Abstract
CYP2D6 genotype is increasingly being integrated into practice to guide prescribing of certain medications. The CYP2D6 drug metabolizing enzyme is susceptible to inhibition by concomitant drugs, which can lead to a clinical phenotype that is different from the genotype-based phenotype, a process referred to as phenoconversion. Phenoconversion is highly prevalent but not widely integrated into practice because of either limited experience on how to integrate or lack of knowledge that it has occurred. We built a calculator tool to help clinicians integrate a standardized method of assessing CYP2D6 phenoconversion into practice. During tool-building, we identified several clinical factors that need to be considered when implementing CYP2D6 phenoconversion into clinical practice. This tutorial shares the steps that the University of Florida Health Precision Medicine Program took to build the calculator tool and identified clinical factors to consider when implementing CYP2D6 phenoconversion in clinical practice.
Keywords: Pharmacogenetics, CYP, Personalized Medicine, Drug-Drug Interactions
Introduction
Cytochrome P450 2D6 isoenzyme (CYP2D6) is a drug metabolizing enzyme responsible for metabolizing approximately 25% of commonly prescribed drugs (e.g., opioids, antidepressants).(1, 2) The CYP2D6 gene is highly polymorphic, and genetic variation confers alleles that have no function, decreased function, or are multiplied, which leads to enzyme activity ranging from absent to greater than normal.(3) Enzyme activity is commonly categorized into phenotypes, which include poor metabolism (PM), intermediate metabolism (IM), normal metabolism (NM), and ultra-rapid metabolism (UM).(4) CYP2D6 is translated from genotype to a genotype-based phenotype using an activity score system.(5) This system assigns individual CYP2D6 alleles an activity value that are added together to provide a genotype-based activity score, which is then translated to a phenotype based on score cut offs.(6)
An individual’s CYP2D6 genotype is increasingly being integrated into clinical care to guide medication prescribing.(7) Results are returned in a laboratory report and include the genotype, often a predicted phenotype, and possibly the activity score. Clinical laboratories may use different terms or phenotypes to describe the enzyme function predicted from the genetic test, although the Clinical Pharmacogenetics Implementation Consortium (CPIC) has recommended standardized terms for laboratory reporting.(4) Laboratories may also differ in how they determine phenotype from activity score as changes were just recently proposed.(8)
CYP2D6 is susceptible to clinically relevant enzyme inhibition (e.g., by drug interactions), which can lead to a clinical phenotype that does not match the genotype-based phenotype, a process called phenoconversion.(9–14) Laboratories typically do not integrate concomitant medications individuals are taking, nor do they typically have access to the individual’s full medication list. For example, an individual who has a genotype-based NM phenotype and is taking bupropion, a strong CYP2D6 inhibitor, would phenoconvert to a PM clinical phenotype. However, the lab report would likely include only the NM phenotype and it would be up to the clinician to recognize the individual has a different clinical phenotype, secondary to drug interaction that should be acted upon.
CYP2D6 inhibitor use is highly prevalent. Specifically, five of the ten CYP2D6 inhibitors classified by the FDA as strong (i.e., bupropion, fluoxetine, paroxetine) or moderate (i.e., duloxetine, mirabegron) are in the 2021 ‘Top 300 Drugs’ list.(15) Recent studies from our group and others indicate that in the clinical settings where CYP2D6 genotype might be used, approximately 20–30% of individuals are also taking an enzyme inhibitor that leads to phenoconversion.(16–19) Integrating CYP2D6 phenoconversion into clinical practice is not yet standard of care as a majority of clinicians have limited knowledge of the drugs that might cause phenoconversion or if knowledgeable, experience on how to integrate into practice, and no tool exists to facilitate integration. At present, phenoconversion is most often integrated into practice manually within clinical consult notes placed by pharmacists into the electronic health record at University of Florida (UF) Health.
UF Health is a site for the upcoming IGNITE II network’s national trial, A Depression and Opioid Pragmatic Trial of Pharmacogenetics (ADOPT-PGx), for which we expect to enroll over 1,000 patients across multiple sites within UF Health and approximately 4,500 patients nationally.(20) The trials will enroll patients with acute pain, chronic pain, or depression for whom CYP2D6 and CYP2C19 test results may be useful to guide opioid or antidepressant therapy. Because of the pragmatic nature of the trial and common use of CYP2D6 inhibitors in the clinical practice setting, patients enrolled in this trial will not be excluded if they are on a medication known to inhibit CYP2D6 and potentially cause phenoconversion. As multiple sites are enrolling for this trial, consistency in phenoconversion implementation is important. Therefore, we needed to create a standardized process to integrate phenoconversion accurately and efficiently into practice to facilitate consistent assignment of clinical phenotypes across clinical trial sites. We opted to create a web-based calculator tool, intended to be used by clinicians to easily integrate CYP2D6 genotype and drug interactions to ensure the correct clinical phenotype is used when making pharmacotherapy decisions. Throughout the process of creating the calculator tool, several clinical factors were identified as important to consider when integrating phenoconversion into practice. Herein we provide a tutorial on the steps the UF Health Precision Medicine Program (PMP) took to implement a standardized method of CYP2D6 phenoconversion into clinical practice by building a CYP2D6 calculator tool.
Step 1: Determine CYP2D6 Phenoconversion Approach
We identified and reviewed three methods for calculating CYP2D6 phenoconversion (Table 1). Borges et al., proposed two methods to account for potential differences in degree of inhibition caused by CYP2D6 inhibitors based on the genotype-based activity score.(21) The first method implemented a standard multiplication factor of 0 or 0.5 based on the presence of a strong inhibitor or a moderate/weak inhibitor, respectively. The other method implemented a multiplication factor based on both the genotype-based activity score and classification of inhibitor present (e.g., 0.25 if activity score was greater than 2 with strong inhibitor present). They found both methods were able to similarly predict phenotype. Mostafa et al., adapted a method similar to the Borges et al., method that accounted for differences in genotype-based activity scores.(22) They considered individuals with a genotype-based activity score between zero and two and taking a strong or moderate inhibitor to have a clinical PM phenotype, while UMs (activity score > 2) taking a moderate or strong inhibitor were considered to have a clinical NM phenotype.
Table 1.
