This study identifies AT2s as the relevant source of GM-CSF for the development and maintenance of alveolar macrophages. During organogenesis, nascent AT2s induce GM-CSF expression in a perfectly timed manner to support the proliferation and differentiation of fetal monocytes that contemporaneously seed the embryonic lungs.
Abstract
Programs defining tissue-resident macrophage identity depend on local environmental cues. For alveolar macrophages (AMs), these signals are provided by immune and nonimmune cells and include GM-CSF (CSF2). However, evidence to functionally link components of this intercellular cross talk remains scarce. We thus developed new transgenic mice to profile pulmonary GM-CSF expression, which we detected in both immune cells, including group 2 innate lymphoid cells and γδ T cells, as well as AT2s. AMs were unaffected by constitutive deletion of hematopoietic Csf2 and basophil depletion. Instead, AT2 lineage-specific constitutive and inducible Csf2 deletion revealed the nonredundant function of AT2-derived GM-CSF in instructing AM fate, establishing the postnatal AM compartment, and maintaining AMs in adult lungs. This AT2-AM relationship begins during embryogenesis, where nascent AT2s timely induce GM-CSF expression to support the proliferation and differentiation of fetal monocytes contemporaneously seeding the tissue, and persists into adulthood, when epithelial GM-CSF remains restricted to AT2s.
Introduction
Proper organ development and homeostasis is reliant not only on the appropriate function of specialized tissue-specific stromal cells but also on the cross talk that occurs between these cells and the stably or transiently integrated tissue-resident immune cell populations. Tissue-resident macrophages (TRMs) are often integral members of these immune cross talk compartments and are best known for their phagocytic activity (Davies et al., 2013). Work over the past years has identified distinct transcriptional programs that are associated with particular tissue-specific functions in TRM subsets (Blériot et al., 2020). Instigation of these programs is particularly critical during the embryonic development of an organ, when TRMs first differentiate from different myeloid progenitors and readily acquire tissue-specific transcriptional signatures (Mass et al., 2016). Although the processes of TRM induction are not well defined, it is presumed that these programs are initiated and maintained by signals derived from the local tissue microenvironment (Guilliams et al., 2020).
Alveolar macrophages (AMs) are a lung-specific type of TRM. They play a central role in maintenance of alveolar homeostasis by removing cellular debris, excess surfactant, and inhaled bacteria, but are also important in preserving lung function during pulmonary viral infections (Hussell and Bell, 2014; Kopf et al., 2015). Found within the lumen of the alveoli, AMs are surrounded by two alveolar epithelial cell types. Alveolar type 1 cells (AT1s) facilitate gas exchange with underlying endothelial cells, while alveolar type 2 cells (AT2s) produce surfactant, act as facultative progenitors in case of alveolar injury, and activate the immune system in response to pathogen-related stimuli (Basil et al., 2020; Whitsett et al., 2019). Notably, this epithelial niche is formed toward the end of embryonic lung development, and its establishment is concomitant with the differentiation of AMs (Guilliams et al., 2013; Schneider et al., 2014b). Work generated over the past several years suggests that differentiating AMs pass through a transitional monocytic stage during their development (Evren et al., 2021; Guilliams et al., 2013; Hoeffel et al., 2015; Liu et al., 2019; Schneider et al., 2014b; Yona et al., 2013), while parabiosis and fate-mapping studies revealed that, similar to other TRM compartments, AMs comprise cells of both fetal and adult monocytic origin (Hashimoto et al., 2013; Schulz et al., 2012; Yona et al., 2013). Novel refined tools enabled the identification of multiple myeloid progenitors that contribute to the pool of AMs and that are produced during partially overlapping successive waves of hematopoiesis (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Liu et al., 2019). Notably, experiments using mixed bone marrow chimeras and adoptive transfers of fetal and adult precursor populations demonstrated that regardless of origin, a major determinant for the generation of a functional AM compartment in mice and humans is signaling via the cytokine GM-CSF (encoded by the gene Csf2; Guilliams et al., 2013; Li et al., 2020; Schneider et al., 2017; Schneider et al., 2014a; Schneider et al., 2014b; Suzuki et al., 2014; van de Laar et al., 2016). Indeed, more than 30 yr ago, Chen et al. (1988) speculated about GM-CSF–mediated AM self-renewal, which is decoupled from bone marrow hematopoiesis. The critical function of GM-CSF for AMs and surfactant homeostasis was later confirmed in knockout mice with deficiencies in the GM-CSF signaling pathway (Dranoff et al., 1994; Nishinakamura et al., 1995; Robb et al., 1995; Stanley et al., 1994). Subsequent findings in patients with rare Csf2 mutations or anti–GM-CSF autoantibodies and in studies with humanized mice demonstrated that this distinctive lung GM-CSF function is fundamentally conserved in humans (Kitamura et al., 1999; Martinez-Moczygemba et al., 2008; Suzuki et al., 2008; Willinger et al., 2011). Mouse studies have demonstrated that the binding of GM-CSF to its receptor (encoded by Csf2ra and Csf2rb) is associated with induction of the transcription factor peroxisome proliferator-activated receptor γ and the corresponding initiation of AM differentiation in lung fetal monocytes during prenatal development (Schneider et al., 2014b). PPARγ is thought to critically regulate an AM-specific transcriptional program, including up-regulation of signature markers CD11c and SiglecF, and to drive terminal AM differentiation in the first 1–2 wk after birth.
Disrupting the mature functional AM population compromises lung homeostasis and in both humans and mice. Loss of GM-CSF signaling and the subsequent deficit of AMs lead to the accumulation of lipoproteinaceous material and cellular debris in the alveolar space, a condition known as pulmonary alveolar proteinosis (PAP; Trapnell et al., 2019). These detrimental phenotypes can be rescued in mice by reestablishing a functional AM compartment via pulmonary overexpression of GM-CSF in Csf2−/− mice (Huffman et al., 1996) or by supplying GM-CSFR–deficient mice with cells that have both intact GM-CSFR and the potential to differentiate into mature AMs (Schneider et al., 2014a; Suzuki et al., 2014). Thus, in addition to its known function in conferring an inflammatory state in myeloid cells under inflammatory and autoimmune conditions (Becher et al., 2016; Hamilton, 2020), GM-CSF also has a dedicated role in shaping lung AM fate (Evren et al., 2020). Indeed, results from early studies published almost 50 yr ago using mouse lung conditioned medium indicated high GM-CSF expression in the lung (Sheridan and Metcalf, 1973); the responsible cells, however, remained elusive. Production of GM-CSF in the lung has since been attributed to multiple hematopoietic and nonhematopoietic lineages, including group 2 innate lymphoid cells (ILC2s), basophils, and epithelial cells, specifically AT2s (Cohen et al., 2018; Guilliams et al., 2013; Schneider et al., 2014b). Basophils were suggested to constitute an important part of this AM niche and to be critical regulators of AM differentiation and function (Cohen et al., 2018). Other sources of GM-CSF may include T cells, B cells, natural killer (NK) cells, innate lymphoid cells (ILCs), fibroblasts, and endothelial cells, as suggested by data obtained from different tissues under inflammatory conditions (Hamilton, 2020). However, it is unclear which cellular sources of GM-CSF are most critical for promoting AM differentiation. It is also not known whether the critical GM-CSF producers differ between embryonic, neonatal, and adult lungs, or if this axis regulates AM maintenance following perinatal AM establishment.
To unambiguously identify the relevant sources of GM-CSF required for the formation of AMs in vivo, we generated novel GM-CSF reporter and conditional knockout mice and proceeded to systematically delete GM-CSF from different hematopoietic and nonhematopoietic compartments. Here, we identify AT2s as a constitutive and dominant nonhematopoietic source of GM-CSF in the lungs of neonatal and adult mice. We further demonstrate that AT2-derived GM-CSF is the necessary signal to initiate and promote pre- and postnatal AM differentiation and subsequent AM maintenance throughout adulthood. GM-CSF is a lineage-defining cytokine in lung epithelial cells, which becomes expressed following AT2 fate specification during late gestation, contemporaneously with AM fate initiation in lung fetal monocytes. Our results unequivocally demonstrate that AMs are critically reliant not on hematopoietic GM-CSF but rather on epithelial GM-CSF derived from AT2s, both for development and for maintenance of the mature AM population.
Results
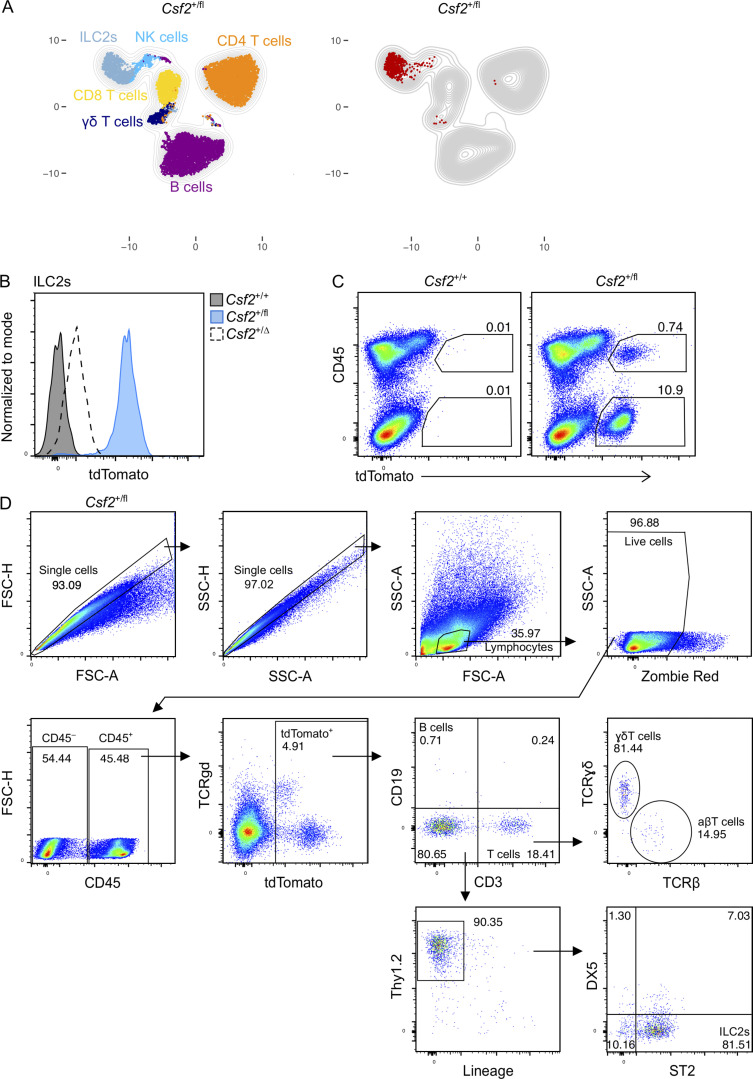
ILC2s are the major hematopoietic source of Csf2 expression in the lungs
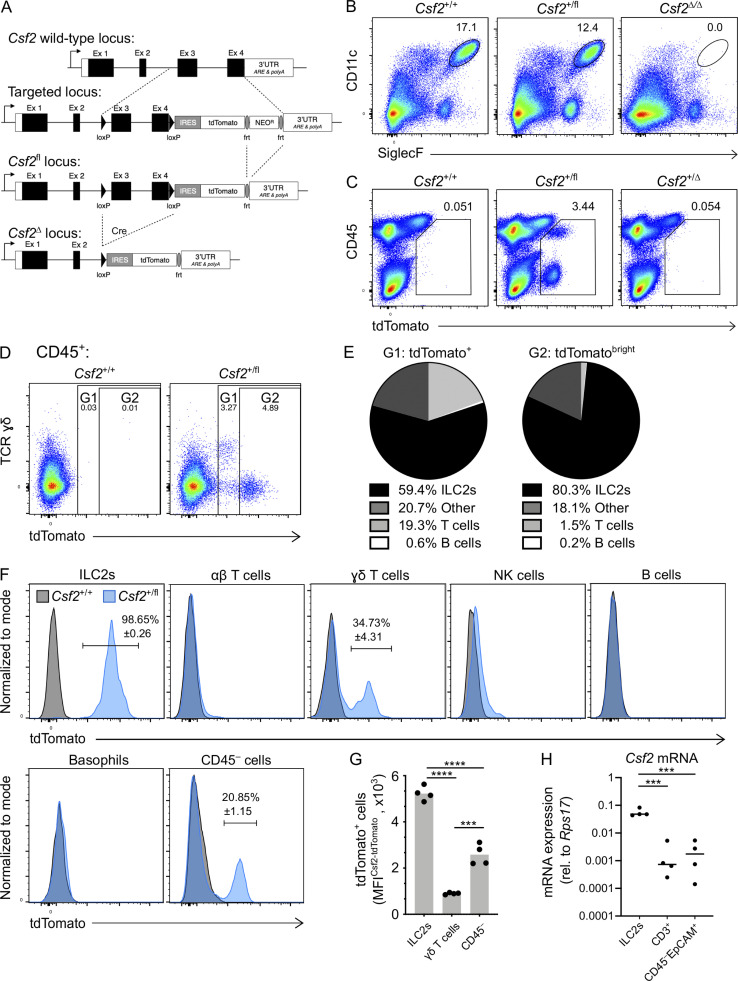
To disentangle the individual contributions of different pulmonary GM-CSF sources, we generated a knock-in mouse model that was designed to report Csf2 expression and to allow Csf2 deletion (Fig. 1 A). In the absence of Cre, transcription of the Csf2flox-tdTomato locus (Csf2fl) results in the production of a bicistronic mRNA encoding GM-CSF and tdTomato, marking GM-CSF–producing cells with fluorescent tdTomato signal while maintaining normal GM-CSF expression. When recombined in the germline to create mice with a global deletion of Csf2 (Csf2Δ), we found that AMs were completely absent from the adult lung, as per other Csf2−/− models; these results validated the loss-of-function property of this novel mouse tool (Fig. 1 B). Evaluation of the reporter function of this tool revealed that tdTomato+ cells were readily detectable in the lungs of Csf2+/fl mice compared with Csf2+/+ littermate controls (Fig. 1 C). As expected, our results suggested the presence of multiple GM-CSF sources, including cells of nonhematopoietic (CD45−) and hematopoietic (CD45+) origin (Fig. 1 C). Subjecting CD45+ lymphoid populations in the adult lung to FlowSOM-guided meta-clustering revealed that the tdTomato+ signal comprised predominantly ILC2s, with small contributions from NK cells and γδ T cells (Fig. S1 A). Notably, deletion of the floxed allele (Csf2+/Δ) resulted in strongly reduced tdTomato expression across all populations (Fig. 1 C), which was >10-fold lower than the signal in cells with the nonrecombined Csf2fl allele (Fig. S1 B). This unexpected reduction in tdTomato expression in the case of Csf2Δ alleles has thus provided us with an endogenous readout for assessing Cre-mediated Csf2 deletion.
Figure 1.
Different hematopoietic cell populations contribute to GM-CSF production in the neonatal lung. (A) Gene-targeting strategy used to engineer Csf2flox-tdTomato (Csf2fl) mice. (B) Flow cytometry analysis of CD11c+SiglecF+ AMs in the lungs of adult Csf2+/+, Csf2+/fl, and Csf2Δ/Δ mice, gated on CD45+ cells. (C) Flow cytometry analysis of tdTomato+ populations in the adult lungs of Csf2+/+, Csf2+/fl, and Csf2+/Δ mice. (D) Flow cytometry of tdTomato expression in Csf2+/+ and Csf2+/fl P10 lungs, gated on CD45+ cells. (E) Percentage contribution from different hematopoietic cells types to the tdTomato+ (left) or tdTomatobright compartment, as gated in D. (F) Expression of tdTomato in the indicated cell populations in lungs of P10 Csf2+/+ (gray) and Csf2+/fl (blue) mice was determined by flow cytometry analysis. Percentages (±SEM) of tdTomato+ cells are indicated. (G) MFI of tdTomato of Csf2+/fl ILC2s, γδ T cells, and CD45− cells in P10 lungs, gated on tdTomato+ cells as indicated in F. (H) Csf2 mRNA expression relative to Rps17 in the three major tdTomato+ cell populations isolated from P10 Csf2+/fl lungs. (B–H) Data are from one experiment representative of two (B and C), five (D-G), or three (H) independent experiments. (G and H) ***, P 0.0001–0.001; ****, P < 0.0001. IRES, internal ribosome entry site; MFI, mean fluorescence intensity; rel., relative; UTR, untranslated region.
