Abstract
Excessive cytokine activity underlies many autoimmune conditions, particularly through the IL-17 and TNFα signaling axes. Both cytokines activate NF-κB, but appropriate induction of downstream effector genes requires coordinated activation of other transcription factors, notably CCAAT/Enhancer Binding Proteins (C/EBPs). Here we demonstrate the unexpected involvement of a post-transcriptional ‘epitranscriptomic’ mRNA modification (N6-methyladenosine, m6A) in regulating C/EBPβ and C/EBPδ in response to IL-17A, as well as IL-17F and TNFα. Prompted by the observation that C/EBPβ/δ-encoding transcripts contain m6A consensus sites, we show that Cebpd and Cebpb mRNAs are subject to m6A modification. Induction of C/EBPs is enhanced by an m6A methylase ‘writer’ and suppressed by a demethylase ‘eraser.’ The only m6A ‘reader’ found to be involved in this pathway was IGF2BP2 (IMP2), and IMP2 occupancy of Cebpd and Cebpb mRNA was enhanced by m6A modification. IMP2 facilitated IL-17-mediated Cebpd mRNA stabilization and promoted translation of C/EBPβ/δ in response to IL-17A, IL-17F and TNFα. RNASeq revealed transcriptome-wide IL-17-induced transcripts that are IMP2-influenced, and RIPSeq identified the subset of mRNAs that are directly occupied by IMP2, which included Cebpb and Cebpd. Lipocalin-2 (Lcn2), a hallmark of autoimmune kidney injury, was strongly dependent on IL-17, IMP2 and C/EBPβ/δ. Indeed, Imp2−/− mice were resistant to autoantibody-induced glomerulonephritis (AGN), showing impaired renal expression of C/EBPs and Lcn2. Moreover, IMP2 deletion initiated only after AGN onset ameliorated disease. Thus, post-transcriptional regulation of C/EBPs through m6A/IMP2 represents a new paradigm of cytokine-driven autoimmune inflammation.
One sentence summary:
The m6A ‘reader’ IMP2 promotes cytokine-induced kidney autoimmunity by posttranscriptional control of C/EBPβ/δ transcription factors.
INTRODUCTION
Over 20 million Americans live with an autoimmune disorder. The estimated economic burden in medical expenses and loss of productivity is enormous, exceeding even cancer care (1, 2). The advent of anti-cytokine antibody drugs three decades ago revolutionized treatment of many autoimmune conditions, beginning with biologics targeting TNF. More recently, IL-17 (IL-17A) and Th17 cells were found to be dysregulated in skin and joint conditions such as psoriasis, psoriatic arthritis and ankylosing spondylitis. IL-17 is also implicated in other autoimmune settings such as multiple sclerosis and autoantibody-induced glomerulonephritis (AGN) (3). In principle, understanding the molecular basis of cytokine-induced autoimmune pathology has potential to inform new therapeutic targets (4).
IL-17 signaling occurs predominantly in non-hematopoietic cell types expressing the IL-17 receptor (5), while nearly all cells express TNF receptors and are responsive to this cytokine. TNFα and IL-17 mediate many overlapping signals, leading to similar, though not identical, downstream gene profiles. Many transcription factors (TF) are activated by one or both of these cytokines, including NF-κB, IκBξ, AP-1, as well as the CCAAT/enhancer binding proteins (C/EBPs). Most known cytokine-induced genes are regulated by combinations of these TFs (6-12).
C/EBPs are leucine zipper TFs described in the 1980’s, yet their regulation remains surprisingly poorly understood, especially compared to other immune TF families such as NF-κB or STATs (13-17). C/EBPs bind to characteristic motifs on target promoters and enhancers and are pleiotropic activators of inflammation in response to numerous immune and microbial stimuli (14). C/EBPs form heterodimers and homodimers and autoregulate their own promoters (14, 16). C/EBPβ exists in multiple, alternatively translated isoforms, and is also subject to inducible phosphorylation that tunes its activity in response to cytokine signals (18-22). Early studies of IL-17 showed that this cytokine induces both C/EBPδ and C/EBPβ, and both are required for induction of downstream effector genes that underlie IL-17 biologic activity (7, 8).
Whereas TNF is a potent activator of new transcription (23), IL-17 is a major activator of post-transcriptional events through a complex network of RNA binding proteins (RBPs) (5). RBPs bind to respective client transcripts at cis-acting elements typically found within 3’ or 5’ untranslated regions (UTRs). A variety of RBPs (Regnase-1, Arid5a, ASF/SF2, HuR, DDX3X, Roquins, Act1) influence downstream RNA stability and/or translation in response to IL-17 (13, 24, 25). Moreover, there is crosstalk between transcriptional and post-transcriptional events, since several mRNAs encoding TFs are subject to post-transcriptional control. This phenomenon is well described for Nfkbiz (encoding the noncanonical NF-κB protein IκBξ) (5, 17, 26) which is regulated by Regnase-1, Arid5a and an Act1/DDX3X complex (26, 27). We recently showed that C/EBPβ, but not C/EBPδ, is regulated in part by Arid5a at the level of translation in the IL-17 pathway (17). Thus, regulation of gene expression by IL-17 and related cytokines involves transcriptional and post-transcriptional circuitry.
An underappreciated determinant of RNA fate is ‘epitranscriptomic’ modification of mRNA (28). N6-methyladenosine (m6A) is added to specific RNAs by methyltransferase enzymes termed ‘writers’, which can be reversed by demethylases (‘erasers’) (29). The consequences of m6A modification are myriad, impacting RNA stabilization, translational efficiency, splicing, nuclear export and phase separation (29). Deletion of the writer METTL3 in T cells disrupts homeostatic expansion and Treg function (30, 31), and viral infections alter m6A modification of host gene expression (32-34). Diverse ‘readers’ bind to m6A directly or to sequence structures influenced by this modification. The best-known m6A readers belong to the YTH-domain family, which typically destabilize their respective client mRNAs. Recent work identified members of the IGF2 mRNA binding protein (IGF2BP) family (known as IMPs) as noncanonical m6A readers that can stabilize target transcripts (35, 36). To date, the m6A pathway has been somewhat overlooked in the immune system (37).
AGN encompasses a heterogenous group of nephritic conditions caused by inappropriate responses to renal autoantigens. The most severe form is characterized by formation of glomerular crescents and tubulointerstitial inflammation (38-40). In humans, treatment with corticosteroids and cyclophosphamide increases AGN survival somewhat, but there is an unmet need to understand events within the kidney to develop better treatment strategies (41). While traditionally considered to be a B-cell-dependent disease, T helper (Th) cells drive renal damage in AGN (42, 43). Tissue-resident memory Th17 cells specific for commensal microbes are overrepresented in human GN nephrectomy samples (44). In mice, these TRM17 cells exacerbate pathology in crescentic GN, which was blocked with anti-cytokine antibodies that inhibit Th17 cell development (44). Lipocalin-2 (Lcn2, 24p3, NGAL) is a biomarker and driver of AGN (7, 45-47) and is potently induced by IL-17 through activation of C/EBPβ and C/EBPδ (6, 19, 26). The present study was prompted by the observation that noncoding sequences in Cebpd and Cebpb contain consensus m6A sites. Indeed, we show that C/EBP mRNAs are subject to m6A modification and are bound to and regulated by IMP2 (35, 48). Imp2−/− mice were resistant to AGN, and loss of IMP2 after induction of AGN ameliorated renal inflammation. Thus, induction of autoimmune nephritis is controlled by an IMP2/m6A epitranscriptomic axis through C/EBP transcription factors.
RESULTS
Cebpd and Cebpb mRNAs are subject to m6A modification
Prototypical genes in the IL-17 target gene signature include Il6, Lcn2 as well as the TFs Cebpd and Cebpb, but regulation of C/EBPs has long been enigmatic. Induction of these mRNAs is seen in numerous cell backgrounds including primary mouse embryonic fibroblasts (MEFs) and human renal epithelial cells (HK-2 (49)) (Fig 1A, fig S1a, b). The proximal promoters of the Il6 and Lcn2 genes contain C/EBP binding elements that are nonredundant for IL-17-dependent activation (6, 7, 19), and knockdown of C/EBPβ and C/EBPδ by siRNA in MEFs abrogated the upregulation of Il6 and Lcn2 by IL-17 as well as regulating one another (Fig 1A, fig S1c). As expected, IL-17 did not activate a Lcn2 promoter with a C/EBP binding site mutation (fig S1d, e) (6). Although C/EBPs were reported to be regulated by NF-κB in response to LPS (50), an IKK inhibitor surprisingly had no impact on induction of Cebpb or Cebpd in response to IL-17, though it abolished expression of Il6 and Lcn2, as expected (fig S1f) (6, 7, 51).
Fig 1. Cebpd and Cebpb mRNAs are subject to m6A modification.

(A) MEFs were transfected with siRNAs targeting Cebpd and Cebpb or control and treated ± IL-17 for 8 h. qPCR of the indicated mRNAs is shown. Data are normalized to untreated samples with control siRNA, from 3 independent experiments ± SEM. (B) Diagram of m6A modification by the m6A methyltransferase (writer) METTL3, and removal by the m6A demethylase (eraser) FTO. (C, D) MEFs were transfected with siRNAs targeting Mettl3 or Fto or non-targeting controls and treated ± IL-17 for 8 h. qPCR of the indicated mRNAs is shown. Data are normalized to untreated samples with control siRNA ± SEM, from 3 independent experiments. (E) MEFs were transfected with the indicated siRNAs and treated ± IL-17 for 6 h. C/EBPδ and C/EBPβ (isoforms LAP*, LAP and LIP (16) were assessed in nuclear extracts by immunoblotting. Blots are representative of 3-4 independent experiments. (F) Band intensity values were quantified from immunoblots. Means ± SEM from all experiments are shown. (G) MEFs transfected with the indicated siRNAs and treated ± IL-17 for 6 h. C/EBPδ and C/EBPβ were assessed in nuclear extracts by immunoblotting. Blots are representative of 4 independent experiments. (H) Band intensity values quantified from immunoblots. Means ± SEM from pooled experiments. (I) MEFs were treated with IL-17 for 3 h and subjected to RIP with m6A or IgG control Abs. qPCR of the indicated mRNAs is presented as % input. Data show mean ± SEM from 4 experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 by ANOVA with post hoc Tukey’s test. N.D, not detected.
