Abstract
Background:
Pancreatic ductal adenocarcinoma (PDAC) has the worst prognosis among common cancers. The genomic landscape of PDAC is defined by four mutational pathways: kirsten rat sarcoma virus (KRAS), cellular tumor antigen p53 (TP53), cyclin dependent kinase inhibitor 2A (CDKN2A), and SMAD family member 4 (SMAD4). However, there is a paucity of data on the molecular features associated with clinical outcomes after surgery or chemotherapy.
Methods:
We performed comprehensive molecular characterization of tumor specimens from 83 patients with PDAC who received surgery, using whole-exome sequencing and ribonucleic acid sequencing on tumor and matched normal tissues derived from patients. We also systematically performed integrative analysis, combining genomic, transcriptomic, and clinical features to identify biomarkers and possible therapeutic targets.
Results:
KRAS (75%), TP53 (67%), CDKN2A (12%), SMAD4 (20%), and ring finger protein 43 (RNF43) (13%) were identified as significantly mutated genes. The tumor-specific transcriptome was classified into two clusters (tumor S1 and tumor S2), which resembled the Moffitt tumor classification. Tumor S1 displayed two distinct subclusters (S1-1 and S1-2). The transcriptome of tumor S1-1 overlapped with the exocrine-like (Collisson)/ADEX (Bailey) subtype, while tumor S1-2 mostly consisted of the classical (Collisson)/progenitor (Bailey) subtype. In the analysis of combinatorial gene alterations, concomitant mutations of KRAS with low-density lipoprotein receptor related protein 1B (LRP1B) were associated with significantly worse disease-free survival after surgery (p = 0.034). One patient (1.2%) was an ultrahypermutant with microsatellite instability. We also identified high protein kinase C lota (PRKCI) expression as an overlapping, poor prognostic marker between our dataset and the TCGA dataset.
Conclusion:
We identified potential prognostic biomarkers and therapeutic targets of patients with PDAC. Understanding these molecular aberrations that determine patient outcomes after surgery and chemotherapy has the potential to improve the treatment outcomes of PDAC patients.
Keywords: KRAS, landscape, LRP1B, pancreatic cancer, PRKCI
Introduction
Pancreatic ductal adenocarcinoma (PDAC) has the worst prognosis among common cancers.1,2 The 5-year survival rate of PDAC is approximately 9%,1,2 despite the recent success with combination chemotherapies and targeted therapy.3–6 The incidence, mortality, and disease burden of PDAC are all rapidly increasing worldwide;1,7,8 PDAC is predicted to become the second leading cause of cancer-related death in the United States by 2030.1,7
The Australian Pancreatic Cancer Genome Initiative (APGI) and the Cancer Genome Atlas (TCGA) Research Network have provided the mutational landscape of PDAC.9,10 The genomic landscape of PDAC is defined by four mutational pathways that are essential drivers of carcinogenesis; kirsten rat sarcoma virus (KRAS), cellular tumor antigen p53 (TP53), cyclin dependent kinase inhibitor 2A (CDKN2A), and SMAD family member 4 (SMAD4). Along with these four major drivers, the additional frequently mutated genes including ring finger protein 43 (RNF43) and AT-rich interactive domain-containing protein 1A (ARID1A) have been identified in the previous systematic genomic profiling of PDAC. The key mutation in PDAC is activated KRAS, which occurs early in carcinogenesis, whereas mutations in SMAD4 occur later and are thought to facilitate the progression of PDAC.9,10 Germline and somatic mutations in the DNA damage repair (DDR) genes, BRCA2, partner and localizer of BRCA2 (PALB2), and ATM, were observed in a minority of cases, representing a class of patients for whom platinum-based chemotherapy and/or poly(ADP-ribose) polymerase (PARP) inhibition may have therapeutic benefit.9,10 Several groups have described transcriptomic subtypes in PDAC, with mostly consistent findings concerning two major lineages that separate PDAC into the squamous (alternatively named basal-like and quasi-mesenchymal) and classical subtypes.11,12
However, there is a paucity of data on the molecular features associated with clinical outcomes after curative surgery and previous studies do not provide any correlative information with respect to chemotherapy response. In the current study, we performed integrative genomics analyses of PDAC patients who received surgery and/or chemotherapy to facilitate identification of biomarkers and possible therapeutic targets.
Materials and methods
Patient enrollment
Among the patients with PDAC who received curative surgery at Samsung Medical Center, Seoul, South Korea, between September 2008 and June 2017, 83 patients with available samples were included in this study. The molecular analysis included whole-exome sequencing and ribonucleic acid (RNA) sequencing on tumor and matched normal tissues derived from patients. Clinical information including age, sex, primary tumor site, histologic type, stage, and treatment outcomes was extracted from hospital records. We also evaluated the outcomes of palliative chemotherapy. Computed tomography scan and/or magnetic resonance imaging were performed every two cycles to evaluate the chemotherapy response according to RECIST 1.1 criteria.13
The collection of specimens and associated clinical data used in this study was approved by the Institutional Review Board of Samsung Medical Center (IRB #2015-10-062 and #2018-03-162). All patients who participated in this study provided written informed consent prior to enrollment and specimen collection. This study was performed in accordance with the principles of the Helsinki Declaration and the Korean Good Clinical Practice guidelines.
