SUMMARY
Emerging studies have highlighted the disproportionate role of Candida albicans in influencing both early community assembly of the bacterial microbiome and dysbiosis during allergic diseases and intestinal inflammation. Nonpathogenic colonization of the human gastrointestinal (GI) tract by C. albicans is common, and the role of this single fungal species in modulating bacterial community reassembly after broad-spectrum antibiotics can be readily recapitulated in mouse studies. One of the most notable features of C. albicans-associated dysbiotic states is a marked change in the levels of lactic acid bacteria (LAB). C. albicans and LAB share metabolic niches throughout the GI tract, and in vitro studies have identified various interactions between these microbes. The two predominant LAB affected are Lactobacillus species and Enterococcus species. Lactobacilli can antagonize enterococci and C. albicans, while Enterococcus faecalis and C. albicans have been reported to exhibit a mutualistic relationship. E. faecalis and C. albicans are also causative agents of a variety of life-threatening infections, are frequently isolated together from mixed-species infections, and share certain similarities in clinical presentation—most notably their emergence as opportunistic pathogens following disruption of the microbiota. In this review, we discuss and model the mechanisms used by Lactobacillus species, E. faecalis, and C. albicans to modulate each other’s growth and virulence in the GI tract. With multidrug-resistant E. faecalis and C. albicans strains becoming increasingly common in hospital settings, examining the interplay between these three microbes may provide novel insights for enhancing the efficacy of existing antimicrobial therapies.
KEYWORDS: Candida albicans, enterococcus, gastrointestinal, lactobacillus, lactic acid bacteria, polymicrobial infection
INTRODUCTION
There is increasing evidence that the fungus Candida albicans plays a disproportionate role in modulating the microbial ecology of the human gut microbiome and subsequent mucosal immune responses. The intestinal microbiota is a multiplex system that is capable of suppressing indigenous pathogens while modulating essential homeostatic functions, such as digestion and immune responses (1). In a recent longitudinal cohort examination of 178 preterm infants, investigators studied the community dynamics of bacteria, fungi, and archaea in the intestine and reported periods of microbial blooms, microbial extinctions, and an inverse correlation between bacterial and fungal loads (2). Most notably, they discovered that interactions with a single fungal species (C. albicans) can influence early microbiome community assembly and inhibit multiple dominant genera of intestinal bacteria. This correlation between Candida levels and a dysbiotic microbiota (including diminished Lactobacillus levels) has also been reported in an atopic cohort of children (3, 4) and parallels our laboratory’s observations in mouse models of antibiotic-mediated dysbiosis and allergic disease (5, 6). Intestinal microbiome-derived Lactobacillus strains can antagonize the opportunistic pathogens Enterococcus faecalis and C. albicans, which are also causative agents of a variety of life-threatening infections and are frequently isolated together from mixed-species infections (7). Studies in which E. faecalis and C. albicans are sole colonizers of the Caenorhabditis elegans gastrointestinal (GI) tract have found that these two microbes promote each other’s growth but actually dampen each other’s virulence, suggesting a complex mutualistic relationship which is likely further influenced by other organisms in their niche (8, 9). Using mouse models, our laboratory has reported an antagonistic relationship between C. albicans and Lactobacillus johnsonii and a positive association between C. albicans colonization and levels of E. faecalis (10–12). These observations, together with others (described in the following sections), provide evidence that C. albicans can play a significant role in modulating mucosal immunity and the intestinal microbiota—particularly the levels of lactic acid bacteria (LAB).
Members of Enterococcus and Candida share certain similarities, including their emergence as opportunistic pathogens after a long nonpathogenic existence in the host microbiome. These microbes can asymptomatically colonize humans, and their pathogenesis generally arises from this asymptomatic colonization (13). Infections involving these organisms are tightly linked to ecological disruption, which may be due to prior antibiotic exposure, treatment with chemotherapeutic agents, medical devices, or anatomic disruption. Consistent with this overlap in clinical presentation, the most severe forms of infection often occur in the same clinical setting—namely, following significant nosocomial exposure, broad-spectrum antibiotic use, and immune defects (14, 15). Furthermore, C. albicans and E. faecalis are common coconstituents in polymicrobial infections, and outgrowth of one of these organisms promotes the outgrowth of the other (7, 16, 17). Multidrug-resistant E. faecalis and C. albicans are becoming increasingly common in hospital settings, so by understanding and exploiting the interplay between these organisms and their antagonistic interacting partners (e.g., Lactobacillus species), we may be able to enhance the efficacy of existing antimicrobials in treating these opportunistic infections.
Unfortunately, research to date on Lactobacillus-Enterococcus-Candida interactions has only examined the cross talk between two of these microbes at a time, and there have been no studies investigating the mechanisms by which C. albicans itself interacts with Enterococcus or Lactobacillus species. In this review, we provide a brief background on each microbe and then examine the existing data on the mechanisms employed by Lactobacillus species, E. faecalis, and C. albicans to modulate each other’s growth and/or virulence. We use this information to construct a mechanism-focused model of the microbial ecology of these three microorganisms in the GI tract, highlighting points where their interactions likely affect human health and disease.
CANDIDA ALBICANS
Candida is a diverse genus of fungi consisting of over 200 species, and yet only a few species are known to cause disease in humans (18). One of the most prevalent of these species is Candida albicans, which asymptomatically colonizes the human intestinal, upper respiratory, and genitourinary tracts as well as the skin. As a polymorphic fungus, specific environmental cues can trigger C. albicans to undergo various morphologic transformations (reviewed in reference 19) that are central to its pathogenesis and elicit different responses from host cells and the surrounding microbiota (20–22). In addition to being one of the most ubiquitous fungal species in the human microbiota, C. albicans is most commonly associated with infection, with Candida glabrata, Candida parapsilosis, Candida krusei, and Candida tropicalis accounting for most of the remaining infections (23). The clinical spectrum of disease caused by these different species is largely indistinguishable, although Candida auris has a greater propensity for localized outbreaks in acute care settings (24). This may be due to direct clonal transmission of C. auris via fomites in these settings, whereas other Candida species are less resilient on abiotic surfaces and are therefore less likely to seed infections and spread in this way (24, 25).
Microbiology
Morphologic and phenotypic plasticity.
These traits enable C. albicans to rapidly adapt to changing environmental conditions and colonize diverse niches. Yeast and hypha are the predominant morphological forms of C. albicans encountered in the GI tract during health and disease (19). Yeast cells are round and proliferate by budding, while hyphal cells are filamentous and exhibit polarized growth whereby polarisomes direct germ tube formation and elongation at the apex of the cell (19). The ability for C. albicans to transition back and forth from yeast to hypha is generally regarded as its most important virulence factor (19, 26, 27).
As a yeast cell, C. albicans may also switch between two distinct phenotypic states named for their colony appearance: “white” and “opaque” (22, 27). Although these phenotypes are not genetically encoded, they are heritable and exhibit distinct transcriptomic profiles (27). Moreover, white and opaque cells preferentially occupy separate niches and differ in their interactions with immune cells, propensity to mate, and conditions in which they can form hyphae (27, 28). In doing so, phenotypic switching also directly impacts C. albicans virulence. Interestingly, passage of C. albicans through the GI tract results in a phenotypic switch into the gastrointestinal-induced transition (“GUT”) phenotype, which possesses a distinct transcriptome from white or opaque cells (29). In contrast to white cells, which are round or oval, GUT cells are elongated and more closely resemble opaque cells in shape; yet they lack many of their defining characteristics, including heat sensitivity, sexual mating capacity, and increased expression of genes encoding secreted aspartyl proteases (29). While white-opaque switching and GUT are the best-studied phenotypic transitions in C. albicans, a number of other phenotypes have been identified (reviewed in references 22 and 30), which may each serve important functions in many aspects of C. albicans biology such as generating variability in commensal and pathogenic populations to promote their adaptability.
C. albicans uses quorum sensing (QS) and pheromone signaling to regulate its morphologic and phenotypic states. Notably, hyphal morphogenesis is inhibited under conditions of high cell density by the QS molecule (QSM) farnesol, which is secreted by C. albicans in a density-dependent manner (31). Farnesol also acts to repress opaque cell formation and C. albicans mating, as it can be produced only by white cells and has even been shown to kill opaque cells (31). In contrast, only opaque cells can secrete pheromones, which promote biofilm formation in white cells as well as sexual mating in opaque cells (31). Host environmental determinants of C. albicans morphological and phenotypic states include pH, temperature, and nutrient availability (22). The combinatorial effects of morphology and phenotype on C. albicans pathogenesis in different physiological contexts are incompletely understood. Nevertheless, it is clear that the transition from commensal to pathogen is multifaceted and not exclusively a consequence of hyphal morphogenesis.
Cell wall composition.
