Abstract
Objective
PE is a pregnancy‐specific syndrome that affects 3%–5% of pregnant women. It often presents as new‐onset hypertension and proteinuria during the third trimester. PE progresses rapidly and may lead to serious complications, including the death of both mother and fetus. In low‐income countries, PE is one of the main causes of maternal and child mortality. While the cause of PE is still debated, clinical and pathological studies suggest that the placenta plays an important role in the pathogenesis of PE.
Materials and Methods
In this single‐cell RNA‐sequencing (RNA‐seq) study, the placenta was taken from the designated position after cesarean section. We compared placental cell subsets and their transcriptional heterogeneity between preeclampsia and healthy pregnancies using the single‐cell RNA‐seq technology. A developmental trajectory of human trophoblasts was shown.
Results
Gene expression in endoplasmic reticulum signaling pathways in syncytiotrophoblast was upregulated in the PE group. The villi cytotrophoblasts (VCT) and extravillous trophoblasts were mainly involved in immune responses.
Conclusion
The placental immune function of patients with PE was altered. Proteasomes, spliceosomes, ribosomes, and mitochondria were abnormally active in the new VCT cell type.
Keywords: extravillous trophoblasts, preeclampsia, single‐cell RNA sequencing, syncytiotrophoblast, villi cytotrophoblast
This information has enriched our understanding of the human placenta and broadened our understanding of the molecular functions of the human placenta in the diseased state. These findings will be helpful in the future discovery of new molecular markers and the discovery of drugs for the treatment of the disease.
![]()
1. INTRODUCTION
The placenta plays a crucial role in anchoring the conceptus, preventing its rejection by the maternal immune system, and delivering effective nourishment for the fetus, including gaseous exchange and transporting nutrients and waste between the fetal and maternal circulatory systems (Burton & Fowden, 2015; Redman & Sargent, 2005; Sibai et al., 2005). The placenta has three main types of epithelial trophoblast cells: the villi cytotrophoblasts (VCTs), the syncytiotrophoblasts (SCTs), and extravillous trophoblasts (EVTs) (Redman & Sargent, 2005; Turco & Moffett, 2019). The monocytic VCTs are located on the basement membrane beneath the SCTs. Unlike other trophoblasts, VCTs express molecular markers related to cell proliferation, such as EGFR, Met, and some members of the Wnt signaling pathway (Jokhi et al., 1994; Leisser et al., 2006; Pijnenborg & Vercruysse, 2013), so it is considered the germinal layer of the trophoblast. SCTs envelop the whole surface area of the villous tree surrounding the pregnancy, consisting of 58 billion nuclei and having a surface area of 12–14 m2 (Napso et al., 2018). SCTs also secrete hormones to support healthy pregnancy that then enter the maternal circulation, such as human chorionic gonadotropin and placental prolactin (Evain‐Brion & Malassine, 2003; Kaufmann et al., 1977; Sonderegger et al., 2007). EVTs are derived from cytotrophoblast cell columns at the distal tips of the villi and have two differentiation pathways (Pijnenborg et al., 1980). Substances released from the placenta into the maternal blood are associated with the clinical symptoms of PE, such as elevated blood pressure and levels of urinary proteins (Mastorakos & Ilias, 2003). When under oxidative stress, SCT release a series of complex factors, including proinflammatory cytokines, exosomes, and antiangiogenic agents, into the maternal circulation. These factors interfere with maternal endothelial function and lead to a systemic inflammatory response (Redman et al., 2014; Velegrakis et al., 2017).
Many studies have examined human placental transcriptome. Tsang et al. identified diverse cellular subtypes in the human placenta and enabled the reconstruction of the trophoblast differentiation trajectory by analyzing over 24,000 nonmarker selected cells from preeclamptic placentas (Tsang et al., 2017). Liu et al. performed single‐cell RNA sequencing on sorted placental cells from first‐and second‐trimester human placentas. New subtypes of trophoblasts, Hofbauer cells, and mesenchymal stromal cells were identified (Liu et al., 2018; Pavlicev et al., 2017).
However, it is still unclear what changes have taken place in gene expression profiles of various cell types in placental tissue under the condition of elevated blood pressure. In this study, we used microfluidic single‐cell digital transcriptomic technology to comprehensively characterize the transcriptomic heterogeneity of the human placenta.
2. MATERIALS AND METHODS
2.1. Sample collection
In this single‐cell RNA‐seqencing (RNA‐seq) study, samples were collected from three pregnant women complicated by PE and three healthy pregnant women from the Weifang Traditional Chinese Hospital, Shandong, China between December 2019 and April 2020. An overview of their clinical data is shown in Table 1. PE was defined as blood pressure ≥140/90 mmHg on at least two occasions, 4 h apart, and developing after a 20‐wk gestation with proteinuria of ≥300 mg in 24 h (Brown et al., 2018). The exclusion criteria consisted of multiple pregnancies, chronic inflammatory, autoimmune, hemolytic and renal diseases, hepatitis, placental ablation, pregnancies with chromosomal aberrations, and maternal diabetes. The study was approved by the Weifang Traditional Chinese Hospital Research and Ethics Committees. Informed consent was obtained from all participants before collection of placental samples. For placental parenchymal biopsy, 1 cm3 of placental tissue was dissected immediately after delivery from a region 2 cm deep and 5 cm away from the umbilical cord insertion point after peeling off the fetal membranes. This study was authorized by the Ethic committe of Weifang Traditional Chinese Hospital.
