Abstract
Background & Aims:
Alterations in the gut microbiome have been associated with the severity of nonalcoholic fatty liver disease (NAFLD). These studies focused exclusively on the bacteria in the microbiome; we investigated changes in the viral microbiome (virome) in patients with NAFLD.
Methods:
In a prospective, cross-sectional, observational study, we extracted RNA and DNA virus-like particles from fecal samples from 73 patients with NAFLD: 29 patients had a NAFLD activity score (NAS) of 0–4, 44 patients had a NAS of 5–8 or cirrhosis (NAS 5–8/LCI), 37 patients had F0–F1 fibrosis, and 36 patients had F2–F4 fibrosis. As controls, 9 subjects without liver disease and 13 patients with mild primary biliary cholangitis were included in the analysis. We performed shotgun metagenomic sequencing of virus-like particles.
Results:
Patients with NAFLD and NAS 5–8/LCI had a significant decrease in intestinal viral diversity compared with patients with NAFLD and NAS 0–4 or controls. The presence of more advanced NAFLD was associated with a significant reduction in the proportion of bacteriophages (phages) compared with other intestinal viruses. Using multivariate logistic regression analysis with leave-1 out cross validation, we developed a model, including a viral diversity index and simple clinical variables, that identified patients with NAS 5–8/LCI with an area under the curve of 0.95 (95% CI, 0.91–0.99) and F2–F4 fibrosis with an area under the curve of 0.88 (95% CI, 0.80–0.95). Addition of data on viral diversity significantly improved multivariate models, including those based on only clinical parameters or bacterial diversity.
Conclusions:
In a study of fecal viromes from patients with NAFLD and controls, we associated histologic markers of NAFLD severity with significant decreases in viral diversity and proportion of bacteriophages. We developed a model based on fecal viral diversity and clinical data that identifies patients with severe NAFLD and fibrosis more accurately than models based only on clinical or bacterial data.
Keywords: microbiota, biomarker, prognostic factor, progression
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is a global health burden with 25% of people in Western countries being affected1. Within NAFLD, patients with a higher NAFLD activity score (NAS) and higher degrees of liver fibrosis on liver histology are at higher risk for disease progression, the development of hepatocellular carcinoma and liver-related mortality2, 3. While several non-invasive approaches have already been developed and validated for prediction of liver fibrosis, liver biopsy is still required for assessing grades of steatosis, inflammation and ballooning4, 5. Currently, there are no approved drug treatments for NAFLD and lifestyle-intervention is the main therapeutic approach. Due to the dramatically increasing incidence of NAFLD, new diagnostic tools to non-invasively assess the degree of histological disease severity as well as new potential treatment targets are urgently needed.
Patients with NAFLD show alterations in the intestinal microbiota, which might contribute to disease development and progression6, and features of NAFLD can even be transmitted by fecal microbiota transfer7, 8. However, microbiome research in NAFLD has almost exclusively focused on intestinal bacteria although immense populations of intestinal viruses reside in the gut and interact with other microorganisms and the human host9, 10. The majority of intestinal viruses are bacteriophages (phages), viruses that can specifically infect bacteria11. Phages are the most abundant but also the least understood microbes in the gut12. Phages may serve as important reservoirs of genetic diversity in the microbiota by acting as vehicles for the horizontal transfer of virulence, antibiotic resistance and metabolic determinants among bacteria13. Bacterial acquisition of phage genes could modify the functional properties of the microbiota, thereby substantially impacting host metabolism and immunity14. Besides phages, mammalian viruses and viruses that infect other microorganisms (archaea, fungi) or plants, are present15, 16. Alterations in the intestinal virome have been described in inflammatory bowel disease17–19, colorectal cancer20 and the development of type 1 diabetes21.
The aim of this study was to characterize the gut virome in a well characterized NAFLD cohort and to explore associations between features of the gut virome with different histological disease stages.
METHODS
Patients.
Patients with NAFLD were prospectively enrolled between March 2015 and December 2018 in the outpatient liver department of the Clinic for Gastroenterology and Hepatology, University Hospital of Cologne, Germany, as previously described22. In this study, a total of 73 patients with NAFLD (64 biopsy-proven patients with NAFLD and 9 patients with NAFLD diagnosed with cirrhosis based on characteristic clinical findings (see criteria below)) were included. Inclusion criteria for patients with NAFLD were the presence of ≥5% fat in histological analysis of liver biopsy, daily alcohol consumption of less than 10g in women and less than 20g in men, absence of steatogenic drugs and absence of other diseases causing secondary steatosis such as human immunodeficiency virus infection, celiac disease, inflammatory bowel disease and absence of other chronic liver diseases, e.g. viral hepatitis, autoimmune hepatitis, toxic liver injury, alcoholic steatohepatitis, cholestatic liver disease, Wilson’s disease and hereditary hemochromatosis.
Exclusion criteria were oral- or intravenous antibiotic treatment within the last 6 months prior to the study, known malignancy, pregnancy, and age <18 years. Further exclusion criteria were ongoing successful lifestyle modifications defined as more than 5% loss of body weight within the last three months prior to enrollment, or current or prior participation in an interventional non-alcoholic steatohepatitis (NASH) study22. Treatment recommendations for study participants did not differ from standard of care, which included overall lifestyle recommendations as indicated in the current European guideline23. All blood samples for laboratory analyses were collected under fasting conditions. Anthropometric measurements were carried out by trained physicians or research assistant nurses.
Type 2 diabetes was defined as glycated hemoglobin (HbA1c) ≥6.5% and/or fasting glucose ≥126 mg/dL and/or use of antidiabetic medications. Metabolic syndrome was defined following the International Diabetes Foundation criteria24. Arterial hypertension was defined as office blood pressure ≥140/90 mmHg on ≥2 measurements during ≥2 occasions or antihypertensive drug treatment.
13 patients with primary biliary cholangitis (PBC), were enrolled in the outpatient liver department of the Clinic for Gastroenterology and Hepatology, University Hospital of Cologne, Germany. PBC diagnosis followed the most recent guidelines (AASLD, EASL) at any time. All patients with PBC were under stable treatment with ursodeoxycholic acid. Nine normal weight subjects without any relevant chronic disease and without increased alcohol consumption (social drinkers who consumed alcohol less than 20 g/day) (controls) were enrolled in Cologne, Germany (n=5) and at St. Luc University Hospital, Université Catholique de Louvain, Brussels, Belgium (n=4). The protocol was approved by the local Ethics Commission and written informed consent was obtained from each patient. The study was performed in accordance with the Declaration of Helsinki.
Liver biopsies.
Liver biopsy was performed in patients with NAFLD with history of persistently elevated serum alanine aminotransferase (ALT) and/or aspartate aminotransferase (AST) of at least 6 months, to rule out potential other liver diseases than NAFLD and if there was clinical suspicion for advanced liver disease. If liver biopsy was performed, samples were evaluated by an experienced liver pathologist who was blinded for all clinical and laboratory patient data. The NASH clinical research network histological scoring system25 was used to evaluate disease activity and severity. Accordingly, the NAS was obtained for each biopsy. This score is defined as the unweighted sum of the scores for steatosis, (0–3), lobular inflammation (0–3), and ballooning (0–2); thus ranging from 0 to 825, 26. Fibrosis was staged from 0–4. Fibrosis stages: 0 none, 1 perisinusoidal or periportal, 2 perisinusoidal and portal/periportal, 3 bridging fibrosis, 4 cirrhosis. Stages 1a, 1b and 1c were summarized as stage 1.
Non-invasive diagnosis of liver cirrhosis.
If the following criteria were present, patients with NAFLD were staged as histological F4 fibrosis without obtaining a liver biopsy: besides hepatic steatosis on liver imaging (ultrasound and/or magnet resonance imaging), liver imaging consistent with liver cirrhosis (e.g. nodular hepatic contour, changes in volume distribution indicating portal hypertension in the absence of portal vein thrombosis, secondary phenomena of portal hypertension such as splenomegaly, enlarged caudate lobe and left lobe lateral segment, regenerative nodules) together with clinical and laboratory signs of portal hypertension/cirrhosis (e.g. low platelets, albumin and prothrombin time, esophageal varices)27.