Figure S1.
The contribution of the hematopoietic compartment to GM-CSF production in the lung and the gating strategy for lymphocytes. (A) Representative UMAP showing the FlowSOM-guided meta-clustering of lymphoid populations in adult lungs of Csf2+/fl mice (left). tdTomato-expressing cells are identified in the corresponding UMAP (right). (B) Expression level of tdTomato in Thy1.2+ST2+ ILC2s from adult lungs of Csf2+/+ (gray), Csf2+/fl (blue), and Csf2+/∆ (dashed line) mice. (C) Flow cytometry analysis of tdTomato expression by CD45+ and CD45− cells in Csf2+/+ and Csf2+/fl P10 lungs. (D) Gating strategy used to identify lymphocytes in Fig. 1, E–G. Lin = CD11b, CD11c, NK1.1, F4/80, Gr-1, FcεRIα, Ter119. (A–D) Data are from one experiment, representative of four (A), two (B), or five (C and D) independent experiments.
Because AMs complete their development shortly after birth (Guilliams et al., 2013; Schneider et al., 2014b), we first characterized the major sources of GM-CSF in the neonatal lung. For this, postnatal day 10 (P10) lungs from Csf2+/+ and Csf2+/fl mice were analyzed by flow cytometry. As observed in the adult lungs (Fig. 1 C), the tdTomato+ cells from neonatal Csf2+/fl mice were composed of CD45− and CD45+ populations, of which the latter showed slightly higher tdTomato expression (Fig. S1 C). Further analysis of the CD45+tdTomato+ cells identified several different cell types, including Lin−Thy1+ST2+ ILC2s and CD3+TCRγδ+ T cells (Fig. 1, D–F; and Fig. S1 D). ILC2s were found to be the major hematopoietic GM-CSF producers by frequency (>80%) and by relative reporter expression, followed by CD3+ T cells; minor contributions to the CD45+ tdTomato signal came from NK cells and B cells (Fig. 1, D–F; and Fig. S1 D). ILC2s produced a strong reporter signal, with nearly all of the Csf2+/fl ILC2s expressing tdTomato (Fig. 1 F). In contrast, only a few of the T cells expressed tdTomato, and the overall tdTomato fluorescent signal was not nearly as strong as was observed with the ILC2s (Fig. 1 F). Indeed, comparing the tdTomato signal intensity across Csf2+/fl ILC2s, γδ T cells, and CD45− cells indicated a significantly higher signal intensity in ILC2s relative to the other two populations (Fig. 1, F and G). To further confirm that the tdTomato signal was faithfully reporting Csf2 expression, quantitative RT-PCR was used to compare the relative Csf2 mRNA in sorted tdTomato+ populations. Overall, higher levels of Csf2 mRNA were detected in the tdTomato+ ILC2s relative to tdTomato+ T cells and tdTomato+CD45−EpCAM+ cells (Fig. 1 H). Together, these results suggest that in the neonatal lung, ILC2s express the overall highest amount of GM-CSF on a per-cell basis and also represent the dominant GM-CSF source within the hematopoietic compartment, which is in agreement with a prior report (Cohen et al., 2018).
Hematopoietic-derived GM-CSF is dispensable for the development of AMs in the neonatal lung
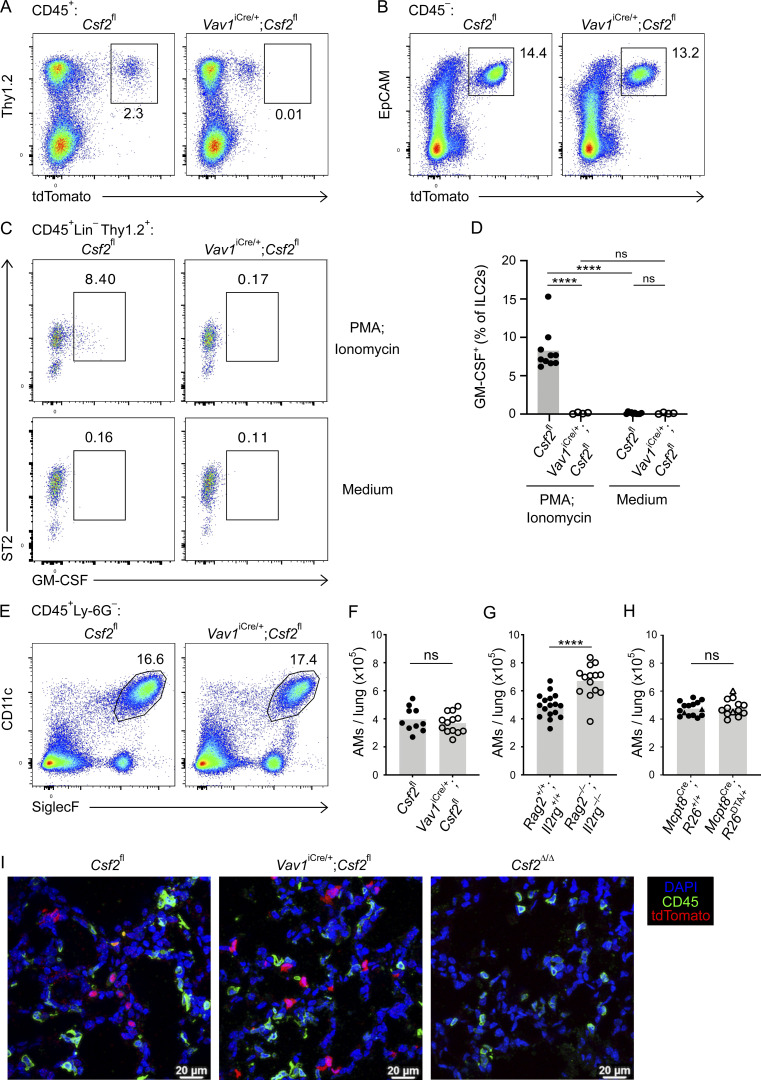
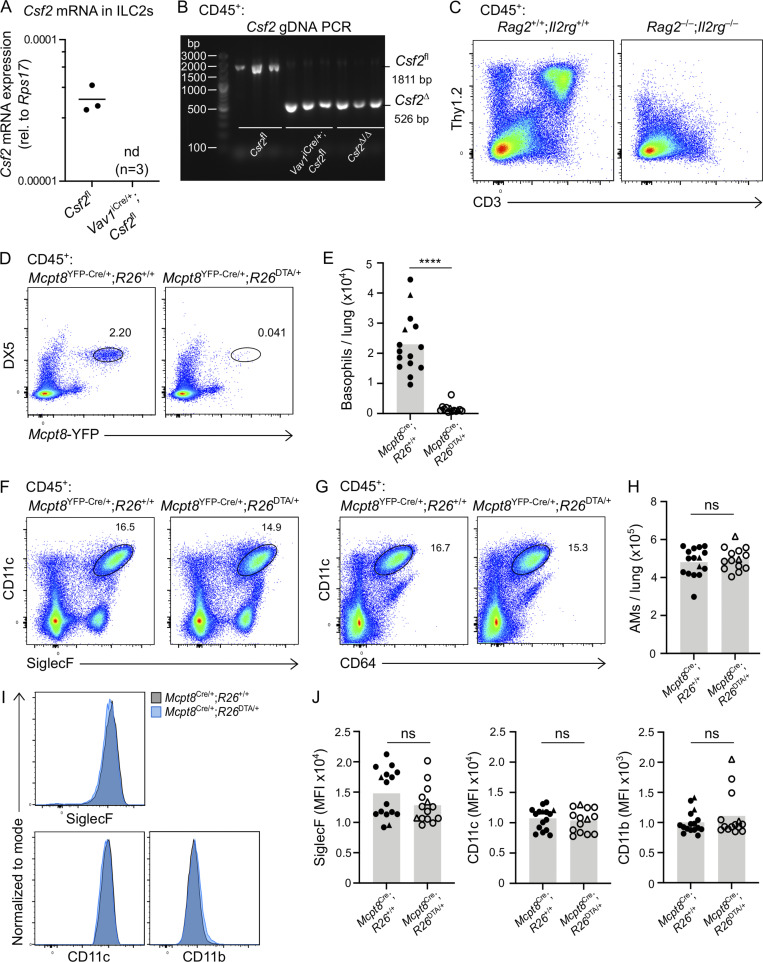
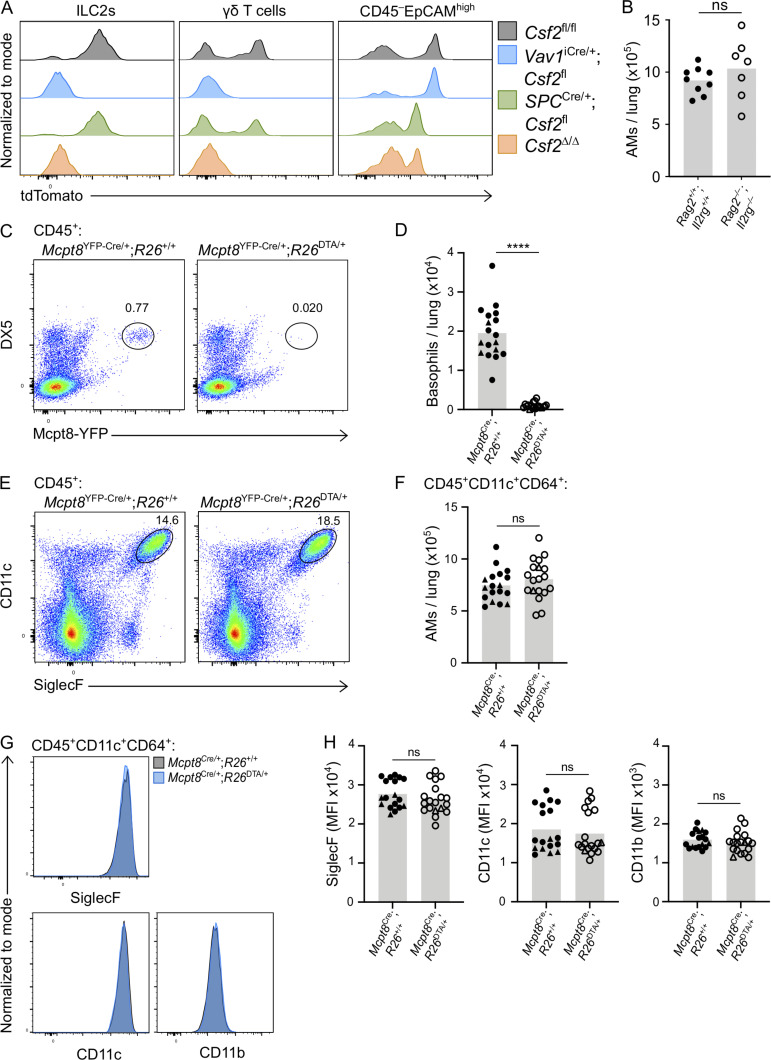
Multiple studies suggest that ILC2s shape the neonatal lung environment via their canonical type 2 effector cytokines, IL-5 and IL-13 (Cohen et al., 2018; de Kleer et al., 2016; Nussbaum et al., 2013; Saluzzo et al., 2017; Schneider et al., 2019; Steer et al., 2017). Indeed, the constitutive Csf2 expression by postnatal lung ILC2s is reminiscent of their production of IL-5, by which they regulate eosinophil homeostasis (Nussbaum et al., 2013). To determine the relative importance of hematopoietic GM-CSF for AM homeostasis, including GM-CSF derived from ILC2s, we deleted Csf2 expression from the hematopoietic compartment by crossing Csf2fl mice with Vav1iCre mice, in which expression of a codon-improved constitutively active Cre is driven by the Vav1 promoter. P10 neonatal lungs from these mice were then analyzed by flow cytometry. For technical reasons and due to occasional germline Cre activity in Vav1iCre mice (see Materials and methods), experimental cohorts included mice with Csf2Δ/fl and Csf2fl/fl alleles (hereafter termed Csf2fl). All conditional knockout groups were compared with their littermate Cre− controls. In comparison to Cre− controls, analysis of P10 lungs from Vav1iCre;Csf2fl mice revealed a reduction in tdTomato signal in CD45+ cells (Fig. 2 A), while the tdTomato signal from the CD45− compartment was unchanged (Fig. 2 B). Within the CD45+ compartment, recombination of Csf2 exons 3 and 4 was verified by quantifying the corresponding region in Csf2 mRNA from sorted ILC2s (Fig. S2 A), as well as by detecting the Csf2Δ allele in sorted CD45+ lung cells by PCR (Fig. S2 B). Both methods demonstrated that the functional Csf2fl allele was deleted specifically from hematopoietic cells in Vav1iCre;Csf2fl mice relative to their littermate controls. Absence of GM-CSF protein expression in ILC2s from Vav1iCre;Csf2fl mice was further validated by in vitro restimulation of lung cells (Fig. 2, C and D).
Figure 2.
Hematopoietic-derived GM-CSF is dispensable for the development of AMs in the neonatal lung. (A–F) Analysis of P10 lungs isolated from Csf2fl and Vav1iCre/+;Csf2fl mice. (A) Flow cytometry analysis of the Thy1.2+tdTomato+ population, gated on CD45+ cells. (B) Flow cytometry analysis of the EpCAM+ tdTomato+ population, gated on CD45− cells. (C) Flow cytometry analysis of GM-CSF production in ILC2s (CD45+Lin−Thy1.2+ST2+) after restimulation with PMA/ionomycin (top row) or incubation with medium only (bottom row). (D) Percentage of GM-CSF+ ILC2s after restimulation or in the presence of medium only. (E) Flow cytometry analysis of CD11c+SiglecF+ AMs, gated on CD45+Ly-6G− cells. (F) Quantification of AMs. (G) Quantification of AMs in P10 lungs of Rag2+/+;Il2rg+/+ and Rag2−/−;Il2rg−/− mice. (H) Quantification of AMs (CD45+SiglecF+CD11c+) in P10 lungs of Mcpt8YFP-Cre;R26+/+ and Mcpt8YFP-Cre;R26DTA/+ mice. Mcpt8YFP-Cre/+ mice are indicated by circles, while Mcpt8YFP-Cre/YFP-Cre mice are indicated by triangles. (I) Representative IF pictures of tdTomato+ cells (red) and CD45+ cells (green) in P10 lungs of Csf2fl, Vav1iCre/+;Csf2fl, and Csf2Δ/Δ mice (scale bars 20 µm). (A–C, E, and I) Data are from one experiment representative of at least two independent experiments. (D and F–H) Data are pooled from two (D), four (F), or three (G and H) independent experiments. ns, P ≥ 0.05; ****, P < 0.0001.
Figure S2.
Validation for effective Cre-recombination in Vav1iCre and that lung hematopoietic GM-CSF contributions, including from lymphocytes and basophils, are dispensable for AM development in the neonatal lung. (A and B) Cells sorted from P10 lungs isolated from Csf2fl and Vav1iCre/+;Csf2fl mice. (A) Csf2 mRNA expression relative to Rps17 in ILC2s (CD45+Lin−Thy1.2+ST2+). nd, not detected; rel., relative. (B) Gel depicting the Csf2 deletion PCR reaction for CD45+ lung cells. The products of the recombined (Csf2∆) and nonrecombined (Csf2fl) allele are indicated. (C) Flow cytometry analysis of P10 lungs of Rag2+/+;Il2rg+/+ and Rag2−/−;Il2rg−/− mice, gated on CD45+ cells. (D–J) Analysis of P10 lungs isolated from Mcpt8YFP-Cre;R26+/+ and Mcpt8YFP-Cre;R26DTA/+ mice. (E, H, and J) Mcpt8YFP-Cre/+ mice are indicated by circles, while Mcpt8YFP-Cre/YFP-Cre mice are indicated by triangles. (D) Flow cytometry analysis of DX5+Mcpt8-YFP+ basophils, gated on CD45+ cells. (E) Quantification of basophils. (F) Flow cytometry analysis of CD45+CD11c+SiglecF+ AMs. (G) Flow cytometry analysis of CD45+CD11c+CD64+ AMs. (H) Quantification of AMs, gated per G. (I) Expression levels of SiglecF, CD11c, and CD11b from AMs, gated per G. Mcpt8YFP-Cre/+;R26+/+ (gray) and Mcpt8YFP-Cre/+;R26DTA/+ (blue) mice. (J) MFI of SiglecF, CD11c, and CD11b for AMs, gated per G. (A–D, F, G, and I) Data are from one experiment (A and B) or one experiment representative of three independent experiments (C, D, F, G, and I). (E, H, and J) Data are pooled from three independent experiments. ns, P ≥ 0.05; ****, P < 0.0001. gDNA, genomic DNA; MFI, mean fluorescence intensity.
