 1C). Subsequently, the loss of Smad3 expression in the double-mutant animals was confirmed by Western analysis of thymus protein extracts, using a Smad3-specific antibody (Fig. 1D).
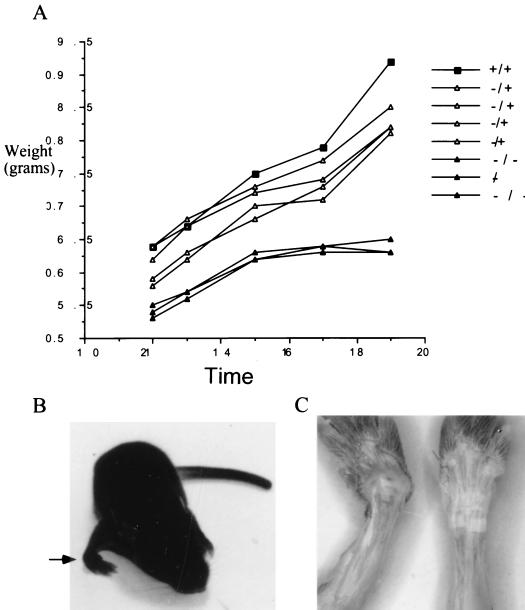
The first noticeable phenotype in these null animals is a decrease in the size and growth rate of young mice. As shown in Fig. 2A, Smad3 null mice are smaller than both wild-type and heterozygous littermates. An additional early phenotype, which occurs in approximately 31% (32 of 103) of null mice, is the presence of medially torqued forepaws (Fig. 2B and C), with a smaller percentage of mice with noticeably torqued hind limbs. Mice with this phenotype can have either one or more limbs affected. In addition, mice with severely affected limbs often develop kyphosis and display marked rib cage malformation often resulting in a concave indentation at the base of the sternum (data not shown). Interestingly, this phenotype is remarkably similar to that of mice expressing a transgenic dominant negative type II TGF-β receptor in bone (50), suggesting that the phenotype described here is intrinsic to the bone. In addition, the similarity between these two phenotypes suggests that the previously described TGF-β effects in bone development are at least partially mediated by Smad3.
FIG. 2.
Smad3 null mice are smaller than wild-type littermates and have an incompletely penetrant forepaw defect. (A) Mouse weights over time (days) in a single, representative litter. (B) Picture of the torqued-wrist defect (arrow) in a 14-day-old null mouse. (C) The skin was removed from the forelimbs of a 14-day-old Smad3 null mouse (left) and a wild-type littermate (right) to better show the severe bending of the forepaw wrist joint of the Smad3 null mouse.
TGF-β-mediated growth inhibition and gene responses are impaired in Smad3 null fibroblasts.
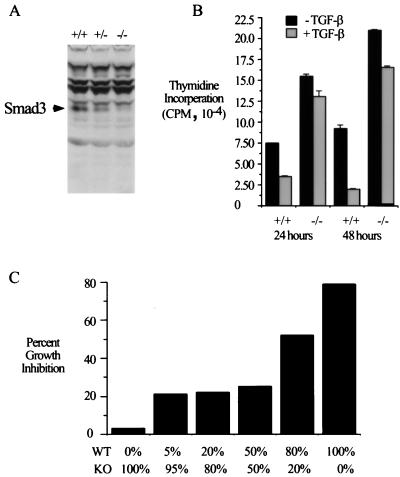
One functional aspect of Smad3 that we hoped to define through the generation of Smad3 null mice is its role in TGF-β-mediated inhibition of cellular proliferation. To test this, we isolated MEFs from both wild-type and Smad3 null mice. As shown in Fig. 3A, Smad3 expression can be detected in wild-type fibroblast lines but not in lines derived from Smad3 null embryos. Using these fibroblasts, we first determined the proliferative responses of these lines to TGF-β. As shown in Fig. 3B, the proliferation of wild-type fibroblasts is inhibited approximately 50% after 24 h and 80% after 48 h of TGF-β treatment. In null fibroblasts, this growth-inhibitory effect of TGF-β is largely lost. In addition, the basal proliferation rate of the null fibroblast lines is approximately twofold higher than that of the wild type. Similar results were obtained for two additional fibroblast lines of each genotype (data not shown). Thus, these results firmly establish an essential role for Smad3 in TGF-β-mediated inhibition of cellular proliferation. Interestingly, none of the known mediators of the growth-inhibitory effect of TGF-β appear to be functioning in fibroblasts. In these cells TGF-β does not alter p21, p15, or CDC25A protein levels, whereas p27 is undetectable (data not shown).
FIG. 3.