Liver stiffness measurement.
Vibration controlled transient elastography (FibroScan, Echosens, Paris, France) was performed in fasting patients by experienced operators, blinded to all clinical patient data. At least 10 valid measurements were performed, and the median value of these measurements was reported in kilopascal (kPa). In accordance with the manufacturer’s protocol, patients were first scanned using the M probe and if indicated by the equipment, patients were re-scanned using the XL probe.
Bacterial DNA extraction and 16S rRNA sequencing.
16S rRNA gene sequencing of human stool samples was performed as previously described28.
Virome preparation and metagenomic sequencing.
Viral nucleic acid was extracted from fecal samples, reverse transcribed and subjected to metagenomic sequencing. For this purpose, we used the NetoVIR protocol with minor modifications29. In brief, fecal samples were resuspended in phosphate-buffered saline and sequentially filtered using 0.8 μm (PES) filter (Sartorius). Any remaining DNA that was not encapsidated was degraded by treating with a mixture of benzonase (EMD Millipore) and micrococcal nuclease (New England Biolabs) followed by EDTA inactivation of DNases. The remaining supernatant was subjected to lysis and viral DNA and RNA were extracted using the QIAamp Viral RNA mini kit without carrier RNA (Qiagen). Amplification was performed using a modified WTA2 (Complete Transcriptome Amplification Kit) protocol from Sigma Aldrich. Library preparation was performed using an adjusted protocol for the Nextera XT DNA Library Preparation kit from Illumina. The size of amplified viral products was determined using a High Sensitivity DNA Kit on a Bioanalyzer (Agilent Technologies), and concentration was measured by High Sensitivity Double Stranded DNA kit on a Qubit Fluorometer (Thermo Fisher Scientific). The sterile water control contained no detectable DNA, indicating no contamination of exogenous DNA during the analysis. Viral DNA from each sample was pooled into equimolar proportions and sequenced on the Illumina platform at the UCSD IGM Genomics Center.
Virome analysis.
Raw sequence reads were deduplicated using Clumpify (https://sourceforge.net/projects/bbmap/) followed by trimming and filtering for low-quality and contaminating human reads using Kneaddata30 with the GRCh38_v25 human genome reference. Reads were aligned and assigned taxonomy using the PathSeq pipeline (distributed in GATK v4.1.3.0)31, 32. Default settings were used, including --min-score-identity and --identity-margin, which were 0.90 and 0.02, respectively. Also, by default, PathSeq will discard alignments if both read pairs do not match the same organism (https://gatk.broadinstitute.org/hc/en-us/articles/360036717051-PathSeqScoreSpark). An inhouse Perl script was made (pathseq2taxsummary.pl) to convert PathSeq concatenated scores.txt files into a MOTHUR33 style .taxsummary file. The perl script is available at https://github.com/JCVenterInstitute/pathseq2taxsummary. Read counts, allowing ambiguity, were imported into R, data were normalized, and richness and diversity were calculated.
Statistical analysis.
Results are expressed as median and range unless stated otherwise. Two groups were compared using the Student’s t-test or Mann-Whitney-Wilcoxon rank-sum test for highly skewed distributions. Three or more groups were compared using one-way ANOVA with Tukey’s post-hoc test or the Kruskal-Wallis test with Dunn’s post-hoc test for highly skewed distributions. Categorical variables were compared using the Fisher’s exact test. All statistical tests were two-sided. Viral diversity and richness were calculated including all detected viruses (phages, mammalian viruses and other viruses, including plant/food derived viruses) using the “phyloseq” package in R34. The average proportion of phages, mammalian viruses and other viruses, including plant/food derived viruses in between groups, was calculated at the family level. Relative abundances were calculated within each virus category (phages vs mammalian viruses vs other viruses) for each taxonomic level. Single phages were analyzed at the species level and summarized according to their hosts. The relative abundance for these summarized phages was calculated within the phages with a known bacterial host. For this purpose, all sequence reads of phage species belonging to one host, e.g. all reads belonging to any of the 47 detected Lactococcus phages were summarized to a new variable “Lactococcus phages”. The presence of a NAS ≥5 and F2–F4 fibrosis have been associated with worse long-term outcomes2, 3, 35. Therefore, patients with NAFLD were grouped according to the NAS in a group with less severe disease (NAS 0–4) versus more severe disease (NAS 5–8 or presence of liver cirrhosis (LCI)). A NAS of 5–8 indicates definitive NASH25. Furthermore, patients with NAFLD and F0–F1 fibrosis and F2–F4 fibrosis were compared. We used Random Forest feature selection to determine features (viral diversity, viral richness and intestinal viruses at species level) that discriminate patients with a NAS of 0–4 (NAS 0–4) from patients with a NAS of 5–8 or cirrhosis (NAS 5–8/LCI) and patients with F0–F1 fibrosis from patients with F2–F4 fibrosis. Multivariate logistic regression analyses were performed to develop prediction models that incorporate clinical features alone or in combination with gut microbial features, to non-invasively predict NAS 5–8/LCI or F2–F4 fibrosis on liver biopsy. Receiver operating characteristic (ROC) analysis was performed with the area under the curve (AUC) to compare all non-invasive approaches36. Further, the likelihood ratio test as implemented in the “rms” package for R37 was used to determine, if adding viral diversity to clinical parameters significantly improves the diagnostic accuracy in multivariate models. Leave-one-out cross validation (LOOCV) was used to validate model accuracy. Under this approach, the prediction model is trained on all the data except for one sample and the prediction is validated in this left-out sample. This procedure is repeated k times (whereas k is the total sample size) and the average error rate is computed for model evaluation38. For all analyses, P values of .05 or less were considered to be statistically significant. Statistical analysis was performed using R statistical software, R version 3.5.139. The R script is available at https://github.com/SchnablLab/ViromeNAFLD.
Data availability.
Raw sequences from 16S rRNA gene sequencing were registered at NCBI under BioProject PRJNA540738. The specific BioSample IDs corresponding to samples used in this study can be found in Supplementary Table 4. Raw virome sequence reads are publicly accessible from the NCBI, through Bioproject number PRJNA622386.
RESULTS
The median age of patients with NAFLD was 55.6 years (range 20.2–79.6 years), 51% were female and the median body mass index (BMI) was 30 kg/m2 (range 22–53 kg/m2). 40% had a NAS of 0–4, and 60% had a NAS of 5–8 or were non-invasively staged as cirrhotic. The main clinical differences between patients with NAS 0–4 and patients with NAS 5–8/LCI were an older age (P = .003), a higher BMI (P = .031), higher proton pump inhibitor (PPI) use (P = .041), and higher AST (P < .001) and ALT levels (P = .002) in patients with NAFLD and NAS 5–8/LCI (Table 1). When we compared patients with NAFLD and F0–F1 fibrosis with those who were staged as F2–F4 fibrosis, we observed an older age (P < .001), higher prevalence of type 2 diabetes (P = .012), higher PPI use (P = .002) and several alterations in laboratory parameters in patients with NAFLD and a higher degree of liver fibrosis (Supplementary Table 1). Normal weight subjects and patients with PBC and mild disease (based on transient elastography) were used as control groups. Eight patients with PBC were overweight and five had steatosis on liver imaging (Supplementary Table 2).
Table 1.