To gain insight into how C/EBPs are regulated in the IL-17 pathway, we examined the noncoding sequences of these intronless genes. According to the RMBase and SRAMP databases (52, 53) Cebpd and Cebpb mRNAs contain several high confidence sites of predicted or empirical N6-methyladenosine (m6A) modification, mainly in the 3’ UTR. In contrast, Ccl20, another mRNA induced by IL-17, contains very few predicted m6A sites (fig S2, table S1). In general, deposition of m6A is mediated by the methyltransferase-like 3 (METTL3) enzyme and can be reversed by demethylases such as α-ketoglutarate-dependent dioxygenase (fat mass and obesity associated protein, FTO) (28) (Fig 1B). In response to IL-17, knockdown of Mettl3 suppressed expression of Cebpd mRNA (Fig 1C) as well as C/EBPδ protein (Fig 1E, F). Mettl3 silencing also showed a trend of reduced C/EBPβ protein expression (Fig 1E, F). Correspondingly, Mettl3 knockdown suppressed expression of the C/EBPβ/δ-dependent mRNAs Il6 and Lcn2, but had no effect on Ccl20 (fig S3). Conversely, knockdown of Fto, an m6A ‘eraser,’ increased Cebpd (Fig 1D) and corresponding C/EBPδ protein levels (Fig 1G, H), and increased Il6 and Lcn2 levels (fig S3). Interestingly, knockdown of Fto only reproducibly impacted the LIP isoform of C/EBPβ, perhaps indicating selective regulation of this TF at the level of alternative translation (Fig 1G, H). These data indicate involvement of the m6A pathway in control of C/EBPs.
To determine if C/EBP mRNAs are subject to m6A modification, we performed RNA immunoprecipitation (RIP) with anti-m6A antibodies (32). There was no enrichment of Cebpd or Cebpb in control IgG RIP samples (Fig 1I). Likewise, Ccl20 was not detected in m6A RIP samples, commensurate with its lack of predicted m6A motifs (fig S2, table S1). However, Cebpd mRNA was substantially increased upon anti-m6A RIP, both at baseline and after IL-17 treatment (Fig 1I). Cebpb mRNA was also constitutively enriched in anti-m6A RIPs, and did not change upon IL-17 signaling (Fig 1I). These data therefore implicate the m6A pathway in regulation of C/EBPs, and consequently the expression of downstream target mRNAs.
The m6A reader IMP2 promotes expression of IL-17 target genes via C/EBPs
Given this evidence that the m6A machinery regulates IL-17-dependent gene expression, we sought to define possible m6A RNA binding proteins (‘readers’) required for this process. Surprisingly, knockdown of none of the canonical YTH m6A readers suppressed Il6, Lcn2 or Cebpd (though knockdown of Ythdf1 increased Cebpd expression, suggesting it may be a negative regulator) (Fig 2A, fig S4). We noted that the predicted m6A motifs in Cebpd and Cebpb corresponded closely to consensus recognition sites for the noncanonical m6A readers of the IGF2BP family (known as IMPs), recently shown to stabilize client mRNAs such as Myc (35, 54). In MEFs, only IMP2 was expressed at substantial levels. To determine if IMP2 participates in the IL-17 pathway, MEFs were transfected with siRNA targeting Imp2, treated with IL-17 for 8 h, and subjected to RNA-Seq and Ingenuity Pathway Analysis (IPA). The top upstream regulators identified by IPA to be IMP2-dependent were TRAF3IP2 (better known as the IL-17-associated adaptor Act1) and IL-17A, as well as closely related pathways including TNFα, LPS, IL-1 family and NF-κB (fig S5a). Strikingly, following Imp2 knockdown, all these pathways were abrogated (Fig 2C).
Fig. 2. IMP2 but not other m6A readers promote expression of IL-17 target genes.
(A) MEFs were transfected with the indicated siRNAs and treated ± IL-17 for 8 h. qPCR of the indicated mRNAs is shown. Data are normalized to untreated samples with control siRNA ± SEM, from 3 independent experiments. (B) MEFs were transfected with the indicated siRNAs. qPCR and immunoblot of IMP2 is shown. Data are normalized to untreated samples with control siRNA ± SEM, from 3 independent experiments. (C) MEFs were transfected with siRNAs targeting Imp2 or non-targeting controls and treated ± IL-17 for 8 h. RNA-seq (n=3) was performed on the Illumina platform. Ingenuity Pathway Analysis of RNASeq showing top 10 predicted upstream regulators. Volcano plots showing the transcriptional response induced by IL-17 ± siImp2. In red are selected transcripts that were significantly changed (P value <0.05 and fold change > 2 or < −2; based on 3 experiments). (D) MEFs were transfected with siRNAs targeting Imp2 or non-targeting control and treated ± IL-17 for 8 h. qPCR of the indicated mRNAs is shown. Data are normalized to untreated samples with control siRNA ± SEM, from 3 independent experiments. (E) Imp2+/+ or Imp2−/− MEFs were treated ± IL-17 for 4 h qPCR of the indicated mRNAs is shown. Data are normalized to untreated samples with control siRNA ± SEM, 3 independent experiments. (F) HK-2 cells were transfected with siRNAs targeting IMP2 or non-targeting control and treated ± IL-17 for 8 h. qPCR of the indicated mRNAs is shown. Data are normalized to untreated samples with control siRNA ± SEM, from 4 independent experiments *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 by ANOVA with post hoc Tukey’s test.
Consistent with this, many characteristic IL-17 signature genes were reduced upon IMP2 knockdown, including Il6, Lcn2, Cebpd and several chemokines (Cxcl1, Cxcl5, Ccl2, Ccl7). (Fig 2C, fig S5b-e). Expression of selected genes was verified by qPCR (Fig 2D). Importantly, not all IL-17-induced genes were impacted by IMP2 deficiency such as Ccl20, revealing a level of specificity in the genes controlled by IMP2 (Fig 2D, fig S5e). This was consistent with absence of Ccl20 in m6A RIP samples (Fig 1I). Further supporting these findings, MEFs derived from Imp2−/− mice showed impaired IL-17 induction of Il6 and Lcn2 but not Ccl20 (Fig 2E). Also consistent with this, IMP2 was required for IL-17-induced expression of IL6 and LCN2 in HK-2 cells, a human renal epithelial cell line (Fig 2F). IMP2 knockdown did not alter expression of proximal signaling intermediates in the IL-17 pathway such as Il17ra, Act 1 (Traf3ip2), Traf2 or Traf6, (fig S5e, f), nor did loss of IMP2 affect expression of any m6A readers/writers/erasers tested (fig S5e, g). Based on IPA predictions (Fig 2C), we evaluated the role of IMP2 in related cytokine pathways. IMP2 deficiency led to reduced IL-17F- and TNFα-mediated upregulation of Il6 and Lcn2 (fig S5h), indicating that the IMP2 pathway is applicable to analogous cytokine pathways.
We next tested the hypothesis that IMP2 controls these downstream genes through regulation of C/EBPs. In MEFs and HK-2 cells, C/EBPδ mRNA and protein were elevated in response to IL-17 in an IMP2-dependent manner, demonstrated by siRNA and in Imp2−/− cells (Fig 3A-C, F, fig S6a). Unlike C/EBPδ, IMP2 deficiency did not impair Cebpb mRNA expression (Fig 3A, B), but protein levels of C/EBPβ were reduced in Imp2−/− cells or following Imp2 silencing. C/EBPβ has three major isoforms generated by alternative translation, LAP*, LAP, LIP; while all were impaired to some degree, LIP appeared to be the most sensitive to IMP2 deficiency (16) (Fig 3C, F, G, fig S6a-d). Likewise, C/EBPδ and C/EBPβ proteins were elevated in response to IL-17F in an IMP2-dependent manner (fig S6e). While TNFα-induced C/EBPβ protein was similarly IMP2-dependent (Fig 3E), TNFα did not induce C/EBPδ protein even at high concentrations, revealing a distinction between the transcription factors induced by IL-17 family cytokines compared to TNFα (Fig 3D).
Fig 3. IMP2 regulates IL-17-induced genes via C/EBPδ and C/EBPβ.

(A, B) MEFs (A) or HK-2 cells (B) transfected with pooled siRNAs targeting Imp2 or non-targeting control and treated ± IL-17 for 8 h. qPCR of the indicated mRNAs is shown. Data are normalized to untreated samples with control siRNA ± SEM, from 3–4 independent experiments. (C) Imp2+/+ or Imp2−/− MEFs treated with IL-17. C/EBPδ, C/EBPβ, IMP2 and YY1 in nuclear lysates were assessed by immunoblotting. Blots are representative of 3 experiments. (D) Imp2+/+ MEFs treated ± indicated doses of IL-17 or TNFα. C/EBPδ and YY1 in nuclear lysates were assessed by immunoblotting. Blots are representative of 2 experiments. (E) Imp2+/+ or Imp2−/− MEFs were treated ± 10 ng/ml TNFα. C/EBPβ and YY1 in nuclear lysates were assessed by immunoblotting. Blots represent 2 experiments. (F) MEFs transfected with siRNAs targeting Imp2 or non-targeting control and treated ± IL-17 for 6 h. C/EBPδ, C/EBPβ, IMP2 and YY1 in nuclear lysates were assessed by immunoblotting. Blots are representative of 4 experiments. (G) Pooled band intensity values with mean ± SEM from 4 experiments. (H) Imp2+/+ or Imp2−/− MEFs were transfected with Luc reporters driven by Il6 or Lcn2 promoters (6, 51). Cells were treated ± IL-17 for 8 h and Luc activity assessed. Data show fold-change relative to untreated Imp2+/+ cells. (I) Imp2−/− MEFs were co-transfected with C/EBPδ or C/EBPβ (7) with Il6- or Lcn2-Luc reporters. Cells were treated ± IL-17 for 8 h and Luc activity assessed. Data show fold-change relative to EV-transfected cells. *P<0.05, **P<0.01, ***P<0.001; ****P<0.0001 by ANOVA with post hoc Tukey’s test.
