Abstract
Major depressive disorder is the most common neuropsychiatric disorder, affecting 11% of veterans. We report results of a large meta-analysis of depression using data from the Million Veteran Program (MVP), 23andMe Inc., UK Biobank, and FinnGen; including individuals of European ancestry (n=1,154,267; 340,591 cases) and African ancestry (n=59,600; 25,843 cases). Transcriptome-wide association study (TWAS) analyses revealed significant associations with expression of NEGR1 in the hypothalamus and DRD2 in the nucleus accumbens, among others. 178 genomic risk loci were fine-mapped, and we identified likely pathogenicity in these variants and overlapping gene expression for 17 genes from our TWAS, including TRAF3. Finally, we were able to show substantial replications of our findings in a large independent cohort (N=1,342,778) provided by 23andMe. This study sheds light on the genetic architecture of depression and provides new insight into the interrelatedness of complex psychiatric traits.
INTRODUCTION
Depression is the most common mental health condition, with lifetime prevalence in the U.S. of more than 20%.1 Over 300 million people, or 4.4% of the world’s population, are estimated to be affected by depression, which imposes substantial costs on individuals and on society at large. Health expenditures exceeded $90 billion for treatment of depression and anxiety disorders in the U.S. in 2013.2 There also is a substantial personal cost to depression; for example, 60% of people who die by suicide have a diagnosed mood disorder. Indeed, depression and mood disorders have been shown to have genetic overlap with suicidal behavior in several recent studies.3–6
Only recently has substantial progress been made in understanding the underlying genetic architecture of depression, led by the Psychiatric Genomics Consortium (PGC) and a large meta-analysis combining results from PGC,7 UKB,8 FinnGen (http://r2.finngen.fi/pheno/F5_MOOD) and 23andMe.9,10 In this article, we describe genome-wide association analysis (GWAS) of ~310,000 participants from the U.S. Department of Veterans Affairs (VA) Million Veteran Program (MVP). MVP is one of the largest and most diverse biobanks in the world with genetic and electronic health record (EHR) data available. Several approaches have previously been taken regarding phenotypes selected for study for a depression GWAS. The PGC2 report7 used a variety of ascertainment methods within the cohorts used for meta-analysis, with a range of case definitions, including expert or clinician ascertainment of formal diagnostic major depressive disorder (MDD) criteria or treatment registers for approximately half of the cohorts, and combinations of self-report and clinical cutoffs on those self-report measures accounting for the other half.7 Other studies 8,10 investigated a broader trait definition of depression, which provided a larger sample size; a greater number of novel loci were discovered, with the potential caveat of less specificity to depression.11 We had available in MVP several potential case definitions, and chose to focus on the definition that provided the highest heritability: the EHR derived International Classification of Diseases (ICD) codes for MDD..
When combined with the prior analysis from PGC, UK Biobank, and 23andMe,10 over 1.2 million participants were available for this study, the largest genetic analysis of depression to date. We identified 178 genetic risk loci and 223 independently significant SNPs. We used the genome-wide association summary statistics from this analysis to investigate genetic correlations between depression and other cohorts with different phenotypic assessments as well as overlap with other related traits. We used genomic structural equation modeling (gSEM) to examine shared genetic architecture and pleiotropy among complex traits. We also investigated functional consequences through fine mapping analysis, transcriptomic enrichment with respect to multiple brain tissues, and functional annotation. The results provide a deep look into the genetic architecture of depression and its underlying complex biology. Finally, we replicated our findings in an entirely independent sample of 1.3 million participants from 23andMe, demonstrating the consistency of GWAS findings once adequate power is achieved.
Results
Primary analysis.
For the ICD code definition of major depressive disorder (see Online Methods for detailed diagnosis definitions), the phenotype with the most available data for the MVP cohort, we conducted a GWAS on 250,215 European individuals (83,810 cases). These MVP data were then included in a meta-analysis in METAL12 using inverse variance weighting with available depression GWAS summary statistics from cohorts of European-ancestry subjects (hereafter, “MDD-META”, Figure 1, Table 1): the PGC and the UK Biobank,10 FinnGen (http://r2.finngen.fi/pheno/F5_MOOD), and 23andMe,9 for a total of 1,154,267 subjects of European ancestry (340,591 cases). We identified 223 independent significant SNPs at 178 genomic risk loci in the primary analysis of European ancestry (Figure 1). We also conducted a GWAS in the African American (AA) sample from MVP in 59,600 participants (25,843 cases). There were no genome-wide significant (GWS) findings from our primary analysis of MDD in African Americans, so we examined overlap with the 223 GWS SNPs from our primary MDD-META meta-analysis of European American ancestry. Of the 223 GWS SNPs from the primary analysis, 206 were available following QC in the AA cohort. 61% (n=125) of the EUR GWS SNPs had the same direction of effect in AAs, with 20 nominally significant (p<0.05) and 1 surviving Bonferroni correction. Finally, we conducted a transancestral meta-analysis of results from the primary GWAS of European and African ancestry. This transancestral analysis of 366,434 cases and 847,433 controls identified 233 independent significant SNPs at 183 genomic risk loci.
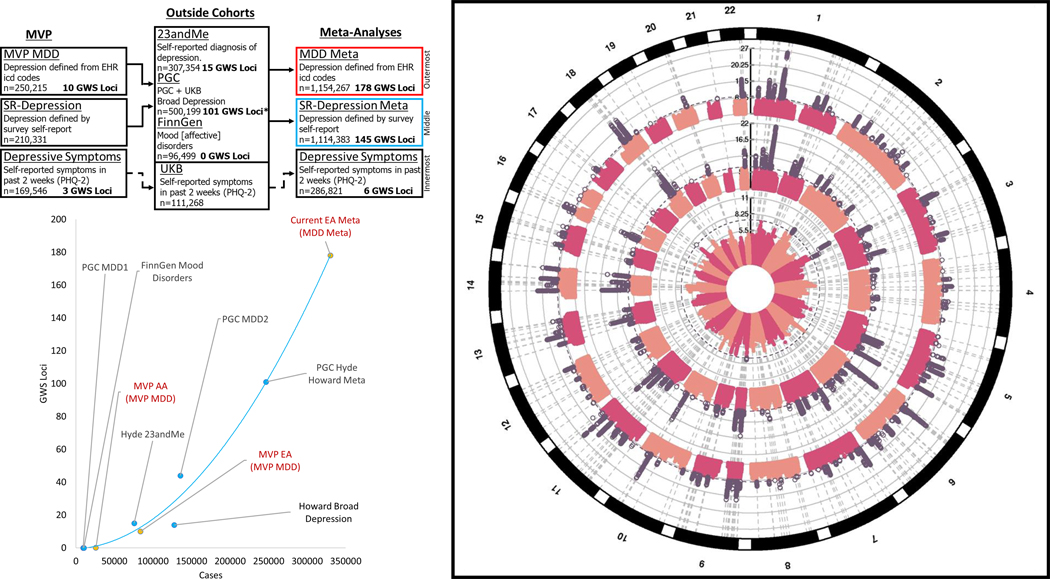
Figure 1. Design of the study and circular Manhattan Plot.
Left Panel: Design of the study (top). Three phenotypes were evaluated within MVP: MDD-META (outermost ring, right panel) which was derived from ICD codes, SR-Depression (middle ring, right panel) which was defined by self-reported diagnosis of depression in the MVP survey, and Depressive symptoms (innermost ring, right panel) which come from the PHQ2 2-item scale found in the MVP survey. MVP-MDD and SR-Depression were each meta-analyzed with depression results from:23andMe, PGC, and FinnGen. MVP PHQ2 was meta-analyzed with results from the PHQ2 2-item scale from UK biobank. Right Panel: Circular Manhattan Plot. Significant results are highlighted in purple. Lower left Panel: Accelerating pace of loci discovery in depression GWAS. Y axis indicates the number of discovered loci in a study, with the X axis showing the number of cases included in each study. Red text and yellow markers indicate original analyses conducted for this study using MVP data for EA, AA and the overall MDD-META meta-analysis of EAs.
Table 1.