Tumor sample collection
Tumor tissues were obtained during surgery. If the tumor content was ⩾40% after pathological assessment, tumor deoxyribonucleic acid (DNA), and RNA were extracted from freshly acquired tissues using a QIAamp Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. For DNA, we used RNase A (cat. #19101; Qiagen). We determined the concentrations and absorbance ratios (OD260/OD280 and OD260/OD230) with an ND1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, MA, USA) and quantified DNA/RNA using a Qubit fluorometer (Life Technologies, CA, USA).
Whole-exome sequencing for tumor tissue
For the generation of standard exome capture libraries, we used the Agilent SureSelect Target Enrichment protocol for Illumina paired-end sequencing library, version B.3, June 2015 (Agilent Technologies, Inc. Santa Clara, CA, USA) with 200 ng of input formaldehyde-fixed, paraffin-embedded DNA. In all cases, the SureSelect Human All Exon V5 probe set was employed. DNA quantity and quality were evaluated using PicoGreen14 and NanoDrop.15 Fragmentation of 1 ng of Inc., MA, USA‘’′ end, and Agilent adapters were ligated to the fragments. Once ligation had been assessed, the adapter-ligated product was amplified by polymerase chain reaction (PCR). Subsequently, the final purified product was quantified using quantitative PCR (qPCR) under the qPCR Quantification Protocol Guide and assessed using the Caliper High Sensitivity DNA LabChip Kit (PerkinElmer Inc., MA, USA). For exome capture, 250 ng of DNA library was mixed with hybridization buffers, blocking mixes, RNase block, and 5 µl of SureSelect All Exon capture library, according to the standard Agilent SureSelect Target Enrichment protocol. Hybridization to capture baits was performed at 65°C using the heated lid option of a thermocycler at 105°C for 24 h on a PCR machine. The captured DNA was amplified. The final purified product was quantified using qPCR according to the qPCR Quantification Protocol Guide and assessed using the TapeStation RNA ScreenTape (Agilent). Finally, we performed sequencing using the HiSeq™ 2500 platform (Illumina, San Diego, CA, USA).
Whole-exome sequencing data analysis
The resulting reads from whole-exome sequencing were mapped to human genome version 19 (hg19) using the Burrows-Wheeler Aligner (version 0.7.12-r1039) with the BWA-MEM algorithm.16 SAMtools sorted the aligned sequences by genomic coordinates (v0.1.19).17 The sorted reads were subjected to the Genome Analysis Toolkit (GATK, v3.6 and v.4.13) for duplicate marking, indel realignment, and base recalibration.18 We performed MuTect2 from GATKv4.13 to detect tumor somatic mutations using the recalibrated BAM files from tumor and matched normal samples. After removing possible germline events (population allele fraction <2.5e−6 in gnomAD), we annotated the called mutations using variant effect predictors (VEP).19 The resulting variant events were converted to MAF format, and then the variants with altered reads <4 were eliminated for further analysis in R. Using MSIsensor, we calculated the microsatellite instability (MSI) score of each tumor by comparing the BAM file of the tumor sample with that of the matched normal sample. We called the tumor ‘MSI’ if the MSI score of the tumor was >3.5.20 Otherwise, the tumors were classified as ‘microsatellite stable (MSS)’. The COSMIC v3 single base substitution (SBS) mutational signatures of each tumor sample were determined by deconstructSigs (v1.8.0, R package).21,22 Significantly mutated genes were identified by MutSigCV (v1.41)23 on MATLAB. To estimate copy number alterations, CNVkit (v0.9.7.b1)24 was used for copy segmentation, followed by GISTIC2.025 to calculate copy number alterations for each gene with log2 threshold = 1.
RNA sequencing
Total RNA concentration was estimated using Quant-IT RiboGreen (Invitrogen, Waltham, MA, USA). To determine the DV200 (% of RNA fragments >200 bp) value, samples were run on the TapeStation RNA ScreenTape (Agilent). Overall, 100 ng of total RNA was subjected to sequencing library construction using a TruSeq RNA Access Library Prep Kit (Illumina) according to the manufacturer’s protocol. Briefly, total RNA was first fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand complementary DNA (cDNA) using SuperScript II reverse transcriptase (Invitrogen, #18064014) and random primers. This was followed by second-strand cDNA synthesis using DNA polymerase I, RNase H, and deoxyuridine 5-triphosphate. The cDNA fragments were subjected to an end-repair process, addition of a single ‘A’ base, and subsequently, ligation of the adapters. Thereafter, the products were purified and enriched with PCR to create the cDNA library. All libraries were normalized, and six were pooled into a single hybridization/capture reaction. Pooled libraries were incubated with a cocktail of biotinylated oligos, corresponding to the coding regions of the genome. Targeted library molecules were captured via hybridized biotinylated oligo probes using streptavidin-conjugated beads. After two rounds of hybridization/capture reactions, the enriched library molecules were subjected to a second round of PCR amplification. The captured libraries were quantified using a KAPA Library Quantification Kit for Illumina Sequencing platforms according to the qPCR Quantification Protocol Guide (KAPA BIOSYSTEMS, #KK4854) and assessed using the TapeStation D1000 ScreenTape (Agilent Technologies, # 5067-5582). Indexed libraries were subsequently submitted to an Illumina HiSeq 2500 platform (Illumina), and paired-end (2 × 100 bp) sequencing was performed by Macrogen Inc. (Seoul, South Korea).
