Abstract
The complexity of human astrocytes remains poorly defined in primary human tissue, requiring better tools for their isolation and molecular characterization. Fluorescence-activated nuclei sorting (FANS) can be used to successfully isolate and study human neuronal nuclei (NeuN+) populations from frozen archival tissue, thereby avoiding problems associated with handling fresh tissue. However, efforts to similarly isolate astroglia from the non-neuronal (NeuN−) element are lacking. A recently developed and validated immunotagging strategy uses three transcription factor antibodies to simultaneously isolate enriched neuronal (NeuN+), astrocyte (paired box protein 6 (PAX6)+NeuN−), and oligodendrocyte progenitor (OLIG2+NeuN−) nuclei populations from non-diseased, fresh (unfixed) snap-frozen postmortem human temporal neocortex tissue.
This technique was shown to be useful for the characterization of cell type-specific transcriptome alterations in primary pathological epilepsy neocortex. Transcriptomic analyses confirmed that PAX6+NeuN− sorted populations are robustly enriched for pan-astrocyte markers and capture astrocytes in both resting and reactive conditions. This paper describes the FANS methodology for the isolation of astrocyte-enriched nuclei populations from fresh-frozen human cortex, including tissue dissociation into single-nucleus (sn) suspension; immunotagging of nuclei with anti-NeuN and anti-PAX6 fluorescently conjugated antibodies; FANS gating strategies and quality control metrics for optimizing sensitivity and specificity during sorting and for confirming astrocyte enrichment; and recommended procurement for downstream transcriptome and chromatin accessibility sequencing at bulk or sn resolution. This protocol is applicable for non-necrotic, fresh-frozen, human cortical specimens with various pathologies and recommended postmortem tissue collection within 24 h.
Introduction
The molecular complexity of human astrocytes remains poorly defined in primary tissue, requiring better tools for their isolation and characterization at high resolution, both in health and disease. Separation of intact human neurons and glia from their niche has proven difficult due to limited access of fresh brain tissue samples, the heavily interconnected nature of glial and neuronal processes, and inevitable cellular activation during processing, all of which limit the molecular characterization of these cell types ex vivo1. Fluorescence-activated nuclei sorting (FANS) has emerged as an alternative to live-cell sorting, enabling the dissociation and immunotagging of nuclei populations from frozen tissue. In the past decade, FANS has become widely used for isolating and molecularly characterizing human neuronal (NeuN+) nuclei populations in a variety of brain specimens and anatomical regions1,2,3,4.
However, similar methods for isolating specific glial nuclei subpopulations from human cortex have been limited, leading to a relative lack of sophistication in the understanding of astrocyte complexity in both normal and diseased tissues. To this end, a previously published protocol was adapted for isolating human neuronal populations using FANS4, and a method was validated to enrich for astrocytes (and for oligodendroglial progenitors) using a triple-antibody combination, capturing astrocytes in both resting and reactive conditions5. To specifically enrich for astrocytes in the NeuN− fraction, antibodies were used against one of two transcription factors known to be differentially expressed across astrocyte populations, PAX6 or SRY-box transcription factor 9 (SOX9)6,7. PAX6 is highly expressed during early fetal development within radial glia-like progenitors in germinal zones and contributes toward both neurogenesis and gliogenesis8,9,10,11 as well as to retinal neuronal specification12. In the adult, PAX6 is differentially overexpressed in resting human astrocytes6 and shows protein co-expression with glial fibrillary acidic protein (GFAP) in human epilepsy tissue astrocytes13.
This protocol describes the simultaneous isolation of neocortical neuronal and astrocyte-enriched nuclei populations by FANS. Fresh (unfixed) snap-frozen (i.e., fresh-frozen) postmortem tissue collected from adult cortex is first mechanically and chemically dissociated. After lysis and ultracentrifugation in a sucrose gradient, the cytoplasmic and extracellular components are discarded while the nuclei are retained. Nuclei are then labeled with fluorescently conjugated nuclear antibodies corresponding to the desired target lineages and sorted using FANS. Following this approach, enrichment of astrocytes is demonstrated in the collected PAX6+NeuN− populations, validated both by a targeted qPCR panel as well as by downstream nuclear RNA sequencing.
Protocol
NOTE: The Program for the Protection of Human Subjects at the Icahn School of Medicine at Mount Sinai (ISMMS) and its Institutional Review Board (IRB) assures the ethical conduct of research and compliance with federal, state, and institutional regulations. In this study, all postmortem specimens used were de-identified, obtained under appropriate consent through the biorepository, and were exempt from “human research” designation by ISMMS’s IRB (HS#14–01007).
1. Buffer preparation
NOTE: If performing downstream RNA sequencing, thoroughly treat all workspaces and tools with RNase Decontamination Solution to prevent mRNA degradation.
- Prepare lysis buffer.
- Dissolve the following in distilled H2O up to 50 mL: 5.47 g of sucrose (0.32 M final), 250 μL of 1 M CaCl2 (5 mM final), 150 μL of 1 M Mg(CH3COO)2 (3 mM final), 10 μL of 500 mM ethylenediaminetetraacetic acid (EDTA) (0.1 mM final), 500 μL of 1 M Tris-HCl (pH 8) (10 mM final), 50 μL of Triton X-100 (0.1% final), and 17 μL of 3 M dithiothreitol (DTT, 1 mM final, add fresh) (see the Table of Materials).