Clinical characteristics of the NAFLD cohort
| Data not available | NAS 0–4 | NAS 5–8/LCI | P value | |
|---|---|---|---|---|
| Total n | 29 | 44 | ||
| Demographics | ||||
| Age, years | 51.9 (28.8–74.2) | 58.9 (20.2–79.6) | .003 | |
| Gender female, n (%) | 13 (44.8) | 24 (54.5) | .478 | |
| Body mass index, kg/m2 | 29.4 (22.5–52.9) | 31.0 (21.9–46.5) | .031 | |
| Waist circumference (cm) | 14 | 106.0 (84.0–130.0) | 113 (81.0–143.0) | .048 |
| Type 2 diabetes, n (%) | 3 (10.3) | 13 (29.5) | .082 | |
| Arterial hypertension, n (%) | 15 (51.7) | 34 (77.3) | .040 | |
| Metabolic syndrome (IDF criteria), n | 9 (31.0) | 24 (54.5) | .057 | |
| (Proton pump inhibitor use, n (%) %) | 1 (3.4) | 10 (22.7) | .041 | |
| Metformin use, n (%) | 2 (6.9) | 11 (25.0) | .063 | |
| Laboratory parameters | ||||
| AST, U/L | 1 | 28.0 (17.0–48.0) | 43.5 (22.0–189.0) | < .001 |
| ALT, U/L | 1 | 36.0 (16.0–97.0) | 54.0 (20.0–239.0) | .002 |
| GGT, U/L | 1 | 79.5 (14.0–732.0) | 71.5 (29.0–334.0) | .876 |
| Alkaline phosphatase, U/L | 1 | 74.5 (43.0–164.0) | 74.5 (43.0–150.0) | .699 |
| Bilirubin, mg/dL | 2 | 0.6 (0.3–1.9) | 0.5 (0.2–2.7) | .593 |
| Albumin, g/L | 2 | 45.0 (40.0–51.0) | 44.0 (34.0–50.0) | .041 |
| Triglycerides, mg/dL | 1 | 120.0 (42.0–1104.0) | 166.0 (55.0–484.0) | .082 |
| Total cholesterol, mg/dL | 1 | 187.0 (104.0–274.0) | 189.5 (104.0–329.0) | .675 |
| HDL cholesterol mg/dL | 6 | 54.0 (16.0–82.0) | 45.0 (27.0–96.0) | .103 |
| LDL cholesterol mg/dL | 8 | 112.0 (47.0–184.0) | 115.0 (42.0–247.0) | .801 |
| Fasting glucose, mg/dL | 1 | 93.5 (80.0–147.0) | 101.0 (63.0–196.0) | .086 |
| HbA1c, % | 7 | 5.3 (4.7–6.6) | 5.6 (4.7–8.3) | .080 |
| Alpha-fetoprotein kU/L | 8 | 2.0 (1.0–10.0) | 3.0 (1.0–85.0) | .055 |
| Creatinine, mg/dL | 1 | 0.8 (0.6–1.4) | 0.8 (0.5–1.3) | .963 |
| Urea, mg/dL | 1 | 28.0 (15.0–45.0) | 29.0 (15.0–48.0) | .655 |
| Uric acid, mg/dL | 1 | 5.9 (2.9–10.5) | 6.2 (2.2–8.7) | .693 |
| Ferritin, μg/L | 1 | 204.0 (19.0–592.0) | 211.5 (16.0–2187.0) | .371 |
| White blood cell count, x1E9/L | 1 | 6.6 (3.7–11.2) | 6.9 (4.0–10.3) | .764 |
| C-reactive protein, mg/L | 2 | 0.0 (0.0–15.5) | 2.1 (0.0–22.9) | .091 |
| Immunoglobulin G, g/L | 3 | 9.9 (6.8–17.3) | 10.9 (6.0–19.2) | .073 |
| Immunoglobulin A, g/L | 3 | 2.4 (0.8–3.9) | 2.5 (0.7–7.4) | .564 |
| Immunoglobulin M, g/L | 3 | 0.9 (0.3–2.6) | 1.0 (0.3–2.6) | .890 |
| Platelet count, x1E9/L | 1 | 232.5 (132.0–386.0) | 213.5 (74.0–373.0) | .181 |
| INR | 1 | 1.0 (0.9–1.1) | 1.0 (0.9–2.4) | .167 |
| Liver histology data of biopsy-proven cohort (n=64) | Scoring | |||
| Total n | 29 | 35 | ||
| Grade of steatosis, n (%) | 0 | 0 (0.0) | 0 (0.0) | |
| 1 | 14 (48.3) | 5 (14.3) | .001 | |
| 2 | 13 (44.8) | 14 (40.0) | ||
| 3 | 2 (6.9) | 16 (45.7) | ||
| Ballooning, n (%) | 0 | 14 (48.3) | 1 (2.9) | < .001 |
| 1 | 14 (48.3) | 16 (45.7) | ||
| 2 | 1 (3.4) | 18 (51.4) | ||
| Grade of inflammation, n (%) | 0 | 9 (31.0) | 0 (0.0) | < .001 |
| 1 | 19 (65.5) | 14 (40.0) | ||
| 2 | 1 (3.4) | 20 (57.1) | ||
| 3 | 0 (0.0) | 1 (2.9) | ||
| Stage of fibrosis, n (%) | 0 | 16 (55.2) | 1 (2.9) | < .001 |
| 1 | 11 (37.9) | 9 (25.7) | ||
| 2 | 2 (6.9) | 11 (31.4) | ||
| 3 | 0 (0.0) | 6 (17.1) | ||
| 4 | 0 (0.0) | 8 (22.9) |
Values are presented as median and range in parentheses or number and percentage. Bold font indicates significance (P value equal or below .05). If liver biopsy was performed (n = 64 patients with NAFLD), samples were evaluated by an experienced liver pathologist who was blinded for all clinical and laboratory patient data. The NASH clinical research network histological scoring system was used to evaluate disease activity and severity. The NAFLD activity score (NAS) was obtained for each biopsy. This score is defined as the unweighted sum of the scores for steatosis, (0–3), lobular inflammation (0–3), and ballooning (0–2); thus ranging from 0 to 8. ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; GGT, gamma-glutamyl-transferase; HbA1c, glycated hemoglobin; INR, international normalized ratio; HDL, High-density lipoprotein; LCI, liver cirrhosis; LDL, Low-density lipoprotein; NAS, NAFLD activity score.
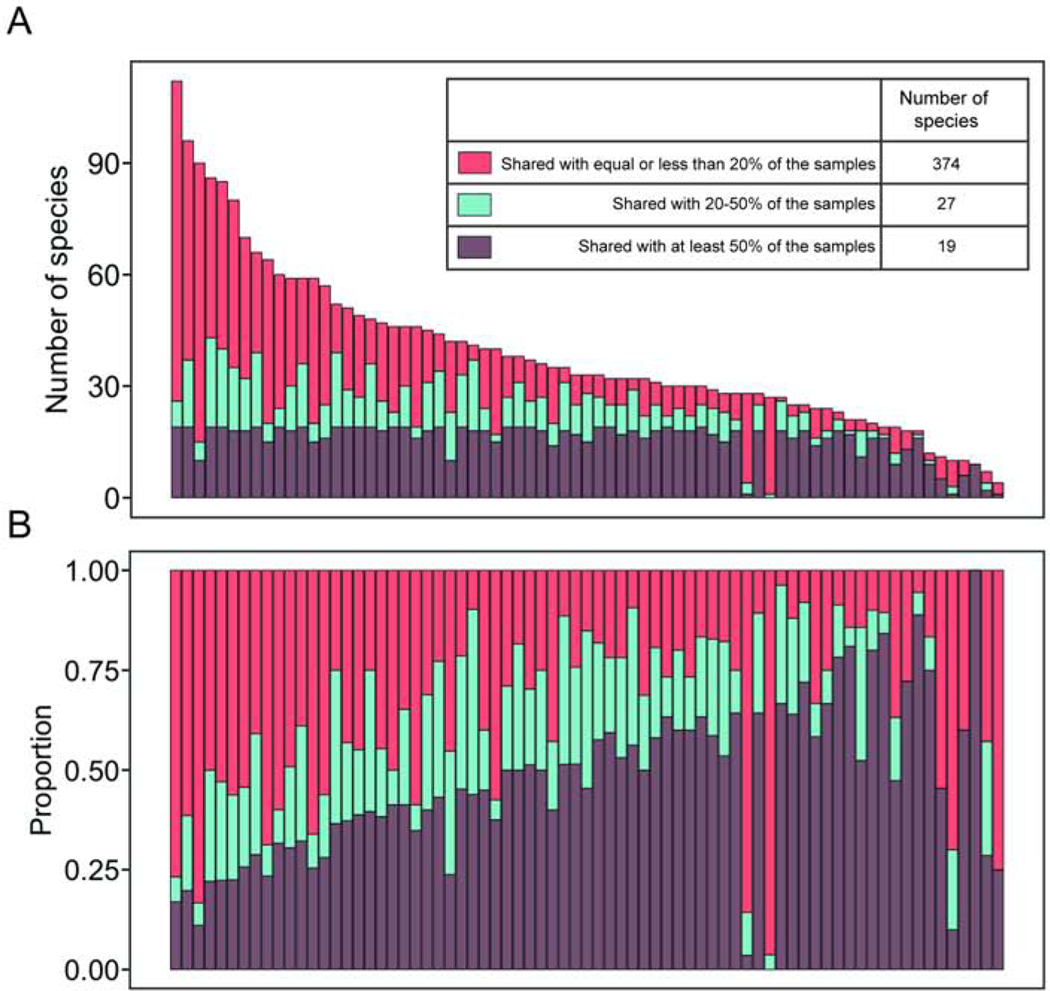
We detected a total of 420 different viral species in the NAFLD cohort. Many of these individual viral species were detected in less than 20% of all fecal samples, while 19 viral species were detected in at least 50% of all samples and 27 species were shared in 20%–50% of the samples (Fig. 1). This indicates that the intestinal virome is individual specific.
