Abstract
Background
Hypoxia within the plateau has a negative effect on skeletal muscle and may play a role in the development of sarcopenia in humans. Tibetans having lived in the Qinghai‐Tibet Plateau for thousands of years, are a high‐risk group for sarcopenia; however, they have a distinctive suite of genetic traits that enable them to tolerate environmental hypoxia and are genetically significantly different from Han Chinese and other lowland populations. Sarcopenia has been consistently found to be associated with single‐nucleotide polymorphisms, but few studies have investigated the role of single‐nucleotide polymorphisms in a range of muscle phenotypes and sarcopenia in Tibetan peoples.
Methods
Our study aimed to investigate the skeletal muscle mass and fat mass of 160 Tibetans (80 men and 80 women) from Lhasa (altitude of 3600 meters) and analyze the association between the polymorphisms of fat mass and obesity protein (FTO) rs9939609, FTO rs9936385, activin type IIB receptor (ACVR2B) rs2276541, insulin receptor substrate 1 (IRS1) 2943656 and sarcopenia.
Result
FTO rs9939609 and rs9936385 polymorphisms were associated with lower limb skeletal muscle mass and sarcopenia for Tibetan women, and TT homozygotes had a higher risk for sarcopenia. But ACVR2B rs2276541 and IRS1 2943656 polymorphisms were unassociated with sarcopenia in Tibetan.
Conclusion
In Tibetans, FTO rs9939609 and rs9936385 polymorphisms were associated with sarcopenia, and ACVR2B rs2276541 and IRS1 2943656 polymorphisms were unassociated with sarcopenia.
Keywords: activin type IIB receptor (ACVR2B), fat mass and obesity‐associated protein (FTO), insulin receptor substrate 1 (IRS1), sarcopenia, single‐nucleotide polymorphism (SNP), Tibetan
Our study aimed to investigate the skeletal muscle mass and fat mass of 160 Tibetans (80 men and 80 women) from Lhasa (altitude of 3600 meters) and analyze the association between polymorphisms of FTO rs9939609, FTO rs9936385 ACVR2B rs2276541, IRS1 2943656 and sarcopenia. We found that FTO rs9939609 and rs9936385 polymorphisms were associated with lower limb skeletal muscle mass and sarcopenia for Tibetan women, and TT homozygotes had a higher risk for sarcopenia. But ACVR2B rs2276541 and IRS1 2943656 polymorphisms were unassociated with sarcopenia in Tibetan.

1. INTRODUCTION
Hypoxia has a negative effect on skeletal muscle, as it can cause hypoxia‐induced muscle loss (Pasiakos et al., 2017). Changes in body composition, such as loss of skeletal muscle mass, frequently occur when humans are exposed to hypoxic environments (Dunnwald et al., 2019; Fusch et al., 1996; Wandrag et al., 2017). Hypoxia can increase the levels of circulating proinflammatory cytokines, leading to inflammation (Eltzschig & Carmeliet, 2011); it also causes an increase in reactive oxygen species, leading to oxidative damage (McGarry et al., 2018). Therefore, it may play a role in the development of sarcopenia in humans. In previous research, the incidences of sarcopenia in Tibetans over 60 were 17.2% in men and 36.0% in women, significantly higher than those in the plain population (Ye et al., 2020). Tibetans are an archaic ethnicity (Lu et al., 2016), have lived at very high altitudes for thousands of years, and have a distinctive suite of genetic traits that enable them to tolerate environmental hypoxia (Simonson et al., 2010; Song et al., 2020), which make them significantly different from the Han and other plain populations genetically (Beall et al., 2010). Recently, the association between single‐nucleotide polymorphisms (SNPs) and sarcopenia was found, and some risk genotypes have been determined (Khanal et al., 2020; Roth et al., 2004), but few studies have investigated the role of SNPs in muscle phenotypes and sarcopenia in Tibetans. Therefore, we investigated whether the identified risk‐genotypes for sarcopenia were equally applicable to Tibetans.
The fat mass and obesity‐associated protein (FTO) gene has been shown to be related to obesity (Loos & Yeo, 2014). Its polymorphisms have been associated with obesity, fat mass, and obesity‐related indicators, such as body mass index (BMI) in adults and children (Hinney et al., 2007; Livshits et al., 2012), which have been studied in various groups of people such as Europeans (Merra et al., 2020; Sällman Almén et al., 2013), Americans (Grant et al., 2008; Wing et al., 2009), and Mexicans (Villalobos‐Comparán et al., 2008). The FTO gene has been associated not only with fat mass but also with lean mass (Ran et al., 2020; Sonestedt et al., 2011). C‐allele carriers at rs9936385 have greater lean mass and the T allele is associated with sarcopenia (Karasik et al., 2019; Zillikens et al., 2017). A‐allele carriers at rs9939609 have lower lean mass and AA homozygotes are more than 3‐fold higher risk for sarcopenia compared to T‐allele carriers (Khanal et al., 2020). ACVR2B codes for a receptor for a negative regulator of skeletal muscle, myostatin, and deletion in ACVR2B causes skeletal muscle hyperplasia (an increase in the number of skeletal muscle fibers) and hypertrophy (an increase in the size of skeletal muscle fibers) (Lee & McPherron, 2001; White & LeBrasseur, 2014). It has previously been identified as a gene of skeletal muscle mass and strength (Klimentidis et al., 2016; Walsh et al., 2007), A‐allele carriers at rs2276541 have greater lean mass. Insulin receptor substrate 1 (IRS1) is a signaling adapter protein, it is downstream of the Insulin‐like growth factor 1 (IGF1) receptor, IGF1 induces skeletal muscle hypertrophy by activating the IGF1/IRS1/PI3K/Akt pathway (Z. Li et al., 2019), and loss of IRS1 can limit the IGF1 pathway and negatively affect skeletal muscle mass (Shi et al., 2011). IRS1 is essential for skeletal muscle growth and protein homeostasis, and polymorphism of rs2943656 is associated with total body lean mass and appendicular lean mass (Zillikens et al., 2017). There is no research which has investigated the association between polymorphisms of FTO rs9939609, FTO rs9936385, ACVR2B rs2276541, IRS1 2943656 and sarcopenia in Tibetans.
2. MATERIALS AND METHODS
2.1. Study population
The survey was carried out via health examinations in August 2016 in Lhasa (altitude of 3600 m), China (Figure 1). Native Tibetans over 40 years of age participated in this study. A total of 1447 subjects (604 men and 843 women) were investigated, including 438 sarcopenia participants (200 men and 238 women) and 1009 healthy participants (404 men and 605 women). We randomly selected 160 adults (80 men and 80 women) from the sarcopenia group and healthy control group with an average age of 53.19 years. The following subjects were excluded from the study: (1) individuals under long‐term bed rest, with sedentary lifestyles, or those who experienced extreme weight loss; (2) individuals with heart, lung, liver, kidney, or brain diseases, inflammatory reaction diseases, malignant tumors, or endocrine diseases; (3) individuals with an absorption disorder, gastrointestinal disease, anorexia, or drug use. The interviewers were trained before the survey. The study was approved by the Research Ethics Committee of Jinzhou Medical University in accordance with the Declaration of Helsinki. Verbal and written informed consent were obtained from all participants.
FIGURE 1.
The map of sample collection place
2.2. Measurement
2.2.1. Height
Height was measured using a portable stadiometer (HM200P, American Charder Company, America) and recorded to the nearest 0.1 cm. Body mass index (BMI) was calculated from weight and height.
2.2.2. Handgrip strength (HS)
HS was measured using a handheld dynamometer based on strain gauge sensors (CAMRY EH101) to the nearest 0.1 kg. Both hands were tested with the participant seated, the elbow flexed at a 110° angle, wrist placed in a neutral position, and the interphalangeal joint of the index finger positioned at a 90° angle. Two readings were obtained for each hand, and the highest value for either hand was used for the analyses.
2.2.3. Gait speed (GS)
According to the criteria of the Asian Working Group for Sarcopenia (AWGS) (Chen et al., 2014), the physical performance was assessed using the usual GS metric. To measure the GS, the participants were asked to walk a 6 m distance at their usual speed. Timing commenced when the participants started foot movement and stopped when the foot contacted the ground after completely crossing the 6 m mark. Canes or walkers were allowed if necessary.
2.2.4. Weight, fat mass, and muscle mass
A bioelectrical impedance analyzer (MC‐180, Bailida) was used to measure weight, fat mass (FM), skeletal muscle mass (SMM), trunk skeletal muscle mass (TSM), left upper limb skeletal muscle mass (LUSM), right upper limb skeletal muscle mass (RUSM), left lower limb skeletal muscle mass (LLSM), right lower limb skeletal muscle mass (RLSM), trunk fat mass (TFM), left upper limb fat mass (LUFM), right upper limb fat mass (RUFM), left lower limb fat mass (LLFM), and right lower limb fat mass (RLFM).
2.3. Sarcopenia diagnosis
According to the recommended diagnostic AWGS algorithm, GS, HS, and skeletal muscle mass index (SMI) were the primary indicators for sarcopenia diagnosis (Chen et al., 2014). The cutoff values for SMI measurements were 8.07 kg/m2 for men and 6.62 kg/m2 for women (Ye et al., 2020). Low physical performance was defined as a GS less than 0.8 m/s (Ye et al., 2020). The cutoff values for HS were <26.7 kg for men and <15.8 kg for women (Ye et al., 2020).
2.4. DNA extraction
First, 2 ml of venous blood was collected from all participants and DNA was extracted from the whole peripheral blood sample using a DNA extraction kit (Blood DNA Extraction Kit, EIibio™). DNA samples were stored at −80°C before genotyping. After DNA extraction, the concentration of the extracted material was obtained using a spectrophotometer (BioPhotometer plus).