- Prepare sucrose buffer
- Dissolve the following in distilled H2O up to 50 mL: 30.78 g of sucrose (1.8 M final), 150 μL of 1 M Mg(CH3COO)2 (3 mM final), 500 μL of 1 M Tris-HCl (pH 8) (10 mM final), 17 μL of 3 M DTT (1 mM final, add fresh).
Materials
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| 10x PBS pH 7.2 | Invitrogen | 70013073 | |
| ANTI-NEUN ANTIBODY CLONE A60 | Millipore | MAB377A5MI | mouse anti-NeuN conjugated to a fluorescent compound AF555 (excitation, 553 nm; emission, 568 nm) |
| ANTI-OLIG2 ANTIBODY CLONE 211 | Millipore | MABN50A4MI | mouse anti-OLIG2 conjugated to a fluorescent compound AF488 (excitation, 499 nm; emission, 520 nm) |
| Bovine Serum Albumin | Fisher | BP9704–100 | |
| Bright-Line Counting Chamber | Hausser Scientific | 3110V | |
| Calcium Chloride Anhydrous | Fisher | C614–3 | |
| Cell Strainers, 40 μM | SP Scienceware | 136800040 | |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Invitrogen | D1306 | |
| DL-Dithiothreitol | Sigma | 43815–1G | |
| DNA Library Kit | Illumina, Nextera | FC-121–1030 | |
| DNAse I | Worthington | LS002139 | |
| Dounce Tissue Grinder | WHEATON | 357542 | |
| FACS Sorter | BD Biosciences | BD FACSAria III | |
| Magnesium Acetate Tetrahydrate | Fisher | M13–500 | |
| PAX6 (PAX6/496) - 100 TESTS | Novus | NBP234705J | |
| RNA Clean & Concentrator | Zymo Research | R1013 | |
| RNaseZap RNase Decontamination Solution | Invitrogen | AM9780 | |
| SMARTer Stranded Total RNA-Seq Kit Pico Input Mammalian | Clontech Laboratories | 635005 | Fragmentation time of 2.5 minutes, as recommended for low RIN RNA values. |
| Sucrose, crystal certified, ACS, 500 mg | Fisher | S5500 | |
| SW 41 Ti Swinging-Bucket Rotor | Beckman Coulter | 331362 | |
| Tris-HCl, 1M Solution, pH 8.0, Molecular Biology Grade, Ultrapure | Thermo Scientific | J22638AE | |
| TritonX-100 | Fisher | BP151–500 | non-ionic surfactant in lysis buffer |
| TRIzol LS Reagent | Invitrogen | 10296028 | |
| TRIzol Reagent | Invitrogen | 15596026 | reagent for isolation of RNA |
| Trypan Blue Solution, 0.4% | Gibco | 15250061 | |
| Ultracentrifuge | Beckman Coulter Optima XE-100 | A94516 | |
| Ultracentrifuge tubes PP 9/16 × 3–1/2 | Beckman Coulter | 331372 | |
| UltraPure Distilled Water (RNAse-, DNAse-free) | Invitrogen | 10977023 | referred to as distilled water |
| Ultrapure EDTA | Life Technologies | 15576–028 |
2. Frozen tissue dissociation into single-nucleus suspension
Add 4 mL of ice-cold lysis buffer to a 7 mL glass tissue douncer (grinder), and keep on ice. Dissect approximately 200–400 mg of fresh-frozen human adult cortex, dounce approximately 50x, and transfer the tissue homogenate to a 12 mL ultracentrifuge polypropylene tube.
-
Using a 5 mL pipette, add 6.5 mL of ice-cold sucrose buffer to the bottom of the ultracentrifuge tube, taking care not to disturb the layer between sucrose and the tissue homogenate.
NOTE: If processing additional samples, carefully balance the tubes by weight by adding additional lysis buffer to the lighter sample. Precise balancing of the ultracentrifuge tubes is essential to prevent damage to the rotor and ensure rotation at the proper speed.
3. Ultracentrifugation
-
Ultracentrifuge the lysate at 24,400 rpm (101,814 × g) for 1 h at 4 °C. Carefully aspirate the supernatant and debris without disturbing the pellet.
NOTE: A pellet may not be visible if starting with less than 200 mg of tissue.
Add 600 μL of phosphate-buffered saline (PBS) (Ca2+, Mg2+ free) to the nuclei pellet and incubate on ice for 10 min before resuspending.
Pipette up and down 50 times to resuspend the nuclei pellet on ice.
Use a hemocytometer to visualize the intact nuclei under a microscope and to ensure the concentration of the resuspension is above 105 nuclei/mL before proceeding to the next step (combine 10 μL of the resuspended nuclei with 10 μL of trypan blue).
4. Antibody incubation
-
To immunotag the sample for FANS, add 500 μL of the resuspended sample pellet, 490 μL of 1x PBS (Ca2+, Mg2+ free), 10 μL of 10% BSA (0.1% final), 1 μL of mouse anti-NeuN antibody conjugated to AF555 (NeuN-AF555, 1:1000 final concentration), and 1 μL of mouse anti-PAX6 antibody conjugated to allophycocyanin (PAX6-APC, 1:1000 final concentration).
NOTE: Alternative antibodies conjugated to other fluorophores may be added (see discussion). Here, mouse anti-OLIG2 conjugated to AF488 (1:1000 concentration) was used with favorable results.
- Perform 4′,6-diamidino-2-phenylindole (DAPI)-only and single-color controls to set up gating parameters, using a small amount of the sample as necessary.