Smad3 is required for TGF-β-mediated growth inhibition in MEFs. (A) Primary MEFs were created from embryonic day 14 mice. Western blotting for Smad3 was performed to determine if these MEFs express Smad3. (B) Smad3 is required for TGF-β-mediated growth inhibition in primary MEFs. MEFs were assayed for TGF-β-mediated growth inhibition after 24 and 48 h of treatment by measurement of [3H]thymidine incorporation. Bars represent the average thymidine incorporation for triplicate wells for each growth condition. (C) TGF-β-mediated growth inhibition in these MEFs is cell autonomous. Various proportions of wild-type (WT) and knockout (KO) MEFs were seeded into single wells as indicated below the bars. Thymidine incorporation assays were performed as for panel B. Data are presented as percent growth inhibition or percent reduction in thymidine incorporation upon TGF-β treatment.
To determine if the growth-inhibitory effect of TGF-β in these cultures is cell autonomous or due to inappropriately regulated production of paracrine factors, growth inhibition by TGF-β of mixed wild-type and null cultures was assayed. As shown in Fig. 3C, different percentages of wild-type and null cells were seeded into the same well, and TGF-β mediated growth inhibition was assayed. The growth-inhibitory effect of TGF-β in these experiments is proportional to the amount of wild-type cells. This suggests that the antiproliferative effect of TGF-β in these cells is most likely cell autonomous and not due to a Smad3-dependent production of growth-inhibitory or inhibition of growth-stimulatory paracrine factors.
As discussed above, the Smads have been characterized as DNA binding transcription factors. To determine the requirement of Smad3 in the activation of specific promoters, we studied the regulation of the widely used TGF-β-responsive promoter 3TP-lux in our model fibroblast system. In wild-type cells, the previously described TGF-β-induced, Smad3-containing DNA binding complex forms on the concatemerized tetradecanoyl phorbol acetate response elements (TREs) present in this promoter. This DNA binding complex is lost in the Smad3 null fibroblasts (Fig. 4A). In addition, transcription from this promoter in wild-type cells is activated 2.4-fold upon TGF-β treatment. This activation is lost in the null fibroblasts and can be restored by cotransfection of a Smad3 expression vector (Fig. 4B). Thus, Smad3 is necessary not only for the growth-inhibitory effects of TGF-β in this system but also for the induction of this specific promoter reporter construct.
To assess the effect of Smad3 loss on the induction of an endogenous gene known to be transcriptionally regulated by TGF-β, we assayed the TGF-β-mediated induction of PAI-1 in primary dermal fibroblasts. As shown in Fig. 4C, the induction of PAI-1 by TGF-β seen in Smad3 heterozygote dermal fibroblasts is greatly reduced in the null cells.
Analysis of the immune cells derived from Smad3 null mice reveals a defect in TGF-β signaling.
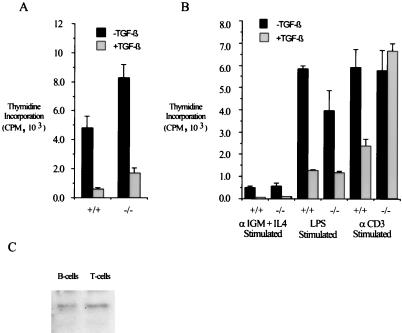
Having defined an essential role for Smad3 in TGF-β signaling in the fibroblast system, we next examined the cell types with highest Smad3 expression, those of lymphoid origin. We first examined the proliferation of splenocytes isolated from wild-type and null animals in the presence and absence of TGF-β. Interestingly, the proliferation of unstimulated primary splenocytes, consisting of a mixed B- and T-cell population, is inhibited by TGF-β regardless of mouse genotype when assayed by tritiated thymidine incorporation (Fig. 5A). Thus, in contrast to the MEF data presented above, Smad3 is not required for TGF-β-mediated inhibition of cellular proliferation in unstimulated splenocytes. In addition, the antiproliferative effects of TGF-β in primary splenocytes stimulated by LPS or αIgM plus IL-4, which specifically stimulate the proliferation of B lymphocytes through the activation of IgM receptor expressed only on the surface of B cells, is largely intact regardless of genotype. However, in primary splenocyte cultures stimulated with αCD3, an activator of the T-cell receptor complex, inhibition of proliferation by TGF-β is seen only in wild-type cultures (Fig. 5B). This difference in Smad3-dependent TGF-β responsiveness of the mixed splenocytes to a specific stimulus is not due to a difference in the expression pattern of Smad3, as demonstrated by Smad3 Western blot analysis of isolated T and B cells (Fig. 5C). Taken together, these data suggest that Smad3 plays a specific role in the inhibition of immune cell proliferation by TGF-β dependent on the nature of stimulus.