TABLE 1.
Clinical characteristics of patients
| Sample | Group | Gestation age, week | Mode of delivery (VD:CS) | Birth weight (g) |
|---|---|---|---|---|
| 2019P1 | PE | 34+5 | CS | 3100 |
| 2019P2 | PE | 35+3 | CS | 2950 |
| 2019P3 | PE | 35+1 | CS | 3020 |
| 2019N1 | HP | 38 | CS | 3500 |
| 2019N2 | HP | 38+2 | CS | 3750 |
| 2019N3 | HP | 38+5 | CS | 3620 |
Abbreviations: CS, cesarean section; HP, healthy pregnancy; PE, preeclampsia; VD, vaginal delivery.
2.2. Tissue dissociation and preparation
GEXSCOPE™ Tissue Preservation Solution (Singleron) was used to store fresh placental tissue. The specimens were washed thrice with Hanks balanced salt solution and cut into 1–2 mm pieces. Tissue pieces were then digested for 15 min with 2 ml of GEXSCOPE™ Tissue dissociation solution (Singleron) at 37°C in a 15 ml centrifuge tube with uninterrupted agitation. After digestion, samples were filtered using 40‐micron sterile strainers and centrifuged at 350 g for 5 min. Then, the supernatant solution was removed and the sediment was resuspended in 1 ml of PBS (HyClone).
2.3. Single‐cell RNA sequencing (scRNA‐seq)
Single‐cell suspensions with 1 × 105 cells/ml in PBS (HyClone) were prepared. The suspensions were then loaded onto microfluidic devices and scRNA‐seq libraries were constructed according to the Singleron GEXSCOPETM protocol using the GEXSCOPE™ Single‐Cell RNA Library Kit (Singleron) (Dura et al., 2019). Individual libraries were diluted to 4 nM and pooled for sequencing. Pools were sequenced on an Illumina HiSeq X with 150 bp paired end reads.
2.4. scRNA‐seq quantifications and statistical analysis
Raw reads were processed to generate gene expression profiles using an internal pipeline. Briefly, after filtering read one without poly T tails, cell barcodes and unique molecular identifiers (UMI) were extracted. Adapters and poly A tails were trimmed (fastp V1) before aligning read two to GRCh38 using Ensemble v.92 gene annotation (fastp 2.5.3a and featureCounts 1.6.2). Reads with the same cell barcode, UMI, and gene were grouped together to calculate the number of UMIs per gene per cell. The UMI count tables of each cellular barcode were used for further analysis. Cell type identification and clustering analysis was performed using the Seurat program. The Seurat program (http://satijalab.org/seurat/, R package, v.3.0.1) was also used for analysis of RNA‐sequencing data. UMI count tables were loaded into R using the read.table function. The parameter resolution was set to 0.8 for the FindClusters function for clustering analyses. The differentially expressed genes (DEGs) were generated by the function FindMarkers of Seurat with logfc.threshold 0.25 and min.pct 0.1; Genes with q_value <0.05 in differential genes list were selected and used for Gene ontology (GO) enrichment analysis by clusterProfiler (Yu et al., 2012); For (Gene Set Enrichment Analysis) GSEA analysis, the differential genes were sorted by the avg_logFC decreasingly. Based on the rearranged genes list, GSEA analysis was performed according to the KEGG database by the ‘gseapy’ python package; Monocle 2 was used for trajectory analysis. 2000 high variable genes generated by FindVariableFeatures function of Seurat were used for genes ordering in trajectory analysis. DDRTree was used for dimensionality reduction of the dataset. And differential genes of pseudotime were screened out by differentialGenesTest function in monocle 2 (Qiu et al., 2017).