- To perform AF555 single color control, add 20 μL of the resuspended sample pellet, 970 μL of 1x PBS (Ca2+, Mg2+ free), 10 μL of 10% BSA (0.1% final), and 1 μL of NeuN-AF555 antibody (1:1000 concentration).
- To perform APC single color control, add 20 μL of the resuspended sample pellet, 970 μL of 1X PBS (Ca2+, Mg2+ free), 10 μL of 10% BSA (0.1% final), and 1 μL of PAX6-APC antibody (1:1000 concentration).
-
To perform DAPI-only control, add 20 μL of the resuspended sample pellet, 970 μL of 1X PBS (Ca2+, Mg2+ free), and 10 μL of 10% BSA (0.1% final) (DAPI will be added later, see 4.4).NOTE: If more than two antibodies are used, performing fluorescence-minus-one (FMO) control is recommended in addition to the single-color controls.
Incubate the samples and the controls with rotation in the dark for 1 h at 4 °C.
Add DAPI at 1:1000 to all the samples and controls, and proceed with sorting.
5. Fluorescence-activated nuclei sorting (FANS)
NOTE: It is recommended to use an institutional flow cytometry facility with assistance from trained personnel unless already proficient in flow cytometry/sorting techniques.
- Gate as shown below.
-
First, and for each sample, gate by forward scatter (FSC-A) vs. side scatter (SSC-A) to include particles in the appropriate size range for nuclei, thus excluding red blood cells and debris (Figure 1A).NOTE: A very clean nuclei preparation can distinguish between a smaller debris cluster and a nuclei cluster.
- Next, and for each sample, gate by FSC-A vs. FSCW (or FSC-A vs. FSC-H) and SSC-A vs. SSC-W (or SSC-A vs. SSC-H) to include only singlet nuclei (Figure 1B).
- Next, and for each sample, gate by DAPI to include intact nuclei singlets (DAPI-high gated population), excluding debris (DAPI-low, on left of gated population) and doublets (on right of gated population) (Figure 1C).
- Set further gating based on fluorescence controls, determined visually as distinct clusters on FACS plots.
- Run NeuN-AF555-only control to determine the cutoff for NeuN+ staining in the channel for AF555 (Figure 2B).
- Run PAX6-APC-only control to determine the cutoff for PAX6+ staining in the APC channel (Figure 2C).
-
NOTE: If using more than two antibodies, an FMO control is recommended to visualize any shifts in populations.
- Once all controls have been run, draw the gates for NeuN+ and PAX6+ collections above the established thresholds.
-
Gate and collect additional glial populations (such as oligodendrocyte progenitor cells, as shown here by OLIG2+) from the NeuN−PAX6−population (Figure 1F).NOTE: In case determining appropriate gating cutoffs is difficult with the flow cytometry software due to indistinct populations, it may be helpful to modify the number of events being visualized (either increasing or reducing the number of events on the FANS plot).
