Abstract
The yeast CTDK-I complex has been implicated in phosphorylation of the carboxy-terminal domain of the RNA polymerase II and in transcription control. It is composed of three polypeptides: Ctk1p and Ctk2p, a cyclin-dependent kinase and a C-type cyclin subunit, respectively; and Ctk3p, a third subunit of unknown function. Cyclins are regulatory proteins whose expression is tightly controlled at the protein level. In this study, we examined the regulation of Ctk2p expression in vivo. Surprisingly, unlike what has been described for cell cycle cyclins, steady-state levels of Ctk2p are composed of two relatively abundant forms, one of them phosphorylated. We show that this phosphorylated form is extremely unstable (half-life, 5 min) and that rapid proteolysis of Ctk2p exhibits growth-related regulation. Furthermore, our data establish that similar to the case for other naturally short-lived proteins, Ctk2p degradation is mediated by the ubiquitin-proteasome pathway. This is the first demonstration that a C-type cyclin is phosphorylated and targeted to the proteasome. Strikingly, neither phosphorylation nor destruction of Ctk2p requires its associated kinase Ctk1p, a feature fundamentally different from that which has been observed for cell cycle cyclins.
Cyclin-dependent protein kinases (CDKs) were first identified as central regulators of the major transitions of the eukaryotic cell division cycle. Their activity is determined by cyclin binding, by both positive and negative regulatory phosphorylation, and by polypeptide CDK inhibitors (40, 41). A single cyclin-dependent kinase subunit associates sequentially with multiple cyclin partners whose abundance fluctuates during the cell cycle (43). Indeed, one aspect of CDK regulation is triggered by the rapid destruction of their cyclin partners (23). Several studies both in yeast and in mammals have demonstrated that cyclins are selectively degraded by the ubiquitin-dependent proteasome pathway (9, 16, 53, 67). Substrates are first marked for degradation by conjugation with several molecules of ubiquitin, a highly conserved 76-amino-acid-long polypeptide that is joined reversibly by a covalent linkage. Usually, ubiquitin molecules are linked tandemly at lysine 48 to form multiubiquitin chains on substrate proteins (47). The resulting ubiquitin chains are in a dynamic state, subject to either further rounds of ubiquitination, ubiquitin removal by deubiquitinating enzymes, or degradation by the 26S proteasome (20). The 26S proteasome is a nuclear and cytosolic multicatalytic proteinase that breaks down targeted substrates to short peptides and recycles the ubiquitin molecules (18).
How proteins are targeted to the ubiquitin-mediated proteasome degradation pathway is not well understood. At least two motifs, which are also found in other ubiquitinated proteins, are necessary for cell cycle cyclin targeting to the ubiquitin proteolytic pathway. In the yeast Saccharomyces cerevisiae, mitotic (Clb1 to Clb4) and S-phase (Clb5 and Clb6) cyclins are targeted by a consensus sequence called the destruction box (16, 24, 42), while targeting of G1 cyclins (Cln1 to Cln3) requires a carboxy-terminal PEST-rich region (49, 51). Regulation of cyclin destruction has also been shown to be dependent on their associated CDKs (7), and several studies indicated that G1 cyclin phosphorylation provides a signal that is necessary for their rapid degradation, suggesting that the relevant aspect of PEST sequences might be their richness in CDK phosphorylation target sites rather than their PEST amino acids per se. Indeed, mutations in the Cdc28p kinase consensus phosphorylation sites of G1 cyclins resulted in their stabilization (28, 68). In summary, phosphorylation of cyclin PEST regions by their associating CDKs triggers their degradation. Thus, expression of a cyclin may be negatively regulated by the same cell cycle machinery that it activates.
C-type cyclins were originally identified in Drosophila melanogaster and humans because they could rescue G1 cyclin function in yeast (33, 34). Since then, cyclins with significant homology to the cyclin box region of cyclin C have also been found in Schizosaccharomyces pombe (14, 39). Except for a recent study carried out in murine cells (36), there has been no evidence supporting a critical role for C-type cyclins in cell cycle progression. Rather, C-type cyclins seem to activate CDKs involved in the regulation of RNA polymerase II transcription. Unlike cell cycle cyclins, their expression does not fluctuate during the cell cycle (6, 39). The S. cerevisiae genome encodes three C-type cyclins that have been identified as subunits of RNA polymerase II carboxy-terminal domain (CTD) kinases. The RNA polymerase II CTD plays an essential role in mRNA synthesis, and its phosphorylation is a key feature of its function (8). The first CTD kinase, Kin28p, is associated with Ccl1p, a C-type cyclin, and with a third factor, Rig2p (12, 61, 62). This complex, like its human counterpart the Cdk7p kinase complex (37), is a component of the general transcription factor TFIIH (44, 58). Kin28p kinase activity is essential, as it is required for basal transcription (62). The second CDK implicated in CTD phosphorylation is the Srb10p/Ume5p/Ssn3p kinase and its cyclin partner Srb11p/Ume3p/Ssn8p, a kinase pair that is a component of the holoenzyme (35). The Srb10/11 complex is structurally related to mammalian Cdk8p kinase and its associated cyclin C (59). It was first characterized as a regulator of meiosis-specific genes in response to glucose (57). Since then, several studies established that Srb10/11 kinase is implicated in transcription repression of sets of genes (1, 27, 65). Cooper et al. (6) showed that Ume3p cyclin is destroyed during meiosis and when cultures are subjected to heat shock. Mutational analysis identified several regions implicated in Ume3p stability: a PEST-rich region, a destruction box-like motif (RXXL), and the highly conserved cyclin box. Strikingly, this process does not appear to be affected in mutants defective for ubiquitin-mediated protein destruction. Furthermore, in contrast to what has been described for yeast cell cycle cyclins, the CDK activated by Ume3p is not required for the rapid degradation of this cyclin.
The third yeast C-type cyclin is a component of the CTDK-I complex, which was isolated in vitro by its ability to phosphorylate CTD-containing fusion proteins (29). It is composed of three subunits: the CTK1 and CTK2 genes encode a kinase and a C-type cyclin, respectively; the third subunit, Ctk3p, shows no similarity to other known proteins (30, 56). Deletion of any of the CTK genes generates cryosensitive cells. CTDK-I is related to the Srb10/11 kinase complex, although it seems to display a distinct role in transcriptional control (26). In vitro studies carried out with HeLa nuclear extracts have shown that CTDK-I can modulate the elongation efficiency of RNA polymerase II (31). In humans, the closest kinase to Ctk1p is Cdk9p, a CTD kinase that was first described as a positive transcription elongation factor (38). A recent study showed that Cdk9p is associated with a C-type cyclin that also interacts specifically with the human immunodeficiency virus type 1 transactivator protein Tat, which acts to enhance the processing efficiency of RNA polymerase II (66, 69). This finding led the authors to propose that this cyclin might be the TAR RNA-binding cofactor for Tat (66).