Identified Phenoconversion application methods
| Current Classification per FDA (23) | Inhibitors | UF Health PMP Adapted method | Reviewed methods | ||
|---|---|---|---|---|---|
| Borges et al., that used standard multiplication factors (21) | Borges et al., That used a multiplication factor dependent on genotype-based AS (21) | Mostafa, et. al., (22)a | |||
| Multiplication factor: | |||||
| Strong | paroxetine | 0 | 0 | if AS >2: 0.25 | if AS ≥ 0–2: 0 |
| if AS = 0.5–2: 0 | |||||
| if AS = 0: 1 | if AS > 2: N/A (convert to NM) | ||||
| quinidine terbinafine |
0 | 1 | 1 | ||
| bupropion fluoxetine |
0 | 0.5 | if AS ≥ 1.75: 0.5 | ||
| if AS = 0.5–1.5: 0.75 | |||||
| if AS =0: 1 | |||||
| Moderate | abiraterone cinacalcet duloxetine lorcaserin mirabegron |
0.5 | 1 | 1 | if AS ≥ 0–2: 0 |
| if AS > 2: N/A (convert to NM) | |||||
| Weak | amiodarone | 1 | 0 | if AS >2: 0.25 | 1 |
| if AS = 0.5–2: 0 | |||||
| if AS = 0: 1 | |||||
| cimetidine clobazam cobicista escitalopram fluvoxamine labetalol ritonavir vemurafenib |
1 | 0.5 | if AS ≥ 1.75: 0.5 | ||
| if AS = 0.5–1.5: 0.75 | |||||
| if AS =0: 1 | |||||
| sertraline celecoxib |
1 | 0.5 | if AS ≥ 1.75: 0.5 | ||
| if AS = 0.5–1.5: 0.75 | |||||
| if AS =0: 1 | |||||
| Not classified | venlafaxine citalopram razadone |
1 | 0.5 | if AS ≥ 1.75: 0.5 | 1 |
| if AS = 0.5–1.5: 0.75 | |||||
| if AS =0: 1 | |||||
| metoclopromide | 1 | 0 | if AS >2: 0.25 | ||
| if AS = 0.5–2: 0 | |||||
| if AS = 0: 1 | |||||
| perhexiline moclonemide flecanide |
1 | 1 | 1 | if AS ≥ 0–2: 0 | |
| if AS > 2: N/A (convert to NM) | |||||
AS: Activity Score; NM: Normal Metabolizer
If multiplication factor = 1 then it was not considered to cause an adjustment
Assessed FDA inhibitors classified as of 2017, changes between 2017 and 2020 were unable to be determined unless identified inhibitors were specifically reported (e.g., perhexiline, moclonemide, flecanide).
CPIC guidelines refer to the Borges et al., method that uses a standard multiplication factor regardless of genotype-based activity score in their guidelines, stating that for patients taking strong inhibitors the CYP2D6 activity score is adjusted to 0 and for patients taking weak or moderate inhibitors the activity score is multiplied by 0.5, with subsequent conversion to the predicted phenotype.(24) As observed in Table 1, the methods differ in their classification of inhibitors. CPIC describes that for phenotype modification, inhibitors are classified as strong, moderate, or weak based on the U.S. Food and Drug Administration’s (FDA) guidance, and others have accepted the FDA’s classifications as a standard of care.(12, 25)
As the CPIC and the FDA are considered authoritative resources, we opted to also adapt the Borges et al., method that uses a standard multiplication factor , and utilize drugs defined as strong or moderate CYP2D6 inhibitors by the FDA (Table 2).(23) The PMP adapted method is described in Figure 1.(21, 24) First, a genotype-based CYP2D6 activity score is obtained, either from the laboratory report or by manual calculation. To calculate the genotype-based activity score, the activity value of each allele must be determined.(6) Then, the activity value for the two alleles is added together, and the sum provides the genotype-based activity score. Next, a genotype-based CYP2D6 phenotype can be assigned based on the genotype-based activity score.(6) Third, the individual’s medications must be assessed to determine if the genotype-based phenotype warrants adjustment. If the individual is taking a medication classified as strong or moderate CYP2D6 inhibitor (Table 2), then the genotype-based activity score is multiplied by 0 or 0.5, respectively. If the patient is taking multiple inhibitors, the stronger inhibitor is accounted for in calculating the activity score. This multiplication step results in an adjusted CYP2D6 activity score. Next, the adjusted activity score is used to determine the clinical CYP2D6 phenotype, using the same activity score to phenotype translation. Finally, if the clinical phenotype is different from the genotype-based phenotype, then phenoconversion has occurred.
Table 2.
CYP2D6 Clinical Inhibitors
| Classification by FDAa(23) | Drug Names |
|---|---|
| Strong | bupropion, fluoxetine, paroxetine, quinidine, terbinafine |
| Moderate | abiraterone, cinacalcet, duloxetine, lorcaserin, mirabegron |
FDA last updated table 3/6/2020
Figure 1: University of Florida Health Precision Medicine Program Phenoconversion Method.
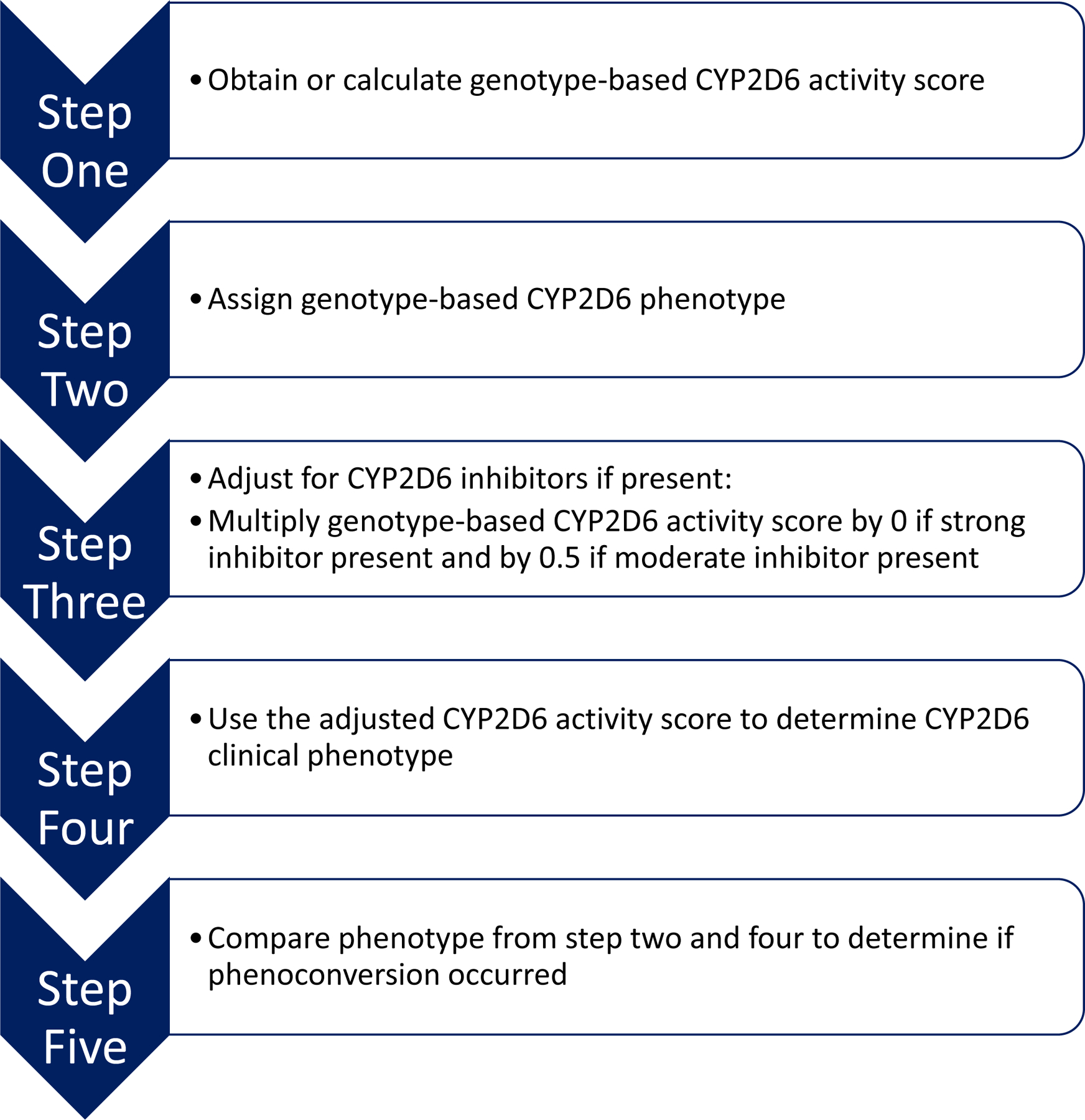
After reviewing the literature for various phenoconversion approaches, the UF Health Precision Medicine Program adapted a method endorsed by the Clinical Pharmacogenetics Implementation Consortium. The steps of this method are outlined in the figure.
