Abstract
The Coronavirus (COVID-19) pandemic impelled several research efforts, from collecting COVID-19 patients’ data to screening them for virus detection. Some COVID-19 symptoms are related to the functioning of the respiratory system that influences speech production; this suggests research on identifying markers of COVID-19 in speech and other human generated audio signals. In this article, we give an overview of research on human audio signals using ‘Artificial Intelligence’ techniques to screen, diagnose, monitor, and spread the awareness about COVID-19. This overview will be useful for developing automated systems that can help in the context of COVID-19, using non-obtrusive and easy to use bio-signals conveyed in human non-speech and speech audio productions.
Keywords: COVID-19, Digital health, Audio processing, Computational paralinguistics
1. Introduction
More than 212 million confirmed cases of coronavirus-induced COVID-19 (C19) infected individuals have been detected in more than 200 countries1 across the world at the time of writing this overview. This pandemic had a wide spectrum of effects on the population, ranging from no symptoms to life-threatening medical conditions and more than four million deaths. The world health organisation (WHO)2 reports as most common symptoms of C19 fever, dry cough, loss of taste and smell, and fatigue; the symptoms of a severe C19 condition are mainly shortness of breath, loss of appetite, confusion, persistent pain or pressure in the chest, and temperature above 38 degrees Celsius.
Monitoring the development of the pandemic and screening the population for symptoms is mandatory. Arguably the procedures mostly used are temperature measurement – e.g., before boarding a plane – and diverse corona rapid tests – e.g., before being allowed to visit a care home. In the clinical test for diagnosing C19 infection, the anterior nasal swabs sample is collected as suggested by Hanson et al. [1]. Amongst alternatives, assessing human audio signals has some advantages: It is non-intrusive, easy to obtain, and both recording and assessment can be done almost instantaneously. It is an open research question whether the human audio signal provides enough ‘markers’ for C19, resulting in good enough performance of classification such that C19 can be told apart from other respiratory diseases and from typical subjects displaying idiosyncrasies in speech production. Note that performance need not necessarily be ‘perfect’: The same way as elevated body temperature can be caused not by C19, it might do to find, out of a larger sample, those persons that have to undergo more detailed medical examination. With other words, taking into account a fair number of false positives might do, given that we obtain a very high number of true positives.
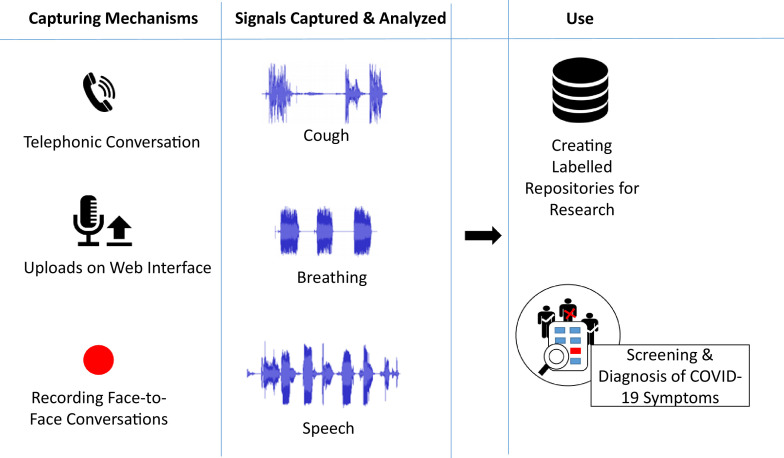
An automated approach to detect and monitor the presence of C19 or its symptoms could be developed using Pattern Recognition – in more general terms, Artificial Intelligence (AI) – based techniques. Although AI techniques are still in the process of reaching a matured stage, they can be used for early detection of the symptoms, especially in the form of a self-care tool in reducing the spread, taking early care, and hence avoiding propagation of the disease; see for overviews [2], [3]. As depicted in Fig. 1 , in this article we are discussing capturing and processing speech and other human audio data for screening and diagnosis of C19. The references included in this overview are searched on google scholar with the keywords ‘COVID’ or ‘Corona virus’ with ‘speech’, ‘audio’, ‘cough’ or ‘breathing’, for the period from January 2020 till 23 March 2021.
Fig. 1.
Capturing and processing audio signals including speech for COVID-19 applications.
The paper is organised as follows. The previous work done for the detection of cough and breath sounds is discussed in the beginning. In the main Section 2, the papers using the detected cough sounds, speech, and breathing signals for the screening and diagnosis of C19 are mentioned. In Section 3, the limitations in identifying C19 status from cough, speech, and breath signals are discussed. Section 4 mentions the challenges and possibilities for future work in the context of using human sounds for identifying C19. Finally, Section 5 concludes the findings and observations from multiple studies attempting to detect C19.
1.1. Previous work in detecting cough & breath
Cough is one of the prominent symptoms of C19; it is thus of interest to know the techniques used in detecting human cough and discriminating it from other similar sounds such as laughter and speech. The motivation of using a microphone-captured signals for the detection of cough events comes from the study by Drugman et al. [4]. They have compared the performance exhibited by several sensors including ECG, thermistor, chest belt, accelerometer, contact, and audio microphones to detect cough events from a database of 32 healthy individuals producing the sounds voluntarily in a confined room. They observed that microphones performed the best in telling apart cough events from other sounds such as speech, laughter, forced expiration, and throat clearing with a sensitivity and specificity, both of 94.5%.
The following studies detailed the audio features used in detecting cough sounds: A study with 38 patients having pertussis cough, croup, bronchiolitis, and asthma is presented by Pramono et al. [5] in which the data are collected from public domain websites such as YouTube and whoopingcough. The cough detection algorithm separates these cough sounds from the non-cough sounds such as speech, laughter, sneeze, throat clearing, wheezing sound, whooping sound, machine noises, and other types of background noise. The non-cough sounds together constitute 1000 separate sound events. The authors report a sensitivity of 90.31%, a specificity of 98.14%, and an F1-score of 88.70%, using logistic regression with three features: (1) the ratio of the median of B-HF to a maximum of B-01, (2) the minimum to a maximum of B-01 contents, and (3) median of lower quantile to a maximum of B-01 contents, where B-01 is between the fundamental frequency (F0) and next harmonic (F1) and B-HF is between 2.5 to 3 kHz. Miranda et al. [6] have shown that Mel Filter Banks (MFBs) performed better than Mel Frequency Cepstral Coefficients (MFCCs) in telling apart cough from other sounds such as speech, sneeze, throat clearing, and other home sounds such as door slams, collisions between objects, toilet flushing, and running engines, on the Google audio set extracts from 1.8 million YouTube videos and the Freesound audio database. The authors report an absolute improvement of 7% in the area under the receiver operating characteristic curve (AUC) by using MFBs over MFCCs.
San et al. [7] conducted in-clinic and outside clinic research to collect speech from individuals with pulmonary disorders and detected pulmonary conditions using two algorithms, one for predicting the obstructive pulmonary disorder and a second one to detect the ratio of a person’s vital capacity to expire in the first second of forced expiration to the full forced vital capacity (FEV1/FVC). The authors conducted this study with 131 participants, in a non-clinical setting, and with 70 participants in a clinical setup. The seven most relevant features identified by the authors are frequency of pause while speaking, shimmer, absolute jitter, relative jitter, maximum of Fast-Fourier Transform (FFT) of inspiratory sound in frequencies from 7.8 kHz to 8.5 kHz, mean of phonation period to inspiratory period ratio, and average phonation time. The authors report a classification accuracy of 0.75% with a RandomForest classifier for the prediction of pulmonary disorders and a mean absolute error of 9.8% for the FEV1/FVC ratio prediction task using an eight dense layered neural network. Yadav et al. [8] used the INTERSPEECH 2013 Computational Paralinguistics Challenge (ComParE) baseline acoustic features [9] for the classification of 47 asthmatic and 48 healthy individuals with a classification accuracy of 75.4% using voiced speech sounds. The authors compared the performance exhibited by these features with that of only MFCCs, and report an absolute improvement of 18.28% over the accuracy given by only MFCCs.