Following confirmation of specific and efficient depletion of hematopoietic-derived GM-CSF in Vav1iCre;Csf2fl mice, the myeloid compartment was analyzed for AMs. Assessment of the AM population indicated that there was no change in the number of AMs in the lung when hematopoietic Csf2 expression was absent (Fig. 2, E and F). Furthermore, AMs were examined in the P10 lungs of Rag2−/−;Il2rg−/− mice, which lack T cells, B cells, NK cells, and ILCs (Fig. S2 C). In the complete absence of these GM-CSF–producing lymphocytes, AM numbers did not decline, but even slightly increased compared with wild-type controls (Fig. 2 G). Together, these findings suggest that hematopoietic GM-CSF is dispensable for AM development.
A previous report, however, suggests that basophils are important regulators of AM development, a function that was in part attributed to the GM-CSF produced by ILC2s and/or basophils themselves (Cohen et al., 2018). Since our results did not support a critical contribution of hematopoietic-derived GM-CSF to AM survival, we further explored the role of basophils in this process by using a validated genetic basophil depletion model (Sullivan et al., 2011), in which basophil-specific Cre expression (Mcpt8YFP-Cre) ablates YFP-tagged basophils through Rosa26-driven expression of the cell-lethal diphtheria toxin A (DTA). The constitutive depletion of basophils in Mcpt8YFP-Cre;R26DTA mice (Fig. S2, D and E), however, did not significantly alter AM numbers in the P10 lung even when using two different gating strategies (Fig. 2 H; and Fig. S2, F–H). Furthermore, we did not detect changes in AM canonical surface marker expression when comparing AMs from basophil-deficient mice with littermate control mice (Fig. S2, I and J).
Overall, these results demonstrate that neither the constitutive loss of hematopoietic Csf2 expression nor the depletion of basophils or ILCs per se compromises the number of AMs in the developing neonatal lung. Indeed, despite a substantial reduction in the Csf2-reporter signal in dissociated lungs from Vav1iCre;Csf2fl mice, in situ analysis by immunofluorescence (IF) revealed that the overall tdTomato signal was still present in these lungs (Fig. 2 I). Furthermore, the lack of CD45 coexpression in the remaining tdTomato+ cells in Vav1iCre;Csf2fl lung sections suggests that these cells exclusively belong to the CD45− compartment and are more numerous than initially anticipated from flow cytometry analysis of dissociated tissue. These studies thus point toward the CD45− compartment as the major orchestrator of GM-CSF–mediated AM differentiation and survival.
Epithelial AT2s are the main source of Csf2 expression in the nonhematopoietic compartment of the neonatal lung
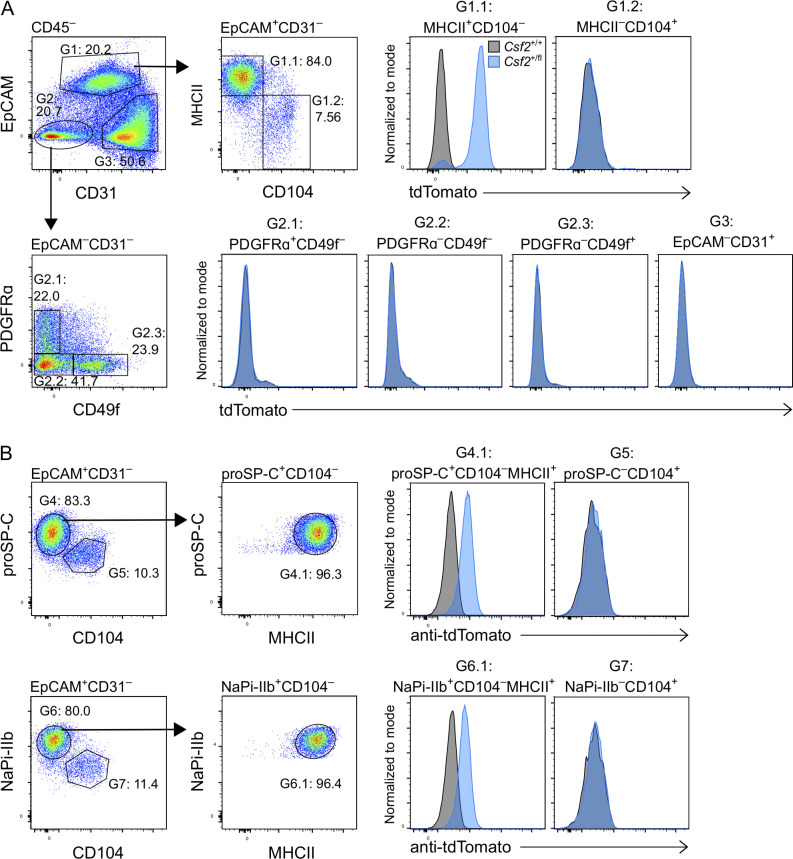
To identify the CD45− cell types that express the Csf2 reporter, neonatal P10 lungs from Csf2+/+ and Csf2+/fl mice were profiled by flow cytometry using a comprehensive panel of markers for epithelial, mesenchymal, and endothelial lineages. tdTomato expression was only detected in EpCAM+ epithelial cells that coexpressed MHCII; no Csf2-reporter signal was detected in CD45−EpCAM− cells, including CD31+ endothelial and PDGFRα+ mesenchymal populations (Fig. 3 A). Airway epithelial cells (EpCAM+CD104+) were also found to be tdTomato− (Fig. 3, A and B). Further characterization of the tdTomato+ epithelial cells revealed that these cells also expressed pro-surfactant protein C (SP-C) and the sodium-dependent phosphate transporter NaPi-IIb (Slc34a2; Fig. 3 B). In the neonatal and adult lung, all three of these markers—MHCII, SP-C, and NaPi-IIb—are very strongly associated with AT2s (Donati et al., 2020; Tighe et al., 2019; Traebert et al., 1999). Taken together, we conclude that AT2s constitutively express Csf2 and that the vast majority of Csf2-expressing epithelial cells are AT2s, at least as assessed using this reporter in dissociated tissue of the neonatal lung.
Figure 3.
AT2s are the main nonhematopoietic source of GM-CSF in the neonatal lung. (A and B) Analysis of P10 lungs isolated from Csf2+/+ (gray) and Csf2+/fl (blue) mice. (A) Flow cytometry analysis of tdTomato expression in the CD45− compartment. Isolated populations include AT2s (G1.1: CD45−CD31−EpCAM+MHCII+CD104−); airway epithelial cells (G1.2: CD45−CD31−EpCAM+MHCII−CD104+); fibroblasts (G2.1: CD45−CD31−EpCAM−PDGFRα+CD49f−); CD45−CD31−EpCAM−PDGFRα−CD49f− cells (G2.2); CD45−CD31−EpCAM−PDGFRα −CD49f+ cells (G2.3); and endothelial cells (G3: CD45−CD31+EpCAM−). (B) Flow cytometry analysis of tdTomato expression in the CD31−EpCAM+ compartment of fixed lungs, gated on CD45− cells. Isolated populations include AT2s (G4.1: proSP-C+CD104−MHCII+; G6.1: NaPi-IIb+CD104−MHCII+), and airway epithelial cells (G5: proSP-C−CD104+; G7: NaPi-IIb−CD104+). (A and B) Data are from one experiment representative of two independent experiments.
Depletion of AT2-derived Csf2 expression abrogates AM development
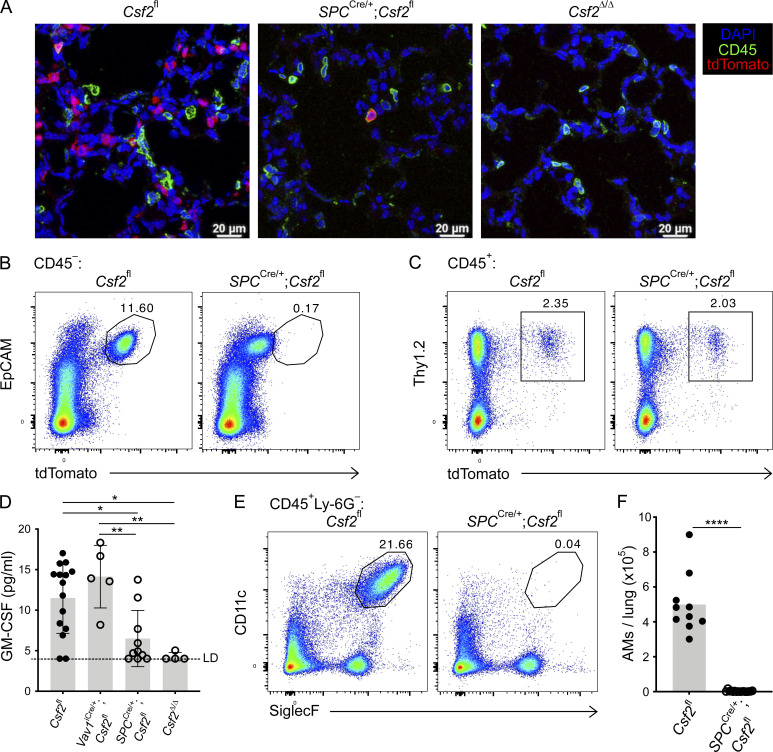
Overall, our data suggest that the major production of nonhematopoietic GM-CSF in the neonatal lung can be specifically attributed to the AT2s. To test the functional relevance of AT2-derived GM-CSF for perinatal AM development, we generated SPCCre;Csf2fl mice, in which Cre expression is under the control of the human SP-C gene promoter (Okubo and Hogan, 2004). Compared with the abundant tdTomato+ cells that were found in lung sections of Csf2fl mice on P10, few tdTomato+ cells were detected in lungs of SPCCre;Csf2fl mice (Fig. 4 A). Notably, these remaining tdTomato+ cells also expressed CD45, identifying them as hematopoietic cells. As expected, tdTomato signal was completely absent in Csf2Δ/Δ lung sections (Fig. 4 A). Flow cytometry analysis of dissociated lungs revealed that Csf2-reporter expression by CD45−EpCAMhigh AT2s was uniformly reduced, whereas no change was detected in tdTomato expression by the CD45+ hematopoietic compartment nor, more specifically, by the ILC2s (Fig. 4, B and C; and Fig. S3, A–C). This suggests that the SPCCre was effective in specifically depleting Csf2 expression in AT2s while leaving hematopoietic Csf2 expression intact. These findings are supported by measurements of total GM-CSF in lung conditioned media, in which GM-CSF levels in conditioned media from P10 SPCCre;Csf2fl and Csf2Δ/Δ lungs were significantly reduced compared with control Csf2fl and Vav1iCre;Csf2fl media samples (Fig. 4 D). The functional consequences of deleting GM-CSF specifically from AT2s were then evaluated by examining the AM compartment in SPCCre;Csf2fl mice and their littermate controls. Remarkably, CD11c+SiglecF+ AMs were completely absent in SPCCre;Csf2fl mice (Fig. 4, E and F). Overall, these results unambiguously demonstrate that Csf2 expression from the epithelial compartment of the neonatal lung, specifically from AT2s, is crucial for AM differentiation and survival.
Figure 4.
AT2-derived GM-CSF is necessary for the development of AMs in the neonatal lung. (A) Representative IF pictures of tdTomato+ cells (red) and CD45+ cells (green) in P10 lungs of Csf2fl, SPCCre/+;Csf2fl, and Csf2Δ/Δ mice (scale bars, 20 µm). (B, C, E, and F) Analysis of P10 lungs isolated from Csf2fl and SPCCre/+;Csf2fl mice. (B) Flow cytometry analysis of the EpCAM+tdTomato+ population, gated on CD45− cells. (C) Flow cytometry analysis of the Thy1.2+tdTomato+ population, gated on CD45+ cells. (D) Total GM-CSF quantification in lung conditioned media from P10 Csf2fl, Vav1iCre/+;Csf2fl, SPCCre/+;Csf2fl, and Csf2Δ/Δ mice, ±SD. LD, limit of detection. (E) Flow cytometry analysis of CD11c+SiglecF+ AMs, gated on live CD45+Ly-6G− cells. (F) Quantification of AMs. (A–E) Data are from one experiment representative of two (A and D) or four (B, C, and E) independent experiments. (F) Data are pooled from two independent experiments representative of four independent experiments. (D and F) *, P 0.01–0.05; **, P 0.01–0.001; ****, P < 0.0001.
Figure S3.
EpCAM+ epithelial cells do not contribute to GM-CSF production in neonatal SPCCre lungs. (A–C) Analysis of P10 lungs isolated from Csf2fl (gray) and SPCCre/+;Csf2fl (blue) mice. (A) Expression level of tdTomato in Thy1.2+ST2+ ILC2s. (B) Gating strategy used to identify CD45−EpCAM+ cells. (C) Expression level of tdTomato in CD45−EpCAMhigh epithelial cells. (A–C) Data are from one experiment, representative of five independent experiments.
