Summary
A myriad of microbes living together with the host constitute microbiota, which possesses very diverse functions in regulation of host physiology. Recently, it has been unequivocally demonstrated that microbiota regulates cancer initiation, progression and responses to therapy. Here we review known pro-tumorigenic and anti-tumorigenic function of microbiota and mechanisms how microbes can regulate cancer cells and immune and stromal cells within the tumor microenvironment.
Keywords: Microbiome, tumor microenvironment, cancer, tumor progression, mechanisms, host-microbiome interactions
Introduction
Unique complex microbial ecosystems are important and irreplaceable components of human body biology and homeostasis. Microbiota of the host appears early after birth through maternal transmission is further shaped through the lifetime by various circumstances and events including host genetics, infections, antibiotic usage, dietary and environmental factors, and presence of various diseases or use of therapies. The presence of large number of live and metabolically active microbes in close proximity to cells represents opportunity for the reciprocal interactions whereby the host modify the microbiota and microbiota derived factors. Indeed, quantitative and qualitative properties of the microbiota can regulate multitude of host processes, including tonic regulation of immune and inflammatory cells, digestion, gastrointestinal motility, cell differentiation and turnover or cognitive and behavioral functions. Likewise, changes in microbiota have been associated or found to be causative of a variety of diseases, including inflammatory conditions [1,2], metabolic [3] and cardiovascular diseases [4], neurological ailments [5] and cancer [6,7]. Since the gastrointestinal tract harbors at least 1014 bacteria [8], many aspects of the “microbiota-host-disease” triad have been studied in the context to intestinal microbiome. However, it has become apparent that other organs, previously thought to be sterile can harbor microbial organisms both in healthy and diseased states or are subject to distant regulation by microbial metabolites produced elsewhere[9,10].
In this review we summarize some of the strategies and mechanisms by which microbiota regulates cancer progression, metastasis and sensitivity to therapies and metastasis development and progression. Importantly, this relation can be both deleterious and beneficial, as the microbiome can exert anti-tumorigenic and pro-tumorigenic effects in the host through distinct molecular and cellular mechanisms.
Protective role of the microbiome in cancer
Live bacteria and their products can accumulate in many tumor types, primary or metastatic [10]. Distant regulation of tumorigenesis and tumor microenvironment is also thought to be a process that can be mediated by microbiota [9,11–15]. While many bacteria and their products were shown to be tumor-promoting [6,16,17], as discussed later, some bacteria clearly exert protective functions during the process of tumorigenesis or facilitate various forms of cancer therapies.
Diverse microbiome community which populates tumor tissue or regulates intra-tumoral processes distantly can exert protective, anti-tumor function by several mechanisms.
First, some bacterial species could initiate and sustain the formation of favorable conditions for growth of beneficial bacteria and together form the niche which suppresses the overgrowth of pathogenic bacteria [18] (Figure 1). Examples of such beneficial bacteria include Lactobacillus or various Clostridium species protecting against pathogenic E.coli or Salmonella colonization in the context of intestinal inflammation [19,20].
Figure 1. Roles of «beneficial» and «pathogenic» bacteria in primary tumor development and metastasis.
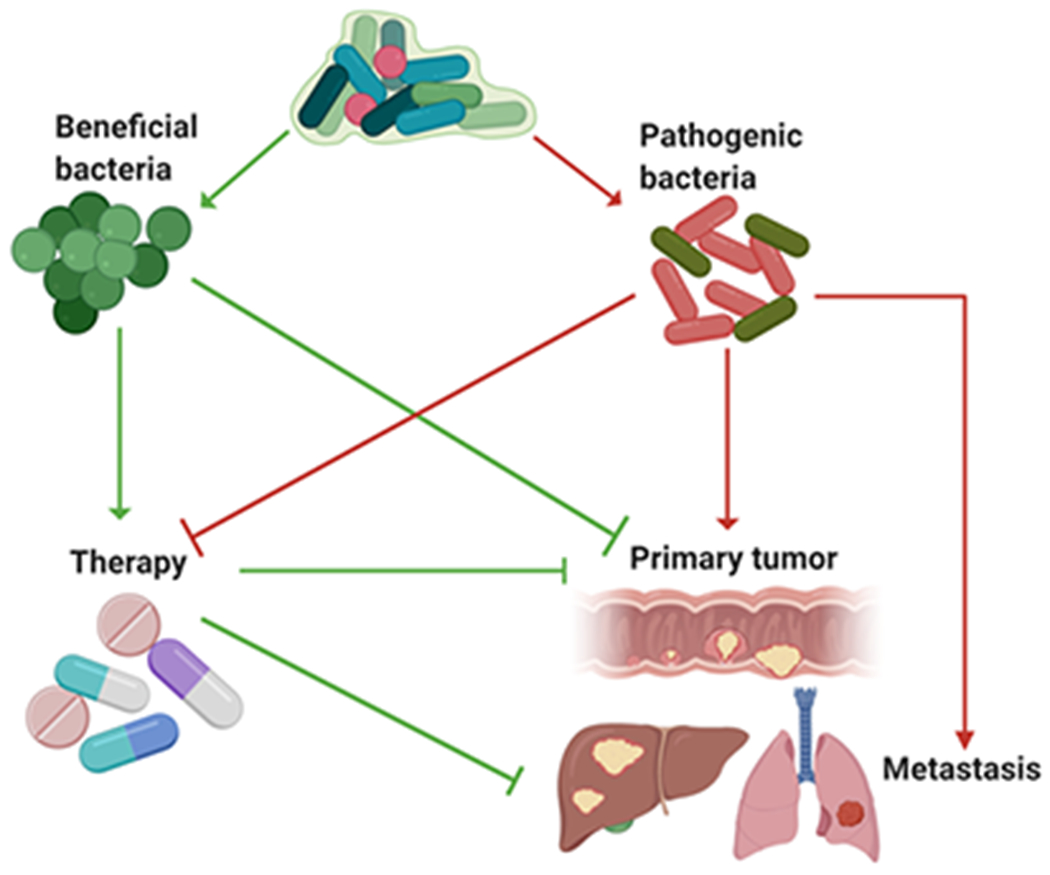
Microbiota contains multiple species with different metabolic activities and other properties, and these species co-exist with the host within the particular niche or tissue. Each relevant bacterial species plays the different role in the primary tumor growth, metastasis and anti-cancer therapy. «Beneficial» bacteria can suppress tumor growth and improve anti-cancer therapies through several direct mechanisms but also can inhibit overgrowth and outcompete «pathogenic» bacteria in the context of cancer. Pathogenic bacteria promote tumor development, metastasis and resistance to therapies. “Green” reflects anti-tumorigenic action, while “Red” is tumor-promoting function. Arrows illustrate active promotion, while blocks illustrate inhibition. Figure was produced using BioRender platform.