2.5. PCR and genotyping
The primer information of DNA fragments containing the FTO rs9939609, FTO rs9936385, ACVR2B rs2276541 and IRS1 rs2943656 was uploaded as supplementary material 1. For all polymorphism samples, the reactions were carried out in a 30 μl volume of 1 μl of DNA, 15 μl of 2XPCR MIX (TAKARA), 12 μl of DDW, and 1 μl of each primer. The PCR conditions were as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 58°C for 20 s and extension at 72°C for 40 s, and then a final extension at 72°C for 5 min. Genotypes for the FTO rs9939609 (TT/AT/AA), FTO rs9936385 (TT/CT/CC), ACVR2B rs2276541(AA/AG/GG), and IRS1 rs2943656 (AA/AG/GG) polymorphism were determined using a 3730 XL Gene Sequencer (ABI).
2.6. Statistical analyses
The Hardy–Weinberg equilibrium (HWE) was calculated to determine the variation in the distribution of alleles and genotypes within the population. The data are expressed as the mean ± standard deviation. The association between genotypes and anthropometric indices and body composition was analyzed via analysis of variance. The chi‐square test was used to analyze the differences in genotype and allele frequency between the sarcopenia group and the control group. Logistic regression analyses were used to analyze the odds ratio (OR) and 95% confidence interval (95% Cl) of sarcopenia among the genotypes. All analyses were performed using SPSS (ver.25.0, IBM Company). The map of China was plotted by QGIS (https://www.qgis.org/).
3. RESULTS
The basic characteristics of the samples are listed in Table 1. There were no significant differences in age and height between the sarcopenia and control groups in men and women (p > 0.05). Comparisons of the FTOs rs9939609, FTO rs9936385, ACVR2B rs2276541, and IRS1 rs2943656 polymorphism between the sarcopenia and healthy groups are given in Tables 2, 3, 4, 5, respectively. The genotypes in the healthy control and sarcopenia cohorts were in HWE. The genotype and allele frequencies of FTO were significantly different between the groups (p < 0.05), particularly for women. The frequency of the T allele was higher in the sarcopenia group than in controls for both SNPs. FTO rs9939609TT homozygotes exhibited a 2.414‐fold higher risk for sarcopenia compared to A‐allele carriers (OR = 2.414, CI: 1.270–4.586, p = 0.007), and FTO rs9936385 TT homozygotes exhibited a 2.414‐fold higher risk for sarcopenia compared to C‐allele carriers (OR = 2.414, CI: 1.270–4.586, p = 0.007). There was no difference of genotype and allele frequencies between health group and sarcopenia group in ACVR2B rs2276541 and IRS1 rs2943656.
TABLE 1.
Anthropometric indices and body composition characteristics of the Tibetan people
| Index | Male (80) | p | Female (80) | p | ||
|---|---|---|---|---|---|---|
| Control (40) | Sarcopenia (40) | Control (40) | Sarcopenia (40) | |||
| Age | 53.73 ± 7.48 | 51.38 ± 7.20 | 0.156 | 52.45 ± 8.53 | 55.20 ± 8.08 | 0.143 |
| Height | 167.59 ± 6.38 | 169.40 ± 7.76 | 0.257 | 159.00 ± 5.08 | 157.38 ± 5.66 | 0.180 |
| Weight | 70.28 ± 9.42 | 57.44 ± 6.54 | 0.000 | 66.40 ± 9.15 | 53.19 ± 8.04 | 0.000 |
| BMI | 24.97 ± 2.67 | 19.98 ± 1.41 | 0.000 | 26.24 ± 3.39 | 21.42 ± 2.73 | 0.000 |
| FFM | 53.64 ± 5.38 | 47.48 ± 5.08 | 0.000 | 42.07 ± 3.25 | 37.51 ± 3.20 | 0.000 |
| FM | 16.68 ± 5.27 | 9.99 ± 3.28 | 0.000 | 24.34 ± 7.18 | 15.70 ± 5.73 | 0.000 |
| SMM | 50.85±5.12 | 45.00 ± 4.82 | 0.000 | 39.56 ± 2.97 | 35.38 ± 2.94 | 0.000 |
| TSM | 28.11 ± 2.56 | 25.57 ± 2.59 | 0.000 | 22.55 ± 1.53 | 21.13 ± 1.87 | 0.000 |
| LUSM | 2.75 ± 0.37 | 2.38 ± 0.31 | 0.000 | 2.05 ± 0.24 | 1.67 ± 0.15 | 0.000 |
| LLSM | 8.56 ± 1.05 | 7.27 ± 0.95 | 0.000 | 6.38 ± 0.69 | 5.42 ± 0.50 | 0.000 |
| RUSM | 2.85 ± 0.38 | 2.48 ± 0.30 | 0.000 | 2.15 ± 0.24 | 1.74 ± 0.17 | 0.000 |
| RLSM | 8.70 ± 1.13 | 7.41 ± 0.95 | 0.000 | 6.54 ± 0.66 | 5.54 ± 0.46 | 0.000 |
| TFM | 9.77 ± 3.30 | 5.74 ± 2.26 | 0.000 | 13.91 ± 4.58 | 8.75 ± 4.04 | 0.000 |
| LUFM | 0.64 ± 0.23 | 0.34 ± 0.13 | 0.000 | 1.17 ± 0.48 | 0.61 ± 0.28 | 0.000 |
| LLFM | 2.89 ± 0.81 | 1.86 ± 0.44 | 0.000 | 4.08 ± 0.88 | 2.90 ± 0.58 | 0.000 |
| RUFM | 0.60 ± 0.21 | 0.33 ± 0.12 | 0.000 | 1.15 ± 0.48 | 0.59 ± 0.28 | 0.000 |
| RLFM | 2.89 ± 0.82 | 1.83 ± 0.43 | 0.000 | 4.18 ± 0.89 | 2.98 ± 0.59 | 0.000 |
| HS | 31.95 ± 8.73 | 30.64 ± 6.88 | 0.460 | 21.41 ± 7.14 | 17.52 ± 4.77 | 0.005 |
Abbreviations: BMI, body mass index; FFM, fat‐free mass; FM, fat mass; HS, handgrip strength; LLFM, left lower limb fat mass; LLSM, left lower limb skeletal muscle mass; LUFM, left upper limb fat mass; LUSM, left upper limb Skeletal muscle mass; RLFM, right lower limb fat mass; RLSM, right lower limb skeletal muscle mass; RUFM, right upper limb fat mass; RUSM, right upper limb skeletal muscle mass; SMM, skeletal muscle mass; TFM, trunk fat mass; TSM, trunk skeletal muscle mass.
TABLE 2.
Comparison of FTO rs9939609 polymorphism between healthy controls and the sarcopenia group
| Group | Genotype (%) | Allele (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | AT | AA | χ 2 | p 1 | A | T | χ 2 | p 2 | |
| Sarcopenia | 54 (67.5) | 23 (28.75) | 3 (3.75) | 7.343 | 0.024 | 29 (18.125) | 131 (81.875) | 6.174 | 0.013 |
| S‐Male | 28 (70.0) | 10 (25.0) | 2 (5.0) | 2.198 | 0.370 | 14 (17.5) | 66 (82.5) | 1.345 | 0.246 |
| S‐Female | 26 (65.0) | 13 (32.5) | 1 (2.5) | 6.143 | 0.046 | 15 (18.75) | 65 (81.25) | 5.375 | 0.020 |
| Control | 37 (46.25) | 38 (47.5) | 5 (6.25) | — | — | 48 (30.0) | 112 (70.0) | — | — |
| C‐Male | 22 (55.0) | 16 (40.0) | 2 (5.0) | — | — | 20 (25.0) | 60 (75.0) | — | — |
| C‐Female | 15 (37.5) | 22 (55.0) | 3 (7.5) | — | — | 28 (35.0) | 52 (65.0) | — | — |
TABLE 3.
Comparison of FTO rs9936385 polymorphism between healthy controls and the sarcopenia group
| Group | Genotype (%) | Allele (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | χ 2 | p 1 | C | T | χ 2 | p 2 | |
| Sarcopenia | 54 (67.5) | 23 (28.75) | 3 (3.75) | 7.343 | 0.024 | 29 (18.125) | 131 (81.875) | 6.174 | 0.013 |
| S‐Male | 28 (70.0) | 10 (25.0) | 2 (5.0) | 2.198 | 0.370 | 14 (17.5) | 66 (82.5) | 1.345 | 0.246 |
| S‐Female | 26 (65.0) | 13 (32.5) | 1 (2.5) | 6.143 | 0.046 | 15 (18.75) | 65 (81.25) | 5.375 | 0.020 |
| Control | 37 (46.25) | 38 (47.5) | 5 (6.25) | — | — | 48 (30.0) | 112 (70.0) | — | — |
| C‐Male | 22 (55.0) | 16 (40.0) | 2 (5.0) | — | — | 20 (25.0) | 60 (75.0) | — | — |
| C‐Female | 15 (37.5) | 22 (55.0) | 3 (7.5) | — | — | 28 (35.0) | 52 (65.0) | — | — |
TABLE 4.