3. RESULTS
3.1. Single‐cell transcriptome profiles distinguished placental cell types
A total of 11,518 cells were obtained from the placental tissues of six women. A graph‐based clustering method was used to identify 13 major cell clusters by uniform manifold approximation and projection (UMAP; Figure 1a). In the PE group, 6434 placental cells included immune cells (3562 cells, 55.36%), trophoblast cells (2507 cells, 38.96%), fibroblasts (157 cells, 2.4%), erythroblasts (142 cells, 2.2%), and endothelial cells (66 cells, 1.0%) (Data file S1; Figure 1b). In the healthy pregnancy group, a similar cell heterogeneity classification was retrieved (Data file S1). Large numbers of macrophages and trophoblast cells were detected in both the PE (GT01) and healthy pregnancy (XG01) groups, with natural killer cells and T cells being higher in the PE group. Trophoblast cells could be further subclassified into EVTs, VCTs, and SCTs by employing sublineage markers such as PARP1, CGA, CYP19A1, HLA‐G, and PAPPA2 (Lee et al., 2016) (Figure 1c). EVTs (1216 cells, 48.5%) were the most abundant type of trophoblast cell, followed by VCTs (1000 cells 39.9%), and SCTs (291 cells 11.6%) in the PE group. (Data file S1). VCTs express the PARP1 gene which encodes a chromatin‐associated enzyme, Poly(ADP‐ribosyl) transferase that modifies various nuclear proteins by Poly(ADP‐ribosyl)ation. CGA, which encodes for a common subunit of the four human glycoprotein hormones chorionic gonadotropin (CG), follicle‐stimulating hormone (FSH), luteinizing hormone (LH), and thyroid‐stimulating hormone (TSH), is primarily expressed by SCTs. EVTs are characterized by the expression of HLA‐G (Fiddes & Goodman, 1981). Diseases associated with HLA‐G include severe preeclampsia and asthma (Hylenius et al., 2004; Nicolae et al., 2005; Sageshima et al., 2003). As presented in Figure 1c, those clusters exhibiting high expression of CD68, APOE, and SPP1 were cells from macrophages. The specific CD37 and NKG7 genes were expressed by T cells and natural killer cells respectively. Clusters with a high expression of IGKC and JCHAIN were plasma cells that participate in the humoral immune response. Clusters with a high expression of PECAM1 were endothelial cells. Erythroblasts specifically expressed hemoglobin subunit genes, such as HBB and HBA1.
FIGURE 1.
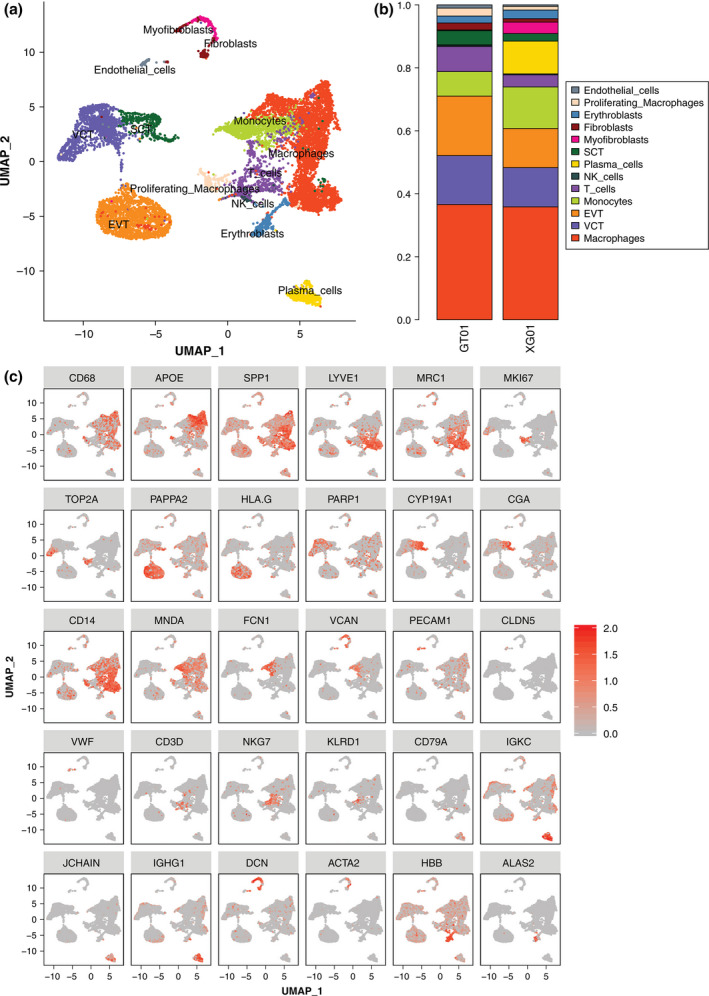
Single‐cell transcriptomic profiling and dissection of the cellular heterogeneity of the human placenta. (a) UMAP distribution of single cells from the 13 defined cell types containing both preeclampsia and normal placental tissue. (b) The proportion of cell clusters in preeclampsia (GT01) and healthy pregnancy (XG01). (c) Biaxial scatter plots showing the expression pattern of specific genes among different subgroups of placental cells
3.2. Identification of differentially expressed genes in trophoblast cells
We compared and analyzed the genes of three types of trophoblast cells in the PE and healthy pregnancy groups. SCTs revealed 610 DEGs (Data file S2). Using the average expression profiles of SCTs, VCTs, and EVTs, we performed unsupervised clustering analysis of the union of the top 40 DEGs including down‐regulated and up‐regulated genes that were expressed (Figure 2a, Figure S1a,b). Using all DEGs, we examined the main signaling pathways involved in SCTs by GSEA. Among them, genes related to protein processing in the endoplasmic reticulum pathway were upregulated in the PE group (Figure 2b). Using upregulated genes, we performed functional annotation with GO analysis. The enriched items of the biological process focused on the regulation of interleukin‐8 production and interleukin‐8 production (Figure 2c). A total of 347 DEGs were identified in VCTs (Data file S3). All these genes were analyzed by GSEA analysis. The expression of genes related to the ribosome pathway was downregulated in the PE group (Figure S1c). The GO enrichment analysis was performed on those genes whose differential gene expression was up‐regulated. The biological process was mainly enriched in the innate immune response and cell chemotaxis (Figure S1d). EVTs revealed 283 DEGs (Data file S4) and GSEA analysis was performed on all these differential genes. Genes related to the herpes simplex infection pathway were upregulated in the PE group (Figure S1e). The GO terms enriched in these cells were associated with the innate immune response and response to cytokines (Figure S1f).