Gene expression calling and analysis
RNA sequence reads were mapped on hg19 by STAR (v2.6.1d)26 and sorted according to genomic coordinates. Cufflinks (v2.2.1) was used to calculate gene expression levels in fragments per kilobase million (FPKM)27 using Ensembl gene annotation. The FPKM values were log2-transformed for further analysis. Based on gene expression profiling of tumor samples, to assign predefined tumor subtypes,11,28–30 nearest template prediction was applied using gene markers of each subtype in R. To identify the differentially expressed genes in tumor samples compared with their matched normal samples, we performed a paired t test and selected genes with adjusted p-values < 0.05 and log2 fold-change >1. As a result, 5012 genes remained that we defined as ‘tumor-specific’ genes. Among these, highly variable genes were selected with a standard deviation (SD) >1 for unsupervised hierarchical clustering (HC). To assume the expression scores of tumor microenvironment-associated cell markers, we used MCPcounter in R.31 Tumor purities based on gene expression profiles were calculated by R package ESTIMATE.32 Expression scores of gene sets were estimated using a single sample gene set enrichment analysis (ssGSEA) algorithm in GSVA (R package).33 GSEA was performed on GSEA-P.34 Gene ontology analysis was performed on DAVID (https://david.ncifcrf.gov/).35,36 Differentially expressed genes (log2 fold change >1 and false discovery rate <0.05) among HC-based tumor groups were identified through DESeq2 in R.37
TCGA PAAD data download
Processed gene expression profiles (in FPKM) and clinical information on pancreatic adenocarcinoma (PAAD) from the TCGA were downloaded through GDC using the R-TCGAbiolinks package.10,38
Survival analysis
All survival analysis was conducted on the R survival package. Disease-free survival (DFS) was defined as the time between surgery and disease recurrence while overall survival (OS) was defined as the time between surgery and death. We performed univariate Cox regression for OS and DFS analysis using age, sex, tumor location, differentiation, adjuvant chemotherapy and radiation therapy, carbohydrate antigen 19-9 (CA19-9) level, and stage. Multivariate Cox regression survival analysis was also conducted with the risk factors (univariate Cox regression analysis p-values < 0.05). To identify tumor-specific gene expression-based poor prognostic factors, univariate Cox regression OS analysis was applied to both the PDAC cohort and to the TCGA PAAD cohort using their gene expression profiles (summarized in log2-transformed FPKM). Genes with a hazard ratio (HR) > 1 and p-values < 0.05 were left and used for multivariate Cox regression analysis with risk clinical factors (i.e., tumor differentiation grade and stage). To reveal gene mutations associated with OS and DFS, we used the R package, maftools.39 The association between genomic alterations and responses to chemotherapy was calculated via Fisher’s exact test in R.
Results
Patient characteristics
The baseline characteristics of the study participants are summarized in Table 1. All 83 patients were pathologically confirmed as PDAC and received curative surgery. Fifty-two patients (62.7%) were male and the median age was 65 years (range, 37–82). There were 26 patients (31.3%) with stage I, 36 (43.3%) with stage II, and 17 (20.5%) with stage III/IV at diagnosis. The most common tumor location was pancreas head (49.4%). CA19-9 levels were higher than the upper limit of normal (ULN, 37 U/ml) in 65 patients (78.3%) at diagnosis.
Table 1.
Baseline characteristics.
| Characteristics | Total (N = 83) |
|---|---|
| Age, years | |
| Median | 65 |
| Range | 37–82 |
| Sex, no. (%) | |
| Male | 52 (62.7) |
| Female | 31 (37.3) |
| Pancreas tumor location, no. (%) | |
| Head | 41 (49.4) |
| Body | 24 (28.9) |
| Tail | 18 (21.7) |
| Differentiation, no. (%) | |
| Well | 6 (7.2) |
| Well to moderately | 1 (1.2) |
| Moderately | 49 (59.3) |
| Moderately to poorly | 5 (6.0) |
| Poorly | 22 (26.5) |
| Stage at diagnosis, no. (%) | |
| IA | 8 (9.6) |
| IB | 18 (21.7) |
| IIA | 5 (6.0) |
| IIB | 31 (37.3) |
| III | 15 (18.1) |
| IV | 2 (2.4) |
| Level of CA19-9 at diagnosis, no. (%) | |
| Normal | 18 (21.7) |
| ULN to ⩽59 × ULN | 60 (72.3) |
| >59 × ULN | 5 (6.0) |
| Adjuvant treatment (N = 42), no. (%) | |
| Chemoradiation | 31 (73.8) |
| Chemotherapy | 10 (12.0) |
| Radiation | 1 (2.4) |
| Palliative first-line chemotherapy (N = 29), no. (%) | |
| Gemcitabine monotherapy | 20 (70.0) |
| Gemcitabine-based combination | 5 (17.2) |
| FOLFIRINOX | 3 (10.3) |
| TS-1/cisplatin | 1 (1.2) |
CA19-9, carbohydrate antigen 19-9; FOLFIRINOX, folinic acid/fluorouracil/irinotecan/oxaliplatin; TS-1, tegafur/gimeracil/oteracil; ULN, upper limit of normal.
We evaluated the clinicopathological factors that are associated DFS and OS. As expected, poorer differentiation and higher stage of cancer resulted in worse prognosis. In addition, a CA19-9 level higher than the ULN was significantly associated with short DFS in univariate analysis (Supplemental Tables 1 and 2).
Genomic landscape of 83 PDAC patients
To characterize genomic and transcriptomic profiles of the 83 PDAC patients, we obtained tumor and matched normal pancreas tissues with blood samples from each PDAC patient. The collected samples were subjected to whole-exome sequencing and RNA sequencing. Whole-exome sequencing data derived from tumor and blood samples were used to estimate somatic mutations and copy number variations.