Comparison of ACVR2B rs2276541 polymorphism between healthy controls and the sarcopenia group
| Group | Genotype (%) | Allele (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | χ 2 | p 1 | A | G | χ 2 | p 2 | |
| Sarcopenia | 24 (30.0) | 36 (45.0) | 20 (25.0) | 0.436 | 0.804 | 84 (52.5) | 76 (47.5) | 0.050 | 0.823 |
| S‐Male | 12 (30.0) | 17 (42.5) | 11 (27.5) | 1.310 | 0.519 | 41 (51.25) | 39 (48.75) | 0.100 | 0.752 |
| S‐Female | 12 (30.0) | 19 (47.5) | 9 (22.5) | 0.857 | 0.651 | 45 (56.25) | 35 (43.75) | 1.601 | 0.206 |
| Control | 21 (26.25) | 40 (50.0) | 19 (23.75) | — | — | 82 (51.25) | 78 (48.75) | — | — |
| C‐Male | 12 (30.0) | 21 (52.5) | 7 (17.5) | — | — | 43 (53.75) | 37 (46.25) | — | — |
| C‐Female | 9 (22.5) | 19 (47.5) | 12 (30.0) | — | — | 37 (46.25) | 43 (53.75) | — | — |
TABLE 5.
Comparison of IRS1 rs2943656 polymorphism between healthy controls and the sarcopenia group
| Group | Genotype (%) | Allele (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | χ 2 | p 1 | A | G | χ 2 | p 2 | |
| Sarcopenia | 2 (2.5) | 26 (32.5) | 52 (65.0) | 0.709 | 0.807 | 30 (18.75) | 130 (81.25) | 0.313 | 0.576 |
| S‐Male | 1 (2.5) | 14 (35.0) | 25 (62.5) | 2.175 | 0.328 | 16 (20.0) | 64 (80.0) | 0.040 | 0.841 |
| S‐Female | 1 (2.5) | 12 (30.0) | 27 (67.5) | 1.600 | 0.670 | 14 (17.5) | 66 (82.5) | 0.954 | 0.329 |
| Control | 4 (5.0) | 26 (32.5) | 50 (62.5) | — | — | 34 (21.25) | 126 (78.75) | — | — |
| C‐Male | 3 (7.5) | 9 (22.5) | 28 (70.0) | — | — | 15 (18.75) | 65 (81.25) | — | — |
| C‐Female | 1 (2.5) | 17 (42.5) | 22 (55.0) | — | — | 19 (23.75) | 61 (76.25) | — | — |
The associations among genotypes of FTO, ACVR2B and IRS1, body composition and anthropometric indices are listed in Tables 6, 7, 8, 9, respectively. Both rs9939609 and rs9936385 were associated with lower limb skeletal muscle mass in Tibetan women, and genotypes AA/CC and AT/CT had higher lower limb skeletal muscle mass compared to TT, but there was no association between FTO genotypes and fat mass. And ACVR2B rs2276541 and IRS1 rs2943656 were unassociated with body composition in Tibetans.
TABLE 6.
Association between FTO rs9939609 genotypes (TT, AT, & AA) and body composition and anthropometric indices
| Male | Genotype (n) | F | p | Female | Genotype (n) | F | p | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | TT (50) | AT (26) | AA (4) | Index | TT (41) | AT (35) | AA (4) | ||||
| Age | 52.06 ± 7.62 | 53.31 ± 7.06 | 53.75 ± 7.89 | 0.294 | 0.746 | Age | 55.05 ± 8.55 | 53.11 ± 8.17 | 47.50 ± 5.80 | 1.739 | 0.183 |
| Height (cm) | 169.70 ± 7.36 | 166.52 ± 6.74 | 166.25 ± 2.63 | 1.965 | 0.147 | Height (cm) | 157.50 ± 5.49 | 158.59 ± 5.46 | 161.75 ± 2.06 | 1.306 | 0.277 |
| Weight (kg) | 63.73 ± 10.53 | 64.69 ± 10.46 | 60.10 ± 7.94 | 0.347 | 0.708 | Weight (kg) | 57.36 ± 9.29 | 62.11 ± 12.21 | 64.48 ± 8.78 | 2.278 | 0.109 |
| BMI (kg/m2) | 22.10 ± 3.22 | 23.29 ± 3.42 | 21.78 ± 3.18 | 1.217 | 0.302 | BMI (kg/m2) | 23.05±3.11 | 24.65 ± 4.60 | 24.68 ± 3.87 | 1.727 | 0.185 |
| FFM (kg) | 50.68 ± 6.34 | 50.47 ± 5.95 | 49.70 ± 3.55 | 0.051 | 0.950 | FFM (kg) | 38.92 ± 3.83 | 40.55 ± 4.09 | 41.95 ± 1.26 | 2.316 | 0.106 |
| FM (kg) | 13.09 ± 5.36 | 14.26 ± 5.90 | 10.40 ± 4.45 | 0.983 | 0.379 | FM (kg) | 18.44 ± 6.29 | 21.58 ± 9.11 | 22.55 ± 7.52 | 1.790 | 0.174 |
| SMM (kg) | 48.03 ± 6.02 | 47.85 ± 5.66 | 47.13 ± 3.39 | 0.048 | 0.953 | SMM (kg) | 36.69 ± 3.52 | 38.16 ± 3.74 | 39.43 ± 1.21 | 2.250 | 0.112 |
| TSM (kg) | 26.90±2.89 | 26.82 ± 3.00 | 26.18 ± 2.00 | 0.116 | 0.89 | TSM (kg) | 21.60 ± 1.86 | 22.04 ± 1.90 | 22.55 ± 0.37 | 0.871 | 0.423 |
| LUSM (kg) | 2.56 ± 0.41 | 2.58±0.36 | 2.58 ± 0.22 | 0.013 | 0.987 | LUSM (kg) | 1.79 ± 0.25 | 1.92 ± 0.29 | 1.93±0.26 | 2.218 | 0.116 |
| LLSM (kg) | 7.94 ± 1.25 | 7.89 ± 1.14 | 7.80 ± 0.98 | 0.035 | 0.965 | LLSM (kg) | 5.70 ± 0.72 | 6.07 ± 0.79 | 6.50 ± 0.62 | 3.731 | 0.028 |
| RUSM (kg) | 2.66 ± 0.42 | 2.66 ± 0.35 | 2.70 ± 0.22 | 0.020 | 0.980 | RUSM (kg) | 1.87 ± 0.27 | 2.01 ± 0.32 | 2.05 ± 0.26 | 2.448 | 0.093 |
| RLSM (kg) | 8.08 ± 1.28 | 8.02 ± 1.19 | 7.95 ± 0.73 | 0.032 | 0.969 | RLSM (kg) | 5.83 ± 0.68 | 6.23 ± 0.81 | 6.53 ± 0.25 | 3.788 | 0.027 |
| TFM (kg) | 7.61 ± 3.35 | 8.33 ± 3.73 | 5.78 ± 2.74 | 1.057 | 0.352 | TFM (kg) | 10.41 ± 4.15 | 12.24 ± 5.86 | 12.75 ± 4.38 | 1.443 | 0.242 |
| LUFM (kg) | 0.48 ± 0.23 | 0.51 ± 0.26 | 0.40 ± 0.18 | 0.439 | 0.646 | LUFM (kg) | 0.78 ± 0.37 | 1.00 ± 0.56 | 1.03 ± 0.54 | 2.154 | 0.123 |
| LLFM (kg) | 2.33 ± 0.82 | 2.52 ± 0.88 | 2.00 ± 0.65 | 0.881 | 0.419 | LLFM (kg) | 3.27 ± 0.76 | 3.69 ± 1.08 | 3.93 ± 1.09 | 2.456 | 0.092 |
| RUFM (kg) | 0.46 ± 0.21 | 0.49 ± 0.23 | 0.40 ± 0.18 | 0.415 | 0.662 | RUFM (kg) | 0.76 ± 0.37 | 0.97 ± 0.57 | 1.00 ± 0.55 | 2.026 | 0.139 |
| RLFM (kg) | 2.31 ± 0.84 | 2.50 ± 0.88 | 2.00 ± 0.68 | 0.820 | 0.444 | RLFM (kg) | 3.36 ± 0.77 | 3.80 ± 1.12 | 3.93 ± 0.97 | 2.353 | 0.102 |
| HS (kg) | 31.08 ± 7.62 | 31.82 ± 7.20 | 30.58 ± 15.31 | 0.092 | 0.912 | HS (kg) | 18.94 ± 6.74 | 19.84 ± 5.82 | 21.58 ± 7.65 | 0.419 | 0.659 |
Abbreviations: BMI, body mass index; FFM, fat‐free mass; FM, fat mass; HS, handgrip strength; LLFM, left lower limb fat mass; LLSM, left lower limb skeletal muscle mass; LUFM, left upper limb fat mass; LUSM, left upper limb skeletal muscle mass; RLFM, right lower limb fat mass; RLSM, right lower limb skeletal muscle mass; RUFM, right upper limb fat mass; RUSM, right upper limb skeletal muscle mass; SMM, skeletal muscle mass; TFM, trunk fat mass; TSM, trunk skeletal muscle mass.
TABLE 7.