C-type cyclins constitute a divergent family of cyclins that display a prime role in RNA polymerase II transcription. Regulation of their expression is poorly documented. The yeast cyclin Ume3p is highly unstable under certain conditions, but how this regulation is achieved remains unknown. To try to understand the regulation of their expression, which probably exhibits specific features, we have studied the expression of Ctk2p, the CTDK-I C-type cyclin subunit. We show that at steady state under standard exponential growth conditions, Ctk2p comprises two forms, one a phosphorylated form that is very unstable. Our data establish that rapid proteolysis of Ctk2p is mediated by the ubiquitin-proteasome pathway. Strikingly, neither phosphorylation nor degradation of Ctk2p requires its activated kinase.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The yeast strains used in this study were WCG4a (MATa ura3 leu2-3,112 his3-11,15) and its isogenic pre1-1 pre2-2 derivative WCG4-11/22a, constructed by Richter-Ruoff et al. (48), and MHY501 (MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1) and its isogenic Δdoa4 derivative MHY623, described by Papa and Hochstrasser (45). The Δctk strains were derived from W303-1B (MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura 3-1). Null mutations were obtained by one-step gene replacement (50) by transformation with PCR fragments of the TRP1 gene, using long oligonucleotides containing 5′ and 3′ sequences of each CTK gene.
Yeast cultures were grown at 30°C in SD (minimal) medium containing 0.67% yeast nitrogen base without amino acids (Difco) supplemented with appropriate nutrients for auxotrophy, plus 2% glucose as a carbon source. For induction of CUP1 promoter-dependent ubiquitin alleles, CuSO4 was added either at 0.2 mM for 5 h or at 0.1 mM overnight. For plasmid loss tests, transformants were grown in YPD (rich) medium (1% yeast extract, 1% Bacto Peptone, 2% glucose) for several generations. Culture dilutions were plated onto nonselective medium prior to replica plating onto selective medium. Yeast colonies unable to grow were scored. Yeast transformations were performed by the LiCl method (22).
Plasmids.
CTK1 and CTK3 genes were obtained as BamHI fragments (2,240 and 1,560 bp, respectively) by PCR on genomic DNA. The CTK2 gene was carried into a BglII cosmid fragment (1,970 bp). To verify the disrupted strains, each gene was cloned into a URA3 CEN plasmid. For overexpression experiments, each gene was cloned on a URA3 2μm plasmid. The epitope-tagged CTK2 gene was constructed via several cloning steps. First, a PCR BamHI fragment containing the CTK2 open reading frame was inserted into the BamHI site of plasmid TL38 (LEU2 2μm) (4). The resulting construct, TL38-CTK2, allowed the expression of a Ctk2p protein fused to two influenza virus hemagglutinin (HA) epitope tags at its N terminus, under the control of the PGK promoter. Second, CTK2 promoter sequences were cloned into pFL38 (URA3 CEN) (3) by insertion of a 940-bp BamHI-NcoI/SalI PCR fragment into the BamHI and SalI sites of the vector, yielding pFL38-PrCTK2. Third, the tagged CTK2 open reading frame was cloned under the control of the CTK2 promoter by insertion of the TL38-CTK2 NcoI-XhoI fragment into the NcoI and XhoI sites of pFL38-PrCTK2. The final construct, CTK2-HA (URA3 CEN), allows expression of a tagged Ctk2p under the control of its own promoter. Plasmid YEp96 (TRP1 2μm Ub) (10) contains a synthetic yeast wild-type ubiquitin gene under the control of the copper-inducible CUP1 promoter. YEp105 (TRP1 2μm Myc-Ub) and YEp110 (TRP1 2μm UbK48R) are identical to YEp96, except that they encode a c-Myc-tagged ubiquitin (11) and a mutant ubiquitin with an arginine instead of lysine at amino acid position 48, respectively (21).
Yeast cell extracts and Western immunoblotting.
Cells were grown to an optical density at 600 nm (OD600) of 0.8 or 3. When indicated, cycloheximide (10 μg/ml; Sigma) was added, and aliquots were frozen after different times of incubation. Cells (10 OD600 units) were resuspended in 100 μl of ice-cold trichloroacetic acid (TCA) buffer (20 mM Tris-HCl [pH 8], 50 mM ammonium acetate, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Cell suspensions were transferred to microcentrifuge tubes containing 100 μl of glass beads (425 to 600 μm in diameter; Sigma) and 100 μl of ice-cold 20% TCA. Cells were disrupted by vigorous vortexing (four times for 1 min each) with 1 min on ice between each vortexing. The beads were washed with 500 μl of an ice-cold 1:1 mixture of 20% TCA and TCA buffer and vortexing twice for 1 min each time. Resulting extracts were centrifuged for 20 min at 4°C at 15,000 rpm, and pellets were resuspended in 100 μl of TCA-Laemmli loading buffer (120 mM Tris base, 3.5% sodium dodecyl sulfate [SDS], 8 mM EDTA, 5% β-mercaptoethanol, 1 mM PMSF, 15% glycerol, 0.01% bromophenol blue). Samples were boiled for 10 min and centrifuged at 15,000 rpm for 10 min. Supernatants were collected, and 10-μl aliquots were immediately loaded on an SDS–13% polyacrylamide gel (acrylamide-bisacrylamide, 33.5:0.3) for polyacrylamide gel electrophoresis (PAGE).
Immunoprecipitation experiments were performed essentially as described in reference 55. Cells were grown to an OD600 of 0.8 or 3 as indicated, and extracts were made from 12 OD600 units. Polyclonal rabbit antibody HA.11 (Babco) was added at 1/125 to the resulting extracts for 1 h at 4°C; 40 μl of 50% protein G immobilized on Sepharose beads (Pharmacia Biotech) was mixed into the samples for 1 h at 4°C. The beads were washed three times with 0.5 ml of extraction buffer without PMSF and Complete protease inhibitor cocktail (Boehringer). Immunoprecipitated proteins were separated by SDS-PAGE as described above.
Proteins were electrotransferred onto nitrocellulose membranes (0.2-μm pore size; Schleicher & Schuell). The membranes were hybridized with HA-specific monoclonal antibody 16B12 or Myc-specific monoclonal antibody 9E10 (Babco) at 1/5,000. After washing, membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Promega). The blots were visualized by enhanced chemiluminescence (Amersham). Ctk2p half-lives in the Δctk2 strain were quantified with a Luminescence Imager (Boehringer).