The C. albicans cell wall is critical for nearly every aspect of its biology and virulence. In addition to providing protection and maintaining its shape, this structure mediates physical interactions with the host and other microorganisms and enables C. albicans to assume various morphologies (32, 33). Not surprisingly, yeast and hyphal cell walls have distinct compositions which are at least partially responsible for their differential recognition by immune cells and the responses they induce (21). Irrespective of its morphological state, the C. albicans cell wall is composed primarily of carbohydrates and heavily glycosylated proteins organized into an inner and outer layer (21, 32). The three major carbohydrates include β-glucans, chitin, and mannose polymers, which may be interspersed or directly associated with cell wall proteins (21, 33). β-Glucans and chitin comprise the inner layer adjacent to the plasma membrane and provide shape and rigidity to C. albicans cells, while mannose polymers covalently linked to proteins (termed mannoproteins or “mannans”) dominate the outer layer and control cell permeability but not overall strength or shape (21, 32). Characterization of yeast- and hypha-specific expression of cell wall structures has proven to be a challenge due to the inherent dynamism of C. albicans in physiological settings. However, certain cell wall components are linked to functions critical for C. albicans virulence, including adhesion, biofilm formation, and interactions with immune cells (reviewed in references 32 and 33).
Metabolism and respiration.
C. albicans requires exogenous sources of carbon, nitrogen, and phosphate for biosynthetic processes as well as pathogenesis. The preferred carbon source of C. albicans is glucose; however, it demonstrates a high degree of metabolic plasticity which allows it to exploit a variety of both fermentable and nonfermentable carbon sources (34, 35). This versatility also enables C. albicans to colonize diverse nutritional niches in the host and outcompete other microorganisms for growth-limiting nutrients (26, 35). For example, when faced with sugar-limiting conditions, C. albicans upregulates genes involved in amino acid transport and metabolism, allowing it to simultaneously assimilate carbon and nitrogen from those amino acids (34). Nitrogen can also be derived from proteins, and several secreted proteases involved in nitrogen acquisition from protein sources are important virulence factors for C. albicans (36–38). Phagosomes are nitrogen-deficient environments, so internalization of fungi by macrophages or neutrophils triggers an upregulation of amino acid biosynthetic pathways to boost nitrogen intake (39–41).
Phenotypic analysis of C. albicans in the presence of different metabolic cues reveals striking differences in optimal growth requirements between white and opaque cells (42), suggesting that these two phenotypes are metabolically specialized for different host niches. In addition, the altered transcriptome of GUT cells reflects an enhanced fitness and commensalism of this phenotype in the mammalian GI tract, as gene expression is skewed toward the metabolism of nutrients encountered there (29). Specific differences in the metabolic pathways of white, opaque, and GUT cells are discussed more in reference 29.
C. albicans metabolism is greatly affected by oxygen levels. As a facultative anaerobe, it grows optimally under aerobic conditions but is capable of both aerobic respiration and anaerobic fermentation (35). The GI tract is largely hypoxic, although steep oxygen gradients exist between the upper and lower GI tracts as well as from the lumen to epithelium (43). This normally restricts microbes to specific regions based on their oxygen tolerances. However, thanks to its metabolic flexibility under fluctuating oxygen levels, C. albicans can thrive in diverse anatomical niches (35). When grown in vitro under microaerophilic conditions, C. albicans exhibits enhanced hyphal growth and biofilm formation as well as β-glucan masking on its cell wall, suggesting that low oxygen promotes its virulence (44–46). In a recent study comparing the metabolomes of C. albicans grown under hypoxic (i.e., 5% oxygen) or aerobic conditions, stark differences were revealed in fundamental metabolic pathways, including glycolysis and cell wall biogenesis (35). These observations suggest that the low oxygen levels encountered in the GI tract promote C. albicans pathogenesis, and yet, C. albicans colonizes most humans without causing infection. Crucially, none of the aforementioned studies account for the effects of surrounding microbial or host cells, which likely alter or interfere with the responses reported in C. albicans monocultures.
Colonization of Mucosal Tissues
Although over 99% of the microbial genes in the human microbiome are bacterial, there has been a growing appreciation for the role of the minor fungal component (“mycobiome”) in human health (47, 48). C. albicans is one of the most abundant fungal species in the mycobiome and is estimated to be present in around 60% of healthy adults (49); although, these colonization rates are reported to be lower in non-Westernized societies (50). C. albicans is also the most common cause of a variety of life-threatening fungal diseases collectively referred to as “candidiasis,” which are typically seeded by indigenous Candida populations (47, 51). Nearly all mucosal surfaces can be colonized by C. albicans, but its primary habitats include the oral cavity, GI tract, and vaginal canal. In a given individual, C. albicans isolates from different anatomical regions are often genetically similar but nonidentical, indicating that they adapt to distinct selective pressures in these regions (52). Its presence at each site is implicated in a number of fungal infections—each with potentially unique target populations and clinical manifestations. However, the focus of this review is on the interactions of C. albicans in the GI tract because that is the major reservoir for disseminated candidiasis and where Enterococcus faecalis and many Lactobacillus species also reside.
Gastrointestinal tract.
The human GI tract can be broken up into the following three major sections: upper (esophagus and stomach), middle (small intestine, which includes the duodenum, jejunum, and ileum), and lower GI tract (colon and rectum). Sampling different regions of the human GI tract for characterization of their microbial communities is a complicated and invasive undertaking. In a 2018 study (53), stool samples from healthy adults were inoculated into bioreactors simulating the different nutritional conditions and oxygen saturation found in these two regions. Bioreactors representing the upper-middle GI tract (i.e., high levels of simple sugars and hypoxic conditions) had reduced bacterial diversity but elevated levels of C. albicans compared with the lower GI tract bioreactors containing low sugar under anoxic conditions (53). Candida abundance in the GI tract is also significantly influenced by short-term diet since a high consumption of carbohydrates within a single week is associated with higher fungal burdens (54). As C. albicans coexists with the bacterial microbiota and diet has been shown to strongly modulate bacterial enterotypes (55), diet-associated changes in C. albicans abundance are likely affected by changes in the levels of certain bacteria and/or their metabolites.
Gnotobiotic mice have been used to evaluate the localization of C. albicans across the mammalian GI tract and understand its spatial organization in the presence or absence of enteric bacteria (56, 57). Strikingly, fungal distribution is dependent on the presence of bacteria. As the sole colonizer, C. albicans was found both in the intestinal lumen and adjacent to the mucus layer along the entire murine GI tract, but the addition of the enteric bacterium Bacteroides thetaiotaomicron led to the formation of a thick outer mucus layer in which C. albicans became embedded (56). B. thetaiotaomicron is known to promote mucin secretion by goblet cells in the GI tract and directly interact with C. albicans cell wall mannans to inhibit fungal growth (56, 58), so its presence likely impacts C. albicans colonization. Indeed, B. thetaiotaomicron and C. albicans were in close association with one another, and B. thetaiotaomicron-processed mucins supported the growth of C. albicans to a greater extent than unprocessed mucins (56). Taken together, these data suggest that C. albicans primarily inhabits upper-middle regions of the GI tract, adjacent to or within the mucus layer. Other studies using gnotobiotic mice (57, 59–61) and piglets (62, 63) have detected high levels of C. albicans in the lower GI tract as well, supporting the notion that C. albicans colonizes throughout the GI tract.
Oral cavity and urogenital tract.
Fungal-bacterial interactions also play an important role in balancing C. albicans commensalism and pathogenesis in the oral and urogenital tracts, so understanding their interactions in these niches may provide insight into their cross talk in the GI tract. C. albicans colonization and infection in the oral and vaginal mucosae are not the focus of this review, but more detail is provided in references 64 and 65.
Establishment of the oral microbiome begins at birth and later expands downstream in the GI tract (66). Crucially, the factors that shape the oral microbiota such as diet and oral hygiene can also impact the microbial composition of the GI tract. Candida species are the most frequently encountered fungi in the oral cavity, appearing in an estimated 75% of healthy individuals (67). C. albicans can grow as a biofilm in both aerobic environments like the mouth and anaerobic environments such as the colon, but its subsequent virulence greatly depends on its interactions with neighboring bacteria; hence, biofilm formation does not necessarily lead to infection. As a matter of fact, C. albicans biofilms in the oral cavity may provide an anoxic niche for many anaerobic bacteria that inhibit its pathogenesis (44).
In addition to the oral cavity, C. albicans is the most frequently identified fungal species in the human urogenital tract—particularly the vagina, where it is present in anywhere from 5% to 20% of healthy individuals (64, 68, 69). Given the pronounced differences between these two mucosae in terms of host defense mechanisms, surrounding microbial communities, and overall architecture (69), this further exemplifies the tremendous adaptability of C. albicans. The mechanisms underlying C. albicans commensalism in the vaginal mucosa may be especially useful in forming hypotheses about its interplay with bacteria in the GI tract. In particular, the healthy human vagina is dominated by Lactobacillus species (70), of which many are also present in the GI tract and have been shown to directly antagonize C. albicans. Reference 71 provides a comprehensive summary of the role of lactobacilli in preventing the pathogenesis of C. albicans and other microbes in the vagina.