-
Collect samples appropriately based on the intended downstream analysis.
Figure 1: Validation of neuron- and astrocyte-enriched isolation by FANS.
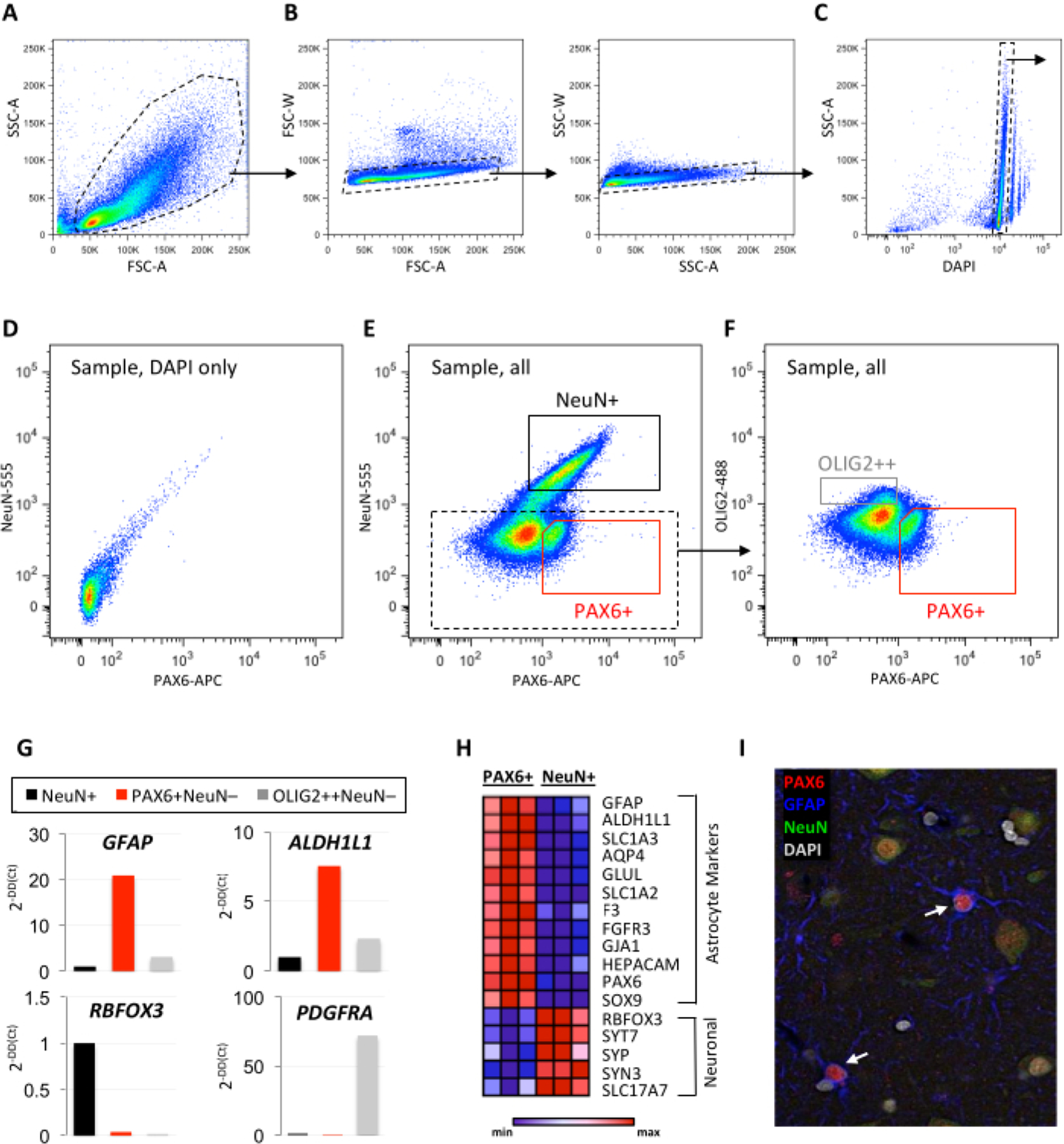
(A–F) Representative examples of sequential FANS gating to exclude debris and doublets (A–C) and define background staining using (D) a DAPI-only control for the collection of (E–F) enriched neuronal (NeuN+), astrocyte (PAX6+NeuN−), and OPC (OLIG2++NeuN−) nuclei populations. (G) Minimal qPCR panel using pan-astrocyte (GFAP, ALDH1L1), neuronal (RBFOX3), and OPC (PDGFRA) markers for quality control of the collected populations (A–G: temporal neocortex autopsy specimen without pathology, 12 h PMI, 100,000 events shown; dotted lines represent positively gated sequential populations; solid lines represent final collection of cell-type enriched populations). (H) Heatmap generated from row-normalized bulk RNA sequencing data showing expression of canonical astrocyte and neuronal markers in PAX6+ and NeuN+ sorted populations (n=3 autopsy cases, temporal neocortex without pathology, PMI 12–21 h). (I) Representative immunofluorescence image of PAX6, GFAP, and NeuN in human neocortex. Arrows indicate astrocytes co-expressing PAX6 and GFAP. Abbreviations: FANS = fluorescence-activate nuclei sorting; DAPI = 4′,6-diamidino-2-phenylindole; NeuN = neuronal nuclei; PAX6 = paired box protein 6; OLIG2 = oligodendrocyte transcription factor 2; OPC = oligodendrocyte progenitor cell; qPCR = quantitative polymerase chain reaction; GFAP = glial fibrillary acidic protein; ALDH1L1 = 10-formyltetrahydrofolate dehydrogenase; RBFOX3 = RNA-binding protein FOX-1 homolog 3; PDGFRA = platelet-derived growth factor alpha; PMI = postmortem interval; SSC-A = side scatter-area; FSC-A = forward scatter area; SSC-W = side scatter width; FSC-W = forward scatter width; NeuN-555 = mouse anti-NeuN conjugated to AF555; PAX6-APC = mouse anti-PAX6 conjugated to allophycocyanin; OLIG2-488 = mouse anti-OLIG2 conjugated to green fluorescent dye for the 488 nm laser line.
Figure 2: Comparative isolation of astrocyte-enriched populations using PAX6 or SOX9 antibodies.
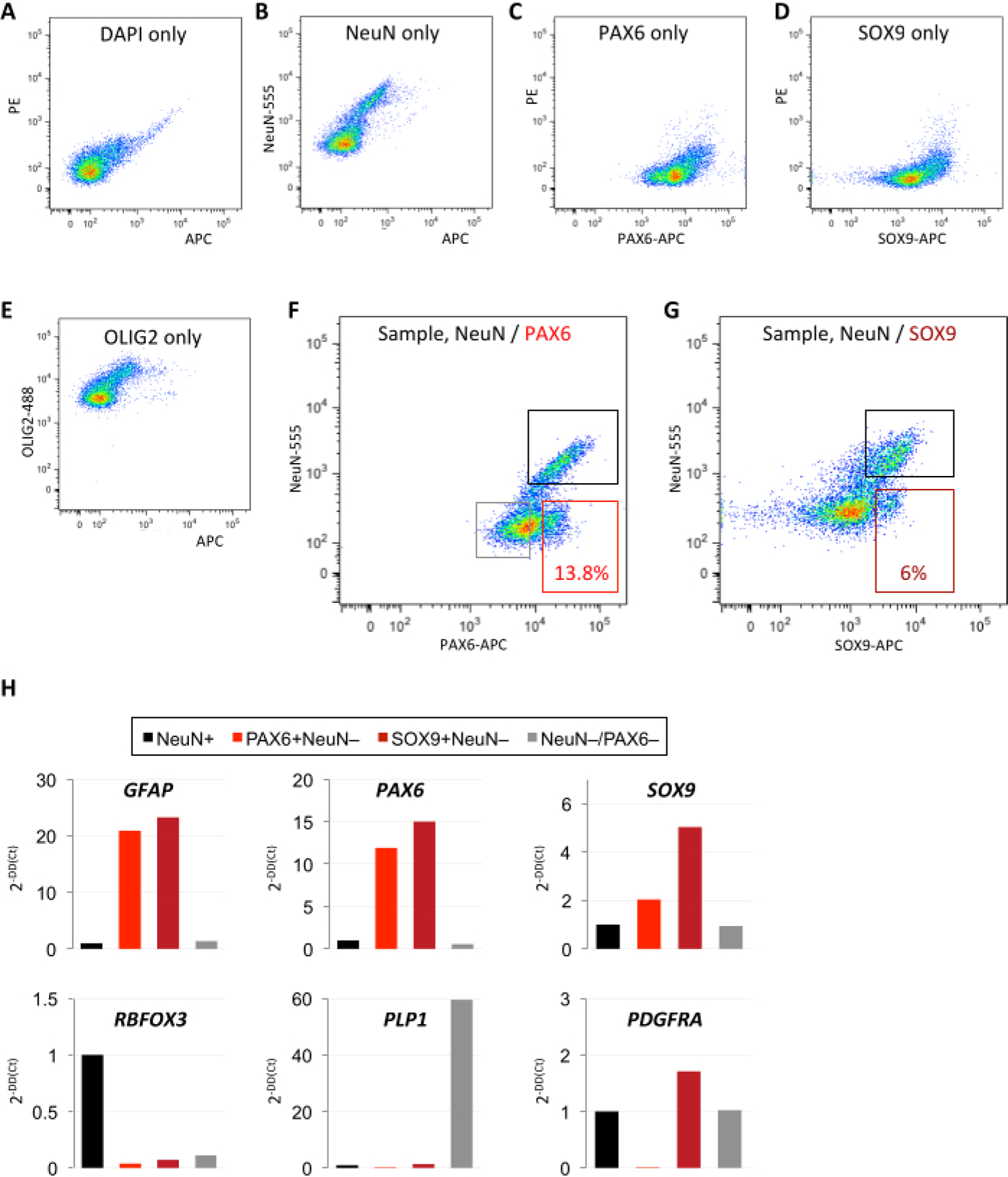
(A-E) Single-color controls for DAPI, NeuN, PAX6, SOX9, and OLIG2 define positive/negative cutoffs in the respective fluorescence channels. (F–G) Comparative FANS method for astrocyte-enriched isolation using (F) NeuN/PAX6 or (G) NeuN/SOX9 antibodies. A greater percentage of events are captured by PAX6 than by SOX9 within the astrocyte-enriched gate. (A–G: identical postmortem non-pathological temporal neocortex specimen used for all