Phosphatase analyses.
Cells (10 OD600 units) were broken with 10% TCA as described above. After centrifugation, pellets were resuspended in 100 μl of 50 mM Tris-HCl (pH 7.5)–2% SDS and cleared by centrifugation. Aliquots of 20 μl were incubated with 2 U of alkaline phosphatase (calf intestinal phosphatase; Pharmacia Biotech) in a final volume of 750 μl of 50 mM Tris-HCl (pH 7.5)–1 mM MgCl2 at 37°C. When indicated, phosphatase inhibitors (100 mM Na2HPO4-NaH2PO4 [pH 7.5], 10 mM EDTA) were added. Proteins were precipitated for 15 min on ice with 750 μl of cold 50% TCA. After centrifugation, pellets were resuspended in 15 μl of TCA-Laemmli loading buffer with heating at 100°C for 10 min. Samples of 10 μl were loaded on an SDS–13% polyacrylamide gel and Western immunoblotted as described above.
RESULTS
Overexpression of Ctk2p cyclin is toxic.
The S. cerevisiae CTDK-I complex is encoded by three genes: CTK1, encoding a CDK subunit; CTK2, encoding a C-type cyclin subunit; and CTK3, encoding a subunit of unknown function. Disruption of any of these genes generates viable cells that display similar growth defects and are unable to grow at low temperatures (56). Cell cycle cyclins expression is tightly regulated at the level of both transcription and protein stability (43). As a consequence, overexpression of cell cycle cyclins usually generates growth defects (9, 60, 68). The CTDK-I complex is implicated not in cell cycle regulation but rather in transcription regulation (26, 31). Similarly to what has been observed for other C-type cyclins implicated in transcription control, CTK2 mRNA levels do not fluctuate during the cell cycle (data not shown). However, because Ctk2p belongs to the cyclin family of proteins, its expression was also expected to be highly regulated. We therefore wanted to know whether its overexpression would generate growth defects. Each protein of the CTDK-I complex was overexpressed by introducing into wild-type cells multicopy plasmids carrying each coding sequence under the control of its cognate promoter. Growth rates were measured for cells grown in minimal medium at 30 and at 15°C (Table 1). Under both conditions, growth rates observed for cells overexpressing either Ctk1p or Ctk3p were identical to that observed for cells carrying a control plasmid, whereas cells overexpressing the Ctk2p cyclin showed significantly slower growth (Table 1). To confirm that overexpression of Ctk2p during exponential growth was toxic for the cells, plasmid loss was quantified. Strains were grown in rich medium for several generations prior to testing for plasmid retention. Whereas similar loss rates were obtained for cells carrying either a plasmid control or cells overexpressing Ctk1p or Ctk3p, cells overexpressing Ctk2p showed a much higher plasmid loss rate (Table 1). Taken together, these results show that overexpressing the C-type cyclin Ctk2p is toxic for the cells. This effect is consistent with a potential tight control of the Ctk2 cyclin C protein expression.
TABLE 1.
Ctk2p overexpression from a multicopy plasmid affects wild-type cell growth
| Plasmid | Doubling time
|
% Plasmid loss (30°C) | |
|---|---|---|---|
| 30°C | 15°C | ||
| 2μm control | 2 h 35 min | 8 h 30 min | 35 |
| 2μm/CTK1 | 2 h 40 min | 9 h 10 min | 25 |
| 2μm/CTK2 | 4 h 50 min | 15 h | >99 |
| 2μm/CTK3 | 2 h 25 min | 8 h 25 min | 26 |
Ctk2p is phosphorylated in vivo.
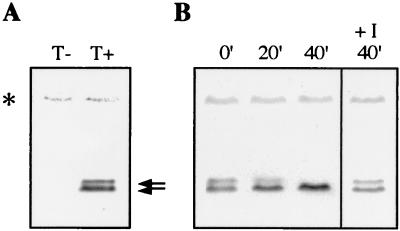
To study Ctk2p expression, its coding sequence was cloned on a single-copy plasmid under the control of the CTK2 promoter sequences. The protein was fused at its N-terminal extremity to influenza virus HA epitope tags. The resulting construct, CTK2-HA, allowed expression of a functional protein because it could restore Δctk2 cell growth at the nonpermissive temperature (data not shown). Western blot analyses permitted detection of two specific bands corresponding to the expected size for Ctk2p (p38), the upper band being present at lower quantities (Fig. 1A). In an attempt to identify these two forms, extracts were treated with alkaline phosphatase (Fig. 1B). Treatment of extracts with alkaline phosphatase in the absence, but not in the presence, of phosphatase inhibitors resulted in the loss of the species with lower mobility. This loss was concomitant with an increase in the levels of the higher-mobility species, showing that it was due to serine and/or threonine dephosphorylation. In early exponentially growing cells, at steady state, Ctk2p occurs as two forms, one corresponding to a phosphorylated form (hereafter called Ctk2p-P).
FIG. 1.
Ctk2p phosphorylation in vivo. Cells were grown in SD medium to an OD600 of 0.8. (A) Extracts from Δctk2 strain transformed with a plasmid control (T−) or with the CTK2-HA construct (T+) were analyzed by Western immunoblotting. Arrows indicate bands corresponding to Ctk2p. (B) Extracts from the Δctk2 strain transformed with the CTK2-HA construct were incubated for different times (minutes) at 37°C in phosphatase buffer with 2 U of alkaline phosphatase in the absence or presence (+I) of phosphatase inhibitors. In all figures, asterisks mark a nonspecific band which remained stable during all experiments, showing that all lanes were equally loaded.
Ctk2p is unstable.