FIGURE 2.
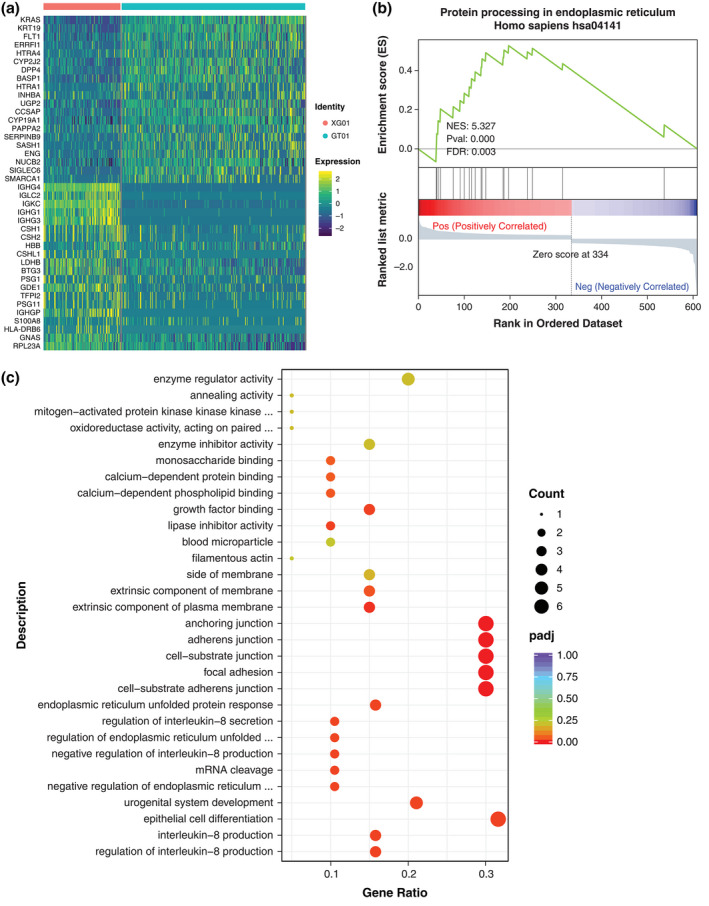
SCT differentially expressed genes and functional changes between the hypertensive group and the healthy pregnancy group. (a) Heat map showing the TOP 40 DEGs, including downregulated and upregulated genes, in SCTs. (b) GSEA result confirming enrichment of the protein processing signaling pathway found in endoplasmic reticulum expression in preeclampsia. (c) GO enrichment term of upregulated genes in SCTs
3.3. Reconstruction of the developmental relationship of the three types of trophoblasts
Our dataset captured a significant number of trophoblastic cells. We observed that VCTs, SCTs, and EVTs were clustered in a continuum (Figure 1a). In fact, it has been proposed that these trophoblast subtypes are developmentally connected (Tsang et al., 2017; Vento‐Tormo et al., 2018). To delineate this relationship and to study the genes that regulate this process, we first identified the highly variable genes from these three clusters using UMAP reclustering analysis. Through the analysis of cell trajectories, we found that VCTs were in the early stage of cell development. We found that both the PE group and the healthy pregnancy group showed a continuous “Y” shaped trajectory of trophoblast development. (Figure 3a,b). FLT1, FN1, and TIMP3 were specifically upregulated along the differentiation pathway of SCTs (Figure 3c).
FIGURE 3.
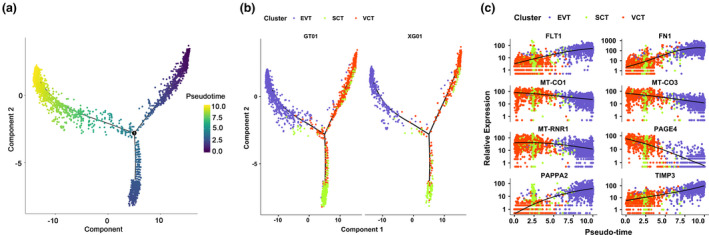
Reconstruction of the developmental relationship of trophoblast cells using pseudotime analysis. (a) The trophoblastic cell developmental trajectory visualization in a biaxial scatter plot. Dark colors indicate early development. (b) Distribution of trophoblast subtypes in trajectory. Trophoblast differentiation tracks are shown separately in the preeclampsia group and the healthy pregnancy group. (c) Dot plot showing the expression of the indicated genes in the indicated subtypes of cells
3.4. Identification of new subtypes of trophoblast cells
Further analysis revealed that the PARP1 positive cells were clustered into three subtypes: VCT‐1, VCT‐2, and VCT‐3 (Figure 4a). The cluster exhibiting the highest expression of CDK1, CCNB1, and RRM2 was VCT‐1. This cluster also had the highest expression of IFIT2 and IFIT3, which were the marker genes identifying VCT‐2 (Figure 4b).