Demographics of European Ancestry Samples for Different Phenotype Definitions
| Cohort | Case | Control | Total (%female) |
|---|---|---|---|
| MVP-MDD | 83,810 | 166,405 | 250,215(7) |
| MVP SR-Depression | 55,228 | 155,103 | 210,331(7) |
| 23andMe self-reported diagnosis of depression | 75,607 | 231,747 | 307,354 (48) |
| UKB/PGC PGC + UKB Broad Depression | 170,756 | 329,443 | 500,199(54) |
| FinnGen Mood [affective] disorders | 10,418 | 86,081 | 96,499 |
|
MDD-META
(MVP MDD + 23andMe + UKB/PGC + FinnGen) |
340,591 | 813,676 | 1,154,267 |
|
SR-Depression Meta
(MVP SR-Depression + 23andMe + UKB/PGC + FinnGen) |
312,009 | 802,374 | 1,114,383 |
| MVP PHQ2 | 175,553(8) | ||
| UKB PHQ2 | 111,268(54) | ||
|
PHQ2 Meta
(MVP PHQ2 + UKB PHQ2) |
286,821 | ||
Replication of primary analysis results.
We performed replication analysis in 1,342,778 independent samples provided by 23andMe, including 455,350 depression cases. 211 variants were available for testing in the 23andMe sample. Of these 211, 2 variants had discordant effect direction (0.9%) but not significantly so (p >= 0.28), 209 variants had concordant effect directions (99.1%), 192 showed at least nominal significance p<0.05 (91%), 144 remained significant after Bonferroni correction for multiple comparisons p<0.05/211=2.37×10–04 (68%), and 81 were genome-wide significant p<5×10–08 (38%). These results are reported in Table S1.
Linkage Disequilibrium Score Regression (LDSC).
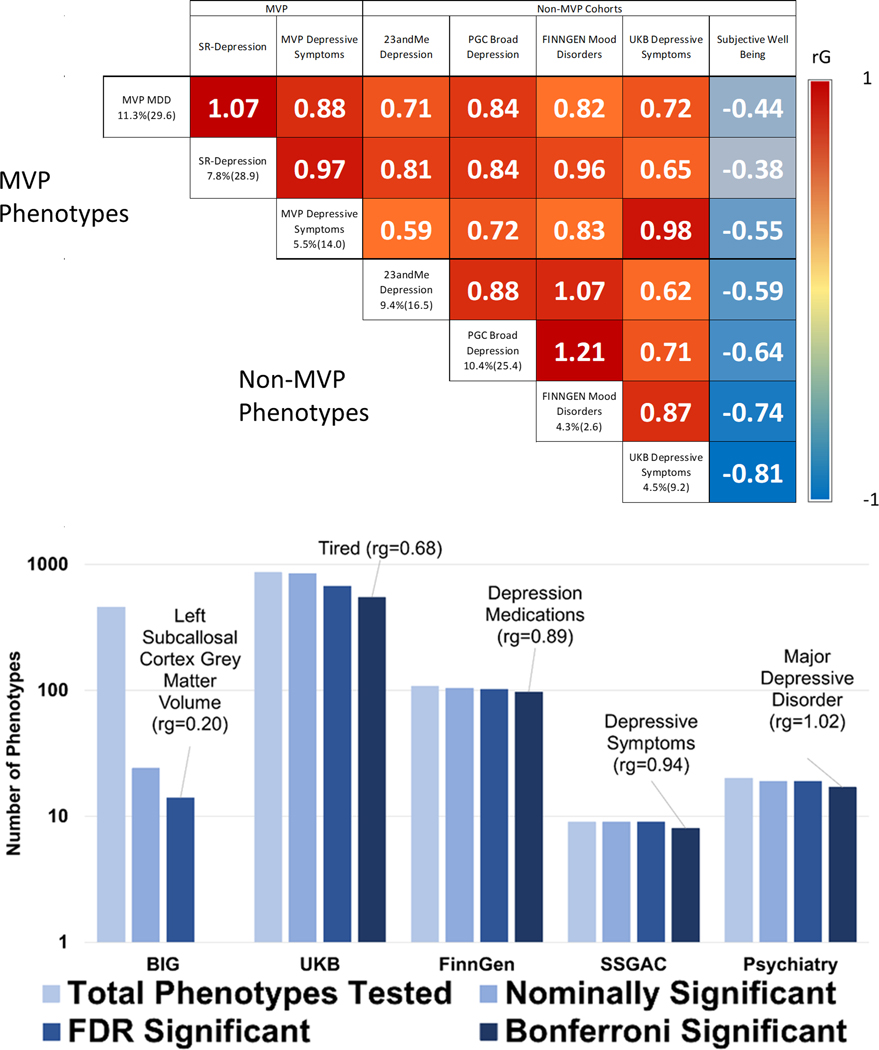
LDSC was used in two ways: 1) to identify genetic correlations and SNP-based heritability within each of the depression cohorts and phenotypes (Table S5); and 2) to identify genetic correlation with other traits based on the primary meta-analysis (MDD-META). Heritability in the primary MDD-META analysis was 11.3% (z= 29.63, sample prevalence 28.6%, population prevalence 20%), while heritability in the secondary analyses of self-reported depression (SR-depression, see online methods) and PHQ-2 were 7.8% (z=28.74, sample prevalence 27.1%, population prevalence 20%) and 5.5% (z=14.0), respectively. Genetic correlation between depression phenotypes ranged from 0.59 to 1.21, with lower rg identified between measures of depressive symptoms and case-control phenotypes (Figure 2 Upper). Some of the genetic correlations from the LD score regression were greater than one; genetic correlation from LDSC does not bound to one,13 and the instances with values higher than one occurred when testing in the same sample with similar phenotype (rg 1.07, SE=0.0343) between MDD and SR-Depression within MVP), or between the somewhat smaller FinnGen sample and the large PGC/UKB broad depression (rg 1.21, SE=0.25) and 23andMe (rg=1.07, SE=0.21) samples. LD-intercept (1.03, SE 0.011) and attenuation ratio (0.0297, SE 0.011) of the LD score regression revealed minimal evidence for inflation or confounding, with 97% of inflation observed due to high polygenicity of depression.
Figure 2. Genetic Correlation.
Upper Panel. Genetic correlations between depression phenotypes, with subjective well-being included as a negative correlation comparator. Heritability (z-score) is given along the left axis of the matrix for each depression phenotype. Values within the matrix represent rg. All correlations are significant following Bonferroni correction for multiple comparisons (0.05/28=p<0.0018). The largest p-value was for the correlation between FinnGen and UKB Depressive symptoms (p=4.06×10–05). P-values and 95% CI are reported in Table S6. Lower Panel. Summary of genetic correlation between MDD-META and 1,457 phenotypes from large-scale genetic studies of mental health and behavior. The Psychiatry category contains phenotypes from the Psychiatric Genomics Consortium, GWAS & Sequencing Consortium of Alcohol and Nicotine use, Million Veteran Program, and International Cannabis Consortium. The labels Tired and left subcallosal cortex grey matter volume represent UKB Field ID 2080 and BIG Field ID 0078, respectively. P-values are two-sided.
Based on significant and robust heritability estimates (h2 z>4), 1,457 traits from available GWAS summary statistics were sufficiently powered to assess genetic correlation with MDD-META. After multiple testing correction (p = 0.05/1,457 trait pairs = 3.43×10−5), 669 phenotypes were significantly genetically correlated with MDD-META (Figure 2 Lower Panel, Supplementary File 1). The most significant phenotypic correlations with MDD-META from each depressive trait category were: (i) depressive symptoms (Social Science Genetic Association Consortium, SSGAC) rg =0.943±0.029, p=1.76×10−228, (ii) depression medications (FinnGen) rg =0.890±0.063, p=6.22×10−45, (iii) major depressive disorder (Psychiatry) rg =1.02±0.017, p<1.39×10-, and (iv) frequency of tiredness/lethargy in last 2 weeks (UKB Field ID 2080) rg =0.684±0.018, p<1.39×10-300. No brain imaging phenotype met corrected significance criteria for genetic correlation with MDD-META; the most significantly genetically correlated brain imaging phenotype, using data provided from the Oxford Brain Imaging Genetics (BIG) project,14 relative to MDD-META was left subcallosal cortex grey matter volume (BIG Field ID 0078) rg =0.205±0.061, p=9.00×10-4.
Transcriptome-Wide Association Study (TWAS).