Next, we assessed the impact of IMP2 deficiency on activation of promoters that require C/EBP binding sites for IL-17/TNFα-mediated induction (6, 7). As shown, induction of a Luciferase (Luc) reporter driven by the Il6 or Lcn2 proximal promoters was largely abrogated in Imp2−/− MEFs (Fig 3H, fig S7a). Activation of the Il6 or Lcn2 promoters in Imp2−/− MEFs could be rescued by ectopic expression of C/EBPδ or C/EBPβ (Fig 3I, fig S7b, c). Together, these data support a role for IMP2 in mediating IL-17 signaling through regulation of C/EBPs.
IMP2 binds Cebpd and Cebpb mRNAs
We next evaluated the binding of IMP2 to selected IL-17-induced transcripts by RIP. Indeed, IMP2 RIP samples contained marked enrichment of both Cebpd and Cebpb mRNA compared to IgG controls, indicating that both transcripts interact directly with IMP2 (Fig 4A). Additionally, IMP2 bound to Ccl7 quite strongly. Il6, Lcn2 and Cxcl1 were also enriched in IMP2 RIP samples, albeit more weakly, and there was no enrichment of Ccl20.
Fig 4. IMP2 binds to Cebpd and Cebpb mRNAs and mediates post-transcriptional regulation.
(A) Imp2+/+ MEFs were treated ± IL-17 for 3 h and subjected to RIP with anti-IMP2 or IgG Abs. qPCR of the indicated mRNAs was normalized to input. Data show mean ± SEM of 4 independent experiments. Inset; IMP2 immunoprecipitates from Imp2+/+ and Imp2−/− MEFs was assessed by immunoblotting. (B) MEFs were treated with IL-17 for 3 h and subjected to RIP-seq with anti-IMP2 Abs. Diagram indicates the intersection between mRNAs that are IMP2-occupied based on RIPSeq compared to mRNAs that are IMP2-influenced at baseline or after IL-17 treatment. (C) Top: Diagram of Cebpd 3’UTR and predicted m6A site mutants (sequence, fig S8). Dashed line indicates sequence in the biotinylated synthetic mRNA. Bottom: HEK293T cells were co-transfected with IMP2-FLAG and a Luc reporter fused to WT Cebpd-3’UTR or sequences in which putative m6A sites were mutated. Lysates were subjected to RIP with anti-FLAG Abs and Luc mRNA assessed by qPCR. Data are normalized to input and show mean ± SEM of 3 independent experiments. (D) Lysates from HEK293T cells transfected with FLAG-tagged IMP2 were incubated with the biotinylated mRNAs corresponding to the Cebpd 3’ UTR (863-982, dashed line in panel C) in which indicated adenosines were m6A-modified or unmodified. RNA pulldowns were performed with streptavidin beads. FLAG-tagged IMP2 analyzed by immunoblotting. Blots are representative of 3 experiments. Pooled band intensity values with mean ± SEM. (E) MEFs were treated with TNFα for 3 h, given actinomycin D ± IL-17 for the indicated times, and Cebpd assessed by qPCR. Left: Data normalized to time=0 (100%), representative of 3 independent experiments. Right: Half-life was estimated by linear regression (17). (F) MEFs were treated ± IL-17 for 3 h and subjected to RIP with anti-eIF4G Abs. mRNAs were assessed by qPCR. Data normalized to untreated Imp2+/+ samples precipitated with IgG. Data are representative of 2 independent experiments. Inset; eIF4G in cytoplasmic RIP fractions from Imp2+/+ MEFs. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 by ANOVA with post hoc Tukey’s test or t-test.
To gain a transcriptome-wide view of the IMP2 clients in the IL-17 pathway, cells were stimulated with IL-17 for 3 h and lysates subjected to RIPSeq with anti-IMP2 Abs or IgG. Results analyzed with Piranha and DESeq2 (55, 56), eliminating non-annotated regions from consideration and using a cutoff of FDR>0.01 and a 2-fold enrichment. We identified 4933 mRNAs with IMP2-interactions. In this analysis we only considered mRNAs with known IMP2 binding motifs, which constituted ~63% of the transcripts in the IMP2-RIP samples (3217 total) (57). Comparison of the RIPSeq and RNASeq datasets distinguished those mRNAs that are direct IMP2 clients (based on RIPSeq) from the entirety of transcripts that are influenced by IMP2 silencing (based on RNASeq) (Fig 4B, Fig S8B). IMP2-bound RNAs identified in RIPSeq included Cebpd, Ccl7, and Il6, thus independently confirming the validity of these mRNAs as authentic IMP2 clients (Fig 4B). Cebpb was also enriched in the RIPSeq dataset, but its abundance was not altered by IMP2 silencing (Fig 1, Fig 4B) (58). Also as expected, there were no detectable interactions between IMP2 and Ccl20, which is upregulated by IL-17 but is neither methylated nor IMP2-dependent. Unexpectedly, Lcn2 was not enriched in the RIPSeq samples, possibly due to its relatively weak association with IMP2 seen in RIP (Fig 4A). These data thus demonstrate that a large subset of IMP2-controlled mRNAs bind to IMP2 directly, and that the rest are likely to be regulated secondarily, for example by IMP2-dependent TFs such as C/EBPβ/δ.
Analysis of Cebpd mRNA revealed consensus IMP2 binding sites within the 3’ UTR that corresponded to high-confidence m6A sites as predicted by the RMBase and SRAMP databases (Fig 4C, fig S8a, c). To better understand interactions between IMP2 and the Cebpd 3’ UTR, we linked this sequence to a luciferase reporter with some or all of the predicted m6A sites mutated to T (Fig 4C). These constructs were co-expressed with FLAG-tagged IMP2 in HEK293 cells, subjected to RIP with anti-FLAG Abs, and Luciferase mRNA enrichment was assessed by qPCR. There was substantial association of IMP2 with a Luc construct fused to the Cebpd 3’ UTR sequence (Fig 4C). In contrast, a Luc-Cebpd 3’ UTR construct in which all putative m6A sites were mutated led to reduced enrichment of Luciferase mRNA upon IMP2-RIP. Mutation of just the two highest confidence m6A sites (940, 977) similarly abolished IMP2 binding, demonstrating that these A sites are necessary for IMP2 binding. Interestingly, they are not sufficient, as a construct in which the remaining five putative m6A sites were mutated also exhibited reduced IMP2 binding (Fig 4C). These results indicate that m6A sites in the Cebpd 3’UTR mediate IMP2 occupancy. This finding is not only in agreement with published work on other m6A-modified 3’ UTRs, but is, to our knowledge, the most detailed mutagenesis of a 3’UTR with respect to m6A usage to date (59-61).
To confirm that IMP2 is a bona fide reader of m6A-modified Cebpd mRNA, we employed an in vitro RNA pulldown assay using synthetic biotinylated ‘bait’ transcripts spanning the Cebpd 3’UTR that either m6A-modified or unmodified (Fig 4D). These mRNAs were incubated with FLAG-tagged IMP2 derived from HEK293T cell overexpression. Samples were precipitated streptavidin-conjugated beads and immunoblotted with anti-FLAG. IMP2 interacted significantly more efficiently with the methylated bait than with the unmethylated controls, although at the highest concentrations IMP2 bound even to a non-modified transcript (Fig 4D). Therefore, m6A modification within the 3’ UTR markedly facilitates the binding of IMP2 to its client Cebpd.
RNA stability is a major mechanism by which inflammatory genes are regulated, so we evaluated the impact of IMP2 on C/EBP mRNA half-life using an mRNA decay assay (17, 62). Imp2+/+ and Imp2−/− MEFs cells were primed with TNFα for 3 h, washed, treated with actinomycin D to stop new transcription, stimulated with IL-17, and mRNA measured over 90 minutes. Transcript half-life (t½) was extrapolated by linear regression, as described (17, 63). IL-17 stabilized the Cebpd transcript in Imp2+/+ cells, increasing its estimated t½ from 86 to 117 minutes (Fig 4E). Cebpd mRNA decayed significantly more rapidly in Imp2−/− than in Imp2+/+ cells. Unlike Cebpd, the stability of Cebpb mRNA was unaffected by Imp2 deficiency or IL-17 treatment (Fig 4E).
IMP2 also regulates protein translation for some client transcripts (64, 65), so we evaluated occupancy of Cebpd and Cebpb mRNAs within the translation initiation complex by performing RIP of eIF4G (17, 66), a scaffolding subunit associated with mRNAs undergoing active translation. In control Imp2+/+ cells, Cebpd and Cebpb but not Gapdh were enriched in eIF4G RIP preparations following IL-17 treatment. However, levels of Cebpd and Cebpb within the eIF4G complex were substantially reduced in Imp2−/− cells (Fig 4F), consistent with reduced C/EBPδ and C/EBPβ translational efficiency. Accordingly, Cebpd and Cebpb are subject to m6A modification, which underlies the capacity of IMP2 to enhance C/EBPδ mRNA half-life and C/EBPδ and C/EBPβ protein expression.
HuR (Elavl1) and Act1 were previously shown to form dimeric complexes that mediate stability and translation of unstable mRNAs in the IL-17 pathway (24, 25). By co-IP, IMP2 associated constitutively and robustly with HuR but not with Act1 (Fig 5A). Like IMP2, knockdown of HuR impaired expression of Cebpd, Il6 and Lcn2 but not Cebpb (Fig 5B). Moreover, HuR deficiency was associated with reduced C/EBPδ and C/EBPβ protein expression (Fig 5C, D), in keeping with prior reports (67-70). These data suggest that IMP2 forms a complex with HuR to regulate C/EBPδ and C/EBPβ and consequently their downstream target genes.
Fig 5. IMP2 forms a complex with HuR to regulate C/EBPs.

(A) Imp2+/+ MEFs were treated ± IL-17 and immunoprecipitated with anti-IMP2 Abs or IgG controls. Blots are representative of 2 independent experiments. (B) MEFs were transfected with pooled siRNAs targeting HuR (Elavl1) or control and treated ± IL-17 for 8 h. Data show mean ± SEM of 3 experiments normalized to untreated. (C) MEFs were transfected with siRNAs targeting HuR or non-targeting control and treated ± IL-17 for 6 h. C/EBPδ, C/EBPβ, HuR and YY1 were assessed by immunoblotting. Blots are representative of 3 experiments. (D) Pooled band intensity values with mean ± SEM from all experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by ANOVA with post hoc Tukey’s test.