Association between FTO rs9936385 genotypes (TT, CT, & CC) and body composition and anthropometric indices
| Male | Genotype (n) | F | p | Female | Genotype (n) | F | p | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | TT (50) | CT (26) | CC (4) | Index | TT (41) | CT (35) | CC (4) | ||||
| Age | 52.06 ± 7.62 | 53.31 ± 7.06 | 53.75 ± 7.89 | 0.294 | 0.746 | Age | 55.05 ± 8.55 | 53.11 ± 8.17 | 47.50 ± 5.80 | 1.739 | 0.183 |
| Height (cm) | 169.70 ± 7.36 | 166.52 ± 6.74 | 166.25 ± 2.63 | 1.965 | 0.147 | Height (cm) | 157.50 ± 5.49 | 158.59 ± 5.46 | 161.75 ± 2.06 | 1.306 | 0.277 |
| Weight (kg) | 63.73 ± 10.53 | 64.69 ± 10.46 | 60.10 ± 7.94 | 0.347 | 0.708 | Weight (kg) | 57.36 ± 9.29 | 62.11 ± 12.21 | 64.48 ± 8.78 | 2.278 | 0.109 |
| BMI (kg/m2) | 22.10 ± 3.22 | 23.29 ± 3.42 | 21.78 ± 3.18 | 1.217 | 0.302 | BMI (kg/m2) | 23.05 ± 3.11 | 24.65 ± 4.60 | 24.68 ± 3.87 | 1.727 | 0.185 |
| FFM (kg) | 50.68 ± 6.34 | 50.47 ± 5.95 | 49.70 ± 3.55 | 0.051 | 0.950 | FFM (kg) | 38.92 ± 3.83 | 40.55 ± 4.09 | 41.95 ± 1.26 | 2.316 | 0.106 |
| FM (kg) | 13.09 ± 5.36 | 14.26 ± 5.90 | 10.40 ± 4.45 | 0.983 | 0.379 | FM (kg) | 18.44 ± 6.29 | 21.58 ± 9.11 | 22.55 ± 7.52 | 1.790 | 0.174 |
| SMM (kg) | 48.03 ± 6.02 | 47.85 ± 5.66 | 47.13 ± 3.39 | 0.048 | 0.953 | SMM (kg) | 36.69 ± 3.52 | 38.16 ± 3.74 | 39.43 ± 1.21 | 2.250 | 0.112 |
| TSM (kg) | 26.90 ± 2.89 | 26.82 ± 3.00 | 26.18 ± 2.00 | 0.116 | 0.89 | TSM (kg) | 21.60 ± 1.86 | 22.04 ± 1.90 | 22.55 ± 0.37 | 0.871 | 0.423 |
| LUSM (kg) | 2.56 ± 0.41 | 2.58 ± 0.36 | 2.58 ± 0.22 | 0.013 | 0.987 | LUSM (kg) | 1.79 ± 0.25 | 1.92 ± 0.29 | 1.93 ± 0.26 | 2.218 | 0.116 |
| LLSM (kg) | 7.94 ± 1.25 | 7.89 ± 1.14 | 7.80 ± 0.98 | 0.035 | 0.965 | LLSM (kg) | 5.70 ± 0.72 | 6.07 ± 0.79 | 6.50 ± 0.62 | 3.731 | 0.028 |
| RUSM (kg) | 2.66 ± 0.42 | 2.66 ± 0.35 | 2.70 ± 0.22 | 0.020 | 0.980 | RUSM (kg) | 1.87 ± 0.27 | 2.01 ± 0.32 | 2.05 ± 0.26 | 2.448 | 0.093 |
| RLSM (kg) | 8.08 ± 1.28 | 8.02 ± 1.19 | 7.95 ± 0.73 | 0.032 | 0.969 | RLSM (kg) | 5.83 ± 0.68 | 6.23 ± 0.81 | 6.53 ± 0.25 | 3.788 | 0.027 |
| TFM (kg) | 7.61 ± 3.35 | 8.33 ± 3.73 | 5.78 ± 2.74 | 1.057 | 0.352 | TFM (kg) | 10.41 ± 4.15 | 12.24 ± 5.86 | 12.75 ± 4.38 | 1.443 | 0.242 |
| LUFM (kg) | 0.48 ± 0.23 | 0.51 ± 0.26 | 0.40 ± 0.18 | 0.439 | 0.646 | LUFM (kg) | 0.78 ± 0.37 | 1.00 ± 0.56 | 1.03 ± 0.54 | 2.154 | 0.123 |
| LLFM (kg) | 2.33 ± 0.82 | 2.52 ± 0.88 | 2.00 ± 0.65 | 0.881 | 0.419 | LLFM (kg) | 3.27 ± 0.76 | 3.69 ± 1.08 | 3.93 ± 1.09 | 2.456 | 0.092 |
| RUFM (kg) | 0.46 ± 0.21 | 0.49 ± 0.23 | 0.40 ± 0.18 | 0.415 | 0.662 | RUFM (kg) | 0.76 ± 0.37 | 0.97 ± 0.57 | 1.00 ± 0.55 | 2.026 | 0.139 |
| RLFM (kg) | 2.31 ± 0.84 | 2.50 ± 0.88 | 2.00 ± 0.68 | 0.820 | 0.444 | RLFM (kg) | 3.36 ± 0.77 | 3.80 ± 1.12 | 3.93 ± 0.97 | 2.353 | 0.102 |
| HS (kg) | 31.08 ± 7.62 | 31.82 ± 7.20 | 30.58 ± 15.31 | 0.092 | 0.912 | HS (kg) | 18.94 ± 6.74 | 19.84 ± 5.82 | 21.58 ± 7.65 | 0.419 | 0.659 |
Abbreviations: BMI, body mass index; FFM, fat‐free mass; FM, fat mass; HS, handgrip strength; LLFM, left lower limb fat mass; LLSM, left lower limb skeletal muscle mass; LUFM, left upper limb fat mass; LUSM, left upper limb skeletal muscle mass; RLFM, right lower limb fat mass; RLSM, right lower limb skeletal muscle mass; RUFM, right upper limb fat mass; RUSM, right upper limb skeletal muscle mass; SMM, skeletal muscle mass; TFM, trunk fat mass; TSM, trunk skeletal muscle mass.
TABLE 8.
Association between ACVR2B rs2276541 genotypes (AA, AG, & GG) and body composition and anthropometric indices
| Male | Genotype (n) | F | p | Female | Genotype (n) | F | p | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | AA (24) | AG (38) | GG (18) | Index | AA (21) | AG (38) | GG (21) | ||||
| Age | 50.00 ± 6.67 | 53.34 ± 7.47 | 54.28 ± 7.65 | 2.205 | 0.117 | Age | 52.29 ± 6.81 | 53.74 ± 9.57 | 55.52 ± 7.43 | 0.786 | 0.459 |
| Height (cm) | 168.92 ± 6.88 | 168.50 ± 7.70 | 167.92 ± 6.45 | 0.099 | 0.906 | Height (cm) | 158.76 ± 5.37 | 157.29 ± 5.50 | 159.24 ± 5.24 | 1.041 | 0.358 |
| Weight (kg) | 65.55 ± 12.51 | 63.17 ± 9.76 | 63.07 ± 8.43 | 0.453 | 0.638 | Weight (kg) | 59.08 ± 10.65 | 58.53 ± 11.84 | 62.80 ± 8.83 | 1.115 | 0.333 |
| BMI (kg/m2) | 22.91 ± 3.83 | 22.24 ± 3.11 | 22.38 ± 3.01 | 0.311 | 0.734 | BMI (kg/m2) | 23.41 ± 3.96 | 23.57 ± 4.29 | 24.71 ± 3.04 | 0.737 | 0.482 |
| FFM (kg) | 51.58 ± 6.46 | 50.31 ± 6.24 | 49.73 ± 5.18 | 0.534 | 0.588 | FFM (kg) | 39.86 ± 4.12 | 39.37 ± 4.04 | 40.48 ± 3.66 | 0.539 | 0.585 |
| FM (kg) | 14.00 ± 6.94 | 12.90 ± 4.97 | 13.36 ± 4.60 | 0.29 | 0.749 | FM (kg) | 19.23 ± 7.57 | 19.17 ± 8.72 | 22.34 ± 5.79 | 1.279 | 0.284 |
| SMM (kg) | 48.90 ± 6.12 | 47.68 ± 5.94 | 47.14 ± 4.92 | 0.535 | 0.588 | SMM (kg) | 37.53 ± 3.78 | 37.09 ± 3.70 | 38.10 ± 3.35 | 0.525 | 0.594 |
| TSM (kg) | 27.26 ± 2.92 | 26.55 ± 3.01 | 26.88 ± 2.52 | 0.447 | 0.641 | TSM (kg) | 21.84 ± 1.86 | 21.62 ± 1.81 | 22.22 ± 1.89 | 0.714 | 0.493 |
| LUSM (kg) | 2.62 ± 0.38 | 2.59 ± 0.42 | 2.45 ± 0.31 | 1.103 | 0.337 | LUSM (kg) | 1.87 ± 0.30 | 1.83 ± 0.27 | 1.89 ± 0.27 | 0.332 | 0.719 |
| LLSM (kg) | 8.13 ± 1.31 | 7.90 ± 1.21 | 7.67 ± 0.96 | 0.796 | 0.455 | LLSM (kg) | 5.94 ± 0.81 | 5.84 ± 0.81 | 5.96 ± 0.69 | 0.196 | 0.822 |
| RUSM (kg) | 2.71 ± 0.39 | 2.68 ± 0.43 | 2.56 ± 0.31 | 0.898 | 0.412 | RUSM (kg) | 1.95 ± 0.30 | 1.91 ± 0.30 | 1.98 ± 0.28 | 0.406 | 0.668 |
| RLSM (kg) | 8.28 ± 1.39 | 8.08 ± 1.18 | 7.69 ± 1.03 | 1.246 | 0.293 | RLSM (kg) | 6.04 ± 0.73 | 5.98 ± 0.79 | 6.14 ± 0.74 | 0.323 | 0.725 |
| TFM (kg) | 8.11 ± 4.21 | 7.44 ± 3.16 | 7.95 ± 3.10 | 0.306 | 0.737 | TFM (kg) | 10.70 ± 4.85 | 10.79 ± 5.67 | 12.94 ± 3.56 | 1.489 | 0.232 |
| LUFM (kg) | 0.51 ± 0.30 | 0.48 ± 0.22 | 0.46 ± 0.18 | 0.263 | 0.77 | LUFM (kg) | 0.86 ± 0.48 | 0.84 ± 0.53 | 1.01 ± 0.38 | 0.929 | 0.399 |
| LLFM (kg) | 2.51 ± 1.08 | 2.33 ± 0.74 | 2.29 ± 0.61 | 0.455 | 0.636 | LLFM (kg) | 3.45 ± 0.93 | 3.38 ± 1.03 | 3.72 ± 0.78 | 0.885 | 0.417 |
| RUFM (kg) | 0.49 ± 0.28 | 0.46 ± 0.19 | 0.46 ± 0.16 | 0.17 | 0.844 | RUFM (kg) | 0.83 ± 0.49 | 0.82 ± 0.53 | 0.98 ± 0.37 | 0.791 | 0.457 |
| RLFM (kg) | 2.50 ± 1.09 | 2.29 ± 0.77 | 2.31 ± 0.61 | 0.476 | 0.623 | RLFM (kg) | 3.52 ± 0.93 | 3.47 ± 1.05 | 3.83 ± 0.81 | 1.011 | 0.369 |
| HS (kg) | 32.61 ± 7.96 | 30.45 ± 8.35 | 31.32 ± 6.64 | 0.555 | 0.577 | HS (kg) | 19.80 ± 5.62 | 19.78 ± 6.95 | 18.55 ± 6.06 | 0.289 | 0.750 |
Abbreviations: BMI, body mass index; FFM, fat‐free mass; FM, fat mass; HS, handgrip strength; LLFM, left lower limb fat mass; LLSM, left lower limb skeletal muscle mass; LUFM, left upper limb fat mass; LUSM, left upper limb skeletal muscle mass; RLFM, right lower limb fat mass; RLSM, right lower limb skeletal muscle mass; RUFM, right upper limb fat mass; RUSM, right upper limb skeletal muscle mass; SMM, skeletal muscle mass; TFM, trunk fat mass; TSM, trunk skeletal muscle mass.