experiments; 12 h PMI; 10,000 events shown). (H) Quality control qPCR analysis confirms enrichment of astrocyte markers and depletion of non-astrocyte markers in PAX6+(NeuN−) and SOX9+(NeuN−) sorted populations from F–G (data normalized to ACTB and quantified as fold change of the NeuN+ population). Abbreviations: DAPI = 4′,6-diamidino-2-phenylindole; NeuN = neuronal nuclei; PAX6 = paired box protein 6; OLIG2 = oligodendrocyte transcription factor 2; SOX9 = SRY-box transcription factor 9; qPCR = quantitative polymerase chain reaction; GFAP = glial fibrillary acidic protein; RBFOX3 = RNA-binding protein FOX-1 homolog 3; PDGFRA = platelet-derived growth factor alpha; PMI = postmortem interval; NeuN-555 = mouse anti-NeuN conjugated to AF555; PAX6-APC = mouse anti-PAX6 conjugated to allophycocyanin; OLIG2-488 = mouse anti-OLIG2 conjugated to green fluorescent dye for the 488 nm laser line.
6. Collection of FANS populations for downstream molecular analyses
- For bulk RNA sequencing, collect 50,000–500,000 nuclei in PBS (see also section 7.1).
- Add 2 mL of sucrose solution, 50 μL of 1 M CaCl2, and 30 μL of 1 M Mg(CH3COO)2, and fill with PBS up to 10 mL.
- Invert and incubate on ice for 15 min, then centrifuge at 900 × g for 10–15 min at 4 °C.
- Aspirate the supernatant, resuspend in 1 mL of RNA-extracting reagent, vortex, freeze on dry ice, and store at −80 °C.
- Alternatively, collect the samples directly into 200 μL of the RNA-extracting reagent. Add the RNA-extracting reagent up to 1 mL after sorting, maintaining a 1:1 ratio of the reagent to sorted sample. Vortex, freeze on dry ice, and store at −80 °C.
For bulk assay for transposase-accessible chromatin using sequencing (ATAC-seq), collect 50,000–75,000 nuclei in PBS in a microcentrifuge tube coated with 5% bovine serum albumin (BSA). Freeze nuclei on dry ice/−80 °C or immediately use them for ATAC preparation.
For sn RNA-seq or ATAC-seq, collect nuclei in 0.04% BSA in PBS (see also section 7.2).
7. Pre-library preparation tips
- Bulk nuclear RNA sequencing library preparation
- After collecting in RNA extraction reagent, perform standard phenol/chloroform RNA extraction by adding phenol/chloroform and precipitating RNA from the upper aqueous layer with ethanol, followed by DNase digestion on tube (15 min).
-
Perform RNA cleanup and concentrate in a final volume of 15 μL of water.NOTE: Using this method, representative recovery from 300 mg of adult cortex sample is ~300,000 NeuN+ nuclei (15–20 ng/μL total RNA after cleanup and concentration) and ~250,000 PAX6+ nuclei (10–12 ng/μL total RNA after cleanup and concentration). This representative yield can vary greatly based on sample quality, gating stringency, and RNA recovery.