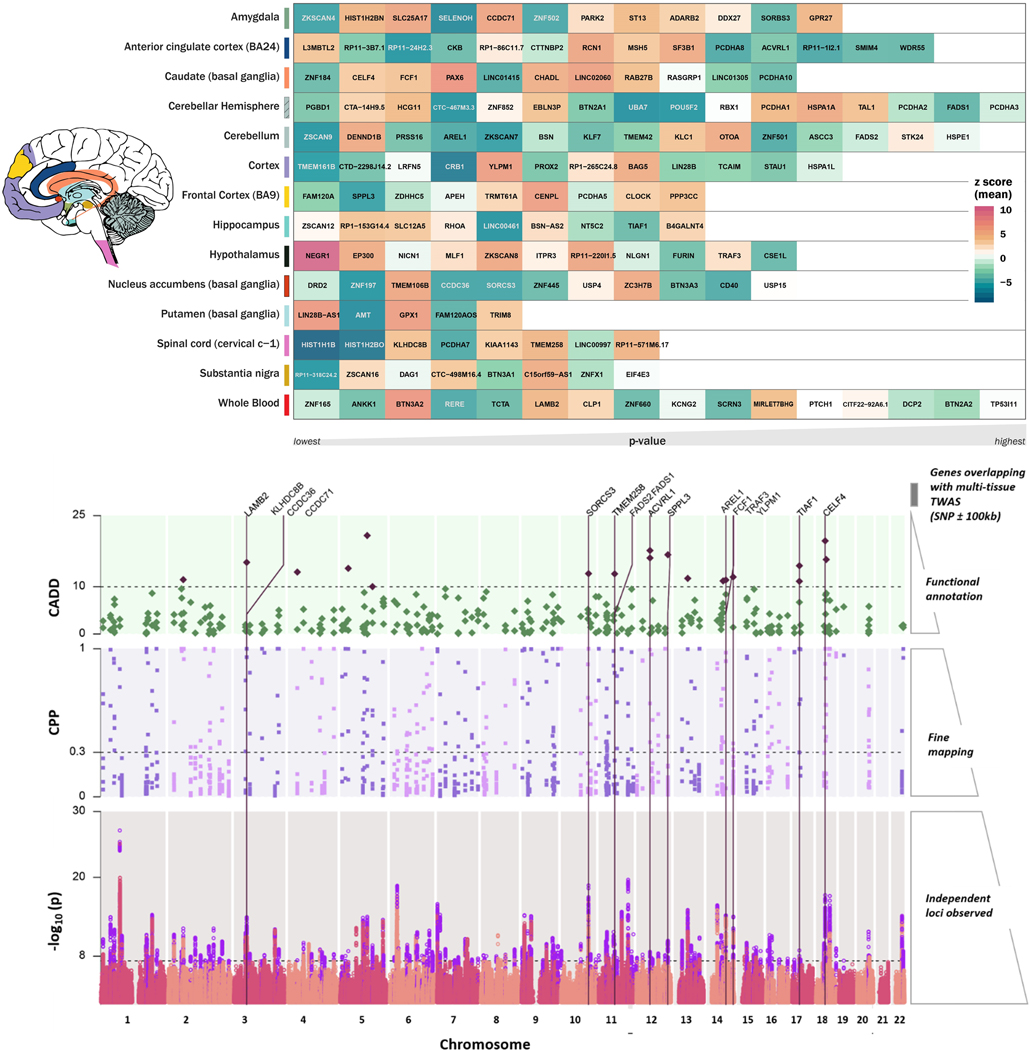
Gene-based association analysis was performed by integrating GWAS association statistics and eQTL data of all brain and whole-blood tissues from GTEx v8. To prioritize target genes further, joint effects of gene expression correlation across tissues was leveraged using S-MultiXcan.15 153 genes and their best representative tissues were below the Bonferroni corrected significance threshold (1.79×10−7) for predicted gene expression in 14 tissues (Figure 3 top; Supplementary file 2). Top genes for each tissue tested were: Amygdala (ZKSCAN4, p=1.65×10−12), anterior cingulate cortex (L3MBTL2, p=1.09×10−14), caudate (ZNF184, p=1.85×10−9), cerebellar hemisphere (PGBD1, p=1.67×10−13), cerebellum (ZSCAN9, p=8.4×10−17), cortex (TMEM161B, p=1.84×10−12), frontal cortex (FAM120A, p=3.25×10−10), hippocampus (ZSCAN12, p=1.14×10−18), hypothalamus (NEGR1, p=3.19×10−25), nucleus accumbens (DRD2, p=1.87×10−20), putamen (LIN28B-AS1, p=2.13×10−12), spinal cord c-1 (HIST1H1B, p=2.90×10−18), substantia nigra (RP11–318C24.2, p=2.41×10−12), and whole blood (ZNF165, p=4.01×10−11).
Figure 3. Top: Tissue-based gene association study (TWAS).
The genes were tested using MetaXcan for 13 brain tissues and whole blood from the GTEx-v8. The genes were compared across tissues to identify best representative tissues for each gene using SMultiXcan. Genes are arranged in order from left to right by respective tissue specific p-value, with the lowest value on the left. The color scale for the gene matrix is based on mean z-score. The values are reported in Supplementary file 2. Bottom: SNP prioritization using fine Mapping and functional scoring. Bottom panel: Manhattan plot showing each genomic risk locus in violet. Middle panel: Each locus was fine mapped, and the causal posterior probability (CPP) on the y-axis is shown for SNPs from the causal set. The SNPs which had CPP ≥0.3 (30%) were annotated using Combined Annotation Dependent Depletion (CADD) scores. Top panel: The SNPs with CADD ≥ 10 are highlighted in purple; these SNPs were positionally mapped to 107 genes within 100kb. Only positional genes overlapping with multi-tissue TWAS results (Supplementary Figure 1) are annotated with vertical lines. Details of the prioritized SNPs are reported in Supplementary file 2.
Variant Prioritization.
All 178 risk loci were fine-mapped (Figure 3 bottom; bottom panel); 1620 SNPs in the causal set out of 14,016 GWS hits have high posterior probability for causal relation with MDD-META (Figure 3 bottom; middle panel). The SNPs with casual posterior probability ≥ 30% were annotated with Combined Annotation Dependent Depletion (CADD) score.16 There were 19 SNPs with CADD scores >10, representing the top 1% of pathogenic variants across the human genome (Figure 3 bottom; top panel). These SNPs were annotated to genes positioned within ±100kb. We found 17 genes overlapping with significant genes identified from cross-tissue TWAS analysis. Each gene-tissue pair was tested for colocalization of the region for eQTL and GWAS. The coloc17 method tests probability of four hypotheses (H0–4). Of these, H4 tests the hypothesis that the same locus is shared between GWAS and tissue-specific eQTL. Loci that were found to have 80% or higher probability for H4 were compared, to understand the LD structure and most prominent variant being shared by GWAS and eQTL. These gene-tissue pairs were CCDC71-Amygdala (H4-PP: 93.1%), FADS1-Cerebellar hemisphere (H4-PP: 96.6%), SPPL3-Frontal Cortex (H4-PP: 83.9%), TRAF3-Hypothalamus (H4-PP: 95.2%) and LAMB2-whole blood (H4-PP: 79.9%) (Supplementary file 2).
Tissue expression analysis and genome-wide gene-based association study (GWGAS).
GWGAS conducted in MAGMA using the MDD-META GWAS meta-analysis identified 426 significant genes after Bonferroni correction for 16,038 protein coding genes. MAGMA tissue expression analysis identified enrichment across all brain tissues and pituitary using data from GTEX v8, with the strongest findings for Brodmann area 9 (p=7.31×10−16), and no enrichment in non-neuronal tissue (Figure S1).
Gene Ontology.
Gene ontology analysis conducted in ShinyGO18 identified 219 biological processes with FDR < 0.05, with top findings involved in nervous system development (q=1.20×10−10), and synapse assembly (q=9.75×10−9) and organization (q=9.75×10−9) (Table S2).
Drug mapping.
The Manually Annotated Targets and Drugs Online Resource (MATADOR)19 database was tested for enrichment for 426 significant genes from the MAGMA analysis. This analysis identified 10 drug annotations with FDR < 0.05 including four drugs that are either estrogen receptor agonists (diethylstilbestrol, Implanon [etonogestril implant]), or anti-estrogens (tamoxifen and raloxifene); in addition to nicotine, cocaine, cyclothiazide, felbamate, and riluzole.
Latent Causal Variable Analysis.
After filtering for suitable trait pairs with LCV-estimated h2 z-scores≥4, 1,667 phenotypes were powered to evaluate causal estimates relative to MDD-META; no statistically significant putatively causal genetic causality proportions (gĉps) were detected.