In the PDAC cohort, the median number of somatic nonsilent coding mutations was 45 (7–10028), which is comparable with previous findings10 [Supplemental Figure 1(a) and (b)]. In addition, including four major drivers in PDAC, five significantly mutated genes (SMGs) were identified; KRAS (75%, q = 4.19e−12), TP53 (67%, q = 1.05e−11), CDKN2A (12%, q = 9.07e−12), SMAD4 (20%, q = 9.07e−12), and RNF43 (13%, q = 1.01e−06), as was also previously reported (Figure 1).10
Figure 1.
Genomic and molecular characteristics of 83 PDAC patients. From top to bottom, shown are the number of mutations per tumor detected by WES (the y-axis indicates the number of mutations in log10 scale), mutational signatures of the COSMIC v3 SBS, treated chemotherapy information and responses to the therapy, clinicopathological features, MSI status, transcriptome subtypes, and genomic alterations in major oncogenic pathways of PDAC. SMGs are indicated by red asterisks.
APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; dMMR–MSI, deficient mismatch repair–microsatellite instability; DSBR, double-strand break repair; gemcitabine-combi, gemcitabine-based combination treatment; germline-NA, not available for MSI status due to lack of normal WES data; MD, moderately differentiated; MSI, microsatellite instable; MSS, microsatellite stable; NE, not evaluable; PDAC, pancreatic ductal adenocarcinoma; PD, progressive disease; PD, poorly differentiated; PR, partial response; S-1/CDDP, TS-1/cisplatin treatment; SBS, single base substitution; SD, stable disease; SMGs, significantly mutated genes; WD, well differentiated; WES, whole-exome sequencing.
Activating KRAS mutations are illustrated in Supplemental Figure 2(a). Multiple oncogenic KRAS alleles were identified, including G12D (n = 23, 27.7%), followed by G12V (n = 22, 26.5%) and G12R (n = 10, 12.0%), as well as another hotspot at codon 12, and 61 mutant alleles at a lower prevalence. KRAS mutations affecting two separate codons were found in one patient (KRASG12D and KRASD92V). Patients who had KRAS mutations showed no significant difference in clinical outcomes, in terms of DFS and response to palliative chemotherapy, compared with patients with KRAS wild-type tumors. However, in the analysis of combinatorial gene alterations, concomitant mutations of KRAS with LRP1B, which is known as a functional tumor suppressor gene, were associated with significantly worse DFS (p = 0.034) [Supplemental Figure 2(b)].
Germline and/or somatic mutations in the DDR system were found in nine patients (10.8%), showing mutations in serine/threonine kinase 11 (STK11) (4.8%), ATM (3.6%), MutL homolog 1 (MLH1) (3.6%), BRCA2 (2.4%), BRCA1 (1.2%), and PALB1 (1.2%). It should be noted that one tumor (#74) was ultrahypermutant with MSI. The mutational signature of this sample showed that 72% of mutations were attributed to a DNA mismatch repair (dMMR)–MSI mechanism with a high frequency of C–T transition [Figure 1 and Supplemental Figure 3(a)]. When we examined its gene expression profiles using RNA sequencing data, this MSI tumor showed high expression scores for immune cell markers, particularly in cytotoxic lymphocytes, natural killer (NK) cells, and monocytic lineage cells. Overexpression of PDCD1 and CD274 was also detected [Supplemental Figure 3(b) and (c)].
Tumor-specific and normal tissue transcriptomic classification of PDAC
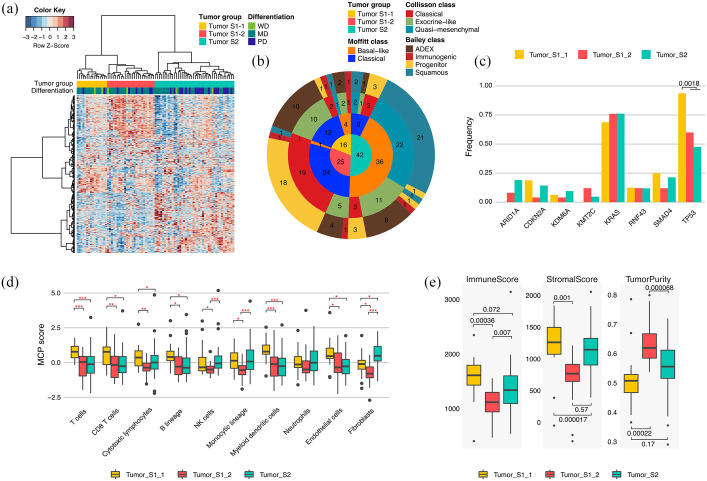
Because molecular characterization of PDAC has been limited by the high level of stromal cell involvement, we conducted tumor-specific and normal tissue transcriptomic analysis. Through differentially expressed gene (DEG) analysis between tumors and their matched normal tissues, we identified 5012 and 413 tumor-specific and normal genes, respectively (absolute log2-fold change >1, adjusted paired t test p-value < 0.05). According to previous studies, PDACs can be classified into several transcriptomic subtypes.11,28,29 To identify tumor-specific and normal subgroups in this cohort and compare them with the previously identified subtypes, we performed unsupervised HC using highly variable tumor-specific and normal genes (standard deviation > 1) [Figure 2(a) and Supplemental Figure 4(a)]. Tumor-specific gene expressing profiling was classified into two clusters (tumor S1 and tumor S2), which resembled the Moffitt tumor classification, and were also generated by tumor-specific genes [Figure 2(a) and (b)]. Tumor S1 displayed two distinct subclusters (S1-1 and S1-2). The transcriptome of tumor S1-1 overlapped with the exocrine-like (Collisson)/ADEX (Bailey) subtype while tumor S1-2 mostly consisted of the classical (Collisson)/progenitor (Bailey) subtype. On the other hand, 88% of basal-like, 96% of quasi-mesenchymal, and 92% of squamous subtypes were clustered as tumor S2 [Figure 2(b)]. We could not find any survival differences between the tumor-specific HC-based PDAC groups, although tumors with poor differentiation were significantly frequent in tumor S2 (p = 0.36, OR = 2.99, 19/27 poorly differentiated versus 23/56 well-differentiated and moderately differentiated) [Figure 2(a) and Supplemental Figure 4(c)]. In addition, we investigated the association between genomic alterations and tumor groups. We found that TP53 mutations occurred with significantly greater frequency in tumor S1-1 than in other groups (p = 0.0018, OR = 13.4) [Figure 2(c)].