Figure 1. Distribution of shared viral species.
420 detected viral species in 73 patients with NAFLD were assigned into three categories based on overlapping characteristics among samples. 19 species were detected in at least 50% of the samples, 27 in 20%–50% of the samples and the remaining 374 species were detected in equal or less than 20% of the samples and therefore more unique to each patient. In panel A and B, one column represents one patient. (A) Absolute number, (B) relative number of detected species in relationship to sharing characteristics.
Decreased viral diversity and compositional alterations in the gut virome of patients with NAFLD and NAS 5–8/LCI
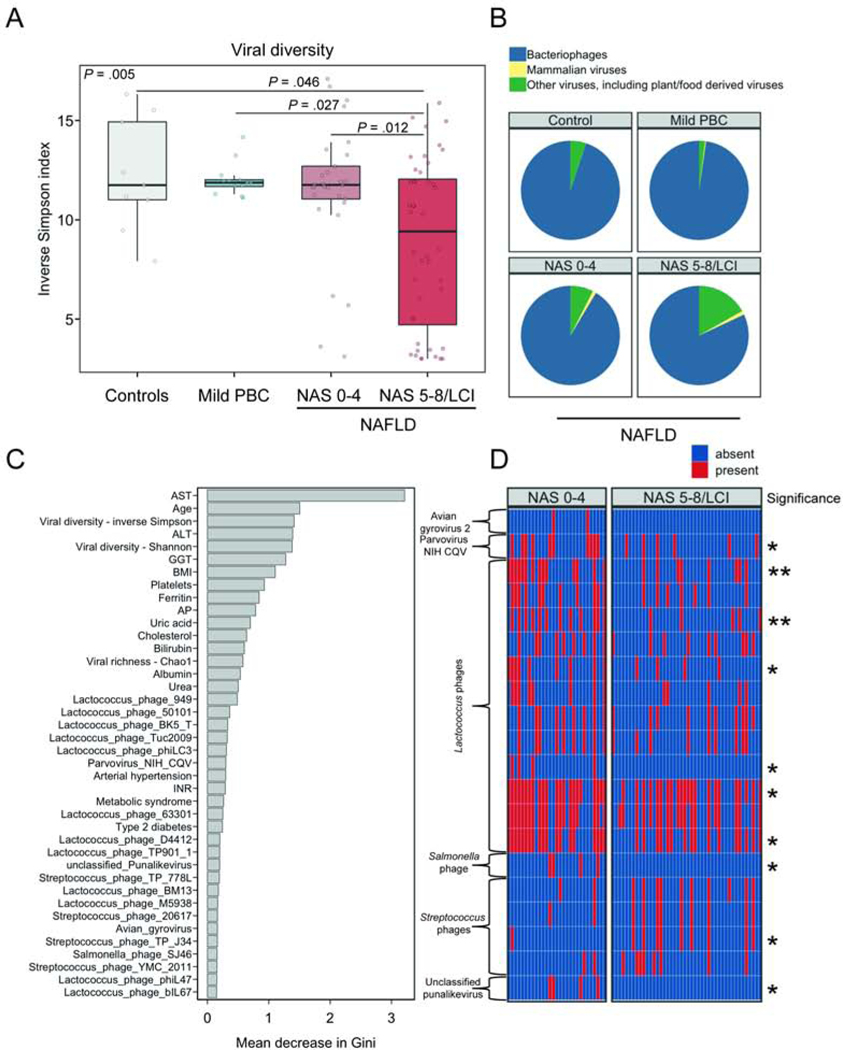
We compared the viral diversity among patients with NAFLD and different disease stages, controls and patients with PBC and mild liver disease. Patients with NAFLD and NAS 5–8/LCI had a significantly lower viral diversity as measured by the inverse Simpson index when compared with patients with NAFLD and NAS 0–4, patients with mild PBC or controls (P = .005, Fig. 2A). Phages were the most abundant viruses in proportion to mammalian viruses, and other viruses (including plant/food derived viruses) (Fig. 2B). Fecal samples from patients with NAFLD and NAS 5–8/LCI contained significantly less phages, in proportion to other viruses, compared with fecal samples from patients with NAFLD and NAS 0–4, patients with mild PBC or controls (phages; NAS 0–4 vs. NAS 5–8/LCI: P = .046, controls vs. NAS 5–8/LCI: P = .038, PBC vs. NAS 5–8/LCI: P = .038, other comparisons not significant, Fig. 2B).
Figure 2. Altered fecal virome composition in patients with NAS 5–8/LCI.
(A) Viral diversity based on the inverse Simpson index. (B) Mean relative abundance of intestinal bacteriophages (phages), mammalian viruses, and other viruses, calculated at the family level. In panel A–B, 29 patients with NAFLD staged as NAS 0–4 and 44 patients with NAFLD staged as NAS 5–8/LCI were compared with 9 controls and 13 patients with mild primary biliary cholangitis (PBC). (C) Random Forest feature selection including the presence/absence of viral taxa at species level together with selected clinical features to discriminate NAS 5–8/LCI from NAS 0–4. (D) Presence/absence heatmap of the relative abundance of viral taxa among the top 40 features identified in Random Forest feature selection. Stars on the right side of the panel indicate significance whereas one star (*) denotes a P value equal or below .05 but higher than .01. Unadjusted and P values adjusted for proton pump inhibitor use can be found in Supplementary Table 3. ALT, alanine aminotransferase; AST, aspartate aminotransferase; AP, alkaline phosphatase; BMI, body mass index; GGT, gamma-glutamyltransferase; INR, international normalized ratio; HDL, High-density lipoprotein; LCI, liver cirrhosis; LDL, Low-density lipoprotein; NAS, NAFLD activity score.
In patients with NAFLD, we observed significant correlations between various metabolic and immunological parameters with the relative abundance of viral taxa. Several Leuconostoc phages were associated with a lower BMI, lower blood glucose and HbA1c levels whereas Escherichia and Enterobacteria phages were associated with increased blood glucose levels (Supplementary Fig. 1).