Shortness of breath is also one of the symptoms of the virus for which smartphone apps can be designed to capture breathing patterns by recording the speech signal. The breathing patterns captured using smartphone microphones are analysed by Azam et al. [10] using wavelet de-noising and Empirical Mode Decomposition for data pre-processing, to detect asthmatic inspiratory cycles. Multiple studies have tried to correlate speech signals with breathing patterns, e.g., Routray et al. [11] using cepstrogram and Nallanthighal et al. [12] using spectrograms. In the Breathing Sub-challenge of Interspeech 2020 ComParE [13], Schuller et al. presented as a baseline a Pearson’s correlation of r = 0.51 on the development, and r = 0.73 on the test data set to correlate speech signals with the breathing signals. They used a piezoelectric respiratory belt for capturing breathing patterns as a reference; more details are given by MacIntyre et al. [14]. In another effort of correlating speech signals with breathing signals, an ensemble system with fusion at both feature and decision level of two approaches is presented by Markitantov et al. [15]. One of the two approaches is a 1D-CNN based end-to-end model having two LSTM layers stacked above it. The other approach uses a pre-trained 2D-CNN ResNet18 with two Gated Recurrent Unit (GRU) layers stacked above it. They combine several deep learning procedures in early/late fusion, reporting r = 0.76 between the speech signal and corresponding breathing values of the test set. Further, Mendonça et al. [16] modified the end-to-end baseline architecture by replacing the LSTMs with Bi-LSTM. They also augmented the challenge data set, with the same data set being modified to emulate Voice over Internet (VoIP) conditions. With the above modifications, they achieved r = 0.728 on the test data set. To explore attention mechanisms, MacIntyre et al. [14] used an end-to-end approach along with a Convolutional RNN (CRNN) for two prediction tasks: the breathing signals captured using a respiratory belt, and the inhalation events. They report a maximum of r = 0.731 in predicting the breathing pattern from the speech signal and a macro averaged F1 value of 75.47% in predicting the inhalation events. The attention step is found to improve the metrics by 0.003 r-value absolute, from r = 0.728 to r = 0.731, and 0.726% F1 value absolute, from 74.743 to 75.469 for the two tasks, respectively. All the three studies mentioned above [14], [15], [16] worked with the data set provided in the Breathing Sub-challenge of Interspeech 2020 ComParE [13].
Outside of the challenge, Nallanthighal et al. [17] attempted to correlate high-quality speech signals captured using an Earthworks microphone M23 at 48 kHz with the breathing signal captured using two NeXus respiratory inductance plethysmography belts over the ribcage and abdomen to measure the changes in the cross-sectional area of the ribcage and abdomen at a sample rate of 2 kHz. They collected data from 40 healthy subjects by making them read a phonetically balanced text (exact text not mentioned in the paper) to train a deep learning model. Using the plethysmography belts, normal quiet breathing is collected for the reference breathing rate. They also asked the participants to produce sustained vowels to estimate their lung capacity. The authors achieved a correlation of 0.42 with the actual breathing signal, a breathing error rate of 5.6%, and a recall of 0.88 for breath event detection.
2. Screening and diagnosing for COVID-19
In this section, we report different algorithms/applications using audio processing developed for the screening and diagnosis of C19. All the efforts are categorised as ‘non-clinical’ and ‘clinical’ as per the clinical validation of the collected and analysed data performed by the authors using gold standard methods such as Reverse Transcription-Polymerase Chain Reaction (RT-PCR) or any similar test.
2.1. Non-clinical analysis
The studies presented in this section have used the data collected from crowd-sourcing platforms. The participants have voluntarily participated by uploading their data along with required metadata including C19 status; the C19 status has not been clinically validated.
2.1.1. Non-clinical cough analysis
Cough detection is about identifying cough sounds and differentiating them from other similar sounds such as speech and laughter; the next step will be identifying C19 specific cough sounds. It requires cough and speech samples from C19 and non-C19 subjects to develop an AI model that can differentiate between them on its own. Fig. 2 shows the number of healthy and C19 positive subjects or data points (items) collected by all the groups having data from more than 100 subjects.
Fig. 2.
Groups (given on the x-axis) that collected and analysed cough, speech, and breathing data as indicated. Although some groups collected all three types of data, they have reported their results based on the analysis of only one of them. The y-axis indicates the frequencies of the healthy and C19 subjects present in the data set. Coughvid, VoiceMed and Spira have reported number of data points; we report here number of subjects. The data sets from Cambridge, Coswara, and Coughvid are publicly available. C & B: Cough & Breath; C, S & B: Cough, Speech & Breath. On the x-axis, reference to bibliography is given in square brackets & number without bracket refers to footnote.
Cambridge University3 provided a web-based platform and an android application to upload three coughs, five breaths, and three speech samples of reading a short sentence, and to report C19 symptoms & status. As explained by Brown et al. [18], the crowdsourced data collected come from more than 10 different countries and comprises samples from 6 613 subjects with 235 C19 positive subjects. Note that in the work presented in Brown et al. [18], only cough and breathing sounds are considered. With a manual examination of each sample, 141 cough and breathing items of 62 as C19 positive tested users and 298 items from 220 non-C19 users are used for building a binary classification model to distinguish between C19 and non-C19 users (Task-1). Similarly, 54 “C19 with cough” samples are distinguished from 32 “non-C19 cough” samples (Task-2), and from 20 non-C19 asthmatic cough samples (Task-3). Further, hand-crafted features, amongst them duration, pitch onset, tempo, and MFCCs, are extracted. Along with the handcrafted features, Brown et al. [18] have used another approach, in which transfer learning using the VGGish model is developed using videos from YouTube. The authors achieved an AUC of 0.8 for distinguishing C19 subjects from non-C19 subjects (Task-1) using logistic regression on VGGish-based feature with a sub-set of the handcrafted features, and again an AUC of 0.8 for distinguishing C19 cough from non-C19 and asthmatic cough (Task-3) using a Support Vector Machine (SVM) on VGGish-based features and all handcrafted features except MFCC and its derivatives. The authors found handcrafted features along with VGGish based features to give the best performance. Together, cough and breathing signals perform best in Task-1. Yet, breathing signals alone are better suited for Task-2. With training data augmentation methods such as amplification, adding white noise, and changing pitch and speed, the authors could improve the classification performance of Task-2 from 0.82 to 0.87 AUC and of Task-3 from 0.80 to 0.88 AUC. Thus, the collection of breathing sounds seems to give more accurate results in classifying individuals having C19 infection. Although it is reported that manual evaluation of the samples has been done to verify the C19 status, how this had been done is not explained in detail.
As described by Sharma et al. [19], another corpus called “Coswara” with 941 subjects and nine different sounds has been created using a web interface developed by IISC Bangalore India. The nine sounds include (1) shallow and (2) deep cough, (3) shallow and (4) deep breathing, the sustained vowels (5) [ey], (6) [i:], and (7) [u:], and one to twenty digits counting in (8) normal and (9) fast speaking rate. The metadata collected from the participants include age, gender, location, current health status, (healthy / exposed / cured / infected) and the presence of co-morbidity. Here, along with the subjects labeling their data, the items were manually assigned to one of the nine categories. The data set comprises audio samples from 104 unhealthy users. After curating, the data set is publicly available at Github.4
Dash et al. [20] generated a new bio-inspired cepstral feature set termed COVID-19 Coefficient (C-19CC) to detect the C19 status. The two datasets Coswara [19] and Cambridge [18] are used for evaluating this new feature set. C-19CC with SVM performs best in detecting C19 from shallow and heavy cough in the Coswara dataset, with an accuracy of 74.1% and 72.3%, respectively. It remains the best performer in identifying C19 from the cough of the Cambridge dataset with an accuracy of 85.7% using SVM. However, MFCCs and their variants have performed better than C-19CC for the speech and breathing samples of both datasets.