Timed induction of Csf2 expression in nascent AT2s initiates AM differentiation in fetal lung monocytes
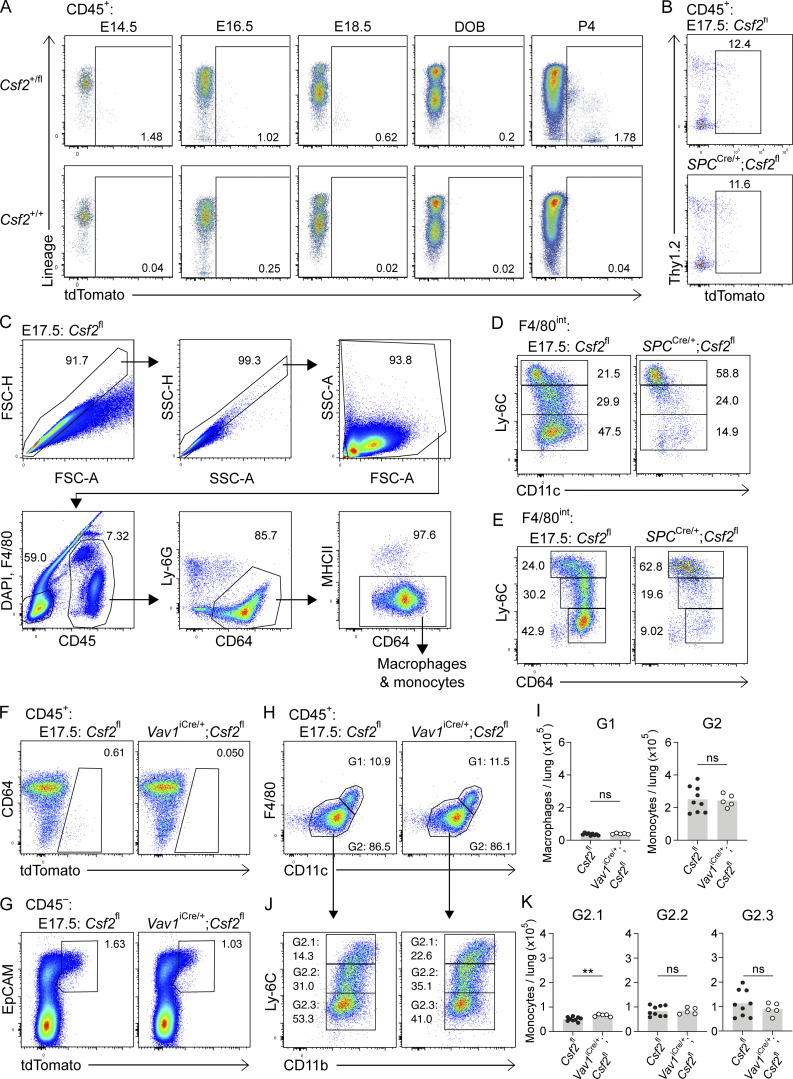
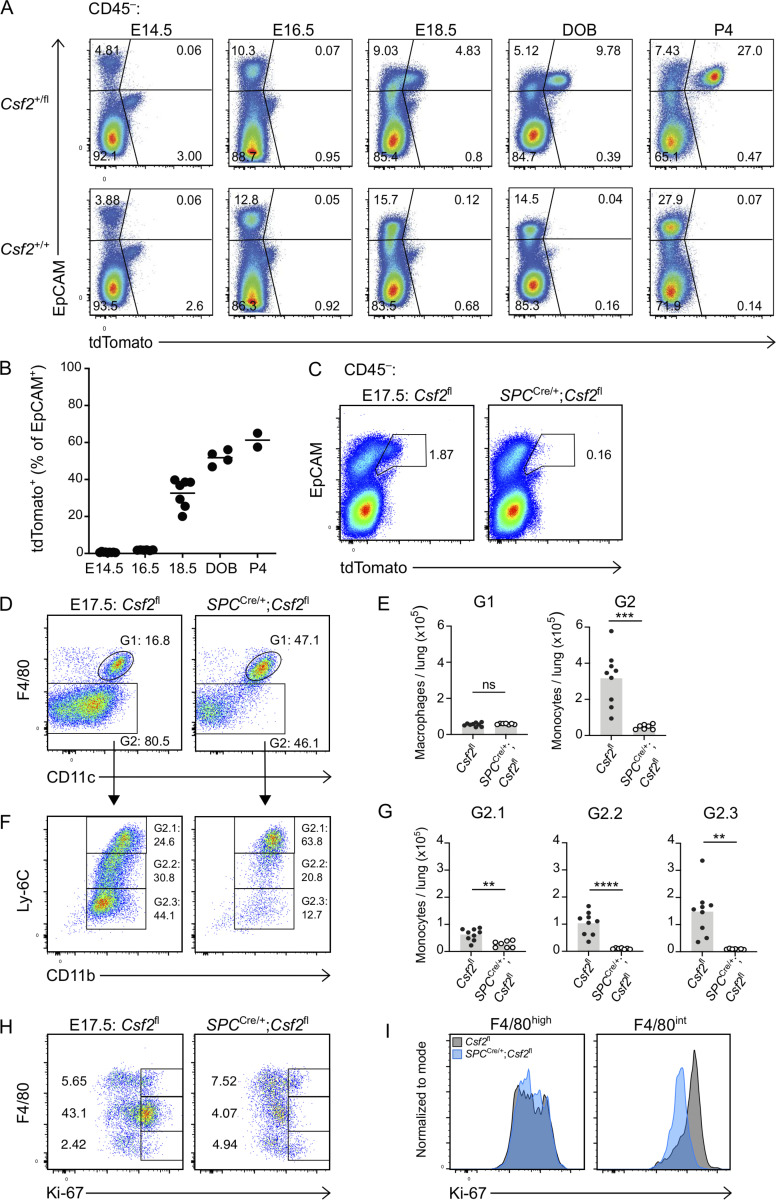
Given the initiation of AM differentiation during fetal development (Guilliams et al., 2013; Schneider et al., 2014b), we speculated that the induction of GM-CSF expression by relevant sources may be timed with the appearance of AM precursors and the initiation of their local differentiation toward an AM fate. To establish a timeline of Csf2 expression, we performed a kinetic analysis of Csf2-tdTomato reporter expression in perinatal mouse lungs. Csf2+/+ and Csf2+/fl embryos and neonates were analyzed at embryonic day (E)14.5, E16.5, E18.5, P0, and P4 to evaluate tdTomato signal in the hematopoietic and epithelial compartments. Few CD45+Linlow/−tdTomato+ cells were detected during embryonic development, though there was a substantial increase following birth (Fig. S4 A). This is consistent with the expansion of lung ILC2s during that time period (de Kleer et al., 2016; Saluzzo et al., 2017; Schneider et al., 2019; Steer et al., 2017). From E14.5 to P4, the vast majority of the tdTomato signal was detected in the CD45−EpCAM+ epithelial compartment. The first tdTomato+ cells were detected at E18.5, followed by a steady increase in the expression level and percentage of Csf2-reporter+ epithelial cells after birth (Fig. 5, A and B). These results imply that epithelial Csf2 expression is first induced in the lung between E16.5 and E18.5, concomitant with the commencement of AT2 differentiation (Treutlein et al., 2014), after which it is maintained constitutively.
Figure S4.
Hematopoietic-derived GM-CSF is not critical for AM fate specification during embryogenesis. (A) Flow cytometry analysis of kinetics of tdTomato expression in perinatal lungs in Csf2+/fl and Csf2+/+ mice ranging from E14.5 to P4, gated on CD45+ cells. DOB, day of birth. (B–E) Analysis of E17.5 lungs isolated from Csf2fl and SPCCre/+;Csf2fl mice. (B) Flow cytometry analysis of tdTomato+ population, gated on CD45+ cells. (C) Gating strategy used to identify fetal macrophage and monocyte populations. (D) Flow cytometry analysis of Ly-6C and CD11c levels in the developing fetal monocyte population (G2 in Fig. 5 D). (E) Flow cytometry analysis of Ly-6C and CD64 levels in the developing fetal monocytes population (G2 in Fig. 5 D). (F–J) Analysis of E17.5 lungs isolated from Csf2fl and Vav1iCre/+;Csf2fl mice. (F) Flow cytometry analysis of the tdTomato+ population, gated on CD45+ cells. (G) Flow cytometry analysis of the EpCAM+tdTomato+ population, gated on CD45− cells. (H) Flow cytometry analysis of F4/80high primitive macrophages (G1) and F4/80int fetal monocytes (G2), gated on CD45+Ly-6G−CD64+MHCII− cells. (I) Quantification of primitive macrophages (G1) and fetal monocytes (G2). (J) Flow cytometry analysis of Ly-6C and CD11b levels in the developing fetal monocytes population (G2). (K) Quantification of Ly-6Chigh (G2.1), Ly-6Cint (G2.2), and Ly-6Clow (G2.3) fetal monocytes. (A) Data are representative of at least two independent experiments per time point. (B–H and J) Data are from one experiment representative of three (B–E) or two (F–H and J) independent experiments. (I and K) Data are pooled from two independent experiments. ns, P ≥ 0.05; **, P 0.01–0.001.
Figure 5.
Timed induction of Csf2 expression in nascent AT2s instructs AM differentiation in fetal lung monocytes. (A) Flow cytometry analysis of kinetics of tdTomato expression in perinatal lungs in Csf2+/fl and Csf2+/+ mice ranging from E14.5 to P4, gated on CD45− cells. DOB, day of birth. (B) Percentage of EpCAM+tdTomato+ cells for time points represented in A. (C–I) Analysis of E17.5 lungs isolated from Csf2fl and SPCCre/+;Csf2fl mice. (C) Flow cytometry analysis of EpCAM+tdTomato+ populations, gated on CD45− cells. (D) Flow cytometry analysis of primitive macrophages (G1; F4/80high) and fetal monocytes (G2; F4/80int), gated on CD45+Ly-6G−CD64+MHCII− cells. (E) Quantification of primitive macrophages (G1) and fetal monocytes (G2). (F) Further flow cytometry analysis of Ly-6C and CD11b levels in the developing fetal monocytes population (G2). (G) Quantification of Ly-6Chigh (G2.1), Ly-6Cint (G2.2), and Ly-6Clow (G2.3) fetal monocytes. (H) Flow cytometry analysis of Ki-67 expression, gated on CD45+ cells. (I) Ki67 expression levels in primitive macrophages (F4/80high) and fetal monocytes (F4/80int) for Csf2fl (gray) and SPCCre/+;Csf2fl (blue) mice. (A and B) Data representative of at least two independent experiments per time point. (B, E, and G) Data are pooled from at least two (B) or three (E and G) independent experiments. (C, D, F, H, and I) Data are from one experiment, representative of three (C, D, and F) or two (H and I) independent experiments. (E and G) ns, P ≥ 0.05; **, P 0.01–0.001; ***, P 0.0001–0.001; ****, P < 0.0001.
Previous reports identified the same time period as the beginning of fetal monocyte differentiation toward an AM identity (Guilliams et al., 2013; Schneider et al., 2014b). To assess if AT2-derived GM-CSF locally initiates these first steps of AM differentiation, we characterized the myeloid compartment in embryonic E17.5 lungs from SPCCre;Csf2fl mice. E17.5 was specifically chosen in order to capture the time point when Csf2-reporter expression first begins to emerge from CD45−EpCAM+ cells (Fig. 5 C). Specific Cre activity was validated by the abrogation of this tdTomato expression (Fig. 5 C), suggesting efficient deletion of Csf2 from the AT2 lineage. Our results were consistent with prior studies reporting activity of the 3.7-kb human SP-C promoter in alveolar epithelial progenitor cells of the primordial lung buds (Wert et al., 1993). The minor Csf2-reporter expression among CD45+ cells was unchanged in the E17.5 lungs of SPCCre;Csf2fl mice (Fig. S4 B). The macrophage/monocyte populations were evaluated based on the canonical surface markers CD64, F4/80, CD11c, CD11b, and Ly-6C (Fig. S4 C and Fig. 5 D). Primitive macrophages were identified as a relatively homogeneous population of F4/80high cells that were equally abundant in SPCCre;Csf2fl and Csf2fl embryos (Fig. 5, D and E). In contrast, the F4/80int cells comprise a heterogeneous compartment consisting of fetal monocytes that adopt a waterfall-shaped distribution of Ly-6C versus CD11b, CD11c, and CD64 on their differentiation trajectory toward preAMs (Fig. 5 F; and Fig. S4, D and E; Guilliams et al., 2013; Schneider et al., 2014b). Notably, there was a significant reduction in these differentiating fetal monocytes in SPCCre;Csf2fl mice, predominantly affecting the Ly-6Cint and Ly-6Clow populations, which were almost completely absent (Fig. 5, D–G; and Fig. S4, D and E). Further assessment of these early F4/80int monocyte and F4/80high macrophage populations for Ki-67 expression showed a significant and specific reduction in proliferation of fetal monocytes but not primitive macrophages in the SPCCre;Csf2fl lungs relative to their littermate controls (Fig. 5, H and I).
To further confirm the dispensability of hematopoietic GM-CSF for the embryonic macrophage/monocyte populations, Vav1iCre embryonic lungs were assessed at E17.5. The Vav1iCre specifically depleted hematopoietic Csf2, leaving the epithelial Csf2 expression intact (Fig. S4, F and G). Examination of the myeloid compartment demonstrated that the differentiation of fetal monocytes toward an AM fate was not impaired in Vav1iCre;Csf2fl embryos relative to their littermate controls (Fig. S4, H–K). Together, our results demonstrate that the intimate relationship between epithelial cells and AMs is already present in the embryonic lung, where timed expression of GM-CSF from developing AT2s is the essential local signal to initiate robust in situ expansion of fetal monocytes and, concomitantly, the first steps of AM differentiation.
GM-CSF ablation specifically in AT2s results in the absence of AMs and the development of PAP
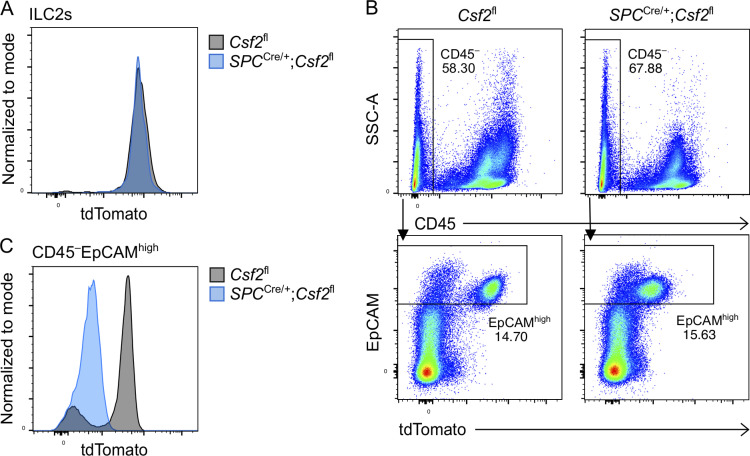
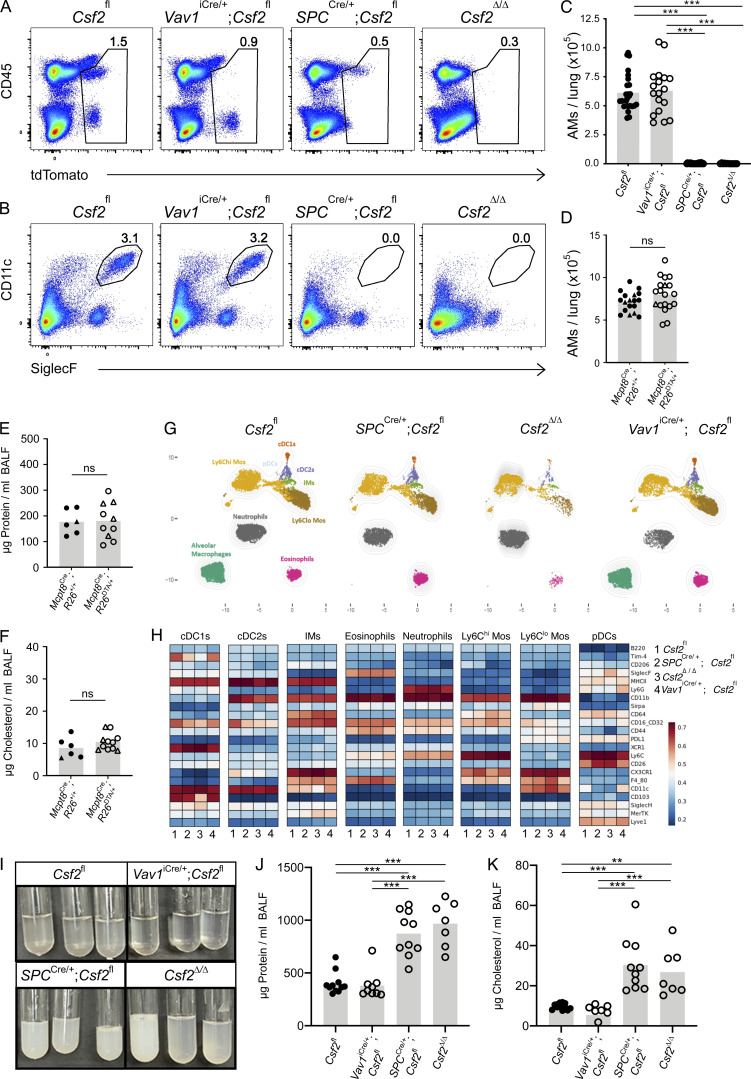
Having identified AT2s as the critical source of GM-CSF for perinatal AM differentiation, we next asked whether this relationship perpetuates throughout adulthood. Given that diverse populations of pulmonary hematopoietic cells are capable of expressing GM-CSF, it is conceivable that their role in AM maintenance is only apparent following AT2-dependent AM development. Many immune cell subsets accumulate over the course of the first few weeks of life and are more numerous in the lungs of adult mice than in the developing lungs of the early postnatal period (de Kleer et al., 2016). This influx of immune cells may remodel postnatal lung niches, affect the relative contribution of GM-CSF from individual sources, or possibly compensate for niche signals that may be missing. To determine whether the accumulation of immune cells in mature lungs modifies the AM–GM-CSF biology observed in neonates and embryos, we profiled myeloid populations in lungs of adult Vav1iCre;Csf2fl, SPCCre;Csf2fl, Csf2Δ/Δ, and littermate control mice. Evaluation of tdTomato expression by flow cytometry demonstrated hematopoietic-specific reduction of tdTomato signal in the presence of Vav1iCre, epithelial-specific reduction of tdTomato signal in the presence of SPCCre, and reduced tdTomato signal in all compartments in the case of Csf2Δ/Δ mice (Fig. 6 A and Fig. S5 A).
Figure 6.
AT2-specific deletion of Csf2 leads to AM depletion in adult lungs. (A–C, G, and H) Analysis of adult lungs isolated from Csf2fl, Vav1iCre/+;Csf2fl, SPCCre/+;Csf2fl, and Csf2Δ/Δ mice. (A) Flow cytometry analysis of tdTomato+ populations. (B) Flow cytometry analysis of CD11c+SiglecF+ AMs, gated on live CD45+Ly-6G− cells. (C) Quantification of AMs. (D) Quantification of AMs (CD45+SiglecF+CD11c+) in adult lungs of Mcpt8YFP-Cre;R26+/+ and Mcpt8YFP-Cre;R26DTA/+ mice. Mcpt8YFP-Cre/+ mice are indicated by circles, while Mcpt8YFP-Cre/YFP-Cre mice are indicated by triangles. (E) Quantification of protein in BALF from mice as in D. (F) Quantification of total cholesterol in BALF from mice as in D. (G) A representative UMAP map showing the FlowSOM-guided meta-clustering of the myeloid compartment. Mos, monocytes; IMs, interstitial macrophages; cDC, conventional DC; and pDC, plasmacytoid DC. (H) Heatmap displaying the median antigen intensity of markers used to generate G. (I) BALF from adult Csf2fl, Vav1iCre/+;Csf2fl, SPCCre/+;Csf2fl, and Csf2Δ/Δ mice. (J) Quantification of protein in BALF as in I. (K) Quantification of total cholesterol in BALF as in I. (A, B, and G–I) Data are from one experiment representative of four (A, B) or two (G–I) independent experiments. (C–F, J, and K) Data are pooled from four (C), three (D), or two (E, F, J, and K) independent experiments. ns, P ≥ 0.05; **, P 0.01–0.001; ***, P 0.0001–0.001.