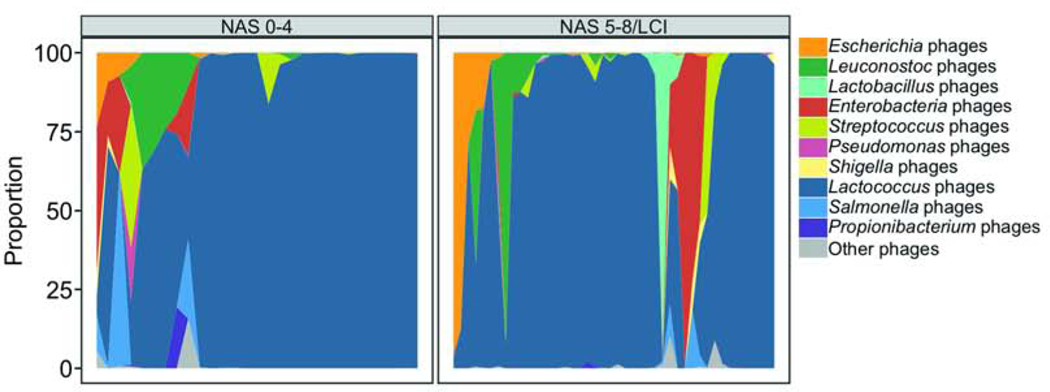
To identify features of the gut virome that discriminate patients with NAFLD and NAS 5–8/LCI from patients with NAFLD and NAS 0–4, we performed Random Forest feature selection under consideration of clinical variables that might be associated with more disease activity as well. Twenty viral taxa were among the top 40 features, and most of them belonged to phages. Overall, most clinical features were of higher importance based on the mean decrease in Gini index for discriminating NAS 5–8/LCI from NAS 0–4 compared with individual viral species. However, viral diversity measures were the third and fifth most important variables following a higher AST and a higher age (Fig. 2C). The 20 most important viral species predominantly belonged to Lactococcus phages whereas several Lactococcus phages were significantly less present in patients with NAFLD and NAS 5–8/LCI (Fig. 2D, Supplementary Table 3). On the other hand, Streptococcus phages TP-J34 were significantly more present in fecal samples from patients with NAFLD and NAS 5–8/LCI (Fig. 2D, Supplementary Table 3). We further summarized phage species according to their known bacterial host. The predominant viral species were Lactococcus phages that were present in almost all samples. Escherichia, Enterobacteria and Lactobacillus phages were more abundant in patients with NAFLD and NAS5–8/LCI but overall, phages were individual specific and several phages were only detected in a few fecal samples (Fig. 3).
Figure 3. Proportion of bacteriophage species summarized by their bacterial host.
Single phages were analyzed at the species level and summarized according to their hosts. The mean relative abundance for these summarized phages was calculated within the phages with a known bacterial host. The x-axis represents individual patients, grouped by the presence of NAS 5–8/LCI. Patients were further grouped according to the relative abundance of specific phages. NAS, NAFLD activity score; LCI, liver cirrhosis.
Although patients with more pronounced disease activity had a higher age, we did not observe significant correlations between age and viral diversity or the proportion of phages, mammalian and plant viruses (Spearman correlation between age and viral diversity (inverse Simpson index): R = −0.14, P = .25, proportion of phages, R = −0.082, P = .49, proportion of mammalian viruses: R = −0.088, P = .46; proportion of plant/other viruses: R = 0.1, P = .40), indicating that age did not have a major impact on the results. There was no significant age difference between controls, patients with PBC and patients with NAFLD (Supplementary Table 2).
NAFLD fibrosis associates with alterations in the gut virome
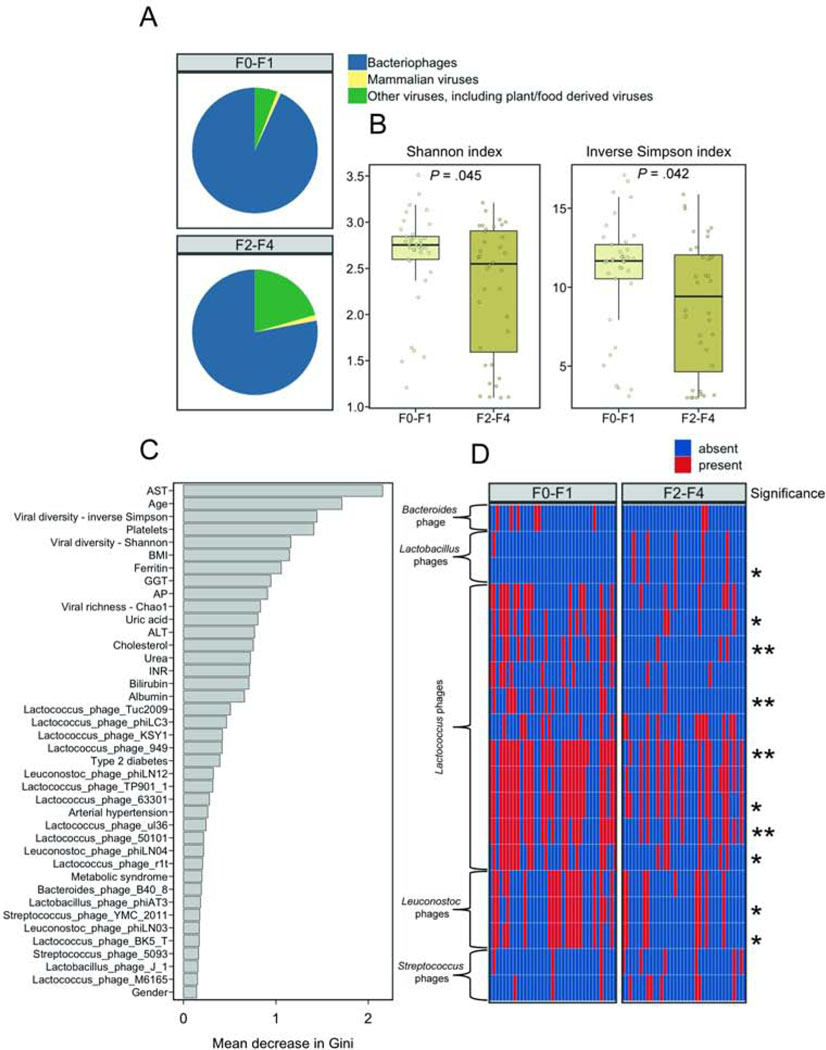
We further investigated if the presence of liver fibrosis is associated with alterations in the gut virome. Patients with NAFLD and F2–F4 fibrosis had a significantly lower proportion of phages compared with patients with NAFLD and no or minimal liver fibrosis (78% mean relative abundance in F2–F4 vs 93% in F0–F1, P < .001; Fig. 4A). The viral diversity was decreased in fecal samples from patients with NAFLD and a higher degree of fibrosis (Fig. 4B).
Figure 4. Altered fecal virome composition in patients with NAFLD and fibrosis.
(A) Mean relative abundance of intestinal bacteriophages (phages), mammalian viruses, and other viruses, calculated at the family level, in fecal samples from patients with NAFLD and F0–F1 fibrosis or patients with NAFLD and F2–F4 fibrosis. (B) Viral diversity based on the Shannon and inverse Simpson indices. (C) Random Forest feature selection including the presence/absence of viral taxa at species level together with selected clinical features to discriminate NAFLD F0–F1 from NAFLD F2–F4. (D) Presence/absence heatmap of the relative abundance of viral taxa among the top 40 features identified in Random Forest feature selection. Stars on the right side of the panel indicate significance whereas one star (*) denotes an adjusted P value equal or below .05 but higher than .01, two stars (**) denote an adjusted P value equal or lower than .01 but higher than .001. Unadjusted and P values adjusted for proton pump inhibitor use can be found in Supplementary Table 3. In panel A–D, 37 patients were staged as F0–F1 fibrosis and 36 patients were staged as F2–F4 fibrosis. ALT, alanine aminotransferase; AST, aspartate aminotransferase; AP, alkaline phosphatase; BMI, body mass index; GGT, gamma-glutamyl-transferase; INR, international normalized ratio; HDL, High-density lipoprotein; LDL, Low-density lipoprotein.