FIG. 5.
Assay of TGF-β’s effects in primary splenocytes reveals both Smad3-dependent and Smad3-independent growth-inhibitory signaling pathways. (A) Smad3 is not required for TGF-β-mediated growth inhibition in primary unstimulated splenocytes. Primary splenocytes were isolated from 8-week-old mice and cultured in the presence or absence of 100 pM TGF-β for 48 h. Cells were incubated with [3H]thymidine for the last 4 h of culture, after which the splenocytes were harvested and 3H incorporation was measured. Bars indicate the average of three identically treated wells for each growth condition; error bars represent the standard deviation. (B) Smad3 is required for TGF-β-mediated growth inhibition of αCD3-stimulated splenocytes. Primary splenocytes were isolated from 8-week-old mice and cultured in the presence of the indicated growth stimuli in the presence or absence of 100 pM TGF-β. Cellular proliferation was assayed by [3H]thymidine incorporation as for panel A. (C) Smad3 is expressed in both B and T cells. Western blotting for Smad3 was performed on purified B and T cells from mature wild-type spleens.
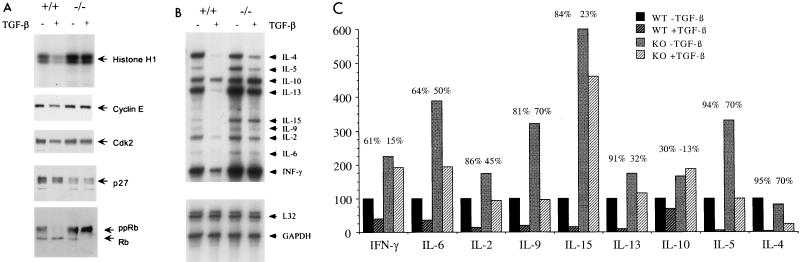
Little is known on the molecular mechanisms through which TGF-β inhibits proliferation of activated B and T cells, making it difficult to predict the role of Smad3 in this system. Concurrent with results obtained for a variety of cell types, TGF-β treatment of αCD3-stimulated wild-type spleen cultures leads to a decrease in Cdk2 kinase activity and a maintenance of Rb in a hypophosphorylated state. These effects occur with minimal change in the levels of Cdk2 and cyclin E and no change in the levels of p27. In contrast, TGF-β-mediated inhibition of Cdk2 kinase activity and maintained activation of Rb do not occur in the Smad3 null spleen cultures, further supporting the different growth properties of wild-type and null immune cells (Fig. 6A). Western analysis of various cell cycle components reveal no TGF-β-mediated change in the levels of the CDC25A phosphatase or the TGF-β-responsive Cdk inhibitors p15 and p21 (data not shown). Thus, the TGF-β growth-inhibitory pathway activated in αCD3-stimulated splenocytes represents a yet to be defined Smad3-dependent mechanism.
FIG. 6.
TGF-β-mediated growth inhibition of αCD3-stimulated splenocytes is associated with a decrease in G1 Cdk activity and cytokine expression. (A) Splenocytes were harvested from wild-type and knockout mice and cultured with αCD3 in the presence or absence of TGF-β. Cell lysates were prepared and subjected to Western blotting for cyclin E, Cdk2, p27, and Rb (lower panels). In addition, Cdk2 kinase activity was assayed by immunoprecipitation of Cdk2 and evaluation of its ability to phosphorylate the exogenous substrate, histone H1 (top panel). (B) Cytokine production was assayed on splenocytes from wild-type and Smad3 null mice treated as for panel A, using an RNase protection assay. The identity of each band is indicated on the right. L32 and GAPDH are controls for mRNA quantity and quality. (C) The intensity of the cytokine RPA bands in panel B was determined by densitometry. Plotted are the relative intensities of each band, with wild-type levels of each cytokine set at 100%.
Subsequently, we examined the effects of TGF-β on cytokine production in αCD3-stimulated primary spleen cultures. In this system, TGF-β prevents the αCD3-mediated increase in the production of a number of different cytokines by the wild-type cells (Fig. 6B). This effect is even more dramatic than the growth-inhibitory effects of TGF-β on these cultures. As shown in Fig. 6B, the production of several cytokines, such as IL-2, IL-4, IL-5, IL-9, IL-13, and IL-15, is more than 80% reduced by TGF-β in wild-type splenocytes (Fig. 6C). In the Smad3 null culture, however, TGF-β clearly does not have the same effect on the levels of these cytokines as seen in the wild-type culture. This loss of TGF-β responsiveness is most marked in gamma interferon (IFN-γ), IL-2, IL-13, and IL-15 production. In addition, the non-TGF-β-treated levels of several cytokines are elevated in the null cultures. These results strongly suggest abnormal regulation of cytokine production in the absence of Smad3-mediated TGF-β signal transduction.