Coughvid5 is another app from EPFL (Ecole Polytechnique Fédérale de Lausanne) to tell apart C19 cough from other cough categories such as normal cold and seasonal allergies. Till date, this data set by Orlandic et al. [21] has more than 20 000 cough samples; all the samples are passed through an open-source cough detection machine learning model to identify the cough segments. More than 2 000 samples are labeled by 3 expert pulmonologists for the respiratory conditions along with the C19 status. Out of the 2 000 labels given by expert 1, 632 C19 positive labels are given; however, there exists no agreement between the three pulmonologists on the C19 diagnosis (Fleiss’ Kappa score 0.00). The data set is publicly available, along with a machine learning model for identifying cough from other sounds. This data set has the samples validated by three pulmonologists, however, they do not agree at all on their evaluation; this shows that just by listening to the samples it is difficult to agree on the C19 status, even for the experts. Both efforts ‘Coughvid’ and ‘Coswara’ are focused on building a data set only and have not been used for further analysis.
The C19 cough data collection at Massachusetts Institute of Technology (MIT) is done using a web app,6 in which each subject gives three prompted cough recordings, diagnosis details, and other demographic metadata. The total data come from 5 320 subjects (2 660 C19 positives). Laguarta et al. [22] used MFCCs with a CNN architecture, and residual-based neural network architecture (ResNets) to build a baseline model using the collected data set, to understand the impact on C19 diagnosis by employing four ‘bio-markers’ (muscle degradation, vocal cords, sentiments, and lung & respiratory tract). The authors used these bio-markers in a pre-training for the baseline model. This baseline model’s performance is then compared with the four variants; it is found that the lung and respiratory tract bio-markers have the most, and the sentiment marker has the least effect on improving the baseline performance of detecting C19 cough samples. The authors report an accuracy of 98.5% in detecting C19 cough; however, the data used for building the models are not clinically validated, hence they intend to work with clinically validated data next. The performance reported is strikingly high and has, therefore, to be scrutinised thoroughly.
VoiceMed7 is another android and web application that captures crowd-sourced speech and cough sounds and returns the C19 infection status on the fly. The different stages in this cloud-based pre-trained CNN based system comprise pre-processing the collected signal, using a cough detector to identify if it is a cough signal, and then a C19 cough detector to further detect if the audio signal is a C19 cough. As explained in a video,8 the authors used 900 coughs and 2 000 non-cough audio samples for building the cough detector. Similarly, the authors employed 165 C19 and 613 non-C19 samples for building the C19 cough detector. The accuracy of the cough classifier is reported to be 83.7% and the accuracy of the C19 classifier is reported to be 89.69% using deep spectrograms. The major challenges mentioned are that, within the entire group of C19 patients, the identification of C19 cough and their separation from the non-C19 cough in elderly individuals and in individuals with respiratory disorders is a further complex problem.
2.1.2. Non-clinical speech analysis
Considering the risky effects of coughing on spreading the infection in the absence of any preventive measures, capturing and analysing speech signals is a dependable alternative. A web interface to capture speech along with cough of C19 patients is developed by Voca.9 The data collected were analysed by Dubnov [23]; they comprise 30 positively diagnosed and 1 811 healthy participants’ speech and cough samples. The candidates are asked to produce [a:], [e:], [o:], counting from 1 to 20, the alphabet from a–z, and to read a segment from a story. When classifying into C19 positive individuals and healthy ones, the author obtained a maximum of 70% accuracy using MFCC features and a CNN architecture. This study uses a rather small portion of C19 diagnosed subjects – only 30. The author has also not described any techniques used for balancing the data set.
2.1.3. Non-clinical breath analysis
As described in Sections 2.1.1 and 2.1.2, multiple attempts are made to analyse respiration along with cough and speech signals. Especially Brown et al. [18] reported that breathing signals are better suited for distinguishing C19 positive users from C19 negative users having asthma and cough. Similarly, Schuller et al. [24] also found breathing signals performing better than coughs in classifying C19 subjects vs. healthy subjects. Employing an ensemble of CNNs for audio and spectrograms, they report an Unweighted Average Recall (UAR, sometimes called ‘macro average’, i.e., per cent mean of the values in the diagonal of the confusion matrix) of 76.1% using breathing sound and 73.7% for coughing sound from the data set collected by Cambridge University [18]. Note that this study uses a sub-set of the Cambridge data. Another analysis using a subset of the data collected by Cambridge University [18] is presented by Coppock et al. [25]. The data comprise of coughing and breathing audio recordings from 62 COVID-19 positives and 293 healthy participants. The authors applied an end-to-end deep network on the joint representation of coughing and breathing audio signals and report an AUC of 0.846.
2.2. Clinical data analysis
The studies presented in this section have used the data collected either from clinical setup or have verified the collected data using clinical tests.
2.2.1. Clinical cough analysis
Another web interface ‘CoughAgainstCovid’10 for collecting C19 cough samples is an initiative by the Wadhwani AI group in collaboration with the Stanford University.11 As described in Bagad et al. [26], the authors collected prompted cough sounds produced by 3 621 individuals using a smartphone microphone in the setups established at testing facilities and isolation wards across India. This data set contains data from 2 001 C19 positive subjects; these subjects’ RT-PCR tests are also employed for confirmation. The authors have also developed a CNN based model for telling apart C19 cough from non-C19 cough sound. Using the features RMSE, tempo, and MFCCs, they obtain a specificity of 31% with a sensitivity of 90%. These efforts from ‘CoughAgainstCovid’ have used clinically validated data which should be more reliable; however, the data are not publicly available.
In the AI4COVID project, the C19 subjects’ validation is done by studying the pathomorphological changes caused by C19 in the respiratory system from their X-rays and Computer Tomography (CT) scans. A cloud-based smartphone app for detecting C19 cough is described by Imran et al. [27]. As a first step, the authors used a CNN based cough detector, which discriminates cough sounds from over 50 environmental sounds. The authors built this detector using the ESC-50 data set [28]. In the next stage, to diagnose a C19 cough, they collected 70 C19, 96 bronchitis, 130 pertussis, and 247 normal cough samples (total 353 non-C19 samples) to train their C19 cough detector model. Using MFCCs for feature representation and t-distributed stochastic neighbour embedding for dimensionality reduction, they trained three models: (1) a deep transfer learning-based multi-class classifier, using a CNN; (2) a classical machine learning-based multi-class classifier, using an SVM; and (3) a deep transfer learning based binary classifier, again using a CNN. These three models reside in the AI4COVID engine, where a decision is made for C19 positive or negative if the output of all the three models’ outputs is the same; else, it declares the test to be inconclusive. With this, the authors report an accuracy of more than 95% in discriminating cough sounds from non-cough sounds. The three engine-based models yield an accuracy of 92.64%, 88%, and 92.85%, respectively, for detecting a C19 cough sound. The overall performance indicates that the app can detect C19 infected individuals with a probability of 77.3%. As seen in Fig. 2, compared to other real-time C19 identifiers such as ‘CoughAgainstCovid’ (2 001 C19 positives), AI4Covid has a much smaller data set comprising of data from 70 C19 positive individuals.
