Abstract
Histones are dynamically modified during chromatin assembly, as specific transcriptional patterns are established, and during mitosis and development. Modifications include acetylation, phosphorylation, ubiquitination, methylation, and ADP-ribosylation, but the biological significance of each of these is not well understood. For example, distinct acetylation patterns correlate with nucleosome formation and with transcriptionally activated or silenced chromatin, yet mutations in genes encoding several yeast histone acetyltransferase (HAT) activities result in either no cellular phenotype or only modest growth defects. Here we report characterization of ESA1, an essential gene that is a member of the MYST family that includes two yeast silencing genes, human genes associated with leukemia and with the human immunodeficiency virus type 1 Tat protein, and Drosophila mof, a gene essential for male dosage compensation. Esa1p acetylates histones in a pattern distinct from those of other yeast enzymes, and temperature-sensitive mutant alleles abolish enzymatic activity in vitro and result in partial loss of an acetylated isoform of histone H4 in vivo. Strains carrying these mutations are also blocked in the cell cycle such that at restrictive temperatures, esa1 mutants succeed in replicating their DNA but fail to proceed normally through mitosis and cytokinesis. Recent studies show that Esa1p enhances transcription in vitro and thus may modulate expression of genes important for cell cycle control. These observations therefore link an essential HAT activity to cell cycle progression, potentially through discrete transcriptional regulatory events.
Cytogenetic and biochemical differences between transcriptionally active and inactive chromatin have long been recognized (for reviews, see references 21, 60, 66, and 71). Particular focus in defining these distinctions has been on the core histones, which in the context of the nucleosome serve as the scaffold for packaging DNA into chromatin. Ultimately, it is chromatin that serves as the in vivo template from which RNA transcription and DNA replication occur, and it has been of central importance to define how histones function in the regulation of these processes.
Previous biochemical and genetic studies and the recent high-resolution structure of the nucleosome (37) point to the amino-terminal tails of the conserved core histones as important effectors for regulation. Extending from the nucleosomal particle, these tails participate in internucleosomal interactions and may be posttranslationally modified to result in local differences in chromatin structure and function by influencing accessibility and activity of polymerases and other regulatory proteins. Understanding these modifications may be crucial for understanding cell- and gene-specific functions. For example, phosphorylation of histone H3 on serine 10 occurs in late G2 and has been proposed to promote binding of factors that may drive chromatin condensation as cells enter mitosis (24).
Among the observed histone modifications, acetylation has been perhaps most thoroughly studied, and it may contribute both to assembly of chromatin in general and to setting transcriptional states for specific loci or entire chromosomes (21, 40, 60). Progress in defining these roles for acetylation came first from mapping and mutating sites on the core histones that were modified and most recently through identification of the enzymes, histone acetyltransferases (HATs) and deacetylases, that catalyze the modifications (for reviews see references 40, 49, and 60). The yeast gene HAT1 was found to encode an enzyme that acetylates primarily newly synthesized histone H4, potentially as part of its assembly into nucleosomes (29, 48). Hat1p associates with Hat2p, an Rba48-like protein that contributes to substrate specificity of the enzyme (48). Although no phenotypes have been observed for yeast cells harboring mutations in these genes, a similar HAT complex exists in human cells (62), suggesting that such conserved activities are functionally important.
The first transcription-linked HAT was identified in Tetrahymena as the homologue of yeast Gcn5p (9). GCN5 is not essential in yeast, but it is necessary for full transcriptional activation of some genes (17), and mutational analyses demonstrate that HAT activity correlates with transcriptional function (33, 65). Although Gcn5p provides the catalytic activity, it is only one component of two large multisubunit HAT complexes that exist in the cell (19). Interestingly, the noncatalytic subunits are also important for in vivo function of the complexes, suggesting that they may be finely regulated for both catalysis and specificity.
Many other proteins with HAT activity have also been identified; these include p300 and the CREB binding protein CBP (4, 43), SRC-1 family proteins (10, 59), and the basal transcription factor TAFII250 (41). The biological significance of the activities of these proteins has not been defined fully, and it is likely that HAT activity is a common feature of both gene-specific and basal regulation.
Sequence analysis identifies a superfamily of other proteins that are either known or predicted acetyltransferases (reviewed in reference 42). For some of these there are no known functions, whereas others have provocative connections to chromatin. We have been especially interested in the SAS (something about silencing) genes of the MYST family (6), which were first identified for their roles in transcriptional silencing in the yeast Saccharomyces cerevisiae (15, 53). Although SAS2 and SAS3 exhibit significant sequence similarity, they have distinct mutant phenotypes suggesting that they can either promote or silence transcription, depending on the locus. Both proteins contain the A motif, a relatively short sequence that may contribute to acetyl coenzyme A binding (42), although neither has been demonstrated to have catalytic activity. We have identified a third yeast SAS family member, ESA1, that encodes an essential HAT. Further, Esa1p functions as the catalytic subunit in the 1.3-MDa NuA4 complex that acetylates both free histones and chromatin and, notably, can promote transcription in vitro (14). Our analysis of recessive conditional alleles reveals that ESA1 is important for progression through the cell cycle, since cells grown under restrictive conditions arrest with a G2/M DNA content and partial depletion of an acetylated form of histone H4. The cell cycle arrest observed upon loss of ESA1 function is dependent on the checkpoint gene RAD9. These observations therefore link specific histone modifications to cell cycle control, potentially mediated through transcriptional regulation.
MATERIALS AND METHODS
Yeast methods.
An esa1Δ::HIS3 null mutation was generated in the S288c diploid strain LPY2553 (MATa/MATα his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 ura3-52/ura3-52) to create LPY2639 (MATa/MATα his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 ura3-52/ura3-52 esa1Δ::HIS3/ESA1) according to standard procedures (5). Temperature-sensitive alleles were generated by amplifying the ESA1 gene by using Taq polymerase in conditions recommended by the manufacturer (Promega), except that deoxynucleoside triphosphates were used at a final concentration of 250 mM. Products were subcloned into a TRP1-CEN vector and tested for complementation of the esa1 null strain LPY2641 (MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1Δ::HIS3 bearing an ESA1-URA3-CEN plasmid) by plasmid shuffle (23). Temperature-sensitive alleles were identified as having complementation defects at 34°C and above. Alleles were sequenced by using an Applied Biosystems automated facility.
Three alleles, esa1-L327S, esa1-L254P, and esa1-414, were integrated according to standard procedures (23) in LPY2639 at the esa1Δ::HIS3 locus with URA3-marked integrating vectors pLP951, pLP949, and pLP952, creating LPY3425, LPY3454, and LPY3443, respectively. Integration events were confirmed by standard methods, and diploids LPY3425 and LPY3454 were sporulated to create the haploids carrying the following alleles for further experiments: esa1-L327S (LPY3430 MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1Δ::HIS3 esa1-L327S::URA3), ESA1 (LPY3431 MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 ESA1), esa1-L254P (LPY3500 MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1Δ::HIS3 esa1-L254P::URA3), and ESA1 (LPY3498 MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 ESA1). Strains for rad9Δ esa1-L254P mutant analysis were generated by crossing LPY3500 and LPY3719 (MATα ade2-101 his3Δ200 lys2-801 trp1Δ1 ura3-52 rad9Δ::HIS3) to create the diploid LPY3779 (MATa/MATα ADE2/ade2-101 his3Δ200/his3Δ200 leu2,3-112/LEU2 LYS2/lys2-801 trp1Δ1/trp1Δ1 ura3-52/ura3-52 esa1Δ::HIS3 esa1-L254P::URA3/ESA1 RAD9/rad9Δ::HIS3); after sporulation and dissection, the haploids with the following mutations were recovered from tetrads with either a 2 HIS:2 his or 4 HIS:0 his segregation pattern: rad9Δ esa1-L254P (LPY3780 MATα ade2-101 his3Δ200 LEU2 lys2-801 trp1Δ1 ura3-52 rad9Δ::HIS3 esa1Δ::HIS3 esa1-L254P::URA3), rad9Δ (LPY3784 MATa ade2-101 his3Δ200 leu2,3-112 LYS2 trp1Δ1 ura3-52 rad9Δ::HIS3), and esa1-L254P (LPY3785 MATa ADE2 his3Δ200 LEU2 lys2-801 trp1Δ1 ura3-52 esa1Δ::HIS3 esa1-L254P::URA3). Additional strains used included LPY2991, a MATa esa1Δ::HIS3 isolate from LPY2639, with a CEN-ESA1 plasmid (pLP795); LPY3291, the same as LPY2991 except that plasmid (pLP863)-borne esa1-414 replaces the ESA1 plasmid; LPY3068, the same as LPY2991 except that ESA1 on a TRP1-2μm plasmid, pLP798, replaces pLP795; LPY4184, the same as LPY2991 but a MATα isolate in which esa1-414 on a TRP1-2μm plasmid, pLP1128, replaces pLP795; a mad3Δ strain (LPY4222 MATα leu2,3-112 his3Δ200 trp1Δ1 ura3-52 mad3Δ::URA3); and a mad3Δ esa1-L254P strain (LPY4223 MATα leu2,3-112 his3Δ200 trp1Δ1 ura3-52 mad3Δ::URA3 esa1Δ::HIS3 esa1-L254P::URA3).