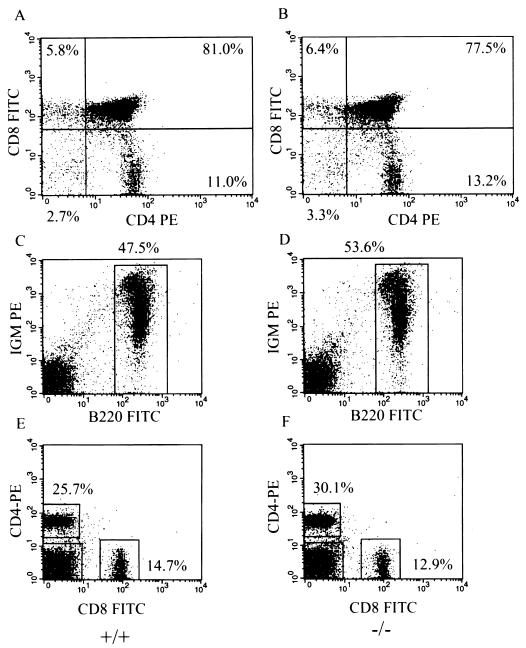
Since lymphocyte proliferation abnormalities are observed in vitro in Smad3 null cells, we next determined whether any abnormalities in the profiles of lymphocyte distribution could be observed in vivo by performing FACS analysis on Smad3 null and wild-type spleens and thymuses. As shown in Fig. 7, the thymuses of Smad3 null mice contain normal proportions of CD4 and CD8 single- and double-positive T cells, suggesting that thymic T-cell maturation is normal in Smad3 null mice. Similarly, the spleens from Smad3 null mice contain normal numbers and percentages of B cells and CD4 and CD8 single-positive T cells, suggesting that there is not an abnormal expansion of lymphocytes in the spleens of Smad3 null mice. In addition, we performed FACS analysis on bone marrow and peripheral lymph nodes for B- and T-cell populations and observed no difference between wild-type and Smad3 null mice (data not shown). Finally, we performed functional analysis of B and T cells by immunizing mice with various antigens and measuring both T-cell-dependent and T-cell-independent antibody production. Again, we did not observe any significant differences in antibody production between wild-type and Smad3 null mice (data not shown).
FIG. 7.
FACs analyses of thymocytes and splenocytes isolated from wild-type and Smad3 null mice demonstrate normal T-cell and B-cell development. (A and B) Representative FACs analysis of wild-type and Smad3 null thymocytes, using αCD4-PE and αCD8-FITC. (C to F) Representative FACS analysis of wild-type and Smad3 null splenocytes, using the indicated conjugated antibodies. All data was gated for viable cells by the absence of 7AAD staining. Percentages represent the proportions of viable cells in each region or quadrant.
DISCUSSION
In an attempt to define the roles of Smad3 in TGF-β-mediated signal transduction, we have created mice harboring a targeted disruption of Smad3. The first striking finding is that Smad3 null mice are viable and survive to adulthood. The analysis of mice deficient in other Smad genes, however, has firmly established the role of this family of proteins in embryonic development. Mice with a targeted disruption of Smad4 display an early embryonic lethal phenotype at embryonic days 6.5 to 8.5. These embryos do not undergo gastrulation or express mesodermal markers, and they show abnormal visceral endoderm development (52, 61). Smad2-deficient mice also die early in development, at embryonic day 9.5, primarily due to a loss of anterior-posterior identity within the embryo. In the absence of anterior-posterior identity, the entire epiblast develops a extraembryonic mesodermal fate, failing to give rise to the three primary germ layers (55). In a separate study, Smad2 was found to play a role in mesoderm formation, left-right patterning, and craniofacial development (43). Additional support for the critical roles of the TGF-β superfamily of ligands and the Smad family of proteins in development has been established in studies of the Xenopus oocyte developmental system (17, 24).
In sharp contrast to mice harboring a targeted disruption of Smad2 and Smad4, the loss of Smad3 function, as we report here, has no discernible effect on embryonic development. It is conceivable that certain functions of Smad3 are redundant with, or compensated for by, that of Smad2. These two proteins are 90% identical at the amino acid level. Both proteins are inducibly phosphorylated by the TGF-β receptors, associate with Smad4, and undergo nuclear accumulation. One main difference is that Smad2 may be expressed as two alternatively spliced variants; one contains two inserts in the MH1 domain of the protein, rendering it unable to bind to DNA (51, 60), whereas the other, without the inserts, is structurally and functionally virtually identical with Smad3 (60). Thus, the molecular functions of Smad2 and Smad3 are most likely overlapping as well as distinct, since functional differences in the DNA binding properties and promoter activation by these molecules have been reported (28, 63, 65). Although we still do not know the expression patterns of the two variants of Smad2, clearly Smad3 cannot fully compensate for the severe defect in Smad2 null mice which may have lost the expression of both forms of Smad2. On the other hand, Smad3 may play a more exclusive role as an effector for TGF-β and possibly activin in adult tissues, whereas Smad2 with its two forms may function more globally in development and possibly in the adult as a signaling mediator of these two ligands.