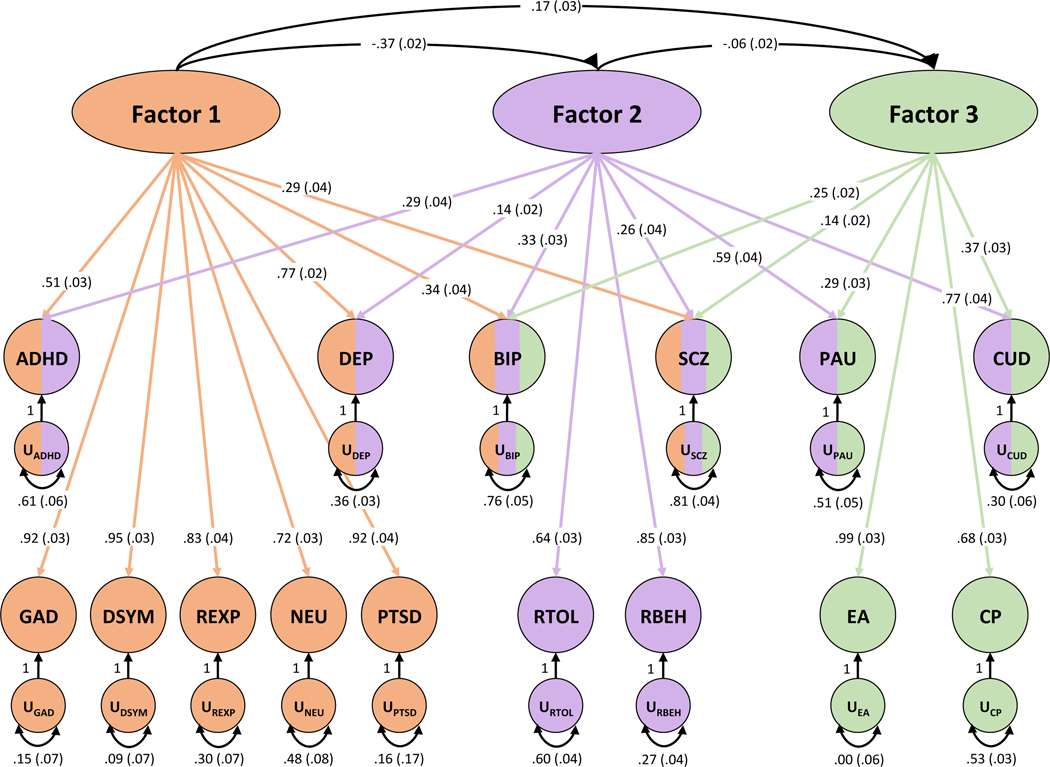
Genomic structural equation modeling (gSEM) was used to evaluate how the MDD-META phenotype relates to 15 previously published large-scale GWAS of mental health and psychiatric phenotypes (See Online Methods and Discussion). Exploratory factor analysis (EFA) was conducted simultaneously on all traits and supported three- (cumulative variance = 0.605) and four-factor models (cumulative variance = 0.624) where each factor contributed over 10% to the cumulative explained variance. Anorexia nervosa did not load onto any factor during EFA and was therefore excluded from confirmatory factor analyses (CFA). CFA did not converge on a four-factor model due to high correlation between two factors. CFA of the three-factor model produced modest fit (comparative fit index = 0.884, X2[83df]=10034.76, AIC=10819.05, standardized root mean square error (SRMR)=0.086; Figure 4, Supplementary File 3). Factor 1 generally represented internalizing phenotypes with major contributions from depressive symptoms (loading = 0.95 ± 0.03), anxiety symptoms (loading = 0.92 ± 0.03), and posttraumatic stress disorder (loading = 0.92 ± 0.04). Factor 2 represented externalizing phenotypes with major contributions from risky behavior (loading = 0.85 ± 0.03) and cannabis use disorder (loading = 0.77 ± 0.04). Factor 3 represented educational attainment (loading = 0.99 ± 0.03) and cognitive performance (loading = 0.68 ± 0.03). MDD-META (DEP, Figure 4, Supplementary File 3) loaded onto factor 1 and, less strongly, on factor 2, independent of its covariance with all other phenotypes (DEP loading on Factor 1 = 0.77 ± 0.02; DEP loading on Factor 2 = 0.14 ± 0.02).
Figure 4. Genomic SEM.
Genomic structural equation modeling of MDD-META (DEP) plus 14 additional traits. Exploratory factor analysis converged on a three-factor model. Arrows represent loading of each phenotype onto a connected factor with loading value and standard error provided for each. Multi-colored phenotypes indicate loading onto more than one factor while monochromatic phenotypes were unique to a single factor. Factor 1 generally represents internalizing symptoms, Factor 2 externalizing behaviors, and Factor 3, education/cognition. The correlation between factors is shown. Phenotype acronyms are: attention deficit hyperactivity disorder (ADHD), MVP MDD-META (DEP), bipolar disorder (BIP), schizophrenia (SCZ), problematic alcohol use (PAU), cannabis use disorder (CUD), anxiety symptoms (GAD), depressive symptoms (DSYM), reexperiencing (REXP), neuroticism (NEU), posttraumatic stress disorder (PTSD), risk tolerance (RTOL), risky behavior (RBEH), educational attainment (EA), and cognitive performance (CP).
Conditional Analysis.
For the mtCOJO analysis (see Online Methods), all eight conditioned versions of the depression GWAS demonstrated substantial similarity to the unconditioned depression GWAS. We observed no changes in h2. All conditioned GWAS had correlation coefficient = 1.00 with the unconditioned GWAS, and genomic control factor and intercepts consistently indicated a lack of population substructure (Figure S2). Though the genome-wide architecture of depression was robust to shared etiology with all other listed comorbid conditions, shared etiology with schizophrenia and anxiety symptoms resulted in substantial loss of GWS SNPs associated with depression when conditioned upon those traits (Figure S2).
Discussion
We present the first genetic study of depression including more than a million informative participants, with new large analyses from the Million Veteran Program meta-analyzed with prior results from the PGC + UK Biobank, 23andMe, and FinnGen, the largest analysis so far in what is a fast-moving field. We investigated genetic correlation between three different definitions (MDD-META, SR-Depression, and PHQ-2) of the depression phenotype within the MVP cohort. We identified 223 independently significant SNPs in 178 genomic loci associated with the primary meta-analysis, using an ICD code derived definition of depression for the MVP sample and GWAS summary statistics from 23andMe, UKB, PGC, and FinnGen. This finding is an increase of 77 loci over the largest previous study that investigated a comparable phenotype.10 As these cohorts used somewhat different definitions for depression (Table 1, Figure 1 Upper Left, Methods), we also used LDSC to examine genetic correlations between MVP depression phenotypes and these differentially defined depression phenotypes in independent cohorts. We investigated genetic correlation with 1,457 traits using available GWAS data, identifying 669 that were significantly correlated. We also used genomic structural equation modeling to evaluate how depression relates to other mental health and psychiatric phenotypes.
The MVP sample added substantially to our ability to discover new loci. Two of the most powerful prior studies conducted to date7,8 had substantial contributions from the UK Biobank. UK Biobank and MVP represent large and non-overlapping samples with consistent phenotypic assessments. This consistency in collection reduces ascertainment heterogeneity within samples and likely increases power to detect new loci. Adding another massive homogenously phenotyped sample here allowed us to discover 77 more loci than previously identified. It also provides a novel large independent cohort for conducting post-GWAS analyses, leveraging the substantial resources already produced by others in the field to improve understanding.
MVP is very informative for depression and related traits with several available measures, so we considered several different diagnosis definitions (Table 1), as follows. In the MVP, we considered (1) an ICD code-based algorithm to determine depression case status based upon diagnosis codes captured in the VA electronic health records (MDD), (2) self-reported diagnosis of depression as reported in the MVP Baseline Survey (SR-Depression), and (3) the 2 item PHQ scale of depressive symptoms in the past 2 weeks, included in the MVP Baseline Survey (depressive symptoms). Genetic correlations between these traits were high (rg 0.81–1.07). We consider the first of these -- MDD-META -- to be our “primary” analysis based on the larger explained heritability and sample size.
For meta-analyses of MDD-META and SR-Depression, we also used available GWAS summary statistics from 23andMe, UKB, PGC, and FinnGen (Table 1). Genetic correlation was conducted between the phenotypes to be meta-analyzed together to quantify potential heterogeneity between the studies to be combined. These studies used a variety of phenotype definitions, with some combining clinical diagnosis of depression based on structured interview and other broader methods,7 such as self-reported treatment,7 or self-reported diagnosis items on questionnaires.9 This analysis is discussed in greater detail in the methods, but the genetic correlations between all traits ranged from 0.71–0.84.