Pal and Sankarasubbu [29] discuss the interpretability of their framework of C19 diagnosis using embeddings for the cough features and symptoms’ metadata. In this study, cough, breathing, and speech with counting from 1 to 10 is collected from 150 subjects; 100 subjects were C19 positive, and 50 were tested negatively during their RT-PCR test. Apart from this, the authors also collected data for bronchitis and asthma cough from online and offline sources. They report an improvement of 5–6% in accuracy, F1-score, recall, specificity, and precision when using both the symptoms’ metadata and cough features for the classification tasks with a 3-layered dense network giving an accuracy of around 96%.
2.2.2. Clinical speech analysis
In the work from Carnegie Mellon University (CMU), features from models of voice production are explored to understand C19 symptoms in speech signals. The data were collected under clinical supervision, while collaborating with a hospital (Merlin Inc., a private firm in Chile), from 512 subjects. Deshmukh et al. [30] used data from only 19 of these 512 subjects, comprising 9 C19 positives and 10 healthy subjects. The method employed is described in Zhao and Singh [31] and is based on the ADLES (Adjoint Least-Squares) algorithm that extracts the features representing the oscillatory nature of the vocal fold for the vowel [a:]. The voice production model is called the “asymmetric body-cover” model that estimates parameters such as glottal pressure, mass, spring, and damping from the left and right vocal folds’ motion speed and acceleration. The authors analysed the differential dynamics of the glottal flow waveform (GFW) during voice production with the recorded speech, as it is too difficult to analyse the GFW of C19 patients. They hypothesise that a greater similarity between the two signals indicates normal voice and a larger difference would mean the presence of anomalies. A CNN based 2-step attention model is used to detect these anomalies from the sustained vowels [a:], [i:], and [u:]. The residual and the phase difference between the two GFWs are reported as the most promising features yielding the best AUC of 0.9 on the sustained vowels [u:] and [i:]. For a larger study, the following details were mentioned in the video published by CMU in a workshop12 ; note that only partial information can be found in the paper. The data were collected from a total of 530 subjects, all of them clinically tested for C19, comprising 299 positively and 231 negatively tested subjects. Each subject provided six recordings: alphabets (no mention of how many and which alphabets), counting 1–20, sustained vowels, and coughs. With these data, classification was done for different train and test partitions; it turned out that with the change in the data partition, the 5-fold cross-validation and AUC values for test data changes. It is mentioned that the results with sustained vowels are better than those obtained with the cough signals, with the vowel [a:] and alphabets giving the best performance, varying with the change in train-test data partition for AUC from 0.73 to 0.95.
A web-based interface for detecting C19 symptoms from the voice is the “Spira Project”.13 This interface asks the participants to record three phrases. Voice samples from C19 patients in the hospital’s COVID wards were collected, with the help of doctors using smartphone microphones. The authors also collected ward noise profiles for performing a noise-robust analysis. In describing their initial results using MFCCs and a CNN architecture, they reported14 an accuracy of 91% in detecting C19 symptoms related to respiratory disorders using 600 samples collected from C19 patients and 6000 control samples.
2.3. Scant data analysis
The studies presented in this section have worked with very small clinical/non-clinical data sets. Some of the studies have not revealed the exact count of the samples they have worked with.
2.3.1. Scant cough data analysis
The following studies have used cough data sets with samples from less than 100 subjects for training a model.
A smartphone-based C19 cough identifier is developed by Pahar et al. [32] using the Coswara data set and another smaller data set collected in South Africa, comprising of clinically validated 8 C19 positive and 13 C19 negative subjects. The authors compared the performance obtained by logistic regression, SVM, multilayer perceptrons, CNN, long-short term memory (LSTM), and Resnet-50. It is found that Resnet-50 performed best in classifying into C19 positive and C19 negative coughs, with an AUC of 0.98, while an LSTM classifier performed best in classifying into C19 positive and C19 negative coughs, with an AUC of 0.94. Yet, such studies with less than 30 subjects can only be seen as indicators; results may vary when analysing more data from the same subjects or data from more subjects. Dunne et al. [33] built a classifier that uses 14 C19 training samples from the Coswara data set and from the Stanford University led Virufy mobile app.15 The authors report an accuracy of 97.5% in classifying the validation set comprising of 38 non-C19 and only 2 C19 instances; note that no independent test set was employed. This is an extremely small data set to draw conclusions for generalising onto real-life settings.
Some efforts towards only data collection comprise ‘Breath for Science’16 : a team of scientists from NYU developed a web-based portal to register the participants where they can enter similar details along with a phone number. On pressing a ‘call me’ button, the participants receive a callback where they have to cough three times after the beep. At the moment, this service is available only for US citizens. The organisers have not published any details about the amount of data collected.
2.3.2. Scant speech data analysis
Following are speech-based efforts with data sets of less than 100 subjects. Some studies have not revealed the exact number of samples that they have worked with.
The Afeka college of engineering developed a mobile application for remote pre-diagnostic assessment of C19 symptoms from the voice and speech signals captured from 29 infected and 59 healthy individuals. All the subjects provided speech, breathing, and cough sounds, and all of them underwent swab tests. The data comprise 70 speakers and 235 items in the training, and 18 speakers and 57 items in the test set. The study presented in a workshop17 focuses on the analysis of the phones [a:] and [z:], cough, and counting from 50 to 80, collected over a period of 14 days. The same study on the cellular call recordings of 88 subjects, with 29 positive and 59 negative C19 clinically tested individuals, is published by Pinkas et al. [34]. The distribution of positive and negative subjects in train and test is not mentioned; yet, the authors point out that they balanced the training data set for C19 status, age, and gender of the subjects. They compared the performance of three deep learning components: an attention-based transformer, a GRU-based expert classifier with aggressive regularisation, and ensemble stacking. [z:] turned out to be a better indicator of laryngeal pathology than [a:]. Among the deep learning techniques, transformer-based experiments gave better F1 scores. They achieved a precision of 0.79 and a recall of 0.78 on the test set.
Speech recordings of TV interviews available on YouTube were collected and analysed by Ritwik et al. [35] for classifying C19 patients vs. healthy individuals. The data set is publicly available18 and comprises 19 speakers with 10 of them tested as C19 positive. The data collected are manually segmented, after which MFB features are calculated for the speech segments. Using the ASpIRE chain model, which is a time-delayed neural network trained on the Fisher English data set described by Ko et al. in Ko et al. [36], Ritwik et al. [35] extracted the posterior probability of phonemes for each frame. When concatenated, this gives a feature vector for each speech utterance. Using an SVM classifier, the authors report an accuracy of 88.6% and an F1-score of 92.7% in classifying C19 patients vs. healthy speakers.
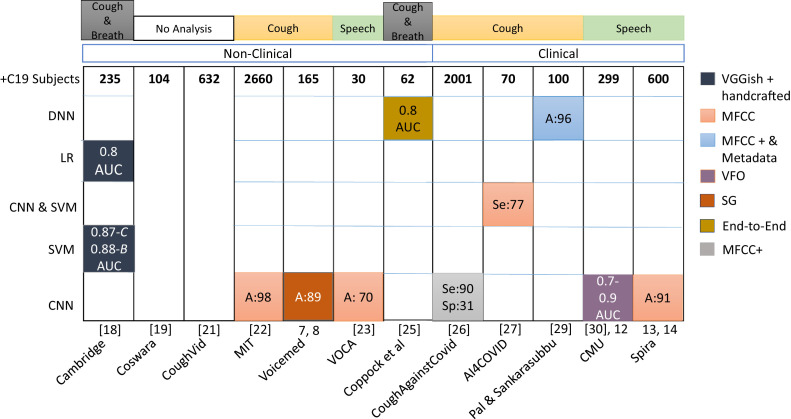
As seen in Fig. 3 , MFCCs are used in more than 50% of the total efforts [18], [19], [22], [23], [26], [27], [33], [37]. However, Alsabek et al. [38] extracted MFCCs from cough, deep breath, and speech signals from seven C19 patients and seven healthy individuals, showing that MFCCs from the speech are not dependable features for this task. Hence, we have to understand those speech-based features that are relevant for differentiating C19 patients from healthy individuals. Bartl-Pokorny et al. [39] studied sustained vowels produced by 11 symptomatic C19 positive and 11 C19 negative German-speaking participants, to assess the 88 eGeMAPS features [40], and report the mean voiced segment length and the number of voiced segments per second as being most important, using a Mann-Whitney U test.