IMP2 deficient mice resist IL-17-driven autoantibody-induced glomerulonephritis (AGN)
Accumulating reports implicate Th17 cells in autoantibody-mediated glomerulonephritis (AGN) (44, 71-76). Here we use an autologous model of rapidly progressive proliferative AGN in which disease is induced by administering rabbit anti-mouse glomerular basement membrane (GBM) serum, leading to deposition of autoantibody complexes in glomeruli (72, 77, 78). The resulting renal pathology is IL-17-driven and shares clinical and pathological features of human crescentic GN (40, 71, 79). Lcn2 is a biomarker and driver of renal damage (80-84). Since Lcn2 is regulated by IMP2, IL-17 and C/EBPs, we postulated that Imp2−/− mice would be resistant to AGN. Imp2+/+ mice but not control Act1−/− mice developed elevated serum blood urea-nitrogen (BUN) and creatinine as well as marked histological renal pathology (Fig 6A) (72). Imp2−/− mice showed reduced BUN and creatinine levels (Fig 6A), reduced numbers of abnormal glomeruli, as well as decreased glomerular sclerosis, hypercellularity, and crescent formation (Fig 6B). Consistent with this, Imp2−/− mice had fewer kidney-infiltrating inflammatory monocytes compared to Imp2+/+ littermates, and a trend to reduced neutrophils (Fig 6C, fig S9). Markers of nephropathy including Lcn2, Il6 and Kim1 (aka, Tim-1 (84)), were also elevated in Imp2+/+ but not Imp2−/− or Act1−/− kidneys (Fig 6D).
Fig 6. IMP2 deficient mice are resistant to IL-17-driven renal autoimmune inflammation.
(A-B) Imp2+/+, Imp2−/− and Act1−/− mice were subjected to AGN (71). Renal dysfunction was assessed at day 14 by ELISA of serum BUN and creatinine. Data pooled from 2 experiments. (B) Left: Representative images of H&E-stained kidney sections at day 14 of AGN (400×). Right: Slides were scored for abnormal glomeruli in Imp2+/+ and Imp2−/− mice (n=3) at day 14 post-AGN. Bars show mean ± SEM. (C) Kidney homogenates were prepared on day 7. Live inflammatory monocytes were determined by staining for CD11b, Ly6C and Ly6G, gated on the live CD45+ population. Graphs show percent of live CD45+CD11b+Ly6ChighLy6G− cells. Data were pooled from 2 independent experiments. (D) The indicated mice were subjected to AGN (Imp2+/+ control n=10; Imp2+/+ AGN n=11, Imp2−/− control n=8, Imp2−/− AGN n=10, Act1−/− control n=6 and Act1−/− AGN n=6). Total RNA was extracted at day 7 and subjected to qPCR. Data were pooled from 2 experiments. (E) Imp2+/+ or Imp2−/− mice were lethally irradiated (900 rad) and reconstituted with BM from the indicated donor mice. After 6 weeks, mice were subjected to AGN. On day 14, BUN and creatinine levels were measured by ELISA. Data pooled from 2 experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, by ANOVA with post-hoc Tukey’s test.
To identify the cellular compartment where IMP2 functions during AGN, Imp2+/+ or Imp2−/− mice were irradiated and adoptively transferred with autologous or reciprocal bone marrow (BM). After 6 weeks of engraftment, chimeric mice were subjected to AGN. Imp2+/+ mice receiving Imp2+/+ or Imp2−/− BM exhibited kidney dysfunction as indicated by elevated serum BUN and creatinine, whereas Imp2−/− mice receiving Imp2+/+ or Imp2−/− BM showed kidney impairment (Fig 6E). Therefore, IMP2 acts in non-hematopoietic cells in AGN (85).
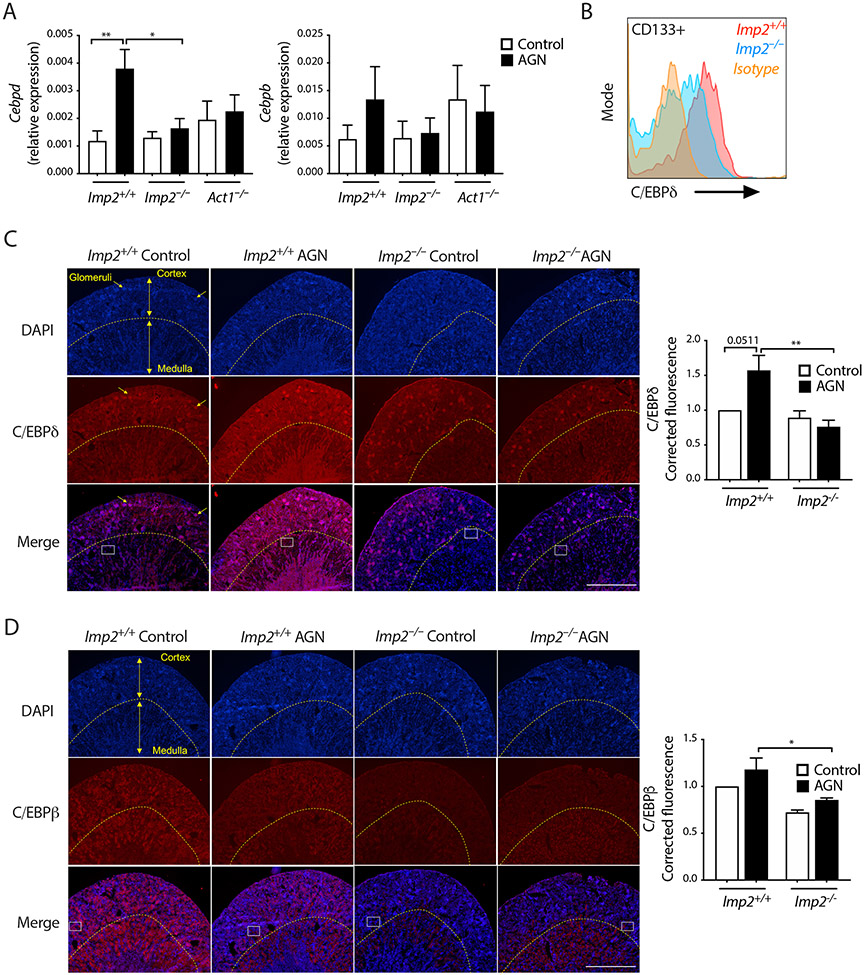
We next determined whether C/EBPs were regulated in the context of AGN and if so, whether this was IMP2-dependent. Indeed, Cebpd mRNA was elevated in total kidney tissue in Imp2+/+ mice during AGN but not Imp2−/− littermates or Act1−/− mice (Fig. 7A). The major IL-17/TNFα-responsive cells implicated in AGN are renal tubular epithelial cells (RTECs) (71, 72). Intracellular staining of C/EBPδ in kidney homogenates showed that C/EBPδ was expressed in CD45−CD133+ RTECs during AGN. Moreover, C/EBPδ staining was decreased in Imp2−/− compared to Imp2+/+ RTECs (Fig 7B; fig S10a). Immunofluorescent staining of Imp2+/+ control kidney sections revealed that C/EBPδ was high in glomeruli and low in RTECs in untreated mice, consistent with prior observations (86). During AGN, C/EBPδ was elevated in both medullary and cortical RTECs in Imp2+/+ but not Imp2−/− mice (Fig 7C; enlarged images: fig S10b), in line with findings in another model of renal injury (87). C/EBPβ expression patterns were strikingly different from C/EBPδ, with far less prominent staining in glomeruli (Fig 7D). Imp2−/− mice exhibited reduced expression of C/EBPβ compared to Imp2+/+ mice, especially in the cortex, regardless of AGN status (Fig 7D, enlarged images: fig S10c). Thus, IMP2 regulates C/EBPδ and C/EBPβ in kidney during AGN.
Fig. 7. Impaired C/EBP in Imp2−/− mice during AGN.
(A) The indicated mice were subjected to AGN (Imp2+/+ control n=10; Imp2+/+ AGN n=11, Imp2−/− control n=8, Imp2−/− AGN n=10, Act1−/− control n=6 and Act1−/− AGN n=6). RNA at day 7 was subjected to qPCR ± SEM. (B) Kidney homogenates from the indicated mice day 7 post-AGN were stained for CD45, CD133 and intracellular C/EBPδ. Data show live CD45−CD133+ cells. Representative FACS plot is shown. (C, D) Left: IF staining of C/EBPδ and C/EBPβ on day 7 post-AGN. Arrows indicate glomeruli. Cortex and medulla in a representative image are indicated. (4X). White boxes indicate sites of 40X images, fig S9. Right: Fluorescence intensity in non-glomerular regions of kidney was quantified by Image J, normalized to Imp2+/+ control. Data are from two regions per slide and 2-3 independent slides (2 mice). *P<0.05, **P<0.01 by ANOVA with post hoc Tukey’s test. Scale bars = 1 mm.
Many of the effects of IMP2 were evident not just after cytokine treatment but also to some extent at baseline (fig S5d). To rule out the possibility that the disease-promoting activity of IMP2 on AGN was due to baseline or developmental effects, we verified that there were no differences in the major immune compartments in Imp2−/− mice spleen or thymus (fig S11). Imp2fl/fl mice (88) were crossed to animals with a constitutive tamoxifen (TAM)-inducible Cre (Rosa26CreERT), which permits gene deletion after administration of TAM. Mice were given TAM starting 1 day after induction of AGN and for the next 5 days (Fig 8A). Deletion of Imp2 was verified by qPCR of kidney (Fig 8B). Mice in which Imp2 was deleted after AGN induction showed a similar improvement of kidney dysfunction as a complete knockout (Fig 8C). These data indicate that IMP2 effects on inflammation are not developmental and that blockade of IMP2 could potentially be targeted to ameliorate AGN.
Fig. 8. Imp2 deletion post AGN induction decreased autoimmune renal dysfunction.