We performed replication analysis in 1,342,778 samples provided by 23andMe (non-overlapping with the 23andMe samples included in our MDD-META), including 455,350 depression cases. 99% of our findings showed concordant direction of effect between these two very large and independent cohorts. Of 211 variants tested, 209 (99%) had the same direction of effect, 192 showed at least nominal significance p<0.05 (91%), 144 remained significant after correction for multiple comparisons p<0.05/211=2.37×10−4 (68%), and 81 were independently genome-wide significant p<5×10−8 (38%). Only 2 SNPs were discordant, both with p>0.05 (0.9%). This very strong replication indicates the consistency of the findings we report herein.
The lead SNP from our primary analysis, rs7531118, (MAF=0.48, p=8.9×10−29) maps close to the neuronal growth regulator 1 gene (NEGR1) and is a brain eQTL for NEGR1. This SNP was at least nominally significant with concordant effect direction in all four studies included in this meta-analysis (MVP p=4.9×10−5, FinnGen p=0.04, PGC+UKB p=1.6×10−17, 23andMe p=2.8×10−8). The S-MultiXcan analysis prioritized hypothalamus as related to NEGR1. Negr −/− mice have shown irregularities in several brain regions, including reduced brain volume in the hippocampus and have also shown abnormalities in social behavior and non-social interest.20 Another study of Negr −/− mice identified a variety of depression-like and anxiety-like features in behavioral assays such as elevated plus maze and forced swim tests.21
The D2 dopamine receptor (DRD2) was another top finding from the TWAS analysis (Figure 3 top), with significant predicted decreased expression in the nucleus accumbens. The mesolimbic dopamine reward circuit, of which nucleus accumbens is a critical part, has long been implicated in depression.22 A recent optogenetic study examining dopaminergic ventral tegmental area (VTA) projections into nucleus accumbens found that dopamine receptors are required for the action of these neurons in depression-related escape behavior.23 Depression-like behavior in animals might be related to depression in humans through links to the reward system and symptoms of anhedonia. A recent randomized proof-of-mechanism trial24 investigated κ-opioid antagonists (KOR) as treatment for anhedonia symptoms. KORs localize within the nucleus accumbens on the terminals of inputs from the mesolimbic dopamine reward circuit. Among the actions of KORs antagonists might be normalization of VTA KOR function and D2 neurons activation, leading to disinhibition of the excitatory circuit they project upon.25 Indeed, the KOR JNJ-67953964 was found to increase VTA activation relative to placebo during reward anticipation, highlighting a potential therapeutic mechanism by which KOR is thought to release inhibition on D2 dopaminergic projections. The group receiving JNJ-67953964 showed reduced anhedonic symptoms relative to controls.24 That this gene and brain tissue emerged from hypothesis-free GWAS and TWAS tissue enrichment is a remarkable finding with respect to known biology, and points to the potential value of other novel findings from this kind of research.
The CUGBP Elav-Like Family Member 4 (CELF4) gene has been highlighted recently in an earlier precursor to this meta-analysis,8 and was our top finding for convergence between functional variant prioritization and multi-tissue TWAS results (Figure 3 bottom, Supplementary File 2). This gene is important in developmental disorders, with deletions of the 18q12.2 region which encompass the gene associated with autism spectrum disorder.26 Celf4 mutant mice show aberrations in sodium channel function, perhaps through increased NAv 1.6 in the axon initial segment of excitatory neurons, and increased susceptibility to seizures.27 We agree with the assertion made in previous studies, now with additional functional and expression evidence, that CELF4 should be a focus of future brain research in depression and depression-like behaviors.
Genetic correlations with available GWAS summary statistics from 1,457 traits were conducted to assess overlap with other traits. There was high genetic correlation between our MDD-META meta-analysis and depression medication prescription in FinnGen (rg=0.89). This could be of value in evaluating depression phenotypes from large cohorts with access to linked pharmacy records; anti-depressant medication prescription may be a viable proxy phenotype for depression diagnosis.
We used ShinyGO18 with the MATADOR19 database to identify overlap between top MAGMA genes and drugs of interest (Figure S3). Riluzole, an NMDA antagonist currently used to treat amyotrophic lateral sclerosis, was one of our top findings. This drug is currently in trials for combination therapy for treatment resistant depression.28 Another drug, cyclothiazide, is an allosteric modulator of AMPA (glutamatergic) receptors. Allosteric modulation of glutamatergic receptors has been considered a mechanistic treatment target for depression.29 This screen also identified an anti-seizure medication, felbamate, which has side effects including increasing depressive symptoms, suicidal ideation, and attempts. These three identified drugs, riluzole, felbamate and cyclothiazide, have been shown to modulate glutamatergic activity.30 Although the exact mechanisms underlying the drugs’ effects on the system remain to be elucidated, it is especially interesting that they were identified in this study considering the emerging evidence of glutamate’s role in the pathophysiology and treatment of mood disorders and the recent US FDA approval of ketamine for treatment-resistant depression. Riluzole has already been identified as a potential antidepressant treatment, with support for its antidepressant properties found in rodent models31 and small clinical studies. However, larger scale clinical trials have not provided clear evidence to support its efficacy. These enrichments, from hypothesis-free association with depression, show converging independent evidence from genetics of existing pharmacological targets based on underlying biological mechanisms.
Genomic SEM was used to investigate relationships between MDD-META and 15 other mental health and neurocognitive phenotypes (Figure 4, Supplementary File 3); summary statistics come from the largest studies available. All traits tested except anorexia nervosa loaded onto at least one factor during exploratory analysis. We identified three factors, with MDD-META loading onto the first two independently of covariance with the other phenotypes. Factor 1 may be thought to represent internalizing phenotypes, with major contributions from MDD-META, anxiety symptoms, and posttraumatic stress disorder. MDD-META also loaded (but less strongly) onto factor 2, which broadly represents externalizing phenotypes and psychosis, with the major contributions coming from risky behavior and cannabis use disorder. MDD-META did not load onto factor 3, which was mostly contributed to by educational attainment and cognitive performance and thus may represent a neurocognitive domain. Many cross-disorder studies using GWAS, this one included, align themselves in ways consistent with existing theories of psychopathology.
We prioritized variants using biologically and statistically informed annotations. To prioritize genes and their target tissues we integrated both transcriptomics and CADD score prioritized variants. This method aided in the identification of shared causal loci for phenotype and tissue-specific eQTLs as evidenced by the high probability for 5 of the 17 genes tested. SNPs at CCDC71 (“Coiled-Coil Domain Containing 71”) have been reported to be associated with depressive symptoms in a multivariate genome wide association meta-analysis, and our prioritized SNP is in strong LD with that study’s lead SNP (current study rs7617480, r2=0.83, D’=1.0).32 The FADS1 protein product, “Fatty Acid Desaturase 1” is involved in fatty-acid regulation and variants in this region have been reported to be associated with depression and substance use disorders. There is consistent evidence in the literature for an association with depleted omega-3 and increased depression risk, though a role for omega-3 supplementation in treatment of depression is still controversial.33 Variants in SPPL3, encoding “Signal Peptide Peptidase Like 3”, were reported to be associated with risk to major depression by Hyde and colleagues.9 The TRAF3 protein product, “TNF Receptor Associated Factor 3”, controls type-1 interferon response,34 and it has been reported that individuals treated with interferon are at high risk to develop depressive symptoms.35 LAMB2 is involved in neuropathic pain and influencing gene expression changes in brain pathways implicated in depression.36
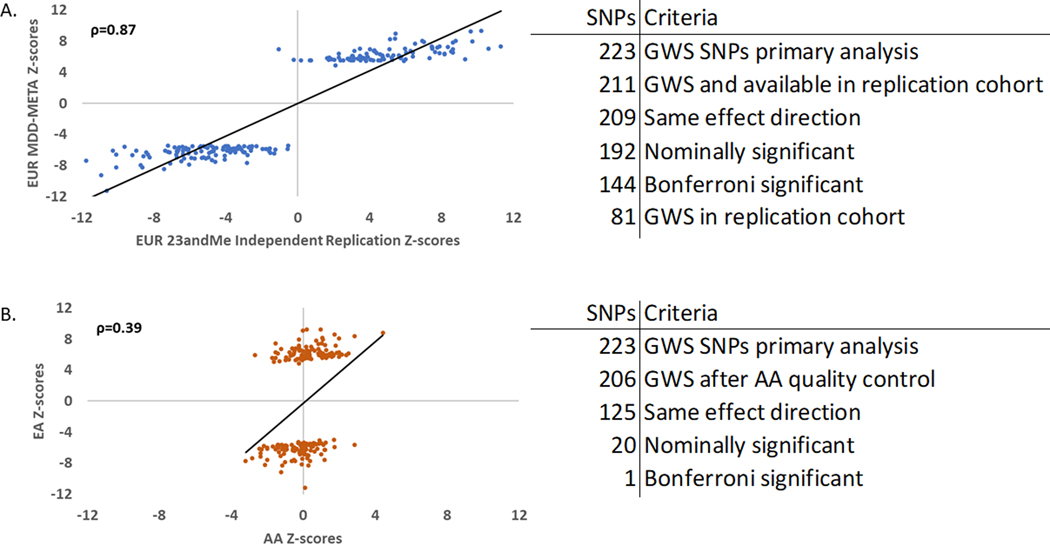
Because no GWS findings were identified in our primary analysis of African ancestry we performed cross ancestry lookups in the summary statistics of European ancestry. Of 223 GWS SNPs from the European ancestry meta-analysis, 206 were available in African ancestry, 61% (n=125) had the same effect direction, 20 were nominally significant (p<0.05), and 1 SNP survived Bonferroni correction (Figure 5). This SNP that survived multiple testing correction (rs1950829 EUR p=7.24Ex10−19, AFR p=9.34×10−6), is in an intron of the “Leucine rich repeat fibronectin type III domain containing 5” (LRFN5) gene. This gene was previously detected in genome-wide gene- and pathway based analyses of depressive symptom burden conducted in three cohorts from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the Health and Retirement Study (HRS), and the Indiana Memory and Aging Study (IMAS).37 As larger samples are collected for more diverse ancestry groups we expect to see more novel loci identified for non-European populations. Finally, we conducted a transancestral meta-analysis by combining studies of African and European ancestries in 1,213,867 participants, thereby identifying 233 independent SNPs and 183 risk loci. For now, transancestral analysis is a way to leverage results from understudied populations.