We decided to exclude weak inhibitors as they only cause a ≥ 1.25 to < 2-fold area under concentration-time curve (AUC)-fold increase of the victim drug, which does not appear to translate to a clinically meaningful amount of inhibition. Additionally, excluding weak inhibitors is well accepted among those considering CYP2D6 inhibitors to determine the clinical phenotype and recent CPIC guidelines now exclude weak inhibitors from their phenotype modification.(22, 25, 26) Taking multiple weak inhibitors are also excluded as limited evidence exists on how to treat this scenario. In contrast to Mostafa et al., we opted to treat genotype-based UMs and NMs the same when on a strong inhibitor. Our rational for this is that the evidence is not compelling enough to stray from the CPIC-endorsed Borges et al., method.(13, 27–32) Specifically, the evidence reviewed suggests that while individuals may not completely phenoconvert from UM to PM per the metabolic ratio phenotyping definition, they had a marked decrease in function.(27, 28, 30) Ultimately, we opted to take a conservative safety-oriented approach. With this approach, it would be recommended that an individual with a genotype-based UM phenotype who is taking a strong inhibitor to be categorized as a clinical PM phenotype and to avoid CYP2D6-mediated opioids. In contrast, if we categorized the individual as a clinical NM phenotype, we would not recommend against use of CYP2D6-mediated opioids, and if increased CYP2D6 enzymatic activity did exist in this patient and they did not phenoconvert, they may be at risk for toxicity with a CYP2D6-mediated opioid.
Step 2: Develop Common Set of Rules to Predict Phenoconversion
Taking a concomitant CYP2D6 inhibitor does not guarantee that phenoconversion will occur. It is possible to stay within the same CYP2D6 phenotype even if the CYP2D6 activity score has changed. In order for the calculator tool to be optimally user friendly, it would need to report out to the user if phenoconversion occurred. To make this happen, we first identified all CYP2D6 genotype-to-phenotype combinations and organized them into a table using the resources on ‘Gene-specific Information Tables for CYP2D6’ from the Pharmacogenomics Knowledgebase (PharmGKB).(6) For each combination, three scenarios were explored; 1) strong inhibitor with or without a moderate inhibitor also present, 2) moderate inhibitor only present, 3) no inhibitor present. The occurrence of phenoconversion was determined for each scenario. At the time of this work, various combinations of genotypes were identified that confer activity scores ranging from zero to ≥ 4.5 by increments of 0.5. Seven possible genotype-based activity scores were examined within each of the three scenarios, and it was identified that phenoconversion could occur for 43% of the possible activity score-CYP2D6 inhibitor use scenarios (n=21) (Table 3).
Table 3:
Scenarios to Determine Occurrence of Phenoconversion
| Starting Activity Score based on Genotype | Phenotype Based on Genotype | Inhibitor present | Adjusted Activity Score | Clinical Phenotype | Phenoconversion Occurrence |
|---|---|---|---|---|---|
| Scenario 1 | |||||
| 0 | PM | strong ± moderate | 0 | PM | no |
| 0.5 | IM | 0 | PM | yes | |
| 1.0 | NMa | 0 | PM | yes | |
| 1.5 | NM | 0 | PM | yes | |
| 2.0 | NM | 0 | PM | yes | |
| 3.0–4.0 | UM | 0 | PM | yes | |
| ≥ 4.5 | UM | 0 | PM | yes | |
| Scenario 2 | |||||
| 0 | PM | moderate only | 0 | PM | no |
| 0.5 | IM | 0.25 | IM | no | |
| 1.0 | NMa | 0.5 | IM | yes | |
| 1.5 | NM | 0.75 | IM | yes | |
| 2.0 | NM | 1.0 | NMa | no | |
| 3.0–4.0 | UM | 1.5–2.0 | NM | yes | |
| ≥ 4.5 | UM | ≥ 2.25 | UM | no | |
| Scenario 3 | |||||
| 0 | PM | none | 0 | PM | no |
| 0.5 | IM | 0.5 | IM | no | |
| 1.0 | NMa | 1.0 | NMa | no | |
| 1.5 | NM | 1.5 | NM | no | |
| 2.0 | NM | 2.0 | NM | no | |
| 3.0–4.0 | UM | 3.0–4.0 | UM | no | |
| ≥ 4.5 | UM | ≥ 4.5 | UM | no | |
PM, poor metabolizer; IM, intermediate metabolizer; NM, normal metabolizer; UM, ultra-rapid metabolizer
Activity score to phenotype translation based on definitions prior to the CYP2D6 genotype to phenotype standardization project. (8)
The scenario results from Table 3 were analyzed to identify common rules to broadly predict phenoconversion. We identified three common rules for predicting phenoconversion. 1) Poor metabolizers do not undergo phenoconversion in presence of a CYP2D6 inhibitor. 2) When a strong inhibitor is present, the clinical phenotype will always be a poor metabolizer. 3) When a moderate inhibitor is present, the clinical phenotype will only change for certain genotype-based activity scores (i.e., 1.0, 1.5, and 3.0 – 4.0). The common rules were utilized to create an algorithm to serve as the foundation to build the automated phenoconversion calculation process (Figure 2).
Figure 2: Phenoconversion Application Algorithm.