Fig. 3.
Acoustic features’ & Machine learning techniques’ usage with the performance reported by different groups (on x-axis) for detecting COVID-19. The first row ‘+C19 subjects’ gives the C19 positive subjects’ count used by the respective groups; sequence of groups same as in Fig. 2. The features used by each group are indicated by the block colour: MFCC; SG: Spectrograms; VFO: Vocal fold Vibrations. Performance reported in the form of A: Accuracy, Se: Sensitivity, Sp: Specificity, and AUC. LR: Logistic regression. ‘Coswara’ and ‘Coughvid’ have not done any analysis with the data set they collected, hence blank blocks are shown for them. The results reported by ’Cambridge’ are: Combined analysis using cough and breath, C: Cough only and B: Breath only. On the x-axis, reference to bibliography is given in square brackets & number without bracket refers to footnote.
2.3.3. Scant breathing data analysis
Following efforts are analysing breath signals from data sets having less than 100 subjects.
In a study by Hassan et al. [41] with 60 healthy and 20 C19 positive subjects, the authors report better accuracy with LSTM based analysis using both breathing (98.2%) and cough (97%) data than for speech (88.2%) with an absolute improvement of 10% and 8.8%, respectively. The feature set used includes spectral centroid, spectral roll-off, zero-crossing rate, MFCCs, and their derivatives. However, analysing breathing signals is less popular than analysing coughs, owing to the challenges in capturing these low amplitude signals in noisy environments.
A preliminary analysis of the sound signals of respiration from nine C19 patients and four healthy volunteers is done by Furman et al. [42] using FFT harmonics. The respiration sounds are recorded using a smartphone microphone. Another such app detecting anomalies from the breathing sound has been developed by the TCS Research team [43]. The authors demonstrated that their app can detect breathing anomalies with an accuracy of 94%; however, the details about the anomalies and their significance in detecting C19 are not described.
3. Discussion and limitations
Although cough, speech, and breathing algorithms obtained sometimes good results on their test data sets, it is essential to validate these systems by using them with larger samples: In the absence of a C19 cure, it will be highly favourable to diagnose the virus symptoms at the earliest with non-invasive and easily available modes such as audio on smartphones and web interfaces. As a prerequisite, every research step, from data collection, annotation, validation of the annotated data to machine learning adopted needs careful analysis. Even though breathing analysis is found to be a promising signal carrying C19 symptoms, lesser attention has been paid to it as compared to cough analysis, maybe because of the challenging and intrusive mechanisms of capturing such low amplitude signals; a solution might be to correlate the speech signals with the breathing patterns and employ the speech signals themselves for classifying. As for the methods used, they seem not to converge; many different methods have been employed, and it is not (yet) possible to disentangle the adequacy of methods from other factors such as phenomenon addressed or reliability of annotation. As for the parameters used, it seems that MFCCs alone will not do; instead, we will have to employ the multitude of acoustic parameters available.
In all, to answer if an AI system can detect C19 when experts cannot do so by simple listening, it is important to consider the three factors: (1) clinically validated ground truth, (2) cough being not (only) an archetypal symptom of C19 and (3) presence of confounding classes such as Chronic Obstructive Pulmonary Disease (COPD), cold, or asthma. In the presence of a clinically validated ground truth, solving the C19 detection problem using machine learning techniques becomes empirical. However, relying on cough as the only human sound for the detection of C19 can be deceptive. Also, understanding the biomarkers that can differentiate C19 from other respiratory disorders seems to be the major challenge.
In the studies conducted for detecting C19 from cough and other human sounds, the analysis is mostly done with a very small number of C19 patients. It is difficult to label the data as C19 or non-C19 when it is collected through crowd-sourcing platforms having no clinical validation. In several studies, subjects donated the data voluntarily and produced the cough sounds in the absence of any ailments. We know from other domains of speech analysis such as emotion detection that acted states have higher intensities than spontaneously expressed emotions. Similarly here, we have to understand whether the prompted cough sounds carry a good enough correlation with spontaneous ones or not. The influence of environmental noises while detecting cough has also been studied by only a few studies. Sometimes, neither partitioning nor stratification of the data is mentioned.
When we relate the number of C19 subjects within studies to the performance measure obtained, then – as expected – a higher number of data points in the training set goes often together with better performance. As seen in Fig. 3, Voca [23] exhibits the lowest performance (70% accuracy) and has the lowest number of C19 subjects – only 30. However, the performance reported by MIT [22] – not although but because it is extremely high – waits for further corroboration.
Certain preventive measures taken by the governmental authorities in many countries have started examining and asking every individual whether they have any C19 symptoms. Such initiatives need a lot of human efforts to be invested. Alternative automation to accomplish such surveys might use a system as described by Lee et al. [44], where, by using speech recognition, synthesis, and natural language understanding techniques, the CareCall system monitors individuals of Korea and Japan who had contact with C19 patients. This monitoring is done over the phone using with and without human-in-the-loop processes for three months. The system has been used with over 13 904 calls; the authors reported 0% false negative (self-reported C19 subjects are not identified by the CareCall system) and 0.92% false-positive rate. A surveillance tool, the FluSense platform [45], has been developed by Al Hossain et al. to detect influenza-like illness from hospital waiting areas using cough sounds. Considering the importance of covering the mouth as a preventive measure against the spread of C19, it is valuable to detect mask-wearing individuals from their voice. The Interspeech 2020 Computational Paralinguistics Challenge (ComParE) [13] featured a mask detection sub-challenge, where the task is to recognise whether the speaker was recorded while wearing a facial mask or not. The winners of this sub-challenge, Szep and Hariri [46], used a deep convolutional neural network-based image classifier on the linear-scale 3-channel spectrograms of the speech segments. They achieved a UAR of 80.1% – 8.3% higher than the baseline, using an ensemble of VGGNet, ResNet, and DenseNet architectures. The caveat has to be made that ensemble methods seem to be highly competitive but might not meet run-time constraints in real-life applications.
4. Next steps and challenges
With the smartphone being the most convenient and available asset that almost every individual carries all the time, more smartphone-based applications for detecting C19 symptoms might help in controlling the spread of the virus. Albes et al. [47] addressed the memory and power consumption issues for importing a deep learning model for detecting a cold from the speech signal. They propose network pruning and quantization techniques to reduce the model size, achieving a size reduction of 95% in Megabytes without affecting recognition performance.
The spread of the disease has equally affected the physical and mental health of individuals. As found by Patel et al. [48], the C19 pandemic has generated unprecedented demand for telehealth based clinical services. It is imperative to study mental health issues such as stress, anxiety, and depression, from speech signals during the C19 period. This demands relevant data. Recently, a study was conducted by Han et al. [49] on the speech signal of C19 diagnosed patients. The behavioural parameters detected from speech include self-reported ratings of sleep quality, fatigue, and anxiety as a reference and achieved an average accuracy of 0.69 in estimating the severity of C19.