Figure 5. Similar-ancestry and Transancestry Replication Analyses.
A. Left panel: Scatter plot for z-score effect sizes for 211 GWS SNPs (Spearman’s ρ=0.87) from the primary MDD-META GWAS on the y axis and the independent 23andMe replication cohort African ancestry (only) GWAS on the x axis. Right panel: Overlap of SNPs from European and African ancestry GWASs. 223 GWS SNPs from the primary analysis, of which 211 were available in the independent 23andMe GWAS. 209 (99%) of the remaining SNPs had the same effect direction, 192 were nominally significant p<0.05 (91%), 144 were Bonferroni significant after correcting for 211 comparisons (68%), and 81 were independently genome-wide significant (38%). B. Left panel: Scatter plot for z-score effect sizes for 206 GWS SNPs (Spearman’s ρ=0.39) from the primary MDD-META GWAS of different ancestries, plotting z-score for European ancestry (only) GWAS on the y axis and African ancestry (only) GWAS on the x axis. Right panel: Overlap of SNPs from European and African ancestry GWASs. 223 GWS SNPs from the primary analysis, of which 206 are available in the AA GWAS following QC. 125 (61%) of the remaining SNPs had the same effect direction, 20 were nominally significant (p<0.05) and one was Bonferroni significant after correcting for 206 comparisons.
We recognize limitations in our study. Maximizing the power available for this analysis comes at the cost of accepting broader biobank phenotyping approaches, which may reduce specificity of findings for the core depression phenotype.11 Nonetheless, strong genetic correlations between the ICD derived MDD with the broader definitions provide confidence in internal consistency, and future studies could look to further refine phenotyping. Although all genetic correlations were significant, there was substantial variance (95% CI= 0.72–1.7) in correlations with the FinnGen sample, probably due to power and heterogeneity in the broad phenotype we used from this sample. Finally, other ancestries remain understudied in relation to Europeans. We hope that the initial results reported here for the MVP African ancestry sample can help advance the field by encouraging additional concerted research in African and other non-European ancestral groups.
In summary, we identified multiple novel loci, and several of these loci serve functions that should prioritize their further study in the pathology of major depression. We examined genetic correlations between depression GWAS and other external phenotypes, largely confirming and strengthening previous observations. We showed substantial enrichments for several brain regions, such as hypothalamus and frontal cortex, known to be important for depression. We also find strong support for the importance of DRD2 in the nucleus accumbens, a finding that is consistent with an emerging role for dopaminergic function in symptoms of anhedonia. Using gene and drug-based enrichments we found overlapping biology with existing drugs – notably those that impact glutamatergic function, but also those that influence the actions of estrogen – that could offer repurposing opportunities. We used genomic structural equation modeling to show how the genetic architecture of depression maps onto the broader genetic structure of mental disorders and cognition, identifying emergent overlap from hypothesis-free GWAS approaches with existing theories of psychopathology with regard to clusters of internalizing and externalizing disorders. Finally, we showed that many of our findings replicate in a large and independent cohort provided by 23andMe, providing evidence for the stability of GWAS findings from adequately powered cohorts.
Online Methods
Participants.
The MVP cohort has been previously described.38–40 GWAS was conducted in each of two tranches of data separately by ancestry, depending upon when the data became available. Ancestry was assigned using 10 principal components (PCs) and the 1000 genomes project phase 3 EUR and AFR reference within each tranche of data. For the analysis of the quantitative phenotype we also performed a GWAS in the UK Biobank sample. Finally, we conducted GWAS meta-analyses of traits related to depression using data from 4 large cohorts (Table 1, Figure 1 Upper Left): the Million Veteran Program (MVP), 34, 41 the PGC/UK Biobank,10 FinnGen, and 23andMe.9 For the ICD definition of depression, the phenotype with the most available data for the MVP cohort, there were 1,154,267 total subjects for primary meta-analysis. For the secondary case control meta-analysis, we performed a similar analysis except we replaced the MDD diagnosis from MVP with the SR-Depression GWAS for a total of 1,114,383 participants. For the secondary analysis of depressive symptoms by PHQ, we included 286,821 total participants from UKB and MVP. We also performed a GWAS in the MVP African American (AA) sample of 59,600 participants. We included these participants in a transancestral meta-analysis with a total sample size of 1,213,867 participants (Figure S5). Cohorts are detailed in Table 1. All data were collected independently and therefore the analysts were blinded to the conditions of the analysis. No randomization was performed. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications. 8,10
Phenotypes.
Within MVP there were three depression phenotypes investigated across five different analyses. We used 1) an ICD code-based algorithm to determine depression case status based upon investigation of the electronic health records (MDD, primary analysis), 2) self-reported physician diagnosis of depression as reported in the MVP baseline survey (SR-Depression), and 3) the 2-item PHQ scale of depressive symptoms in the past 2 weeks, included in the MVP baseline survey (depressive symptoms). Phenotypes in outside cohorts for UKB-PGC and 23andMe have been previously described.7–10 See Table 1 and Figure 1 for summary. For the ICD code-based algorithm in MVP, codes used to assess case status are presented in Table S3. Cases included people with at least one inpatient diagnosis code or two outpatient diagnosis codes for Major Depressive Disorder (MDD). Controls include only those without any inpatient or outpatient depression diagnosis codes for depression.
Secondary phenotype definitions.
A similar meta-analysis was conducted using self-reported (SR)-Depression (see Methods) from MVP, conducted on 210,331 individuals who completed survey items on self-reported diagnosis of depression by a medical professional; the total meta-analysis with the traits from PGC, UK Biobank and FinnGen included 1,114,383 subjects. A third analysis considered depressive symptoms from the Patient Health Questionniare-2 (PHQ-2),42 a 2-item scale which assesses depressive symptoms within the prior two weeks (Table S4). For this phenotype, data were only available from MVP and UK Biobank, with a total sample of 286,821 European participants.
GWASs and meta-analyses.
GWAS analysis was carried out in the MVP cohorts by logistic regression for MDD and SR-Depression and by linear regression for PHQ2 within each ancestry group and tranche using PLINK 2.0 on dosage data, covarying for age, sex, and the first 10 PCs. A similar GWAS was performed using linear regression in the UK Biobank samples, also using age, sex, and the first 10 PCs for PHQ2.
In individuals of European ancestry for MDD-META and SR-Depression, meta-analysis was performed using METAL with inverse variance weighting for: MVP tranche 1, MVP tranche 2, the PGC-UKB MDD-META meta-analysis,10 23andMe,9 and FinnGen Mood [affective] disorders ( http://r2.finngen.fi/pheno/F5_MOOD). For the PHQ2 meta-analysis, the procedures were the same for the following samples: MVP tranche 1, MVP tranche 2, and UK Biobank. Meta-analysis in the African American participants was carried out only between tranche 1 and 2 of the MVP data due to absence of data in the other samples. Our results depend on contributions from many sources over many years. Some of the contributory studies and historical context of GWASes for MDD are presented in Table S5.