Protein expression.
Exponentially growing Escherichia coli BL21(DE3)pLysS cells transformed with either pLP820 (pRSETc vector [Invitrogen]), pLP831 (pRSETc-ESA1), pLP1138 (pRSETc-esa1-L254P), pLP942 (pRSETc-esa1-414), or pLP832 (pRSETc-esa1-L327S) were induced by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.5 mM. Cells were harvested after 3 h at 25°C, and lysate was prepared by a freeze-thaw cycle followed by sonication in buffer (20% sucrose, 30 mM Tris [pH 8.0], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 100 μl per optical density at 600 nm of 0.25). Supernatants from total cell equivalent lysates that had been clarified by centrifugation were used in HAT assays.
Acetyltransferase activity assays.
Fifty micrograms of calf thymus histones (Sigma), 20 μg of yeast histones (a gift of J. Pilon and P. Laybourn), or 500 μg of unacetylated bovine serum albumin (BSA; Amresco) was combined in reaction buffer (32) with 15 μg of recombinant Esa1p, Esa1-L327Sp, or pRSETc total soluble cell lysate and 0.1 μCi of 3H-acetyl coenzyme A (2.3 Ci/mmol; ICN). Following incubation at 30°C for 10 min, samples were processed as described elsewhere (7) prior to scintillation counting. Approximately equivalent as well as sevenfold more Esa1-L327Sp than Esa1p was used in HAT assays based on quantitation of Coomassie blue-stained gels, using the Alpha Digital Imaging System 2000 program. For fluorography, aliquots of the HAT assay reactions were separated on a sodium dodecyl sulfate (SDS)–18% polyacrylamide gel and stained with Coomassie brilliant blue. The gel was then treated for fluorography with Intensify enhancer (NEN) and processed according to the manufacturer’s instructions.
Microsequencing.
HAT assays were performed essentially as described above except that 3 μg of synthetic peptide corresponding to the N terminus of histone H4, H3, or H2A was combined with 3 μg of recombinant Esa1p total soluble cell lysate and 1.0 μCi of 3H-acetyl coenzyme A (2.58 Ci/mmol; NEN). Reaction mixes were incubated for 30 min at 22°C (H4 peptide) or for 3 h at 15°C (H3 and H2A peptides). Individual reactions were microsequenced as described elsewhere (32, 58) at the Protein Chemistry Core Facility at the Baylor School of Medicine. H4 and H2A peptides were gifts of M. Parthun and D. Gottschling. The H3 peptide was synthesized on an Applied Biosystems instrument, using standard chemistry and manufacturer’s recommendations.
Flow cytometry and microscopic analyses.
Flow cytometry of 10,000 cells/sample was performed as described elsewhere (69). For 4′,6-diamidino-2-phenylindole (DAPI) staining, cells were fixed in 70% acetone–30% methanol on dry ice for at least 1 h, washed twice in phosphate-buffered saline (PBS), and then stained with DAPI (Boehringer Mannheim) at a final concentration of 0.25 μg/ml. For indirect immunofluorescence, 37% formaldehyde was added to cultures to a final concentration of 5%. Samples were agitated for 5 min at their relevant experimental temperatures and then fixed for an additional 45 min at 22°C. Cells were washed once in PBS, washed twice in 1.2 M sorbitol in PBS (SPBS), and then resuspended in SPBS with 24 mM β-mercaptoethanol and Zymolyase 70000 (ICN) at a final concentration of 29 μg/ml. After 30 min at 30°C, samples were washed once in SPBS and twice in PBS. Staining was performed on poly-l-lysine-coated multiwell slides. Samples were preblocked (PBS with 10 mg of BSA per ml and 0.5% Nonidet P-40). Rabbit anti-Kar2p serum was used at a dilution of 1:1,000, and rat antitubulin serum (YOL1/34; Serotec, Inc.) at was used 1:2,000. Primary incubations for 1 h at 37°C were followed by six PBS washes. Secondary antibodies, fluorescein isothiocyanate-conjugated goat anti-rabbit (Jackson Laboratories) at 1:300 and Texas red-conjugated donkey anti-rat (Jackson Laboratories) at 1:50, were incubated at 22°C for 30 min. After six PBS washes, DAPI was added as described above. After washing, slides were mounted in Citifluor (Ted Pella, Inc.), staining was visualized with a Zeiss Axiophot fluorescence microscope, and images were collected with a COHU charge-coupled device camera using the Scion Image 1.57 program. For electron microscopy in collaboration with Tom Giddings at the University of Colorado, samples were prepared as described elsewhere (44). Sections of 80 nm were cut and examined on a Philips CM10 microscope at 80 kV with a magnification of either ×10,500 or ×25,000. Images were collected by using a Gatan BioScan camera and Gatan Digital Micrograph 2.5 (PPC) software.
Immunoblotting analysis.
Exponentially growing cells were split, pelleted, and resuspended into 28 or 37°C medium and incubated at the appropriate temperature for 4 h (except LPY3430, which was incubated for 8 h). Protein from 5 × 108 cells was recovered by glass bead lysis. Briefly, cells were washed in lysis buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, 10 mM Tris [pH 7.5]), and the pellets were frozen at −20°C and then resuspended in 0.4 to 0.8 ml of lysis buffer containing 1 μg each of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK), leupeptin, pepstatin A, aprotinin per ml and 0.2 mM phenylmethylsulfonyl fluoride. Acid-washed glass beads were added, and the cells were vortexed six times in 1-min bursts. Extracted protein was quantitated by the Bio-Rad protein assay, diluted into sample buffer, boiled for 5 min, and loaded onto an SDS–18% polyacrylamide gel. The separated proteins were then transferred to nylon membrane, blocked in TBSTM (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 20, 2% dry milk) for 1 h at room temperature, and then incubated with antiserum directed against the histone H4-acetylated Lys5 residue (1:500 in TBSTM; Serotec) or antiserum directed against total histones (0.5 μg/ml in TBSTM; Boehringer-Mannheim) for 8 h at 4°C. Blots were washed, incubated with horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse antiserum (1:10,000 or 1:2,500, respectively; Promega) for 1 h at room temperature, washed, and processed for chemiluminescence as described elsewhere (56).
RESULTS
ESA1 is an essential gene in the MYST family with similarity to acetyltransferases.
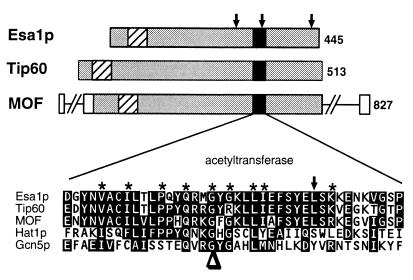
During analysis of the yeast SAS2 and SAS3 silencing genes (53), we discovered that another, very similar but previously uncharacterized open reading frame (ORF) (YOR244w) existed on yeast chromosome XV. This ORF is predicted to encode a 445-amino-acid protein (Fig. 1) with sequence features that include an acetyltransferase motif and an amino-terminal chromo domain, a motif found in many proteins thought to regulate transcription via their chromatin association (1, 31, 47). YOR244w is most similar [BLASTP (3) scores of P = 10−100 or better] to the human MYST family proteins Tip60 and MOZ and the Drosophila melanogaster MOF protein (6, 25, 26). To investigate the function of YOR244w, we constructed a null allele by deleting one copy of the ORF (5) in a diploid yeast strain. This diploid was sporulated, and of the four spores from each tetrad, only two formed colonies. Microscopic analysis demonstrated that all four spores germinated with approximately the same kinetics, but cells from two of the spores in every tetrad stopped growing after three to four rounds of division. Genotyping revealed that the surviving spores were all wild type for YOR244w, and a plasmid-borne copy of the wild-type gene rescued the lethality associated with the deletion. Thus, the YOR244w gene product is essential for cellular viability, and we have named it ESA1, for essential SAS family acetyltransferase.