A role for Smad3 in TGF-β-mediated growth inhibition.
Our initial goal in these studies was to define the role of Smad3 in the regulation of cellular proliferation by TGF-β. Since previous studies on this topic have involved overexpression of Smads and the use of various tumor lines which likely harbor additional mutations, a role for the Smads in the regulation of proliferation remained uncertain. To this end, we have demonstrated that Smad3 is required for TGF-β-mediated growth inhibition in at least two cellular contexts: αCD3-stimulated primary splenocytes and primary MEFs.
The results from primary splenocyte cultures are particularly interesting in that TGF-β-mediated growth inhibition is dependent on Smad3 only under certain stimulated growth conditions. The proliferation of unstimulated, LPS-stimulated, and αIgM–IL-4-stimulated splenocytes is inhibited in response to TGF-β treatment in wild-type cells and to a nearly identical extent in Smad3 null cells. In contrast, a large reduction in TGF-β-mediated growth inhibition is seen in the Smad3 null splenocytes specifically when they are stimulated by αCD3. Thus, there appear to be both Smad3-dependent and Smad3-independent growth-inhibitory signaling pathways for TGF-β. We have also observed a similar defect in TGF-β-mediated growth inhibition in MEFs derived from Smad3 null mice. In these cells, the growth-inhibitory effect of TGF-β is largely absent, and this lack of TGF-β effect is most likely cell autonomous.
The molecular nature of the growth-inhibitory effects of TGF-β is one of its most studied properties. Through the work of a number of groups, a model has been put forward in which TGF-β regulates proliferation by inhibiting the activity of Cdk complexes. This function of TGF-β is likely due, in part, to its ability to increase the expression of the Cdk inhibitors p21 and p15, decrease the expression of a number of different cyclins, Cdks, the phosphatase CDC25A, and c-Myc, as well as regulate the activity of p27 (reviewed in reference 20). The signaling mechanisms of TGF-β-mediated growth inhibition vary significantly from one cell type to another. Unfortunately, none of the previously described TGF-β-mediated growth-inhibitory pathways appear to be functioning in wild-type MEFs or αCD3-stimulated splenocytes. Specifically, MEFs and αCD3-stimulated splenocytes down regulate G1 cyclin-Cdk complex activity without significant changes in the levels of p21, p15, p27, cyclin E, or Cdk2. Thus, Smad3 does not act through these defined downstream effectors to mediate the growth-inhibitory effects of TGF-β in these cells. Consequently, these findings suggest a novel Smad3-dependent growth-inhibitory pathway for TGF-β.
The work presented here is complemented by a recent report by Zhu et al., characterizing the phenotype of an independently created mouse line with a targeted insertion into the second exon of Smad3 (68). This group describes a high prevalence of colon tumor in the 129sv mouse background, and a lower prevalence of a less aggressive tumor phenotype in the 129-C57BL/6 hybrid mouse background. Although not experimentally addressed, it is an attractive hypothesis that this tumor formation occurs due to defects in TGF-β-mediated growth inhibition of the sort that we describe here. It remains to be determined, however, if these tumors arise from some other TGF-β–Smad3-dependent cellular effect or through a mechanism unrelated to TGF-β signaling. It is intriguing that we have not yet observed the 30% prevalence of colon tumors in our 129-C57BL/6 hybrid lines as in the reported study. This discrepancy may be due to differences in genetic background of the Smad3 null animals or even targeting strategies. It is also possible that a higher prevalence of tumors may still occur in our lines with longer time or when the mice with mixed genetic background are inbred into a pure 129 mouse line.
In addition to its antiproliferative role in the context of tumor suppression, TGF-β is a well-documented global inhibitor of immune system function. This function of TGF-β is evidenced by the phenotype of TGF-β1 null mice (9, 26). These mice present with a multifocal inflammatory disease, with lymphocyte infiltration into multiple organs and production of autoimmune antibodies (9, 13). The phenotype of these mice may be attributed to a loss of the antiproliferative effect of TGF-β1 on both B and T cells (22, 23). Given the fact that Smad3 is most highly expressed in the spleen and thymus, and the accumulating evidence that Smad3 is regulated by TGF-β, the development of an overactive inflammatory phenotype similar to that of the TGF-β1 knockout mice may have been expected in the Smad3 null mice. This phenotype, however, is not observed.