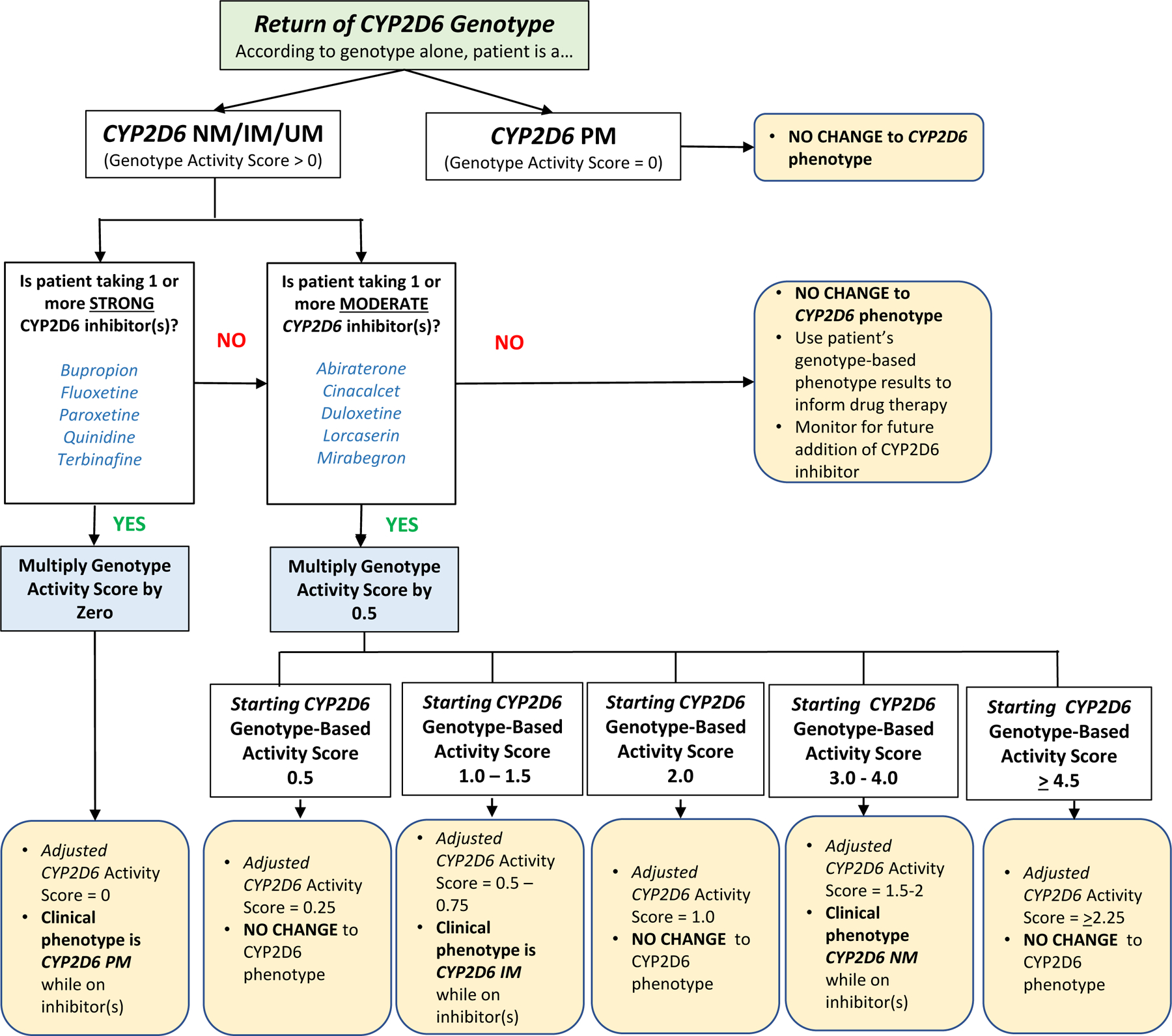
The three common rules identified for predicting phenoconversion [ 1) Poor metabolizers do not undergo phenoconversion in presence of a CYP2D6 inhibitor. 2) When a strong inhibitor is present, the clinical phenotype will always be a poor metabolizer. 3) When a moderate inhibitor is present, the clinical phenotype will only change for certain genotype-based activity scores] were utilized to create an algorithm that served as the foundation to build the automated calculator tool.
Step 3: Build and Refine Automated Calculator Tool Based on Internal Testing
We worked with the UF College of Pharmacy’s web development team to build off the algorithm to create an online-automated calculator. This was an iterative process that included testing to ensure internal validity, improvements based on feedback, and integrating solutions for identified complicating clinical factors. Specifically, internal reviews were conducted by trainees and 10 faculty members within UF’s Center for Pharmacogenomics and Precision Medicine. The reviews identified five clinical factors that impacted the calculator functionality along with other aesthetic modifications that resulted in ten revisions to date. The original and final versions of the alpha calculator are shown in Figure 3. The five clinical factors identified are related to updates from the pharmacogenetics community (n=2), to laboratory testing and/or resulting that are unique to the CYP2D6 gene (n=3). Each factor is described below, and further details can be found in Table 4.
Figure 3: Original and Final Versions of the CYP2D6 Calculator (Alpha version).
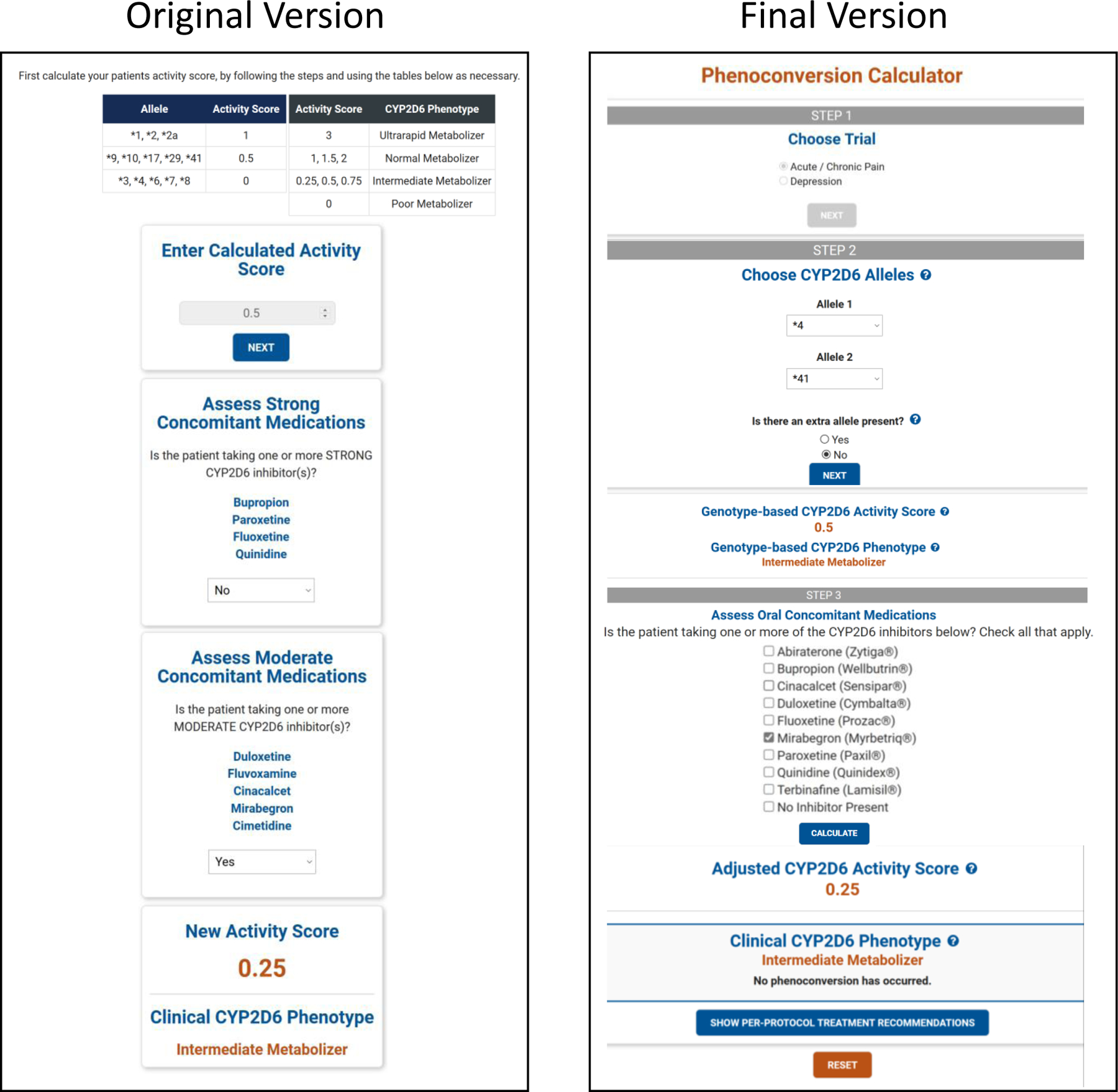
The calculator tool underwent several revisions during the build process to incorporate complicating clinical factors and user-feedback. The left side of the figure shows the original version, where the user had to calculate and enter their own genotype-based activity score. The right side of the figure shows the final version of the calculator, where the user selects their patient’s CYP2D6 alleles and then the calculator determines the genotype-based activity score. This is reflective of the alpha version that will be used in the IGNITE II trials.