FIG. 1.
Esa1p has sequence similarity (gray boxes) with human Tip60 and D. melanogaster MOF. A region of strong sequence similarity to acetyltransferases including yeast Hat1p and Gcn5p is indicated (black boxes). Similar and identical residues in this region are highlighted. Residues most conserved among known and predicted acetyltransferases in the A motif as defined by Neuwald and Landsman (42) are noted with asterisks. Positions of temperature-sensitive esa1 alleles (see text) are noted by arrows above the Esa1p cartoon; the position of the esa1-L327S allele is also shown by an arrow in the sequence expansion. The conserved G→E mutation in mof (25) is marked with an arrowhead below the sequence. An N-terminal chromo domain (1, 25, 31, 47) is found in these three SAS family members (diagonal stripes). The number of amino acids in each protein is indicated at the right.
Esa1p acetylates histones with a distinct specificity pattern.
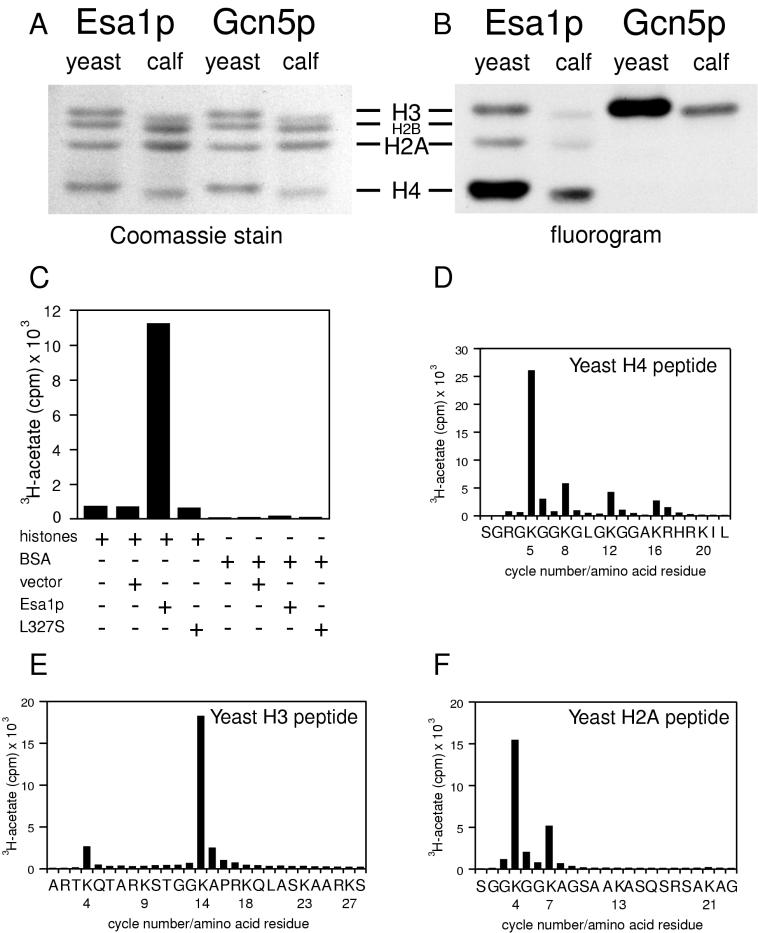
A clue for the cellular role of ESA1 came from the motif that it shares with other yeast proteins demonstrated to have in vitro catalytic acetyltransferase activity, such as Hat1p, thought to function in chromatin assembly, and Gcn5p, which functions in transcription (9, 29, 48). To test if Esa1p had HAT activity in vitro, we cloned the gene into a vector allowing its inducible expression in E. coli. Extracts prepared from bacteria expressing Esa1p or Gcn5p were tested for HAT activity in a standard assay (7, 32) by mixing the extracts with 3H-acetyl coenzyme A and histones from either yeast or calf thymus. These reaction mixtures were resolved by using SDS–18% polyacrylamide gels that were stained (Fig. 2A) and subjected to fluorography (Fig. 2B). Recombinant Esa1p acetylated histone substrates, but the pattern of incorporation was distinct from that of Gcn5p. Esa1p acetylated primarily histone H4 and to a lesser extent H3 and H2A. In contrast, Gcn5p was most active on H3, although upon longer exposures, as in published reports (32), weak H4 acetylation was observed. In parallel liquid HAT assays performed with extracts prepared from bacteria transformed with vector alone, or without histones or with BSA substituted for histones, no acetylation was observed (Fig. 2C). Therefore, Esa1p’s activity is specific for histones under these reaction conditions. To identify the lysines acetylated by recombinant Esa1p, we used synthetic peptides corresponding to the lysine-rich amino-terminal tails of yeast H4, H3, and H2A as substrates in standard reactions. The labeled peptides were then subjected to microsequencing coupled with the direct determination of radioactivity incorporated into each position of the peptide (Fig. 2D to F). The results show clearly that Lys5 was the major site of Esa1p acetylation on the yeast H4 peptide. Lysines at positions 8, 12, and 16 were also acetylated but less efficiently, demonstrating that each of the acetylatable lysines in the H4 tail is a potential target for Esa1p. For H3, Lys14 was the major target, with modest acetylation of Lys4. The yeast H2A peptide was acetylated predominantly at Lys4 and moderately at Lys7. This pattern of substrate specificity is distinct from those previously defined for yeast HATs and agrees with independent results for Esa1p obtained in assays using purified histones, including nonyeast proteins, as substrates (57). For example, the deposition-related Hat1p acetylates H4, predominantly on Lys12, although under conditions of reduced specificity Lys5 is also acetylated as is H2A (29, 48). The transcription-linked HAT, Gcn5p, can acetylate H4 on Lys16 but has significant preference for Lys14 of histone H3 in vitro (32). H2A is not a substrate for Gcn5p. However, Lys4 of H2A was identified previously as the sole in vivo site of acetylation in murine leukemia L1210 cells (46), although to date the relevant L1210 HAT has not been reported. Thus, it is clear that ESA1 encodes a HAT with distinct substrate specificities, which may indicate novel or perhaps multiple cellular functions. To begin to understand these functions, we identified conditional alleles so that loss-of-function phenotypes could be examined.
FIG. 2.
Recombinant Esa1p acetylates histones. Products of HAT assays using either yeast or calf histones as substrates were resolved on an SDS–18% polyacrylamide gel that was stained with Coomassie brilliant blue (A) and treated for fluorography (B). The activities of recombinant Esa1p were compared with that of recombinant Gcn5p; whereas Esa1p preferentially acetylates H4, Gcn5p preferentially acetylates H3 (32). (C) Bacterial extracts containing equivalent amounts of recombinant Esa1p, vector control, or recombinant Esa1-L327Sp were incubated with 3H-acetyl coenzyme A and either calf histones or BSA. Esa1p acetylates calf histones but not BSA. No activity was observed with vector control or Esa1-L327Sp lysates or with two other mutant proteins tested (data not shown). The preferred targets of Esa1p are Lys5 of histone H4 (D), Lys14 of histone H3 (E), and Lys4 and Lys7 of histone H2A (F). Note that lysines 8, 12, and 16 in H4 were also acetylated. When repetitive yields for the cycles of Edman degradation were calculated, the extent of acetylation at each of these sites was equivalent and significantly less than that of Lys5.
Temperature-sensitive esa1 mutants lack HAT activity in vitro and deplete cellular pools of histone H4 acetylated on Lys5 in vivo.