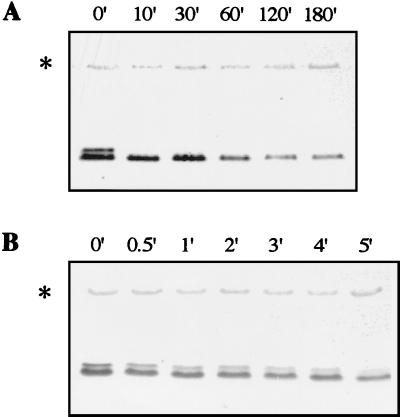
Protein stability is known to play an important role in regulation (19). In yeast, G1 and mitotic cyclins as well as CDK inhibitors have been shown to be very unstable (23). Cyclin abundance is often rate limiting for CDK activity, and control of cyclin degradation rate is important in regulating cell cycle transitions. Examination of the yeast C-type cyclin Ume3p regulation showed that this cyclin is subjected to rapid destruction (6). To study Ctk2p stability, cycloheximide, an inhibitor of cytoplasmic translation, was added to cultures grown at 30°C in SD medium. Ctk2p levels were monitored by immunodetection. The lower band corresponding to the nonphosphorylated form of the cyclin was relatively unstable (half-life, 60 min; Fig. 2A). Furthermore, the upper band corresponding to the phosphorylated form was highly unstable, as we could not detect it 10 min after cycloheximide addition (Fig. 2A). To get a better insight into Ctk2p-P turnover, its levels were followed just after cycloheximide addition (Fig. 2B). Only 5 min after blocking protein synthesis, we could detect half of the initial levels of Ctk2p-P (half-life, 5 min; Fig. 2B). The same experiments conducted with a Myc-tagged Ctk2p protein generated identical results (data not shown). We conclude that Ctk2p is unstable in exponentially growing cells.
FIG. 2.
Ctk2p turnover in early exponential cells. Δctk2 cells transformed with the CTK2-HA construct were grown in SD medium to an OD600 of 0.8. Aliquots were taken at different times (minutes) of incubation at 30°C after the addition of cycloheximide. Resulting cellular extracts were analyzed by Western immunoblotting. After the addition of cycloheximide, Ctk2p levels were monitored for 180 min (A) and 5 min (B) to analyze turnover of the nonphosphorylated and phosphorylated species, respectively.
Ctk2p turnover exhibits growth-related regulation.
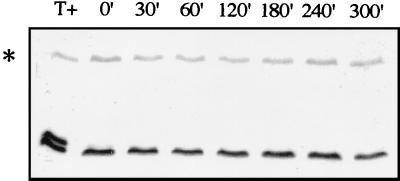
A recent study showed growth-related changes in phosphorylation of the yeast RNA polymerase II CTD (46). Also, Cooper et al. (6) reported that Ume3p turnover is dependent on environmental changes. For these reasons, we examined Ctk2p expression in cells grown under different conditions. Extracts were made from late exponentially growing cells (OD600 = 3). Western blot analyses permitted detection of only one band whose migration corresponded to that of the nonphosphorylated form (Fig. 3). Ctk2p stability was determined after cycloheximide addition. In contrast to our previous observations made from cells grown to an OD600 of 0.8, Ctk2p levels did not vary for 4 h after inhibition of protein synthesis. This result shows that in late exponentially growing cells, Ctk2p turnover is dramatically reduced, indicating that the regulation of Ctk2p turnover is dependent on growth conditions. Concomitant with this process, we could not detect phosphorylated Ctk2p species, suggesting that either Ctk2p is not phosphorylated or Ctk2p-P is extremely rapidly dephosphorylated.
FIG. 3.
Ctk2p turnover in late exponential cells. Δctk2 cells transformed with the CTK2-HA construct were grown in SD medium to an OD600 of 3. Aliquots were taken at different times (minutes) of incubation at 30°C after the addition of cycloheximide. Resulting cellular extracts were analyzed by Western immunoblotting. T+, protein extracts from Δctk2 cells transformed with the CTK2-HA construct grown to an OD600 of 0.8.
Taken together these results indicate a correlation between Ctk2p phosphorylation state and cyclin stability, suggesting that Ctk2p phosphorylation is a signal for its rapid degradation. This is reminiscent of previous data showing that phosphorylation of yeast cell cycle cyclins can trigger their selective degradation (28, 67, 68).
Ctk2p-P degradation is impaired in a mutant defective in proteasome activity.
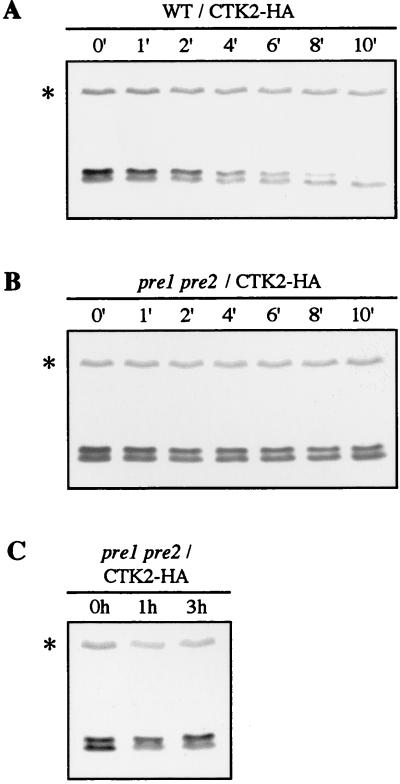
Eukaryotic cells contain two distinct systems for protein degradation: intravacuolar proteolysis (63) and a ubiquitin-mediated proteolysis that involves the proteasome and occurs in the cytoplasm as well as in the nucleus (18). Biochemical and genetic evidence indicates that the ubiquitin-proteasome pathway is involved in the degradation of abnormal proteins and regulatory proteins with naturally short half-lives such as cell cycle cyclins (20). Ctk2p turnover was not affected in the vacuolar protease-deficient pra1 prb1 prc1 cps1 mutant strain (17) (data not shown). To establish whether Ctk2p was a substrate for the proteasome degradation pathway, we examined Ctk2p stability in the yeast pre1 pre2 mutant strain, which is defective in two of the proteasome catalytic subunits (48). The CTK2-HA construct was introduced into pre1 pre2 and wild-type isogenic cells. Cultures were subjected to a shift to the nonpermissive temperature (37°C) prior to addition of cycloheximide to impose the block in proteasome function. In wild-type cells, immunodetection of Ctk2p-P revealed a turnover of a few minutes, similar to what was previously observed (Fig. 4A). In contrast, in pre1 pre2 cells, Ctk2p-P levels remained unchanged 10 min after cycloheximide addition (Fig. 4B). We note that before cycloheximide addition, the Ctk2p-P/Ctk2p ratio was different from that observed previously, indicating that it is dependent on genetic background and/or temperature. In conclusion, Ctk2p-P is dramatically stabilized in a mutant strain that exhibits defects in the activity of the proteasome, showing that its degradation is mediated by the 26S proteasome pathway.
FIG. 4.
Ctk2p turnover in cells that have impaired catalytic proteasome subunits. WCG4a (wild type [WT]; A) and isogenic WCG4-11/22a (pre1 pre2; B and C) cells transformed with the CTK2-HA construct were grown in SD medium to an OD600 of 0.8. For panels A and B, cultures were then shifted to the nonpermissive temperature (37°C) for 40 min prior to the addition of cycloheximide, and aliquots were taken at different times (minutes) of incubation at 37°C. (C) Cultures were shifted to the nonpermissive temperature for 1 or 3 h. Resulting cellular extracts were analyzed by Western immunoblotting.