Similar to what we observed in distinguishing patients with NAFLD and NAS 0–4 from patients with NAFLD and NAS 5–8/LCI, clinical features played a more important role than viral species, based on the mean decrease in Gini index, to discriminate patients with NAFLD and at least significant fibrosis from patients with NAFLD and low fibrosis. However, viral diversity and richness indices were among the top discriminating features. When we compared the top viral species, detected by Random Forest feature selection, we found that several Lactococcus and Leuconostoc phages were significantly decreased in patients with NAFLD and higher degrees of liver fibrosis whereas Lactobacillus phage phiAT3 was significantly increased (Fig. 4D, Supplementary Table 3). When we summarized all reads belonging to 47 detected different Lactococcus phages, the relative Lactococcus phage abundance was significantly decreased in patients with F2–F4 fibrosis compared with patients with F0–F1 fibrosis (P = .047), but did not reach significance when comparing patients with NAS 5–8/LCI vs. NAS 0–4 (P = .063).
Taken together, this data indicates that more advanced liver disease in NAFLD is associated with a lower intestinal viral diversity and compositional alterations of specific gut viral taxa.
Proton pump inhibitor use associated with changes in the virome
Among several drugs, PPI use has the strongest influence on the gut bacterial microbiome40. Since we observed significantly more patients with NAS 5–8/LCI or F2–F4 fibrosis using PPI, we further investigated associations between PPI use and alterations in the intestinal virome. Patients with NAS 5–8/LCI and PPI use (n = 10/44 = 22.7%) had a significantly (P = .015) less proportion of phages and a significantly (P = .049) higher proportion of other viruses (including plant/food derived viruses) compared with patients with NAS 5–8/LCI that did not take PPI (Supplementary Fig. 2). Similar trends were observed for patients with NAFLD and F2–F4 fibrosis, however, these changes were not significant (P = .08 for phages, P = .15 for other viruses) (Supplementary Fig. 2). This indicates, that observed differences in proportion of phages and other viruses between patients with NAS 0–4 and patients with NAS 5–8/LCI as well as patients with F0–F1 versus F2–F4 fibrosis might be in part attributable to the use of PPI. On the other hand, PPI use was not significantly associated with a lower viral diversity and importantly, adjusting the association between NAS 5–8/LCI or F2–F4 fibrosis with viral diversity for PPI use in a multivariate logistic regression model, did not affect significance (outcome: NAS 5–8/LCI, dependent: inverse Simpson index: OR .80 (95% CI 0.68–0.92), adjusted P = .004, outcome: F2–F4, OR 0.84 (95% CI 0.73–0.95), adjusted P = .010). We further adjusted the P values corresponding to heatmaps (Fig. 2D, Fig. 4D) for PPI use in multivariate logistic regression models. Most of the associations that were significantly associated with more disease severity in the univariate analysis, remained significant after adjustment for PPI (Supplementary Table 3).
Metformin use, on the other hand, was not associated with differences in the viral diversity or the proportion of phages/mammalian viruses/other viruses (data not shown).
Viral diversity improves non-invasive prediction of histological disease severity
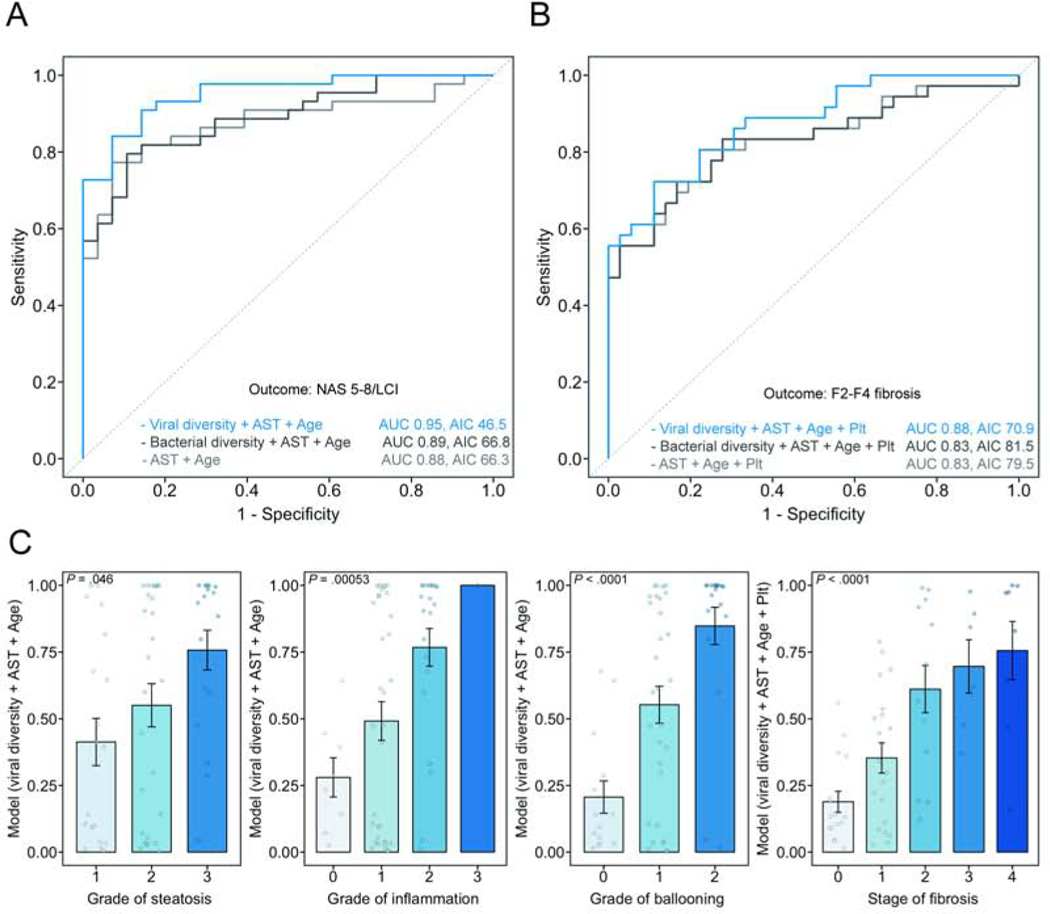
We next performed multivariate logistic regression analyses with the aim to noninvasively detect the presence of NAS 5–8/LCI and F2–F4 fibrosis. We selected the top predictors determined by Random Forest feature selection. Age and AST alone had an AUC of 0.88 (95% CI 0.80–0.96, LOOCV error rate: 0.15) to predict the presence of NAS 5–8/LCI. Whereas adding bacterial diversity (inverse Simpson index) did not improve the model accuracy, adding the viral diversity (inverse Simpson index) significantly improved the diagnostic accuracy as compared with a model computed with age and AST alone (likelihood ratio P value < .001). This model (viral diversity + age + AST) had an AUC of 0.95 (95% CI 0.91–0.99, LOOCV error rate: 0.10) for the prediction of NAS 5–8/LCI (Fig. 5A).
Figure 5. Prediction of more severe liver disease using clinical features and intestinal viral diversity.
Receiver operating curves (ROC) were performed based on multivariate models to predict the presence of (A) NAS 5–8/LCI and (B) F2–F4 fibrosis. The most important variables detected by Random Forest feature selection were included in the models, with or without viral diversity (inverse Simpson index) and bacterial diversity (inverse Simpson index). Likelihood ratio test for the detection of NAS 5–8/LCI, model comparison age + aspartate aminotransferase (AST), versus viral diversity + age + AST, P < .001; detection of F2–F4 fibrosis, model comparison age + AST + platelet counts (Plt), versus viral diversity + age + AST + platelet counts, P = .001. (C) Association between the calculated model value for each individual patient and the individual components of the NAFLD activity score and the stage of fibrosis. AUC, area under the curve; AIC Akaike information criterion; NAS, NAFLD activity score; LCI, liver cirrhosis.