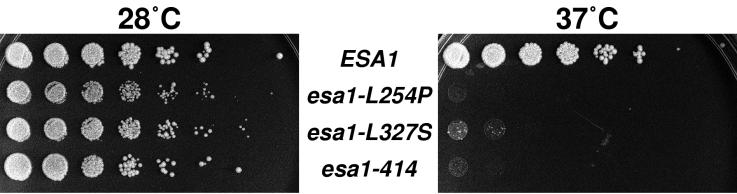
We screened a library of plasmids encoding ESA1 that had been amplified by using standard procedures. We recovered those plasmids that fully complemented lethality of strains with the esa1Δ allele at permissive temperatures (28°C or below) but that failed to complement at restrictive temperatures (34°C or above). Twenty-two esa1 alleles met these criteria, and growth phenotypes of strains harboring three recessive esa1 mutants are shown in Fig. 3. These three mutant alleles have been integrated into the normal chromosomal locus of ESA1. The esa1-414 phenotype is most severe in that haploid strains with this allele are viable only when the mutated gene is plasmid borne and presumably present at slightly increased gene dosage. In esa1-414, deletion of a single nucleotide at position 1887 leads to a frameshift mutation in codon 414, altering 10 amino acids before terminating the ORF 22 amino acids prematurely. The esa1-L254P and esa1-L327S mutations substitute proline or serine for conserved leucines, and notably the esa1-L327S substitution lies within the acetyltransferase domain itself (Fig. 1). All three mutant strains grew well at permissive temperatures but poorly at elevated temperatures (Fig. 3), suffering 60 to 80% lethality within the first 4 h of incubation. When recombinant protein encoded by esa1 mutant alleles was tested in standard liquid HAT assays using either equivalent or excess amounts of mutant compared to wild-type protein, no activity was detected (Fig. 2C and data not shown) at any temperature tested from 1 to 37°C.
FIG. 3.
esa1 mutants are temperature sensitive. Temperature sensitivity was observed by plating wild-type ESA1 (LPY3498) and esa1 mutant (esa1-L254P [LPY3500], esa1-L327S [LPY3430], and esa1-414 [LPY3291]) strains in fivefold serial dilutions at 28°C (left) and 37°C (right). All three alleles are recessive mutations.
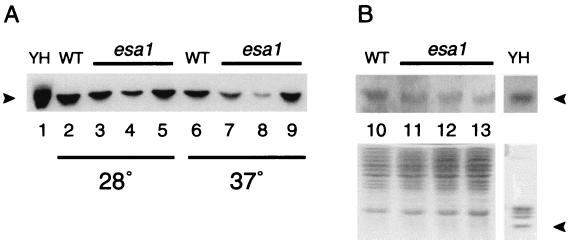
To determine whether the loss of HAT activity in vitro correlated with changes in histone acetylation in vivo, we assayed protein extracts prepared from mutant and control strains by immunoblotting. In particular, we used an antiserum specific for the acetylated Lys5 residue of histone H4, the preferred site for recombinant Esa1p activity, to compare relative levels of acetylation of wild-type and mutant strains at permissive and restrictive temperatures (Fig. 4). Each of the three esa1 mutants had detectable levels of the acetylated isoform at the permissive temperature, although there were modest reductions relative to the wild-type control. Under restrictive conditions, each of the mutant strains had decreased levels of acetylation, with the most severely affected mutant, the esa1-414 strain, showing the greatest reduction. This reduction was not simply due to loss of viability, because extracts prepared from a dam1 temperature-sensitive mutant that also suffers significant loss of viability under these conditions (25a) had wild-type levels of Lys5 acetylation (data not shown). The change in Lys5-acetylated H4 does not simply reflect a decrease in cellular pools of H4. When the same samples of wild-type and mutant proteins prepared at 37°C were probed with an antibody to evaluate total histones, levels of H4-immunoreactive material were comparable. Although this loss of Lys5 acetylation is correlated with loss of viability of the esa1 mutant strains, it is likely that other sites on H4, other histone targets, or the cumulative effects of loss of acetylation on all of Esa1p’s targets together contribute to the essential function of Esa1p. These possibilities may be distinguished in future studies, but it is clear from Fig. 2 and 4 that the esa1 temperature-sensitive alleles encode proteins that are defective for HAT activity both in vitro and in vivo.
FIG. 4.
esa1 mutants display conditional decreases in the level of histone H4-Lys5 acetylation in vivo. Whole-cell lysates from wild-type (WT; lanes 2, 6, and 10) or esa1 temperature-sensitive (lanes 3 to 5, 7 to 9, and 11 to 13) strains grown at 28°C (lanes 2 to 5) or 37°C (lanes 6 to 13) were separated on an SDS–18% polyacrylamide gel and either transferred to nylon and probed with an antiserum directed against Ly5-acetylated H4 (A) or probed with a control antiserum directed against histones (B; upper panel shows H4 immunoreactive band) or stained with Coomassie brilliant Blue (B, lower panel). Purified yeast histones (YH; approximately 6 μg; lane 1) were run in parallel. esa1-L254P (LPY3500; lanes 3, 7, and 11), esa1-414 (LPY3291; lanes 4, 8, and 12), and esa1-L327S (LPY3430; lanes 5, 9, and 13) strain displayed decreased Lys5-acetylated immunoreactivity at the nonpermissive temperature of 37°C. Arrowheads denote migration of histone H4.
esa1 mutant strains are defective in cell cycle control.
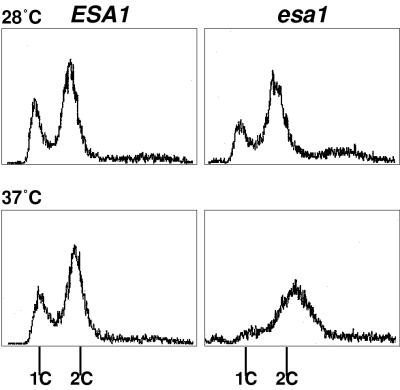
To explore further the nature of the striking loss of viability that correlates with loss of HAT activity, we used a variety of cell biological techniques to compare wild-type ESA1 and esa1 temperature-sensitive strains. Mutant and wild-type cells were first grown at the permissive temperature and then split so that a portion of each was shifted to the restrictive temperature. At multiple time points after the shift to 37°C, we compared the cells for morphology and DNA content. The mutant cells became distinctly blocked in the cell cycle at the restrictive temperature. As shown in Fig. 5, 4 h after the temperature shift, ESA1 cells were dividing normally and two peaks, indicative of G1 and G2/M positions in the cycle, were easily distinguished by flow cytometric analysis. In contrast, the mutant cells stopped dividing after being shifted to 37°C and accumulated at G2/M. This terminal arrest phenotype was observed for every esa1 temperature-sensitive strain examined, although the time required to achieve the arrest varies between alleles (data not shown). Because the mutant cells arrest with G2/M DNA content, loss of ESA1 function apparently does not interfere with DNA synthesis, but the esa1 mutants are unable to proceed through mitosis and cytokinesis.
FIG. 5.
esa1 temperature-sensitive mutants arrest in the G2/M phase of the cell cycle after DNA replication. Wild-type ESA1 (LPY3498) (left) and mutant esa1-L254P (LPY3500) (right) cells were incubated at 28 and 37°C for 4 h and analyzed for DNA content by flow cytometry. The x and y axes represent relative fluorescence intensity and number of cells, respectively. esa1 mutants arrested with G2/M DNA content after DNA replication at the restrictive temperature of 37°C. This mutant phenotype was also observed for esa1-L327S, esa1-414, and four other temperature-sensitive alleles tested (data not shown). The time to reach the arrest varied somewhat depending on the allele of ESA1.