Table 4.
Complicating Clinical Factors leading to Calculator Revisions
| Complicating clinical factor | Revision | Rationale |
|---|---|---|
| Results of the CYP2D6 genotype to phenotype standardization project. | Instead of having the user calculate and/or enter the genotype-based CYP2D6 activity score, the user can now enter the CYP2D6 alleles and the calculator will compute a discrete genotype-based activity score | PharmGKB has annotations of all CPIC guidelines, which allows a user to enter the alleles for a gene and be presented with key information from the guideline (e.g., activity score, metabolizer status, recommendations).(6) Initially, we thought it would be best for the users of the calculator to go to PharmGKB to obtain the genotype-based activity score if needed. However, after the CYP2D6 genotype to phenotype standardization project,(8) the activity scores that populate are based on the consensus definition, which is a range. For example, when a user enters CYP2D6 *4/*41 into PharmGKB, the resulting activity score is “0 < x < 1.25”. The true activity score for this combination is 0.5, which indeed falls within that range. However, the range is not useful when a discrete number is required for multiplication adjustment purposes. |
| Laboratory limitations in detecting copy number variation (e.g., duplicated allele is unknown and exact number of copies is either known or unknown) | Calculator asks user to enter if an extra allele is present (yes or no), and if yes, to enter total alleles (unknown, 3, 4, or ≥ 5) |
Unknown copies: To continue utilizing a discrete number for the genotype-based activity score, we used a minimum value approach. Specifically, if the user enters that a duplication was present, we would assume at minimum, the allele with the lower activity value had two copies. An upper range activity was not calculated, instead the upper range was only determined when translating to phenotype. For example, if the adjusted activity score was 0.25+, then the resulting clinical phenotype was “Intermediate-to-Ultrarapid metabolizer”. Known Copies: The calculator will determine what the genotype-based activity score would be if allele 1 is the duplicated allele, and if allele 2 is the duplicated allele. For example, if the CYP2D6 genotype is *4/*41 with three duplication copies, or a total of 4 copies, the genotype-based activity score is 0.5–1.5. If the *4 was the duplicated allele, the genotype-based activity score would be 0.5, and if *41 was duplicated then it would be 1.5, producing a range of 0.5–1.5. This results in a ranged genotype-based phenotype of “Intermediate-to-Normal metabolizer”. |
| Difference in reporting CYP2D6 copy number variation and lack of a standard | Instead of asking “how many duplicates” were present, the calculator now asks “how many total alleles” | Laboratories often report copy number variation as (*1/*2)x3 or (*1/*2) N3, meaning a total of three copies of CYP2D6 alleles are present, with either *1 or *2 being duplicated. To avoid confusion with how laboratories were reporting the calculator was updated to align with reports. If a user wants to enter that a total of 2 alleles is present, then they should answer that “no” extra alleles are present, as there is no copy number variation in that scenario. |
| CYP2D6*10 allele downgraded from activity value of 0.5 to 0.25 | CYP2D6*10 allele was reassigned a new activity value (0.25) and additional starting genotype possibilities were built | To ensure all CYP2D6 alleles as defined by CPIC are in the calculator and the activity values match the literature. |
| Ranged CYP2D6 phenotypes | Per-protocol recommendations account for range phenotype and include recommendations as if the individual had the more extreme phenotype | The standard of care at both Mayo Clinic and UF Health was to err on the side of caution and treat the individual as if they had the more extreme phenotype. |
The first complicating clinical factor was a direct result of the CYP2D6 genotype to phenotype standardization project.(8) Initially, the user had to calculate and/or enter the genotype-based CYP2D6 activity score. Directions were provided on how to calculate the activity score if needed (i.e., using the activity values listed in the CYP2D6 allele functionality table from PharmGKB).(6) The CYP2D6 genotype to phenotype standardization project provides ranges of activity scores (e.g., 0 < x <1.25), rather than discrete numbers, to correlate to different phenotypes. As the calculator depends on a discrete activity score to calculate the phenotype appropriately, we updated the calculator to calculate the genotype-based activity score based on the alleles selected by the user to ensure consistency.
Laboratories are often limited in their ability to detect which allele is multiplicated and the number of copies present, leading to ranged genotype-based activity scores.(7) Once the calculator was responsible for calculating the genotype-based activity score, we needed to accommodate for these possible ranged activity scores. Certain laboratories can detect that a copy number variation is present, but are not able to determine the number of copies nor which allele is duplicated or multiplicated. Other laboratories within the IGNITE II network are able to determine the number of copies (up to four), but not which allele is multiplicated. When it is unknown how many copies are present, a minimum value approach (a discrete number) is used, and the upper range of the genotype-based CYP2D6 activity score is not calculated. When it is known how many copies are present, a minimum and maximum value are calculated. Both scenarios often result in a ranged CYP2D6 clinical phenotype being displayed.
Initially when the multiplications were built in the calculator, it asked the user how many copies were present (e.g., if three copies of one allele was present and one copy of the other allele, you would need to enter 3). It was identified that most labs that were able to detect the number of copies would report the total number of alleles present (e.g., 4 from previous example). While there is no standard on how to report CYP2D6 copy number variation,(33) based on the laboratories in the network, we revised the calculator to ask the user to enter the total alleles to align with the majority of laboratory reports.
An additional complicating clinical factor that resulted from the CYP2D6 genotype-to- phenotype standardization project was that the CYP2D6 *10 allele had its activity value downgraded from 0.5 to 0.25.(8) This increased the number of genotype-based activity score combinations for “starting total activity score from genotype” that were handed off to the development team to build from seven to ten possible genotype-based activity scores. Each possibility was built for the three previously described scenarios (strong inhibitor, moderate inhibitor, no inhibitor). Adding in copy number variations and accounting for genotype-based activity scores with CYP2D6 *10 made the calculator build more intricate. Once the calculator had to account for these additions, the total genotype-based activity score combinations increased from seven to 40 and were provided to the development team to build for each of the three scenarios. Specifically, it was more complex when adjusting for moderate inhibitors (Table S1). When the initial scenarios were investigated (Table 3), moderate inhibitors caused phenoconversion in 43% of the moderate inhibitor only scenarios (3/7), which increased to 58% with the additional combinations (23/40).