(A-C) Indicated mice were administered 2 mg tamoxifen (TAM) i.p. for 5 days starting day 1 after AGN induction. (B) Imp2 was assessed by qPCR at day 14. (C) BUN and creatinine were assessed at day 14. Data pooled from 2 experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, by ANOVA with post-hoc Tukey test. (D) Model of signaling through IMP2. Cebpb and Cebpd transcripts are subject to m6A modifications, which facilitate IMP2 mediated-post-transcriptional regulation. Imp2 deficiency impaired mRNAs whose expression is dependent on C/EBPs. Imp2-deficient mice are resistant to IL-17-driven renal inflammation. Diagram created on Biorender.com.
DISCUSSION
It has long been appreciated that blocking cytokines such as TNFα is an effective treatment for many, though not all, autoimmune conditions. The contribution of the Th17/IL-17 pathway to autoimmunity has become appreciated with the success of biologic drugs targeting IL-17, the IL-17 receptor, or upstream regulators of Th17 cells (89-91). However, the cost of antibody-based drugs is often prohibitive, so there is an unmet need for therapies that are more universally applicable (4). In principle, any molecule in cytokine signal transduction could serve as a pharmacological target, but since our understanding of cytokine signaling is incomplete, there are hurdles to rational drug design (5).
Specificity in the RNA methylation pathway is particularly driven by m6A readers (29), and in this regard IMP-family proteins were only recently recognized to be classified as such (35, 36). Moreover, IMP proteins have not been linked to autoimmunity or cytokine signal transduction (48), but rather to metabolism, stem cell maintenance, type 2 diabetes and cancer (37, 64, 85). Although one report described an IGF2BP2 polymorphism in a psoriasis cohort, a condition that is strongly IL-17- and TNFα-driven, expression of IGF2BP2 was linked only to altered triglycerides, not to psoriasis per se (92).
Glomerulonephritis (GN) causes mortality and morbidity in Goodpasture disease and ANCA vasculitis among other conditions (93). In mice, IL-17A, IL-17F and TNFα drive AGN pathology, and anti-IL-17 and anti-TNF blocking antibodies ameliorate disease in mouse models of crescentic GN (39, 44, 71-75, 94-97). Tissue-resident Th17 cells are implicated in human GN, and a recent report shows that these T cells recognize commensal microbes, such as Staphylococcus aureus and Candida albicans (39, 44, 94, 98, 99). In renal epithelial cells, IL-17 and TNFα signal cooperatively to activate inflammatory genes (7, 45, 71, 100, 101). Hence dual blockade of these cytokines is under consideration for treating autoimmunity (102). Autoimmunity is potentiated by multifactorial interacting gene networks, and decades of research in both humans and animal models of disease has underscored this complexity. Several genes controlled by IMP2 help explain its contribution to cytokine-mediated AGN, particularly Lcn2 and Il6 (81, 82, 103). Though IMP2 regulates lamininβ2 (Lamb2), a glomerular basement membrane component that promotes renal barrier permeability, Lamb2 is not regulated by IL-17 in murine AGN (104). In a model of anti-Thy1-induced GN, loss of C/EBPδ protected from disease, though its connections to IL-17 were unknown at the time that study was conducted (86). Conversely, C/EBPδ-deficiency caused worse pathology in the unilateral ureteral obstruction (UUO) model of kidney fibrosis (87). Strikingly, IL-17 signaling similarly attenuates pathology in UUO (105) and RTECs are the dominant cell type expressing C/EBPδ and responding to IL-17 in the kidney (87)(106, 107).
We originally linked C/EBPβ and C/EBPδ to IL-17 signaling in 2004, finding that C/EBPδ in particular contributes to IL-17 activation of downstream genes and to signaling cooperativity between IL-17 and TNFα (7). The role of C/EBPβ in the IL-17 pathway is not always straightforward, as it serves as a transcriptional activator (7, 8, 108) but can also be a repressor, for example when subject to phosphorylation (19, 109). The present work led to the unexpected observation that C/EBPβ and C/EBPδ expression is controlled at a post-transcriptional level by the noncanonical m6A reader IMP2. Genes regulated by C/EBPs (e.g., Il6, Lcn2) are consequently IMP2-dependent. Interestingly, IMP2 was the only m6A reader seen to increase cytokine-induced mRNAs in these studies, whereas the more common YTH-domain readers did not, thus revealing considerable specificity in terms of how the m6A pathway operates in response to cytokines. Here we confirm that TNFα does not upregulate C/EBPδ (46), representing a divergence in the mechanisms by which inflammatory cytokines trigger downstream gene expression. Thus, the m6A/IMP2/C/EBP axis represents a previously-unrecognized avenue of signal activation that is likely to apply to many cytokines, probably in distinct and specific modalities.
In recent years, IL-17 has become appreciated as a potent regulator of post-transcriptional gene expression (5, 110-112). Moreover, post-transcriptional regulation pathways activated by IL-17 exhibit crosstalk with transcriptional signaling in intricate ways. For example, IκBξ, a noncanonical member of the NF-κB family, is required for many IL-17-induced genes (113). The mRNA encoding IκBξ (Nfkbiz) is intrinsically unstable, subject to both positive and negative regulation by IL-17-activated RBPs. For example, the endoribonuclease Regnase-1 (MCPIP1) degrades Nfkbiz upon binding the 3’ UTR (26). Regnase-1 mRNA (Zc3h12a) is subject to autoregulation as well as control by the DDX3X RNA helicase (27, 114). Additionally, Regnase-1 activity is counteracted by Arid5a, which binds to Nfkbiz and thereby enhances translation of IκBξ (17), which in turn triggers a cascade of downstream IL-17-dependent genes. Arid5a controls translation of C/EBPβ, but surprisingly does not impact C/EBPδ (17). In contrast, IMP2 did not impact Nfkbiz or Zc3h12a expression. Although IL-17 leads to inducible phosphorylation of C/EBPβ by ERK and GSK3β (19, 20), IMP2 did not impact IL-17-dependent activation of MAPK pathways (fig S12). In these studies, only a subset of IL-17-dependent transcripts are regulated by IMP2. Collectively, these data suggest that IMP2 operates in a distinct and specific post-transcriptional pathway (Fig. 8D).
HuR is an RBP that influences many facets of mRNA fate, including transcript stability and translation. Upon IL-17 signaling, HuR binds to Act1, a multifunctional signaling protein required for all known IL-17-dependent signals. Act1, in conjunction with TRAF2, has the capacity to bind to target mRNAs and control their stability as well as direct translation (24, 25). Here we show that IMP2 interacts with HuR but not with Act1, in agreement with findings that HuR facilitates IMP2 function (35). HuR has been shown to regulate C/EBPβ and C/EBPδ mRNA nuclear export, translation, and stability (67-70). Hence, there is remarkable complexity, specificity and crosstalk among the transcriptional and post-transcriptional regulators that determine the output of cytokine-derived signals (5).
The concept that IMP2 may exert distinct effects depending on its client mRNA has precedent (54, 88, 115, 116), but the basis of its binding selectivity was elusive until the discovery that IMP2 recognizes N6-methyladenosine (m6A) (35). This RNA modification is mediated by core methyltransferases (e.g., METTL3/14) and is regulated dynamically by demethylase enzymes (e.g., FTO). Although there are only limited studies interrogating the functional significance m6A in the immune system, this modification occurs in innate immunity in response to viral infection (32, 33), and loss of METTL3 in T cells was reported to impair T cell homeostasis through SOCS-mediated inhibition of IL-7/STAT5 signaling (30). METTL3 in T cells also controlled IL-2/STAT5 and thus Treg function (31). By evaluating the upstream methylase METTL3, those studies implicated m6A broadly in immune system control, but as yet the specific readers recognizing the m6A RNA mark have not been described in these settings.
The establishment of a rapid inflammatory response through activation of post-transcriptional events requires a tonic level of mRNA expression (112). IMP2 has potent effects on RNAs induced by cytokines, but also influences their baseline expression (54, 58, 115). In agreement with this, in the absence of a stressor, the immune system appeared normal in Imp2−/− mice. Moreover, IMP2-dependent effects on AGN were not caused by aberrant developmental issues, since deletion of IMP2 after AGN onset resulted in a similar suppression of renal damage as a full IMP2 knockout. Therefore, IMP2 is not merely a rheostat that alters the ability of cells to respond to all signals, but rather exerts selective effects, many of which rely on the C/EBP pathway.
In summary, these findings uncover a new role for m6A epitranscriptomic RNA marks and the noncanonical m6A reader IMP2 in cytokine-driven autoimmunity. The capacity of IMP2 to direct posttranscriptional regulation of Cebpd and Cebpb potentiates expression of genes reliant on these TFs, which in turn underlie pathology in autoimmune inflammation of the kidney (Fig 8D). Exploiting RNA therapeutically is attractive given its potential for exquisite specificity and the possibility of targeting otherwise “un-druggable” molecules. Accordingly, strategies are in development that target RNA or RBPs (117, 118). In the IL-17 pathway, a preclinical study using aptamers to target the RNA-binding properties of Act1 elegantly illustrated proof-of-principle for this concept (24). Still, taking full advantage of RNA requires a complete understanding of the molecular players involved, especially RBPs, and the relevant client transcript mRNAs regulated within pertinent target cells.
Materials and Methods
Study design
The objective of this study was to determine how C/EBPs are regulated in the IL-17 signal pathway. We used cell culture studies and a model of AGN to define the role of m6A pathway components on IL-17 signaling. Sample sizes were determined by power analyses from pilot or previously published data. Mice of both sexes were assigned randomly to experimental cohorts. Unless noted, experiments were done 3 or more independent times. Data from multiple experiments were pooled unless noted. Investigators were not blinded to groups except for immunofluorescent image analysis. No data were excluded. Endpoints were selected based on prior studies. The details of the reagents used are provided in Table S2.
Mice
All mice were on the C57BL/6 background. Cohorts were age- and sex-matched. Act1−/− mice were from NIH and Imp2−/− mice were obtained under MTA from Oxford, UK. Wild type (WT) controls were usually derived from breeding of heterozygote mice to generate littermate controls. In some instances, WT mice were from Taconic Farms or The Jackson Laboratory, and were co-housed with experimental cohorts for at least 3 weeks to normalize microbiota. Mice were housed in SPF conditions. Protocols were approved by the University of Pittsburgh IACUC.