FIGURE 4.
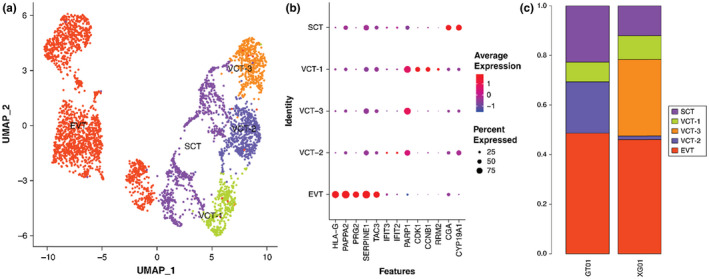
Trophoblast subtypes are present. (a) Biaxial scatter plot showing single‐cell transcriptomic clustering of the VCT subtype using UMAP analysis. (b) Biaxial scatter plots showing the expression pattern of specific genes among different trophoblast cell subgroups. (c) The histogram shows the proportion of VCT subtypes in the preeclampsia group and the healthy pregnancy group. VCT‐2 is mainly distributed in the preeclampsia group, while VCT‐3 is mainly distributed in the healthy pregnancy group
VCT‐2 cells were mainly distributed in the PE group, while VCT‐3 cells were mainly distributed in the healthy pregnancy group (Figure 4c). To further understand the gene functions of VCT‐2 and VCT‐3, correlation analysis was performed. The expression of VCT‐2 was compared with other trophoblast genes, and 1151 DEGs were obtained (Data file S5). GSEA analysis of all differential genes showed that the expression of genes related to the proteasome, spliceosome, and ribosome pathways was upregulated, but the genes related to antigen processing and presentation were downregulated (Figure 5a, Figure S2a). We found that proteasome related genes, such as PSMA7 and PSMC1, were upregulated to varying degrees in the VCT‐2 differential geneset (Data file S6). The enriched GO terms in VCT‐2 were associated with cellular respiration, energy derivation by oxidation of organic compounds, and generation of precursor metabolites and energy (Figure 5b). Genes whose expression levels were upregulated included mitochondrial coding genes related to the cellular respiratory chain (Data file S5).
FIGURE 5.
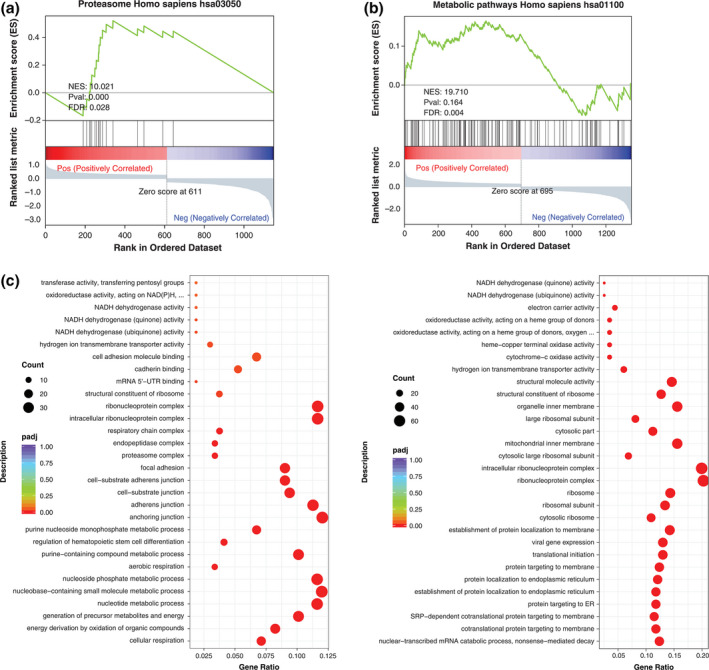
Functional enrichment and signal pathway analysis of new subtypes of trophoblast cells. (a) GSEA result confirming enrichment of the proteasome signaling pathway in VCT‐2. Gene sets from VCT‐2 are compared with other trophoblast cells. (b) Dot plots show the GO enrichment term of upregulated genes in VCT‐2. (c) GSEA result confirming enrichment of metabolic pathways in VCT‐3. Gene sets from VCT‐3 are compared with other trophoblast cells. (d) GO enrichment term of upregulated genes in VCT‐3
VCT‐3 was compared with other trophoblast cells to obtain 1353 DEGs (Data file S7). GSEA analysis of all the differential genes showed that the expression of genes related to metabolic pathways and the Alzheimer's disease pathway were upregulated (Figure 5c, Figure S2b, Data file S8). The expression of genes related to natural killer cell‐mediated cytotoxicity and rheumatoid arthritis were downregulated (Figure S2b). Based on GO analysis it appears from the highly enriched terms that VCT‐3 participates in the nuclear‐transcribed mRNA catabolic process, and cotranslational protein targeting to the membrane (Figure 5d). Related genes are: RPL23A and RPS14, etc. (Data file S7).