TABLE 9.
Association between IRS1 rs2943656 genotypes (AA, AG, & GG) and body composition and anthropometric indices
| Male | Genotype (n) | F | p | Female | Genotype (n) | F | p | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | AA (4) | AG (23) | GG (53) | Index | AA (2) | AG (29) | GG (49) | ||||
| Age | 53.25 ± 7.46 | 50.48 ± 6.81 | 53.40 ± 7.58 | 1.278 | 0.284 | Age | 55.00 ± 14.14 | 52.79 ± 7.82 | 54.39 ± 8.63 | 0.345 | 0.709 |
| Height (cm) | 167.75 ± 3.50 | 170.37 ± 7.71 | 167.74 ± 7.00 | 1.125 | 0.330 | Height (cm) | 154.50 ± 3.54 | 159.19 ± 4.49 | 157.74 ± 5.89 | 1.133 | 0.328 |
| Weight (kg) | 72.83 ± 14.58 | 63.24 ± 10.21 | 63.45 ± 9.96 | 1.614 | 0.206 | Weight (kg) | 59.70 ± 20.51 | 62.56 ± 10.99 | 58.16 ± 10.33 | 1.524 | 0.224 |
| BMI (kg/m2) | 25.95 ± 5.36 | 21.76 ± 3.09 | 22.52 ± 3.10 | 2.911 | 0.060 | BMI (kg/m2) | 24.80 ± 7.50 | 24.64 ± 3.97 | 23.31 ± 3.74 | 1.127 | 0.329 |
| FFM (kg) | 53.83 ± 3.82 | 50.15 ± 6.30 | 50.49 ± 6.09 | 0.632 | 0.535 | FFM (kg) | 37.95 ± 5.87 | 40.58 ± 4.04 | 39.40 ± 3.83 | 1.040 | 0.358 |
| FM (kg) | 19.03 ± 11.05 | 13.13 ± 4.81 | 12.99 ± 5.16 | 2.328 | 0.104 | FM (kg) | 21.80 ± 14.71 | 22.00 ± 7.65 | 18.78 ± 7.55 | 1.640 | 0.201 |
| SMM (kg) | 51.05 ± 3.65 | 47.53 ± 5.98 | 47.86 ± 5.79 | 0.641 | 0.530 | SMM (kg) | 35.80 ± 5.37 | 38.19 ± 3.71 | 37.11 ± 3.50 | 1.028 | 0.363 |
| TSM (kg) | 28.30 ± 0.48 | 26.58 ± 2.96 | 26.84 ± 2.92 | 0.608 | 0.547 | TSM (kg) | 20.50 ± 1.13 | 21.98 ± 1.69 | 21.81 ± 1.94 | 0.614 | 0.544 |
| LUSM (kg) | 2.80 ± 0.26 | 2.53 ± 0.41 | 2.57 ± 0.38 | 0.850 | 0.431 | LUSM (kg) | 1.85 ± 0.49 | 1.92 ± 0.31 | 1.81 ± 0.24 | 1.465 | 0.237 |
| LLSM (kg) | 8.53 ± 1.45 | 7.90 ± 1.21 | 7.88 ± 1.17 | 0.550 | 0.579 | LLSM (kg) | 5.65 ± 1.63 | 6.09 ± 0.86 | 5.80 ± 0.67 | 1.443 | 0.243 |
| RUSM (kg) | 2.85 ± 0.30 | 2.61 ± 0.41 | 2.67 ± 0.39 | 0.650 | 0.525 | RUSM (kg) | 1.95 ± 0.49 | 2.01 ± 0.33 | 1.90 ± 0.26 | 1.478 | 0.234 |
| RLSM (kg) | 8.68 ± 1.54 | 8.01 ± 1.33 | 8.02 ± 1.16 | 0.540 | 0.585 | RLSM (kg) | 5.90 ± 1.56 | 6.28 ± 0.80 | 5.90 ± 0.68 | 2.408 | 0.097 |
| TFM (kg) | 11.23 ± 6.80 | 7.64 ± 2.97 | 7.54 ± 3.29 | 2.187 | 0.119 | TFM (kg) | 12.15 ± 8.56 | 12.46 ± 4.79 | 10.63 ± 5.02 | 1.247 | 0.293 |
| LUFM (kg) | 0.73 ± 0.50 | 0.47 ± 0.20 | 0.47 ± 0.22 | 2.247 | 0.113 | LUFM (kg) | 1.10 ± 0.99 | 1.02 ± 0.49 | 0.80 ± 0.44 | 2.187 | 0.119 |
| LLFM (kg) | 3.28 ± 1.68 | 2.33 ± 0.76 | 2.32 ± 0.76 | 2.590 | 0.082 | LLFM (kg) | 3.75 ± 2.05 | 3.78 ± 0.98 | 3.30 ± 0.85 | 2.471 | 0.091 |
| RUFM (kg) | 0.70 ± 0.45 | 0.47 ± 0.19 | 0.45 ± 0.19 | 2.665 | 0.076 | RUFM (kg) | 1.05 ± 0.92 | 1.00 ± 0.50 | 0.78 ± 0.44 | 1.980 | 0.145 |
| RLFM (kg) | 3.28 ± 1.68 | 2.33 ± 0.75 | 2.30 ± 0.78 | 2.592 | 0.081 | RLFM (kg) | 3.90 ± 2.12 | 3.87 ± 1.00 | 3.39 ± 0.87 | 2.411 | 0.096 |
| HS (kg) | 39.68 ± 9.17 | 30.93 ± 7.74 | 30.82 ± 7.58 | 2.499 | 0.089 | HS (kg) | 21.55 ± 9.55 | 19.89 ± 7.60 | 19.13 ± 5.49 | 0.238 | 0.788 |
Abbreviations: BMI, body mass index; FFM, fat‐free mass; FM, fat mass; HS, handgrip strength; LLFM, left lower limb fat mass; LLSM, left lower limb skeletal muscle mass; LUFM, left upper limb fat mass; LUSM, left upper limb skeletal muscle mass; RLFM, right lower limb fat mass; RLSM, right lower limb skeletal muscle mass; RUFM, right upper limb fat mass; RUSM, right upper limb skeletal muscle mass; SMM, skeletal muscle mass; TFM, trunk fat mass; TSM, trunk skeletal muscle mass.
4. DISCUSSION
Skeletal muscle strength and mass are strongly determined by genetics (Tan et al., 2012), and inter‐individual variation in muscle phenotype may be attributed to genetic factors, environmental factors, and/or gene–environment interactions (Pratt et al., 2019). Previous studies have reported that skeletal muscle development is impaired and lean mass is significantly reduced in FTO‐deficient mice (McMurray et al., 2013; Wang et al., 2017). Changes in FTO gene expression are associated with changes in skeletal muscle percentage (Doaei et al., 2019), but it has remained unclear how FTO affects skeletal muscle mass. FTO gene polymorphism in China has only been found in the Han (Huang et al., 2011) and Mongolian (Zhang et al., 2018) Chinese, but no research has involved the Tibetans. We found that the FTO rs9939609 and rs9936385 polymorphisms of Tibetan women were associated with lower limb skeletal muscle mass and sarcopenia. TT homozygotes were at higher risk for sarcopenia (OR = 2.414, CI: 1.270–4.586, p = 0.007) (Figures 2 and 3). This risk genotype is in line with the results of Sonestedt et al., (2011), Karasik et al., (2019), Zillikens et al., (2017), and Khanal et al., (2020), but different from those of Khanal et al., (2020) and Heffernan et al. (2017). These apparent differences in genotypic associations could reflect genetic and environmental differences. The minor allele (A) frequency of rs9939609 was 24.06% for the total Tibetan population, 18.1% in the sarcopenia group, and 30.0% in the healthy control group. It was significantly diverse among different populations, being the highest in Turks (40.2%) and lowest in Mongolians (12.0%) (Table 10). Few studies have considered the FTO rs9936385 polymorphism, and the differences in allele frequency among populations are still unknown. In this study, the C allele frequency was 18.1% in the Tibetan sarcopenia group and 30.0% for healthy controls.