These findings may be explained by the fact that under several conditions for assay of B- and T-cell cultures in vitro, the antiproliferative effect of TGF-β is intact in Smad3 null cells. Thus, under in vivo conditions, the proliferation of B and T cells may be appropriately inhibited under most circumstances by endogenous TGF-β. Since this is likely the case, a more subtle or incompletely penetrant inflammatory phenotype may still emerge in the Smad3 null mice. These findings also support a model in which although Smad3 is important in regulating the antiproliferative effects of TGF-β under certain conditions, TGF-β can also activate or use other Smad3-independent pathways to exert a growth-inhibitory effect.
A role for Smad3 in TGF-β-mediated gene responses.
TGF-β can affect the expression of a number of different genes of diverse functions (46). The identification of Smads as sequence-specific DNA binding transcription factors supports the notion that the regulation of specific genes by TGF-β may be through the functions of Smad2, Smad3, and Smad4. Both 3TP-lux, a well-studied promoter reporter used for the analysis of TGF-β signaling, and the promoter of PAI-1, a highly TGF-β inducible extracellular matrix protein, contain Smad3-Smad4 DNA binding sites (11, 21, 63). Although the Smads have been implicated in the TGF-β-mediated induction of 3TP-lux and PAI-1, these studies are based largely on Smad overexpression and dominant negative studies, leaving the question of the physiological role for Smad3 in TGF-β-mediated gene activation unresolved (29, 35, 65). In addition, we have shown in a previous study that the Smad-DNA interaction is dispensable for the activation of 3TP-lux by TGF-β, bringing into question the role of Smad3 in the regulation of this promoter (63). Here we demonstrate that Smad3 is integral for transactivation of 3TP-lux and PAI-1, as their induction by TGF-β is reduced in the absence of Smad3. Interestingly, although Smad2 has been shown in the context of overexpression to activate 3TP-lux and PAI-1 (29), no compensation by Smad2 is observed in the Smad3 null fibroblasts.
In addition to the studies of 3TP-lux and PAI-1, we have investigated the role of Smad3 in the regulation of other genes by TGF-β. Specifically we provide evidence that the TGF-β-mediated down regulation of αCD3-stimulated cytokine production is Smad3 dependent. This suggests that Smad3 may play an important role in both the activation as well as the repression of gene expression. We have also evaluated the role of Smad3 in the regulation of several additional promoters by TGF-β in our Smad3 null model system. Specifically, we describe the role of Smad3 in the induction of c-Jun by TGF-β in a separate study (56). In this study we find that the c-Jun promoter contains a Smad3-Smad4 complex binding site and that Smad3 is required for the induction of c-Jun in MEFs. Thus, Smad3 may be specifically required for the activation of transcription from a subset of TGF-β-responsive promoters. In this regard, the Smad3 null MEF system provides a useful tool to define these genes which are regulated by TGF-β through Smad3. In doing so, we may be able to define novel genes or pathways which are at the root of the Smad3-dependent, TGF-β antiproliferative effects which we have described.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Cheryl Bock and the Duke Transgenic Mouse Facility for help in the generation of Smad3-deficient mice, Michael Cook and Lynn Martinek for flow cytometry services; Rik Derynk for Smad2 and Smad4 expression constructs, Allan Balmain and Sheelagh Frame for providing the dermal fibroblast isolation protocol, Yong Yu for technical help, and the members of the Wang lab for helpful scientific discussion.
This work was supported by grants from the NIH to X.-F.W. (DK45746 and CA75368) and to Y.Z. (CA72433). J.P.F. was supported by a fellowship (DAMD17-98-1-8067) from the Department of Defense Breast Cancer Research Program. X.-F.W. is a Leukemia Scholar, and Y.Z. is a Whitehead Fellow.