Intriguingly, C. albicans pathogenesis does not occur at the same rates or under the same circumstances between the oral cavity and urogenital tract. Oropharyngeal candidiasis (OPC) occurs much less frequently in humans than vulvovaginal candidiasis (VVC), which is likely due to microbiological differences in C. albicans itself as well as differences in surrounding microbial communities and host defense. Indeed, despite higher colonization rates in the oral cavity, OPC occurs almost exclusively in immunocompromised individuals, whereas 75% of all women experience at least one episode of VVC in their lifetime, regardless of immune status (69). While infections of the oral and vaginal mucosae are not the topic of this review, it is important to remember the variable nature of C. albicans in different niches when making generalizations about its pathogenesis.
Pathogenesis
Prevalence and risk factors.
C. albicans ranks among the top four most common causes of health care-associated bloodstream infections (BSIs) in the United States, with an estimated mortality rate of 40% to 55% (72). In addition to BSIs (termed “candidemia”), infections can be localized—as in OPC and VVC—or invasive, with invasive candidiasis (IC) accounting for the majority of deaths (73, 74). Notably, over 60% of IC cases across the globe are attributable to C. albicans derived from indigenous populations in the GI tract (73, 75). Thus, C. albicans represents a significant threat to human health.
There are many well-established risk factors for C. albicans infections. Candidiasis is especially prevalent in intensive care units (ICUs), where high doses of antibiotics and immunosuppressive drugs are regularly administered to patients (72). Using a conditional logistic regression model, Wenzel and Gennings (76) projected that ICU patients exposed to antibiotics have a 10% to 30% chance of succumbing to a C. albicans BSI, and by adding just one other risk factor (e.g., prior colonization with C. albicans), that likelihood jumps to 50%. According to this study, simply being admitted to an ICU carries an inherent risk of over 33%. Antibiotics can sufficiently reduce the levels of antagonistic bacteria to enable C. albicans outgrowth and have been successfully used to colonize mice with stable levels of C. albicans (11, 12, 77) since mice typically lack indigenous populations of this fungus (78). Similarly, immunodeficiencies due to chemotherapy or other immunosuppressive drugs (79), underlying neutropenia (80), and/or infection with human immunodeficiency virus (HIV) (65) are considerable risk factors in the development of candidiasis, although the latter is primarily associated with OPC. These and other predictors for C. albicans infections are extensively reviewed in reference 73.
Host innate immunity.
Phagocytes are responsible for the elimination of C. albicans and critical for preventing systemic candidiasis. Toll-like receptors (TLRs) and C-type lectins on the surface of neutrophils and macrophages recognize β-glucans and mannans on the fungal cell wall, leading to either pro- or anti-inflammatory responses (81, 82). Stimulation of TLR2 by C. albicans can trigger the production proinflammatory cytokines, including tumor necrosis factor alpha (TNF-α), interleukin-1α (IL-1α), IL-1β, and IL-6, while stimulation of TLR4 results in chemokine production and thus enhanced neutrophil recruitment (82, 83). Moreover, the innate immune receptors TLR2, TLR4, Dectin-1, and mannose recognize different structures in the C. albicans cell wall and can elicit different responses as a result (82). Phagocytosis of C. albicans by neutrophils is mediated by β-glucans (84), although β-glucans can also trigger residual cytokine production in other cells (82). The β-glucan layer is typically “masked” by the outer mannan layer, enabling C. albicans to evade recognition by host cells (85). Cell wall remodeling that results in the masking and unmasking of β-glucans is therefore an important mechanism by which C. albicans modulates its survival and virulence in a host. β-Glucan expression is considerably reduced on hyphal cells, which is associated with enhanced virulence and reduced induction of proinflammatory cytokine or chemokine release compared with yeast cells (81). Cell wall chitin is recognized by NOD2, TLR9, and mannose receptor, leading to the release of the anti-inflammatory cytokine IL-10 (86). This response likely represents the restoration of the immune balance once a C. albicans infection has been controlled. Strains of C. albicans isolated from active disease often have considerably higher chitin content than commensal isolates, and any damage to β-glucans may expose more chitin to host receptors, thereby dampening the host immune response (86, 87). As such, immune responses to C. albicans are greatly influenced by its morphology. Taken together, innate immunity to C. albicans is largely context dependent and determined by both the host cell type/receptor and fungal morphology.
Virulence factors.
Some of the most widely recognized factors contributing to C. albicans virulence include adhesion, hyphal morphogenesis, biofilm formation, and phenotypic switching (88). Specialized cell wall proteins called adhesins mediate adherence to host cells and other microorganisms, as well as abiotic surfaces, such as intravenous (i.v.) catheters. The agglutinin-like sequence (ALS) and hypha-associated GPI-linked proteins are among the best studied of these adhesins and are both significantly upregulated during infection (89–91). Following initial adherence to biotic or abiotic surfaces, C. albicans may proliferate and form biofilms enclosed by a protective extracellular matrix that enhances its resistance to host and microbial defense mechanisms (88). Hyphal morphogenesis is also essential for C. albicans pathogenesis, as is evidenced by the fact that mutants unable to form hyphae are avirulent (92, 93). However, the propensity for C. albicans to undergo hyphal morphogenesis is in large part determined by its phenotypic state and vice versa (22). While both white and opaque cells undergo filamentation in vivo, white cells more readily establish lethal systemic infections in a host, which is predominantly temperature dependent (28). Furthermore, white cells are more susceptible to phagocytosis by macrophages than opaque cells, indicating that these phenotypes interact differently with immune cells (28). Interestingly, the observed hypervirulence of white cells is also dependent on their ability to undergo filamentation, as deletion of genes regulating hyphal morphogenesis significantly reduces the lethality of infection (28).
A recent review by Kumamoto and colleagues (94) highlights several studies in which expression of hypha-specific genes was associated with attenuated colonization fitness in the mouse GI tract. Of note, serial passage of C. albicans in antibiotic-treated mice promoted C. albicans competitive fitness by selecting for mutations in FLO8 and EFG1, which are required for hyphal morphogenesis (95). Similarly, C. albicans mutants with deletions in the hyphal-associated genes EFG1, BRG1, TEC1, ROB1, and UME6 were able to outcompete the wild-type strain in the mouse GI tract, whereas a UME6-overexpressing mutant had significantly reduced fitness (96). Surprisingly, the UME6-knockout strain displayed a hyperfit phenotype but retained a normal ratio of yeast and hyphae, indicating that morphology alone does not dictate commensal fitness. Although these data suggest that hyphae have a competitive disadvantage, C. albicans typically colonizes the GI tract as a mixture of yeast and hyphal cells (94, 96), so the ability to transition between yeast and hyphal forms is likely important for commensal colonization.
The proximal GI tract has been reported to be dominated by yeast while hyphae predominate the lower GI tract (96), suggesting that tissue-specific signals are involved in the control of hyphal transformation. This may also reflect differences in antihyphal signals from bacteria (especially LAB) in those regions. Together, these observations raise the intriguing possibility that inhibition of hyphal morphogenesis by Lactobacillus species actually enhances its fitness rather than reducing its virulence. However, it is important to keep in mind that lactobacilli affect many aspects of C. albicans biology which can outweigh the effect of hyphal suppression on colonization, so the net impact of their interactions may be a decreased growth of C. albicans in the GI tract.
Among the lesser-known virulence factors are secreted molecules involved in host cell penetration and pH sensing and regulation and various proteins involved in environmental stress responses, which are discussed at length in reference 88. Importantly, C. albicans virulence factors can be antagonized by various components of the host immune system and surrounding microbiota and represent potential targets for antifungal drugs. However, because C. albicans virulence factors are so interconnected, it is critical to include readouts such as host survival or inflammation in any in vivo infection model when making claims about the impact of a specific condition or organism on C. albicans pathogenesis.
Prostaglandins and mucosal immunity.
The production of bioactive lipid mediators is only one of many mechanisms by which C. albicans directly interacts with and alters the surrounding immune milieu to promote its survival. Prostaglandins can be synthesized by host cell cyclooxygenases that act on arachidonic acid released from the plasma membrane by phospholipases (97). Host-derived prostaglandin E2 (PGE2) is one of the signals for hyphal transformation in C. albicans (92, 98–100). Interestingly, C. albicans is capable of producing authentic PGE2 as well as functionally related cross-reactive oxylipins from host-derived arachidonic acid (97–101) via an unconventional pathway involving a fatty acid desaturase (Ole2) and multicopper oxidase (Fet3) (101). Both host and fungal PGE2 shift the adaptive immune response in favor of fungal survival by promoting Th2 responses and thus inhibiting anti-Candida Th1 responses, lymphocyte proliferation, and phagocytosis (97).