Second, some of the metabolites produced by bacteria may be essential tools in their interactions with epithelial and cancer cells for tumor growth suppression. Short chain fatty acids (SCFA), which are produced by different commensals from fiber fermentation, inhibit myeloid cell driven pro-tumorigenic inflammation and regulate proliferation of epithelial and stem cell compartment [21,22]. Several vitamins, including biotin, cobalamin, folate, niacin, pantothenate, pyridoxine and others have also been linked to anti-tumor activities [23]. Phytochemicals such as polyphenols, which are widely present in fruits and vegetables, are metabolized by certain gut bacterial species into the active forms. Polyphenols metabolites may prevent cancer progression through the impact on cell cycle arrest and apoptosis and also through inhibition of inflammatory cytokines production [24].
Third, distinct bacterial species or “aggregate microbiota” are essential for maturation and tonic stimulation of the immune system, which then can exert immunosurveillance functions at different stages of tumor development [25,26]. Classical Coley experiments and anti-tumor effect of Coley toxins where bacterial infections led to regression of established tumors served as a foundation for studies in this direction [27]. On the other hand, germ free mice lack fully matured immune system and also depletion of microbiota with broad spectrum antibiotics can reduce the efficacy of immunesurveillance in mouse models [28]. This function is further extended to the ability of microbes to stimulate activation of various immune cells subsets; for example, members of Lactobacilla phylum activate antitumor immune responses by dendritic cell (DC) maturation [29] and subsequently for acquisition of cytotoxic properties by T cells, NK cells, NK T cells and anti-tumorigenic myeloid cells [30].
Fourth mechanism stems from the aforementioned ability of bacteria to drive activation of immune system and is related to the ability of some bacteria to improve anti-cancer therapies [6]. Germ-free or antibiotic-treated tumor bearing mice do not properly respond to standard chemotherapeutic treatment with oxaliplatin [31] and gut bacteria are needed for the effect of cyclophosphamide, an anti-cancer immunomodulatory agent, which acts in particular through an increase in the intestinal permeability and translocation of immunostimulatory bacteria into secondary lymphoid organs, such as lymph nodes, for proper stimulation of anticancer immune responses [32]. Microbiota is also essential for efficiency of and sensitivity to various immunotherapies, such as combination of CpG-oligodeoxynucleotides (ODN), a ligand of Toll-like receptor 9 (TLR9), and inhibitory interleukin-10 (IL-10) receptor antibodies (anti-IL-10R) therapy [31]. It was shown that some commensal bacteria such as Alistipes and Ruminococcus are directly responsible for production of anti-tumorigenic TNF and regulation of reactive oxygen species essential for tumor restriction in this model [31]. Another facet of microbiota action is its ability to induce and heighten the responses to immunotherapies based on the usage of immune checkpoint blockade approaches. Responses to anti-CTLA4 or anti-PD-1/PD-L1 or combinational therapies positively correlated with overall diversity of microbiota as well as with the presence of particular species, such as Bacteroides and others [25,33–35] . These findings have led to trials of probiotics or fecal transplants from responders to non-responders in order to understand the mechanisms which underline these effects of microbiota. Mechanistically, the microbiota can regulate the production of cytokines with anti-tumorogenic and immunostimulatory properties, such as IL-12, TNF and others [31]. As the microbial diversity associated with improved responses, it is tempting to hypothesize that microbial genes provide an excellent basis for molecular mimicry of cancer neoantigens, i.e. when the same peptides present as cancer neoantigen in the tumor are present within one of the bacterial proteins. In this scenario, the presence of bacteria provides both a signal (so called “signal 1”) for stimulation of innate immunity, and an additional signal (“signal 2”) as the MHC- peptide complex, which is identical to that on cancer cells; and the presence of checkpoint blocker alleviates the exhaustion or repression of neoantigen/tumor specific T cells. This has been demonstrated in particular for Bacteroides fragilis which encodes for the peptide found in melanoma [33] or for a bacteriophage infecting Enterococcus species associated with enhanced response to anti-PD-1 immunotherapy [36].