If Ctk2p phosphorylation is a signal for its degradation, affecting this latter process might lead to the accumulation of phosphorylated species. Western blots analyses were performed with extracts made from pre1 pre2 cells carrying the CTK2-HA construct, grown to the permissive temperature and shifted to the nonpermissive temperature (Fig. 4C). After incubation at the nonpermissive temperature, we observed a significant increase in Ctk2p-P steady-state levels correlated with a decrease in that of Ctk2p. This result shows that a block in Ctk2p destruction results in an important increase in Ctk2p-P/Ctk2p ratio, supporting the idea that phosphorylated Ctk2p species are indeed those that are rapidly degraded by the proteasome pathway.
Ctk2p-P degradation is an ubiquitin-mediated process.
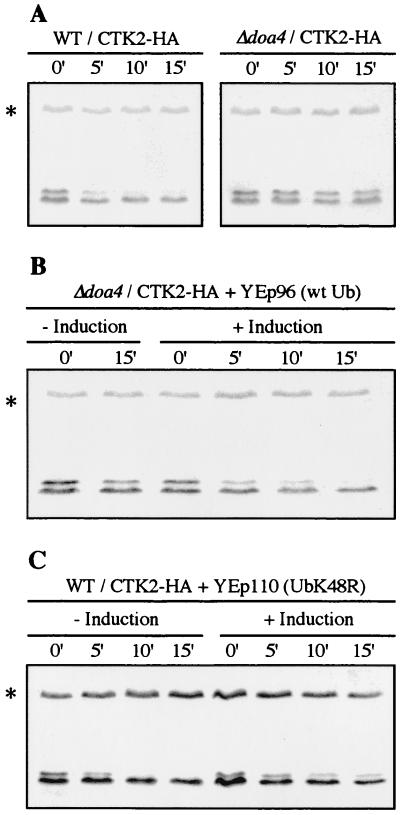
Although the 26S proteasome degrades an unknown number of proteins that are recognized without undergoing ubiquitination, the ubiquitin system constitutes the major targeting process leading to selective degradation (47). Ubiquitin is covalently attached to the protein substrate through an isopeptide bond in a multistep reaction, and repeated ubiquitination reactions culminate in attachment of a multiubiquitin chain that marks the target protein for rapid degradation by the 26S proteasome (20). Ubiquitin hydrolases act to remove ubiquitin from conjugated forms during either protein degradation or signalling. Cells disrupted for the DOA4 gene encoding a deubiquitinating enzyme are disturbed for normal metabolism of ubiquitin and display a strong inhibition of the turnover of several soluble substrates of the ubiquitin pathway (45). To determine if Ctk2p degradation was mediated by the ubiquitin pathway, we studied Ctk2p turnover in cells disrupted for the DOA4 gene. We compared the rates of Ctk2p turnover in Δdoa4 and isogenic wild-type cells after inhibition of protein synthesis. As previously observed, Ctk2p-P turnover was very fast in wild-type cells, whereas the amounts of immunodetected Ctk2p-P in Δdoa4 cells decreased only slightly after 15 min (Fig. 5A). Similar to what was observed in a proteasome mutant strain, Ctk2p-P degradation was strongly affected in Δdoa4 mutant cells. Several studies have suggested that the amount of intracellular ubiquitin available for ubiquitination is limiting in Δdoa4 cells (15, 45, 54). To find out whether an increase in ubiquitin levels could restore Ctk2p instability in the Δdoa4 mutant strain, wild-type and Δdoa4 cells were transformed with the multicopy plasmid YEp96, encoding a synthetic ubiquitin gene under the control of a CUP1-inducible promoter (10). CuSO4 was added for 5 h to induce the CUP1 promoter, and Ctk2p levels were monitored by immunodetection after cycloheximide addition. Overexpression of wild-type ubiquitin did not affect Ctk2p-P turnover in wild-type cells (data not shown). In contrast, overexpression of ubiquitin rescued Ctk2p-P turnover in Δdoa4 cells (half-life, 5 to 10 min; Fig. 5B). In summary, we observed a significant stabilization of Ctk2p-P in Δdoa4 cells. This effect was partially reversed by overproduction of wild-type ubiquitin, suggesting that an ubiquitin-mediated process is required for Ctk2p-P rapid degradation.
FIG. 5.
Ctk2p degradation is a ubiquitin-dependent process. Cells were grown in SD medium to an OD600 of 0.8, and aliquots were taken at different times (minutes) of incubation at 30°C after the addition of cycloheximide. Resulting cellular extracts were analyzed by Western immunoblotting. (A) MHY501 (wild type [WT]) and isogenic MHY623 (Δdoa4) cells transformed with the CTK2-HA construct. (B) Δdoa4 cells cotransformed with the CTK2-HA and YEp96 (wild-type ubiquitin [wt Ub]) plasmids. (C) MHY501 (wild type) cells cotransformed with the CTK2-HA and YEp110 (UbK48R) plasmids. In the right portions (+ Induction) of panels B and C, cells were grown for 5 h with CuSO4 before they reached an OD600 of 0.8 in order to induce ubiquitin expression from the CUP1 promoter.
The targeting of substrates of the ubiquitin pathway to the proteasome is predominantly accomplished by the ligation of a polyubiquitin chain assembled through isopeptide bonds connecting the carboxyl-terminal Gly76 of one ubiquitin moiety to the ɛ-amino group of Lys48 of the adjacent ubiquitin molecule (5, 64). A modified ubiquitin carrying an arginine at position 48 instead of lysine (UbK48R) can still be conjugated to proteins but fails to function as an acceptor within the polyubiquitin chain, thus acting as a multiubiquitin chain terminator (2, 5, 13, 21, 52). If Lys48-linked multiubiquitination is a prerequisite for Ctk2p degradation, the presence of UbK48R should affect this process. The multicopy plasmid YEp110, carrying a gene encoding the variant UbK48R under the control of the CUP1 promoter (21), was introduced into wild-type cells. Expression of UbK48R was induced for 5 h, and Ctk2p fate after cycloheximide addition was monitored by immunodetection. When UbK48R expression was induced, we could observe only a slight decrease in Ctk2p-P levels 15 min after cycloheximide addition, indicating a significant increase in Ctk2p-P stability (Fig. 5C). Since in all of these experiments cells also contained considerable amounts of wild-type ubiquitin, and because the mutant ubiquitin can be removed from the multiubiquitin chains (13), the inhibition of Ctk2p degradation by UbK48R was expected to be leaky. For that reason, we also analyzed the effect of UbK48R expression in Δdoa4 cells that are thought to contain lower levels of free endogenous ubiquitin. Overproduction of UbK48R in Δdoa4 cells resulted in a further and complete stabilization of Ctk2p-P (data not shown). In conclusion, Ctk2p-P destruction is substantially impaired by overexpression of UbK48R, demonstrating that this process requires the formation of Lys48-linked polyubiquitin chains.