4. DISCUSSION
By analyzing the transcripts of 11,518 cells, the difference between the PE and healthy pregnancy groups was revealed at the single‐cell level. We obtained 1240 DEGs in total. Through further analysis of the signal pathway of SCTs, we found that the expression of protein processing in endothelial reticula was upregulated, indicating that endoplasmic reticulum function was active in the diseased state. This was thought to be related to the intermittent hypoxia of the placenta caused by narrow spiral arteries. During hypoxia, protein folding by the endoplasmic reticula is suspended, resulting in a cessation of cell proliferation and subsequent severe apoptosis (Huppertz et al., 2006). Our findings were consistent with previous studies. The release of apoptotic trophoblast particles into the maternal blood will stimulate inflammation in blood vessels (Redman & Sargent, 2007). The Go enrichment analysis of VCTs and EVTs showed that the cellular function of the PE group focused on the immune response, suggesting that the placental immune function of patients with PE was altered. On the one hand, EVTs need to remodel the uterine spiral arteries, and at the same time to invade the decidual tissue of the uterus and anchor the placenta to the uterine wall. In these processes, different types of maternal cells, such as decidual macrophages, uterine NK cells, and stromal cells, are encountered. To adapt to each other, reasonable immune regulation is necessary (Moffett et al., 2017; Pollheimer et al., 2018). Through our experiments, and for the first time, we identified three subtypes of VCTs. After GSEA analysis of VCT‐2, we found that the proteasomes, spliceosomes, ribosomes, and mitochondria were abnormally active in this cell. Proteasomes are a type of protein complex; their main function is to degrade proteins that are not needed or that have been damaged in cells through the breaking of peptide bonds. The proteasome degradation pathway is essential for many cell processes, including the cell cycle, gene expression regulation, and oxidative stress response, etc. (Shang & Taylor, 2011). Plasma proteasome and immunoproteasome are significantly upregulated in PE and hemolysis, elevated liver enzymes, and thrombocytopenia syndrome (Berryman et al., 2019). Sun et al. analyzed the gene expression profile of peripheral blood of patients with PE using gene microarray technology, and found that the expression of PSMC5 was increased (Sun et al., 2008). Our findings confirmed many previous conclusions on PE, such as stress to endoplasmic reticula and mitochondrial dysfunction. Different from previous transcriptome studies, our results were accurate to the single‐cell level.
There are some limitations of the current study. First, only three pregnant women were reruited in each group. Our results should be validated in large‐scale studies. Secondly, the interactions among these active signaling pathways in PE warrant further investigation.
5. CONCLUSION
This study has enriched the understanding of human placenta and its molecular functions in the diseased state. These findings will contribute to the future discovery of new molecular markers and the discovery of drugs for the treatment of the disease.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
T Z, H Z, QQ B, and YC C were involved in study design. SH L, SW Y, P J, L L, and SJ D prepared the samples for the experiments. T Z and XL W carried out data analysis. QQ B and M S drafted the manuscript. All the authors contributed to critical discussions and approved the final version of the manuscript.
ETHICS STATEMENT
This study was authorized by the Ethic committee of Weifang Traditional Chinese Hospital.
Supporting information
Fig S1‐S2
Data S1‐S8
ACKNOWLEDGEMENTS
We acknowledge everyone for their helpful contributions on this paper.
Zhang, T., Bian, Q., Chen, Y., Wang, X., Yu, S., Liu, S., Ji, P., Li, L., Shrestha, M., Dong, S., Guo, R., & Zhang, H. (2021). Dissecting human trophoblast cell transcriptional heterogeneity in preeclampsia using single‐cell RNA sequencing. Molecular Genetics & Genomic Medicine, 9, e1730. 10.1002/mgg3.1730
Funding information
Key Project of Shandong Province Higher Educational Science and Technology Program (Grant No. J18KZ013); Support Program for Youth Innovation technology in Colleges and Universities of Shandong Province (2019KJK004).
Contributor Information
Rong Guo, Email: 406422461@qq.com.
Hong Zhang, Email: Hong_Z@21cn.com.
DATA AVAILABILITY STATEMENT
The data generated in this study are available on request to the corresponding author.