Figure 2.
Tumor-specific transcriptomic subgroups in PDAC. (a) Unsupervised HC of 83 PDAC tumor tissues based on gene expression profiling; 399 tumor-specific genes with SD > 1 were used in this clustering. Bars at the top of a heat map represent HC-based tumor groups (top) and tumor differentiation grade (bottom). (b) Pie chart showing the number of samples in HC-based PDAC groups (inner circle), Moffitt subtypes (second inner ring), Collisson subtypes (third inner ring), and Bailey subtypes (outer ring). (c) Alteration frequency of each HC-based tumor group for genes altered in >4 PDAC patients (p-value: Fisher’s exact test). (d) Z-normalized MCPcounter scores among HC-based tumor groups. Z-normalization was performed across the tumor samples for each cell marker in the tumor microenvironment. Statistically significant differences in scores among the tumor groups are indicated by red asterisks (*p < 0.05, **p < 0.01, ***p < 0.001, Student’s t test). (e) Immune scores (left panel), stromal scores (middle panel), and tumor purities (right panel) in HC-based tumor groups (p-value: Student’s t test). ESTIMATE calculated the scores and tumor purities based on gene expression profiling.
HC, hierarchical clustering; MD, moderately differentiated; PDAC, pancreatic ductal adenocarcinoma; PD, poorly differentiated; SD, standard deviation; WD, well differentiated.
To dissect the cellular composition of the tumor microenvironment (TME) according to HC-based tumor group, we applied ESTIMATE and MCPcounter algorithms [Figure 3(d) and (e)]. With lower tumor purities, higher scores for immune cell and endothelial markers were found in tumor S1-1 than in other types. By comparison, markers for fibroblasts were upregulated in tumor S2, which showed high mesenchymal traits. ESTIMATE and MCPcounter results demonstrated the low infiltration levels of TME in tumor S1-2. Collectively, we identified three clusters using tumor-specific genes: tumor S1-1 was characterized by high stromal/immune cell infiltration and high frequency of TP53 mutations; tumor S1-2 displayed high tumor purities, resembling the conventional classical subtype, and; tumor S2, which is similar to the basal-like/quasi-mesenchymal/squamous subtype, showed increased expression levels for fibroblast markers and enrichment of tumors with poor differentiation.
Figure 3.
Identification of PRKCI as a poor survival predictor in PDAC. (a) Workflow for identifying poor prognostic markers in both SMC (Samsung Medical Center, this study cohort) and the TCGA PAAD cohort. (b–d) Kaplan–Meier curves showing the overall (b) and disease-free (c) survival differences between patients with relatively high and relatively low PRKCI expression levels in the SMC (b, c) and the TCGA (d) dataset. (e, f) PRKCI expression levels in tumor and normal tissues in the SMC (e) (p-value, paired t test) and the TCGA (f) dataset (p-value, unpaired t test).
DEG, differentially expressed genes; FC, log2-fold change; FDR, false discovery rate; HZ, hazard ratio; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; PRKC1, protein kinase C iota type.
Unsupervised HC with normal genes also revealed two clusters depending on the expression levels of overall normal genes [Supplemental Figure 4(a)]. The first cluster (normal S1) showed an absence of expression of normal genes, but tumors belonging to the other cluster (normal S2) expressed normal genes. Gene ontology analysis revealed that expressed normal genes in normal S2 were involved in proteolysis and digestion [Supplemental Figure 4(b)]. In addition, 13 of 16 tumor S1-1 samples were classified as normal S2, consistent with TME analysis. Tumor S1-1 showed transcriptomic similarity with the exocrine-like or ADEX subtype; it is controversial whether the exocrine-like/ADEX subtype is a result of normal tissue contamination or is a bona fide tumor subtype. Although tumor S1-1 had low tumor cellularity, there were PDAC tumor-specific genes present such as PROM1 (a well-known cancer stem cell marker40), the expression levels of which were significantly upregulated in tumor S1-1 (Supplemental Table 3). This suggests that tumor S1-1 is one of the PDAC tumor types rather than the result of simple contamination by normal tissue.