-
Perform quantitative polymerase chain reaction (qPCR) prior to sequencing to confirm enrichment of astrocytes based on high differential expression of canonical astrocyte markers (GFAP, SOX9), 10-formyltetrahydrofolate dehydrogenase (ALDH1L1)) and depletion of neuronal and other cell lineage markers (Figure 1G).NOTE: Collection of the double-negative population (NeuN−PAX6−) allows for a more accurate, multitiered qPCR quality control analysis for the relative enrichment of astrocytes (Figure 2H).
- Generate RNA-seq libraries using a kit recommended for low RNA integrity number values, as expected from postmortem samples.
- Single-nucleus sequencing library preparation
- Perform sn sequencing through an institutional sequencing core.
- For nanofluidics-based processing and sequencing, use the recommended concentration of 1 × 106 cells/mL in at least 60 μL of 1x PBS + 0.04% BSA for single-nucleus RNA-sequencing (snRNA-seq) and 3 × 106 cells/mL in at least 20 μL of 1x PBS + 0.04% BSA for single-nucleus Assay for Transposase-Accessible Chromatin using sequencing (snATAC-seq).
Representative Results
Nuclei were collected from fresh (unfixed) snap-frozen temporal neocortex tissue with a postmortem collection time of 12 h. After tissue dissociation into nuclei suspension, samples were incubated with antibodies against NeuN, PAX6, and OLIG2, and sorted according to the gating shown in Figure 1 and Figure 2. Nuclei were collected from NeuN+, PAX6+NeuN−, and OLIG2+NeuN− sorted populations (Figure 1E,F and Figure 2F). A targeted qPCR panel revealed enrichment for the pan-astrocyte markers, GFAP and ALDH1L1, in the PAX6+(NeuN−) population (Figure 1G and Figure 2H). Additionally, the OLIG2+ population was enriched for the oligodendrocyte progenitor cell (OPC) marker PDGFRA (Figure 1G). Bulk RNA sequencing of the collected populations showed comparative enrichment of astrocyte markers and depletion of neuronal markers in the PAX6+(NeuN−) population (Figure 1H). Immunofluorescence confirmed the colocalization of PAX6 with GFAP in adult human cortical astrocytes (Figure 1I).
Examining single-color controls reveals distinct positive and negative populations for each marker, which enabled the setting of accurate cutoffs for collection (Figure 2A–G). Furthermore, astrocyte-enriched FANS isolation was compared with two different antibodies against astrocyte nuclear markers, SOX9 and PAX6. A higher percentage of nuclei events was captured by FANS using PAX6 (~13.8%) than by FANS using SOX9 (~6%) within the astrocyte-enriched gate (Figure 2F,G). A targeted qPCR panel revealed enrichment for GFAP, PAX6, and SOX9 in both SOX9+(NeuN−) and PAX6+(NeuN−) populations (Figure 2H). Retroactively comparing the dissociation and staining of several samples with varying postmortem interval (PMI) of tissue collection revealed that a shorter PMI was associated with greater intact nuclei recovery (Figure 3A). Frozen tissue with a PMI of up to 24 h yielded a high rate of intact nuclei, up to 90%; at 30 h PMI, however, very few intact nuclei could be recovered (Figure 3A). Including an antibody-wash step in the standard protocol (which typically omits this step to optimize recovery) did not reveal significant shifts in the separation of distinct NeuN+/− or PAX6+/− populations (Figure 3B).
Figure 3: Dependent and independent metrics for successful FANS experiments.
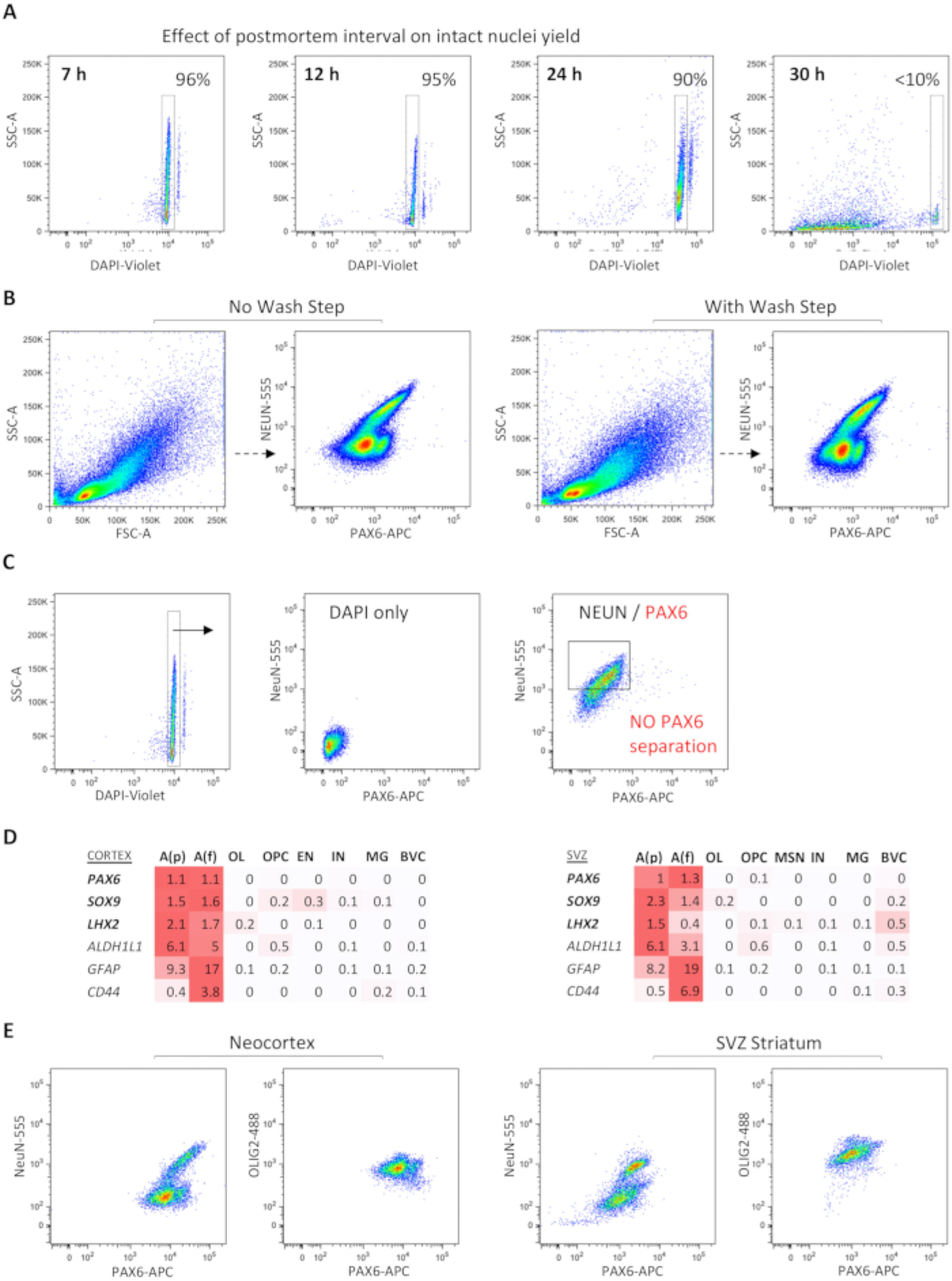