AGN
AGN was induced by i.p. injection of rabbit IgG (0.1 mg/ml) in CFA (2.5 mg/ml; Sigma, St. Louis, MO) on day -3. Heat-inactivated rabbit anti-mouse GBM serum (Lampire Biological Laboratories, Pipersville, PA) was injected (100 μl) i.v. (71). BUN and creatinine levels were measured on days 7 or 14 by a BUN ELISA kit (Bioo Scientific, Austin, TX) or a QuantiChrom Creatinine Assay Kit (BioAssay Systems, DICT-500). Kidney sections were stained with H&E and images acquired on an EVOS FL Auto Imaging System.
Radiation Chimeras
On day −1, recipients (WT, CD45.1) or Imp2−/− (CD45.2) were given sulfamethoxazole and trimethoprim in drinking water for 10 days. On day 0, mice were irradiated (900 rad). On day 1, 5 ×106 donor femoral BM cells were injected i.v. into recipients. After 6 weeks, BM reconstitution was verified by flow cytometry.
Cell Culture
MEFs, HK-2 and HEK293T cells were cultured in α-minimum essential medium (α-MEM; Sigma-Aldrich, St. Louis MO) with L-glutamine, antibiotics and 10–15% FBS. HK-2 cells (ATCC) were cultured in DMEM/F12 (Gibco), antibiotics and 10% FBS. IL-17, IL-17F and TNFα (PeproTech, Rocky Hill, NJ) were used at 200 and 10 ng/ml. Actinomycin D (Sigma-Aldrich) was used at 10 μg/ml.
siRNA, Plasmids and Luciferase assays
ON-TARGETplus SMARTpool siRNAs were from Dharmacon (Lafayette, CO). MEFs and HK-2 cells were seeded overnight in antibiotic-free media and transfected 18 h later with 50 nM siRNA in DharmaFECT Reagent 1. Culture media was replaced after 24 h, and cytokines administered 24 h later. The Lcn2 and Il6 promoter constructs were described (6, 51). MEFs were transfected with FuGENE HD (Promega). HEK293T cells were transfected using CaPO4. Constructs containing the 5’UTR, coding sequences and 3’UTR of murine Cebpb was synthesized by Bon Opus Biosciences (Millburn, NJ). Luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega) and normalized to a Renilla luciferase control (17).
qPCR, RNA-Seq, RIP-Seq
RNA was isolated with RNeasy Mini Kits (Qiagen), and cDNA was synthesized by SuperScript III First Strand Kits (Thermo Fisher Scientific, Waltham MA). Real-time qPCR was performed with SYBR Green FastMix ROX (Quanta Biosciences) on a 7300 Real-Time instrument (Applied Biosystems), normalized to Gapdh. Primers were from QuantiTect Primer Assays (QIAGEN). RNASeq libraries were prepared from MEF mRNA (Nextera XT Kit). RNASeq was performed on Illumina NextSeq 500 by the Health Sciences Sequencing Core at the University of Pittsburgh.
For RIP-Seq, libraries were generated using SMART-Seq v4 Ultra Low Input RNA kit (Takara Biosciences) and sequenced on a NextSeq500. Raw reads were aligned to mm10 using STAR (119) followed by identification of global binding of IMP2 using Piranha (55). Differentially enriched IMP2 interactions were identified with DESeq2 (56) by comparing 3 replicates of IMP2-RIP-seq data and control IgG. Identification of IMP2 motifs (35, 120) was performed using MDS2 (57) and viewed using Integrative Genomics Viewer (Broad Institute).
Immunoblotting, Immunoprecipitation
Western blotting and IPs were performed as described (17). Immunoblotting Abs: METTL3 (Proteintech, 1:1000), FTO (Santa Cruz Biotechnology, 1:1000), IMP2 (MBL, 1:1000; Cell Signaling, 1:1000), C/EBPδ (Cell Signaling, 1:1000; Santa Cruz Biotechnology, 1:1000), C/EBPβ (Biolegend, 1:500; Santa Cruz Biotechnology, 1:1000), YY1 (Santa Cruz Biotechnology, 1:1000), β-actin (Abcam, 1:25,000), phospho-p38 MAPK, p38 MAPK, phospho-JNK , JNK, phospho-p44/42, p44/42 (Cell Signaling, 1:1000), eIF4G (Cell Signaling, 1:1000). Blots viewed with a FluorChem E imager (Protein Simple, Santa Clara CA).
RNA immunoprecipitation (RIP) and in vitro RNA pulldown
For IMP2 RIPs, extracts were isolated with lysis buffer [100 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.0), 0.5% NP-40, 1 mM dithiothreitol] with RNAseOUT (100 U/ml; Invitrogen). For eIF4G RIP, extracts were isolated with lysis buffer [0.3% CHAPS, 40 mM HEPES (pH 7.5), 120 mM NaCl, 1 mM EDTA, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 50 mM NaF, 1.5 mM sodium orthovanadate, 1 mM DTT]. Buffers included a protease inhibitor cocktail (Sigma-Aldrich). Lysates were precleared protein A agarose (Roche) or protein G–conjugated magnetic Dynabeads (ThermoFisher) and immunoprecipitated with Abs against IMP2 (MBL), eIF4G (Cell Signaling) or FLAG (Sigma). Beads were washed with NT2 buffer and digested with DNase I (Roche Applied Science) and protease K (Sigma). Total RNA was extracted with acid phenol or TRIzol.
For RNA pulldowns, biotinylated RNA corresponding to the Cebpd 3’UTR (bases 863-982) was synthesized with specified adenosines subject to m6A modification (Dharmacon). RNAs were incubated with lysates from HEK293T cells transfected with FLAG-IMP2, precipitated with streptavidin Dynabeads M-280 (ThermoFisher), isolated by magnetic separation and subjected to immunoblotting.
For m6A RIP (MeRIP) (32), 20–50 μg RNA was fragmented and purified by ethanol precipitation. 0.1 fmol of a control m6A-modified Gaussia luciferase RNA or unmodified Cypridina luciferase RNA (supplied with the EpiMark N6-methyladenosine Enrichment kit) were spiked in each sample. For RIP, Protein G Dynabeads (Thermo Fisher) were washed in MeRIP buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.5], 0.1% NP-40), and incubated with anti-m6A Abs for 2 h at 4°C. After washing, anti-m6A conjugated beads were incubated with mRNA for 4 h in RNasin (Promega). Up to 3% of mRNA was used for input. Beads were washed with MeRIP buffer, low salt wash buffer (50 mM NaCl, 10 mM Tris-HCl [pH 7.5], 0.1% NP-40), and wash buffer (500 mM NaCl, 10 mM Tris-HCl [pH 7.5], 0.1% NP-40). m6A-modified RNA was eluted in MeRIP buffer containing 5 mM m6A salt (Santa Cruz Biotechnology). Eluates were pooled and concentrated by ethanol precipitation. Input and IP fractions were reverse transcribed using the iScript cDNA synthesis kit (BioRad) and subjected to qPCR.
Immunofluorescence, flow cytometry
Frozen sections (8 μm) were fixed in 100% methanol and blocked with 5% goat serum (500622, Life Technologies) in Triton X. Primary antibodies were incubated overnight: C/EBPδ (Abcam, 1:200), normal rabbit IgG (Abcam, 1:200), C/EBPβ (Abcam, 1:200) and rabbit monoclonal IgG (Abcam, 1:200). Slides were incubated with goat anti-Rabbit Cy3 antibody (A15020, Thermo Fisher) or DAPI. Slides were visualized on an EVOS FL microscope (Life Technologies).
For flow cytometry, kidneys were harvested following perfusion with PBS. Cells were digested with collagenase IV (1mg/mL) in HBSS. Antibodies: anti-CD45 (clone 30-F11, Thermo Fisher), anti-Ly6G (clone 1A8, BD Biosciences), anti-Ly6C (clone HK1.4, eBioscience), anti-CD11b (clone M1/70, BioLegend), anti-CD133 (clone 13A4, Thermo Fisher), anti-C/EBPδ (Abcam, 1:500), rabbit polyclonal Abs (Abcam, 1:500), and secondary goat anti-Rabbit Alexa Fluor 488 Abs (Thermo Fisher). Dead cells were excluded using Ghost Dye (eBioscience). C/EBPδ intracellular staining was performed with the FOXP3 staining kit (eBioscience). Data acquired with LSR Fortessa and analyzed using FlowJo software (TreeStar).
RNA decay assays
MEFs were primed with TNFα (10 ng/ml) for 3 h and treated with 10 μg/ml actinomycin D (ActD; Sigma-Aldrich) ± 200 ng/ml IL-17 as described (17). For each mRNA, quantity (%) was calculated by normalizing ΔΔCt to the ΔΔCt of samples primed with TNFα.
Statistics
One-way ANOVA with post hoc Tukey’s analysis or Student’s t-test was used to assess significance, P < 0.05 considered significant. Data were analyzed on GraphPad Prism. Each symbol represents one mouse.
Supplementary Material
Data file S1. Raw data file (Excel spreadsheet)
Fig S1. C/EBP proteins regulate Lcn2 promoter activity
Fig S2. Predicted m6A sites in IL-17 target gene transcripts
Fig S3. Mettl3/FTO axis modulates IL-17-induced Il6 and Lcn2 expression
Fig S4. YTH family m6A readers in IL-17 signaling
Fig S5. IMP2 promotes expression of a subset of IL-17-, IL-17F- and TNFα-induced target genes
Fig S6. IMP2 promotes expression of C/EBPδ and C/EBPβ in HK-2 cells and MEFs
Fig S7. Impaired activation of C/EBP-dependent promoters in Imp2−/− cells
Fig S8. IMP2 clients by RIPSeq and IMP2/m6A binding sites in Cebpd mRNA
Fig S9. IMP2-deficient mice show reduced inflammatory monocyte infiltration during AGN
Fig S10. C/EBP expression in kidney during AGN
Fig S11. Characterization of the immune compartment in spleen and thymus of Imp2−/− mice at baseline
Fig S12. IMP2 is not required for MAPK activation
Table S1. Frequencies of putative m6A methylation sites in three IL-17-induced genes
Table S2. Reagents used in this study
MDAR checklist.