Figure S5.
Lung hematopoietic GM-CSF contributions, including from lymphocytes and basophils, are dispensable for AM survival in the adult lung. (A) tdTomato signal from Thy1.2+ST2+ ILC2s, CD3+TCRγδ+ γδ T cells, and CD45−EpCAMhigh cells in adult lungs of Csf2fl/fl (gray), Vav1iCre/+;Csf2fl (blue), SPCCre/+;Csf2fl (green), and Csf2∆/∆ (orange) mice. (B) AM quantification from adult lungs of Rag2+/+;Il2rg+/+ and Rag2−/−;Il2rg−/− mice. (C–H) Analysis of adult lungs isolated from Mcpt8YFP-Cre;R26+/+ and Mcpt8YFP-Cre;R26DTA/+ mice. (D, F, and H) Mcpt8YFP-Cre/+ mice are indicated by circles, while Mcpt8YFP-Cre/YFP-Cre mice are indicated by triangles. (C) Flow cytometry analysis of DX5+Mcpt8-YFP+ basophils, gated on CD45+ cells. (D) Quantification of basophils. (E) Flow cytometry analysis of CD45+CD11c+SiglecF+ AMs. (F) Quantification of CD45+CD11c+CD64+ AMs. (G) Expression levels of SiglecF, CD11c, and CD11b by CD45+CD11c+CD64+ AMs. Mcpt8YFP-Cre/+;R26+/+ (gray) and Mcpt8YFP-Cre/+;R26DTA/+ (blue) mice. (H) MFI of SiglecF, CD11c, and CD11b for CD45+CD11c+CD64+ AMs. (A, C, E, and G) Data are from one experiment representative of four (A) or three (C, E, and G) independent experiments. (B, D, F, and H) Data are pooled from two (B) or three (D, F, and H) independent experiments. ns, P ≥ 0.05; ****, P < 0.0001. MFI, mean fluorescence intensity.
Analysis of the adult lung myeloid compartment across the different genotypes duplicated the results found in neonatal lungs. The AMs in Vav1iCre;Csf2fl lungs were present in numbers comparable to those of control lungs; this contrasted strongly with the lungs of both SPCCre;Csf2fl and Csf2Δ/Δ mice, in which the AMs were completely absent (Fig. 6, B and C). Analysis of AMs in the absence of lymphocytes using adult Rag2−/−;Il2rg−/− mice also revealed no significant changes in AM numbers relative to wild-type controls (Fig. S5 B). Notably, we again examined the effects of constitutive basophil depletion on AM homeostasis. Similar to our observations in neonatal mice, basophils were absent from lungs of adult Mcpt8YFP-Cre;R26DTA mice (Fig. S5, C and D), whereas the number and surface marker profile of their AMs were indistinguishable from those of littermate control lungs (Fig. 6 D; and Fig. S5, E–H). Protein and cholesterol concentrations in the bronchioalveolar lavage fluid (BALF) were also comparable between the two groups, suggesting functional AMs in the absence of basophils (Fig. 6, E and F).
To further fortify the results observed with Vav1iCre and SPCCre-mediated Csf2 deletion, we performed comprehensive spectral flow cytometry profiling and high-dimensional analysis of the myeloid compartments in these mice. The results showed normal representation of the AM cluster in control and Vav1iCre;Csf2fl lungs, whereas this cluster in the lungs of both the SPCCre;Csf2fl and the Csf2Δ/Δ mice was completely absent (Fig. 6 G). Other myeloid populations could be identified based on their expression of signature markers and were largely unperturbed (Fig. 6 H).
It has been well documented in both humans and mice that missing or malfunctioning AMs, arising due to defects in GM-CSF signaling, result in the accumulation of cellular and surfactant debris within the lumen of the alveoli, a pathological condition known as PAP (Trapnell et al., 2019). To further verify the importance of different GM-CSF sources for the homeostatic function of AMs, BALF was collected from the lungs of Vav1iCre;Csf2fl and SPCCre;Csf2fl mice and analyzed with that from Csf2Δ/Δ and pooled littermate control mice for signs of PAP. From both the flow cytometry plots and visual inspection of the extracted BALF (Fig. 6 I), excess cellular debris was evident in the SPCCre and Csf2Δ/Δ samples but was not detectable in the Vav1iCre or the control samples. Quantification of total protein and cholesterol content of the BALF further indicated significantly higher levels of both protein and cholesterol in SPCCre and Csf2Δ/Δ lungs relative to Vav1iCre and control lungs (Fig. 6, J and K). Together, these results suggest that the loss of epithelial Csf2 expression not only results in loss of the entire AM population but also leads to PAP in the lungs of these mice. The presence of PAP further demonstrates that the absent AM population has not been functionally replaced. Furthermore, it suggests that hematopoietic sources are neither able to compensate for the loss in GM-CSF production by AT2s nor have a tangible effect on homeostatic AM function.
Inducible AT2-specific ablation of Csf2 expression in adult lungs results in AM population atrophy
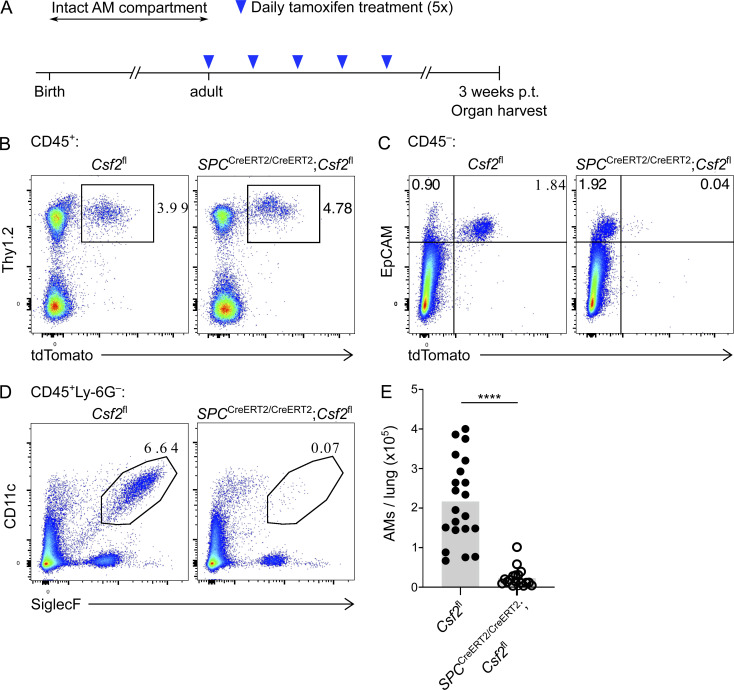
Despite the clear reliance of developing AMs on AT2-derived GM-CSF, it was still not certain whether fully developed AMs remain dependent on GM-CSF for their survival. Furthermore, constitutive absence of AMs and the resulting presence of PAP could impact Csf2 expression by non-AT2s and thereby impede a definitive assessment of the critical GM-CSF source during adulthood. Our data thus demonstrated a requirement for assessing how mature AMs are maintained in the lungs. To address this query, we generated a conditional inducible mouse model in which Csf2 expression from SP-C+ AT2 cells could be selectively depleted in adulthood upon treatment with tamoxifen. This was achieved by crossing Csf2fl reporter mice with a tamoxifen-inducible SPCCreERT2 mouse line. SPCCreERT2/CreERT2;Csf2fl and Csf2fl control mice were treated with tamoxifen, and their lungs were analyzed by flow cytometry 3 wk later (Fig. 7 A). Evaluation of the tdTomato signal from CD45+Lin−Thy1.2+ cells and epithelial CD45−EpCAM+ cells revealed that there was no change in the hematopoietic-derived tdTomato signal in the presence of tamoxifen-activated SPCCreERT2, but a specific decrease in the tdTomato signal from AT2s (Fig. 7, B and C). This suggests that the tamoxifen treatment results in a specific deletion of Csf2 expression from AT2s but does not affect Csf2 expression from the hematopoietic compartment. 3 wk after tamoxifen treatment, AMs were found to be absent in the presence of activated SPCCreERT2 (Fig. 7, D and E). Together, these results suggest that even after the postnatal development of AMs is complete, AT2-derived GM-CSF continues to be a critical niche factor for the maintenance of AM in the adult alveoli. Furthermore, the crucial function of nourishing and maintaining AMs via GM-CSF production appears to be unique to the AT2 lineage.
Figure 7.
Inducible AT2-specific ablation of Csf2 expression in adult lungs results in AM population atrophy. (A) Timeline of tamoxifen treatment and organ harvest. p.t., post-treatment. (B–E) Analysis of adult lungs isolated from Csf2fl and SPCCreERT2/CreERT2;Csf2fl mice 3 wk after tamoxifen treatment as in A. (B) Flow cytometry analysis of the Thy1.2+tdTomato+ population, gated on CD45+ cells. (C) Flow cytometry analysis of the EpCAM+tdTomato+ population, gated on CD45− cells. (D) Flow cytometry analysis of CD11c+SiglecF+ AMs, gated on live CD45+Ly-6G− cells. (E) Quantification of AMs. (B–D) Data are from one experiment representative of three independent experiments. (E) Data are pooled from three experiments. ****, P < 0.0001.
Discussion
Using a novel Csf2-reporter allele and multiple conditional knockout strains, our study generated findings critical to the understanding of the AM niche in the developing and adult lung. First, we defined hematopoietic and nonhematopoietic sources of GM-CSF in the perinatal lung. Second, we demonstrated that the hematopoietic compartment, including ILC2s and basophils, is neither a necessary source of GM-CSF nor a critical compartment for AM development. Third, using multiparameter flow cytometry, we identified AT2s as the major, if not exclusive, nonhematopoietic pulmonary compartment with constitutive GM-CSF expression. Lastly, constitutive and inducible deletion of Csf2 from the AT2 lineage revealed that AT2-derived GM-CSF plays a necessary and nonredundant function in promoting AM fate in fetal monocytes, in establishing the postnatal AM compartment, and in regulating AM maintenance in the adult lung.
Transgenic reporter alleles and single-cell RNA sequencing analysis have spurred major developments in the understanding of cytokine biology, enabling the identification of cytokine sources in tissues and guiding further in vivo validation. Here, we report the generation of a new GM-CSF reporter line suitable to investigating GM-CSF expression in situ and ex vivo. Furthermore, this Csf2fl reporter can be readily converted to a conditional knockout allele by crossing the reporter mice to a Cre-expressing mouse line. The decrease in tdTomato signal after Cre-mediated recombination of the loxP-flanked Csf2 exons, which we speculate occurs due to deletion of the associated splice acceptor sites and thus reduced expression of the truncated bicistronic transcript, enables a convenient readout of the recombination efficiency. Using this novel tool to study GM-CSF expression in the neonatal mouse lung, we identified multiple hematopoietic sources, including ILC2s, T cells, and NK cells, among which ILC2s produced the most GM-CSF on a per-cell level. Our data are in agreement with prior reports, whose findings were based on Csf2 fate-mapping and single-cell RNA sequencing analysis (Cohen et al., 2018; Komuczki et al., 2019).
Despite the significant homeostatic expression of GM-CSF by various pulmonary immune cells, our studies using Vav1iCre-mediated Csf2 deletion clearly demonstrate that hematopoietic-derived GM-CSF is dispensable for the postnatal differentiation and the subsequent maintenance of AMs. This finding was unexpected given the high Csf2 expression by ILC2s and the reports suggesting that GM-CSF is included in the ILC2 effector-cytokine repertoire that is induced perinatally (Cohen et al., 2018; de Kleer et al., 2016; Saluzzo et al., 2017; Schneider et al., 2019) and that led to speculation about ILC2 functions in shaping homeostatic myeloid cell compartments beyond the regulation of eosinophils (Nussbaum et al., 2013). Furthermore, there are reports of important ILC–GM-CSF interactions in other contexts. Notably, ILC3-derived GM-CSF in the colon facilitates dendritic cell (DC)– and macrophage-mediated regulatory T cell (T reg cell) differentiation and thereby contributes to oral tolerance (Mortha et al., 2014). Under conditions of tissue perturbation, ILC-mediated cross talk via GM-CSF can amplify inflammation through activation of monocytes and eosinophils (Griseri et al., 2015; Song et al., 2015). Although not addressed by our studies, similar cellular interaction modules might also be active in the lung, including in allergic inflammation (Behrens et al., 2015; Nobs et al., 2021). The biological function of constitutive Csf2 expression by pulmonary ILC2s, which we validated on an mRNA and protein level, however, remains unclear. It is possible that ILC2s create local tissue microenvironments that are enriched for GM-CSF and provide context-dependent support for particular myeloid compartments, such as eosinophils or DCs. Indeed, bone marrow ILC2s were recently reported to increase GM-CSF expression and to support hematopoietic recovery under conditions of stress hematopoiesis following treatment with 5-fluorouracil (Sudo et al., 2021). Further studies are thus required to reveal the more subtle consequences of lung hematopoietic Csf2 deficiency with the necessary spatial resolution.
The dispensability of hematopoietic GM-CSF for developing AMs was also surprising given a recent report that highlighted basophils as a critical regulator of AM development. In the developing lung, basophils were reported to express GM-CSF and to rely on GM-CSF receptor signaling; these cells were also suggested to critically regulate AM differentiation and numbers (Cohen et al., 2018). Our own investigations into basophil-AM biology using a genetic model of constitutive basophil depletion, however, did not confirm any changes in AM numbers nor any obvious defects in AM development (as determined by expression of signature surface markers) in the lungs of neonatal and adult mice. We can only speculate about the reason for these conflicting results. In addition to a genetic model of constitutive basophil depletion, Cohen et al. also employed an antibody-mediated basophil depletion strategy to functionally test the role of basophils in vivo. Newborn mice were treated intranasally with a relatively large amount of MAR-1 antibody, which was followed by lung cell analysis the following day. This may be a somewhat short time window to study basophil depletion–mediated consequences on AM development, though it does fall within the time window of fetal monocyte-to-AM differentiation. The MAR-1 antibody itself is known to bind and deplete FcεRIα-expressing cells including basophils, but concomitant antibody-mediated FcεRIα cross-linking has also been demonstrated to potently activate basophils, causing their degranulation and release of bioactive mediators in vitro and in vivo (Hübner et al., 2011; Pellefigues et al., 2019). Furthermore, a recent report demonstrated binding of MAR-1 to FcγRI and FcγRIV (Tang et al., 2019). Such off-target effects could alter gene expression in other cells, warranting caution when critically relying on the use of MAR-1 treatment. Although our studies using a genetic basophil-depletion strategy lack the high resolution needed to detect subtle transcriptional alterations in the AM compartment, our results cast doubt on the suggested importance of basophils for AM development.
Using comprehensive flow cytometry profiling, we identified AT2 cells as the major, if not exclusive, nonhematopoietic source of pulmonary GM-CSF under homeostatic conditions. Our embryonic analysis revealed that the production of epithelial GM-CSF, first detectable during late gestation and maintained across the entire life span, is acquired concomitantly with AT2 lineage commitment. The time point around which Csf2 expression is first detected (E16.5–E18.5) corresponds to the transition from the canalicular stage of lung organogenesis (E15–E17) to the saccular stage of development (E17–P0; Swarr and Morrisey, 2015). During this period, epithelial cells, which may derive from bipotent or precommitted progenitors, begin to express transcriptional signatures reminiscent of mature AT1s and/or mature AT2s while they progressively differentiate toward nascent AT1 and AT2 lineages (Desai et al., 2014; Frank et al., 2019; Gonzalez et al., 2019; Treutlein et al., 2014; Zepp et al., 2021). Although our Csf2 reporter is first detectable by flow cytometry in E17.5–E18.5 lungs, assigning the onset of fetal Csf2 expression to a precise developmental stage is complicated by not only the dynamic epithelial compartment differentiation and the coexpression of AT1 and AT2 markers by progenitors (including SP-C), but also the differences in maturation kinetics between GM-CSF and tdTomato, the latter being a slow-maturing fluorescent protein (Balleza et al., 2018; Gehart et al., 2019). Further studies are needed to definitively identify the first Csf2-expressing epithelial cells in the fetal lung and to determine whether these early GM-CSF producers are already committed to the AT2 lineage.