In addition to prostaglandin production, C. albicans colonization of the GI tract can modulate mucosal immunity and barrier function. In antibiotic-treated mice, GI colonization by C. albicans can promote the development of a CD4+ Th2 cell-mediated allergic airway response to mold spore challenge (5, 6). Subsequent studies have shown that M2 macrophage polarization in the lung is a critical component of the response, and suppression of PGE2 synthesis blocks this polarization, resulting in decreased allergic airway inflammation (102). C. albicans colonization of mice also significantly enhances GI epithelial leak of orally delivered OVA and drives an increase in the number of mast cells in the intestinal mucosa (103, 104). IL-9 production and mast cell activation are critical steps in the process whereby C. albicans induces epithelial barrier leak (104). Additional evidence of a positive feedback loop between C. albicans colonization and intestinal inflammation has also been reported in a mouse model of dextran sulfate sodium (DSS)-induced colitis in which C. albicans colonization was enhanced by inflammation-inducing injury to the intestinal epithelium (105).
Clinical Significance
The clinical spectrum of candidiasis is associated with the immune status of the patient. In immunologically intact patients, the most common manifestation is VVC, which is generally well treated with localized therapy (106). However, even in immunocompetent hosts, recurrent VVC impacts an estimated 138 million women each year (107). OPC and other superficial mycoses can last longer in patients with defects in cellular immunity, such as those with AIDS or those receiving high doses of corticosteroids (108, 109). C. albicans can also cause severe invasive infection, which is tightly linked to neutropenia and ICU exposure (110, 111). In these patients, IC can be indistinguishable from bacterial sepsis (112). Invasive focal infections, such as endocarditis and endophthalmitis, are due to seeding by indigenous C. albicans following systemic infection or introduction from devices such as intravenous (i.v.) catheters and ventricular shunts or i.v. drug use (113, 114). Additionally, chemotherapeutic agents which lead to disruption of mucous membranes increase the risk for systemic infections, presumably because they also facilitate entry of organisms from the GI tract into the bloodstream. Interestingly, Candida pneumonia is extraordinarily rare (115), as is pneumonia due to Enterococcus species (116). Candida pneumonia has historically been attributed to contamination of the airways by oral C. albicans, but positive sputum cultures and bronchoalveolar lavage cultures are not uncommon (117, 118). More recently, recognition of the continuity of the microbiome between the upper and lower airways has raised the significant possibility that Candida isolation in those sites may not represent contamination (118).
LACTIC ACID BACTERIA
Broadly speaking, the term lactic acid bacteria (LAB) refers to a group of bacteria that produce lactic acid as the primary by-product of carbohydrate metabolism. More precisely, LAB belong to the order Lactobacillales and actually constitute an extremely diverse group in terms of metabolic pathways, tolerances to environmental stresses, pathogenic potential, and ecological niches (119). While LAB are often equated with Lactobacillus species or other generally harmless or even beneficial bacterial strains, this group also includes common human pathogens—most notably E. faecalis.
Microbiology
LAB are Gram-positive, non-spore-forming, aerotolerant anaerobes that ferment a variety of carbohydrates to lactic acid and are generally oxidase, catalase, and cytochrome negative (120). Most LAB are fastidious organisms and require complex nutrients for growth, including amino acids, vitamins, and minerals (121). Although lacking heme enzymes involved in oxidative stress response (119), LAB can use alternative radical scavenging metals such as manganese (122), zinc (123), and selenium (124). The absence of cytochromes in LAB means that they cannot use oxygen to generate ATP by means of an electron transport chain. Instead, they rely on sugar fermentation, which generates lactic acid either exclusively (i.e., homolactic fermentation) or in conjunction with other end products (i.e., heterolactic fermentation) (125). However, in addition to fermentation, LAB can generate ATP from noncarbohydrate substrates through pathways that also serve to counteract the acidification caused by lactic acid accumulation (119).
The LAB group encompasses many well-known genera including Lactobacillus, Enterococcus, Lactococcus, and Streptococcus (119, 126). Subgroups of LAB that were originally proposed by Orla-Jensen in the early 20th century are still used to this day (126). The classification criteria include morphology (rods versus cocci), fermentation pathway (homofermentative versus heterofermentative), optimal growth temperatures, carbon dioxide production (from glucose versus gluconate), requirement for thiamine, ability to reduce fructose to mannitol, and ability to hydrolyze arginine. Reference 126 provides a more complete overview of these subgroups.
Colonization and Distribution
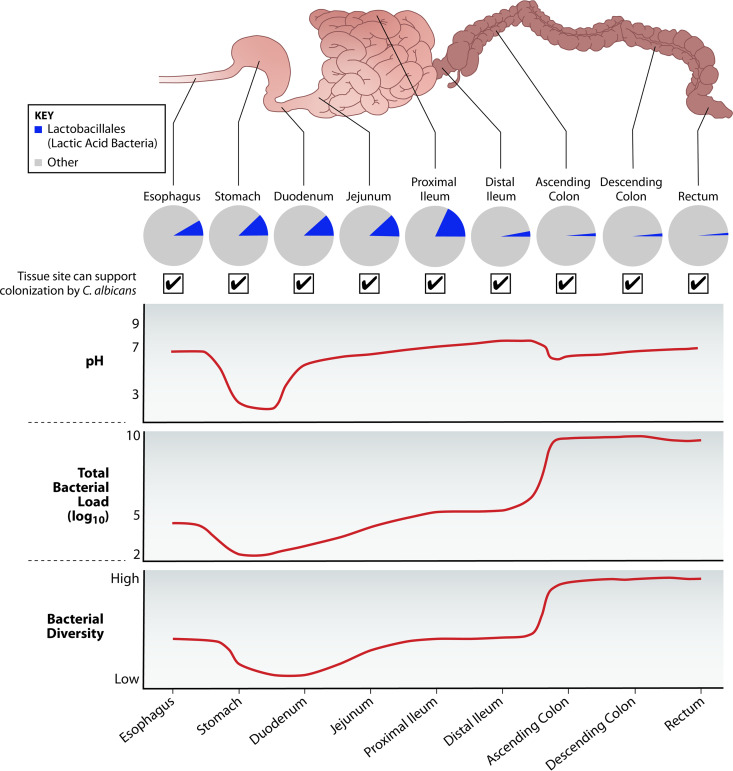
The heterogeneity of LAB as a group is evidenced by their presence in diverse niches, including the human GI and vaginal tracts, dairy and meat products, soil and decomposing plant material, and sewage water (127). LAB comprise a staggering 90% to 100% of the vaginal microbiota (which is predominantly Lactobacillus species [128]) and inhabit every major region of the GI tract (Fig. 1).
FIG 1.
Distribution of LAB and C. albicans along the human GI tract. LAB including Lactobacillus and Enterococcus species belong to the order Lactobacillales, which are found in proportionally higher numbers in the upper GI tract. The predominance of LAB in proximal regions such as the stomach and small intestine is a result of their tolerance to the extremely low pH, bile salts, and oxygen gradients within those niches. Although the total number of LAB is relatively stable throughout the GI tract, the total bacterial load and diversity increase dramatically starting at the ileocecal junction, so their relative abundance is much lower in the large intestine (131, 132). The fact that LAB abound in microbially sparse regions suggests that whatever interactions they have with other cells in those regions play a significant role in their functioning as well as host physiology. Importantly, C. albicans is also found in every region of the GI tract, including these low-biodiversity niches, so the interplay between LAB and C. albicans is expected to have a profound impact on host health.
In humans, microbial levels in the upper regions of the GI tract are restricted by extremely low pH, toxic bile salts, oxygen tension, and fast-flowing digesta (129). Consequently, bacterial load is relatively low (101 to 103 microbes per gram in the stomach and duodenum [130]), as is the overall microbial diversity (131, 132), making the proportion of LAB in these regions comparatively high (Fig. 1). The colon is home to the highest microbial diversity and abundance, with numbers reaching an estimated 1011 per gram of contents (130). Although absolute numbers of LAB are relatively stable from the upper to lower GI tract (131), they represent a much smaller fraction of the total bacteria in the distal regions of the small intestine to the rectum (Fig. 1).
Given the high proportion of LAB in areas with extremely low bacterial abundance and diversity, their interactions with other microorganisms in these niches are likely to have a significant influence on human health. Indeed, the genera Lactobacillus and Enterococcus have received considerable attention for their roles in colonization resistance, microbial carriage, opportunistic infection, and host immunity.
Lactobacillus Species
Microbiology.
Lactobacillus species can be either aerotolerant or anaerobic, exist as rods or coccobacilli, and have moderate to high tolerance to salt and acid. Their unusually high resistance to low pH is thought to be mediated by the F0F1-ATPase, which establishes a proton gradient across their membrane (133, 134). Furthermore, many lactobacilli carry bile salt hydrolase (BSH) proteins that enable them to tolerate high concentrations of bile acids in the small intestine (135, 136). As LAB, they have complex nutritional requirements and produce lactic acid as a by-product of sugar fermentation. More precisely, lactobacilli can be either homofermentative (i.e., generate at least 85% lactic acid from fermentation) or heterofermentative (i.e., generate equal amounts of lactic acid, ethanol and/or acetic acid, and CO2 from fermentation) (137).
Colonization of the gastrointestinal tract.