(A) Effect of sample collection PMI on the quality of nuclei: lower PMI enables the recovery of a greater number of intact nuclei with adequate results up to 24 h. (B) The inclusion of a wash step after antibody incubation does not visually appear to shift FANS populations. (C) Example of a failed FANS experiment with lack of distinct PAX6+(NeuN−) population (on right) despite the presence of intact nuclei and low background (on left) (postmortem temporal neocortex, non-pathological, 7 h PMI). (D) Heatmap representation of adult human sn RNA-seq data showing differential expression of PAX6 and two other astrocyte nuclear factors, SOX9 and LHX2, across protoplasmic and fibrous astrocyte subpopulations in both cortex and SVZ, compared to other cell types (red color gradient represents spectrum of log-normalized average gene expression values; n=3 distinct autopsy cases, 43,619 total nuclei. (E) Distinct brain regions (temporal neocortex vs. SVZ and subjacent striatum) show different patterns of NeuN/PAX6/OLIG2 separation. Abbreviations: FANS = fluorescence-activate nuclei sorting; DAPI = 4′,6-diamidino-2-phenylindole; NeuN = neuronal nuclei; PAX6 = paired box protein 6; OLIG2 = oligodendrocyte transcription factor 2; GFAP = glial fibrillary acidic protein; ALDH1L1 = 10-formyltetrahydrofolate dehydrogenase; LXH2 = LIM homeobox 2; PMI = postmortem interval; SSC-A = side scatter-area; FSC-A = forward scatter area; NeuN-555 = mouse anti-NeuN conjugated to AF555; PAX6-APC = mouse anti-PAX6 conjugated to allophycocyanin; OLIG2–488 = mouse anti-OLIG2 conjugated to green fluorescent dye for the 488 nm laser line; SVZ = subventricular zone; A(p) = protoplasmic (gray matter) astrocytes; A(f) = fibrous (white matter) astrocytes; OL = oligodendrocytes; OPC = oligodendrocyte progenitor cells; EN = excitatory neurons; MSN = medium spiny neurons; MG = microglia; BVC = blood vessel cells.
Occasionally, poor separation of PAX6+NeuN− populations could be seen, even in samples with a high percentage of viable nuclei (Figure 3C), necessitating the repetition of the tissue dissociation and the FANS protocol. Single-nucleus RNA-seq studies further prioritized PAX6 as a top differentially expressed nuclear transcription factor across both protoplasmic and fibrous adult astrocyte subpopulations, not only in the neocortex, but also in the subventricular zone (SVZ) and adjacent striatum (Figure 3D). Comparison of NEUN/PAX6/OLIG2 triple FANS between the neocortex and striatum derived from the same brain sample showed region-specific differences in the separation of PAX6+ nuclei (Figure 3E). This protocol is currently validated for the neocortex only.
Discussion
Experimental design following the outlined protocol should be finalized after considering several biological and technical factors. Starting tissue samples are fresh-frozen, without having been fixed, and preferably have a short postmortem collection interval to maximize nuclei recovery. Based on experience, a PMI of up to 24 h allows for adequate nuclei recovery; however, a PMI of 12 h or less is preferable to optimize intact nuclei recovery. Additional factors apart from PMI, including temperature of body storage and pH levels of the tissue, may also affect nuclei yield, but were not accounted for in these studies14. From anecdotal experience, FANS recovery has been found to be comparable with the same tissue being stored up to five years at −80 °C, although controlled experiments were not performed to assess the variability of storage on cell-type specific expression. Washing the sample after antibody incubation is another variable that may need consideration. In general, a wash step decreases the overall yield, but may improve the separation of distinct immunolabeled populations. In these studies, no difference was observed in the separation of NEUN/PAX6 populations when including or excluding a post-antibody wash step, but this step may be beneficial when using other primary and secondary antibodies, especially if they are not preconjugated.