Our results also raise interesting questions about the regulation of this cytokine, which is constitutively expressed across vastly different cell types including AT2s, ILC2s, and T cells. Regulation of Csf2 expression in epithelial cells is particularly intriguing, as it may be suggestive of tissue-immune cross talk. Recent studies reported increased Csf2 expression in alveolar and bronchial epithelial cells downstream of IL-1R signaling following Legionella pneumophila infection and house dust mite treatment (Liu et al., 2020; Willart et al., 2012). Furthermore, reduced NF-κB signaling in alveolar epithelial cell cultures was found to be associated with their decreased Csf2 expression (Sturrock et al., 2018). It will be interesting to explore whether similar pathways are involved in the onset of Csf2 expression in early AT2s and to investigate whether homeostatic regulation of epithelial GM-CSF production is altered after birth.
Unexpectedly, given prior reports, we found that AMs are critically reliant not on hematopoietic GM-CSF but on epithelial GM-CSF derived from AT2s. Using genetic models of constitutive SPCCre-mediated and inducible SPCCreERT2-mediated deletion of Csf2, lungs from fetal, neonatal, and adult time points demonstrate that regulation of epithelial GM-CSF–AM-AT2 biology occurs on multiple levels. GM-CSF from early SPCCre-expressing epithelial cells or their progeny (likely bipotent epithelial progenitors or nascent AT2s) supports the proliferation and local expansion of fetal monocytes. These c-Myb–dependent myeloid progenitors can first be detected in lungs around E13 (Hoeffel et al., 2015; Schulz et al., 2012), several days before the first signs of GM-CSF expression based on our kinetic analysis. The mechanism of this anticipatory positioning of AM progenitors is unclear, and our findings suggest that additional pathways may exist to ensure their survival until lung epithelial–derived GM-CSF drives local expansion and instructs differentiation toward an AM fate. We found that this process is blocked in SPCCre/+;Csf2fl mice, which replicate the phenotype previously reported for global Csf2−/− mice (Schneider et al., 2014b), and that completion of AM differentiation postnatally is solely dependent on GM-CSF from nascent and mature AT2s. Thus, our studies provide further evidence for the existence of discrete tissue niches that critically regulate cell fate of developing tissue-resident immune cells (Guilliams and Scott, 2017). Additionally, constitutive GM-CSF production from AT2s is required for the maintenance of a mature AM compartment; our results reveal that AMs disappear within a few weeks of ablating AT2-specific Csf2 expression in adult mice. Indeed, a role for GM-CSF in regulating AM self-renewal was suggested based on the capacity to stimulate the proliferation of isolated AMs in vitro (Chen et al., 1988). It remains to be determined if this loss of AMs occurs due to abrogated self-renewal or enhanced cell death, the latter possibly caused by decreased expression of anti-apoptotic factors as suggested by studies using isolated AMs (Draijer et al., 2019).
Our studies unequivocally demonstrate the strict requirement for AT2-derived GM-CSF in AM development and maintenance. They also reveal the limitations of inferring ligand-receptor pairs and interaction modules primarily based on transcriptional data from dissociated organs lacking spatial context. However, emerging high-resolution, spatially resolved transcriptomics, which enables the mapping of gene expression data to protein and structural information, will likely prove very useful for predicting cellular interactions in complex tissues with better precision via an unbiased systems-level approach (Marx, 2021). In the lung, the demand for a niche factor produced by a discrete source, despite its even higher expression in other cell types that fail to compensate, is remarkable. For GM-CSF and AMs, this may in part be explained simply by the higher abundance of AT2s compared with ILC2s. The total number of AT2s in an adult mouse lung is estimated to be ∼107 (Dzhuraev et al., 2019), while that of ILC2s may be <105 (Ricardo-Gonzalez et al., 2018). In addition, AT2s are ideally positioned for intimate interaction with AMs, and organization of the AT2 secretion machinery might favor apical release of GM-CSF directly into the alveolar lumen where AMs reside. In contrast, ILC2s are found in collagen-rich adventitial niches that localize to bronchovascular areas, and they are rarely found in the alveolar parenchyma (Dahlgren et al., 2019). Therefore, ILC2-derived GM-CSF may be spatially segregated from AMs and consumed locally by other cell types. The processes that drive such niche organization and positioning of differentiating tissue-resident immune cells during organogenesis are largely unknown. It is intriguing to speculate that circulating fetal monocytes are recruited to the luminal side of the saccule by gradients of GM-CSF, where the highest GM-CSF concentration exists in what will later become the luminal side of mature alveoli and the niche environment for AMs.
Collectively, our study unveils AT2s as a constitutive and dominant source of GM-CSF in the alveolar niche, nurturing AMs in prenatal as well as postnatal stages of lung development. This AT2–GM-CSF–AM relationship persists from last gestation throughout adulthood, such that consistent AT2-derived GM-CSF is crucial for both AM development and maintenance.
Materials and methods
Mice
SftpcCre (SPCCre; Tg(Sftpc-Cre)1Blh; Okubo and Hogan, 2004) mice were generously provided by J. Rock via R. Locksley. SftpcCreERT2 (SPCCreERT2) knock-in mice (Sftpctm1(Cre/ERT2,rtTA)Hap; Chapman et al., 2011) were generously provided by H. Chapman via R. Locksley. Vav1iCre (B6.Cg-Commd10Tg(Vav1-iCre)A2Kio/J; 008610) mice were purchased from The Jackson Laboratory. Mcpt8YFP-Cre;R26DTA mice were previously described (Sullivan et al., 2011) and provided by R. Locksley. Rag2−/−;Il2rg−/− mice were provided by B. Becher via S. Tugues and were compared with in-house–bred wild-type controls.
Csf2fl mice were generated by homologous gene targeting in C57BL/6 embryonic stem cells using a previously described targeting strategy and reporter cassette that was modified to remove the BGHpA (von Moltke et al., 2016). Briefly, a pKO915-DT (Lexicon Genetics)–based targeting vector was generated with the following components encoded downstream of the DTA-SV40pA: a 2.6-kb 5′ homology arm; a loxP site and the complete third and fourth exon of Csf2, a reporter cassette; and a 3.2-kb 3′ homology arm beginning in the 3′ untranslated region of Csf2. The reporter cassette encoded (in order from 5′ to 3′) a loxP site, encephalomyocarditis virus internal ribosome entry site, tdTomato, and an frt-flanked neomycin resistance cassette. The final construct was linearized with NotI and transfected by electroporation into C57BL/6 embryonic stem cells. Cells were grown on irradiated feeders with the aminoglycoside G418 in the media, and neomycin-resistant clones were screened for 5′ and 3′ homologous recombination by PCR. Positive clones were selected and screened by PCR using the Expand Long Range dNTPack (Sigma), PCR buffer supplemented with 2% DMSO, and the primer pairs for the 5′ region (LR 5′ forward [fw]: 5′-AATCCAGTTCACGCAGCTTGTACG-3′; LR 5′ reverse [rev]: 5′-CCTTCAGCCCCTTGTTGAATACGC-3′) and the 3′ region (LR 3′ fw: 5′-TCGCCTTCTATCGCCTTCTTGACG-3′; LR 3′ rev: 5′-GATTTCCATCCATCTGAGACGGTG-3′) of the modified locus. Clones were injected into albino C57BL/6 blastocysts to generate chimeras, and the male pups with highest ratios of black-to-white coat color were selected to breed with homozygous Gt(ROSA26)FLP1/FLP1 females (The Jackson Laboratory; 009086) to excise the neomycin resistance cassette. Deletion of neomycin was confirmed by PCR using fw 5′-GGCTACTACTACGTGGACAC-3′ and rev 5′-CCAAGTTCCTGGCTCATTAC-3′. Offspring with the germline-transmitted and neo-deleted Csf2fl allele were backcrossed to C57BL/6 mice to cross out the FLP1 allele. Csf2fl genotyping was done using primers loxP fw: 5′-GCTTTTGAAATAGTGCTTCCCCAC-3′ and loxP rev: 5′-AGGTTCCCAGTTCCAAGTGCTGTC-3′, yielding a 163-bp wild-type band and a 131-bp floxed band, respectively. Deletion of the floxed allele (Csf2Δ) was detected by PCR using primers loxP fw (as above) and LR 5′ rev: 5′-CCTTCAGCCCCTTGTTGAATACGC-3′, yielding a 526-bp recombined band or a 1,811-bp floxed band.
Csf2Δ/Δ mice with global Csf2 deletion were generated from SPCCre/+;Csf2fl mice in which Cre is active with high frequency in the male germline, resulting in transmission of a recombined allele.
Germline Cre activity was found not only in the SPCCre line but occasionally also in the Vav1iCre line. Because of repeated germline Cre activity in both male and female Vav1iCre mice, it is possible to have transmission of a Csf2Δ allele to offspring. The germline-deleted allele, however, could not be readily distinguished from the Vav1iCre recombined allele in biopsy samples due to the presence of hematopoietic cells in the biopsies. Thus, all conditional knockout groups for both Vav1iCre and SPCCre were compared with their littermate Cre− controls and included both Csf2fl/fl and Csf2Δ/fl mice. Of note, the lungs of both Csf2Δ/fl and Csf2fl/fl mice contained mature AM populations and were free from any overt lung pathology (data not shown), as previously demonstrated by Dranoff et al. (1994).
All mice were generated on or previously backcrossed to the C57BL/6 background. Mice were bred and housed at the University of Zurich Laboratory Animal Sciences Center in Zurich, Switzerland, in a specific and opportunistic pathogen–free barrier facility. Experimental animals were internally transferred to a specific pathogen-free barrier facility and were housed in individually ventilated cage units. Animal experiments were reviewed and approved by the cantonal veterinary office of Zurich (permit numbers ZH134/2019 and ZH117/2019). Experiments were performed using age- and sex-matched groups whenever possible.
Mouse treatments
Adult SPCCreERT2;Csf2fl mice were treated with tamoxifen dissolved at 20 mg/ml in corn oil (Sigma). 20 µg tamoxifen per gram body weight was administered by oral gavage once a day over the course of 5 d. Mice were then sacrificed 3 wk after the last tamoxifen treatment.
Tissue processing for flow cytometry
Unless indicated otherwise, all adult mice were euthanized by lethal i.p. pentobarbital injection; pups were euthanized by either live decapitation or lethal i.p. pentobarbital injection. Lungs were perfused through the right cardiac ventricle with cold PBS and incubated in ice-cold RMPI-1640 until time of dissociation. Lungs underwent an initial mechanical dissociation using the m_lung_01_02 program on the gentleMACS dissociator (Miltenyi Biotec). Lung tissues were then digested with 50 µg/ml Liberase TM (Roche) and 25 µg/ml DNase I (Roche) in prewarmed RPMI-1640 for 35 min at 37°C with gentle rocking. This was followed by further dissociation using the m_lung_02_01 program on the gentleMACS dissociator. Single-cell suspensions were obtained after passing the homogenized samples through 70-µm cell strainers. Following red blood cell lysis (10× BD Pharm Lyse), cells were washed, filtered, and stained for flow cytometry.
Embryonic lungs were harvested from timed-pregnant dams at various time points ranging from E14.5 to E18.5. After euthanasia of the dam by CO2 inhalation, embryos were removed and placed in ice-cold PBS until time of dissection. For E16.5–E18.5 embryos, lungs were perfused intracardially with cold PBS before removal. In all cases, dissected lungs were incubated in cold FACS buffer (2% FBS and 0.05% sodium azide in PBS) until time of dissociation. Tissue was manually dissociated with scissors and digested with 50 µg/ml Liberase TM (Roche) and 25 µg/ml DNase I (Roche) in prewarmed RPMI-1640 for 15–20 min at 37°C with gentle rocking. Partially digested tissue was triturated with a P1000 pipette and further digested for 15–20 min at 37°C with gentle rocking. Upon completion of the digestion, tissue was triturated and filtered through 50-µm cell strainers. Following red blood cell lysis (10× BD Pharm Lyse), cells were washed, filtered, and stained for flow cytometry.
Where indicated, total cell numbers were enumerated with CountBright absolute counting beads (Thermo Fisher Scientific; C36950).
Flow cytometry
Fc block (anti-mouse CD16/32, 2.4G2) was purchased from BioXCell. Anti-mouse CD45 (30-F11) and rat anti-mouse SiglecF (E50-2440) were purchased from BD Biosciences; anti-mouse I-A/I-E (M5/114.15.2), anti-mouse CD31 (390), anti-mouse CD104 (346-11A), anti-mouse CD140a (APA5), anti-mouse CD326 (G8.8), anti-mouse CD49f (GoH3), anti-mouse Podoplanin (8.1.1), anti-mouse Ly-6A (D7), anti-mouse CD11c (N418), anti-mouse XCR1 (ZET), anti-mouse CD11c (M1/70), anti-mouse CD103 (2E7), anti-mouse Ly-6G (1A8), anti-mouse Ly-6C (HK1.4), anti-mouse CD64 (X54-5/7.1), anti-mouse F4/80 (BM8), anti-mouse GR-1 (RB6-8C5), anti-mouse NK1.1 (PK136), anti-mouse CD8a (53–6.7), anti-mouse CD196 (29-2L17), anti-mouse CD19 (6D5), anti-mouse CD4 (RM4-5), anti-mouse CD90.2 (30-H12), anti-mouse FcER1a (MAR-1), anti-mouse CD3 (17A2), anti-mouse TCRb (H57-597), anti-mouse CD49b (DX5), anti-mouse TCRgd (GL3), and anti-mouse SIRPɑ (P84) were purchased from BioLegend; anti-mouse NKp46 (29A1.4) was purchased from Invitrogen; and anti-mouse ST2 (DJ8) was purchased from MD Bioproducts. Dead cells were excluded using DAPI, fixable violet Live/Dead (Thermo Fisher Scientific), or Zombie Red (Biolegend). For intracellular staining of proteins, cells were fixed using either 4% paraformaldehyde (PFA; Electron Microscopy Science) or, in the case of Ki-67 staining, the FoxP3 fixation kit (Invitrogen). Following fixation, cells were washed and stained in permeabilization buffer (Thermo Fisher Scientific; 00–8333-56) with either anti-mouse Ki-67 (Thermo Fisher Scientific; SolA15); anti-mouse GM-CSF (MP1-22E9, Biolegend); or primary anti-dsRed (Sicgen; AB8181-200, 1:200) together with primary rabbit anti–proSP-C (Sigma; AB3786, 1:1,000) or primary rabbit anti–NaPi-IIb (provided by C. Wagner; Hilfiker et al., 1998; 1:400), followed by a wash and secondary donkey anti-goat IgG-Alexa Fluor 555 (Thermo Fisher Scientific; A-32816, 1:4,000) and donkey anti-rabbit IgG-Alexa Fluor 488 (Thermo Fisher Scientific; A-21206, 1:4,000).
Samples were analyzed on a FACSymphony (BD Biosciences) with five lasers (355 nm, 405 nm, 488 nm, 561 nm, and 639 nm). For doublet exclusion, samples were gated by forward scatter (FSC)–H and FSC-A, followed by side scatter (SSC)–H and SSC-A; FSC-A and SSC-A gating was used to exclude debris, followed by dead cell exclusion. Unless otherwise indicated, lineage (Lin)− cells were defined as lacking CD3, CD4, CD8, CD19, DX5, CD11b, CD11c, NK1.1, F4/80, Gr-1, FcεRIα, and Ter119. AMs were gated as CD45+Ly-6G−CD11c+SiglecF+CD64+. Data analysis was performed using FlowJo (Tree Star).