It would be interesting to evaluate whether multi-modal analysis helps to improve the accuracy of C19 detection: Image analysis is providing novel solutions using X-ray [50], [51], [52], [53], [54], [55] and chest CT images [56], [57], [58], [59], [60], [61], [62], [63]. Some of them [50], [52], [55], [58], [60], [64] have discriminated C19 from another pulmonary disorder (pneumonia). Hryniewska et al. [65] present a checklist for the development of an ML model for lung image analysis, pointing out the urgent need for better quality and quantity of image data. One of the topics in the checklist is data augmentation, which includes image visibility, the inclusion of areas of interest, and sensible transformations. Li et al. [64] demonstrated the use of simple auxiliary tasks on both 4758 CT and 5821 X-ray images using CNN-based deep networks for improving the network performance. Considering that C19 primarily affects the respiratory system – by thus being a genuine object of speech and voice analysis, both modalities might complement each other, yielding better performance.
5. Conclusion
Speech and human audio analysis are found to be promising for C19 analysis. As shown in Fig. 3, the early results exhibited by the studies performed by different groups indicate the feasibility of C19 detection from audio signals. As seen in Fig. 2, those groups which have collected all three types of audio data (cough, speech, and breathing) have not yet analysed them completely and together. Several initiatives towards identifying cough sounds and distinguishing C19 cough from other illnesses are currently being pursued. Such detectors, when integrated with chat-bots, can enhance the screening, diagnosing, and monitoring efforts while reducing human interventions. Further research is required for cough, breathing, and speech signal-based C19 analysis, where it is more important to identify the exact bio-markers. Moreover, exact benchmarking with strictly identical constellations such as identical databases and partitioning is highly needed to tell apart random from systematic factors; first initiatives are the forthcoming challenges at Interspeech 2021, see [66], [67].
With increasing correlations established between speech and breathing signals, detecting breathing disorders from the speech signals will be useful. Many elderly individuals have been inside home for almost the entire year. The past research on the detection of stress needs to be taken forward in the C19 context for the elderly population. Besides, promising applications using language processing and other signal analyses have been shown. In sum, we are positive that the combination of intelligent audio, speech, language, and other signal analysis can help make an important contribution in the fight against the C19 and oncoming similar pandemics – alone, or in combination with other methods.
Although the technology makes it feasible to monitor individuals for wearing a mask, coughing, sneezing and also for a healthy mental wellbeing, the privacy of an individual stands above it; since audio signals can enable de-anonymisation, it is essential to store and maintain such information in an anonymous way for further analysis. Applying a responsible AI in this context is described in Leslie [68]. As seen from the studies discussed in Section 2, the data in the context of C19 are sparse and in need of validation by performing gold standard test such as RT-PCR or Chest X-ray analysis by experts. Although it is crucial to have a speech based C19 screening, there is a greater need of having close to zero false negative rates of such a tool. The pure breathing studies have been promising, but of course, they suffer from sparse data as well.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank all researchers, health supporters, and others helping in this crisis. Our hearts are with those affected and their families and friends. We acknowledge funding from the German BMWi by ZIM grant no. 16KN069402 (KIrun), and from the European Community’s Seventh Framework Programme under grant agreement no. 826506 (sustAGE ).
Biographies
Gauri Deshpande is working as senior scientist at the behavioural business and social sciences lab of Tata Consultancy Services (TCS) Research Center. She is an external Ph.D. student at the University of Augsburg, Germany under the guidance of Prof. Björn Schuller. Her research interest includes speech signal processing, affective computing, and behavioural signal processing.
Anton Batliner received his doctoral degree in Phonetics in 1978 at LMU Munich. He is now with the Chair of Embedded Intelligence for Health Care and Wellbeing at University of Augsburg, Germany. He is co-editor/author of two books and author/co-author of more than 300 technical articles, with an h-index of 48 and 11.000 citations. His main research interests are all (cross-linguistic) aspects of prosody and (computational) paralinguistics.
Björn W. Schuller is full professor and head of the chair of Embedded Intelligence for Health Care and Wellbeing, at the University of Augsburg, Germany. He is full professor of Artificial Intelligence and Head of GLAM - Group on Language, Audio & Music, at Imperial College London. He is Chief Scientific Officer (CSO) and Co-Founding CEO of audEERING GmbH, Gilching/Germany. He is visiting professor at school of Computer Science and Technology, at Harbin Institute of Technology, Harbin/P.R. China amongst other Professorships and Affiliations.
Footnotes
https://www.worldometers.info/coronavirus/, retrieved August 24, 2021.
Virufy, http://archive.is/hbrfE
References
- 1.Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Lee M.J., Loeb M., Patel R., El Alayli A., Kalot M.A., Falck-Ytter Y., Lavergne V., Morgan R.L., Murad M.H., Sultan S., Bhimraj A., Mustafaand R.A. Infectious diseases society of america guidelines on the diagnosis of COVID-19. Clin. Infect. Dis. 2020:1–27. doi: 10.1093/cid/ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.B. W. Schuller, D. M. Schuller, K. Qian, J. Liu, H. Zheng, X. Li, COVID-19 and computer audition: an overview on what speech & sound analysis could contribute in the SARS-cov-2 corona crisis, arXiv:2003.11117 (2020). [DOI] [PMC free article] [PubMed]
- 3.G. Deshpande, B. Schuller, An overview on audio, signal, speech, & language processing for COVID-19, arXiv:2005.08579 (2020).
- 4.Drugman T., Urbain J., Bauwens N., Chessini R., Valderrama C., Lebecque P., Dutoit T. Objective study of sensor relevance for automatic cough detection. IEEE J. Biomed. Health Inform. 2013;17(3):699–707. doi: 10.1109/jbhi.2013.2239303. [DOI] [PubMed] [Google Scholar]
- 5.Pramono R.X.A., Imtiaz S.A., Rodriguez-Villegas E. Proceedings of the 41st Annual International Conference of the Engineering in Medicine and Biology Society (EMBC) IEEE; Berlin, Germany: 2019. Automatic cough detection in acoustic signal using spectral features; pp. 7153–7156. [DOI] [PubMed] [Google Scholar]
- 6.Miranda I.D.S., Diacon A.H., Niesler T.R. Proceedings of the 41st Annual International Conference of the Engineering in Medicine and Biology Society (EMBC) IEEE; Berlin, Germany: 2019. A comparative study of features for acoustic cough detection using deep architectures; pp. 2601–2605. [DOI] [PubMed] [Google Scholar]
- 7.San Chun K., Nathan V., Vatanparvar K., Nemati E., Rahman M.M., Blackstock E., Kuang J. 2020 IEEE International Conference on Pervasive Computing and Communications (PerCom) IEEE; 2020. Towards passive assessment of pulmonary function from natural speech recorded using a mobile phone; pp. 1–10. [Google Scholar]
- 8.Yadav S., Keerthana M., Gope D., Maheswari U.K., Ghosh P.K. Proceedings of the 45th International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; Barcelona, Spain: 2020. Analysis of acoustic features for speech sound based classification of asthmatic and healthy subjects; pp. 6789–6793. [Google Scholar]
- 9.Schuller B., Steidl S., Batliner A., Vinciarelli A., Scherer K., Ringeval F., Chetouani M., Weninger F., Eyben F., Marchi E., Mortillaro M., Salamin H., Polychroniou A., Valente F., Kim S. Proceedings of the 14th Annual Conference of the International Speech Communication Association. INTERSPEECH; Lyon, France: 2013. The INTERSPEECH 2013 computational paralinguistics challenge: social signals, conflict, emotion, autism; pp. 148–152. [Google Scholar]