Outside the direct functions of the calculator, ranged phenotypes were another complicating clinical factor that had to be addressed. Specifically, how does one treat an individual clinically with a ranged phenotype? We conducted a literature review and found limited guidance on how to treat patients with this scenario. We identified that the standard of care at both Mayo Clinic and UF Health was to err on the side of caution and treat the individual as if they had the more extreme phenotype.(34) For example, an individual with a range clinical phenotype of CYP2D6 “NM to UM” (activity score of ≥ 1.5) would get the same recommendation to avoid CYP2D6-mediated opioids as a CYP2D6 UM. Since the calculator is being used in a trial setting by multiple sites, we added in per-protocol recommendations based on the clinical phenotype to ensure clinical application consistency among sites. Adding in the recommendations resulted in an additional build for CYP2C19 genotype-based phenotypes as the trial will be providing recommendations for selective serotonin reuptake inhibitors for depression patients that depend on both CYP2D6 and CYP2C19 results. Of note, no phenoconversion adjustments are made for CYP2C19 as no standardization on how to adjust the CYP2C19 phenotype based on genotype and inhibitor use is available at present.(35)
Step 4: Perform External Usability Testing
Usability testing was conducted via a standardized form to gather feedback about calculator accessibility, intuitiveness, visual appearance, functionality, and ease of interpreting results. An email was distributed to the IGNITE II sites requesting a minimum of two participants per site to conduct usability testing. In addition, trainees in pharmacogenetic programs at UF who were not involved in the development of the calculator were also recruited to conduct testing. A case accompanied each form to walk the user through the calculator and allowed for complicated scenarios to be tested. Each user who tested the calculator completed Likert scale questions, which were on a scale of one to five where one was “strongly disagree” and five was “strongly agree”. Additional feedback was documented in the comments section. Upon return of the results, the averages were calculated for the questions, and the comments were reviewed.
Twenty-nine participants completed the usability testing. These users were healthcare providers (i.e., pharmacists, physicians) (31%), research coordinators (6.9%), program managers (13.8%), and pharmacogenetic trainees (i.e., graduate students, post-docs) (48.3%). The results of the Likert-scale questions are summarized in Table 5. Most notably, the statement “resulting terms can benefit from additional explanatory text” received an average score of 3.51 indicating the calculator could benefit from additional explanatory text. Upon reviewing the comments, the team was able to identify ways to expand on the language and create explanatory text that will appear when the user hovers their mouse over specific sections. Additionally, each step is now explained more thoroughly by having an information section for the user. The addition of the information section addressed the testing participants’ concerns about understanding the relevance of the patient’s score in comparison to other scores.
Table 5:
Usability Test Results
| Questions | Mean (SD) |
|---|---|
| Arrangement of content is well organized; sequentially and logically ordered | 4.93 (0.25) |
| Calculator is easy to navigate and use; steps are intuitive | 4.90(0.30) |
| Instructions are clear | 4.97(0.18) |
| Resulting terms are clear | 4.77(0.67) |
| Resulting terms can benefit from additional explanatory text | 3.51(1.45) |
| Calculator’s functioned technically in the expected way | 4.77(0.76) |
| Calculator is visually appealing | 4.57(0.72) |
Step 5: Develop Strategy for Ongoing Maintenance Calculator
The calculator requires continuous ongoing maintenance to ensure it is operating correctly. Specifically, new literature on phenoconversion will need to be reviewed to determine if the standard of care approach should be updated. Currently, the alleles in the calculator are those that were listed as having a defined function and corresponding activity value in the ‘CYP2D6 Allele Functionality Table’ on PharmGKB as of February 1st 2020.(6) Changes in allele function and corresponding activity value as well as new alleles will need to be monitored to update the calculator accordingly.
Discussion
The use of CYP2D6 genotyping in clinical practice is growing and is increasingly being used to guide prescribing of common medications, such as opioids and antidepressants.(7) Since CYP2D6 is susceptible to clinically relevant enzyme inhibition, it is important to consider concomitant CYP2D6 inhibitors. Determining whether a person has phenoconverted is key when prescribing medications as utilizing the correct clinical phenotype is vital to avoiding undesired clinical outcomes. Having a simple method to implement phenoconversion into clinical practice is crucial to its uptake. We believe our phenoconversion calculator tool is that solution. The calculator was created in order to be used in the upcoming IGNITE II trials.(20) This automation replaces the manual process used in our previous chronic and acute pain trials and provides standardization among sites.(17, 19) We recognized the value this calculator may have for the public and also created a beta version, called Propelling Clinical Pharmacogenomics into Practice (PROP™). This version is not trial-specific and can be accessed via the web (https://precisionmedicine.ufhealth.org/phenoconversion-calculator/).
CYP2D6 is a complex gene, and as such, creating the phenoconversion calculator tool was multifaceted. We encountered several complicating clinical factors that led us to describe key lessons for implementing CYP2D6 phenoconversion in clinical practice.
First, there are multiple resources for CYP2D6 inhibitors, and they do not have consistency between them. Most notable of the information sources are the FDA table of inhibitors, Indiana University’s Drug Interactions Flockhart Table™, and University of Washington’s Drug Interaction Database, Drugbank.(23, 36–38) The classification of inhibitors into strong, moderate, or weak classifications based on increases in substrate AUC, put forth by the FDA, is well accepted. However, results can vary from study to study, and several limitations exist that can impact the magnitude of the pharmacokinetic interaction, including but not limited to multiple pathways of metabolism, genetic variation, dose of inhibitor or substrate given.(12, 39) As Stout et al., points out, over-categorization of inhibitors may lead to potentially harmful actions. As such, it is important to integrate a reputable resource when considering CYP2D6 inhibitors in clinical practice. The FDA is an authoritative resource that maintains their evidence-based table of inhibitors, which was last updated in March 2020.(40) This makes the FDA table of inhibitors a trustworthy resource to use when assessing for the presence of CYP2D6 inhibitors.
Pharmacogenetics is a field with constant emerging evidence. During the development of this phenoconversion calculator tool, CYP2D6*10 was downgraded from an activity value of 0.5 to 0.25.(8) In order to implement the impact of moderate inhibitors, the tool is based off a calculated starting total genotype-based activity score, which is a discrete numbers between zero and two, without considering duplication or multiplication. The starting total genotype-based activity score is then used to calculate the true genotype-based activity score if warranted based on the presence of duplications or multiplications. (Table S1). Prior to this change we had seven starting CYP2D6 activity score numbers, and this increased to ten after the change. This created a more complex build. The tool is currently working from allele functionality tables that define the activity value for each allele. Knowing these activity values can change in the future and to avoid manual updates, we intend to take advantage of PharmGKB application programming interface (API) resources in the future as an enhancement update to the beta version, PROP™. The CYP2D6 genotype to phenotype translation was also recently updated, where an activity score of 1.0 is now categorized as an IM, rather than NM.(8) As our alpha calculator tool is being implemented in the upcoming IGNITE II trials where an individual with an activity score of 1.0 will be treated the same as NMs, we did not update the phenotype translation. However, the publicly available beta version, PROP™, aligns with current CPIC phenotypes. Some laboratories within the IGNITE network may be updating their lab reporting to align with new translation. For this reason, along with the annotations on PharmGKB reporting the consensus ranges, the user must now enter the CYP2D6 alleles in the calculator. This allows the tool to calculate the activity score, adjust based on concomitant CYP2D6 inhibitors if necessary, and then output the per-protocol recommendations based on the adjusted activity score, thus providing consistency among IGNITE network sites. The beta version, PROP™, does not include drug therapy recommendations at this time, we hope to enhance it in the future to align with CPIC guidelines recommendations. The tool uses activity scores to predict phenotype, as endorsed by CPIC, however, it should be noted that evidence is emerging suggesting that activity score may be an oversimplification. Instead, a continuous scale method may be more accurate in predicting the enzyme activity or using next-generation sequencing to capture structural variation in CYP2D6.(41, 42) The ADOPT PGx trial will have the opportunity to compare patient-reported outcomes between NM and IM phenotypes, as determined by the calculator.