REFERENCES
- 1.Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana J L. TβRI phosphorylation of Smad2 on Ser 465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 2.Arai T, Akiyama Y, Okabe S, Ando M, Endo M, Yuasa Y. Genomic structure of the human Smad3 gene and its infrequent alterations in colorectal cancers. Cancer Lett. 1998;122:157–163. doi: 10.1016/s0304-3835(97)00384-4. [DOI] [PubMed] [Google Scholar]
- 3.Attisano L, Wrana J L. Signal transduction by members of the transforming growth factor-β superfamily. Cytokine Growth Factor Rev. 1996;7:327–339. doi: 10.1016/s1359-6101(96)00042-1. [DOI] [PubMed] [Google Scholar]
- 4.Baker J C, Harland R M. From receptor to nucleus: the Smad pathway. Curr Opin Genet Dev. 1997;7:467–473. doi: 10.1016/s0959-437x(97)80072-x. [DOI] [PubMed] [Google Scholar]
- 5.Balazovich K J, Fernandez R, Hinkovska-Galcheva V, Suchard S J, Boxer L A. TGF-β stimulates degranulation and oxidant release by adherent human neutrophils. J Leukoc Biol. 1996;60:772–777. doi: 10.1002/jlb.60.6.772. [DOI] [PubMed] [Google Scholar]
- 6.Barrett M T, Schutte M, Kern S E, Reid B J. Alletlic loss and mutational analysis of the DPC4 gene in esophageal adenocarcinoma. Cancer Res. 1996;56:4351–4353. [PubMed] [Google Scholar]
- 7.Chen X, Rubock M J, Whitman M. A transcriptional partner for MAD proteins in TGF-β signaling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 9.Dang H, Geiser A G, Letterio J J, Nakabayashi T, Kong L, Fernandes G, Talal N. SLE-like autoantibodies and Sjogren’s syndrome-like lymphoproliferation in TGF-β knockout mice. J Immunol. 1995;155:3205–3212. [PubMed] [Google Scholar]
- 10.Datto M B, Li Y, Panus J F, Howe D J, Xiong Y, Wang X-F. Transforming growth factor β induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanim. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J-M. Direct binding of Smad3 and Smad4 to critical TGF-β inducible elements in the promoter of human plasminogen activator inhibitor 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derynck R, Zhang Y. Intracellular signalling: the mad way to do it. Curr Biol. 1996;7:1226–1229. doi: 10.1016/s0960-9822(96)00702-6. [DOI] [PubMed] [Google Scholar]
- 13.Diebold R J, Eis M J, Yin M, Ormsby I, Boivin G P, Darrow B J, Saffitz J E, Doetschman T. Early-onset multifocal inflammation in the transforming growth factor β1-null mouse is lymphocyte mediated. Proc Natl Acad Sci USA. 1995;92:12215–12219. doi: 10.1073/pnas.92.26.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L W, Bapat B, Gallinger S, Andrulis I L, Thomsen G H, Wrana J L, Attisano L. MADR2 maps to 18q21 and encodes a TGFβ-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 15.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 16.Hata A, Lo R S, Wotton D, Lagna G, Massague J. Mutations increasing autoinhibition inactivate tumor suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 17.Heasman J. Patterning the Xenopus blastula. Development. 1997;124:4179–4191. doi: 10.1242/dev.124.21.4179. [DOI] [PubMed] [Google Scholar]
- 18.Heldin C H, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through Smad proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 19.Howe J R, Roth S, Ringold J C, Summers R W, Jarvinen H J, Sistonen P, Tomlinson I P M, Houlston R S, Bevan S, Mitros F A, Stone E M, Aaltonen L A. Mutations in the Smad4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 20.Hu PP-C, Datto M B, Wang X-F. Molecular mechanism of transforming growth factor-β signaling. Endocrine Rev. 1998;19:349–363. doi: 10.1210/edrv.19.3.0333. [DOI] [PubMed] [Google Scholar]
- 21.Hua X, Liu X, Ansari D O, Lodish H F. Synergistic cooperation of TFE3 and Smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kehrl J H, Roberts A B, Wakefield L M, Jakowlew S, Sporn M B, Fauci A S. Transforming growth factor β is an important immunomodulatory protein for human B lymphocytes. J Immunol. 1986;137:3855–3860. [PubMed] [Google Scholar]
- 23.Kehrl J H, Wakefield L M, Roberts A B, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn M B, Fauci A S. Production of transforming growth factor β by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med. 1986;163:1037–1050. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessler D S, Melton D A. Vertebrate embryonic induction: mesoderm and neural patterning. Science. 1994;266:596–604. doi: 10.1126/science.7939714. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Johnson K, Chen H J, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediate activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 26.Kulkarni A B, Huh C G, Becker D, Geiser A, Lyght M, Flanders K C, Roberts A B, Sporn M B, Ward J M, Karlsson S. Transforming growth factor β1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkarni A B, Karlsson S. Inflammation and TGF-β1: lessons from the TGF-β1 null mouse. Res Immunol. 1997;148:453–456. doi: 10.1016/s0923-2494(97)82669-7. [DOI] [PubMed] [Google Scholar]
- 28.Labbe E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF-β-dependent transcription through the forkhead DNA-binding protein Fast2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 29.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and Smad proteins in TGF-β signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 30.Lechleider R J, de Caestecker M P, Dehejia A, Polymeropoulos M H, Roberts A B. Serine phosphorylation, chromosomal localization, and transforming growth factor-β signal transduction by human bsp-1. J Biol Chem. 1996;271:17617–17620. doi: 10.1074/jbc.271.30.17617. [DOI] [PubMed] [Google Scholar]