REFERENCES
- Berryman, K., Buhimschi, C. S., Zhao, G., Axe, M., Locke, M., & Buhimschi, I. A. (2019). Proteasome levels and activity in pregnancies complicated by severe preeclampsia and hemolysis, elevated liver enzymes, and thrombocytopenia (HELLP) syndrome. Hypertension, 73(6), 1308–1318. 10.1161/HYPERTENSIONAHA.118.12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M. A., Magee, L. A., Kenny, L. C., Karumanchi, S. A., McCarthy, F. P., Saito, S., Hall, D. R., Warren, C. E., Adoyi, G., & Ishaku, S. (2018). The hypertensive disorders of pregnancy: ISSHP classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens, 13, 291–310. [DOI] [PubMed] [Google Scholar]
- Burton, G. J., & Fowden, A. L. (2015). The placenta: a multifaceted, transient organ. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 370(1663), 20140066. 10.1098/rstb.2014.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura, B., Choi, J. Y., Zhang, K., Damsky, W., Thakral, D., Bosenberg, M., Craft, J., & Fan, R. (2019). scFTD‐seq: freeze‐thaw lysis based, portable approach toward highly distributed single‐cell 3’ mRNA profiling. Nucleic Acids Research, 47(3), e16. 10.1093/nar/gky1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evain‐Brion, D., & Malassine, A. (2003). Human placenta as an endocrine organ. Growth Horm IGF Res, 13, S34–S37. 10.1016/S1096-6374(03)00053-4. [DOI] [PubMed] [Google Scholar]
- Fiddes, J. C., & Goodman, H. M. (1981). The gene encoding the common alpha subunit of the four human glycoprotein hormones. Journal of Molecular and Applied Genetics, 1(1), 3–18. [PubMed] [Google Scholar]
- Huppertz, B., Kadyrov, M., & Kingdom, J. C. (2006). Apoptosis and its role in the trophoblast. American Journal of Obstetrics and Gynecology, 195(1), 29–39. 10.1016/j.ajog.2005.07.039 [DOI] [PubMed] [Google Scholar]
- Hylenius, S., Andersen, A. M., Melbye, M., & Hviid, T. V. (2004). Association between HLA‐G genotype and risk of pre‐eclampsia: A case‐control study using family triads. Molecular Human Reproduction, 10(4), 237–246. 10.1093/molehr/gah035 [DOI] [PubMed] [Google Scholar]
- Jokhi, P. P., King, A., & Loke, Y. W. (1994). Reciprocal expression of epidermal growth factor receptor (EGF‐R) and c‐erbB2 by non‐invasive and invasive human trophoblast populations. Cytokine, 6(4), 433–442. 10.1016/1043-4666(94)90068-X [DOI] [PubMed] [Google Scholar]
- Kaufmann, P., Stark, J., & Stegner, H. E. (1977). The villous stroma of the human placenta. I. The ultrastructure of fixed connective tissue cells. Cell and Tissue Research, 177(1), 105–121. 10.1007/BF00221122 [DOI] [PubMed] [Google Scholar]
- Lee, C. Q., Gardner, L., Turco, M., Zhao, N., Murray, M. J., Coleman, N., Rossant, J., Hemberger, M., & Moffett, A. (2016). What is Trophoblast? A combination of criteria define human first‐trimester trophoblast. Stem cell reports, 6(2), 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisser, C., Saleh, L., Haider, S., Husslein, H., Sonderegger, S., & Knöfler, M. (2006). Tumour necrosis factor‐alpha impairs chorionic gonadotrophin beta‐subunit expression and cell fusion of human villous cytotrophoblast. Molecular Human Reproduction, 12(10), 601–609. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Fan, X., Wang, R., Lu, X., Dang, Y. L., Wang, H., Lin, H. Y., Zhu, C., Ge, H., Cross, J. C. & Wang, H. (2018). Single‐cell RNA‐seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Research, 28(8), 819–832. 10.1038/s41422-018-0066-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastorakos, G., & Ilias, I. (2003). Maternal and fetal hypothalamic‐pituitary‐adrenal axes during pregnancy and postpartum. Annals of the New York Academy of Sciences, 997, 136–149. 10.1196/annals.1290.016 [DOI] [PubMed] [Google Scholar]
- Moffett, A., Chazara, O., & Colucci, F. (2017). Maternal allo‐recognition of the fetus. Fertility and Sterility, 107(6), 1269–1272. 10.1016/j.fertnstert.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Napso, T., Yong, H. E. J., Lopez‐Tello, J., & Sferruzzi‐Perri, A. N. (2018). The role of placental hormones in mediating maternal adaptations to support pregnancy and lactation. Frontiers in Physiology, 9, 1091. 10.3389/fphys.2018.01091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae, D., Cox, N. J., Lester, L. A., Schneider, D., Tan, Z., Billstrand, C., Kuldanek, S., Donfack, J., Kogut, P., Patel, N. M., Goodenbour, J., Howard, T., Wolf, R., Koppelman, G. H., White, S. R., Parry, R., Postma, D. S., Meyers, D., Bleecker, E. R., … Ober, C. (2005). Fine mapping and positional candidate studies identify HLA‐G as an asthma susceptibility gene on chromosome 6p21. American Journal of Human Genetics, 76(2), 349–357. 10.1086/427763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicev, M., Wagner, G. P., Chavan, A. R., Owens, K., Maziarz, J., Dunn‐Fletcher, C., Kallapur, S. G., Muglia, L., & Jones, H. (2017). Single‐cell transcriptomics of the human placenta: inferring the cell communication network of the maternal‐fetal interface. Genome Research, 27(3), 349–361. 10.1101/gr.207597.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnenborg, R., Dixon, G., Robertson, W. B., & Brosens, I. (1980). Trophoblastic invasion of human decidua from 8 to 18 weeks of pregnancy. Placenta, 1(1), 3–19. 10.1016/S0143-4004(80)80012-9 [DOI] [PubMed] [Google Scholar]