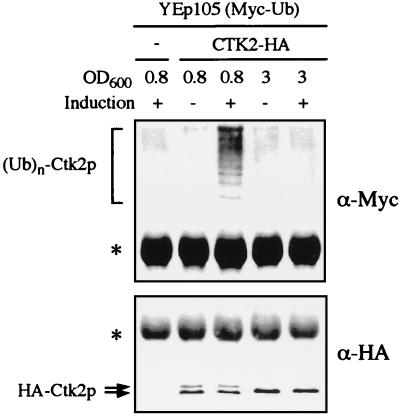
One difficulty in establishing whether the degradation of a short-lived protein requires its conjugation to the ubiquitin system stems from the fact that depending on the relative rates of ubiquitination, deubiquitination, and degradation of the ubiquitin-containing protein, the steady-state levels of ubiquitinated species may range from negligible to readily detectable. Despite the fact that Ctk2p was readily observed by immunodetection, we did not observe any ubiquitinated forms. In an attempt to detect such species, Ctk2p was immunoprecipitated from wild-type cells overexpressing a Myc-tagged variant of ubiquitin from the CUP1 promoter (11). Ctk2p species were further analyzed by Western immunoblotting (Fig. 6). Hybridization with Myc antibodies allowed detection of high-molecular-weight bands specific of the CTK2-HA construct only when Myc-ubiquitin expression was induced (Fig. 6, upper panel). Furthermore, these ubiquitin conjugates could not be observed when extracts were made from late exponential cells (OD600 = 3) (Fig. 6). Hybridization with HA antibodies (Fig. 6, lower panel) confirmed the presence of Ctk2p-ubiquitin conjugates only when phosphorylated Ctk2p forms were detected (OD = 0.8). On the contrary, we observed no ubiquitin conjugates when the cyclin was stable and its steady-state comprised only the unphosphorylated species (OD = 3). Altogether, these results demonstrate that Ctk2p is a substrate for the ubiquitin-proteasome pathway.
FIG. 6.
Ctk2p ubiquitin conjugates. W303-1B (wild type) cells cotransformed with plasmid YEp105 (Myc-tagged ubiquitin [Myc-Ub]) and either a control plasmid (−) or the CTK2-HA construct were grown in SD medium to an OD600 of 0.8 or 3 as indicated. CuSO4 was (+) or was not (−) added overnight to the cultures in order to induce expression of the Myc-tagged ubiquitin. After immunoprecipitation with HA polyclonal antibodies, proteins were analyzed by Western immunoblotting with monoclonal Myc (upper panel) and monoclonal HA (lower panel) antibodies successively. Arrows and asterisks indicate bands corresponding to Ctk2p and nonspecific products, respectively.
Ctk2p is phosphorylated in cells lacking the Ctk1p kinase.
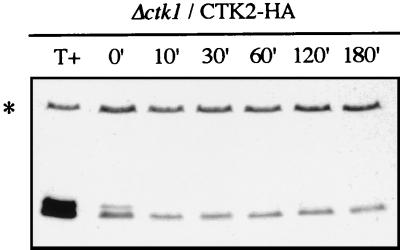
Cell cycle cyclins have been shown to be rapidly degraded via the ubiquitin-proteasome pathway, and in several cases, mutagenesis analyses revealed that phosphorylation by their associated kinase provides a signal that promotes this process (28, 68). The fact that degradation of a cyclin is dependent on its cognate CDK subunit renders the cyclin-activated CDK activity self-limiting. To determine whether Ctk2p phosphorylation was dependent on its cognate kinase subunit, CTK2-HA construct was introduced into cells disrupted for the CTK1 gene. Western blots analyses allowed detection of two bands corresponding to Ctk2p (Fig. 7), showing that the cyclin Ctk2p was phosphorylated in the absence of its activated kinase. Moreover, after cycloheximide addition, we observed a similar turnover for the phosphorylated form in Δctk1 cells compared to wild-type cells (half-life, 5 min; Fig. 7 and data not shown). We note that Ctk2p abundance was lower in Δctk1 mutant cells than in wild-type cells, consistent with Northern blots analyses showing that CTK2 mRNA steady-state levels were substantially diminished in Δctk1 cells (data not shown). Also, the Ctk2p-P/Ctk2p ratio was slightly lower in Δctk1 cells than in wild-type cells, this effect being correlated with a significant stabilization of the unphosphorylated form. If Ctk2p phosphorylation is not as efficient in Δctk1 cells as in wild-type cells, as a consequence one would expect to observe its stabilization. To obtain a better insight into the regulation of Ctk2p turnover in Δctk1 cells, we analyzed Ctk2p expression in Δctk1 cells grown to an OD of 3. As previously observed in wild-type cells (Fig. 3), Ctk2p was composed of only one band corresponding to the unphosphorylated form (data not shown). In conclusion, if there is a slight difference in Ctk2p turnover in Δctk1 cells, it is probably nonspecifically affected due to the fact that Ctk2p is not an active protein, as it cannot be part of the CTDK-I complex.
FIG. 7.
Ctk2p phosphorylation in Δctk1 cells. Δctk1 cells transformed with the CTK2-HA construct were grown in SD medium to an OD600 of 0.8. Aliquots were taken at different times (minutes) of incubation at 30°C after the addition of cycloheximide. Resulting cellular extracts were analyzed by Western immunoblotting. As a control (T+), protein extracts from Δctk2 cells transformed with the CTK2-HA construct grown to an OD600 of 0.8 were loaded on the gel.
We conclude that neither Ctk2p phosphorylation nor degradation is affected in Δctk1 cells, showing that these processes require neither CTDK-I complex assembly nor its activity.
DISCUSSION
The C-type cyclin Ctk2p is phosphorylated in vivo.