Pro-tumorigenic effects of microbiota in tumorigenesis and tumor progression
A concept of “infection-driven” cancers is highly plausible, as HPV, HCV, HBV, Helicobacter pylori and other infectious organisms have been shown to aid to directly promote transformation and tumor growth and can be considered as bona fide “infectious carcinogens” [37]. However, recent evidence also suggests that pathobionts, commensal organisms that can sometimes become pathogenic, can also drive tumorigenesis. The underlying mechanisms for microbiota driving tumorigenesis are somehow overlapping with the case of anti-tumorigenic mechanisms discussed above, and include the induction of pro-tumorigenic inflammation and immunosuppression, aberrant tissue repair program, direct genotoxic effects, as well as metabolic effects on cancer cells and cells of the tumor microenvironment (Figure 2).
Figure 2. Mechanisms of microbiota-host interactions promoting tumorigenesis.
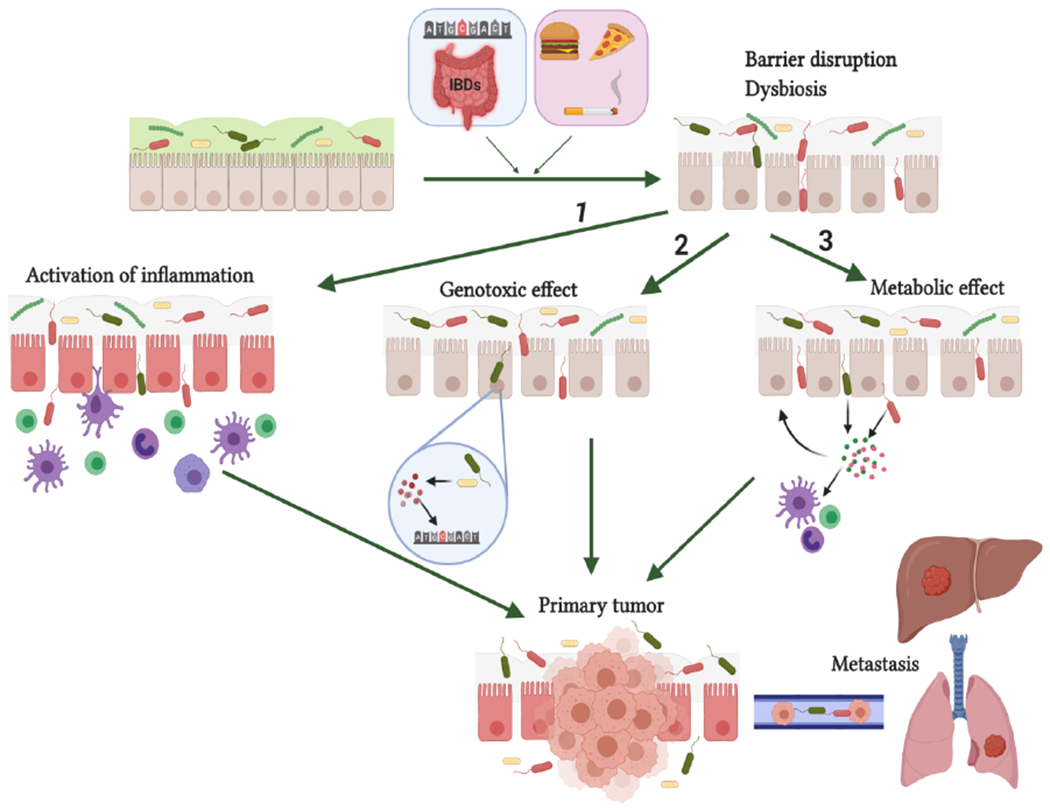
Non-modifiable (genetic) and environmental factors promote intestinal epithelial barrier disruption and dysbiosis. Barrier dysfunction causes bacterial penetration through the epithelium and their interaction with host cells by several mechanisms: (1) immune cell activation and triggering of pro-tumorigenic inflammation; (2) Epithelial cell DNA damage by bacterial toxins or bacteria-induced genotoxic products (e.g. ROS) ; (3) modulation of immune, stromal and cancer cell function through the effect of bacterial metabolites acting locally or distantly. Figure was produced using BioRender platform.