The 23andMe phenotype was based on responses to 4 questions: “Have you ever been diagnosed by a doctor with any of the following psychiatric conditions?”, “Have you ever been diagnosed with clinical depression?”, “Have you ever been diagnosed with or treated for any of the following conditions? (Depression)”, and “In the last 2 years, have you been newly diagnosed with or started treatment for any of the following conditions? (Depression)”. Cases were defined as having responded “Yes” to any of the above questions, and controls, when not a case and at least 1 “No” response to the above questions.
The FinnGen diagnosis is defined by the F5 Mood category and was downloaded from Freeze 2 of the database (http://r2.finngen.fi/pheno/F5_MOOD). This phenotype is broad and contains manic episodes, bipolar disorders, depression, persistent mood disorders, and other unspecified mood [affective] disorders. Data from UKB8 is a broad depression phenotype based on affirmative responses to either of the questions: “Have you ever seen a general practitioner for nerves, anxiety, tension or depression?”, and “Have you ever seen a psychiatrist for nerves, anxiety, tension or depression?”. PGC data also has been previously reported,7 and come from meta-analysis of 35 cohorts with a spectrum of depression phenotypes, including some with clinical diagnosis from structured interviews and others with broader definitions. LD-intercept (1.03, SE 0.011) and attenuation ratio (0.0297, SE 0.011) of the LD score regression revealed minimal evidence for inflation or confounding, with 97% of inflation observed due to high polygenicity of depression (Figure S4). Data distribution was assumed to be normal but this was not formally tested.
Replication of primary analysis.
An independent GWAS was run at 23andMe using logistic regression assuming an additive model for allelic effects, while covarying for age, sex, 4 principal components and array platform, followed by SNP lookups of our 221 independent GWS SNPs. The phenotype was identical to that reported from9 (discussed in detail in the section above) but consisting of an entirely independent sample of 455,350 cases and 887,428 (n=1,342,778) not previously included in any reported primary analysis.
Post-GWAS analysis
Linkage Disequilibrium Score Regression.
For post-GWAS analysis, FinnGen was removed a priori due to potential for increased heterogeneity in the phenotype definition due to the broad nature of inclusion in the F5 Mood phenotype. Genetic correlation analyses were performed using LDSC to assess the degree of genetic overlap between phenotypes and across the cohorts included in the analysis. Per-trait observed-scale SNP-based heritability estimates were calculated via LDSC using the 1000 Genomes Project European linkage disequilibrium reference panel. 13 Heritability estimates were calculated for 1,468 phenotypes from FinnGen, 4,083 phenotypes from UKB, 3,143 brain image derived phenotypes from the Oxford Brain Imaging Genetics (BIG) project, and phenotypes from the Psychiatric Genomics Consortium (PGC), the Social Science Genetic Association Consortium (SSGAC), and the Genetics of Personality Consortium (GPC). Heritability z-scores were calculated by dividing the heritability estimate per phenotype by its associated standard error. Phenotypes with heritability z-scores ≥ 4 were considered suitable for genetic correlation against MDD-META. 13 For continuous UKB phenotypes we restricted our analyses to use inverse-rank normalized phenotypes instead of untransformed phenotypes. Genetic correlations are summarized by total phenotypes tested, nominally significant (p<0.05), and after application of 5% false discovery rate and Bonferroni thresholds (Figure 2 Lower Panel).
Latent Causal Variable (LCV).
The LCV model was used to infer genetic causal relationships between trait pairs using the 1000 Genomes Project European linkage disequilibrium reference panel. MDD-META was subjected to LCV with all traits described above for genetic correlation analysis. Due to differences in heritability calculation method and the number of SNPs used by LCV versus LDSC, genetic correlation results were not used to inform LCV trait pair selection. Genetic causality proportions (gĉp) were interpreted only when the heritability z-score of both traits was ≥ 7, as determined by LCV, not LDSC.43 Fully causal relationships were deduced for significant trait pairs with gĉp estimates ≥ 0.70; otherwise gĉp estimates were considered evidence for partial causality.43
Genomic structural equation modeling (SEM).
Genomic SEM was performed using GWAS summary statistics in the genomicSEM and lavaan R packages.44 Exploratory factor analyses (EFA) were performed on 16 traits simultaneously (MDD-META [the main phenotype of interest for this study], attention deficit hyperactivity disorder, anorexia nervosa, bipolar disorder, cannabis use disorder, cognitive performance, depressive symptoms, educational attainment, anxiety symptoms, neuroticism, posttraumatic stress disorder, problematic alcohol use, reexperiencing, risk tolerance, risky behavior, and schizophrenia). EFAs were performed for 1 through N factors until the addition of factor N contributed less than 10% explained variance to the model. Confirmatory factor analysis was performed using the diagonally-weighted least squares estimator and a genetic covariance matrix of munged GWAS summary statistics for all 16 phenotypes based on the 1000 Genome Project Phase 3 European linkage disequilibrium reference panel.
Transcriptome-Wide Association Study (TWAS).
We performed transcriptome-wide association study using MetaXcan for 13 brain tissues and whole blood using GTEx v8. The MetaXcan framework consists of two prediction models for GTEx v8; elastic net and MASHR-based model for deriving eQTL values. The MASHR model is biologically informed, with Deterministic Approximation of Posteriors (DAP-G) based fine mapped variables and recommended by the developers.45 Since the eQTL effect is shared across several tissues, the joint effect of eQTL in 14 tissues was tested using the S-MultiXcan, developed under the MetaXcan toolkit.15 We applied Bonferroni correction (corrected p-value threshold = 1.79×10−7) for all gene-tissue pair tested.
Variant prioritization.
Each of the risk loci, determined from FUMA (default LD = 0.6), were fine-mapped using CAVIAR.46 The set of causal SNPs were annotated with CADD16 scores followed by positional gene mapping within ±100kb. The genes that overlapped with significant gene cross-tissue eQTL analysis were further tested for colocalization. Coloc17 was used to test colocalization between specific gene eQTL tissue pairs (GTEx v8). The LocusCompareR R package was used to generate regional plots of tissue-specific eQTL and GWAS p-values.
Genome-wide gene-based association study (GWGAS) and Enrichment Analysis.
Summary statistics from the primary MDD-META meta-analysis were loaded into Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA GWAS) to test for gene-level associations using Multi-Marker Analysis of GenoMic Annotation (MAGMA).47 Input SNPs were mapped to 17,927 protein coding genes. The GWS threshold for the gene-based test was therefore determined to be p = 0.05/17,927 = 2.79×10-6. Genes from MAGMA’s gene-based association were used for gene ontology and drug-set enrichment using the ShinyGO18 web tool.
Conditional Analysis
To evaluate whether the genetic signal of depression was independent of signals from comorbid conditions, we employed multi-trait-based conditional and joint analysis (mtCOJO) in GCTA.48 With mtCOJO, per-SNP effect estimates and association statistics of MDD-META were adjusted for the causal effects between MDD and seven comorbid conditions estimated by Mendelian randomization. We required at least 2 genome-wide significant SNPs after Heidi outlier testing with which to estimate causality between phenotypes. MDD was conditioned eight times: once each for alcohol use disorder, digestive disorders, educational attainment, fibromyalgia, neuroticism (SSGAC), schizophrenia, and subjective well-being; and once using all seven correlates simultaneously. In this experimental design, we generated 8 new versions of depression GWAS summary statistics termed “conditioned” GWAS to analyze for heritability, genetic correlation versus the original unconditioned depression GWAS, SNP effects, and p-value survival. These analyses are described in Methods under Post-GWAS analysis: Linkage Disequilibrium Score Regression. Conditioned GWAS generated from mtCOJO are free of collider biases when estimate causal relationship between depression and each comorbid condition.49
Due to SNP-matching procedures to condition depression with other phenotypes, some GWS SNPs for depression were not found in the conditioned depression GWAS. Where necessary, we selected proxy SNPs for each depression GWS SNP using SNPsnap50 with default settings. For each conditioned version of the depression GWS a subset of SNPs could not be matched using direct or proxy SNP matching.
Supplementary Material
Acknowledgements
We want to acknowledge the participants and investigators of the FinnGen study, 23andMe, the UK Biobank, PGC and the Million Veteran Program. We would like to thank the research participants and employees of 23andMe for making this work possible. We thank the veterans who participate in the Million Veteran Program.