- 10.Azam M.A., Shahzadi A., Khalid A., Anwar S.M., Naeem U. Proceedings of the 40th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC) IEEE; Honolulu, Hawaii: 2018. Smartphone based human breath analysis from respiratory sounds; pp. 445–448. [DOI] [PubMed] [Google Scholar]
- 11.Routray A., Mohamed Ismail Yasar A.K. Proceedings of the 27th European Signal Processing Conference (EUSIPCO) IEEE; A Coruña, Spain: 2019. Automatic measurement of speech breathing rate; pp. 1–5. [Google Scholar]
- 12.Nallanthighal V.S., Strik H. Proceedings of the 16th Annual Conference of the International Speech Communication Association. INTERSPEECH; Graz, Austria: 2019. Deep sensing of breathing signal during conversational speech; pp. 4110–4114. [Google Scholar]
- 13.Schuller B.W., Batliner A., Bergler C., Messner E.-M., Hamilton A., Amiriparian S., Baird A., Rizos G., Schmitt M., Stappen L., Baumeister H., MacIntyre A.D., Hantke S. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. The INTERSPEECH 2020 computational paralinguistics challenge: elderly emotion, breathing & masks; pp. 2042–2046. [Google Scholar]
- 14.MacIntyre A.D., Rizos G., Batliner A., Baird A., Amiriparian S., Hamilton A., Schuller B.W. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. Deep attentive end-to-end continuous breath sensing from speech; pp. 2082–2086. [Google Scholar]
- 15.Markitantov M., Dresvyanskiy D., Mamontov D., Kaya H., Minker W., Karpov A. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. Ensembling end-to-end deep models for computational paralinguistics tasks: compare 2020 mask and breathing sub-challenges; pp. 2072–2076. [Google Scholar]
- 16.Mendonça J., Teixeira F., Trancoso I., Abad A. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. Analyzing breath signals for the interspeech 2020 compare challenge; pp. 2077–2081. [Google Scholar]
- 17.Nallanthighal V.S., Härmä A., Strik H. Proceedings of the 45th International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; Barcelona, Spain: 2020. Speech breathing estimation using deep learning methods; pp. 1140–1144. [Google Scholar]
- 18.Brown C., Chauhan J., Grammenos A., Han J., Hasthanasombat A., Spathis D., Xia T., Cicuta P., Mascolo C. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2020. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data; pp. 3474–3484. [Google Scholar]
- 19.Sharma N., Krishnan P., Kumar R., Ramoji S., Chetupalli S.R., Nirmala R., Ghosh P.K., Ganapathy S. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. Coswara - a database of breathing, cough, and voice sounds for COVID-19 diagnosis; pp. 4811–4815. [Google Scholar]
- 20.Dash T.K., Mishra S., Panda G., Satapathy S.C. Detection of COVID-19 from speech signal using bio-inspired based cepstral features. Pattern Recognit. 2021;117:107999. doi: 10.1016/j.patcog.2021.107999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.L. Orlandic, T. Teijeiro, D. Atienza, The COUGHVID crowdsourcing dataset: a corpus for the study of large-scale cough analysis algorithms, arXiv:2009.11644 (2020). [DOI] [PMC free article] [PubMed]
- 22.Laguarta J., Hueto F., Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. Open J. Eng. Med. Biol. 2020;1:275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubnov T. Signal Analysis and Classification of Audio Samples From Individuals Diagnosed With COVID-19. UC San Diego; 2020. Ph.D. thesis. [Google Scholar]
- 24.B.W. Schuller, H. Coppock, A. Gaskell, Detecting COVID-19 from breathing and coughing sounds using deep neural networks, arXiv:2012.14553 (2020).
- 25.Coppock H., Gaskell A., Tzirakis P., Baird A., Jones L., Schuller B.W. End-2-end COVID-19 detection from breath & cough audio. BMJ Innov. 2021;7:8. doi: 10.1136/bmjinnov-2021-000668. [DOI] [PubMed] [Google Scholar]; to appear.
- 26.P. Bagad, A. Dalmia, J. Doshi, A. Nagrani, P. Bhamare, A. Mahale, S. Rane, N. Agarwal, R. Panicker, Cough against COVID: evidence of COVID-19 signature in cough sounds, arXiv:2009.08790 (2020).
- 27.Imran A., Posokhova I., Qureshi H.N., Masood U., Riaz M.S., Ali K., John C.N., Hussain M.D.I., Nabeel M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform. Med. Unlocked. 2020;20:100378. doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piczak K.J. Proceedings of the 23rd International Conference on Multimedia. ACM; Brisbane, Australia: 2015. ESC: dataset for environmental sound classification; pp. 1015–1018. [Google Scholar]
- 29.Pal A., Sankarasubbu M. Proceedings of the 36th Annual ACM Symposium on Applied Computing. 2021. Pay attention to the cough: early diagnosis of COVID-19 using interpretable symptoms embeddings with cough sound signal processing; pp. 620–628. [Google Scholar]
- 30.Deshmukh S., Al Ismail M., Singh R. Proceedings of the 46th International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; Virtual: 2021. Interpreting glottal flow dynamics for detecting COVID-19 from voice; pp. 1055–1059. [Google Scholar]
- 31.Zhao W., Singh R. Proceedings of the 45th International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; Barcelona, Spain: 2020. Speech-based parameter estimation of an asymmetric vocal fold oscillation model and its application in discriminating vocal fold pathologies; pp. 7344–7348. [Google Scholar]
- 32.Pahar M., Klopper M., Warren R., Niesler T. COVID-19 cough classification using machine learning and global smartphone recordings. Comput. Biol. Med. 2021;135:104572. doi: 10.1016/j.compbiomed.2021.104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunne R., Morris T., Harper S. High accuracy classification of COVID-19 coughs using Mel-frequency cepstral coefficients and a convolutional neural network with a use case for smart home devices. Res. Square Prepr. 2020:1–14. [Google Scholar]
- 34.Pinkas G., Karny Y., Malachi A., Barkai G., Bachar G., Aharonson V. SARS-CoV-2 detection from voice. IEEE Open J. Eng. Med. Biol. 2020;1:268–274. doi: 10.1109/OJEMB.2020.3026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.K.V.S. Ritwik, S.B. Kalluri, D. Vijayasenan, COVID-19 patient detection from telephone quality speech data, arXiv:2011.04299 (2020).
- 36.Ko T., Peddinti V., Povey D., Khudanpur S. Proceedings of the 16th Annual Conference of the International Speech Communication Association. INTERSPEECH; Dresden, Germany: 2015. Audio augmentation for speech recognition; pp. 3586–3589. [Google Scholar]
- 37.Bansal V., Pahwa G., Kannan N. Proceedings of the 3rd International Conference on Computing, Power and Communication Technologies (GUCON) IEEE; Greater Noida, (NCR New Delhi) India: 2020. Cough classification for COVID-19 based on audio MFCC features using convolutional neural networks; pp. 604–608. [Google Scholar]
- 38.Alsabek M.B., Shahin I., Hassan A. Proceedings of the International Conference on Communications, Computing, Cybersecurity, and Informatics (CCCI) IEEE; Sharjah, UAE: 2020. Studying the similarity of COVID-19 sounds based on correlation analysis of MFCC; pp. 1–5. [Google Scholar]
- 39.K.D. Bartl-Pokorny, F.B. Pokorny, A. Batliner, S. Amiriparian, A. Semertzidou, F. Eyben, E. Kramer, F. Schmidt, R. Schönweiler, M. Wehler, B.W. Schuller, The voice of COVID-19: acoustic correlates of infection, arXiv:2012.09478 (2020). [DOI] [PMC free article] [PubMed]
- 40.Eyben F., Scherer K.R., Schuller B.W., Sundberg J., André E., Busso C., Devillers L.Y., Epps J., Laukka P., Narayanan S., Truong K.P. The Geneva minimalistic acoustic parameter set (GeMAPS) for voice research and affective computing. IEEE Trans. Affect. Comput. 2016;7(2):190–202. [Google Scholar]
- 41.Hassan A., Shahin I., Alsabek M.B. Proceedings of the International Conference on Communications, Computing, Cybersecurity, and Informatics (CCCI) IEEE; Sharjah, UAE: 2020. COVID-19 detection system using recurrent neural networks; pp. 1–5. [Google Scholar]
- 42.Furman E.G., Charushin A., Eirikh E., Malinin S., Sheludko V., Sokolovsky V., Furman G. The remote analysis of breath sound in COVID-19 patients: a series of clinical cases. medRxiv. 2020 [Google Scholar]
- 43.Harini S., Deshpande P., Rai B. Proceedings of the 7th International Electronic Conference on Sensors and Applications. Sciforum, MDPI, Online; 2020. Breath sounds as a biomarker for screening infectious lung diseases. [Google Scholar]; 1–1
- 44.S.-W. Lee, H. Jung, S. Ko, S. Kim, H. Kim, K. Doh, H. Park, J. Yeo, S.-H. Ok, J. Lee, S. Lim, M. Jeong, S. Choi, S. Hwang, E.-Y. Park, G.-J. Ma, S.-J. Han, K.-S. Cha, N. Sung, J.-W. Ha, Carecall: a call-based active monitoring dialog agent for managing COVID-19 pandemic, arXiv:2007.02642 (2020).