Identification of poor prognosis-related genes
As PDAC remains one of the most fatal malignancies and displays low tumor purity, the identification of tumor-specific prognostic markers and therapeutic targets is important for PDAC patient treatment. To discover tumor-specific genes that predict prognosis of PDAC, we performed Cox regression survival analysis using gene expression levels [Figure 3(a)]. Univariate Cox regression analysis with OS was applied to the 5012 tumor-specific genes. This analysis revealed 164 genes that met the criteria (HR > 1, p-value < 0.05). Of these 164 genes, 82 were identified as worse prognosis-related genes via multivariate Cox regression analysis with the prognostic factors (tumor stage and differentiation). The same procedure was performed with TCGA PAAD data to identify the prognostic genes that can be validated in an independent dataset. As a result, only PRKCI overlapped with the TCGA PAAD dataset. PDAC patients with higher expression levels of PRKCI showed worse OS and worse PFS [Figure 3(b)–(d)]. A significant upregulation of PRKCI in tumors compared with normal controls was confirmed [Figure 3(e) and (f)].
Response to palliative first-line chemotherapy
Among 83 patients, 29 received palliative first-line chemotherapy due to disease progression [Figure 4(a)]. Twenty patients were treated with gemcitabine monotherapy and five were treated with gemcitabine-based combination therapy (2, gemcitabine/nab-paclitaxel; 2, gemcitabine/erlotinib; 1, gemcitabine/capecitabine). FOLFIRINOX treatment was performed for another three patients. As a result, one, 16, and 10 patients achieved partial response (PR), stable disease (SD), and progressive disease (PD), respectively, and two patients were not evaluable [Figure 4(b)]. The overall clinical benefit ratio (PR + SD) was 58.6% [95% confidence interval (CI), 3.3–30.7].
Figure 4.
Genomic and molecular correlates of clinical benefit of chemotherapy. (a) Swimmer plots illustrating overall survival from surgery for patients whose clinical response to palliative first-line chemotherapy was available (top panel) and patients who did not receive palliative first-line chemotherapy (bottom panel). (b) Genomic and molecular characteristics of 29 PDAC patients who received chemotherapy. From top to bottom, progression-free survival in months, tumor mutation burden (log10 scale), mutational signatures of the COSMIC v3 SBS, treated chemotherapy information and responses to the therapy, clinicopathological features, and genomic alterations in major oncogenic pathways of PDAC. (c) GSEA plot showing the upregulation of the KEGG_ABC_TRANSPORTERS gene set in tumors derived from patients who achieved PD to chemotherapy compared with patients with clinical benefit of chemotherapy. (d) ssGSEA scores of the KEGG_ABC_TRANSPORTERS gene set were significantly higher in PD than in PR/SD patients (p-value, Wilcoxon rank sum test).
APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; dMMR–MSI, deficient mismatch repair–microsatellite instability; DSBR, double-strand break repair; gemcitabine-combi, gemcitabine-based combination treatment; GSEA, gene set enrichment analysis; MD, moderately differentiated; NE, not evaluable; PDAC, pancreatic ductal adenocarcinoma; PD, progressive disease; PD, poorly differentiated; PR, partial response; S-1/CDDP, TS-1/cisplatin treatment; SBS, single base substitution; SD, stable disease; ssGSEA, single sample GSEA; WD, well differentiated.
When we examined genomic correlates of clinical benefit of chemotherapy, none of the genomic alterations of core pathways were associated with clinical benefit of chemotherapy [Figure 4(b)]. Therefore, we conducted GSEA to explore whether there are distinct molecular gene sets among patients with clinical benefit of chemotherapy. We found ATP-binding cassette (ABC) transporter-associated genes were upregulated in patients who did not have clinical benefit of chemotherapy [Figure 4(c) and (d)].
Discussion
Using whole-exome and transcriptome sequencing, we performed integrative molecular characterization of 83 patients with PDAC receiving surgery and/or chemotherapy. The genomic landscape of PDAC, including four major driving mutations (KRAS, TP53, CDKN2A, and SMAD4), was comparable with the previous APGI and TCGA datasets9,10 and we also found that a substantial proportion of patients harbored germline and somatic mutations in their DDR systems. In addition, we identified factors that affect clinical outcomes, such as concomitant mutational status of KRAS and LRP1B, the expression level of PRKCI, and the overexpression of ABC transporter proteins.
KRAS, which plays an essential role in the signaling pathways that regulate cell growth and differentiation, is the most frequently mutated oncogene in PDAC. However, the association between KRAS mutational status and clinical outcome after surgery has not been clearly established. One recent study showed that KRAS G12D-mutant tumors were associated with worse DFS after surgery, compared with KRAS wild-type tumors.41 That study also showed that multiple alterations (⩾3) among the four major driver mutations (KRAS, CDKN2A, SMAD4, and TP53) are associated with worse DFS. Our study did not find a significant association between KRAS mutational status and clinical outcomes after surgery. Our study population had a relatively lower proportion of head cancer (49% versus 72~79%) and a relatively higher proportion of T1-2 disease (81.9% versus 8–26%). The previous study also included a majority of white ethnicity and included only 21% of Asian populations. It should be noted that our study included a significantly higher proportion of patients who received adjuvant concurrent chemoradiotherapy according to the institutional protocol (73.8% versus 22–40%), even though the proportion of node-positive disease is similar or slightly lower (58.8 % versus 67–77%). These differences in baseline tumor characteristics and adjuvant treatment strategy may contribute to the discordance of the association between KRAS mutational status and disease-free survival after surgery.