Laboratory limitations regarding copy number variation introduced range phenotypes, and through this process we had to think through how to treat individuals with ranged phenotypes in clinical practice. Since there is limited evidence in this regard, we opted to err on the side of caution and treat as if the individual has the most extreme phenotype. Deciding this upfront also allows for consistency among sites as we were able to build this decision into the calculator tool. Additionally, variability in reporting copy number variation can be a source of confusion and requires explanation and/or education in order to have consistency in interpretation. There is currently no standard in how to report CYP2D6 copy number variation, but hopefully in the future there will be.(33)
Applying phenoconversion in practice allows clinicians to act on the estimated drug metabolizing capacity of the individual based on genetics and interacting medications. It is limited as it is just an estimation, and it is possible, for example in CYP2D6 UMs, that some increased activity remains even with a strong CYP2D6 inhibitor. However, in the absence of routine therapeutic drug monitoring of the active drug/metabolite, which is not realistic in typical routine clinical practice for these chronic medications, the estimating approach is appropriate. Additionally, for CYP2D6-mediated opioids UM and PMs have the same drug therapy recommendation to avoid, in contrast to a NM where the medication would be okay to use. After reviewing and analyzing the relevant available evidence, we are comfortable providing recommendations that UMs do indeed phenoconvert.(27, 28, 30) As with use of genotype data in clinical practice, incorporation of phenoconversion should be implemented with clinical judgment; the calculator is not intended to replace this. Patient medications are dynamic, and thus phenoconversion can be reversed when the interacting drug is discontinued. Similarly, for a patient not on a CYP2D6 inhibitor at the time of initial clinical phenotype assignment, the addition of an inhibitor can alter the phenotype. Clinicians should be cognizant that discontinuation or addition of a CYP2D6 inhibitor may result in different recommendations and will require the healthcare provider to re-consult the calculator. Additionally, some medications may be both metabolized by CYP2D6 while also causing CYP2D6 enzyme inhibition (e.g., paroxetine). In these scenarios, we do not consider the CYP2D6 self-inhibition when making clinical recommendations for that specific drug.
Phenoconversion appears more common in patients with chronic pain (29%) as compared to depression (4%), as observed from two pragmatic implementations conducted at UF Health.(18) Patients with chronic pain often are treated with antidepressants, either for depression comorbidity or for adjunct agent to treat pain. These agents make up a large portion of the CYP2D6 inhibitors (e.g., duloxetine, paroxetine, bupropion).
It must be noted that phenoconversion can be caused by other nongenetic factors such as age, cancer, smoking, pregnancy and inflammation and a limitation of the calculator is that it does not account for these factors.(14) Our focus has been on drugs causing phenoconversion, also referred to as drug-drug-gene interactions (DDGIs) as this is where the evidence and guidance for phenoconversion exists.(43) This concept is similar to drug-drug interactions (DDIs), which are very common. In fact, recently 19% of 30 potentially clinically significant interactions were shown to occur as DDGIs.(43, 44) For example, mirabegron is a moderate CYP2D6 inhibitor that interacts with tramadol and is a common drug alert in electronic health systems. Genotyping for CYP2D6 brought more attention to DDIs like this as the interaction is exaggerated if the individual is a CYP2D6 IM as compared to NM. Alerts based on drug-gene interactions are becoming more commonplace as genotyping becomes integrated into standard clinical care. There is however no accepted method of translating DDGI into prescribing decisions (e.g., CYP2D6 IM and taking mirabegron and tramadol). A potential future application for our calculator would be embedding it in electronic health record to help mitigate DDGIs. This would require reliance upon the active medication list in the medical record, which is known to be inaccurate.(45, 46)
In summary, our CYP2D6 calculator tool is applicable to a broad audience as more institutions are implementing CYP2D6 in their healthcare systems.(7) The tool meets an important need, to aid clinicians in easily implementing phenoconversion, a very common phenomenon. The steps provided in this tutorial can be used by other sites when implementing phenoconversion whether they opt to use our calculator or not as the phenoconversion method and clinical factors to consider are clearly described.
Supplementary Material
1. Supplemental Material
Acknowledgements:
We would like to acknowledge the internal and external testers of the calculator for their valuable time spent testing the calculator functionality and for providing feedback for improvement including the individuals within the NIH IGNITE Network. Additionally, we thank all of the students and trainees who worked on various parts of the calculator development.
Funding information: This work was supported by grants from the National Institutes of Health (U01 HG007269 [as part of the IGNITE Network); Clinical Translational Science Institute (CTSI) program, which is supported by the NIH National Center for Advancing Translational Sciences (NCATS) (UL1TR001427)
Footnotes
Disclaimer: As Editor-in-Training of Clinical Pharmacology & Therapeutics, D. Max Smith was not involved in the review or decision process for this paper.
Conflict of interest statement: The authors declared no competing interests for this work.
References:
- (1).Wilkinson GR Drug metabolism and variability among patients in drug response. The New England journal of medicine 352, 2211–21 (2005). [DOI] [PubMed] [Google Scholar]
- (2).Taylor C, Crosby I, Yip V, Maguire P, Pirmohamed M & Turner RM A Review of the Important Role of CYP2D6 in Pharmacogenomics. Genes 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gaedigk A Complexities of CYP2D6 gene analysis and interpretation. International review of psychiatry (Abingdon, England) 25, 534–53 (2013). [DOI] [PubMed] [Google Scholar]
- (4).Caudle KE et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genetics in medicine : official journal of the American College of Medical Genetics 19, 215–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ & Leeder JS The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clinical pharmacology and therapeutics 83, 234–42 (2008). [DOI] [PubMed] [Google Scholar]
- (6).Whirl-Carrillo M et al. Pharmacogenomics knowledge for personalized medicine. Clinical pharmacology and therapeutics 92, 414–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cavallari LH et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genetics in medicine : official journal of the American College of Medical Genetics 21, 2255–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Caudle KE et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clinical and translational science 13, 116–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Shah RR & Smith RL Addressing phenoconversion: the Achilles’ heel of personalized medicine. British journal of clinical pharmacology 79, 222–40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Owen RP, Sangkuhl K, Klein TE & Altman RB Cytochrome P450 2D6. Pharmacogenetics and genomics 19, 559–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gardiner SJ & Begg EJ Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacological reviews 58, 521–90 (2006). [DOI] [PubMed] [Google Scholar]
- (12).Cicali EJ, Smith DM, Duong BQ, Kovar LG, Cavallari LH & Johnson JA A Scoping Review of the Evidence Behind Cytochrome P450 2D6 Isoenzyme Inhibitor Classifications. Clinical pharmacology and therapeutics, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Storelli F, Matthey A, Lenglet S, Thomas A, Desmeules J & Daali Y Impact of CYP2D6 Functional Allelic Variations on Phenoconversion and Drug-Drug Interactions. Clinical pharmacology and therapeutics 104, 148–57 (2018). [DOI] [PubMed] [Google Scholar]
- (14).Klomp SD, Manson ML, Guchelaar HJ & Swen JJ Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review. J Clin Med 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).ClinCalc LLC. The Top 300 of 2021 <https://clincalc.com/DrugStats/Top300Drugs.aspx> Accessed 1 April 2021.