- Pijnenborg, R., & Vercruysse, L. (2013). A.A.W. Hubrecht and the naming of the trophoblast. Placenta, 34(4), 314–319. 10.1016/j.placenta.2013.01.002 [DOI] [PubMed] [Google Scholar]
- Pollheimer, J., Vondra, S., Baltayeva, J., Beristain, A. G., & Knöfler, M. (2018). Regulation of placental extravillous trophoblasts by the maternal uterine environment. Frontiers in Immunology, 9, 2597. 10.3389/fimmu.2018.02597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Mao, Q., Tang, Y., Wang, L., Chawla, R., Pliner, H. A., & Trapnell, C. (2017). Reversed graph embedding resolves complex single‐cell trajectories. Nature Methods, 14(10), 979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman, C. W., & Sargent, I. L. (2005). Latest advances in understanding preeclampsia. Science, 308(5728), 1592–1594. 10.1126/science.111172 [DOI] [PubMed] [Google Scholar]
- Redman, C. W., & Sargent, I. L. (2007). Microparticles and immunomodulation in pregnancy and pre‐eclampsia. Journal of Reproductive Immunology, 76(1–2), 61–67. 10.1016/j.jri.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Redman, C. W., Sargent, I. L., & Staff, A. C. (2014). IFPA Senior Award Lecture: making sense of pre‐eclampsia – Two placental causes of preeclampsia? Placenta, 35(Suppl), S20–25. 10.1016/j.placenta.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Sageshima, N., Ishitani, A., Omura, M., Akasaki, M., Umekage, H., Katabuchi, H., Okamura, H., & Hatake, K. (2003). Necrotic feature of the trophoblasts lacking HLA‐G expression in normal and pre‐eclamptic placentas. American Journal of Reproductive Immunology, 49(3), 174–182. 10.1034/j.1600-0897.2003.00010.x [DOI] [PubMed] [Google Scholar]
- Shang, F., & Taylor, A. (2011). Ubiquitin‐proteasome pathway and cellular responses to oxidative stress. Free Radical Biology and Medicine, 51(1), 5–16. 10.1016/j.freeradbiomed.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibai, B., Dekker, G., & Kupferminc, M. (2005). Pre‐eclampsia. Lancet, 365(9461), 785–799. 10.1016/S0140-6736(05)71003-5 [DOI] [PubMed] [Google Scholar]
- Sonderegger, S., Husslein, H., Leisser, C., & Knöfler, M. (2007). Complex expression pattern of Wnt ligands and frizzled receptors in human placenta and its trophoblast subtypes. Placenta, 28, S97–S102. 10.1016/j.placenta.2006.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C. J., Zhang, W. Y., Yu, S., & Cui, M. H. (2008). Microarray analysis of differentially expressed genes in peripheral leucocytes derived from severe preeclampsia and normotensive pregnancies. Zhonghua Fu Chan Ke Za Zhi, 43(9), 651–656. [PubMed] [Google Scholar]
- Tsang, J. C. H., Vong, J. S. L., Ji, L., Poon, L. C. Y., Jiang, P., Lui, K. O., Ni, Y. B., To, K. F., Cheng, Y. K. Y., Chiu, R. W. K. & Lo, Y. M. D. (2017). Integrative single‐cell and cell‐free plasma RNA transcriptomics elucidates placental cellular dynamics. Proceedings of the National Academy of Sciences of the United States of America, 114(37), E7786–E7795. 10.1073/pnas.1710470114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco, M. Y., & Moffett, A. (2019). Development of the human placenta. Development, 146, 22. 10.1242/dev.163428 [DOI] [PubMed] [Google Scholar]
- Velegrakis, A., Sfakiotaki, M., & Sifakis, S. (2017). Human placental growth hormone in normal and abnormal fetal growth. Biomedical Reports, 7(2), 115–122. 10.3892/br.2017.930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento‐Tormo, R., Efremova, M., Botting, R. A., Turco, M. Y., Vento‐Tormo, M., Meyer, K. B., Park, J. E., Stephenson, E., Polański, K., Goncalves, A., Gardner, L., Holmqvist, S., Henriksson, J., Zou, A., Sharkey, A. M., Millar, B., Innes, B., Wood, L., Wilbrey‐Clark, A., … Teichmann, S. A. (2018). Single‐cell reconstruction of the early maternal‐fetal interface in humans. Nature, 563(7731), 347–353. 10.1038/s41586-018-0698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang, L. G., Han, Y., & He, Q. Y. (2012). ClusterProfiler: an R package for comparing biology theme among gene cluster. Omics, 16(5), 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2
Data S1‐S8
Data Availability Statement
The data generated in this study are available on request to the corresponding author.