Interestingly, we found that concomitant mutation of KRAS and LRP1B is associated with worse PFS after surgery. LRP1B encodes an endocytic low-density lipoprotein (LDL)-family receptor and the receptor binds to multiple extracellular ligands, including fibrinogen and apolipoprotein (apoE)-carrying lipoproteins.42 LRP1B is known as a functional tumor suppressor gene; inactivating mutations of LRP1B are frequently observed in diverse solid cancers, including melanoma, lung cancer, and gastric cancer.43–45 Owing to its large coding sequence (16 kbp), LRP1B is often missed as significantly mutated on gene analysis, whereas its mutation could still have a functional consequence in tumorigenesis and heterogeneity.46 Our study identified nine patients (10.8%) harboring LRP1B mutations and found that LRP1B was significantly downregulated in tumor tissues, compared with normal tissues. We also conducted GSEA and found that proliferation/cell cycle-associated gene sets were enriched in LRP1B and KRAS mutant samples, compared with tumors with only KRAS mutations [Supplemental Figure 2(c)]. Interestingly, a recent report found that down-regulation of LRP1B in colon cancer promoted the growth and migration of cancer cells via aberrant activation of beta-catenin/T-cell factor signaling.47 These findings might indicate that LRP1B loss of function may accelerates tumor cell proliferation in RAS-activated circumstances and promotes metastasis of tumor cells.
Our study also showed that ~10% of tumor samples harbored germline or somatic mutations in at least one of the DDR genes, including STK11 (4.8%), ATM (3.6%), MLH1 (3.6%), BRCA2 (2.4%), BRCA1 (1.2%), and PALB1 (1.2%), all of which are potential predictive biomarkers sensitive to PARP inhibition.48 Of note, one patient was ultrahypermutated (267 mutations/Mb) and showed high MSI. For PDAC, the frequency of ultrahypermutation and high MSI is not well known and varies considerably between studies. A recent systematic review reported that high MSI in PDAC is rare but exists in 1–2% of cases. That study also showed that high MSI in PDAC is strongly associated with medullary and mucinous/colloid histology and is usually KRAS-TP53 wild type.49,50 In the context of precision oncology, our study also confirmed that MSI status should be determined as part of a first-line analysis in PDAC with typical histology, despite its low prevalence, for all potential therapeutics, such as anti-programmed cell death protein 1 (PD1) immunotherapies.
Molecular subtypes guide preclinical and clinical therapeutic development and treatment in many cancer types. In pancreatic cancer, several transcriptomic subtypes have been described.11,28,29 In our transcriptomic analysis, tumors were classified into two clusters (S1 and S2), with tumor S1 displaying two distinct subclusters (S1-1 and S1-2). Our transcriptomic subtypes of S1 and S2 resembled the Moffitt tumor subtypes. Subclusters of tumor S1-1 overlapped with the exocrine-like subtype described by Collisson et al.,12 and the ADEX subtype described by Bailey et al., 11while tumor S1-2 mostly consisted of the classical subtype of Collisson et al.,12 and the progenitor subtype of Bailey et al.11 On the other hand, 88% of the basal-like, 96% of the quasi-mesenchymal, and 92% of the squamous subtypes were clustered as tumor S2. It was previously shown that the squamous subtype was significantly associated with poorer prognosis than the ADEX, progenitor, and immunogenic subtypes. However, the present study could not identify survival differences between the tumor-specific, HC-based PDAC groups. Our study included patients who received surgical resection plus mainly adjuvant chemoradiotherapy and analyzed DFS after surgery. In contrast, the previous study analyzed the OS according to the data from the Central Cancer Registry and treating clinicians.11 The difference in the survival endpoint may cause the discordance between the current study and the previous report regarding the association between molecular subtypes and the survival outcomes between the current study and the previous study. In the context of precision medicine, more research is needed on the molecular subtypes of PDAC for optimization of current therapeutics, and the discovery of novel therapeutic targets.
In this study, we identified high PRKCI expression levels as an overlapping, poor prognostic marker between our dataset and the TCGA dataset. PRKCI (encoding for PKCι) is an oncogene that is frequently overexpressed and associated with poor outcomes in lung cancers.51,52 In KRAS-mutant lung adenocarcinoma, PKCι establishes and maintains an aggressive stem-like, tumor-initiating cell phenotype by activating a PKCι–ELF3–NOTCH3 signaling axis that drives transformed growth and tumor initiation.53 Together with previous lung adenocarcinoma studies, our study supports the notion that pharmacologic blockade of PKCι signaling can be a potential therapeutic option in KRAS-mutant PDAC.
Overexpression of ABC transporter proteins is responsible for drug efflux, which attenuates the efficacy of chemotherapy by protecting tumor cells against chemotherapeutic agents.54,55 In the precision oncology era, cytotoxic chemotherapy, such as gemcitabine, gemcitabine/nab-paclitaxel, and FOLFIRINOX, is still the mainstay of treatment for metastatic pancreatic cancer.4,5 As a result, our results highlight that ABC transporters could be important therapeutic targets for overcoming cytotoxic chemotherapy resistance in PDAC patients.
Conclusions
We performed integrative genomics analyses of PDAC patients who received surgery and/or chemotherapy, and reconfirmed genomic landscapes, major driver mutations, and DDR pathway mutations associated with PDAC. We also discovered potential prognostic biomarkers and therapeutic targets, such as concomitant mutations of KRAS and LRP1B, the expression level of PRKCI, and the overexpression of ABC transporter proteins. In the future, understanding the molecular aberrations that determine patient outcomes after surgery and chemotherapy has the potential to improve treatment outcomes of PDAC patients.