Loss of epithelial barrier function and increased bacterial colonization of transformed tissues
Epithelial and mucosal surfaces of the body as well as tumors are colonized by bacteria at different degrees ranging from modest (lung, pancreas) to high bacterial loads (colon). Indeed, colonic epithelium and lumen are rich in bacterial content, which requires the presence of a strong dedicated barrier including fortified cell-to-cell contacts, mucus coverage and continuous secretion of anti-bacterial peptides and immunoglobulins A (IgA) [38]. Because of this barrier, in healthy state bacterial transfer into systemic circulation and transport into other tissue is a rare and inefficient process. Even local interaction of bacteria with epithelial cells via bacteria-derived metabolites, such as SCFA, secondary bile acids, vitamins and polyamines may be resctricted by a functional barrier [39]. Barrier function however can be deteriorated by autoimmunity and inflammation, diets characterized by high fat or sugar content, the presence of invasive species or dysbiosis of different etiologies, including antibiotics overuse [40–44]. However, neoplastic transformation itself can lead to dysregulation of the epithelial barrier, directly linking early transformation with the loss of epithelial integrity and barrier against microbiota [45,46]. For example, in colorectal cancer, early transformation caused by dysregulation of tumor suppressor APC leads to the loss of proper cell junction organization and tight junctions as well as to cellular hyperproliferation and a loss of terminally differentiated subset of mucus-producing Goblet cells [45,46]. Altogether, this causes an increase in direct contact of commensal with neoplastic epithelial cells and other cells of tumor microenvironment and facilitate the access of microbially-derived products to the neoplastic tissue[45,46]. In addition, as inflamed or normal tissue progresses to adenoma and to carcinoma, microbiota is disrupted, probably due to changes in the barrier and epithelial compartment itself [47,48]. Among the bacteria enriched in CRC patients is Fusobacterium nucleatum [17,49,50] that encodes surface receptor FadA which binds to E-cadherin. Such binding and accumulation in transformed tissues is likely aided by the loss of barrier function and by direct physical FadA-E-cadherin interaction. Loss or alteration in barrier function and changes in metabolic state of epithelial cells can also lead to accumulation of other species, such as biofilm-forming Escherichia coli [51] and enterotoxigenic Bacteroides fragilis [52]. Such biofilms initially found in CRC would be probably detected in other epithelial cancers, such as lung and pancreatic cancers. Overgrowth of bacterial biofilms in the direct proximity of transformed tissue is likely to enforce stronger and persistent interactions between microbes and transformed tissue surface [53] and could facilitate reciprocal loss of epithelial barrier function at the tumor site. Furthermore, some bacteria like B. fragilis are especially efficient at establishment of biofilms and their presence initiates a cascade of local colonization with other bacteria such as E.coli [54]. The presence of biofilms is not obligatory for the bacteria to be associated with tumors or to drive tumor promoting functions though, as pro-tumorigenic bacteria and components of microbiota have been detected in lung [55–57], pancreatic [58,59] and other tumors without formal demonstration that they are organized in biofilms.
Once increased interaction of bacteria and epithelial-tumor microenvironment compartment is established due to the loss of barrier function, increased colonization or formation of biofilms, bacteria can drive tumor progression by several mechanisms as it is described next.
Genotoxic effect of bacteria and induction of tumor-driving mutations
Distinct components of microbiota can release or induce synthesis of genotoxic compounds to affect host DNA by inducing mutations. Arguably, these processes are critical for tumor initiation and progression. Toxins, genotoxic compounds and other bacteria-induced metabolites have been directly implicated tumorigenesis by mutations or otherwise altering the expression levels of key oncogenes and tumor suppressors [60]. Helicobacter pylori gene product CagA inhibit the function of p53 tumor suppressor protein in gastric epithelial cells [61], while some Staphylococcus strains secrete toxic hemolysins and enterotoxins, which may contribute to the accumulation of mutations in malignant T cells [62]. Salmonella typhi produces a protein toxin virulence factor A that directly increases the expression of oncogenic β-catenin expression [63]. Genotoxic strains of E. coli harbor PKS “pathogenicity island” which encodes biosynthetic machinery for production of colibactin- a toxin which alkylates DNA at adenine residues [51] and causes mutations in the intestinal epithelial cells [64,65]. By genomic sequencing, a similar mutational signature was found in a subgroup of over 5800 human cancer genomes, mainly in colorectal cancers and these tumors were specifically associated with E.coli expressing colibactin [65]. Monocolonization with PKS+ but not with colibactin/PKS negative E. coli NC101 increase cancer development in Il10−/− mice treated with azoxymethane [64] and other models of colon cancer, and mutation and PKS-specific mutational signature can be directly recapitulated in vitro using intestinal organoids colonized with E. coli [65]. Enterotoxic strains of B. Fragilis produce BFT toxin, previously implicated into exacerbation of colitis and colon cancer tumorigenesis. BFT binds to a specific colon epithelial receptor, activates the Wnt and NF-κB pathways, promotes proliferation and accelerates tumorigenesis [66]. Secondary bile acids, which are converted from primary bile acids by various members of Firmicutes family, possess DNA–damaging capabilities and therefore can aid cancer induction [67,68]. Another mechanism by which microbiota can induce DNA damage is the induction of oxidative stress and nitric oxide production through the activation of innate immune cells. Such oxidative DNA damage is characterized by presence of 8-oxoguanine and double-stranded DNA breaks and can affect key tumor suppressors, such as p53 [69]. During chronic inflammation, such genetic lesions can be present in visibly normal areas of epithelium and subsequently give rise to cancer through the process of «field cancerization» [70]. Prolonged IBD also triggers ROS-dependent, mutation-induced colitis associated cancer in animal models without further addition of exogenous carcinogens, suggesting an instrumental role for inflammation in induction of cancer-driving mutations [71]. As discussed above, changes in barrier function may be again facilitate of closer and persistent contact between bacteria and target population of cells acquiring mutations.