High-dimensional analysis
For spectral cytometry data, samples were analyzed on a Cytek Aurora (Cytek Biosciences) with five lasers (355 nm, 405 nm, 488 nm, 561 nm, and 640 nm). The compensation matrix was corrected using FlowJo software (Tree Star). Compensated samples were gated on live cells, singlets, and CD45+CD3−CD19−NK1.1−Ter119− cells (Fig. 6, G and H) or CD45+CD11b−CD11c−GR-1−Ter119−F4/80−FcεR1α− cells (Fig. S1 A), exported, and then imported into R Studio. Before automated high-dimensional data analysis, spectral cytometry data were transformed using an inverse hyperbolic sine (arcsinh) function with a cofactor in the range of 1,200 and 3,500. Additionally, all spectral cytometry data were normalized between 0 and 1 to the 99.999th percentile of the merged sample.
Automated population identification
For automated population identification, we first performed a step of FlowSOM clustering to generate a starting point of 100 nodes on preprocessed and combined flow cytometry datasets (Hartmann et al., 2016; Van Gassen et al., 2015). This was then followed by expert-guided manual meta-clustering using the parameters listed in each figure legend. The respective k-value was manually chosen (in the range of 7 and 12), and identified clusters were then annotated and merged based on a similarity of antigen expression in order to uphold the biological relevance of the dataset. Heatmaps display median expression levels of all markers per merged population and were plotted using the R package ‘‘pheatmap’’ (Kolde, 2019). For data visualization, we used Uniform Manifold Approximation and Projection (UMAP; McInnes et al., 2020 Preprint). To create a UMAP, we pooled 50,000 equally proportioned cells from each genotype.
Restimulation
For restimulation experiments, P10 lungs from Vav1iCre/+;Csf2fl and littermate controls were collected and processed as above. In a 96-well plate, lung cells were incubated for 4 h at 37°C and 5% CO2 in 200 μl of either complete RPMI media (RPMI-1640, 10% FBS, 1 mM Hepes, 100 U/ml/100 μg/ml Pen/Strep, and 55 µM 2-mercaptoethanol) or complete RPMI media supplemented with 1× Protein Transport Inhibitor Cocktail (Thermo Fisher Scientific; 00–4980-03), 0.05 μg/ml PMA (Sigma; P1585), and 0.5 μg/ml ionomycin (Sigma; I0634). Upon completion of the incubation, cells were washed, stained, and analyzed by flow cytometry.
Cell sorting
For quantitative PCR (qPCR) analysis of major Csf2-expressing cell populations (Fig. 1 H), Csf2-tdTomato+ cells were sorted from homogenized lungs that were pooled from two or three individual Csf2+/fl P10 mice. The three cell populations collected were ILC2s (tdTomato+CD45+Lin−CD3−Thy1.2+ST2+), T cells (tdTomato+CD45+CD3+), and epithelial cells (tdTomato+CD45−EpCAM+). Cells were sorted directly into RLT Lysis Buffer (Qiagen) using a FACSAria III (BD Biosciences). Each channel was loaded with 5,000–100,000 cells from each sample.
To verify recombination of the Csf2fl allele in ILC2s by qPCR (Fig. S2 A), ILC2s (CD45+Lin−CD3−Thy1.2+ST2+) were sorted from homogenized P10 lungs of Csf2fl and Vav1iCre/+;Csf2fl mice. Cells were sorted directly into RLT Lysis Buffer (Qiagen) using a FACSAria III (BD Biosciences). Each channel was loaded with 15,000–30,000 cells per sample.
For PCR analysis of the Csf2Δ allele (Fig. S2 B), CD45+ cells were sorted from homogenized P10 lungs of Csf2fl and Vav1iCre/+;Csf2fl mice. 600,000 cells were sorted into RPMI-1640 using a FACSAria III (BD Biosciences). The cells were then digested, and the recombination state of the Csf2fl allele was detected by standard PCR (Kapa Hotstart Mouse Genotyping Mix; Sigma; KK7352) using the same primer combination as for Csf2Δ genotyping listed above.
Quantitative RT–PCR
RNA was isolated from FACSorted cells using the Quick-RNA Microprep kit (Zymo Research; R1050) and reverse transcribed using the SuperScript IV VILO Master Mix with ezDNase Enzyme (Thermo Fisher Scientific; 11766050). The resulting cDNA was used as a template for qPCR with the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific; A25918) on a 7500 Fast Real-Time PCR System (Applied Biosystems). Transcripts were normalized to Rps17 (40S ribosomal protein S17) expression, and relative expression was shown as 2−ΔCt. Primer sequences were Rps17, 5′-CGCCATTATCCCCAGCAAG-3′, 5′-TGTCGGGATCCACCTCAATG-3′; Csf2 5′-ACATGACAGCCAGCTACTAC-3′, 5′-TCAAAGGGGATATCAGTCAG-3′.
Bronchoalveolar lavage
Mice were euthanized by lethal i.p. pentobarbital injection. The trachea was exposed and cannulated with a 20-G catheter (Surflo). 1 ml of cold PBS was instilled intratracheally; without removing the catheter or syringe, the lung was flushed three times with ∼0.7 ml of the instilled PBS. Total recovered volume was ∼0.8 ml/mouse. The BALF was then centrifuged at 1,500 rpm for 5 min at 4°C. The supernatant was collected and stored at −80°C until further use. The cell pellets were washed with cold FACS buffer and stored on ice until time of staining. Total protein in BALF was quantified using the Pierce BCA protein assay kit (Thermo Fisher Scientific; 23225) per manufacturer’s instructions. Total cholesterol in BALF was quantified using the Cholesterol/Cholesteryl Ester Assay Kit (Abcam; ab65359) per manufacturer’s instructions.
Conditioned lung medium for cytokine detection
P10 pups were euthanized by lethal i.p. pentobarbital injection; lungs were then removed under aseptic conditions and incubated in a 24-well plate with 1.5 ml of complete RPMI media at 37°C and 5% CO2. Lung conditioned media was harvested after 24 h and stored at −80°C until time of analysis. Cytokine concentration in lung conditioned media was measured using the Biolegend Legendplex Assay (740134). The assay was performed according to the manufacturer’s instruction, with the exception that 8 µl of sample, standard, assay buffer, beads, detection antibodies, and Streptavidin-PE were used instead of the recommended 25 µl.
Immunofluorescence
Mice were euthanized by lethal i.p. pentobarbital, and the trachea was exposed. Lungs were instilled intratracheally with 1 ml of ice-cold 4% PFA followed by prewarmed 0.5% low-melting agarose. Lungs were covered with ice until the agarose solidified, removed, and then incubated in cold 4% PFA for 3 h at 4°C in the dark. Samples were dehydrated in 30% sucrose for 24 h at 4°C, embedded in OCT Cryo Media (Medite Inc.) on dry ice, and stored at −80°C.
Frozen lungs embedded in OCT Cryo Media (Medite Inc.) were cut at the Cryostat (Leica CM3050) into 8-µm-thick sections and placed on glass slides. Before staining, sections were washed for 5 min in PBS, circled with a hydrophobic PAP pen (Dako pen), and blocked with blocking buffer for 1 h at room temperature. After blocking, samples were washed for 5 min in PBS and stained with primary antibodies rabbit anti-dsRed (1/250; Rockland; 600–401-379) and goat anti-CD45 (1/200; R&D Systems; AF114) for 1 h at room temperature. Sections were washed 3 × 5 min in PBS and stained with secondary antibodies donkey anti-rabbit Alexa Fluor 555 (1/1,000; Invitrogen; A-31572) and donkey anti-goat Alexa Fluor 488 (1/1,000; Invitrogen; A-11055) for 1 h at room temperature. Thereafter, samples were washed 3 × 5 min in PBS and stained with DAPI for 10 min. After a last wash with PBS for 5 min, tissues were mounted using Vectashield Antifade Mounting Medium (Vecta Laboratories). Fluorescence was detected with a Leica confocal microscope (Leica SP8 inverse; Leica Camera AG). Leica LAS X software was used to acquire and process the images.
Quantification and statistical analysis
All experiments were performed using randomly assigned mice without investigator blinding, with an aim of at least n = 3 per group per biological replicate. No statistical methods were used to predetermine sample size. All data points and n values reflect biological replicates. No data were excluded unless there was a major experimental error that justified the exclusion. Statistical analyses of two experimental groups were performed using unpaired, two-tailed Student’s t tests; if the P value was <0.05, results were deemed statistically significant. When statistical analyses of more than two experimental groups were required, ordinary one-way ANOVAs were employed; further analyses of statistically significant ANOVA results (P < 0.05) were performed using a Tukey’s multiple comparisons test. In the case of two independent variables (condition and genotype), a two-way ANOVA was employed; further analyses of statistically significant ANOVA results (P < 0.05) were performed using a Tukey’s multiple comparisons test. Intragroup variation was not assessed, except in the case of Mcpt8YFP-Cre/+ and Mcpt8YFP-Cre/YFP-Cre mice, which allowed for these two genotypes to be pooled and experimental groups to be assigned solely on the status of the Rosa26DTA allele. All statistical analyses were performed using Prism 8 (GraphPad Software).
Online supplemental material
Fig. S1 shows the contribution of the hematopoietic compartment to GM-CSF production in the lung and the gating strategy for lymphocytes. Fig. S2 shows a validation for effective Cre-recombination in Vav1iCre and that lung hematopoietic GM-CSF contributions, including from lymphocytes and basophils, are dispensable for AM development in the neonatal lung. Fig. S3 shows that EpCAM+ epithelial cells do not contribute to GM-CSF production in neonatal SPCCre lungs. Fig. S4 shows that hematopoietic-derived GM-CSF is not critical for AM fate specification during embryogenesis. Fig. S5 shows that lung hematopoietic GM-CSF contributions, including from lymphocytes and basophils, are dispensable for AM survival in the adult lung.
Acknowledgments
We thank Brigitte Herzog (University of Zurich [UZH], Zurich, Switzerland) for technical expertise; M. Kopf (Eidgenössische Technische Hochschule Zurich, Zurich, Switzerland) for helpful discussion and support with mouse rederivation; J. Rock (Genentech, South San Francisco, CA), H. Chapman (University of California, San Francisco, San Francisco, CA), and S. Tugues (UZH, Zurich, Switzerland) for providing mice; C. Wagner (UZH, Zurich, Switzerland) for providing the anti–NaPi-IIb; and members of the Schneider laboratory for helpful discussions.
This work was supported by grants from the University of California San Francisco Diabetes Research Center, US National Institutes of Health, the Howard Hughes Medical Institute, and the Sandler Asthma Basic Research Center at the University of California San Francisco to R.M. Locksley; the Swiss National Science Foundation (310030_188450) to B. Becher; and the Peter Hans Hofschneider Professorship for Molecular Medicine to C. Schneider.
Author contributions: J. Gschwend and S. Sherman conceived the study, designed and performed experiments, analyzed data, and wrote the manuscript with C. Schneider. F. Ridder performed additional experiments and assisted with data analysis. X. Feng helped with experiments. H.-E. Liang designed the original tdTomato reporter cassette, performed the ES cell work for Csf2fl mice, and provided scientific input. R.M. Locksley acquired funding and provided resources for Csf2 targeting and provided scientific input. B. Becher assisted with data analysis and provided scientific input. C. Schneider generated Csf2fl mice together with H.-E. Liang, directed the study, acquired funding, and wrote the paper with J. Gschwend and S. Sherman.
References
- Balleza, E., Kim J.M., and Cluzel P.. 2018. Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods. 15:47–51. 10.1038/nmeth.4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basil, M.C., Katzen J., Engler A.E., Guo M., Herriges M.J., Kathiriya J.J., Windmueller R., Ysasi A.B., Zacharias W.J., Chapman H.A., et al. 2020. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell. 26:482–502. 10.1016/j.stem.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher, B., Tugues S., and Greter M.. 2016. GM-CSF: From Growth Factor to Central Mediator of Tissue Inflammation. Immunity. 45:963–973. 10.1016/j.immuni.2016.10.026 [DOI] [PubMed] [Google Scholar]
- Behrens, F., Tak P.P., Østergaard M., Stoilov R., Wiland P., Huizinga T.W., Berenfus V.Y., Vladeva S., Rech J., Rubbert-Roth A., et al. 2015. MOR103, a human monoclonal antibody to granulocyte-macrophage colony-stimulating factor, in the treatment of patients with moderate rheumatoid arthritis: results of a phase Ib/IIa randomised, double-blind, placebo-controlled, dose-escalation trial. Ann. Rheum. Dis. 74:1058–1064. 10.1136/annrheumdis-2013-204816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blériot, C., Chakarov S., and Ginhoux F.. 2020. Determinants of Resident Tissue Macrophage Identity and Function. Immunity. 52:957–970. 10.1016/j.immuni.2020.05.014 [DOI] [PubMed] [Google Scholar]
- Chapman, H.A., Li X., Alexander J.P., Brumwell A., Lorizio W., Tan K., Sonnenberg A., Wei Y., and Vu T.H.. 2011. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J. Clin. Invest. 121:2855–2862. 10.1172/JCI57673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.D., Mueller M., and Chou T.H.. 1988. Role of granulocyte/macrophage colony-stimulating factor in the regulation of murine alveolar macrophage proliferation and differentiation. J. Immunol. 141:139–144. [PubMed] [Google Scholar]
- Cohen, M., Giladi A., Gorki A.D., Solodkin D.G., Zada M., Hladik A., Miklosi A., Salame T.M., Halpern K.B., David E., et al. 2018. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell. 175:1031–1044.e18. 10.1016/j.cell.2018.09.009 [DOI] [PubMed] [Google Scholar]
- Dahlgren, M.W., Jones S.W., Cautivo K.M., Dubinin A., Ortiz-Carpena J.F., Farhat S., Yu K.S., Lee K., Wang C., Molofsky A.V., et al. 2019. Adventitial Stromal Cells Define Group 2 Innate Lymphoid Cell Tissue Niches. Immunity. 50:707–722.e6. 10.1016/j.immuni.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, L.C., Jenkins S.J., Allen J.E., and Taylor P.R.. 2013. Tissue-resident macrophages. Nat. Immunol. 14:986–995. 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kleer, I.M., Kool M., de Bruijn M.J., Willart M., van Moorleghem J., Schuijs M.J., Plantinga M., Beyaert R., Hams E., Fallon P.G., et al. 2016. Perinatal Activation of the Interleukin-33 Pathway Promotes Type 2 Immunity in the Developing Lung. Immunity. 45:1285–1298. 10.1016/j.immuni.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Desai, T.J., Brownfield D.G., and Krasnow M.A.. 2014. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 507:190–194. 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati, Y., Blaskovic S., Ruchonnet-Métrailler I., Lascano Maillard J., and Barazzone-Argiroffo C.. 2020. Simultaneous isolation of endothelial and alveolar epithelial type I and type II cells during mouse lung development in the absence of a transgenic reporter. Am. J. Physiol. Lung Cell. Mol. Physiol. 318:L619–L630. 10.1152/ajplung.00227.2019 [DOI] [PubMed] [Google Scholar]
- Draijer, C., Penke L.R.K., and Peters-Golden M.. 2019. Distinctive Effects of GM-CSF and M-CSF on Proliferation and Polarization of Two Major Pulmonary Macrophage Populations. J. Immunol. 202:2700–2709. 10.4049/jimmunol.1801387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranoff, G., Crawford A.D., Sadelain M., Ream B., Rashid A., Bronson R.T., Dickersin G.R., Bachurski C.J., Mark E.L., Whitsett J.A., et al. 1994. Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science. 264:713–716. 10.1126/science.8171324 [DOI] [PubMed] [Google Scholar]
- Dzhuraev, G., Rodríguez-Castillo J.A., Ruiz-Camp J., Salwig I., Szibor M., Vadász I., Herold S., Braun T., Ahlbrecht K., Atzberger A., et al. 2019. Estimation of absolute number of alveolar epithelial type 2 cells in mouse lungs: a comparison between stereology and flow cytometry. J. Microsc. 275:36–50. 10.1111/jmi.12800 [DOI] [PubMed] [Google Scholar]
- Evren, E., Ringqvist E., and Willinger T.. 2020. Origin and ontogeny of lung macrophages: from mice to humans. Immunology. 160:126–138. 10.1111/imm.13154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evren, E., Ringqvist E., Tripathi K.P., Sleiers N., Rives I.C., Alisjahbana A., Gao Y., Sarhan D., Halle T., Sorini C., et al. 2021. Distinct developmental pathways from blood monocytes generate human lung macrophage diversity. Immunity. 54:259–275.e7. 10.1016/j.immuni.2020.12.003 [DOI] [PubMed] [Google Scholar]
- Frank, D.B., Penkala I.J., Zepp J.A., Sivakumar A., Linares-Saldana R., Zacharias W.J., Stolz K.G., Pankin J., Lu M., Wang Q., et al. 2019. Early lineage specification defines alveolar epithelial ontogeny in the murine lung. Proc. Natl. Acad. Sci. USA. 116:4362–4371. 10.1073/pnas.1813952116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehart, H., van Es J.H., Hamer K., Beumer J., Kretzschmar K., Dekkers J.F., Rios A., and Clevers H.. 2019. Identification of Enteroendocrine Regulators by Real-Time Single-Cell Differentiation Mapping. Cell. 176:1158–1173.e16. 10.1016/j.cell.2018.12.029 [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero, E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F., and Rodewald H.R.. 2015. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 518:547–551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, R., Leaffer D., Chapin C., Gillespie A.M., Eckalbar W., and Dobbs L.. 2019. Cell fate analysis in fetal mouse lung reveals distinct pathways for TI and TII cell development. Am. J. Physiol. Lung Cell. Mol. Physiol. 317:L653–L666. 10.1152/ajplung.00503.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griseri, T., Arnold I.C., Pearson C., Krausgruber T., Schiering C., Franchini F., Schulthess J., McKenzie B.S., Crocker P.R., and Powrie F.. 2015. Granulocyte Macrophage Colony-Stimulating Factor-Activated Eosinophils Promote Interleukin-23 Driven Chronic Colitis. Immunity. 43:187–199. 10.1016/j.immuni.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams, M., and Scott C.L.. 2017. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17:451–460. 10.1038/nri.2017.42 [DOI] [PubMed] [Google Scholar]
- Guilliams, M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., Deswarte K., Malissen B., Hammad H., and Lambrecht B.N.. 2013. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210:1977–1992. 10.1084/jem.20131199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams, M., Thierry G.R., Bonnardel J., and Bajenoff M.. 2020. Establishment and Maintenance of the Macrophage Niche. Immunity. 52:434–451. 10.1016/j.immuni.2020.02.015 [DOI] [PubMed] [Google Scholar]
- Hamilton, J.A. 2020. GM-CSF in inflammation. J. Exp. Med. 217:e20190945. 10.1084/jem.20190945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, F.J., Bernard-Valnet R., Quériault C., Mrdjen D., Weber L.M., Galli E., Krieg C., Robinson M.D., Nguyen X.H., Dauvilliers Y., et al. 2016. High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J. Exp. Med. 213:2621–2633. 10.1084/jem.20160897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., et al. 2013. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 38:792–804. 10.1016/j.immuni.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker, H., Hattenhauer O., Traebert M., Forster I., Murer H., and Biber J.. 1998. Characterization of a murine type II sodium-phosphate cotransporter expressed in mammalian small intestine. Proc. Natl. Acad. Sci. USA. 95:14564–14569. 10.1073/pnas.95.24.14564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel, G., Chen J., Lavin Y., Low D., Almeida F.F., See P., Beaudin A.E., Lum J., Low I., Forsberg E.C., et al. 2015. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 42:665–678. 10.1016/j.immuni.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner, M.P., Larson D., Torrero M.N., Mueller E., Shi Y., Killoran K.E., and Mitre E.. 2011. Anti-FcεR1 antibody injections activate basophils and mast cells and delay Type 1 diabetes onset in NOD mice. Clin. Immunol. 141:205–217. 10.1016/j.clim.2011.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman, J.A., Hull W.M., Dranoff G., Mulligan R.C., and Whitsett J.A.. 1996. Pulmonary epithelial cell expression of GM-CSF corrects the alveolar proteinosis in GM-CSF-deficient mice. J. Clin. Invest. 97:649–655. 10.1172/JCI118461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell, T., and Bell T.J.. 2014. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 14:81–93. 10.1038/nri3600 [DOI] [PubMed] [Google Scholar]
- Kitamura, T., Tanaka N., Watanabe J., Uchida K., Kanegasaki S., Yamada Y., and Nakata K.. 1999. Idiopathic pulmonary alveolar proteinosis as an autoimmune disease with neutralizing antibody against granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 190:875–880. 10.1084/jem.190.6.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde, R. 2019. pheatmap: Pretty Heatmaps. https://rdrr.io/cran/pheatmap/ (accessed May 26, 2021).