- 45.Al Hossain F., Lover A.A., Corey G.A., Reich N.G., Rahman T. Proceedings of the Interactive, Mobile, Wearable and Ubiquitous Technologies. Vol. 4. ACM; 2020. Flusense: a contactless syndromic surveillance platform for influenza-like illness in hospital waiting areas; pp. 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szep J., Hariri S. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. Paralinguistic classification of mask wearing by image classifiers and fusion; pp. 2087–2091. [Google Scholar]
- 47.Albes M., Ren Z., Schuller B., Cummins N. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. Squeeze for sneeze: compact neural networks for cold and flu recognition; pp. 4546–4550. [Google Scholar]
- 48.Patel P.D., Cobb J., Wright D., Turer R., Jordan T., Humphrey A., Kepner A.L., Smith G., Rosenbloom S.T. Rapid development of telehealth capabilities within pediatric patient portal infrastructure for COVID-19 care: barriers, solutions, results. J. Am. Med. Inform. Assoc. (JAMIA) 2020;27(7):1116–1120. doi: 10.1093/jamia/ocaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J., Qian K., Song M., Yang Z., Ren Z., Liu S., Liu J., Zheng H., Ji W., Koike T., Li X., Zhang Z., Yamamoto Y., Schuller B.W. Proceedings of the 21st Annual Conference of the International Speech Communication Association. INTERSPEECH; Shanghai, China: 2020. An early study on intelligent analysis of speech under COVID-19: severity, sleep quality, fatigue, and anxiety; pp. 4946–4950. [Google Scholar]
- 50.Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., Liu X. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest XZ-rays. Pattern Recognit. 2021;110:107613. doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan Y., Liu J., Yao R., Yuan X. COVID-19 detection from X-ray images using multi-kernel-size spatial-channel attention network. Pattern Recognit. 2021;119:108055. doi: 10.1016/j.patcog.2021.108055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vieira P., Sousa O., Magalhes D., Rablo R., Silva R. Detecting pulmonary diseases using deep features in X-ray images. Pattern Recognit. 2021;119:108081. doi: 10.1016/j.patcog.2021.108081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guarrasi V., DAmico N.C., Sicilia R., Cordelli E., Soda P. Pareto optimization of deep networks for COVID-19 diagnosis from chest X-rays. Pattern Recognit. 2021;121:108242. doi: 10.1016/j.patcog.2021.108242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malhotra A., Mittal S., Majumdar P., Chhabra S., Thakral K., Vatsa M., Singh R., Chaudhury S., Pudrod A., Agrawal A. Multi-task driven explainable diagnosis of COVID-19 using chest X-ray images. Pattern Recognit. 2021:108243. doi: 10.1016/j.patcog.2021.108243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shorfuzzaman M., Hossain M.S. MetaCOVID: a siamese neural network framework with contrastive loss for n-shot diagnosis of COVID-19 patients. Pattern Recognit. 2021;113:107700. doi: 10.1016/j.patcog.2020.107700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oulefki A., Agaian S., Trongtirakul T., Laouar A.K. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recognit. 2021;114:107747. doi: 10.1016/j.patcog.2020.107747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., Shi Y., Shi F., Gao Y., Liu J., Zhang J., Shen D. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. Pattern Recognit. 2021;113:107828. doi: 10.1016/j.patcog.2021.107828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou J., Xu J., Jiang L., Du S., Feng R., Zhang Y., Shan F., Xue X. Periphery-aware COVID-19 diagnosis with contrastive representation enhancement. Pattern Recognit. 2021;118:108005. doi: 10.1016/j.patcog.2021.108005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J., Xu H., Zhang S., Li X., Chen J., Zheng J., Gao Y., Tian Y., Liang Y., Ji R. Joint segmentation and detection of COVID-19 via a sequential region generation network. Pattern Recognit. 2021;118:108006. doi: 10.1016/j.patcog.2021.108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao C., Xu Y., He Z., Tang J., Zhang Y., Han J., Shi Y., Zhou W. Lung segmentation and automatic detection of COVID-19 using radiomic features from chest CT images. Pattern Recognit. 2021;119:108071. doi: 10.1016/j.patcog.2021.108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Carvalho Brito V., Dos Santos P.R.S., de Sales Carvalho N.R., de Carvalho Filho A.O. Covid-index: a texture-based approach to classifying lung lesions based on CT images. Pattern Recognit. 2021;119:108083. doi: 10.1016/j.patcog.2021.108083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mu N., Wang H., Zhang Y., Jiang J., Tang J. Progressive global perception and local polishing network for lung infection segmentation of COVID-19 CT images. Pattern Recognit. 2021;120:108168. doi: 10.1016/j.patcog.2021.108168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X., Yao L., Zhou T., Dong J., Zhang Y. Momentum contrastive learning for few-shot COVID-19 diagnosis from chest CT images. Pattern Recognit. 2021;113:107826. doi: 10.1016/j.patcog.2021.107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J., Zhao G., Tao Y., Zhai P., Chen H., He H., Cai T. Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19. Pattern Recognit. 2021;114:107848. doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hryniewska W., Bombiński P., Szatkowski P., Tomaszewska P., Przelaskowski A., Biecek P. Checklist for responsible deep learning modeling of medical images based on COVID-19 detection studies. Pattern Recognit. 2021;118:108035. doi: 10.1016/j.patcog.2021.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.B.W. Schuller, A. Batliner, C. Bergler, C. Mascolo, J. Han, I. Lefter, H. Kaya, S. Amiriparian, A. Baird, L. Stappen, S. Ottl, M. Gerczuk, P. Tzirakis, C. Brown, J. Chauhan, A. Grammenos, A. Hasthanasombat, D. Spathis, T. Xia, P. Cicuta, L.J.M. Rothkrantz, J. Zwerts, J. Treep, C. Kaandorp, The INTERSPEECH 2021 computational paralinguistics challenge: COVID-19 cough, COVID-19 speech, escalation & primates, arXiv:2102.13468 (2021).
- 67.A. Muguli, L. Pinto, N. Sharma, P. Krishnan, P.K. Ghosh, R. Kumar, S. Ramoji, S. Bhat, S.R. Chetupalli, S. Ganapathy, V. Nanda, DiCOVA challenge: dataset, task, and baseline system for COVID-19 diagnosis using acoustics, arXiv:2103.09148 (2021).
- 68.D. Leslie, Tackling COVID-19 through Responsible AI Innovation: Five Steps in the Right Direction, 2020, arXiv:2008.06755.