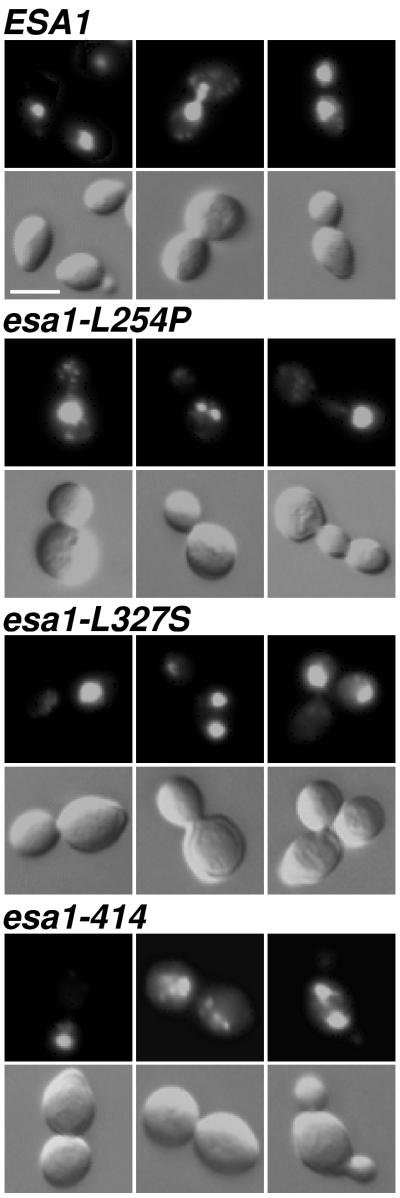
To evaluate the morphology of the mutant arrest, cells from the wild-type strain and the three mutants shown in Fig. 3 were fixed and stained with DAPI to visualize DNA (Fig. 6). The wild-type ESA1 cells were observed at all positions in the cell cycle, with normal populations of 37% unbudded, 27% small-budded, and 36% large-budded cells. Apparently normal mitotic cells, in which DAPI-staining material extended between mother and daughter buds, were seen in only 8% of the mutant cells examined. More than 70% of the mutant cells were arrested with a large bud. Of these, 75% contained a single region of DAPI-stained chromatin; the remaining 25% contained what appeared to be fragmented chromatin. Interestingly, the chromatin could be in either the mother or the daughter bud (data not shown) as determined by Calcofluor staining (51), indicating that normal nuclear migration occurring early in mitosis (45, 74) was not inhibited. Variants of the large-bud arrested mutant cells, in which an additional bud has formed, were also observed as <10% of the population (Fig. 6, right panels for mutants). In these cases, DAPI staining was sometimes seen in more than one of the buds, but one bud usually remained unstained. Together, the large-budded and variant large-budded forms comprised approximately 80% of the esa1 cells at the restrictive temperature. For all of the mutants, increased incubation time under restrictive conditions led to increased cell volume. We interpret the continued growth of the arrested mutant cells to indicate that Esa1p is not required for cell growth but instead is necessary for cell cycle progression. The observation that the mutant cells undergo cell cycle arrest with replicated DNA after the shift to the restrictive temperature reveals that Esa1p function may be required during G2 or mitosis.
FIG. 6.
esa1 mutants display improper DNA segregation. ESA1 (LPY3498), esa1-L254P (LPY3500), and esa1-414 (LPY3291) cells were incubated at 37°C for 4 h, and esa1-L327S (LPY3430) cells were incubated at 37°C for 8 h, fixed, and stained with DAPI. esa1 mutant cells were predominantly large budded as visualized by Nomarski optics (left and center columns, bottom), with some triple-budded cells (right column, bottom). DAPI staining indicates that esa1 mutant cells either failed to segregate or abnormally segregated their chromatin in comparison to wild-type cells. Bar, 5 μm.
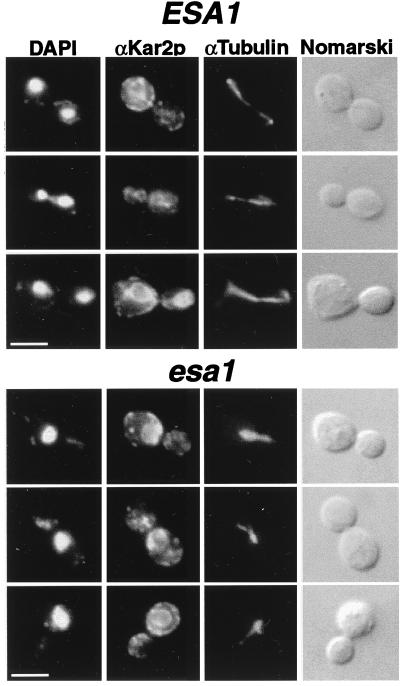
To establish whether the failure was in mitosis, we examined cells by indirect immunofluorescence microscopy using antitubulin (27, 28) and anti-Kar2p (54) staining as markers for mitotic and cytoplasmic microtubules and the nuclear envelope-endoplasmic reticulum (ER) membranes (Fig. 7). In wild-type cells at both temperatures and in esa1 cells at the permissive temperature, the staining appeared normal and consistent with the cell’s position in the cell cycle as inferred by DAPI staining. At the restrictive temperature, among the more than 80% abnormal cells in the esa1 population shown here, the anti-Kar2p staining encircled the DAPI-stained chromatin (Fig. 7) and occasionally extended in fingers throughout the cytoplasm and even into the bud of the cell pair lacking DAPI staining. This staining pattern is consistent with that expected for the nuclear envelope and ER (54). The microtubular structures in the mutant cells were generally oriented with the long axis of the cell and usually had one region of more intense staining within the DAPI-stained nucleus, along with somewhat lighter, occasionally forked structures extended into the other bud of the cell. We interpret these to be spindle and cytoplasmic microtubules, respectively, that are comparable to microtubule structures in other mutants with G2/M arrests (18). For wild-type large-budded cells, the nucleus had migrated between mother and daughter buds, with long mitotic spindles stretched between them, and occasionally short bundles of cytoplasmic microtubules were visible extended from the cytoplasmic faces of the spindle pole bodies. The mutant cells were never observed to have long spindles, consistent with the failure of the nucleus to migrate normally between mother and daughter buds.
FIG. 7.
esa1 mutants disrupt mitotic cell cycle progression. ESA1 (LPY3498) and esa1-L254P (LPY3500) cells were incubated at 37°C for 4 h and prepared for indirect immunofluorescence microscopy. esa1 mutant cells exhibited aberrant nuclear and short spindle morphologies at the restrictive temperature. As visualized by double-label immunofluorescence microscopy, antisera directed against Kar2p detected the nuclear envelope and ER and α-tubulin detected microtubules. Both the nuclear envelope and the microtubules extended only partially through the bud neck in comparison to the wild type but coincided with the DAPI-stained region. Bars represent 5 μm.
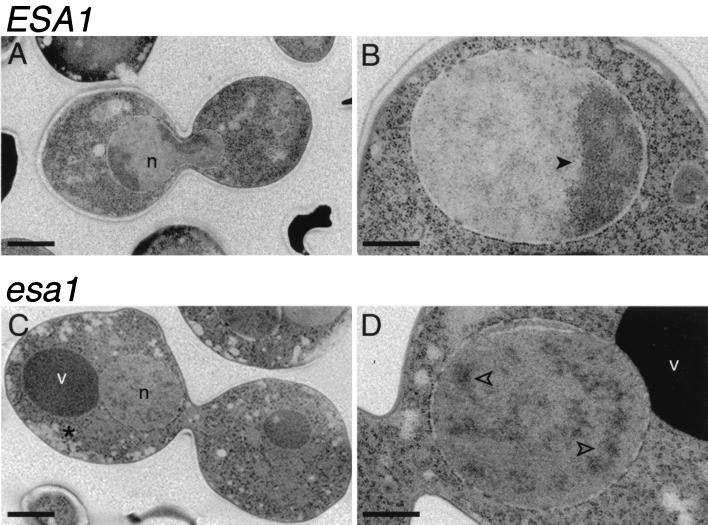
To evaluate the morphology of arrested esa1 mutant cells with greater resolution, samples from a temperature shift experiment were prepared for electron microscopy using a rapid-freeze protocol (44). Clear differences were observed in thin sections of the wild-type and mutant cells (Fig. 8). Images of large-budded cells are shown here for comparison, although they represent only about 36% of the ESA1 population. The cytoplasm of the esa1 mutant cells was consistently observed to be more highly vesiculated than that of wild-type cells (Fig. 8C), but striking differences were also observed in the substructure of the nuclei. In wild-type cells, the nucleus is of relatively uniform density, with increased density found restricted to the fairly compact heterochromatic nucleolus (Fig. 8A and B). In the dividing cell, this more electron-dense material was seen at both poles of the lobed nucleus extending between mother and daughter cells. However, in the esa1 cells, the nucleus was generally restricted to one side of the mother-daughter pair (Fig. 8C and D), confirming our results with light microscopy. The mitotic spindles appeared structurally normal, including short bipolar spindles with connecting microtubules and normally nucleated cytoplasmic microtubules. However, in some instances, the spindle was oriented aberrantly and was even observed to lie horizontally across the bud neck rather than extending through it. The nuclei themselves were also distinct in that the nucleolus was often exaggerated, and electron-dense material was consistently scattered throughout the nucleus (Fig. 8D) rather than remaining restricted to the nucleolar compartment. This dispersal was detected with various degrees of severity in 96% of the 65 mutant nuclei examined, but it was observed in only <1% of wild-type nuclei and has not been seen in other ultrastructural analyses of comparably prepared cdc mutants that arrest with large buds (43, 70a).