- 31.Letterio J J, Roberts A B. Molecule of the month. TGF-β: a critical modulator of immune cell function. Clin Immunol Immunopathol. 1997;84:244–250. doi: 10.1006/clin.1997.4409. [DOI] [PubMed] [Google Scholar]
- 32.Liberati, N. T., M. B. Datto, J. P. Frederick, X. Shen, C. Wong, E. M. Rougier-Chapman, and X.-F. Wang. Smads bind directly to the Jun family of AP-1 transcription factors. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 33.Liu F, Hata A, Baker J C, Doody J, Carcamo J, Harland R M, Massague J. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, Pouponnot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGF-β-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Sun Y, Constantinescu S N, Karam E, Weinberg R A, Lodish H F. Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. MADR2 is a substrate of the TGF-β receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 37.Markowitz S D, Roberts A B. Tumor suppressor activity of the TGF-β pathway in human cancers. Cytokine Growth Factor Rev. 1996;7:93–102. doi: 10.1016/1359-6101(96)00001-9. [DOI] [PubMed] [Google Scholar]
- 38.Massague J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 39.Massague J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 40.Nagatake M, Takagi Y, Osada H, Uchida K, Mitsudomi T, Saji S, Shimokata K, Takahasi T. Somatic in vivo alterations of the DPC4 gene at 18q21 in human lung cancers. Cancer Res. 1996;56:2718–2720. [PubMed] [Google Scholar]
- 41.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C H, Miyazono K, ten Dijke P. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5252–5262. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakao A, Roijer E, Imamura T, Souchelnytskyi S, Stenman G, Heldin C H, ten Dijke P. Identification of Smad2, a human Mad-related protein in the transforming growth factor β signaling pathway. J Biol Chem. 1997;272:2896–2900. doi: 10.1074/jbc.272.5.2896. [DOI] [PubMed] [Google Scholar]
- 43.Nomura M, Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 44.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Mad-related genes in the human. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 45.Riggins G J, Kinzler K W, Vogelstein B, Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997;57:2578–2580. [PubMed] [Google Scholar]
- 46.Roberts A B, Sporn M B. The transforming growth factor-β. In: Sporn M B, Roberts A B, editors. Handbook of experimental pharmacology, peptide growth factors and their receptors. Heidelberg, Germany: Springer; 1990. pp. 419–472. [Google Scholar]
- 47.Savage C, Das P, Finelli A L, Townsend S R, Sun C Y, Baird S E, Padgett R W. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor β pathway components. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schutte M, Hruban R H, Hedrick L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, Sidransky D, Casero R A, Meltzer P S, Hahn S A, Kern S E. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 49.Sekelsky J J, Newfeld S J, Raftery L A, Chartoff E H, Gelbart W M. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serra R, Johnson M, Filvaroff E H, LaBorde J, Sheehan D M, Derynck R, Moses H L. Expression of truncated, kinase-defective TGF-β type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteroarthritis. J Cell Biol. 1997;139:541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y, Wang Y-F, Jayaraman L, Yang H, Massague J, Pavletich N P. Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 52.Sirard C, de la Pompa J L, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern S E, Rossant J, Mak T W. The tumor suppressor gene Smad4/DPC4 is required for the gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin C H. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 54.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 55.Waldrip W R, Bikoff E K, Hoodless P A, Wrana J L, Robertson E J. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- 56.Wong C, Rougier-Chapman E M, Frederick J P, Datto M B, Liberati N T, Li J-M, Wang X-F. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol Cell Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X-F, Massague J. TGF-β signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 58.Wrana J L, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 59.Wu R-Y, Zhang Y, Feng X-H, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. Alternatively-spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- 61.Yang X, Li C, Xu X, Deng C. The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proc Natl Acad Sci USA. 1998;95:3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yingling J M, Das P, Savage C, Zhang M, Padgett R W, Wang X-F. Mammalian dwarfins are phosphorylated in response to TGF-β and are implicated in control of cell growth. Proc Natl Acad Sci USA. 1996;93:8940–8944. doi: 10.1073/pnas.93.17.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X-F. The tumor suppressor Smad4 is a transforming growth factor β-inducible DNA binding protein. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–618. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Feng X-H, Wu R-Y, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Feng X-H, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 68.Zhu Y, Richardson J A, Parada L F, Graff J M. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]