The exact numbers and species of Lactobacillus present in the human GI tract at any given moment have been highly variable across studies and cohorts (128). This lack of consistency makes it difficult to distinguish stably colonizing species from transient species, which are predominantly derived from the consumption of Lactobacillus-containing foods such as yogurt, cheese, and pickles. Nevertheless, there are some species that are more commonly detected in human samples and not generally present in food (e.g., L. acidophilus, Lactobacillus crispatus, Lactobacillus salivarius, Lactobacillus gasseri, L. johnsonii, and Lactobacillus reuteri) (138), suggesting that they may be indigenous to the human GI tract. In addition, 16S rRNA and terminal restriction fragment length polymorphism (T-RFLP) analyses repeatedly demonstrate that Lactobacillus species constitute an appreciable fraction of the microbial population in those regions. This includes the identification of lactobacilli in gastric (139–141) as well as jejunal and ileal (142, 143) biopsy specimens from healthy humans. Considerably higher numbers of lactobacilli are found to persist in the GI tracts of other animals (138), which has been proposed to be due to differences in the epithelium that potentiate Lactobacillus adhesion (144).
Impact on human health.
Of the LAB, lactobacilli are most closely associated with health benefits. Moreover, despite the plethora of strains present in both humans and the environment, very few have been proven to have any pathogenic potential (145). A report drafted by the joint FAO/WHO Working Group (146) notes that Lactobacillus is one of two bacterial genera most commonly exhibiting probiotic attributes—i.e., conferring health benefits on the host. The ability of lactobacilli to not only withstand the hostile environments of the stomach and proximal small intestine but also persist in the lower GI tract make them viable for orally administered probiotics. More importantly, many Lactobacillus species have been shown to have antagonistic effects on a variety of pathogens both in vitro and in vivo (for a comprehensive review, see reference 130).
Reduced levels of Lactobacillus in the GI tract are correlated with a number of human diseases, including irritable bowel syndrome (IBS), type 1 diabetes, HIV, multiple sclerosis, colon cancer, and allergies (128, 147). It is difficult to ascertain the relative contribution of (i) disease onset that leads to an inhospitable environment for lactobacilli or (ii) Lactobacillus depletion by some other means that promotes disease onset. In either case, administration of probiotic Lactobacillus species can sometimes significantly improve symptoms (148–151). This suggests that the intrinsic metabolic activities or properties of Lactobacillus can benefit their host by modulating the microbiota and/or mucosal immune responses. For example, oral administration of L. johnsonii can protect mice in a murine model of cockroach antigen-induced allergic airway disease (4). Lactobacilli produce butyrate and other short-chain fatty acids (SCFAs) following pyruvate fermentation (152, 153), which play important roles in promoting colonic mucosal function and regulating inflammatory responses (154, 155). Importantly, butyrate isolated from Lactobacillus cultures can inhibit C. albicans hyphal morphogenesis (92). Given that lactobacilli and C. albicans are present in all regions of the human GI tract, including the low-biodiversity niches of the stomach and small intestine, Lactobacillus species may be central to preventing the outgrowth of C. albicans and other similarly resilient opportunistic pathogens.
Enterococcus Species
Microbiology.
Enterococcus species are broadly characterized as non-spore-forming, ovoid, facultative anaerobes (156). In contrast to lactobacilli, they are exclusively homofermentative and can be either harmless or pathogenic (156). Enterococci can survive under an exceptionally wide range of environmental stresses, including temperature (5 to 50°C), pH (4.5 to 10), oxygen (aerobic or anaerobic), and bile salts (up to 40% [wt/vol]) (157). Importantly, pathogenic strains possess highly plastic genomes and are able to transfer genetic material from their environment via pheromone-responsive conjugative plasmids and can therefore acquire exogenous antimicrobial resistance genes and/or other genes that might increase their virulence or propensity to cause infection (156, 157). Together, these attributes account for the broad distribution of Enterococcus species in fermented food and dairy products, aquatic and soil environments, plants, and GI tracts of mammals as well as insects and reptiles (158).
Colonization of the gastrointestinal tract.
Enterococci are core members of the human microbiome and are estimated to be 10 to 100 times more abundant than lactobacilli in the GI tract (158). Enterococcus species have been isolated from every major region of the GI tract, including the largely inhospitable stomach environs, but are predominantly found in the jejunum, ileum, cecum, and rectosigmoid junction (142). The remarkable hardiness of enterococci, combined with their genomic plasticity and ability to produce antimicrobial compounds, can impart a considerable competitive advantage to these bacteria within the microbiota. Moreover, pathogenic species such as E. faecalis can exploit their intrinsic and acquired antibiotic resistance to proliferate and dominate the GI tract in the event of an antibiotic-induced disruption of the surrounding microbiota (11, 12, 159). Given that some Enterococcus species are opportunistic pathogens that can cause serious infections (156), there is a critical need to better understand their interactions with microbes that share their niches and antagonize their growth and/or pathogenesis.
Clinical significance.
Similar to C. albicans, Enterococcus species can cause a broad range of clinical infections when the opportunity presents itself. Notably, although the genus Enterococcus is comprised of 34 species, the vast majority of infections are caused by just two species, namely, E. faecalis and Enterococcus faecium (156, 160). Their clinical presentations are similar, but their epidemiology and antimicrobial resistance patterns differ, leading to distinct clinical challenges (161). Both are common causative agents of a variety of nosocomial infections, with E. faecalis being more common in clinical infections following antibiotic regimens but having more favorable clinical outcomes relative to E. faecium (162–164). These species have attributes that facilitate their persistence in the health care environment, including tolerance to desiccation and acidic environments, and have a particular proclivity to acquire antibiotic resistance. E. faecalis is generally susceptible to ampicillin, whereas E. faecium is more frequently resistant. Both species can harbor vancomycin resistance through the acquisition of a transposon-encoded vancomycin resistance cassette (156, 160, 165).
Localized infection can result from inoculation of organisms from the gastrointestinal tract into the urinary tract, wounds, or peritoneum and may subsequently lead to systemic infection (156, 160–170). Both E. faecalis and E. faecium can cause endocarditis, and although E. faecalis is more frequently the instigator, E. faecium also presents a significant therapeutic challenge—particularly ampicillin- and vancomycin-resistant isolates (168). The microbiological attributes that promote enterococcal persistence along with their intrinsic and acquired resistance to many first-line antibiotics have led E. faecalis and E. faecium to become significant problems in critical care settings, especially for patients with hematological malignancy, extensive antibiotic exposure, and/or prolonged hospitalization (169).
INTERACTIONS BETWEEN LACTOBACILLI AND C. ALBICANS
Exopolysaccharides
Bacterial surface polysaccharides, including lipopolysaccharides (LPSs), capsular polysaccharides (CPSs), and exopolysaccharides (EPSs), are key mediators of adherence, biofilm formation, and evasion of host defenses (171). In addition to their role in pathogenesis, surface polysaccharides are thought to be involved in the probiotic activity of certain Lactobacillus strains. Lactobacillus rhamnosus GG harbors galactose-rich EPS molecules on its surface which contribute to its stable colonization in the GI tract and consequently improve its capacity to antagonize pathogens (171).
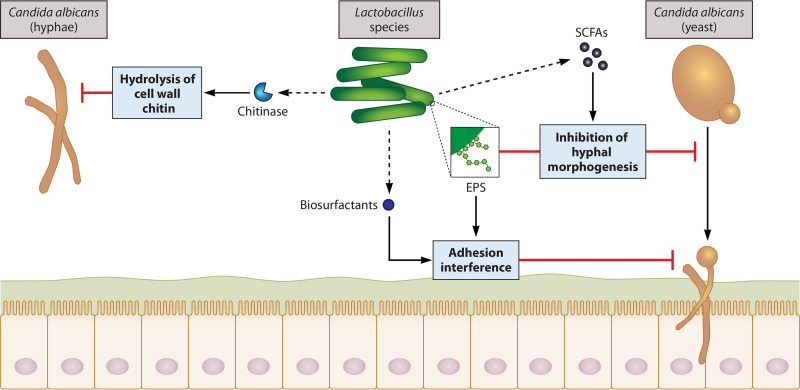
Tissue adhesion and hyphal morphogenesis are two requisite events in C. albicans pathogenesis, so many of the identified probiotic interference mechanisms involve inhibition of one or both of these processes. Specifically, the galactose-rich EPSs of L. rhamnosus GG were found to block adhesion of C. albicans to several epithelial cell lines despite low adhesion of L. rhamnosus GG itself to these cells (172). Given that C. albicans has lectin-like adhesins that can bind bacterial surface polysaccharides such as EPSs, the antiadhesive effects of L. rhamnosus GG are likely due to coaggregation with C. albicans rather than competitive exclusion. Furthermore, L. rhamnosus GG EPSs reduced the ratio of hyphae to yeast cells in a dose-dependent manner (172). Interestingly, this study found that EPS-deficient L. rhamnosus GG mutants that harbored other galactose-rich molecules were also able to inhibit hyphal morphogenesis, indicating its antihyphal activity is due to the presence of galactose-rich polymers and may not be dependent on EPSs per se. A mutant lacking SpaCBA pili was still able to reduce the number of hyphal cells, demonstrating that pili are not critical for colonization resistance against C. albicans like they are for enterococci (172, 173). Galactose-rich surface EPSs thus constitute one of the mechanisms by which Lactobacillus species can antagonize C. albicans in the GI tract (Fig. 2).