Due to the variability in the sensitivity of the sorting equipment, it is necessary to perform appropriate controls at each sorting event. If excessive extracellular debris is seen in the resuspended sample when visualizing on a hemocytometer, the sample can be passed through a 40 μm filter to retain only the nuclei. If sample is limited, beads tagged with the appropriate fluorophore may also be used for single-color and FMO controls. If the samples appear to not be enriched for the desired astrocyte populations, as determined either by qPCR or sequencing results, it may be helpful to gate all NeuN− as well as PAX6+(NeuN−) populations more strictly, leaving more room between the gate cutoffs for positive and negative populations.
Most of the validation studies described here were performed using OLIG2 in addition to NeuN and PAX6, to simultaneously enrich for neuronal, astrocyte, and oligodendrocyte progenitor (OPCs) nuclei populations. It is recommended to include OLIG2 as a third lineage marker to enrich for cortical astrocytes more effectively within the NeuN− fraction. The presence or absence of FITC-conjugated OLIG2 does not appear to shift the PAX6+ population; therefore, the assumption is that experiments excluding OLIG2 will result in similar astrocyte enrichment. Of note, it is essential that the PAX6+ population be gated from the NeuN− population, as some neuronal populations express both NeuN and PAX615. Astrocytes can also be sorted according to SOX9 expression; however, staining and sorting with SOX9 leads to less robust enrichment of astrocytes compared to PAX6. Although clear separation was observed between neuronal, astrocytic, and OPC lineages in the adult cortex, a similar separation was not observed between PAX6+ and OLIG2+ populations in the adult SVZ and adjacent striatum.
While it is possible that some astrocytes from the SVZ express low levels of OLIG2, the collected populations were only validated by qPCR (data not shown); hence, downstream RNA sequencing may be helpful in further defining the purity of these populations. Additionally, this protocol can be adapted to isolate nuclei from frozen fetal brain tissue and brain tumor tissue using population-specific nuclear markers. Because these tissues are considerably more cellular, it may be helpful to start with less tissue to minimize the aggregation of nuclei. When sorting aneuploid nuclei from neoplastic tissue, DAPI gating is adjusted to account for potentially higher fluorescence. It is strongly recommended that users perform a careful validation prior to performing FANS on neoplastic tissue, as the current protocol is validated for non-neoplastic samples only. Overall, the above-described protocol puts forward a validated strategy for isolating enriched astrocyte populations, adapted from previously established protocols to enrich for neurons. This methodology can be used to effectively isolate astrocyte nuclei populations from both normal and diseased cortical brain tissues for use in further molecular analyses.
Acknowledgments
We like to thank members in Pathology and Neurosurgery at the Icahn School of Medicine at Mount Sinai for help with the procurement of de-identified brain tissue and ISMMS’s Flow Cytometry CORE for expert advice. The study was partially funded by NIH RF1DA048810, R01NS106229, R03NS101581 (to N.M.T.), and R61DA048207 (to S.A.).
Footnotes
Disclosures
The authors have nothing to disclose.
References
- 1.Mitchell A, Roussos P, Peter C, Tsankova N, Akbarian S The future of neuroepigenetics in the human brain. Progress in Molecular Biology and Translational Science. 128, 199–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang Y, Matevossian A, Huang HS, Straubhaar J, Akbarian S Isolation of neuronal chromatin from brain tissue. BMC Neuroscience. 9, 42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akbarian S et al. The PsychENCODE project. Nature Neuroscience. 18 (12), 1707–1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matevossian A, Akbarian S Neuronal nuclei isolation from human postmortem brain tissue. Journal of Visualized Experiments: JoVE. (20), 914 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tome-Garcia J et al. Cell type-specific isolation and transcriptomic profiling informs glial pathology in human temporal lobe epilepsy. BioRxiv. doi: 10.1101/2020.12.11.421370 (2020). [DOI] [Google Scholar]
- 6.Zhang Y et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 89 (1), 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W et al. SOX9 Is an astrocyte-specific nuclear marker in the adult brain outside the neurogenic regions. Journal of Neuroscience. 37 (17), 4493–4507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manuel MN, Mi D, Mason JO, Price DJ Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Frontiers in Cellular Neuroscience. 9, 70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakurai K, Osumi N The neurogenesis-controlling factor, Pax6, inhibits proliferation and promotes maturation in murine astrocytes. Journal of Neuroscience. 28 (18), 4604–4612 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong S, et al. Decoding the development of the human hippocampus. Nature. 577 (7791), 531–536 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Pollen A et al. Molecular identity of human outer radial glia during cortical development. Cell. 163 (1), 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cvekl A, Callaerts P PAX6: 25th anniversary and more to learn. Experimental Eye Research. 156, 10–21 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Goc J, Liu JY, Sisodiya SM, Thom M A spatiotemporal study of gliosis in relation to depth electrode tracks in drug-resistant epilepsy. European Journal of Neuroscience. 39 (12), 2151–2162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monoranu CM et al. pH measurement as quality control on human post mortem brain tissue: a study of the BrainNet Europe consortium. Neuropathology and Applied Neurobiology. 35 (3), 329–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ninkovic J et al. The transcription factor Pax6 regulates survival of dopaminergic olfactory bulb neurons via crystallin αA. Neuron. 68 (4), 682–694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]