FIGURE 2.
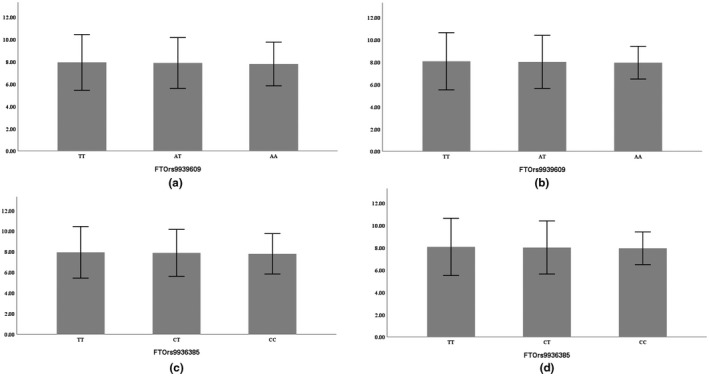
Comparison of the lower limb muscle mass (in men) among three genotypes (a,c: left lower limb, b,d: right lower limb, mean ± SD)
FIGURE 3.
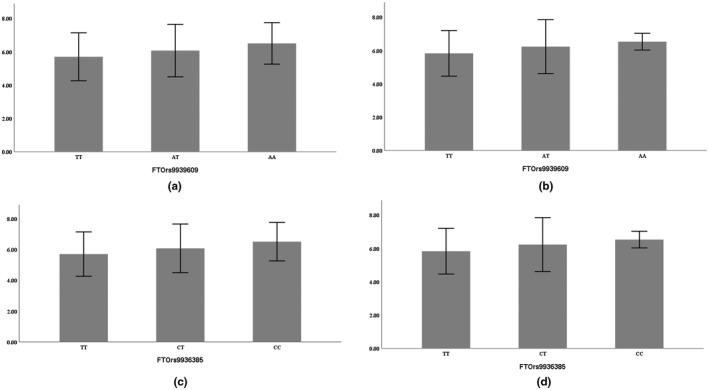
Comparison of lower limb muscle mass (in women) among three genotypes (a,c: left lower limb, b,d: right lower limb, mean ± SD)
TABLE 10.
Comparison of FTO rs9939609 polymorphism among different populations
| Group | Reference | n | TT | AT | AA | A | T |
|---|---|---|---|---|---|---|---|
| Tibetan (Control) | — | 80 | 37 (46.25) | 38 (47.5) | 5 (6.25) | 48 (30.0) | 112(70.0) |
| Tibetan (Sarcopenia) | — | 80 | 54 (67.5) | 23 (28.75) | 3 (3.75) | 29 (18.1) | 131 (81.9) |
| Han (Hubei) | Huang et al., (2011) | 1200 | 450 (37.5) | 545 (45.4) | 205 (17.1) | 955 (39.8) | 1445 (60.2) |
| Mongolians | Zhang et al., (2018) | 200 | 156 (78.0) | 40 (20.0) | 4 (2.0) | 48 (12.0) | 352 (88.0) |
| Taiwanese | Chang et al. (2008) | 1525 | 1158 (75.9) | 347 (22.8) | 20 (1.3) | 387 (12.7) | 2663 (87.3) |
| Kazakh | Sikhayeva et al. (2017) | 767 | 436 (56.8) | 274 (35.7) | 57 (7.4) | 388 (25.3) | 1146 (74.7) |
| Mexican | Villalobos‐Comparán et al., (2008) | 424 | 297 (70.0) | 114 (26.9) | 13 (3.1) | 140 (16.5) | 708 (83.5) |
| Russian | Guilherme et al. (2019) | 754 | 320 (42.4) | 333 (44.2) | 101 (13.4) | 535 (35.5) | 973 (64.5) |
| Brazilian | Guilherme et al. (2019) | 652 | 261 (40.0) | 297 (45.6) | 94 (14.4) | 485 (37.2) | 819 (62.8) |
| Indian | Ningombam et al. (2018) | 521 | 381 (73.1) | 138 (26.5) | 2 (0.4) | 142 (13.6) | 900 (86.4) |
| Swede | Gustavsson et al. (2014) | 4402 | 1629 (37.0) | 2066 (46.9) | 707 (16.1) | 3480 (39.5) | 5324 (60.5) |
| Japanese | Karasawa et al. (2010) | 2639 | 1680 (63.7) | 837 (31.7) | 122 (4.6) | 1081 (20.5) | 4197 (79.5) |
| Turkish | Ağagündüz and Gezmen‐Karadağ (2019) | 200 | 77 (38.5) | 85 (42.5) | 38 (19.0) | 161 (40.2) | 239 (59.8) |
FTO is an upstream regulator of the mammalian target of rapamycin (mTOR) pathway, FTO linked amino acid availability and mammalian target of rapamycin complex 1 (mTORC1) signaling, which regulates growth and translation; cells lacking FTO display decreased activation of the mTORC1 pathway and increased autophagy (Gulati et al., 2013; Loos & Yeo, 2014). According to Wang et al., (2017), the expression of PGC‐1α and mitochondrial biogenesis with mTOR dependency is regulated by FTO, and skeletal muscle differentiation and development are influenced by the mTOR‐PGC‐1α‐mitochondria axis. In skeletal muscle, FTO mRNA expression is negatively associated with lipid accumulation and positively correlated with glucose oxidation rates and the expression of genes involved in oxidative phosphorylation including PGC‐1α (Grunnet et al., 2009; Wu et al., 2017). FTO mRNA abundance in skeletal muscle is significantly reduced in elderly subjects compared to younger ones (Grunnet et al., 2009). In the elderly, the decrease in FTO mRNA may reduce mTOR signaling, leading to impairment of skeletal muscle differentiation and development, and increasing autophagy; it may also reduce the expression of PGC‐1α, impair mitochondrial function, accelerate muscle atrophy, increase lipid accumulation in skeletal muscle cells, and have a negative impact on muscle mass, leading to sarcopenia. According to Mehrdad et al. (2020), the FTO rs9939609 polymorphism is associated with serum levels of leptin, and the homozygotes for the FTO rs9939609 minor allele (A) have higher levels than the TT genotype. Appendicular skeletal muscle mass is negatively correlated with leptin, and compared to non‐sarcopenic subjects, serum levels of leptin are significantly higher in sarcopenic patients (C. W. Li et al., 2019; Waters et al., 2008). The increase in leptin may play a key role in the development of age‐related sarcopenia (Morley, 2001; Ng et al., 2018). Therefore, further investigation is needed to determine the association between FTO polymorphisms with serum levels of leptin in Tibetans.
Previous studies have reported that the polymorphisms of ACVR2B rs2276541 and IRS1 rs2943656 were associated with lean mass, but we found there was no association between polymorphisms of ACVR2B rs2276541 and IRS1 rs2943656 and sarcopenia and body composition in Tibetans, it may be attributed to genetic differences. This proves that the identified risk genes cannot be applied to all populations. Both ACVR2B and IRS1 code for a receptor, and further investigation is needed to research the association between polymorphisms of upstream gene (MSTN, IGF1) and sarcopenia in Tibetans. Interestingly, we found that the rs9939609 polymorphism was associated with the rs9936385 polymorphism in Tibetans. The A allele at rs9939609 and C allele at rs9936385 appeared concurrently, and further investigation is needed to determine the relationship between the two SNPs. And the previous study has found that the FTO mRNA level correlated positively with gene expression levels of hypoxia‐inducible factor‐1a(HIF‐1a) related to hypoxia in adipose tissue (Lappalainen et al., 2010). We speculate that the simultaneous mutation of the two SNPs in Tibetan people was caused by long‐term exposure to the plateau environment, and it may have a protective effect on their skeletal muscle mass. The main limitation of our study was the limited sample size, which may have led to a certain deviation in the association between sarcopenia and gene polymorphisms.
5. CONCLUSION
To the best of our knowledge, this is the first study to investigate the association between FTO rs9939609, FTO rs9936385, ACVR2B rs2276541, and IRS1 rs2943656 polymorphisms with sarcopenia in the Tibetan population. Our study provides evidence for an association between FTO polymorphisms and lower limb muscle mass and sarcopenia in Tibetan women, TT homozygotes had a higher risk for sarcopenia. And we found there is no association between polymorphisms of ACVR2B rs2276541, IRS1 rs2943656 and sarcopenia and body composition in Tibetans.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
6. ETHICS STATEMENT
The study was approved by the Research Ethics Committee of Jinzhou Medical University in accordance with the Declaration of Helsinki. Verbal and written informed consent were obtained from all participants.
AUTHOR CONTRIBUTIONS
Xin Li, Ying Chen, Yaqiong Jiang, and Wenhui Li carried out the investigation of the Tibetan population. Liping Ye guided and helped perform the experiments. Youfeng Wen guided the investigation, and modified and reviewed the manuscript. Xianpeng Zhang carried out the experiments, and conceived and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Lhasa Tama Community Health Service Center for their help.