For the detection of F2–F4 fibrosis, adding viral diversity to a model calculated based on clinical variables (age + AST + platelet counts) with or without bacterial diversity significantly increased the diagnostic accuracy, however with a higher LOOCV error rate (AUC 0.88, 95% CI 0.80–0.95, LOOCV 0.17, likelihood ratio P value (compared with the clinical model) = .001, Fig. 5B).
The prediction models, calculated based on viral diversity and clinical parameters, were evenly distributed over all individual histological parameters (grade of steatosis, inflammation, ballooning and fibrosis stage) (Fig. 5C).
Overall, this data indicates that assessing the viral diversity in fecal samples from patients with NAFLD might be helpful to non-invasively detect the presence of more severe liver disease.
Associations between phages and their bacterial host
We next investigated relationships between phages and bacterial genera in patients with NAFLD. An abundance heatmap visualizes the log10 relative abundance of the 20 most abundant phages in direct relationship with their bacterial host in each individual patient.
Phage/bacteria relationship was characterized by high abundance of some phages and decreased abundances of their bacterial host and vice versa (Fig. 6). For example, Lactococcus phages were the most abundant phages, but the level of Lactococcus was comparably low. Bacteroides was high abundant, whereas the corresponding Bacteroides phages were low abundant and detected in only a few patients. On the other hand, a less stringent relationship was seen for Streptococcus and Streptococcus phages (Fig. 6). No obvious relationship was observed for several other phages with the abundance of the bacterial host. Also, viral diversity did not correlate with bacterial diversity (Spearman correlation between viral and bacterial Shannon index: R=0.13, P=0.262, and inverse Simpson index: R=0.10, P=0.393).
Figure 6. Association between bacteriophages and bacteria.
Abundance heatmap showing the log10 relative abundance of the 20 most abundant phages in direct relationship with their bacterial host in each individual patient (x-axis).
DISCUSSION
In this study, we show that a more pronounced disease activity in NAFLD is associated with specific alterations in the intestinal virome. We found that patients with NAFLD and NAS 5–8/LCI or F2–F4 fibrosis had a significantly lower gut viral diversity and significantly fewer phages in proportion to other viruses. We further observed that specific viral taxa, such as several Lactococcus phages were less frequently present in patients with more severe disease. A logistic regression model using simple clinical parameters and the gut viral diversity had a high diagnostic performance for the non-invasive detection of more severe liver disease. In our study, a decreased viral diversity was associated with NAS 5–8/LCI and fibrosis. Since there is no non-invasive biomarker available to accurately detect patients with NAS 5–8/LCI, viral diversity together with simple clinical parameters might help in the future to predict more advanced disease non-invasively.
Analyzing the viral species level, we observed an individual specific composition of the virome. The more species we detected in one patient, the higher the proportion of taxa that were shared in only up to 20% of the samples, consistent with other publications19, 41, 42. Despite high individual differences in viral composition, longitudinal studies have shown a high stability of an individual’s gut virome over time42, 43. The high individuality might limit the utility of individual viral species as potential diagnostic or prognostic biomarker but assessing the overall viral diversity might be a broader applicable and therefore a more robust biomarker.
NAFLD is a multifactorial disease with well-known and less well understood risk factors; an older age, the presence of type 2 diabetes, increased BMI, genetic variants like the common PNPLA3 (encoding patatin like phospholipase domain containing protein 3) p.I148M polymorphism and dietary factors are well known factors for disease development and progression1. It has been demonstrated in several studies that patients with NAFLD, particularly those with advanced disease stages have a reduced bacterial diversity and compositional changes in the gut microbiome44–47. There are several mechanisms of how alterations in the gut bacterial microbiota might modulate disease. Gut barrier dysfunction observed in patients with NAFLD can lead to microbial translocation to the liver, where microbial components such as lipopolysaccharide can induce an inflammatory cascade. Further, intestinal bacteria are the rate-limiting step in deconjugation of bile acids, produced by the liver, and to convert primary bile acids into secondary bile acids. Bile acids act as signaling molecules through binding to host nuclear and G-protein-coupled receptors, which impacts several host metabolic functions. Other potential mechanistic pathways of how the gut bacterial microbiome might affect NAFLD include synthesis of short-chain fatty acids, an increased energy harvest by gut bacteria as well as endogenous ethanol production48.
In this interplay, the gut virome might play a role by either affecting the host directly (for instance through causing host immune responses) or by influencing the bacterial microbiota. One can speculate that a reduced viral diversity could lead to a quantitative increase of certain bacteria in patients with NAFLD, which then might contribute to modulation of disease. Similar to patients with NAFLD and more pronounced disease, patients with Clostridium difficile infections have a significantly lower viral diversity as compared with controls49. The treatment response in fecal microbiota transfer performed in these patients, was also associated with the donor viral diversity, in which a high viral diversity in the donor feces was associated with favorable effects49. Transfer of the fecal virome from mice with a lean phenotype into mice fed a high-fat diet, resulted in reduced weight gain and amelioration of metabolic parameters, which was accompanied by shift in the gut bacterial microbiota composition50. Although this data indicates an active role of phages shaping the bacterial microbiome, causal relationships in the complex human situation are elusive. Alterations in the gut virome and particularly in the gut “phageome” might also govern the taxonomic composition of the bacterial microbiota or might be primarily driven by the host disease. It is unclear what factors are driving a reduced viral diversity. Similar to the bacterial microbiome, several factors, such as environmental, dietary, genetic as well as medications might play a role in virome composition. Longitudinal studies in humans are needed to further characterize the interaction between bacteria and phages in patients with NAFLD and preclinical studies are required to determine the contribution of phages to disease progression.
The strength of our study is the very well characterized study cohort with available liver histology. Assessing the intestinal virome is challenging and we cannot rule out that a possible DNA amplification bias and bacterial DNA contamination might have resulted in a distorted taxonomic profiling of fecal samples. An additional shortcoming of this study is the low number of known viral genomes in public databases43. Improvements in sequencing, bioinformatics and probably the use of phage culturing and phage proteomics techniques are needed to further investigate this viral “dark matter”43.
In conclusion, we show that histological disease severity of NAFLD is associated with changes in the gut virome. While mechanistic studies are needed to investigate a potential causal role in disease progression, the intestinal virome could contain information to identify patients with NAFLD at risk for future liver-related complications.
Supplementary Material
What you need to know:
Background and Context:
Alterations in the gut microbiome have been associated with the severity of nonalcoholic fatty liver disease (NAFLD), but these studies focused only on change in bacteria; little is known changes in the viral microbiome (virome) in patients with NAFLD.
New Findings:
In a study of fecal viromes from patients with NAFLD and controls, the authors associate histologic markers of NAFLD severity with significant decreases in viral diversity and proportion of bacteriophages. They developed a model based on fecal viral diversity and clinical data that identifies patients with severe NAFLD and fibrosis.
Limitations:
This was an association study—further studies are needed to determine whether changes in intestinal viruses are a cause or effect of NAFLD, and how these changes might come about during development of fatty liver.
Impact:
Fecal viromes might be analyzed to identify patients at risk for severe disease and also to identify therapeutic targets.
Lay Summary:
This study identified changes in the population of viruses in fecal samples of patients with fatty liver that might contribute to development of disease or be used to identify patients with more severe disease.
Acknowledgements:
This study was in part supported by a DFG fellowship (LA 4286/1–1) (to S.L.), by the “Marga und Walter Boll-Stiftung”, project number 210–03-2016, and the “Köln Fortune” research pool, Faculty of Medicine, University of Cologne, Germany, project number 160/2014 (to M.D.), NIH grants R01 AA020703, R01 AA24726, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.), and services provided by P30 DK120515 and P50 AA011999. R.L. receives funding support from NIEHS (5P42ES010337), NCATS (5UL1TR001442), NIDDK (R01DK106419), and DOD PRCRP (CA170674P2). P.S. received funding support from the Fond National de Recherche Scientifique Belgium (J.0146.17 and T.0217.18) and ARC 2018 grant, Université Catholique de Louvain, Brussels, Belgium
Conflicts of interest: B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics and Patara Pharmaceuticals. B.S.’s institution UC San Diego has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company and Axial Biotherapeutics. R.L. serves as a consultant or advisory board member for Arrowhead Pharmaceuticals, AstraZeneca, Boehringer Ingelheim, Bristol-Myer Squibb, Cirius, CohBar, Galmed, Gemphire, Gilead, Glympse bio, Intercept, Ionis, Merck, Metacrine, Inc., NGM Biopharmaceuticals, Novo Nordisk, Pfizer, and Viking Therapeutics. In addition, his institution has received grant support from Allergan, Boehringer-Ingelheim, Bristol-Myers Squibb, Eli Lilly and Company, Galmed Pharmaceuticals, Genfit, Gilead, Intercept, Janssen, Madrigal Pharmaceuticals, NGM Biopharmaceuticals, Novartis, Pfizer, pH Pharma, and Siemens. He is also co-founder of Liponexus, Inc.