FIG 2.
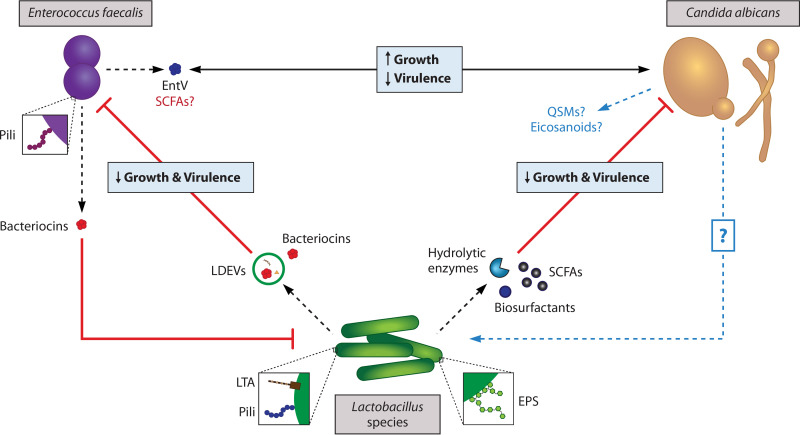
Mechanisms of interaction between Lactobacillus species and C. albicans. Lactobacilli antagonize both the growth and pathogenesis of C. albicans in the GI tract via secreted and cell surface molecules. Some Lactobacillus species secrete hydrolytic enzymes that can degrade components of the C. albicans cell wall that are critical for immune evasion and pathogenesis. Notably, L. rhamnosus GG and L. johnsonii can produce proteins with chitinase activity that hydrolyze cell wall chitin and thus reduce C. albicans growth and hyphal morphogenesis. Exopolysaccharides (EPSs) present on the cell surface of lactobacilli mediate adherence to mucosal surfaces and sustained colonization of the GI tract, which enhances their capacity to antagonize pathogens. While some forms of EPSs may prevent C. albicans biofilm formation by blocking potential mucosal binding sites, the antiadhesive effect of L. rhamnosus GG is due to coaggregation with C. albicans, as its EPS has a low affinity for epithelial cells but can bind lectin-like adhesins on C. albicans. Galactose-rich EPSs on L. rhamnosus GG are also thought to inhibit C. albicans hyphal morphogenesis, although the exact mechanism still needs to be resolved. Lactobacilli produce short-chain fatty acids (SCFAs) from the fermentation of carbohydrates, which interfere with hyphal morphogenesis and biofilm formation. Finally, some Lactobacillus species secrete amphipathic biosurfactants which block C. albicans adherence to the intestinal epithelium by altering the electrochemical properties of the host-microbial interface.
Short-Chain Fatty Acids
An essential role of lactobacilli in the gut microbiota is to metabolize partially and nondigestible carbohydrates into short-chain fatty acids (SCFAs). The three major SCFAs are butyrate, acetate, and propionate, which have a number of health-promoting effects, including maintaining the structure, integrity, and function of the intestinal epithelium; inhibiting proinflammatory immune responses; and stimulating the production of mucin (reviewed in reference 174). Crucially, many of these volatile fatty acids also exhibit antimicrobial activities against C. albicans and other pathogens (92, 175–177).
While attempting to identify specific factors in serum involved in C. albicans hyphal morphogenesis, it was observed that coculture of C. albicans with live—but not heat killed—lactobacilli significantly reduced germ tube formation, which suggested that metabolic activity is necessary for their inhibitory activity (92). Interestingly, spent culture supernatants from each of the Lactobacillus strains used in that study (L. casei, L. paracasei, and L. rhamnosus GG) also ablated C. albicans germ tube formation, indicating that this inhibitory effect is mediated by a secreted metabolic by-product. Of the three commercially available SCFAs tested, only butyrate was found to have a similar effect when added to C. albicans cultures at physiologically relevant concentrations (92). Butyrate is therefore the most potent of these SCFAs in blocking hyphal morphogenesis and may be at least partially responsible for the antihyphal activity of many Lactobacillus species in the GI tract (Fig. 2). But how exactly do these compounds interfere with hyphal morphogenesis? Given that butyrate and other SCFAs have wide-ranging effects on both epithelial and immune cells and that hyphal morphogenesis is a complex process involving many pathways, the answer to this question is likely to involve more than one mechanism.
Butyrate can act as a histone deacetylase inhibitor (HDACi) (178, 179), which is noteworthy because HDACs are thought to control the transcription of genes involved in C. albicans yeast-hyphal transitions (180). In one study, synthetic HDACis were shown to significantly reduce C. albicans germ tube formation as well as adhesion to human epithelial cells (181). Another group found that butyrate inhibited C. albicans growth and filamentation but also enhanced the production of nitric oxide by macrophages and thus their ability to kill C. albicans cells (176). While this group did not implement any experiments to directly link the effects of butyrate on C. albicans virulence to its function as a HDACi, they hypothesized that that was the likely mechanism (176). The observation that butyrate enhances the antifungal activity of macrophages is indication that SCFAs may also act indirectly by modulating the host immune response to C. albicans.
In addition to macrophages and other innate immune cells, SCFAs can influence CD4+ T cells during fungal infection. Stimulation of CD4+ T cells by dendritic cells (DCs) with a C. albicans antigen can either promote or dampen inflammatory responses, depending on the CD4+ T cell subset (182). For example, activation of Foxp3+ regulatory T cells (Tregs) triggers an anti-inflammatory response that reduces C. albicans pathology, while Th17 cell activation leads to the release of chemokines and proinflammatory cytokines (e.g., IL-17A), which augments tissue damage (182). Interestingly, although Tregs suppress Th17 responses in the GI tract, they enhance protective Th17 responses during OPC infections (183). In a mouse model of OPC, antibiotic-induced depletion of the bacterial microbiota resulted in reduced numbers of Tregs and Th17 cells, corresponding to increased fungal loads and oral pathology (177). Treatment with SCFAs reversed the pathology caused by C. albicans and concomitantly increased the numbers of Tregs and IL-17A-producing cells (177). Assuming that similar interactions occur in the GI tract, Lactobacillus-derived SCFAs may promote protective CD4+ T cell responses to C. albicans. In support of this hypothesis, Guinan and colleagues (184) showed that microbial-derived SCFAs protect against C. albicans colonization of the mouse GI tract and can inhibit processes associated with its virulence. More specifically, mice treated with cefoperazone had higher fungal loads in their cecal contents, which correlated with significantly lower levels of SCFAs. They also found that physiological concentrations of SCFAs significantly reduced C. albicans growth, biofilm formation, and hyphal morphogenesis in vitro. Together, these results underscore the importance of SCFA-producing lactobacilli in preventing C. albicans infections.
Biosurfactants
A variety of microorganisms, including lactobacilli, secrete amphipathic molecules called biosurfactants, which accumulate at interfaces and are especially important in mediating cell adherence and desorption from surfaces (185). Lactobacillus and Streptococcus species have been shown to use biosurfactants to displace uropathogenic E. faecalis from glass in parallel-plate flow chambers (186). In addition to their effects on pathogenic bacteria, Lactobacillus-derived biosurfactants can inhibit fungal adhesion and biofilm formation by altering the hydrophobicity and electrical properties of interfaces.
Interestingly, planktonic growth of a nosocomial C. albicans strain was stimulated by high concentrations of a L. brevis-derived biosurfactant, while adherent growth and biofilm formation on medical-grade surfaces have consistently been reduced by biosurfactants from a variety of lactobacilli (187, 188). This suggests that biosurfactants do not affect cell viability but rather derive their antifungal activity from their antiadhesive properties. Recently, a biosurfactant from a vaginal L. crispatus isolate was shown to inhibit C. albicans adhesion to a human epithelial cell line, demonstrating its potential functionality within the GI tract (189). Together, these data illustrate that the secretion of biosurfactants is a mechanism by which Lactobacillus species can suppress C. albicans adhesion to the epithelium and thus its pathogenesis in the GI tract (Fig. 2).
Hydrolytic Enzymes
The fungal cell wall is a unique and essential structure that not only provides protection and cell rigidity but also mediates interactions with the external environment. Antifungal drugs typically target cell wall structures such as β-glucans, mannans, and chitin because these components are not present in human cells and are capable of modulating the host immune response to promote fungal dissemination (190). Importantly, C. albicans harbors a highly transmutable cell wall that enables it to switch morphologies and transition from a commensal to a pathogen. Compared with the yeast form, hyphae are associated with elevated cell wall chitin (191), which is important for immune evasion and as well as resistance to echinocandin antifungals such as caspofungin (87). Echinocandins target the catalytic subunit of the β(1,3)-glucan synthase Fks1, leading to exposure of the underlying layer of chitin, and so resistance is most often acquired through point mutations in FKS1, as illustrated in a study by Lee and colleagues (87). In this study, C. albicans that was induced to produce excess chitin showed reduced susceptibility to caspofungin in vitro and in vivo, and some of these clones had acquired point mutations in FKS1 even without exposure to caspofungin. Thus, cell wall chitin constitutes an important C. albicans virulence factor and may be targeted by bacteria with antifungal activity.