The following members of the 23andMe Research Team contributed to this study:
Michelle Agee, Stella Aslibekyan, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah K. Clark, Sarah L. Elson, Kipper Fletez-Brant, Pierre Fontanillas, Nicholas A. Furlotte, Pooja M. Gandhi, Karl Heilbron, Barry Hicks, David A. Hinds, Karen E. Huber, Ethan M. Jewett, Yunxuan Jiang, Aaron Kleinman, Keng-Han Lin, Nadia K. Litterman, Marie K. Luff, Jennifer C. McCreight, Matthew H. McIntyre, Kimberly F. McManus, Joanna L. Mountain, Sahar V. Mozaffari, Priyanka Nandakumar, Elizabeth S. Noblin, Carrie A.M. Northover, Jared O’Connell, Aaron A. Petrakovitz, Steven J. Pitts, G. David Poznik, J. Fah Sathirapongsasuti, Anjali J. Shastri, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Robert J. Tunney, Vladimir Vacic, Xin Wang, Amir S. Zare.
From the Yale Department of Psychiatry, Division of Human Genetics, we would like to thank and acknowledge the efforts of Ann Marie Lacobelle, Christa Robinson, and Chelsea Tyrell.
Funding
Supported by funding from the Veterans Affairs Office of Research and Development Million Veteran Program grant MVP025 and VA Cooperative Studies Program CSP575B. DFL was supported by a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation.
Footnotes
Conflict of Interest
Dr. Stein reports receiving consulting fees in the past 3 years from Acadia Pharmaceuticals, Aptinyx, Bionomics, Clexio Biosciences, EmpowerPharm, Genentech/Roche, GW Pharmaceuticals, Janssen, Jazz Pharmaceuticals, and Oxeia Biopharmaceuticals.
In the last 12 months Dr. Sanacora has provided consulting services to Allergan, Axsome Therapeutics, Biohaven Pharmaceuticals, Boehringer Ingelheim International GmbH, Bristol-Myers Squibb, Clexio Biosciences, Epiodyne, Intra-Cellular Therapies, Janssen, Lundbeck, Minerva pharmaceuticals, Navitor Pharmaceuticals, NeuroRX, Noven Pharmaceuticals, Otsuka, Perception Neuroscience, Praxis Seelos Pharmaceuticals and Vistagen Therapeutics. He has received funds for contracted research from Janssen Pharmaceuticals, Merck, and Usona Institute. He holds equity in Biohaven Pharmaceuticals and has received royalties from Yale University paid from patent licenses with Biohaven Pharmaceuticals.
Jingchunzi Shi and Suyash Shringarpure are employed by and hold stock or stock options in 23andMe, Inc.
Dr. Gelernter is named as co-inventor on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018.
All other authors declare that they have no conflict of interest. No other conflicts are reported.
Data Availability
The GWAS summary statistics generated during and/or analyzed during the current study are available via dbGaP; the dbGaP accession assigned to the Million Veteran Program is phs001672.v1.p. The website is: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?studyid=phs001672.v1.p1.
The full GWAS summary statistics for the 23andMe discovery data set will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset-access/ for more information and to apply to access the data.
Code Availability
No custom code was used in this study. Software and R packages used were discussed in text.
Ethics statement
The Central VA Institutional Review Board (IRB) and site-specific IRBs approved the MVP study. All relevant ethical regulations for work with human subjects were followed in the conduct of the study, and written informed consent was obtained from all participants. For 23andMe, participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (E&I Review). Participants were included in the analysis on the basis of consent status as checked at the time data analyses were initiated.
Disclaimer
‘This publication reflects the views of the authors and should not be construed to represent the views or policies of the Department of Veterans Affairs, the Substance Abuse and Mental Health Services Administration, the Food and Drug Administration, the Department of Health and Human Services, or the U.S. government.
References
- 1.Hasin DS et al. Epidemiology of Adult DSM-5 Major Depressive Disorder and Its Specifiers in the United States. JAMA Psychiatry 75, 336–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roehrig C. Mental Disorders Top The List Of The Most Costly Conditions In The United States: $201 Billion. Health Affairs 35, 1130–1135 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Mullins N. et al. GWAS of Suicide Attempt in Psychiatric Disorders and Association With Major Depression Polygenic Risk Scores. Am J Psychiatry 176, 651–660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strawbridge RJ et al. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine 41, 517–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levey DF et al. Genetic associations with suicide attempt severity and genetic overlap with major depression. Transl Psychiatry 9, 22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Docherty AR et al. Genome-Wide Association Study of Suicide Death and Polygenic Prediction of Clinical Antecedents. Am J Psychiatry 177, 917–927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wray NR et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard DM et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat Commun 9, 1470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyde CL et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 48, 1031–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard DM et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22, 343–352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai N. et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat Genet 52, 437–447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willer CJ et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40, 161–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bycroft C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbeira AN et al. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet 15, e1007889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rentzsch P, Witten D, Cooper GM, Shendure J & Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giambartolomei C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge SX, Jung D & Yao R. ShinyGO: a graphical enrichment tool for animals and plants. Bioinformatics (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunther S. et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res 36, D919–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh K. et al. Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci Rep 9, 5457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noh K. et al. Negr1 controls adult hippocampal neurogenesis and affective behaviors. Mol Psychiatry 24, 1189–1205 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Nestler EJ & Carlezon WA Jr. The mesolimbic dopamine reward circuit in depression. Biol Psychiatry 59, 1151–9 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Tye KM et al. Dopamine neurons modulate neural encoding and expression of depression-related behaviour. Nature 493, 537–541 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krystal AD et al. A randomized proof-of-mechanism trial applying the ‘fast-fail’ approach to evaluating kappa-opioid antagonism as a treatment for anhedonia. Nat Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlezon WA Jr., Beguin C, Knoll AT & Cohen BM Kappa-opioid ligands in the study and treatment of mood disorders. Pharmacol Ther 123, 334–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilling M. et al. A 3.2 Mb deletion on 18q12 in a patient with childhood autism and high-grade myopia. Eur J Hum Genet 16, 312–9 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Sun W. et al. Aberrant sodium channel activity in the complex seizure disorder of Celf4 mutant mice. J Physiol 591, 241–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakurai H. et al. Longer-term open-label study of adjunctive riluzole in treatment-resistant depression. J Affect Disord 258, 102–108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alt A, Nisenbaum ES, Bleakman D & Witkin JM A role for AMPA receptors in mood disorders. Biochem Pharmacol 71, 1273–88 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Pittenger C. et al. Riluzole in the treatment of mood and anxiety disorders. CNS Drugs 22, 761–86 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Chowdhury GM et al. Transiently increased glutamate cycling in rat PFC is associated with rapid onset of antidepressant-like effects. Mol Psychiatry 22, 120–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baselmans BML et al. Multivariate genome-wide analyses of the well-being spectrum. Nat Genet 51, 445–451 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wani AL, Bhat SA & Ara A. Omega-3 fatty acids and the treatment of depression: a review of scientific evidence. Integr Med Res 4, 132–141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hacker H, Tseng PH & Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol 11, 457–68 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Chiu WC, Su YP, Su KP & Chen PC Recurrence of depressive disorders after interferon-induced depression. Transl Psychiatry 7, e1026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Descalzi G. et al. Neuropathic pain promotes adaptive changes in gene expression in brain networks involved in stress and depression. Sci Signal 10(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nho K. et al. Comprehensive gene- and pathway-based analysis of depressive symptoms in older adults. J Alzheimers Dis 45, 1197–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelernter J. et al. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat Neurosci (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Harrington KM et al. Gender Differences in Demographic and Health Characteristics of the Million Veteran Program Cohort. Womens Health Issues 29, S56–S66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levey DF et al. Reproducible Genetic Risk Loci for Anxiety: Results From approximately 200,000 Participants in the Million Veteran Program. Am J Psychiatry, appiajp201919030256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kroenke K, Spitzer RL & Williams JB The Patient Health Questionnaire-2: validity of a two-item depression screener. Med Care 41, 1284–92 (2003). [DOI] [PubMed] [Google Scholar]
- 43.O’Connor LJ & Price AL Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet 50, 1728–1734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grotzinger AD et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav 3, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbeira AN et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 9, 1825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B & Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe K, Taskesen E, van Bochoven A & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu ZH et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nature Communications 9(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL & Kraft P. Adjusting for Heritable Covariates Can Bias Effect Estimates in Genome-Wide Association Studies. American Journal of Human Genetics 96, 329–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pers TH, Timshel P & Hirschhorn JN SNPsnap: a Web-based tool for identification and annotation of matched SNPs. Bioinformatics 31, 418–420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.