Abbreviations:
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- AUC
area under the curve
- BMI
body mass index
- HbA1c
glycated hemoglobin
- LCI
liver cirrhosis
- LOOCV
leave one out cross validation
- NAFLD
non-alcoholic fatty liver disease
- NAS
NAFLD activity score
- phages
bacteriophages
- PBC
primary biliary cholangitis
- PPI
proton pump inhibitor
- ROC
receiver operating curve
Footnotes
Author names in bold designate shared co-first authorship
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Diehl AM, Day C. Cause, Pathogenesis, and Treatment of Nonalcoholic Steatohepatitis. N Engl J Med 2017;377:2063–2072. [DOI] [PubMed] [Google Scholar]
- 2.Ekstedt M, Hagström H, Nasr P, et al. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology 2015;61:1547–1554. [DOI] [PubMed] [Google Scholar]
- 3.Ajmera V. Nonalcoholic fatty liver disease activity score and mortality: Imperfect but not insignificant. Hepatology 2016;64:309–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vilar-Gomez E, Chalasani N. Non-invasive assessment of non-alcoholic fatty liver disease: Clinical prediction rules and blood-based biomarkers. J Hepatol 2018;68:305–315. [DOI] [PubMed] [Google Scholar]
- 5.Balakrishnan M, Loomba R. The Role of Noninvasive Tests for Differentiating NASH From NAFL and Diagnosing Advanced Fibrosis Among Patients With NAFLD. Journal of clinical gastroenterology 2020;54:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayakumar S, Loomba R. Review article: emerging role of the gut microbiome in the progression of nonalcoholic fatty liver disease and potential therapeutic implications. Aliment Pharmacol Ther 2019;50:144–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Roy T, Llopis M, Lepage P, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 2013;62:1787–94. [DOI] [PubMed] [Google Scholar]
- 8.Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143:913–6.e7. [DOI] [PubMed] [Google Scholar]
- 9.Jiang TT, Shao TY, Ang WXG, et al. Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host Microbe 2017;22:809–816.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carding SR, Davis N, Hoyles L. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther 2017;46:800–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmelcher M, Loessner MJ. Bacteriophage endolysins: applications for food safety. Curr Opin Biotechnol 2016;37:76–87. [DOI] [PubMed] [Google Scholar]
- 12.Mirzaei MK, Maurice CF. Menage a trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 2017;15:397–408. [DOI] [PubMed] [Google Scholar]
- 13.Canchaya C, Fournous G, Chibani-Chennoufi S, et al. Phage as agents of lateral gene transfer. Curr Opin Microbiol 2003;6:417–24. [DOI] [PubMed] [Google Scholar]
- 14.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol 2013;14:654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell 2009;138:30–50. [DOI] [PubMed] [Google Scholar]
- 16.Virgin HW. The virome in mammalian physiology and disease. Cell 2014;157:142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo T, Lu XJ, Zhang Y, et al. Gut mucosal virome alterations in ulcerative colitis. Gut 2019;68:1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clooney AG, Sutton TDS, Shkoporov AN, et al. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host Microbe 2019;26:764–778.e5. [DOI] [PubMed] [Google Scholar]
- 20.Nakatsu G, Zhou H, Wu WKK, et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 2018;155:529–541.e5. [DOI] [PubMed] [Google Scholar]
- 21.Vehik K, Lynch KF, Wong MC, et al. Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nat Med 2019;25:1865–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang S, Demir M, Duan Y, et al. Cytolysin-positive Enterococcus faecalis is not increased in patients with non-alcoholic steatohepatitis. Liver International 2020;40:860–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol 2016;64:1388–402. [DOI] [PubMed] [Google Scholar]
- 24.International Diabetes Federation. Recommendations For Managing Type 2 Diabetes In Primary Care, 2017. www.idf.org/managing-type2-diabetes.
- 25.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313–21. [DOI] [PubMed] [Google Scholar]
- 26.Brunt EM, Kleiner DE, Wilson LA, et al. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology 2011;53:810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeom SK, Lee CH, Cha SH, et al. Prediction of liver cirrhosis, using diagnostic imaging tools. World journal of hepatology 2015;7:2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang S, Martin A, Farowski F, et al. High Protein Intake Is Associated With Histological Disease Activity in Patients With NAFLD. Hepatology Communications 2020;n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conceicao-Neto N, Yinda KC, Van Ranst M, et al. NetoVIR: Modular Approach to Customize Sample Preparation Procedures for Viral Metagenomics. Methods Mol Biol 2018;1838:85–95. [DOI] [PubMed] [Google Scholar]
- 30.McIver LJ, Abu-Ali G, Franzosa EA, et al. bioBakery: a meta’omic analysis environment. Bioinformatics 2018;34:1235–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostic AD, Ojesina AI, Pedamallu CS, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol 2011;29:393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker MA, Pedamallu CS, Ojesina AI, et al. GATK PathSeq: a customizable computational tool for the discovery and identification of microbial sequences in libraries from eukaryotic hosts. Bioinformatics 2018;34:4287–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMurdie P, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013;8(4), pp. e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angulo P, Kleiner DE, Dam-Larsen S, et al. Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2015;149:389–97.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalski J, Tu XM. Modern Applied U-Statistics: Wiley, 2008. [Google Scholar]
- 37.Harrell FE. rms: Regression Modeling Strategies. R package version 4.3–1. 2015. [Google Scholar]
- 38.Elisseeff A, Pontil M. Leave-one-out error and stability of learning algorithms with applications Stability of Randomized Learning Algorithms Source. International Journal of Systems Science 2002. [Google Scholar]
- 39.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.2018. [Google Scholar]
- 40.Imhann F, Vich Vila A, Bonder MJ, et al. The influence of proton pump inhibitors and other commonly used medication on the gut microbiota. Gut Microbes 2017;8:351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manrique P, Bolduc B, Walk ST, et al. Healthy human gut phageome. Proceedings of the National Academy of Sciences of the United States of America 2016;113:10400–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shkoporov AN, Clooney AG, Sutton TDS, et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell host & microbe 2019;26:527–541.e5. [DOI] [PubMed] [Google Scholar]
- 43.Shkoporov AN, Hill C. Bacteriophages of the Human Gut: The “Known Unknown” of the Microbiome. Cell Host Microbe 2019;25:195–209. [DOI] [PubMed] [Google Scholar]
- 44.Loomba R, Seguritan V, Li W, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab 2017;25:1054–1062.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rau M, Rehman A, Dittrich M, et al. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J 2018;6:1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Chierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017;65:451–464. [DOI] [PubMed] [Google Scholar]
- 47.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tripathi A, Debelius J, Brenner DA, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 2018;15:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo T, Wong SH, Lam K, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 2018;67:634–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasmussen TS, Mentzel CMJ, Kot W, et al. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 2020:gutjnl-2019–320005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences from 16S rRNA gene sequencing were registered at NCBI under BioProject PRJNA540738. The specific BioSample IDs corresponding to samples used in this study can be found in Supplementary Table 4. Raw virome sequence reads are publicly accessible from the NCBI, through Bioproject number PRJNA622386.