Zhang, X., Ye, L., Li, X., Chen, Y., Jiang, Y., Li, W., & Wen, Y. (2021). The association between sarcopenia susceptibility and polymorphisms of FTO, ACVR2B, and IRS1 in Tibetans. Molecular Genetics & Genomic Medicine, 9, e1747. 10.1002/mgg3.1747
Funding information
This study was supported by grants from the National Natural Science Foundation of China (No. 31571233) and the scientific research project from the Education Department of Liaoning Province (No. JYTJCZR2020074).
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES
- Beall, C. M., Cavalleri, G. L., Deng, L., Elston, R. C., Gao, Y., Knight, J., Li, C., Li, J. C., Liang, Y., McCormack, M., Montgomery, H. E., Pan, H., Robbins, P. A., Shianna, K. V., Tam, S. C., Tsering, N., Veeramah, K. R., Wang, W., Wangdui, P., … Zheng, Y. T. (2010). Natural selection on EPAS1 (HIF2) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences, 107(25), 11459–11464. 10.1073/pnas.1002443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐K., Liu, L.‐K., Woo, J., Assantachai, P., Auyeung, T.‐W., Bahyah, K. S., Chou, M.‐Y., Chen, L.‐Y., Hsu, P.‐S., Krairit, O., Lee, J. S. W., Lee, W.‐J., Lee, Y., Liang, C.‐K., Limpawattana, P., Lin, C.‐S., Peng, L.‐N., Satake, S., Suzuki, T., … Arai, H. (2014). Sarcopenia in Asia: Consensus report of the Asian Working Group for Sarcopenia. Journal of the American Medical Directors Association, 15(2), 95–101. 10.1016/j.jamda.2013.11.025 [DOI] [PubMed] [Google Scholar]
- Doaei, S., Kalantari, N., Mohammadi, N. K., Izadi, P., Gholamalizadeh, M., Eini‐Zinab, H., Salonurmi, T., Jarrahi, A., Rafieifar, S., Janipoor, R., Sadeghypor, M., Tabesh, G., & Goodarzi, M. (2019). Up‐regulation of FTO gene expression was associated with increase in skeletal muscle mass in overweight male adolescents. Archives of Medical Science, 15(5), 1133–1137. 10.5114/aoms.2019.87239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünnwald, T., Gatterer, H., Faulhaber, M., Arvandi, M., & Schobersberger, W. (2019). Body composition and body weight changes at different altitude levels: A systematic review and meta‐analysis. Frontiers in Physiology, 10. 10.3389/fphys.2019.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig, H. K., & Carmeliet, P. (2011). Hypoxia and inflammation. New England Journal of Medicine, 364(7), 656–665. 10.1056/NEJMra0910283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusch, C., Gfrorer, W., Koch, C., Thomas, A., Grunert, A., & Moeller, H. (1996). Water turnover and body composition during long‐term exposure to high altitude (4,900‐7,600 m). Journal of Applied Physiology, 80(4), 1118–1125. 10.1152/jappl.1996.80.4.1118 [DOI] [PubMed] [Google Scholar]
- Grant, S. F. A., Li, M., Bradfield, J. P., Kim, C. E., Annaiah, K., Santa, E., Glessner, J. T., Casalunovo, T., Frackelton, E. C., Otieno, F. G., Shaner, J. L., Smith, R. M., Imielinski, M., Eckert, A. W., Chiavacci, R. M., Berkowitz, R. I., & Hakonarson, H. (2008). Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS One, 3(3), e1746. 10.1371/journal.pone.0001746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunnet, L. G., Nilsson, E., Ling, C., Hansen, T., Pedersen, O., Groop, L., Vaag, A., & Poulsen, P. (2009). Regulation and function of FTO mRNA expression in human skeletal muscle and subcutaneous adipose tissue. Diabetes, 58(10), 2402–2408. 10.2337/db09-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati, P., Cheung, M. K., Antrobus, R., Church, C. D., Harding, H. P., Tung, Y.‐C.‐ L., Rimmington, D., Ma, M., Ron, D., Lehner, P. J., Ashcroft, F. M., Cox, R. D., Coll, A. P., O'Rahilly, S., & Yeo, G. S. H. (2013). Role for the obesity‐related FTO gene in the cellular sensing of amino acids. Proceedings of the National Academy of Sciences, 110(7), 2557–2562. 10.1073/pnas.1222796110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan, S. M., Stebbings, G. K., Kilduff, L. P., Erskine, R. M., Day, S. H., Morse, C. I., McPhee, J. S., Cook, C. J., Vance, B., Ribbans, W. J., Raleigh, S. M., Roberts, C., Bennett, M. A., Wang, G., Collins, M., Pitsiladis, Y. P., & Williams, A. G. (2017). Fat mass and obesity associated (FTO) gene influences skeletal muscle phenotypes in non‐resistance trained males and elite rugby playing position. BMC Genetics, 18(1). 10.1186/s12863-017-0470-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinney, A., Nguyen, T. T., Scherag, A., Friedel, S., Brönner, G., Müller, T. D., Grallert, H., Illig, T., Wichmann, H.‐E., Rief, W., Schäfer, H., & Hebebrand, J. (2007). Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS One, 2(12), e1361. 10.1371/journal.pone.0001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Sun, Y., & Sun, J. (2011). Combined effects of FTO rs9939609 and MC4R rs17782313 on obesity and BMI in Chinese Han populations. Endocrine, 39(1), 69–74. 10.1007/s12020-010-9413-6 [DOI] [PubMed] [Google Scholar]
- Karasik, D., Zillikens, M. C., Hsu, Y.‐H., Aghdassi, A., Akesson, K., Amin, N., Barroso, I., Bennett, D. A., Bertram, L., Bochud, M., Borecki, I. B., Broer, L., Buchman, A. S., Byberg, L., Campbell, H., Campos‐Obando, N., Cauley, J. A., Cawthon, P. M., Chambers, J. C., … Ohlsson, C. (2019). Disentangling the genetics of lean mass. The American Journal of Clinical Nutrition, 109(2), 276–287. 10.1093/ajcn/nqy272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal, P., He, L., Stebbings, G., Onambele‐Pearson, G. L., Degens, H., Williams, A., Thomis, M., & Morse, C. I. (2020). Prevalence and association of single nucleotide polymorphisms with sarcopenia in older women depends on definition. Scientific Reports, 10(1). 10.1038/s41598-020-59722-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimentidis, Y. C., Bea, J. W., Thompson, P., Klimecki, W. T., Hu, C., Wu, G., Nicholas, J. S., Ryckman, K. K., & Chen, Z. (2016). Genetic variant in ACVR2B is associated with lean mass. Medicine & Science in Sports & Exercise, 48(7), 1270–1275. 10.1249/mss.0000000000000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen, T., Kolehmainen, M., Schwab, U., Pulkkinen, L., de Mello, V. D. F., Vaittinen, M., Laaksonen, D. E., Poutanen, K., Uusitupa, M., & Gylling, H. (2010). Gene expression of FTO in human subcutaneous adipose tissue, peripheral blood mononuclear cells and adipocyte cell line. Journal of Nutrigenetics and Nutrigenomics, 3(1), 37–45. 10.1159/000320732 [DOI] [PubMed] [Google Scholar]
- Lee, S.‐J., & McPherron, A. C. (2001). Regulation of myostatin activity and muscle growth. Proceedings of the National Academy of Sciences, 98(16), 9306–9311. 10.1073/pnas.151270098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.‐W., Yu, K., Shyh‐Chang, N. G., Li, G.‐X., Jiang, L.‐J., Yu, S.‐L., Xu, L.‐Y., Liu, R.‐J., Guo, Z.‐J., Xie, H.‐Y., Li, R.‐R., Ying, J., Li, K., & Li, D.‐J. (2019). Circulating factors associated with sarcopenia during ageing and after intensive lifestyle intervention. Journal of Cachexia, Sarcopenia and Muscle, 10(3), 586–600. 10.1002/jcsm.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Cai, B., Abdalla, B. A., Zhu, X., Zheng, M., Han, P., & Zhang, X. (2019). LncIRS1 controls muscle atrophy via sponging miR‐15 family to activate IGF1‐PI3K/AKT pathway. Journal of Cachexia, Sarcopenia and Muscle, 10(2), 391–410. 10.1002/jcsm.12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits, G., Malkin, I., Moayyeri, A., Spector, T. D., & Hammond, C. J. (2012). Association of FTO gene variants with body composition in UK twins. Annals of Human Genetics, 76(5), 333–341. 10.1111/j.1469-1809.2012.00720.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos, R. J. F., & Yeo, G. S. H.(2014). The bigger picture of FTO—The first GWAS‐identified obesity gene. Nature Reviews Endocrinology, 10(1), 51–61. 10.1038/nrendo.2013.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D., Lou, H., Yuan, K., Wang, X., Wang, Y., Zhang, C., Lu, Y., Yang, X., Deng, L., Zhou, Y., Feng, Q., Hu, Y. A., Ding, Q., Yang, Y., Li, S., Jin, L. I., Guan, Y., Su, B., Kang, L., & Xu, S. (2016). Ancestral origins and genetic history of Tibetan highlanders. The American Journal of Human Genetics, 99(3), 580–594. 10.1016/j.ajhg.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, T., Biniecka, M., Veale, D. J., & Fearon, U. (2018). Hypoxia, oxidative stress and inflammation. Free Radical Biology and Medicine, 125, 15–24. 10.1016/j.freeradbiomed.2018.03.042 [DOI] [PubMed] [Google Scholar]