FIG. 8.
esa1 mutants have aberrant ultrastructural morphology. Wild-type ESA1 (LPY3498) (A and B) and esa1-L254P (LPY3500) (C and D) cells were incubated at 37°C for 4 h and prepared by rapid freezing for electron microscopy. In wild-type cells, the nucleolar electron-dense material was at both poles of the dividing nucleus (n) (A) or in a crescent shape in a nondividing cell (B, closed arrowhead). However, in the esa1 mutant (C and D, open arrowheads), the electron-dense material was dispersed throughout the nucleus. The majority of the mutant large-budded cells showed the nucleus on one side of the bud neck, in contrast to wild-type cells, where the nucleus extended equally through the bud neck. The cytoplasm of the esa1 mutant (C, asterisk) was also more highly vesiculated than in wild-type cells. Each panel shows a different cell, and scale bars shown represent 1 μm (A and C) and 0.5 μm (B and D). v, vacuoles.
Considered together, the enzymatic and cell biological analyses of the esa1 mutants reveal important consequences for loss of ESA1 function. We were especially interested in the defect in cell cycle progression because previous research demonstrated that the acetylatable amino-terminal H3 and H4 tails can be essential for cell cycle control. In particular, if cells are depleted of normal H3 and H4 histones and are left only with H3 and H4 lacking amino-terminal tails (36), they rapidly assume an arrest comparable to that which we observed in esa1 cells. In considering the mechanisms leading to the arrest, it was relevant to consider the role of checkpoint functions (reviewed in references 16 and 68).
The esa1 cell cycle arrest is dependent on the RAD9 checkpoint gene.
Cell cycle checkpoint functions are ordinarily defined as those functions that perceive either damaged structures or failure to complete processes necessary for normal fidelity of cell duplication (16, 68). Frequently, cell cycle arrests are triggered by checkpoint functions allowing time for repair or completion of the lagging step. We wanted to determine whether the cell cycle arrest observed in the esa1 mutants was dependent on checkpoint functions. It seemed possible that, depending on the precise role(s) of the Esa1p HAT activity, the cells could trigger checkpoints registering failure to replicate chromatin or failure in cell cycle transcriptional control. It should be noted that DNA checkpoints defined to date, such as that controlled by the RAD9 gene (39, 67), appear to register DNA damage, not necessarily chromatin damage. Further, the esa1 cell cycle arrest observed is distinct from that ordinarily seen at checkpoints in that there is a substantial loss of cell viability for the esa1 mutants, whereas in classically defined checkpoint arrests, viability remains high upon return to permissive conditions.
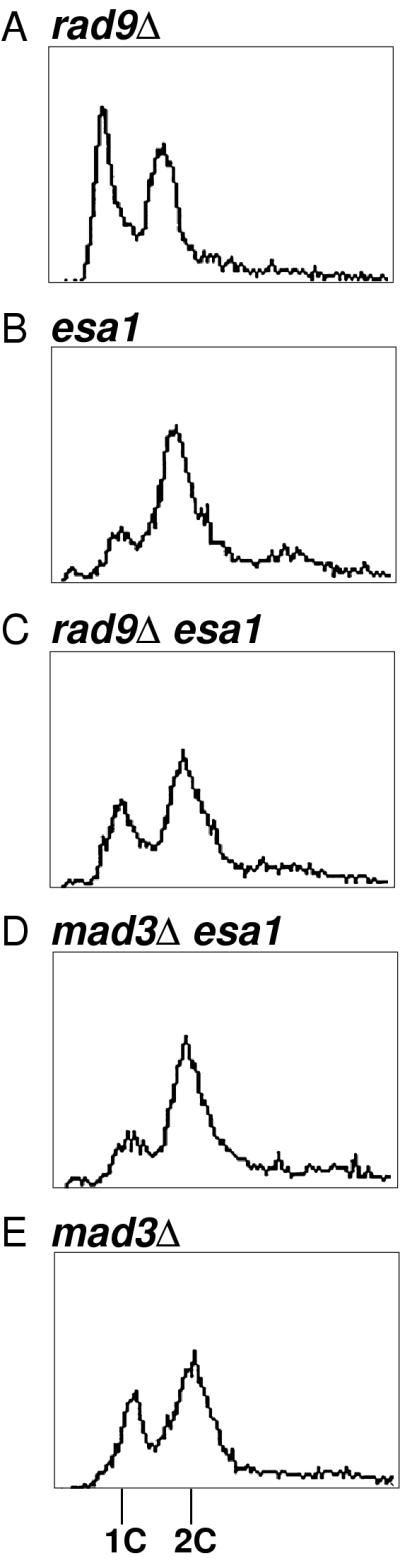
To assess the role of checkpoint control in the esa1 cell cycle arrest, we constructed isogenic rad9Δ esa1 double- and single-mutant strains for temperature shift experiments as described above. At 4 h after the shift to the restrictive temperature, the rad9Δ esa1 cells had severe losses in viability comparable to the esa1 single mutants, whereas the rad9Δ cells remained fully viable (data not shown). Flow cytometry also revealed that the rad9Δ single-mutant strain was unaffected by growth at 37°C (Fig. 9A), whereas the esa1 strain arrested with 2C DNA content (Fig. 9B). The rad9Δ esa1 double-mutant strain, however, no longer arrested with a simple 2C DNA content (Fig. 9C). Instead, both 1C and 2C peaks were apparent, indicative of cells in both G1 and G2/M phases of the cell cycle. Budding indices confirmed and extended the flow cytometry data. Rather than arresting with >80% large-budded and abnormal cells, the rad9Δ esa1 strain was 46% unbudded, 25% small budded, and 26% large budded. In this population, 64% of the cells were clearly abnormal, often appearing exaggerated in size.
FIG. 9.
The esa1 mutant G2/M arrest is dependent on the RAD9 checkpoint gene. (A) rad9Δ (LPY3784), (B) esa1-L254P (LPY3785), (C) rad9Δ esa1-L254P (LPY3780), (D) mad3Δ esa1-L254P (LPY4223), and (E) mad3 (LPY4222) mutant cells were incubated at 37°C and then analyzed for DNA content by flow cytometry. The x and y axes represent relative fluorescence intensity and number of cells, respectively. The esa1-L254P and mad3Δ esa1-L254P mutants arrested after DNA replication with approximately 80% of the cells containing 2C DNA content, whereas the rad9Δ and mad3 cells have normal DNA content for dividing cells. The rad9Δ esa1-L254P double mutant had both 1C and 2C peaks, indicating that esa1-L254P cell cycle arrest is dependent on RAD9 but not on MAD3.
To determine if the cell cycle arrest was dependent on the mitotic checkpoint, we constructed an esa1 strain also deleted for the MAD3 gene (34, 70). No change was observed in cell cycle arrest at the restrictive temperature assayed by flow cytometry compared to the esa1 mutant alone (Fig. 9D and E). Therefore, the cell cycle arrest observed in the esa1 mutant strain is dependent on the checkpoint function of RAD9 but not on the MAD3 mitotic checkpoint.