- (16).Knisely MR et al. CYP2D6 drug-gene and drug-drug-gene interactions among patients prescribed pharmacogenetically actionable opioids. Applied nursing research : ANR 38, 107–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Smith DM et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genetics in medicine : official journal of the American College of Medical Genetics 21, 1842–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cicali EJ et al. Challenges and lessons learned from clinical pharmacogenetic implementation of multiple gene-drug pairs across ambulatory care settings. Genetics in medicine : official journal of the American College of Medical Genetics 21, 2264–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Thomas CD et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genetics in medicine : official journal of the American College of Medical Genetics, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Implementing Genomics in Practice Pragmatic Clinical Trials Network <https://www.genome.gov/Funded-Programs-Projects/Implementing-Genomics-in-Practice-IGNITE-2-Pragmatic-Clinical-Trials-Network> Accessed 1 April 2021.
- (21).Borges S et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. Journal of clinical pharmacology 50, 450–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mostafa S, Kirkpatrick CMJ, Byron K & Sheffield L An analysis of allele, genotype and phenotype frequencies, actionable pharmacogenomic (PGx) variants and phenoconversion in 5408 Australian patients genotyped for CYP2D6, CYP2C19, CYP2C9 and VKORC1 genes. Journal of neural transmission (Vienna, Austria : 1996) 126, 5–18 (2019). [DOI] [PubMed] [Google Scholar]
- (23).Drug Development and Drug Interactions: Table of Substrates, Inhibitors, and Inducers U.S. Food & Drug Adminsitration. <https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers#table3-2> Accessed 1 April 2021.
- (24).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clinical pharmacology and therapeutics 95, 376–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lopes GS et al. Sex Differences in Associations Between CYP2D6 Phenotypes and Response to Opioid Analgesics. Pharmacogenomics and personalized medicine 13, 71–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clinical pharmacology and therapeutics, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Borges S et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clinical pharmacology and therapeutics 80, 61–74 (2006). [DOI] [PubMed] [Google Scholar]
- (28).Dalén P, Dahl M, Andersson K & Bertilsson L Inhibition of debrisoquine hydroxylation with quinidine in subjects with three or more functional CYP2D6 genes. British journal of clinical pharmacology 49, 180–4 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kiss Á et al. Phenoconversion of CYP2D6 by inhibitors modifies aripiprazole exposure. European archives of psychiatry and clinical neuroscience 270, 71–82 (2020). [DOI] [PubMed] [Google Scholar]
- (30).Laine K et al. Inhibition of cytochrome P4502D6 activity with paroxetine normalizes the ultrarapid metabolizer phenotype as measured by nortriptyline pharmacokinetics and the debrisoquin test. Clinical pharmacology and therapeutics 70, 327–35 (2001). [PubMed] [Google Scholar]
- (31).Lam YW, Gaedigk A, Ereshefsky L, Alfaro CL & Simpson J CYP2D6 inhibition by selective serotonin reuptake inhibitors: analysis of achievable steady-state plasma concentrations and the effect of ultrarapid metabolism at CYP2D6. Pharmacotherapy 22, 1001–6 (2002). [DOI] [PubMed] [Google Scholar]
- (32).Preskorn SH et al. Cytochrome P450 2D6 phenoconversion is common in patients being treated for depression: implications for personalized medicine. The Journal of clinical psychiatry 74, 614–21 (2013). [DOI] [PubMed] [Google Scholar]
- (33).Nofziger C et al. PharmVar GeneFocus: CYP2D6. Clinical pharmacology and therapeutics 107, 154–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nassan M, Nicholson WT, Elliott MA, Rohrer Vitek CR, Black JL & Frye MA Pharmacokinetic Pharmacogenetic Prescribing Guidelines for Antidepressants: A Template for Psychiatric Precision Medicine. Mayo Clinic proceedings 91, 897–907 (2016). [DOI] [PubMed] [Google Scholar]
- (35).Lima JJ et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clinical pharmacology and therapeutics 109, 1417–23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Drug Interactions Flockhart Table™ Indiana University. <https://drug-interactions.medicine.iu.edu/MainTable.aspx> Accessed 1 April 2021.
- (37).Drug Bank <https://www.drugbank.ca/> Accessed 1 April 2021.
- (38).Hachad H, Ragueneau-Majlessi I & Levy RH A useful tool for drug interaction evaluation: the University of Washington Metabolism and Transport Drug Interaction Database. Human genomics 5, 61–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Stout SM et al. Interpretation of Cytochrome P-450 Inhibition and Induction Effects From Clinical Data: Current Standards and Recommendations for Implementation. Clinical pharmacology and therapeutics, (2020). [DOI] [PubMed] [Google Scholar]
- (40).Yang X, Pfuma Fletcher E, Huang SM, Zineh I & Madabushi R Regulatory Efforts to Facilitate Evaluation and Clinical Management of Drug-Drug Interaction Risks. Clinical pharmacology and therapeutics, (2020). [DOI] [PubMed] [Google Scholar]
- (41).van der Lee M et al. A unifying model to predict variable drug response for personalised medicine 2020.03.02.967554 (2020). [Google Scholar]
- (42).Dalton R et al. Interrogation of CYP2D6 Structural Variant Alleles Improves the Correlation Between CYP2D6 Genotype and CYP2D6-Mediated Metabolic Activity. Clinical and translational science 13, 147–56 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Verbeurgt P, Mamiya T & Oesterheld J How common are drug and gene interactions? Prevalence in a sample of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping. Pharmacogenomics 15, 655–65 (2014). [DOI] [PubMed] [Google Scholar]
- (44).Malki MA & Pearson ER Drug-drug-gene interactions and adverse drug reactions. The pharmacogenomics journal 20, 355–66 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Coleman EA, Smith JD, Raha D & Min SJ Posthospital medication discrepancies: prevalence and contributing factors. Arch Intern Med 165, 1842–7 (2005). [DOI] [PubMed] [Google Scholar]
- (46).Bedell SE et al. Discrepancies in the use of medications: their extent and predictors in an outpatient practice. Arch Intern Med 160, 2129–34 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Supplemental Material