2. Metabolic effect and regulation of epithelial and stem cell compartment
Collectively, microbiota harbors at least 100 times more genes than the host, with substantial numbers of these genes encoding for the production of various metabolites, some of them identical to the host metabolites and some very unique. Diets modulate cancer risk and some do so through the metabolic activity of microbiota [72–74] involved in the regulation of lipid or cholesterol metabolism [75,76] or fructose content [77,78]. Short chain fatty acids (SCFA), such as butyrate, acetate and propionate, are produced during the fermentation of fibers by various bacteria including Bacteroides and Firmicutes [79]. SCFA has anti-inflammatory effects [7,79] and can act on both epithelial and immune cells [78] within the tumor microenvironment, with its effects on CRC still being studied. Microbial-driven metabolism of bile acids is important in the development of cancer [80]. Deoxycholic acid (DCA) produced by intestinal microbiota promotes CRC development by modulating signaling through muscarinic 3 and Wnt receptors pathways [81,82]. Bacterial metabolism can further affect and promote tumorigenesis through additional mechanisms including metabolic activation of pro-carcinogenic compounds, regulation of hormone metabolism; and modification of inflammatory pathways. Moreover, during therapy, microbiota can participate in metabolic dismantling of active therapeutic compounds, including in metastatic cancers [83–85], resulting in toxic secondary byproducts, lowering the tolerable dose and effectively weakening the therapy [86,87]. Since such metabolites or their secondary metabolites are absorbable and go into systemic circulation, they can affect many other cancers beyond the GI tract. For instance, the intestinal microbiome secretes bioactive metabolites (reactivated estrogens, short chain fatty acids, amino acid metabolites, or secondary bile acids) that modulate breast cancer [88].
Induction of pro-tumorigenic inflammation and bacteria-driven tissue repair.
Chronic inflammation promotes cancer progression and metastasis through a variety of mechanisms and microbes and their metabolites are one of the most prominent inducers of inflammation. If barrier function of transformed epithelium is weakened or if tumor or metastatic seed is permanently colonized with bacteria, chronic exposure to bacterial metabolites leads to chronic inflammation. Inflammation can then drive tumor initiation, growth, progression and metastasis via mechanisms related to regulation of cancer stem cells compartments, immunosuppression, induction of mutations, modulation of tumor microenvironment and stromal cells [89,90]. One of the most prominent mechanisms is the ability of bacteria to activate innate and adaptive immune cells within the TME via recognition through Toll-like receptor, NOD- like receptor and other PRR to stimulate the production of pro-inflammatory cytokines, which both regulate immunity and microenvironment , but also serve as a tissue protective and repair responses in epithelial and cancer cells [45,46]. Indeed, the presence of some of the cytokines (IL-17A, IL-6, IL-11) correlate with poor prognosis in CRC and other types of cancer [91]. Expression of these cytokines can be induced by intratumoral or barrier-seeding microbes. Alternatively, the microbiota can also induce specific types of inflammation, for example through the activation of Th17 cell differentiation in response to bacterial infected apoptotic innate immune cells [92].
Microbiota and inflammation can be also linked to the activation of stem cell/cancer stem cell compartments, which is essential for cancer growth , progression, metastasis and resistance to therapy [93,94]. The plasticity of cells in response to microbe- driven inflammation allows the transition of cells from one stage of differentiation to another, which in turn promote the epithelial–mesenchymal transition and metastasis [95,96]. Survival of metastasis can be regulated by microbiota and microbial products locally (e.g. in the liver, which is generally rich in microbe-derived signals) [97] or systemically, for example in the case of metastasis to the lung. Lipopolysaccharide (LPS) was shown to induce survival and outgrowth of cancer cells metastasizing to the lung by activation of inflammation-driven NF-kB signaling [98,99]. LPS also promoted metastatic spread by increasing integrin-mediated adhesion of cancer cells to the endothelial cells in the vessels [100].
Recent studies of microbiome, in particular associated with tumor development and therapy, only started to uncover the modalities and the mechanisms of host-microbe interactions during the disease progression and therapy. It is likely that further insights will come from cataloguing new and unique microbial metabolic functions followed by mechanistic experiments in animal models of cancer. These mechanisms will be further validated in patients to establish common functions and physiological denominators for the effects of microbes on cancer, particularly using existing wealthy collections of normal and cancer tissues Next, additional diagnostic, preventive or therapeutic approaches in cancer can be developed based on the presence of specific intra-tumoral or surface microbes, links between microbiota and diet, pro- and pre-biotic approaches, ability to introduce or deplete specific microbial species or their mutant versions, or approaches to bypass live microbiota function(s) with distinct set of bacteria-derived metabolites.
Table 1.
Summary: roles of microbiota in tumorigenesis and anti-cancer therapy
| Anti-tumorigenic | Pro-tumorigenic |
|---|---|
| «Beneficial» bacteria > «pathogenic» bacteria competition | Activation of Tumor elicit inflammation (TEI) |
| Tumor suppressive metabolites | Genotoxic effect on epithelial and stromal cells |
| Immune system maturation and tonic stimulation for effective cancer immunosurveillance | Metabolic effect via the metabolic activation of carcinogenic compounds |
| Immunostimulation and improvement of responses to chemo-, radio- and immunotherapies of cancer | Åctivation of cancer stem cell compartments |
| Activation of cell plasticity in the tumor microenvironment | |
| Inhibition of cytotoxic immune responses (immunosuppression) |
Highlights.
Microbiota is important in various aspects of host physiology including regulation of cancer development and progression
Microbiota can play negative or positive effects on tumorigenesis and efficacy of therapies
Microbiota regulates tumorigenesis via few distinct mechanisms including interactions with cancer cells and tumor microenvironment
Pro-tumorigenic microbiota stimulates tumor elicited inflammation, alters metabolism, hinder therapies and can promote genotoxicity
Anti-tumorigenic microbiota outcompetes «pathogenic» microbes, metabolically suppresses cancer, facilitates immunity and therapies.