We have studied the expression of the yeast C-type cyclin Ctk2p, a cyclin subunit of the CTDK-I complex that is implicated in phosphorylation of the RNA polymerase II CTD. We show that steady-state levels of Ctk2p in pseudo-wild-type exponential-phase cells (a functional tagged version of the cyclin was expressed from its cognate promoter carried by a single-copy plasmid introduced into Δctk2 cells) are composed of two forms, one a phosphorylated form. This is the first description of a C-type cyclin phosphorylation. Examination of Ctk2p protein sequence did not reveal the presence of a strong PEST-rich motif, and as yet no phosphorylated residues have been identified. One difficulty in studying cell cycle cyclins at the protein levels is that yeast cells contain very little of these proteins. In contrast, phosphorylated as well as nonphosphorylated species of Ctk2p are relatively abundant during exponential growth. This striking feature might reflect a role for Ctk2p in regulating transcription rather than cell cycle progression. Indeed, cell cycle CDKs are known to interact sequentially with several distinct cyclins whose abundance fluctuates during the cell cycle (43). Yeast CDKs activated by C-type cyclins seem to be an exception from this principle, as only one cyclin has been characterized per kinase subunit, indicating clearly that different features govern regulation of CDKs that are implicated in transcription control.
That Ctk2p steady state is composed of nonphosphorylated as well as phosphorylated species raises the question of which species are functional. Because in the absence of Ctk2p-P the cyclin is stable, it is likely that the CTDK-I complex is active. Indeed, under growth conditions where Ctk2p steady state is mainly composed of the unphosphorylated form, preliminary experiments suggest that the CTDK-I complex is active (unpublished results). Several studies have shown that cell cycle cyclin phosphorylation is required for the control of their degradation but not for their capacity to activate their associated kinase (9, 28). It is thus likely that, similar to the case for cell cycle cyclins, Ctk2p phosphorylation is not required for its function.
Ctk2p degradation is mediated by the ubiquitin-proteasome pathway.
We have blocked cytoplasmic translation with cycloheximide to study Ctk2p stability. We show that under these conditions, Ctk2p is a moderately unstable protein (half-life, 60 min) whereas its phosphorylated form is extremely unstable (half-life, 5 min). Because C-type cyclin expression may vary upon diverse stress conditions, pulse-chase experiments would be required to determine whether Ctk2p turnover is affected by cycloheximide.
In eukaryotic cells, protein degradation is mediated by two distincts systems: the nonspecific intravacuolar proteolysis system, which is rather implicated in the stress response (63); and the proteasome ubiquitin-mediated pathway, which has been implicated in specific degradation of abnormal proteins as well as short-lived proteins (18). Ctk2p turnover is not affected in cells deficient in the major vacuolar peptidases (data not shown), whereas it is markedly affected in a mutant strain deficient for two of the proteasome catalytic subunits (pre1 pre2) (48), indicating that Ctk2p destruction is mediated by the proteasome pathway. Although other cases have been described, targeting to the proteasome usually requires attachment of ubiquitin chains to substrate proteins (47). These chains are highly dynamic, with rapid addition or removal of ubiquitin by deubiquitinating enzymes. Ctk2p degradation is substantially impaired in cells lacking the DOA4 gene, which encodes one of the 16 deubiquitinating enzymes characterized in yeast (20, 45). This impairment is partially rescued by overexpression of wild-type ubiquitin, confirming previous observations indicating that levels of free ubiquitin are limiting in Δdoa4 cells (15, 45, 54). Finally, we could observe Ctk2p-ubiquitin conjugates by using tagged ubiquitin. As expected, these forms were detected under conditions where Ctk2p displays a short turnover but not under conditions where Ctk2p is stable. Moreover, consistent with a function of the ubiquitin system in Ctk2p degradation, expression of a mutant ubiquitin (UbK48R) that prevents the formation of polyubiquitin chains (2, 13, 21, 52) generated an increase in Ctk2p stability. Taken together, these results demonstrate the role of K48-linked polyubiquitination in the rapid degradation of Ctk2p by the proteasome.
The yeast C-type cyclin Ume3p has also been reported to be down-regulated but there was no evidence for the involvement of the ubiquitin pathway in its degradation (6). The authors suggested that Ume3p might be destroyed through a mechanism different from those described for other cyclins. On the contrary, we show that similar to the case for other short-lived proteins, rapid destruction of Ctk2p is mediated by the specific ubiquitin-proteasome pathway.
Control of Ctk2p turnover.
Patturajan et al. (46) reported recently that the pattern of CTD phosphorylation in yeast varies in response to growth conditions and environmental stress. Among the three C-type cyclins identified in yeast, Ume3p and Ctk2p are associated with nonessential CTD kinases that are implicated in transcription regulation. Ume3p is rapidly destroyed when cells enter the meiotic pathway or are exposed to heat shock (6). Ctk2p is expected to be stabilized whenever CTDK-I kinase activity is required. In this report, we show that Ctk2p is very stable in late-exponential-phase cells, demonstrating that this regulatory protein is not constitutively short-lived.
Several lines of evidence suggest that Ctk2p phosphorylation is a signal for its rapid destruction: (i) as discussed above, Ctk2p phosphorylation does not seem to be required for CTDK-I function, indicating that this event must be implicated in another process; (ii) in late-exponential-phase cells, Ctk2p stabilization is, as expected, correlated with the absence of ubiquitin conjugates but also with the absence of Ctk2p phosphorylation; (iii) Ctk2p-P is extremely unstable in wild-type cells, whereas it is very stable in a proteasome mutant strain; (iv) finally, we observe an important increase in the Ctk2p-P/Ctk2p ratio when the cyclin destruction is blocked. Taken together, these data strongly suggest that similar to the case for G1 cyclins, Ctk2p degradation is induced by phosphorylation. It has been shown that the degradation of cell cycle cyclins is dependent on their associated kinase activity (7, 28, 68). Once the cyclin is expressed, it activates its kinase, which in return phosphorylates its cyclin, a signal inducing its rapid and specific ubiquitin-mediated degradation through the proteasome pathway. Unexpectedly, neither phosphorylation nor turnover of Ctk2p is significantly affected in cells disrupted for the gene encoding the kinase subunit of the CTDK-I complex. We cannot exclude the possibility that the Ctk1p kinase plays a role in Ctk2p phosphorylation in wild-type cells; however, if such phosphorylation events take place, they are unlikely to play a role in the control of Ctk2p turnover.
Similarly, it has been shown that the rapid degradation of Ume3p was not dependent on the presence of the CDK that it activates (6). Although Ume3p phosphorylation has not been reported, it is worth noting that integrity of the Ume3p cyclin box, i.e., its kinase binding domain (25, 32), is required for its breakdown (6). This peculiar feature raises the interesting possibility that the Ume3p cyclin box is required for an interaction with another CDK. It thus appears that unlike turnover of cell cycle cyclins, turnover of C-type cyclins is not controlled by their cognate kinases, and it will be important to determine whether Ctk2p phosphorylation is dependent on another CDK that could be activated by CTD kinase cyclins or could phosphorylate monomeric C-type cyclins.