Acknowledgments:
We thank the HSSC@CHP for RNAseq support. We thank L. Minichiello, University of Oxford, and J. Avruch, Harvard, for Imp2−/− mice and valuable suggestions. S. Majumder and MJ McGeachy provided helpful input. We are grateful to Ulrich Siebenlist (deceased) for Act1−/− mice.
Funding:
NIH supported SLG (DE022550, AI107825, AI147383), PSB (DK104680, AI142354) and SMH (AI125416). SLG and PSB were supported by the Rheumatology Research Foundation. SMH was supported by the Burroughs Wellcome Fund. Work was also supported in part by the University of Pittsburgh Center for Research Computing.
Footnotes
Competing Interests: The authors declare no competing interests.
Data and Materials Availability: The RNASeq and RIPSeq data are available through the NCBI GEO resource under accession number GSE178710. Mutant mouse strains used in this study are available by MTA (Act1−/−, NIH; Il17ra−/−, Amgen; Imp2−/−, Oxford University). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Helmick CG et al. , Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum 58, 15–25 (2008). [DOI] [PubMed] [Google Scholar]
- 2.A. C. o. Rheumatology. (American College of Rheumatology, 2012). [Google Scholar]
- 3.McGeachy MJ, Cua DJ, Gaffen SL, The IL-17 Family of Cytokines in Health and Disease. Immunity 50, 892–906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slivka PF et al. , Small Molecule and Pooled CRISPR Screens Investigating IL17 Signaling Identify BRD2 as a Novel Contributor to Keratinocyte Inflammatory Responses. ACS Chem Biol 14, 857–872 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Li X, Bechara R, Zhao J, McGeachy MJ, Gaffen SL, Interleukin 17 receptor-based signaling and implications for disease. Nature Immunology 20, 1594–1602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen F, Hu Z, Goswami J, Gaffen SL, Identification of common transcriptional regulatory elements in interleukin-17 target genes. J Biol Chem 281, 24138–24148 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Ruddy MJ et al. , Functional cooperation between interleukin-17 and tumor necrosis factor-a is mediated by CCAAT/enhancer binding protein family members. J Biol Chem 279, 2559–2567 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Patel DN et al. , Interleukin-17 stimulates C-reactive protein expression in hepatocytes and smooth muscle cells via p38 MAPK and ERK1/2-dependent NF-kappaB and C/EBPbeta activation. J Biol Chem 282, 27229–27238 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsen JR, Borregaard N, Cowland JB, Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/EBP-beta nor C/EBP-delta. J Biol Chem 285, 14088–14100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slowikowski K et al. , CUX1 and IkappaBzeta (NFKBIZ) mediate the synergistic inflammatory response to TNF and IL-17A in stromal fibroblasts. Proc Natl Acad Sci U S A 117, 5532–5541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Y et al. , The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol 8, 247–256 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Chang SH, Park H, Dong C, Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem 281, 35603–35607 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Amatya N, Garg AV, Gaffen SL, IL-17 Signaling: The Yin and the Yang. Trends Immunol 38, 310–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukada J, Yoshida Y, Kominato Y, Auron PE, The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine 54, 6–19 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Ko CY, Chang WC, Wang JM, Biological roles of CCAAT/Enhancer-binding protein delta during inflammation. J Biomed Sci 22, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramji DP, Foka P, CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J 365, 561–575. (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amatya N et al. , IL-17 integrates multiple self-reinforcing, feed-forward mechanisms through the RNA-binding protein Arid5a. Science Signaling 11, eaat4617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen F, Gaffen SL, Structure-function relationships in the IL-17 receptor: Implications for signal transduction and therapy. Cytokine 41, 92–104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen F et al. , IL-17 Receptor Signaling Inhibits C/EBPbeta by Sequential Phosphorylation of the Regulatory 2 Domain. Sci Signal 2, ra8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang QQ et al. , Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U S A 102, 9766–9771 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pulido-Salgado M, Vidal-Taboada JM, Saura J, C/EBPbeta and C/EBPdelta transcription factors: Basic biology and roles in the CNS. Prog Neurobiol 132, 1–33 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Huber R, Pietsch D, Panterodt T, Brand K, Regulation of C/EBPbeta and resulting functions in cells of the monocytic lineage. Cell Signal 24, 1287–1296 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Shi JH, Sun SC, Tumor Necrosis Factor Receptor-Associated Factor Regulation of Nuclear Factor kappaB and Mitogen-Activated Protein Kinase Pathways. Front Immunol 9, 1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herjan T et al. , IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat Immunol 19, 354–365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herjan T et al. , HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J Immunol 191, 640–649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg AV et al. , MCPIP1 Endoribonuclease Activity Negatively Regulates Interleukin-17-Mediated Signaling and Inflammation. Immunity 43, 475–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somma D et al. , CIKS/DDX3X Interaction Controls the Stability of the Zc3h12a mRNA Induced by IL-17. J Immunol 194, 3286–3294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H, Wei J, He C, Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell 74, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaccara S, Ries RJ, Jaffrey SR, Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20, 608–624 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Li HB et al. , m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong J et al. , m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res 28, 253–256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gokhale NS et al. , Altered m(6)A Modification of Specific Cellular Transcripts Affects Flaviviridae Infection. Mol Cell, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y et al. , N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365, 1171–1176 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Paramasivam A, Vijayashree Priyadharsini J, Novel insights into m6A modification in circular RNA and implications for immunity. Cell Mol Immunol, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H et al. , Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20, 285–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H, Weng H, Chen J, m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 37, 270–288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shulman Z, Stern-Ginossar N, The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat Immunol 21, 501–512 (2020). [DOI] [PubMed] [Google Scholar]
- 38.McAdoo SP, Pusey CD, Anti-Glomerular Basement Membrane Disease. Clin J Am Soc Nephrol 12, 1162–1172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas P, IL-17 in renal immunity and autoimmunity. J Immunol 201, 3153–3159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurts C, Panzer U, Anders HJ, Rees AJ, The immune system and kidney disease: basic concepts and clinical implications. Nat Rev Immunol 13, 738–753 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Lahmer T, Heemann U, Anti-glomerular basement membrane antibody disease: a rare autoimmune disorder affecting the kidney and the lung. Autoimmun Rev 12, 169–173 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Ernandez T, Mayadas TN, The Changing Landscape of Renal Inflammation. Trends Mol Med 22, 151–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shlomchik MJ, Activating systemic autoimmunity: B’s, T’s, and tolls. Curr Opin Immunol 21, 626–633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krebs CF et al. , Pathogen-induced tissue-resident memory TH17 (TRM17) cells amplify autoimmune kidney disease. Sci Immunol 5, (2020). [DOI] [PubMed] [Google Scholar]
- 45.Shen F, Ruddy MJ, Plamondon P, Gaffen SL, Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J Leukoc Biol 77, 388–399 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Sonder SU et al. , IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem 286, 12881–12890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansen C et al. , IkappaBzeta is a key driver in the development of psoriasis. Proc Natl Acad Sci U S A 112, E5825–5833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai N, The Diverse Functions of IMP2/IGF2BP2 in Metabolism. Trends Endocrinol Metab 31, 670–679 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Ryan MJ et al. , HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int 45, 48–57 (1994). [DOI] [PubMed] [Google Scholar]
- 50.Litvak V et al. , Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol 10, 437–443 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eickelberg O et al. , Transforming growth factor-beta1 induces interleukin-6 expression via activating protein-1 consisting of JunD homodimers in primary human lung fibroblasts. J Biol Chem 274, 12933–12938. (1999). [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q, SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res 44, e91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xuan JJ et al. , RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res 46, D327–D334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hafner M et al. , Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uren PJ et al. , Site identification in high-throughput RNA-protein interaction data. Bioinformatics 28, 3013–3020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao T, Shu J, Cui J, A systematic approach to RNA-associated motif discovery. BMC Genomics 19, 146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai N et al. , IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and Other mRNAs encoding mitochondrial proteins. Cell Metab 21, 609–621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McFadden MJ et al. , Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep 34, 108798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gokhale NS et al. , Altered m(6)A Modification of Specific Cellular Transcripts Affects Flaviviridae Infection. Mol Cell 77, 542–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Regue L et al. , RNA m6A reader IMP2/IGF2BP2 promotes pancreatic beta-cell proliferation and insulin secretion by enhancing PDX1 expression. Mol Metab, 101209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henness S et al. , IL-17A augments TNF-alpha-induced IL-6 expression in airway smooth muscle by enhancing mRNA stability. J Allergy Clin Immunol 114, 958–964 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Masuda K et al. , Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc Natl Acad Sci U S A, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christiansen J, Kolte AM, Hansen T, Nielsen FC, IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol 43, 187–195 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Dai N et al. , mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev 25, 1159–1172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lahr RM et al. , La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cherry J, Jones H, Karschner VA, Pekala PH, Post-transcriptional control of