Acknowledgements
We apologize to colleagues whose important original work could not be cited due to the citation limits. Figures were prepared using BioRender platform. The work was supported by Pew Scholar in Biomedical Sciences award, NIH R01CA227629 and CA218133, AACR-Landon Innovator Award and US-Israel BSF grant, as well as by NIH P30 CA-006927 to Fox Chase Cancer Center.
Footnotes
Declarations of Interest: none
References
- 1.Tran HQ, Ley RE, Gewirtz AT, Chassaing B: Flagellin-elicited adaptive immunity suppresses flagellated microbiota and vaccinates against chronic inflammatory diseases. Nat Commun 2019, 10:5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koh JH, Kim WU: Dysregulation of gut microbiota and chronic inflammatory disease: from epithelial defense to host immunity. Exp Mol Med 2017, 49:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chittim CL, Martinez Del Campo A, Balskus EP: Gut bacterial phospholipase Ds support disease-associated metabolism by generating choline. Nat Microbiol 2019, 4:155–163. [DOI] [PubMed] [Google Scholar]
- 4.Fatkhullina AR, Peshkova IO, Dzutsev A, Aghayev T, McCulloch JA, Thovarai V, Badger JH, Vats R, Sundd P, Tang HY, et al. : An Interleukin-23-Interleukin-22 Axis Regulates Intestinal Microbial Homeostasis to Protect from Diet-Induced Atherosclerosis. Immunity 2018, 49:943–957 e949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer EA, Knight R, Mazmanian SK, Cryan JF, Tillisch K: Gut microbes and the brain: paradigm shift in neuroscience. J Neurosci 2014, 34:15490–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA: The microbiome, cancer, and cancer therapy. Nat Med 2019, 25:377–388. [DOI] [PubMed] [Google Scholar]
- 7.Garrett WS: Cancer and the microbiota. Science 2015, 348:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Integrative HMPRNC: The Integrative Human Microbiome Project. Nature 2019, 569:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. : The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368:973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, et al. : Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Saeedi BJ, Liu KH, Owens JA, Hunter-Chang S, Camacho MC, Eboka RU, Chandrasekharan B, Baker NF, Darby TM, Robinson BS, et al. : Gut-Resident Lactobacilli Activate Hepatic Nrf2 and Protect Against Oxidative Liver Injury. Cell Metab 2020, 31:956–968 e955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zagato E, Pozzi C, Bertocchi A, Schioppa T, Saccheri F, Guglietta S, Fosso B, Melocchi L, Nizzoli G, Troisi J, et al. : Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol 2020, 5:511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zitvogel L, Daillere R, Roberti MP, Routy B, Kroemer G: Anticancer effects of the microbiome and its products. Nat Rev Microbiol 2017, 15:465–478. [DOI] [PubMed] [Google Scholar]
- 14.Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S, Becker J, Gonzalez A, McKinstry D, Hasso M, et al. : Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep 2020, 31:107471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan JL, Wu S, Geis AL, Chan GV, Gomes TAM, Beck SE, Wu X, Fan H, Tam AJ, Chung L, et al. : Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol 2019, 12:164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD: The gut microbiome modulates colon tumorigenesis. MBio 2013, 4:e00692–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW: Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/beta-Catenin Signaling via its FadA Adhesin. Cell host & microbe 2013, 14:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng M, Tabashsum Z, Patel P, Bernhardt C, Biswas D: Linoleic Acids Overproducing Lactobacillus casei Limits Growth, Survival, and Virulence of Salmonella Typhimurium and Enterohaemorrhagic Escherichia coli. Front Microbiol 2018, 9:2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litvak Y, Mon KKZ, Nguyen H, Chanthavixay G, Liou M, Velazquez EM, Kutter L, Alcantara MA, Byndloss MX, Tiffany CR, et al. : Commensal Enterobacteriaceae Protect against Salmonella Colonization through Oxygen Competition. Cell Host Microbe 2019, 25:128–139 e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Hegazy WA, Guo L, Gao X, Courtney AN, Kurbanov S, Liu D, Tian G, Manuel ER, Diamond DJ, et al. : Effective cancer vaccine platform based on attenuated salmonella and a type III secretion system. Cancer Res 2014, 74:6260–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. : Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504:446–450. [DOI] [PubMed] [Google Scholar]
- 22.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, et al. : The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50:432–445 e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galluzzi L, Vitale I, Senovilla L, Olaussen KA, Pinna G, Eisenberg T, Goubar A, Martins I, Michels J, Kratassiouk G, et al. : Prognostic impact of vitamin B6 metabolism in lung cancer. Cell Rep 2012, 2:257–269. [DOI] [PubMed] [Google Scholar]
- 24.Cardona F, Andres-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuno MI: Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem 2013, 24:1415–1422. [DOI] [PubMed] [Google Scholar]
- 25.Elinav E, Garrett WS, Trinchieri G, Wargo J: The cancer microbiome. Nat Rev Cancer 2019, 19:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy KD, Ronchi F, Geuking MB: Host-microbiota interactions and adaptive immunity. Immunol Rev 2017, 279:63–69. [DOI] [PubMed] [Google Scholar]
- 27.Nauts HC, Fowler GA, Bogatko FH: A review of the influence of bacterial infection and of bacterial products (Coley’s toxins) on malignant tumors in man; a critical analysis of 30 inoperable cases treated by Coley’s mixed toxins, in which diagnosis was confirmed by microscopic examination selected for special study. Acta Med Scand Suppl 1953, 144:1–103. [PubMed] [Google Scholar]
- 28.Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, Shen G, Hu S: Microbiota modulate tumoral immune surveillance in lung through a gammadeltaT17 immune cell-dependent mechanism. Cancer Res 2014, 74:4030–4041. [DOI] [PubMed] [Google Scholar]