Supplemental Material
Supplemental material, sj-tif-1-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-tif-2-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-tif-3-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-tif-4-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-5-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Acknowledgments
A subset of the biospecimens analyzed in this study was provided by Samsung Medical Center Biobank.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI14C2640 & HR20C0025).
ORCID iDs: Jung Yong Hong  https://orcid.org/0000-0003-1363-9332
https://orcid.org/0000-0003-1363-9332
Joon Oh Park  https://orcid.org/0000-0001-6502-2612
https://orcid.org/0000-0001-6502-2612
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Jung Yong Hong, Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
Hee Jin Cho, Innovative Therapeutic Research Center, Precision Medicine Research Institute, Samsung Medical Center, Seoul, Korea; Department of Biomedical Convergence Science and Technology, Kyungpook National University, Daegu, Korea.
Seung Tae Kim, Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
Young Suk Park, Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
Sang Hyun Shin, Department of Surgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
In Woong Han, Department of Surgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
Jeeyun Lee, Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
Jin Seok Heo, Department of Surgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
Joon Oh Park, Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, 81 Irwon-ro, Gangnam-gu, Seoul, 06351, Korea.
References
- 1.GBD 2017 Pancreatic Cancer Collaborators. The global, regional, and national burden of pancreatic cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2019; 4: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 3.Conroy T, Hammel P, Hebbar M, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med 2018; 379: 2395–2406. [DOI] [PubMed] [Google Scholar]
- 4.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011; 364: 1817–1825. [DOI] [PubMed] [Google Scholar]
- 5.Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013; 369: 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019; 381: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
- 8.Henley SJ, Ward EM, Scott S, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer 2020; 126: 2225–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017; 32: 185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531: 47–52. [DOI] [PubMed] [Google Scholar]
- 12.Collisson EA, Bailey P, Chang DK, et al. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2019; 16: 207–220. [DOI] [PubMed] [Google Scholar]
- 13.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 14.Singer VL, Jones LJ, Yue ST, et al. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal Biochem 1997; 249: 228–238. [DOI] [PubMed] [Google Scholar]
- 15.Desjardins P, Conklin D. NanoDrop microvolume quantitation of nucleic acids. J Vis Exp 2010; 45: 2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol 2016; 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014; 30: 1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016; 17: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020; 578: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talevich E, Shain AH, Botton T, et al. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016; 12: e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011; 12: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011; 17: 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015; 47: 1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One 2010; 5: e15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016; 17: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013; 4: 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013; 14: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Kuehn H, Gould J, et al. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 2007; 23: 3251–3253. [DOI] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016; 44: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018; 28: 1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glumac PM, LeBeau AM. The role of CD133 in cancer: a concise review. Clin Transl Med 2018; 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian ZR, Rubinson DA, Nowak JA, et al. Association of alterations in main driver genes with outcomes of patients with resected pancreatic ductal adenocarcinoma. JAMA Oncol 2018; 4: e173420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolaev SI, Rimoldi D, Iseli C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 2011; 44: 133–139. [DOI] [PubMed] [Google Scholar]
- 44.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda H, Rust AG, Ward JM, et al. Sleeping beauty transposon mutagenesis identifies genes that cooperate with mutant Smad4 in gastric cancer development. Proc Natl Acad Sci U S A 2016; 113: E2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Chong W, Wu Q, et al. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol 2019; 10: 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Sun P, Gao C, et al. Down-regulation of LRP1B in colon cancer promoted the growth and migration of cancer cells. Exp Cell Res 2017; 357: 1–8. [DOI] [PubMed] [Google Scholar]
- 48.Pilie PG, Tang C, Mills GB, et al. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol 2019; 16: 81–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luchini C, Brosens LAA, Wood LD, et al. Comprehensive characterisation of pancreatic ductal adenocarcinoma with microsatellite instability: histology, molecular pathology and clinical implications. Gut 2021; 70: 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Humphris JL, Patch AM, Nones K, et al. Hypermutation in pancreatic cancer. Gastroenterology 2017; 152: 68–74.e2. [DOI] [PubMed] [Google Scholar]
- 51.Regala RP, Davis RK, Kunz A, et al. Atypical protein kinase Cι is required for bronchioalveolar stem cell expansion and lung tumorigenesis. Cancer Res 2009; 69: 7603–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin N, Liu Y, Khoor A, et al. Protein kinase Ciota and Wnt/β-catenin signaling: alternative pathways to Kras/Trp53-driven lung adenocarcinoma. Cancer Cell 2019; 36: 156–167.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali SA, Justilien V, Jamieson L, et al. Protein kinase ciota drives a NOTCH3-dependent stem-like phenotype in mutant KRAS lung adenocarcinoma. Cancer Cell 2016; 29: 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2002; 2: 48–58. [DOI] [PubMed] [Google Scholar]
- 55.Lage H. An overview of cancer multidrug resistance: a still unsolved problem. Cell Mol Life Sci 2008; 65: 3145–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-tif-1-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-tif-2-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-tif-3-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-tif-4-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-5-tam-10.1177_17588359211038478 for Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer by Jung Yong Hong, Hee Jin Cho, Seung Tae Kim, Young Suk Park, Sang Hyun Shin, In Woong Han, Jeeyun Lee, Jin Seok Heo and Joon Oh Park in Therapeutic Advances in Medical Oncology