DISCUSSION
Histone acetylation is a posttranslational modification that historically has been correlated with both assembly and distinct transcriptional states of chromatin (8, 21). Here we report identification of the yeast ESA1 gene that is essential for cell viability and encodes a protein with HAT activity. The enzymatic activity of Esa1p has a unique pattern of substrate specificity among yeast HATs, and intriguingly, the essential role appears tied to progression through the cell cycle after DNA replication is complete. ESA1 is one of a family of highly conserved genes which were first identified for their transcriptional regulatory functions in yeast, Drosophila, and humans (6, 15, 25, 26, 53). It is likely that at least part of Esa1p’s essential functions are transcriptional since it is the catalytic component of NuA4, a large, multisubunit complex that can activate transcription (14, 61). These results therefore connect a specific HAT to cell cycle progression and raise the possibility that targets of HAT activity feed into checkpoint controls or contribute to their regulation through modulation of downstream transcriptional events.
Esa1p acetylates histones with a distinct pattern of substrate specificity.
Both recombinant Esa1p and native protein purified in the NuA4 complex (14) have significant HAT activity directed toward histone H4. With synthetic peptides or purified histones as substrates, Esa1p can acetylate each of the four lysines in the N-terminal tail that are most significant for H4 function (this report and references 14 and 57). The pronounced preference toward Lys5 of H4 is consistent with a role in the deposition of newly synthesized histones (8), although the comparable site is acetylated in the transcriptionally active macronuclei of rapidly growing Tetrahymena (12). Esa1p’s pattern of acetylation observed on H3 is also seen for HAT activity of Gcn5p and TAFII250 (32, 41), both transcriptional activators. Although H2A’s acetylation has been less thoroughly studied, it can be acetylated in vivo in animal cells (46) at the same position as we observe for Esa1p’s activity in vitro. These same specificities have also been observed independently and recently reported, including using purified Drosophila H4 and chicken H2A histones as substrates (57). Further, a catalytically active recombinant fragment of the human protein Tip60 acetylates H4, H3, and H2A free histones (72), revealing that not only are the SAS family proteins highly related, but the specificities of their activities may be as well. Although in vitro specificities of HATs should be interpreted cautiously (see, e.g., references 48 and 75), our data along with those of others (57, 72) do identify Esa1p as a candidate H4 HAT, distinguished comparatively by its targets, of which Lys5 of H4 appears relevant in vivo (Fig. 4). Because the Lys5-acetylated isoform of H4 is also depleted in gcn5Δ mutants (75), there likely must be other sites and other substrates that, in concert, contribute to the essential nature of ESA1’s function. Like gcn5 strains which display impaired HAT activity in vivo when residues either within the A box or outside this region are mutated (33, 65), we recovered esa1 mutants with lesions within the A box (esa1-L327S) as well as outside it (esa1-L254P and esa1-414) that exhibit decreased H4 Lys5 acetylation in vivo. Interestingly, the esa1-414 mutant shows the strongest decrease, suggesting a role for the carboxy terminus in maintaining a functional complex (14).
The Esa1p-containing NuA4 complex, identified for its acetylation of nucleosomal histones H2A and H4, is both physically and genetically distinct from the other large HAT complexes such as Ada and SAGA (14, 19) that appear most active toward H3. The NuA4 complex in esa1-414 strains retains HAT activity at permissive temperature, but this activity is lost upon a 4-h shift to the restrictive temperature. It is possible that H2A and H4 are relevant substrates in vivo of Esa1p and that the H3 acetylation results from relaxed stringency in vitro with the recombinant protein. Reduced site specificity was observed for recombinant Hat1p in the absence of its binding partner, the Rba48-like protein Hat2, which appears to function as a specificity factor (48). In contrast, broader activity in vivo than originally predicted has recently been suggested for Gcn5p (75). Thus, Esa1p may acetylate H2A and H4 in the context of the NuA4 complex but may act on H3 as the catalytic subunit of a different complex, influenced by different specificity factors. Determining whether Esa1p is catalytically active in more than one complex and whether it has distinct substrate specificities (including nonhistone proteins) depending on its cellular localization or its targeted genomic loci will be critical to understanding its function. Previously proposed models predict that HAT complexes may be regulated through subunit exchange (55). Indeed, in early studies, a single Tetrahymena HAT activity was shown to have both deposition- and transcription-related activities (11). In light of the current molecular identification of HATs, it would be revealing to know the detailed molecular composition of that potentially bifunctional activity.
Connecting HATs and checkpoint control through ESA1’s role in cell cycle progression.
The cell cycle arrest of temperature-sensitive esa1 mutant cells is rapid and is accompanied by loss of viability. These effects suggest that appropriate histone acetylation is critical for cell cycle progression. Interestingly, recent results indicate that gcn5 mutants, which ordinarily have only modest growth phenotypes, die in combination with subsets of H3 and H4 mutants and also suffer G2/M effects (75). Previous studies revealed that mutations of H4 N-terminal lysines can lead to G2/M delays that are RAD9 dependent (39). Similarly, deletion of N-terminal tails of either H3 or H4 also delays the cell cycle, and deletion of both kills cells in G2/M (36). Our results provide an explanation for these observations by demonstrating that modification of N-terminal histone tails may be essential and may couple acetylation events to cell cycle control, independent of structural changes resulting from mutation of the histones. Because the N-terminal tail of either H3 or H4 alone can support viability (36), it is possible that Esa1p may acetylate both histones in vivo and that these particular acetylation patterns are important for chromatin structure. It is also possible that Esa1p may modify some key nonhistone target(s). Alternatively, Esa1p’s HAT activity may function to regulate transcription of a gene or group of genes, which are themselves required for proper cell cycle control, although ESA1 itself is not transcriptionally regulated through the cell cycle (13). The closely related genes SAS2 and SAS3 can influence transcriptional regulation of the silent mating-type loci and telomeric reporter genes (15, 53). Preliminary studies indicate that ESA1 does not contribute to regulation of these loci, furthering the likelihood that its functions are distinct.
Insight into the nature of the esa1 cell cycle arrest comes from flow cytometric and microscopic analyses. These studies reveal that the arrest occurs after the cells have completed DNA synthesis but before the successful completion of mitosis. Compared to overall bud and cell size, the migration of the nucleus and extension of the mitotic spindle appear abnormal. Ultrastructural analysis suggests that there may be irregularities in the electron-dense chromatin itself. The significance of this observation is not yet understood. It remains possible that Esa1p functions in multiple capacities, some directly related to chromatin structure and assembly in mitosis and others transcriptionally focused.
The observation that the esa1 temperature-sensitive cell cycle arrest is dependent on the RAD9 checkpoint function but not on the MAD3 checkpoint function deepens the potential significance of understanding ESA1’s role in the cell cycle. RAD9 participates in sensing DNA damage and the execution of functions necessary for repair. Our results raise the possibility that incompletely or inappropriately acetylated chromatin may be recognized as damage, although the precise mechanisms for monitoring such damage remain to be defined. It is formally possible that the very end stages of DNA replication are not completed in the esa1 mutants, to degrees not detectable at the limits of flow cytometric resolution, but we prefer the interpretation that the arrest occurs after S phase because of the nuclear positioning and mitotic spindle phenotypes. The high loss of viability observed at the arrest distinguishes it from normal checkpoint-regulated processes. Perhaps the damage in the esa1 mutant cells is so severe as to be irreparable. Alternatively, Esa1p may itself contribute to checkpoint functions akin to the essential RAD53 gene (2, 68), potentially by activating transcriptional targets required at cell cycle transition points, thereby regulating the synthesis of factors required for return to normal cell cycle progression.
Essential roles for essential HATs.
GCN5 and HAT1 were the first yeast genes determined to encode transcription- and deposition-related HAT activities (9, 29, 48). Surprisingly, strains in which only these genes are deleted have modest growth phenotypes under special conditions or no detectable cellular phenotypes. The TFIID component TAFII250, an essential factor of the basal transcriptional machinery (50, 52), has intrinsic HAT activity (41), although whether this is its essential function is not known. It is interesting that mutants with temperature-sensitive alleles of the yeast TAFII250 gene homologue, TAF145 also arrest the cell cycle at their restrictive temperature (63). This arrest appears to be due to loss of transcriptional activation of G1 cyclins and not loss of global RNA polymerase II-directed transcription (64), although whether these mutant alleles are specifically defective in HAT activity remains to be determined. These results, considered together with those for the esa1 cell cycle arrest, raise the possibility that gene-specific or global chromatin modification provides an additional control for transcriptional pathways such as those activated by DNA damage or other cell cycle perturbations.