CCAAT/enhancer-binding protein beta (C/EBPbeta) expression: formation of a nuclear HuR-C/EBPbeta mRNA complex determines the amount of message reaching the cytosol. J Biol Chem 283, 30812–30820 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gantt K, Cherry J, Tenney R, Karschner V, Pekala PH, An early event in adipogenesis, the nuclear selection of the CCAAT enhancer-binding protein {beta} (C/EBP{beta}) mRNA by HuR and its translocation to the cytosol. J Biol Chem 280, 24768–24774 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Hsiao YW et al. , CCAAT/enhancer binding protein delta in macrophages contributes to immunosuppression and inhibits phagocytosis in nasopharyngeal carcinoma. Sci Signal 6, ra59 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Li B, Si J, DeWille JW, Ultraviolet radiation (UVR) activates p38 MAP kinase and induces post-transcriptional stabilization of the C/EBPdelta mRNA in G0 growth arrested mammary epithelial cells. J Cell Biochem 103, 1657–1669 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Ramani K et al. , An essential role of interleukin-17 receptor signaling in the development of autoimmune glomerulonephritis. J Leukoc Biol 96, 463–472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pisitkun P et al. , Interleukin-17 cytokines are critical in development of fatal lupus glomerulonephritis. Immunity 37, 1104–1115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krohn S et al. , IL-17C/IL-17 Receptor E Signaling in CD4(+) T Cells Promotes TH17 Cell-Driven Glomerular Inflammation. J Am Soc Nephrol 29, 1210–1222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riedel JH et al. , IL-17F Promotes Tissue Injury in Autoimmune Kidney Diseases. J Am Soc Nephrol 27, 3666–3677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner JE et al. , IL-17A production by renal gammadelta T cells promotes kidney injury in crescentic GN. J Am Soc Nephrol 23, 1486–1495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mohamed R et al. , Low-Dose IL-17 Therapy Prevents and Reverses Diabetic Nephropathy, Metabolic Syndrome, and Associated Organ Fibrosis. J Am Soc Nephrol 27, 745–765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du Y, Fu Y, Mohan C, Experimental anti-GBM nephritis as an analytical tool for studying spontaneous lupus nephritis. Arch Immunol Ther Exp (Warsz) 56, 31–40 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Fu Y, Du Y, Mohan C, Experimental anti-GBM disease as a tool for studying spontaneous lupus nephritis. Clin Immunol 124, 109–118 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Krebs CF, Schmidt T, Riedel JH, Panzer U, T helper type 17 cells in immune-mediated glomerular disease. Nat Rev Nephrol 13, 647–659 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Viau A et al. , Lipocalin 2 is essential for chronic kidney disease progression in mice and humans. J Clin Invest 120, 4065–4076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang CC et al. , Urinary neutrophil gelatinase-associated lipocalin is a potential biomarker for renal damage in patients with systemic lupus erythematosus. J Biomed Biotechnol 2012, 759313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torres-Salido MT et al. , Neutrophil gelatinase-associated lipocalin as a biomarker for lupus nephritis. Nephrol Dial Transplant 29, 1740–1749 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Haase M, Haase-Fielitz A, Bellomo R, Mertens PR, Neutrophil gelatinase-associated lipocalin as a marker of acute renal disease. Curr Opin Hematol 18, 11–18 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Satirapoj B, Tubulointerstitial Biomarkers for Diabetic Nephropathy. J Diabetes Res 2018, 2852398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao J, Mu Q, Huang H, The Roles of Insulin-Like Growth Factor 2 mRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Stem Cells Int 2018, 4217259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takeji M et al. , CCAAT/Enhancer-binding protein delta contributes to myofibroblast transdifferentiation and renal disease progression. J Am Soc Nephrol 15, 2383–2390 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Duitman J et al. , CCAAT-enhancer binding protein delta (C/EBPdelta) attenuates tubular injury and tubulointerstitial fibrogenesis during chronic obstructive nephropathy. Lab Invest 94, 89–97 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Degrauwe N et al. , The RNA Binding Protein IMP2 Preserves Glioblastoma Stem Cells by Preventing let-7 Target Gene Silencing. Cell Rep 15, 1634–1647 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Lubberts E, The IL-23-IL-17 axis in inflammatory arthritis. Nat Rev Rheumatol 11, 415–429 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Gaffen SL, Jain R, Garg A, Cua D, IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat Rev Immunol 14, 585–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koenders MI, van den Berg WB, Novel therapeutic targets in rheumatoid arthritis. Trends Pharmacol Sci 36, 189–195 (2015). [DOI] [PubMed] [Google Scholar]
- 92.Kargun K et al. , IGF2BP2 gene polymorphism in patients with psoriasis. Biomedical Res 28, 3619–3622 (2017). [Google Scholar]
- 93.Lahmer T et al. , Mineralocorticoid receptor antagonism and aldosterone synthesis inhibition do not improve glomerulosclerosis and renal interstitial fibrosis in a model of chronic kidney allograft injury. Kidney Blood Press Res 35, 561–567 (2012). [DOI] [PubMed] [Google Scholar]
- 94.Paust HJ et al. , The IL-23/Th17 axis contributes to renal injury in experimental glomerulonephritis. J Am Soc Nephrol 20, 969–979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khan SB et al. , Antibody blockade of TNF-alpha reduces inflammation and scarring in experimental crescentic glomerulonephritis. Kidney Int 67, 1812–1820 (2005). [DOI] [PubMed] [Google Scholar]
- 96.Le Hir M, Haas C, Marino M, Ryffel B, Prevention of crescentic glomerulonephritis induced by anti-glomerular membrane antibody in tumor necrosis factor-deficient mice. Lab Invest 78, 1625–1631 (1998). [PubMed] [Google Scholar]
- 97.Karkar AM, Smith J, Pusey CD, Prevention and treatment of experimental crescentic glomerulonephritis by blocking tumour necrosis factor-alpha. Nephrol Dial Transplant 16, 518–524 (2001). [DOI] [PubMed] [Google Scholar]
- 98.Krebs CF et al. , Autoimmune Renal Disease Is Exacerbated by S1P-Receptor-1-Dependent Intestinal Th17 Cell Migration to the Kidney. Immunity 45, 1078–1092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hunemorder S et al. , TH1 and TH17 cells promote crescent formation in experimental autoimmune glomerulonephritis. J Pathol 237, 62–71 (2015). [DOI] [PubMed] [Google Scholar]
- 100.Noack M, Beringer A, Miossec P, Additive or Synergistic Interactions Between IL-17A or IL-17F and TNF or IL-1beta Depend on the Cell Type. Front Immunol 10, 1726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Griffin GK et al. , IL-17 and TNF-alpha sustain neutrophil recruitment during inflammation through synergistic effects on endothelial activation. J Immunol 188, 6287–6299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen F et al. , Combined Blockade of TNF-alpha and IL-17A Alleviates Progression of Collagen-Induced Arthritis without Causing Serious Infections in Mice. J Immunol 202, 2017–2026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gijbels K, Brocke S, Abrams JS, Steinman L, Administration of neutralizing antibodies to interleukin-6 (IL-6) reduces experimental autoimmune encephalomyelitis and is associated with elevated levels of IL-6 bioactivity in central nervous system and circulation. Mol Med 1, 795–805 (1995). [PMC free article] [PubMed] [Google Scholar]
- 104.Schaeffer V, Hansen KM, Morris DR, LeBoeuf RC, Abrass CK, RNA-binding protein IGF2BP2/IMP2 is required for laminin-beta2 mRNA translation and is modulated by glucose concentration. Am J Physiol Renal Physiol 303, F75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramani K et al. , IL-17 Receptor Signaling Negatively Regulates the Development of Tubulointerstitial Fibrosis in the Kidney. Mediators Inflamm 2018, 5103672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li D.-d. et al. , Antibody-induced glomerulonephritis pathology is amplified by RTEC-intrinsic IL-17 signaling and restrained by the endoribonuclease Regnase-1. BioRXiv 2021/425972, (2021). [Google Scholar]
- 107.Ramani K et al. , Unexpected kidney-restricted role for IL-17 receptor signaling in defense against systemic Candida albicans infection. JCI Insight 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ruddy MJ, Shen F, Smith J, Sharma A, Gaffen SL, Interleukin-17 regulates expression of the CXC chemokine LIX/CXCL5 in osteoblasts: Implications for inflammation and neutrophil recruitment. J. Leukoc. Biol 76, 135–144 (2004). [DOI] [PubMed] [Google Scholar]
- 109.Maekawa T et al. , Antagonistic effects of IL-17 and D-resolvins on endothelial Del-1 expression through a GSK-3beta-C/EBPbeta pathway. Nat Commun 6, 8272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Song X, Qian Y, The activation and regulation of IL-17 receptor mediated signaling. Cytokine 62, 175–182 (2013). [DOI] [PubMed] [Google Scholar]
- 111.Iwakura Y, Ishigame H, Saijo S, Nakae S, Functional specialization of interleukin-17 family members. Immunity 34, 149–162 (2011). [DOI] [PubMed] [Google Scholar]
- 112.Hao S, Baltimore D, The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10, 281–288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun SC, The non-canonical NF-kappaB pathway in immunity and inflammation. Nat Rev Immunol 17, 545–558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fu M, Blackshear PJ, RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol 17, 130–143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dai N et al. , IGF2 mRNA binding protein-2 is a tumor promoter that drives cancer proliferation through its client mRNAs IGF2 and HMGA1. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Conway AE et al. , Enhanced CLIP Uncovers IMP Protein-RNA Targets in Human Pluripotent Stem Cells Important for Cell Adhesion and Survival. Cell Rep 15, 666–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Crooke ST, Witztum JL, Bennett CF, Baker BF, RNA-Targeted Therapeutics. Cell Metab 27, 714–739 (2018). [DOI] [PubMed] [Google Scholar]
- 118.Sahin U, Kariko K, Tureci O, mRNA-based therapeutics--developing a new class of drugs. Nat Rev Drug Discov 13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 119.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dominguez D et al. , Sequence, Structure, and Context Preferences of Human RNA Binding Proteins. Mol Cell 70, 854–867 e859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1. Raw data file (Excel spreadsheet)
Fig S1. C/EBP proteins regulate Lcn2 promoter activity
Fig S2. Predicted m6A sites in IL-17 target gene transcripts
Fig S3. Mettl3/FTO axis modulates IL-17-induced Il6 and Lcn2 expression
Fig S4. YTH family m6A readers in IL-17 signaling
Fig S5. IMP2 promotes expression of a subset of IL-17-, IL-17F- and TNFα-induced target genes
Fig S6. IMP2 promotes expression of C/EBPδ and C/EBPβ in HK-2 cells and MEFs
Fig S7. Impaired activation of C/EBP-dependent promoters in Imp2−/− cells
Fig S8. IMP2 clients by RIPSeq and IMP2/m6A binding sites in Cebpd mRNA
Fig S9. IMP2-deficient mice show reduced inflammatory monocyte infiltration during AGN
Fig S10. C/EBP expression in kidney during AGN
Fig S11. Characterization of the immune compartment in spleen and thymus of Imp2−/− mice at baseline
Fig S12. IMP2 is not required for MAPK activation
Table S1. Frequencies of putative m6A methylation sites in three IL-17-induced genes
Table S2. Reagents used in this study
MDAR checklist.