In an effort to elucidate the mechanisms by which certain lactobacilli block C. albicans hyphal morphogenesis, an assortment of Lactobacillus strains was evaluated for their relative ability to block C. albicans hyphal morphogenesis and biofilm formation. These experiments identified the closely related species L. rhamnosus GG, L. casei, and L. paracasei as being the most potent inhibitors (192). Cell-free supernatants from L. rhamnosus GG cultures had the strongest inhibitory activity, which was attributed to the presence of lactic acid and the peptidoglycan hydrolase major secreted protein 1 (MspI). MspI has chitinase activity, suggesting that L. rhamnosus GG uses C. albicans cell wall chitin as a substrate to simultaneously enhance its own growth and block C. albicans hyphal morphogenesis. Additionally, L. rhamnosus GG MspI functions optimally at acidic pH, supporting the synergistic role of lactic acid (192).
In a more recent study, L. johnsonii and B. thetaiotaomicron, whose populations are significantly reduced during colitis and Candida overgrowth, were shown to directly interact with C. albicans and induce cell wall degradation (58). More specifically, L. johnsonii significantly reduced C. albicans growth through its chitinase-like activity, which correlated with improvements in clinical outcomes and restoration of microbial balance in a DSS-induced colitis mouse model challenged with C. albicans. Overall, these findings indicate that some Lactobacillus species produce hydrolytic enzymes that degrade chitin structures in C. albicans cell walls and thus prevent hyphal morphogenesis (Fig. 2).
INTERACTIONS BETWEEN LACTOBACILLI AND ENTEROCOCCUS FAECALIS
Mucus-Binding Pili
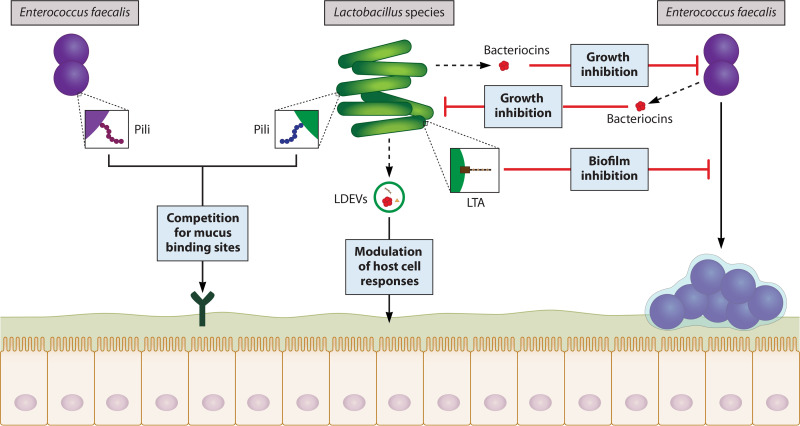
Broadly speaking, a microbial strain is considered competitive if its activity reduces the fitness of another strain with which it shares resources. Competition between two ecologically stable microbes can result in either domination of one strain, establishment of separate metabolic niches (i.e., a divergence in resource requirements), or separation into distinct “territorial niches” from an originally mixed population (193). Given that E. faecalis establishes invasive infection by forming biofilms (wherein adherence to mucus is a necessary first step) and lactobacilli are known to have mucus-binding structures on their surface (194, 195), competition for mucus-binding sites is a potential means by which certain Lactobacillus strains inhibit E. faecalis pathogenesis.
This possibility was substantiated by a study in which a vancomycin-resistant Enterococcus (VRE) strain of E. faecium was observed to have increased mucus-binding capacity compared with nonpathogenic enterococci (173). Moreover, the VRE isolate had undergone mutations such that its pili more closely resembled the mucus-binding SpaCBA pili of L. rhamnosus GG, presumably improving its ability to adhere. In the absence of L. rhamnosus GG cells, E. faecium adherence was significantly reduced by antibodies raised against the mucus-binding domain of SpaCBA, suggesting cross-reactivity between the mucus-binding pili of the two strains. Based on these observations, lactobacilli may normally outcompete enterococci for adherence to the mucus layer and thus prevent biofilm formation and subsequent pathogenesis.
In agreement with these results, administration of L. rhamnosus GG to VRE-positive patients has been demonstrated to decrease and even clear VRE colonization in several randomized trials (196, 197). While it should be noted that E. faecium and E. faecalis strains of VRE have distinct clinical features and likely interact differently with lactobacilli in the GI tract, as members of the genus Enterococcus, they have similar requirements for pathogenesis (198). First and foremost, they must bind the mucosal layer in sufficient numbers to form a biofilm whereby they become more resistant to host and microbial defense mechanisms. Competitive exclusion via mucus-binding pili is therefore predicted to be an important strategy used by certain strains of Lactobacillus to inhibit E. faecalis biofilm formation as well (Fig. 3).
FIG 3.
Mechanisms of interaction between Lactobacillus species and E. faecalis. Lactobacilli and enterococci compete with one another for binding sites along the GI tract via mucus-binding pili on their cell surface. Lactobacillus species generally have high-affinity mucus-binding pili and thereby block E. faecalis adherence. Initial adherence to host cells is required for the establishment of biofilms, which are associated with enhanced resistance to antimicrobials and host immune responses; thus, competitive exclusion via surface pili can either dampen or facilitate E. faecalis pathogenesis, depending on their relative affinity for binding sites. Lipoteichoic acid (LTA) structures found in the cell wall of Lactobacillus species are also capable of antagonizing E. faecalis biofilm formation and are speculated to achieve this effect by regulating the expression of E. faecalis quorum-sensing molecules (QSMs) critical to biofilm formation and integrity. Both lactobacilli and E. faecalis produce bacteriocins with antimicrobial activity against the other. The bacteriocin CRL 1238 secreted by vaginal and intestinal L. salivarius strains reduces the growth of E. faecalis, while the E. faecalis-derived bacteriocins enterocin 1071 and enterolysin A target Lactobacillus species. Bacteriocins can be secreted on their own or within extracellular vesicles, which offer protection from proteolytic enzymes and can deliver their cargo to more distal sites without triggering an adverse immune response. More specifically, Lactobacillus-derived extracellular vesicles (LDEVs) from L. acidophilus and L. plantarum have been shown to reduce the growth and virulence of E. faecalis by delivering bacteriocins to E. faecalis and inducing an upregulation of host defense genes.
Lipoteichoic Acids
Lipoteichoic acid (LTA) is an important cell wall component unique to Gram-positive bacteria, including lactobacilli and enterococci. There are five different types of LTA which vary in structure and elicit different immune responses following recognition by Toll-like receptors (TLRs) (199). For example, LTA from Lactobacillus plantarum weakly stimulates TLR2 and thus poorly induces inflammatory mediators, while LTA from pathogenic E. faecalis acts as a virulence factor and mediates biofilm formation in addition to triggering production of proinflammatory cytokines (200, 201). A study investigating the effects of L. plantarum LTA (LpLTA) on the Gram-positive pathogen Staphylococcus aureus found that LpLTA interferes with S. aureus biofilm formation by inhibiting production of the QSM autoinducer-2 (AI-2). As a result, S. aureus had reduced expression of intracellular adhesion (ica) locus genes which are essential for synthesis of the major exopolysaccharide component of biofilms (202). Lactobacillus-derived LTA is anticipated to have a similar effect on E. faecalis biofilms.
Recently, LTAs from several Lactobacillus species, including L. plantarum, L. acidophilus, L. casei, and L. rhamnosus GG, were found to inhibit E. faecalis biofilm formation and even disrupt preformed biofilms (203, 204). Interestingly, although crystal violet staining showed dose-dependent decreases in adherent E. faecalis cells after incubation with LTA, the optical density at 600 nm was unchanged. This result implies that Lactobacillus LTA does not interfere with E. faecalis planktonic growth—only its ability to form biofilms (Fig. 3). Scanning electron microscopy revealed that LpLTA could reduce the size of 3-week-old E. faecalis biofilms and also enhance antibiotic-induced biofilm disruption (204). Based on the LpLTA and S. aureus data (202), it is likely that LTAs similarly regulate expression of E. faecalis QS molecules, such as sortase A and enterococcal surface protein (Esp), which are known to be important for its biofilm formation and integrity (205, 206).
Lactobacillus-Derived Bacteriocins
Bacteriocins are a diverse group of antimicrobial peptides that are produced by bacteria and can provide them with a competitive advantage (207). The rising concern of antibiotic resistance has stimulated an interest in bacteriocins as an alternative and/or supplement to antibiotics, with the notion that physical interference by intact cells might not be required for decolonization of competing bacteria. In particular, many Lactobacillus strains are capable of generating a variety of bactericidal agents, including bacteriocins, hydrogen peroxide, and organic acids (208), which could represent interaction mechanisms used by commensal lactobacilli to inhibit pathogens such as E. faecalis.