- 29.Cai S, Kandasamy M, Rahmat JN, Tham SM, Bay BH, Lee YK, Mahendran R: Lactobacillus rhamnosus GG Activation of Dendritic Cells and Neutrophils Depends on the Dose and Time of Exposure. J Immunol Res 2016, 2016:7402760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marinelli L, Tenore GC, Novellino E: Probiotic species in the modulation of the anticancer immune response. Semin Cancer Biol 2017, 46:182–190. [DOI] [PubMed] [Google Scholar]
- 31.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. : Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. : The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342:971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. : Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350:1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. : Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. : Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fluckiger A, Daillere R, Sassi M, Sixt BS, Liu P, Loos F, Richard C, Rabu C, Alou MT, Goubet AG, et al. : Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 2020, 369:936–942. [DOI] [PubMed] [Google Scholar]
- 37.Grivennikov SI, Greten FR, Karin M: Immunity, inflammation, and cancer. Cell 2010, 140:883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakshani CR, Morales-Garcia AL, Althaus M, Wilcox MD, Pearson JP, Bythell JC, Burgess JG: Evolutionary conservation of the antimicrobial function of mucus: a first defence against infection. NPJ Biofilms Microbiomes 2018, 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy M, Blacher E, Elinav E: Microbiome, metabolites and host immunity. Curr Opin Microbiol 2017, 35:8–15. [DOI] [PubMed] [Google Scholar]
- 40.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. : A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167:1339–1353 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao Y, Wu K, Mehta R, Drew DA, Song M, Lochhead P, Nguyen LH, Izard J, Fuchs CS, Garrett WS, et al. : Long-term use of antibiotics and risk of colorectal adenoma. Gut 2018, 67:672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert AM, Sinani H, Schloss PD: Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. MBio 2015, 6:e00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nieuwenhuis EE, Blumberg RS: The role of the epithelial barrier in inflammatory bowel disease. Adv Exp Med Biol 2006, 579:108–116. [DOI] [PubMed] [Google Scholar]
- 44.Abraham C, Medzhitov R: Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140:1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. : Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491:254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greten FR, Grivennikov SI: Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tilg H, Adolph TE, Gerner RR, Moschen AR: The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33:954–964. [DOI] [PubMed] [Google Scholar]
- 48.Sears CL, Pardoll DM: The intestinal microbiome influences checkpoint blockade. Nat Med 2018, 24:254–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kostic AD, Howitt MR, Garrett WS: Exploring host-microbiota interactions in animal models and humans. Genes Dev 2013, 27:701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. : Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carra A, Brennan CA, Chun E, Ngo L, Samson LD, et al. : The human gut bacterial genotoxin colibactin alkylates DNA. Science 2019, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, et al. : The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015, 60:208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomkovich S, Dejea CM, Winglee K, Drewes JL, Chung L, Housseau F, Pope JL, Gauthier J, Sun X, Muhlbauer M, et al. : Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest 2019, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, et al. : Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359:592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, Polley EC, Bowman ED, Khan MA, Robles AI, et al. : Interaction between the microbiome and TP53 in human lung cancer. Genome Biol 2018, 19:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, Ameh S, Sandel D, Liang XS, Mazzilli S, et al. : Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 2019, 176:998–1013 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, Li Y, Yie TA, Lhakhang T, Olsen E, et al. : Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am J Respir Crit Care Med 2018, 198:1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McAllister F, Khan MAW, Helmink B, Wargo JA: The Tumor Microbiome in Pancreatic Cancer: Bacteria and Beyond. Cancer Cell 2019, 36:577–579. [DOI] [PubMed] [Google Scholar]
- 59.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, et al. : The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018, 8:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irrazabal T, Belcheva A, Girardin SE, Martin A, Philpott DJ: The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell 2014, 54:309–320. [DOI] [PubMed] [Google Scholar]
- 61.Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, Piazuelo MB, Correa P, Washington MK, El-Rifai W, et al. : Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology 2010, 139:1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dinges MM, Orwin PM, Schlievert PM: Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 2000, 13:16–34, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogan MR, Patterson LL, Wang JY, McBride JW: Bacterial Manipulation of Wnt Signaling: A Host-Pathogen Tug-of-Wnt. Front Immunol 2019, 10:2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. : Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338:120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, et al. : Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 2020, 580:269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chung L, Orberg ET, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, et al. : Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wahlstrom A, Sayin SI, Marschall HU, Backhed F: Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab 2016, 24:41–50. [DOI] [PubMed] [Google Scholar]
- 68.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F: Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 2013, 17:225–235. [DOI] [PubMed] [Google Scholar]