In conclusion, we show in this report that similar to expression of cell cycle cyclins, Ctk2p expression is regulated by rapid proteolysis mediated by the ubiquitin-proteasome pathway. However, it is apparent that regulation of the turnover of C-type cyclins exhibits specific features that are likely to be the signature of their function in transcription control, as indeed another kinase activity can phosphorylate Ctk2p, thereby probably regulating its turnover.
ACKNOWLEDGMENTS
We are very grateful to C. Jacq for scientific and financial support during this work. We thank M. Hochstrasser and D. Wolf for providing strains. Special thanks are due to R. Haguenauer-Tsapis for stimulating discussions and critical reading of the manuscript. We also thank members of C. Jacq’s laboratory for many helpful discussions and to S. Moore for helpful comments and for proofreading the manuscript.
G.H. was supported by a French Ministère de la Recherche et des Technologies fellowship.
REFERENCES
- 1.Balciunas D, Ronne H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995;23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- 3.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 4.Chardin P, Camomis J H, Gale N W, Van Aelst L, Schlessinger J, Wigler M, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to Grb2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 5.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 6.Cooper K F, Mallory M J, Smith J B, Strich R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross F R, Blake C M. The yeast Cln3 protein is an unstable activator of Cdc28. Mol Cell Biol. 1993;13:3266–3271. doi: 10.1128/mcb.13.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 9.Deshaies R J, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ecker D J, Khan M I, Marsh J, Butt T R, Crooke S T. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem. 1987;262:3524–3527. [PubMed] [Google Scholar]
- 11.Ellison M J, Hochstrasser M. Epitope-tagged ubiquitin. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 12.Faye G, Simon M, Valay J-G, Fesquet D, Facca C. Rig2, a RING finger protein that interacts with the Kin28/Cc11 CTD kinase in yeast. Mol Gen Genet. 1997;255:460–466. doi: 10.1007/s004380050518. [DOI] [PubMed] [Google Scholar]
- 13.Finley D, Sadis S, Monia B P, Boucher P, Ecker D J, Crooke S T, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furnari B A, Russell P, Leatherwood J. pch1+, a second essential C-type cyclin gene in Schizosacchromyces pombe. J Biol Chem. 1997;272:12100–12106. doi: 10.1074/jbc.272.18.12100. [DOI] [PubMed] [Google Scholar]
- 15.Galan J M, Haguenauer-Tsapis R. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 17.Heinmeyer W, Kleinschmidt J, Saidowsky J, Escher C, Wolf D. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilt W, Wolf D H. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 19.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 21.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito H, Fukuda Y, Murata K, Kumura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King R W, Deshaies R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 24.King R W, Glotzer M, Kirschner M W. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Cell Biol. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi H, Stewart E, Poon R, Adamczewski J P, Gannon J, Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 29.Lee J M, Greenleaf A L. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci USA. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J M, Greenleaf A L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J M, Greenleaf A L. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- 32.Lees E M, Harlow E. Sequences within the conserved cyclin box of human cyclin A are sufficient for binding to and activation of cdc2 kinase. Mol Cell Biol. 1993;13:1194–1201. doi: 10.1128/mcb.13.2.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Léopold P, O’Farrell P H. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell. 1991;66:1207–1216. doi: 10.1016/0092-8674(91)90043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 35.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z-J, Ueda T, Miyazaki T, Tanaka N, Mine S, Tanaka Y, Taniguchi T, Yamamura H, Minami Y. A critical role for cyclin C in promotion of the hematopoietic cell cycle by cooperation with c-Myc. Mol Cell Biol. 1998;18:3445–3454. doi: 10.1128/mcb.18.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 38.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 39.Molz L, Beach D. Characterization of the fission yeast mcs2 cyclin and its associated protein kinase activity. EMBO J. 1993;12:1723–1732. doi: 10.1002/j.1460-2075.1993.tb05817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 41.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 42.Murray A W, Solomon M J, Kirschner M W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 43.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 44.Nigg E A. Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control? Curr Opin Cell Biol. 1996;8:312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- 45.Papa F R, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 46.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 47.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 48.Richter-Ruoff B, Wolf D H, Hochstrasser M. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 1994;354:50–52. doi: 10.1016/0014-5793(94)01085-4. [DOI] [PubMed] [Google Scholar]
- 49.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 50.Rothstein R J. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 51.Salama S R, Hendricks K B, Thorner J. G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol Cell Biol. 1994;14:7953–7966. doi: 10.1128/mcb.14.12.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schork S M, Thumm M, Wolf D H. Catabolite inactivation of Fructose-1,6-biphosphatase of Saccharomyces cerevisiae. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- 53.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 54.Singer J D, Manning B M, Formosa T. Coordinating DNA replication to produce one copy of the genome requires genes that act in ubiquitin metabolism. Mol Cell Biol. 1996;16:1356–1366. doi: 10.1128/mcb.16.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song W, Carlson M. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 1998;17:5757–5765. doi: 10.1093/emboj/17.19.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sterner D E, Moon Lee J, Hardin S E, Greenleaf A L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin–cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Svejstrup J Q, Vichi P, Egly J-M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 59.Tassan J-P, Jaquenoud M, Léopold P, Schultz S J, Nigg E A. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc Natl Acad Sci USA. 1995;92:8871–8875. doi: 10.1073/pnas.92.19.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valay J-G, Dubois M-F, Bensaude O, Faye G. Ccl1, a cyclin associated with protein kinase Kin28, controls the phosphorylation of RNA polymerase II largest subunit and mRNA transcription. C R Acad Sci (Paris) 1996;319:183–189. [PubMed] [Google Scholar]
- 62.Valay J-G, Simon M, Dubois M-F, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 63.Van Den Hazel H B, Kielland-Brandt M C, Winther J R. Review: biosynthesis and function of yeast vacuolar proteases. Yeast. 1996;12:1–16. doi: 10.1002/(sici)1097-0061(199601)12:1<1::aid-yea902>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 64.Van Nocker S, Vierstra R D. Multiubiquitin chains linked through lysine 48 are abundant in vivo and are competent intermediates in the ubiquitin proteolytic pathway. J Biol Chem. 1993;268:24766–24773. [PubMed] [Google Scholar]
- 65.Wahi M, Johnson A D. Identification of genes required for α2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei P, Garber M E, Fang S-M, Fischer W H, Jones K A. A novel Cdk9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 67.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 68.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]