Essential roles for other HATs are revealed by the male-specific lethality resulting from mutation of the closely related Drosophila mof gene. In this case, the increased transcription of the X chromosome that is correlated with H4 Lys16 acetylation fails, and death results because dosage compensation is not achieved (25). Female flies with mof mutations appear completely normal (25), suggesting that mof may encode a sex-specific HAT or that there are sex-specific requirements for histone acetylation.
Whether the vertebrate MYST family genes are essential is not yet known. Among the human homologues (6), there may be genes encoding both essential and nonessential functions, as do the yeast genes. By comparison, it has been recently reported that the murine p300 and CBP genes are both essential, and loss of either gene results in multiple developmental defects and early embryonic lethality (73). Although it has not been possible to separate p300’s transcriptional activation properties from its HAT activity (38), CBP and p300’s essential roles are likely to be transcriptional ones, mechanistically dependent on HAT activity. CBP is known to interact with p53 to potentiate the transcriptional activation functions of p53 in inhibiting entry into G1 (22, 35). Furthermore, p53 is also activated in response to DNA damage (for a review see reference 30). Thus, regulated transcriptional activities of HAT complexes may participate at several levels in coordinating cell cycle regulation and responses to DNA or chromatin damage.
In considering the contribution of chromatin modification to biological regulation, it is clear that histone acetylation can facilitate fine-tuning of transcription as well as more profound changes in chromatin structure. By analogy to Gcn5p (20), Esa1p may participate catalytically in more than one complex. Whether its catalytic activity is strictly transcription related (14, 61), or whether it has other roles, even in chromatin assembly, will be an important issue to address. The fact that the three closely related genes SAS2, SAS3, and ESA1 appear to have distinct, nonredundant functions in unicellular yeast implies that the homologous genes in vertebrates may also have distinct functions. For example, it is not known how loss or misregulation of Tip60 may contribute to human immunodeficiency virus type 1 Tat transcription (26) or how MOZ, when fused to CBP, may lead to acute myeloid leukemia (6). Understanding the specific functions of these proteins, and how they promote changes in chromatin structure and transcriptional control, will ultimately depend on identifying their in vivo targets and the complexes in which they act.
ACKNOWLEDGMENTS
A.S.C. and J.E.L. contributed equally to this work.
We thank our colleagues M. Parthun, D. Gottschling, M. Jones, J. Pilon, P. Laybourn, J. Rine, M. Rose, S. Roth, F. Solomon, R. Sternglanz, and T. Weinert for providing reagents and advice and D. Allis and J. Lucchesi for communicating results prior to publication. D. Wuttke and W. Bishop provided invaluable advice and assistance with peptide synthesis. T. Giddings made the electron microscopic analysis possible. We thank R. Cook and his colleagues at the Baylor Protein Chemistry Core Facility for their excellent service. We thank K. Grischuk, S. Mohr, and M. Winey for assistance with microscopy and flow cytometry and Y. Han for assistance with sequencing the esa1 alleles. We appreciate comments on the manuscript by L. Freeman-Cook, J. Heilig, M. Klymkowsky, L. Leinwand, J. Sherman, E. Stone, and M. Winey.
This work was initiated with support from the National Science Foundation and the University of Colorado Graduate School Council for Research and Creative Work and has been continued with research and training grant funding from the National Institutes of Health.
REFERENCES
- 1.Aasland R, Stewart A F. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein, HP1. Nucleic Acids Res. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Freiberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Baudin A, Ozier-Kalogeropoulos O, Denouel A, LaCroute F, Cullen C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;14:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrow J, Stanton V P, Andresen J M, Becher R, Behm F G, Chaganti R S K, Civin C I, Disteche C, Dube I, Frischauf A M, Horsman D, Mitelman F, Watmore A E, Housman D E. The t(8;16)(p11;p13) translocation of acute myeloid leukemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 9.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a transcriptional co-activator linking gene expression to histone acetylation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 11.Chicoine L G, Richman R, Cook R G, Gorovsky M A, Allis C D. A single histone acetyltransferase from Tetrahymena macronuclei catalyzes deposition-related acetylation of free histones and transcription-related acetylation of nucleosomal histones. J Cell Biol. 1987;105:127–135. doi: 10.1083/jcb.105.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chicoine L G, Schulman I G, Richman R, Cook R G, Allis C D. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. J Biol Chem. 1986;261:1071–1076. [PubMed] [Google Scholar]
- 13.Cho R J, Campbell M J, Winzler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 14.Côté, J., R. T. Utley, S. Allard, A. Clarke, P. Grant, J. Savard, L. Pillus, and J. L. Workman. The NuA4 transcription activation/histone H4 acetyltransferase complex contains the essential Esa1 protein as the catalytic subunit and the essential ATM-related cofactor Tra1p. Submitted for publication.
- 15.Ehrenhofer-Murray A, Rivier D, Rine J. The role of Sas2, an acetyltransferase homolog, in silencing and ORC function in Saccharomyces cerevisiae. Genetics. 1997;145:923–934. doi: 10.1093/genetics/145.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 17.Georgakopoulos T, Thieros G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 19.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 20.Grant P A, Sterner D E, Duggan L J, Workman J L, Berger S L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 21.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 22.Gu W, Shi X-L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 24.Hendzel M J, Wei Y, Mancini M A, VanHooser A, Ranalli T, Brinkley B R, Bazett-Jones D O, Allis C D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 25.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi J C. mof, a putative acetyltransferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Jones, M. Personal communication.
- 26.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 27.Kilmartin J V, Adams A E M. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilmartin J V, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new non-secreting cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–25677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 30.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 31.Koonin E V, Zhou S, Lucchesi J C. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo M-H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 33.Kuo M-H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of Gcn5p is required for the activation of targeted genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R, Murray A W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–31. doi: 10.1016/0092-8674(81)90015-5. . (Erratum, 79: following 388, 1994.) [DOI] [PubMed] [Google Scholar]
- 35.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 36.Ling X, Harkness T A A, Schultz M G, Fisher-Adams G, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 37.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Balbás M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Megee P C, Morgan B A, Smith M M. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 40.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 42.Neuwald A F, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 43.Ogryzko V O, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 44.O’Toole E T, Mastronarde D N, Giddings T H, Winey M, Burke D J, McIntosh J R. Three-dimensional analysis and ultrastructural design of mitotic spindles from the cdc20 mutant of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1–11. doi: 10.1091/mbc.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer R E, Koval M, Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989;109:3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pantazis P, Bonner W M. Quantitative determination of histone modification. H2A acetylation and phosphorylation. J Biol Chem. 1981;256:4669–4675. [PubMed] [Google Scholar]
- 47.Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- 48.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 49.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 50.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pringle J R, Preston R A, Adams A E, Stearns T, Drubin D G, Haarer B K, Jones E W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 52.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 53.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 54.Rose M D, Mistra L M, Vogel J P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 55.Roth S Y, Allis C D. The subunit exchange model of histone acetylation: HAT modules and ‘transient’ nucleosomes. Trends Cell Biol. 1996;6:371–375. doi: 10.1016/0962-8924(96)20032-7. [DOI] [PubMed] [Google Scholar]
- 56.Schneppenheim R, Budde U, Dahlmann N, Rautenberg P. Luminography—a new, highly sensitive visualization method for electrophoresis. Electrophoresis. 1991;12:367–372. doi: 10.1002/elps.1150120508. [DOI] [PubMed] [Google Scholar]
- 57.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sobel R E, Cook R G, Allis C D. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 59.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 60.Turner B M. Histone acetylation as an epigenetic determinant of long-term transcriptional regulation. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Utley R T, Ikeda K, Grant P A, Côté J, Steger D L, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 62.Verrault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1997;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 63.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 64.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 65.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiler K S, Wakimoto B T. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 67.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 68.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 69.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wells W A, Murray A W. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70a.Winey, M., and T. Giddings. Personal communication.
- 71.Wolffe A. Chromatin structure and function. New York, N.Y: Academic Press; 1992. [Google Scholar]
- 72.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 73.Yao T-P, Oh S P, Fuchs M, Zhou N-D, Ch’ing L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–371. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 74.Yeh E, Skibbens R V, Cheng J W, Salmon E D, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang W, Bone J R, Edmonson D G, Turner B M, Roth S Y. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]