- McMurray, F., Church, C. D., Larder, R., Nicholson, G., Wells, S., Teboul, L., Tung, Y. C. L., Rimmington, D., Bosch, F., Jimenez, V., Yeo, G. S. H., O'Rahilly, S., Ashcroft, F. M., Coll, A. P., & Cox, R. D. (2013). Adult onset global loss of the Fto gene alters body composition and metabolism in the mouse. PLoS Genetics, 9(1), e1003166. 10.1371/journal.pgen.1003166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrdad, M., Doaei, S., Gholamalizadeh, M., Fardaei, M., Fararouei, M., & Eftekhari, M. H. (2020). Association of FTO rs9939609 polymorphism with serum leptin, insulin, adiponectin, and lipid profile in overweight adults. Adipocyte, 9(1), 51–56. 10.1080/21623945.2020.1722550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merra, G., Gualtieri, P., Cioccoloni, G., Falco, S., Bigioni, G., Tarsitano, M. G., Capacci, A., Piccioni, A., Costacurta, M., Franceschi, F., & Di Renzo, L. (2020). FTO rs9939609 influence on adipose tissue localization in the Italian population. European Review for Medical and Pharmacological Sciences, 24(6), 3223–3235. 10.26355/eurrev_202003_20689 [DOI] [PubMed] [Google Scholar]
- Morley John, E. (2001). Anorexia, sarcopenia, and aging. Nutrition, 17(7–8), 660–663. 10.1016/s0899-9007(01)00574-3 [DOI] [PubMed] [Google Scholar]
- Ng, T. P., Lu, Y., Choo, R. W. M., Tan, C. T. Y., Nyunt, M. S. Z., Gao, Q. I., Mok, E. W. H., & Larbi, A. (2018). Dysregulated homeostatic pathways in sarcopenia among frail older adults. Aging Cell, 17(6), e12842. 10.1111/acel.12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasiakos, S. M., Berryman, C. E., Carrigan, C. T., Young, A. J., & Carbone, J. W. (2017). Muscle protein turnover and the molecular regulation of muscle mass during hypoxia. Medicine & Science in Sports & Exercise, 49(7), 1340–1350. 10.1249/mss.0000000000001228 [DOI] [PubMed] [Google Scholar]
- Pratt, J., Boreham, C., Ennis, S., Ryan, A. W., & De Vito, G. (2019). Genetic associations with aging muscle: A systematic review. Cells, 9(1), 12– 10.3390/cells9010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, S., Jiang, Z.‐X., He, X., Liu, Y. U., Zhang, Y.‐X., Zhang, L., Pei, Y.‐F., Zhang, M., Hai, R., Gu, G.‐S., Liu, B.‐L., Tian, Q., Zhang, Y.‐H., Wang, J.‐Y., & Deng, H.‐W. (2020). Replication of FTO Gene associated with lean mass in a Meta‐Analysis of Genome‐Wide Association Studies. Scientific Reports, 10(1). 10.1038/s41598-020-61406-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, S. M., Zmuda, J. M., Cauley, J. A., Shea, P. R., & Ferrell, R. E. (2004). Vitamin D receptor genotype is associated with fat‐free mass and sarcopenia in elderly men. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 59(1), B10–B15. 10.1093/gerona/59.1.b10 [DOI] [PubMed] [Google Scholar]
- Sällman Almén, M., Rask‐Andersen, M., Jacobsson, J. A., Ameur, A., Kalnina, I., Moschonis, G., Juhlin, S., Bringeland, N., Hedberg, L. A., Ignatovica, V., Chrousos, G. P., Manios, Y., Klovins, J., Marcus, C., Gyllensten, U., Fredriksson, R., & Schiöth, H. B. (2013). Determination of the obesity‐associated gene variants within the entire FTO gene by ultra‐deep targeted sequencing in obese and lean children. International Journal of Obesity, 37(3), 424–431. 10.1038/ijo.2012.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Luo, L., Eash, J., Ibebunjo, C., & Glass, D. J. (2011). The SCF‐Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling. Developmental Cell, 21(5), 835–847. 10.1016/j.devcel.2011.09.011 [DOI] [PubMed] [Google Scholar]
- Simonson, T. S., Yang, Y., Huff, C. D., Yun, H., Qin, G., Witherspoon, D. J., Bai, Z., Lorenzo, F. R., Xing, J., Jorde, L. B., Prchal, J. T., & Ge, R. (2010). Genetic evidence for high‐altitude adaptation in Tibet. Science, 329(5987), 72–75. 10.1126/science.1189406 [DOI] [PubMed] [Google Scholar]
- Sonestedt, E., Gullberg, B., Ericson, U., Wirfält, E., Hedblad, B., & Orho‐Melander, M. (2011). Association between fat intake, physical activity and mortality depending on genetic variation in FTO. International Journal of Obesity, 35(8), 1041–1049. 10.1038/ijo.2010.263 [DOI] [PubMed] [Google Scholar]
- Song, D., Navalsky, B. E., Guan, W., Ingersoll, C., Wang, T., Loro, E., Eeles, L., Matchett, K. B., Percy, M. J., Walsby‐Tickle, J., McCullagh, J. S. O., Medina, R. J., Khurana, T. S., Bigham, A. W., Lappin, T. R., & Lee, F. S. (2020). TibetanPHD2, an allele with loss‐of‐function properties. Proceedings of the National Academy of Sciences, 117(22), 12230–12238. 10.1073/pnas.1920546117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L. J., Liu, S. L., Lei, S. F., Papasian, C. J., & Deng, H. W. (2012). Molecular genetic studies of gene identification for sarcopenia. Human Genetics, 131(1), 1–31. 10.1007/s00439-011-1040-7 [DOI] [PubMed] [Google Scholar]
- Villalobos‐Comparán, M., Teresa Flores‐Dorantes, M., Teresa Villarreal‐Molina, M., Rodríguez‐Cruz, M., García‐Ulloa, A. C., Robles, L., & Canizales‐Quinteros, S. (2008). The FTO gene is associated with adulthood obesity in the Mexican population. Obesity (Silver Spring), 16(10), 2296–2301. 10.1038/oby.2008.367 [DOI] [PubMed] [Google Scholar]
- Walsh, S., Metter, E. J., Ferrucci, L., & Roth, S. M. (2007). Activin‐type II receptor B (ACVR2B) and follistatin haplotype associations with muscle mass and strength in humans. Journal of Applied Physiology, 102(6), 2142–2148. 10.1152/japplphysiol.01322.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandrag, L., Siervo, M., Riley, H. L., Khosravi, M., Fernandez, B. O., Leckstrom, C. A., Martin, D. S., Mitchell, K., Levett, D. Z. H., Montgomery, H. E., Mythen, M. G., Stroud, M. A., Grocott, M. P. W., & Feelisch, M. (2017). Does hypoxia play a role in the development of sarcopenia in humans? Mechanistic insights from the Caudwell Xtreme Everest Expedition. Redox Biology, 13, 60–68. 10.1016/j.redox.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Huang, N., Yang, M., Wei, D., Tai, H., Han, X., Gong, H., Zhou, J., Qin, J., Wei, X., Chen, H., Fang, T., & Xiao, H. (2017). FTO is required for myogenesis by positively regulating mTOR‐PGC‐1α pathway‐mediated mitochondria biogenesis. Cell Death & Disease, 8(3), e2702. 10.1038/cddis.2017.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, D. L., Qualls, C. R., Dorin, R. I., Veldhuis, J. D., & Baumgartner, R. N. (2008). Altered growth hormone, cortisol, and leptin secretion in healthy elderly persons with sarcopenia and mixed body composition phenotypes. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 63(5), 536–541. 10.1093/gerona/63.5.536 [DOI] [PubMed] [Google Scholar]
- White, T. A., & LeBrasseur, N. K. (2014). Myostatin and sarcopenia: Opportunities and challenges ‐ A mini‐review. Gerontology, 60(4), 289–293. 10.1159/000356740 [DOI] [PubMed] [Google Scholar]
- Wing, M. R., Ziegler, J., Langefeld, C. D., Ng, M. C. Y., Haffner, S. M., Norris, J. M., Goodarzi, M. O., & Bowden, D. W. (2009). Analysis of FTO gene variants with measures of obesity and glucose homeostasis in the IRAS Family Study. Human Genetics, 125(5–6), 615–626. 10.1007/s00439-009-0656-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W., Feng, J., Jiang, D., Zhou, X., Jiang, Q., Cai, M., Wang, X., Shan, T., & Wang, Y. (2017). AMPK regulates lipid accumulation in skeletal muscle cells through FTO‐dependent demethylation of N6‐methyladenosine. Scientific Reports, 7(1). 10.1038/srep41606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L., Wen, Y., Chen, Y., Yao, J., Li, X., Liu, Y., Song, J., & Sun, Z. (2020). Diagnostic reference values for sarcopenia in Tibetans in China. Scientific Reports, 10(1). 10.1038/s41598-020-60027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Xia, X., Fang, S., & Yuan, X. (2018). Relationship between fat mass and obesity‐associated (FTO) gene polymorphisms with obesity and metabolic syndrome in ethnic Mongolians. Medical Science Monitor, 24, 8232–8238. 10.12659/msm.910928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillikens, M. C., Demissie, S., Hsu, Y. H., Yerges‐Armstrong, L. M., Chou, W. C., Stolk, L., Livshits, G., Broer, L., Johnson, T., Koller, D. L., Kutalik, Z., Luan, J., Malkin, I., Ried, J. S., Smith, A. V., Thorleifsson, G., Vandenput, L., Hua Zhao, J., Zhang, W., … Kiel, D. P. (2017). Large meta‐analysis of genome‐wide association studies identifies five loci for lean body mass. Nature Communications, 8(1), 1–3. 10.1038/s41467-017-00031-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