- 69.Irrazabal T, Thakur BK, Kang M, Malaise Y, Streutker C, Wong EOY, Copeland J, Gryfe R, Guttman DS, Navarre WW, et al. : Limiting oxidative DNA damage reduces microbe-induced colitis-associated colorectal cancer. Nat Commun 2020, 11:1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curtius K, Wright NA, Graham TA: An evolutionary perspective on field cancerization. Nat Rev Cancer 2018, 18:19–32. [DOI] [PubMed] [Google Scholar]
- 71.Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, Rickman BH, Rogers AB, Moroski-Erkul CA, McFaline JL, et al. : DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J Clin Invest 2008, 118:2516–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schulz MD, Atay C, Heringer J, Romrig FK, Schwitalla S, Aydin B, Ziegler PK, Varga J, Reindl W, Pommerenke C, et al. : High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 2014, 514:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mihaylova MM, Cheng CW, Cao AQ, Tripathi S, Mana MD, Bauer-Rowe KE, Abu-Remaileh M, Clavain L, Erdemir A, Lewis CA, et al. : Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell Stem Cell 2018, 22:769–778 e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. : High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016, 531:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK: Specialized metabolites from the microbiome in health and disease. Cell Metab 2014, 20:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Araujo JR, Tazi A, Burlen-Defranoux O, Vichier-Guerre S, Nigro G, Licandro H, Demignot S, Sansonetti PJ: Fermentation Products of Commensal Bacteria Alter Enterocyte Lipid Metabolism. Cell Host Microbe 2020, 27:358–375 e357. [DOI] [PubMed] [Google Scholar]
- 77.Goncalves MD, Lu C, Tutnauer J, Hartman TE, Hwang SK, Murphy CJ, Pauli C, Morris R, Taylor S, Bosch K, et al. : High-fructose corn syrup enhances intestinal tumor growth in mice. Science 2019, 363:1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bu P, Chen KY, Xiang K, Johnson C, Crown SB, Rakhilin N, Ai Y, Wang L, Xi R, Astapova I, et al. : Aldolase B-Mediated Fructose Metabolism Drives Metabolic Reprogramming of Colon Cancer Liver Metastasis. Cell Metab 2018, 27:1249–1262 e1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F: From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165:1332–1345. [DOI] [PubMed] [Google Scholar]
- 80.Ocvirk S, O’Keefe SJ: Influence of Bile Acids on Colorectal Cancer Risk: Potential Mechanisms Mediated by Diet - Gut Microbiota Interactions. Curr Nutr Rep 2017, 6:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, Chrysovergis K, Wade PA: An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol 2018, 19:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zou S, Fang L, Lee MH: Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol Rep (Oxf) 2018, 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stojanovska V, McQuade RM, Fraser S, Prakash M, Gondalia S, Stavely R, Palombo E, Apostolopoulos V, Sakkal S, Nurgali K: Oxaliplatin-induced changes in microbiota, TLR4+ cells and enhanced HMGB1 expression in the murine colon. PLoS One 2018, 13:e0198359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chiang NJ, Tsai KK, Hsiao CF, Yang SH, Hsiao HH, Shen WC, Hsu C, Lin YL, Chen JS, Shan YS, et al. : A multicenter, phase I/II trial of biweekly S-1, leucovorin, oxaliplatin and gemcitabine in metastatic pancreatic adenocarcinoma-TCOG T1211 study. Eur J Cancer 2020, 124:123–130. [DOI] [PubMed] [Google Scholar]
- 85.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. : Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koppel N, Maini Rekdal V, Balskus EP: Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hitchings R, Kelly L: Predicting and Understanding the Human Microbiome’s Impact on Pharmacology. Trends Pharmacol Sci 2019, 40:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miko E, Kovacs T, Sebo E, Toth J, Csonka T, Ujlaki G, Sipos A, Szabo J, Mehes G, Bai P: Microbiome-Microbial Metabolome-Cancer Cell Interactions in Breast Cancer-Familiar, but Unexplored. Cells 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Canli O, Nicolas AM, Gupta J, Finkelmeier F, Goncharova O, Pesic M, Neumann T, Horst D, Lower M, Sahin U, et al. : Myeloid Cell-Derived Reactive Oxygen Species Induce Epithelial Mutagenesis. Cancer Cell 2017, 32:869–883 e865. [DOI] [PubMed] [Google Scholar]
- 90.Robles AI, Traverso G, Zhang M, Roberts NJ, Khan MA, Joseph C, Lauwers GY, Selaru FM, Popoli M, Pittman ME, et al. : Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology 2016, 150:931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, Church SE, Lafontaine L, Fischer M, Fredriksen T, et al. : Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44:698–711. [DOI] [PubMed] [Google Scholar]
- 92.Torchinsky MB, Garaude J, Martin AP, Blander JM: Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature 2009, 458:78–82. [DOI] [PubMed] [Google Scholar]
- 93.Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Munoz P, et al. : The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8:511–524. [DOI] [PubMed] [Google Scholar]
- 94.de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, et al. : A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 2017, 543:676–680. [DOI] [PubMed] [Google Scholar]
- 95.Blanpain C, Fuchs E: Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 2014, 344:1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oskarsson T, Batlle E, Massague J: Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell 2014, 14:306–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, et al. : Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Luo JL, Maeda S, Hsu LC, Yagita H, Karin M: Inhibition of NF-kappaB in cancer cells converts inflammation-induced tumor growth mediated by TNFalpha to TRAIL-mediated tumor regression. Cancer Cell 2004, 6:297–305. [DOI] [PubMed] [Google Scholar]
- 99.Cai Z, Tchou-Wong KM, Rom WN: NF-kappaB in lung tumorigenesis. Cancers (Basel) 2011, 3:4258–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsu RY, Chan CH, Spicer JD, Rousseau MC, Giannias B, Rousseau S, Ferri LE: LPS-induced TLR4 signaling in human colorectal cancer cells increases beta1 integrin-mediated cell adhesion and liver metastasis. Cancer Res 2011, 71:1989–1998. [DOI] [PubMed] [Google Scholar]