- Komuczki, J., Tuzlak S., Friebel E., Hartwig T., Spath S., Rosenstiel P., Waisman A., Opitz L., Oukka M., Schreiner B., et al. 2019. Fate-Mapping of GM-CSF Expression Identifies a Discrete Subset of Inflammation-Driving T Helper Cells Regulated by Cytokines IL-23 and IL-1β. Immunity. 50:1289–1304.e6. 10.1016/j.immuni.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Kopf, M., Schneider C., and Nobs S.P.. 2015. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 16:36–44. 10.1038/ni.3052 [DOI] [PubMed] [Google Scholar]
- Li, F., Okreglicka K.M., Pohlmeier L.M., Schneider C., and Kopf M.. 2020. Fetal monocytes possess increased metabolic capacity and replace primitive macrophages in tissue macrophage development. EMBO J. 39:e103205. 10.15252/embj.2019103205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Gu Y., Chakarov S., Bleriot C., Kwok I., Chen X., Shin A., Huang W., Dress R.J., Dutertre C.A., et al. 2019. Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell. 178:1509–1525.e19. 10.1016/j.cell.2019.08.009 [DOI] [PubMed] [Google Scholar]
- Liu, X., Boyer M.A., Holmgren A.M., and Shin S.. 2020. Legionella-Infected Macrophages Engage the Alveolar Epithelium to Metabolically Reprogram Myeloid Cells and Promote Antibacterial Inflammation. Cell Host Microbe. 28:683–698.e6. 10.1016/j.chom.2020.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moczygemba, M., Doan M.L., Elidemir O., Fan L.L., Cheung S.W., Lei J.T., Moore J.P., Tavana G., Lewis L.R., Zhu Y., et al. 2008. Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRalpha gene in the X chromosome pseudoautosomal region 1. J. Exp. Med. 205:2711–2716. 10.1084/jem.20080759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx, V. 2021. Method of the Year: spatially resolved transcriptomics. Nat. Methods. 18:9–14. 10.1038/s41592-020-01033-y [DOI] [PubMed] [Google Scholar]
- Mass, E., Ballesteros I., Farlik M., Halbritter F., Günther P., Crozet L., Jacome-Galarza C.E., Händler K., Klughammer J., Kobayashi Y., et al. 2016. Specification of tissue-resident macrophages during organogenesis. Science. 353:aaf4238. 10.1126/science.aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, L., Healy J., and Melville J.. 2020. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. https://arxiv.org/abs/1802.03426 (Preprint posted September 18, 2020)
- Mortha, A., Chudnovskiy A., Hashimoto D., Bogunovic M., Spencer S.P., Belkaid Y., and Merad M.. 2014. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 343:1249288. 10.1126/science.1249288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinakamura, R., Nakayama N., Hirabayashi Y., Inoue T., Aud D., McNeil T., Azuma S., Yoshida S., Toyoda Y., Arai K., et al. 1995. Mice deficient for the IL-3/GM-CSF/IL-5 beta c receptor exhibit lung pathology and impaired immune response, while beta IL3 receptor-deficient mice are normal. Immunity. 2:211–222. 10.1016/1074-7613(95)90046-2 [DOI] [PubMed] [Google Scholar]
- Nobs, S.P., Pohlmeier L., Li F., Kayhan M., Becher B., and Kopf M.. 2021. GM-CSF instigates a dendritic cell-T-cell inflammatory circuit that drives chronic asthma development. J. Allergy Clin. Immunol. 147:2118–2133.e3. 10.1016/j.jaci.2020.12.638 [DOI] [PubMed] [Google Scholar]
- Nussbaum, J.C., Van Dyken S.J., von Moltke J., Cheng L.E., Mohapatra A., Molofsky A.B., Thornton E.E., Krummel M.F., Chawla A., Liang H.E., and Locksley R.M.. 2013. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 502:245–248. 10.1038/nature12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo, T., and Hogan B.L.. 2004. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J. Biol. 3:11. 10.1186/jbiol3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellefigues, C., Mehta P., Prout M.S., Naidoo K., Yumnam B., Chandler J., Chappell S., Filbey K., Camberis M., and Le Gros G.. 2019. The Basoph8 Mice Enable an Unbiased Detection and a Conditional Depletion of Basophils. Front. Immunol. 10:2143. 10.3389/fimmu.2019.02143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez, R.R., Van Dyken S.J., Schneider C., Lee J., Nussbaum J.C., Liang H.E., Vaka D., Eckalbar W.L., Molofsky A.B., Erle D.J., and Locksley R.M.. 2018. Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol. 19:1093–1099. 10.1038/s41590-018-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb, L., Drinkwater C.C., Metcalf D., Li R., Köntgen F., Nicola N.A., and Begley C.G.. 1995. Hematopoietic and lung abnormalities in mice with a null mutation of the common beta subunit of the receptors for granulocyte-macrophage colony-stimulating factor and interleukins 3 and 5. Proc. Natl. Acad. Sci. USA. 92:9565–9569. 10.1073/pnas.92.21.9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluzzo, S., Gorki A.D., Rana B.M.J., Martins R., Scanlon S., Starkl P., Lakovits K., Hladik A., Korosec A., Sharif O., et al. 2017. First-Breath-Induced Type 2 Pathways Shape the Lung Immune Environment. Cell Rep. 18:1893–1905. 10.1016/j.celrep.2017.01.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., Nobs S.P., Heer A.K., Kurrer M., Klinke G., van Rooijen N., Vogel J., and Kopf M.. 2014a. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 10:e1004053. 10.1371/journal.ppat.1004053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., and Kopf M.. 2014b. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15:1026–1037. 10.1038/ni.3005 [DOI] [PubMed] [Google Scholar]
- Schneider, C., Nobs S.P., Heer A.K., Hirsch E., Penninger J., Siggs O.M., and Kopf M.. 2017. Frontline Science: Coincidental null mutation of Csf2rα in a colony of PI3Kγ-/- mice causes alveolar macrophage deficiency and fatal respiratory viral infection. J. Leukoc. Biol. 101:367–376. 10.1189/jlb.4HI0316-157R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., Lee J., Koga S., Ricardo-Gonzalez R.R., Nussbaum J.C., Smith L.K., Villeda S.A., Liang H.E., and Locksley R.M.. 2019. Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity. 50:1425–1438.e5. 10.1016/j.immuni.2019.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Sheridan, J.W., and Metcalf D.. 1973. A low molecular weight factor in lung-conditioned medium stimulating granulocyte and monocyte colony formation in vitro. J. Cell. Physiol. 81:11–23. 10.1002/jcp.1040810103 [DOI] [PubMed] [Google Scholar]
- Song, C., Lee J.S., Gilfillan S., Robinette M.L., Newberry R.D., Stappenbeck T.S., Mack M., Cella M., and Colonna M.. 2015. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 212:1869–1882. 10.1084/jem.20151403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley, E., Lieschke G.J., Grail D., Metcalf D., Hodgson G., Gall J.A., Maher D.W., Cebon J., Sinickas V., and Dunn A.R.. 1994. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc. Natl. Acad. Sci. USA. 91:5592–5596. 10.1073/pnas.91.12.5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer, C.A., Martinez-Gonzalez I., Ghaedi M., Allinger P., Mathä L., and Takei F.. 2017. Group 2 innate lymphoid cell activation in the neonatal lung drives type 2 immunity and allergen sensitization. J. Allergy Clin. Immunol. 140:593–595.e3. 10.1016/j.jaci.2016.12.984 [DOI] [PubMed] [Google Scholar]
- Sturrock, A., Woller D., Freeman A., Sanders K., and Paine R. III. 2018. Consequences of Hypoxia for the Pulmonary Alveolar Epithelial Cell Innate Immune Response. J. Immunol. 201:3411–3420. 10.4049/jimmunol.1701387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo, T., Motomura Y., Okuzaki D., Hasegawa T., Yokota T., Kikuta J., Ao T., Mizuno H., Matsui T., Motooka D., et al. 2021. Group 2 innate lymphoid cells support hematopoietic recovery under stress conditions. J. Exp. Med. 218:e20200817. 10.1084/jem.20200817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.M., Liang H.E., Bando J.K., Wu D., Cheng L.E., McKerrow J.K., Allen C.D., and Locksley R.M.. 2011. Genetic analysis of basophil function in vivo. Nat. Immunol. 12:527–535. 10.1038/ni.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Sakagami T., Rubin B.K., Nogee L.M., Wood R.E., Zimmerman S.L., Smolarek T., Dishop M.K., Wert S.E., Whitsett J.A., et al. 2008. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. J. Exp. Med. 205:2703–2710. 10.1084/jem.20080990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Arumugam P., Sakagami T., Lachmann N., Chalk C., Sallese A., Abe S., Trapnell C., Carey B., Moritz T., et al. 2014. Pulmonary macrophage transplantation therapy. Nature. 514:450–454. 10.1038/nature13807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarr, D.T., and Morrisey E.E.. 2015. Lung endoderm morphogenesis: gasping for form and function. Annu. Rev. Cell Dev. Biol. 31:553–573. 10.1146/annurev-cellbio-100814-125249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X.Z., Jung J.B., and Allen C.D.C.. 2019. A case of mistaken identity: The MAR-1 antibody to mouse FcεRIα cross-reacts with FcγRI and FcγRIV. J. Allergy Clin. Immunol. 143:1643–1646.e6. 10.1016/j.jaci.2018.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe, R.M., Redente E.F., Yu Y.R., Herold S., Sperling A.I., Curtis J.L., Duggan R., Swaminathan S., Nakano H., Zacharias W.J., et al. 2019. Improving the Quality and Reproducibility of Flow Cytometry in the Lung. An Official American Thoracic Society Workshop Report. Am. J. Respir. Cell Mol. Biol. 61:150–161. 10.1165/rcmb.2019-0191ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traebert, M., Hattenhauer O., Murer H., Kaissling B., and Biber J.. 1999. Expression of type II Na-P(i) cotransporter in alveolar type II cells. Am. J. Physiol. 277:L868–L873. [DOI] [PubMed] [Google Scholar]
- Trapnell, B.C., Nakata K., Bonella F., Campo I., Griese M., Hamilton J., Wang T., Morgan C., Cottin V., and McCarthy C.. 2019. Pulmonary alveolar proteinosis. Nat. Rev. Dis. Primers. 5:16. 10.1038/s41572-019-0066-3 [DOI] [PubMed] [Google Scholar]
- Treutlein, B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., and Quake S.R.. 2014. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 509:371–375. 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Laar, L., Saelens W., De Prijck S., Martens L., Scott C.L., Van Isterdael G., Hoffmann E., Beyaert R., Saeys Y., Lambrecht B.N., and Guilliams M.. 2016. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity. 44:755–768. 10.1016/j.immuni.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Van Gassen, S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., and Saeys Y.. 2015. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 87:636–645. 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- von Moltke, J., Ji M., Liang H.E., and Locksley R.M.. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 529:221–225. 10.1038/nature16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wert, S.E., Glasser S.W., Korfhagen T.R., and Whitsett J.A.. 1993. Transcriptional elements from the human SP-C gene direct expression in the primordial respiratory epithelium of transgenic mice. Dev. Biol. 156:426–443. 10.1006/dbio.1993.1090 [DOI] [PubMed] [Google Scholar]
- Whitsett, J.A., Kalin T.V., Xu Y., and Kalinichenko V.V.. 2019. Building and Regenerating the Lung Cell by Cell. Physiol. Rev. 99:513–554. 10.1152/physrev.00001.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willart, M.A., Deswarte K., Pouliot P., Braun H., Beyaert R., Lambrecht B.N., and Hammad H.. 2012. Interleukin-1α controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. J. Exp. Med. 209:1505–1517. 10.1084/jem.20112691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willinger, T., Rongvaux A., Takizawa H., Yancopoulos G.D., Valenzuela D.M., Murphy A.J., Auerbach W., Eynon E.E., Stevens S., Manz M.G., and Flavell R.A.. 2011. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc. Natl. Acad. Sci. USA. 108:2390–2395. 10.1073/pnas.1019682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona, S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp, J.A., Morley M.P., Loebel C., Kremp M.M., Chaudhry F.N., Basil M.C., Leach J.P., Liberti D.C., Niethamer T.K., Ying Y., et al. 2021. Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science. 371:eabc3172. 10.1126/science.abc3172 [DOI] [PMC free article] [PubMed] [Google Scholar]