In one study, the in vitro antagonistic activity of several Lactobacillus strains isolated from curd samples was tested against some common human pathogens (209). Of these lactobacilli, isolates identified as L. fermentum had the most potent activity against E. faecalis. Treatment of the bacterial supernatants with protease led to a loss of inhibitory activity, but growth inhibition was still observed when supernatants were treated with catalase and organic acid-neutralizing agents, ruling out hydrogen peroxide and organic acids as the relevant antimicrobial products. Although the Lactobacillus-derived bacteriocins themselves were not characterized, this study identifies several environmental strains of Lactobacillus that produce bacteriocins with inhibitory activity against E. faecalis.
Probiotic Lactobacillus strains from environmental samples have been the focus of most bacteriocin-related research to date. However, in order to understand the interactions of indigenous lactobacilli with other microbes in the GI tract, it is important to also consider strains derived from mucosal sites. Notably, L. salivarius isolated from the human vagina and GI tract have also been shown to antagonize major pathogens, including E. faecalis, through the production of several bacteriocins. Of these bacteriocins, CRL 1238 has been shown to specifically inhibit E. faecalis, while Abp118 is active against other Gram-positive pathogens such as Listeria monocytogenes (210–212). It remains to be determined whether Abp118 targets E. faecalis as well. Taken together, these data indicate that bacteriocins from many lactobacilli—either environmental or stable colonizers in the human microbiota—can antagonize E. faecalis (Fig. 3).
Enterococcus-Derived Bacteriocins
E. faecalis and other enterococci use pheromone-responsive plasmids (PRPs) to transfer virulence, antibiotic resistance, and bacteriocin genes to other enterococcal cells (213). PRPs facilitate inter- and intraspecies gene transfer, making them likely contributors to the widespread antibiotic resistance observed in E. faecalis and E. faecium strains. The ability to transfer bacteriocin genes to other enterococci may be a mechanism by which E. faecalis can outcompete lactobacilli and other niche competitors.
pPD1 is an E. faecalis PRP system that carries the bacteriocin determinant bac21 (214). Notably, E. faecalis strains harboring pPD1 have resistance to bacteriocin 21 (Bac-21) and are able to outcompete closely related enterococci lacking pPD1 or lacking functional bac21 within this plasmid in the mouse GI tract (215). Conjugative plasmid-mediated transfer of pPD1 confers resistance to Bac-21 in addition to spreading its genetic material and thus the ability to produce Bac-21. As a result, E. faecalis strains lacking pPD1 are sensitive to Bac-21 and displaced from the GI tract. Although Bac-21 was not found to significantly affect levels of lactobacilli in the mouse GI tract, other E. faecalis bacteriocins, including plasmid-encoded enterocin 1071 and chromosome-encoded enteroylsin, are active against Lactobacillus species (216, 217). E. faecalis-derived bacteriocins can therefore impart a competitive advantage over lactobacilli and other enterococci, which likely promotes E. faecalis pathogenesis (Fig. 3).
Extracellular Vesicles
Bacterial extracellular vesicles (EVs) are membrane-bound structures that can enclose a variety of proteins, lipids, and nucleic acids and are capable of reshaping the microbial environment. Importantly, they can traverse and be taken up by epithelial cells and thus deliver their cargo to host cells in distal sites to modulate host physiology (218). More specifically, Lactobacillus-derived EVs (LDEVs) suppress proinflammatory signaling and enhance expression of tight junction proteins (219) (Fig. 3). Supporting this notion, EVs isolated from gut-associated L. plantarum were found to prolong the survival of C. elegans worms previously infected with a vancomycin-resistant strain of E. faecium (220). While this effect was attributed solely to upregulation of host defense genes in both C. elegans and mammalian cells, it is also possible that EVs from this and/or other Lactobacillus strains carry bacteriocins to pathogenic enterococci, leading to enhanced protection from infection.
Recently, L. acidophilus-derived EVs were found to contain bacteriocins and deliver them to niche competitors by fusing with target cell membranes (221). Considering that the use of bacteriocins as therapeutics for antibiotic-resistant infections has been hampered by a lack of reliable delivery methods, EVs could be a promising solution in that they offer protection from proteolytic enzymes and may not induce adverse immune reactions. Furthermore, bacteriocins derived from lactobacilli have been shown to specifically antagonize E. faecalis (209, 212), so although their transportation has not explicitly been characterized, LDEVs are a conceivable mechanism by which E. faecalis-targeting bacteriocins can be delivered (Fig. 3).
INTERACTIONS BETWEEN E. FAECALIS AND C. ALBICANS
Despite them being prominent nosocomial pathogens, the interplay between C. albicans and E. faecalis in the GI tract is vastly understudied. Consequently, with the exception of a few studies, the mechanisms underlying their interactions have not been explored, much less elucidated. Here, we report what is currently understood on this topic but would like to emphasize that this lack of knowledge should be considered an opportunity for future research, as C. albicans and E. faecalis each pose significant threats to human health.
Induction of White-Opaque Regulatory Switch
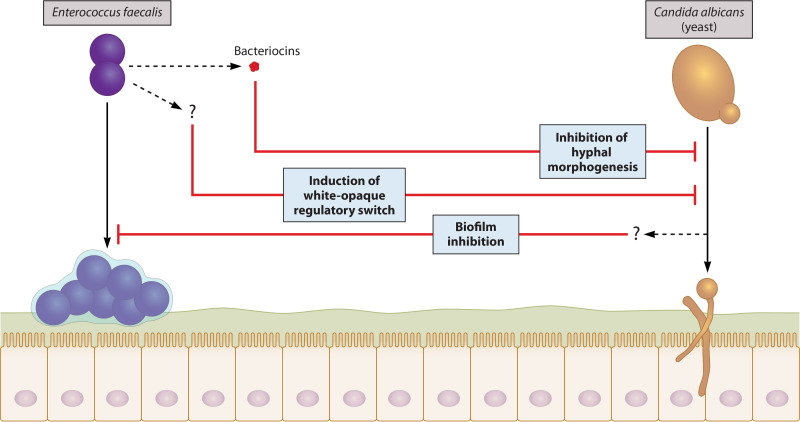
Interactions between E. faecalis and C. albicans are especially significant due to the fact that they are both opportunistic pathogens that occupy the same niches and are frequently coisolated from polymicrobial infections. C. albicans and E. faecalis form mixed-species biofilms in vitro, making it possible to study their interaction mechanisms. The presence of E. faecalis can significantly reduce overall biofilm thickness without affecting the growth of E. faecalis itself, suggesting that E. faecalis antagonizes C. albicans biofilm formation (44). Gene expression analysis showed a strong upregulation of the white-opaque master regulator WOR1 in C. albicans cells. The opaque phenotype is less prone to hyphal morphogenesis than the white phenotype under physiological conditions encountered in the GI tract and is therefore generally considered less virulent (222). However, other genes involved in the white-opaque regulatory switch (i.e., WOR2, WOR3, and CZF1) were not upregulated, indicating that E. faecalis only partially induces this switch and is therefore more likely repressing white cell formation than promoting a switch to opaque. Nevertheless, these results demonstrate that E. faecalis can directly interact with C. albicans in biofilms and alter its gene expression to inhibit pathogenesis (Fig. 4).
FIG 4.
Mechanisms of interaction between E. faecalis and C. albicans. Interestingly, these two opportunistic pathogens enhance each other’s growth but dampen each other’s virulence in vivo. The mechanisms underlying this unusual interplay are largely unknown and require additional research—particularly in regard to how C. albicans interacts with E. faecalis. E. faecalis induces the upregulation of the white-opaque master regulator WOR1 in C. albicans. Notably, other genes involved in the white-opaque regulatory switch are not upregulated by E. faecalis, but even partial induction of the less virulent opaque phenotype leads to a significant reduction in C. albicans biofilm thickness. The specific signals and pathways involved in this outcome have yet to be determined. A better characterized interaction mechanism involves the production of a bacteriocin by E. faecalis. E. faecalis uses its Fsr quorum-sensing system not only to regulate its own virulence but also to sense C. albicans and release the bacteriocin EntV, which inhibits C. albicans hyphal morphogenesis and biofilm formation. It is unclear whether EntV itself is also responsible for the observed reduction in E. faecalis virulence or if C. albicans produces molecules that suppress E. faecalis biofilm formation, either directly or by modulating other cells’ activities.
Enterococcus-Derived Bacteriocins
E. faecalis virulence is regulated by its Fsr QS system. In this system, three genes (fsrA, fsrB, and fsrC) respond to extracellular accumulation of a peptide lactone by producing the proteases gelatinase (GelE) and serine protease (SprE), whose enzymatic activity is required for E. faecalis biofilm formation (223). Surprisingly, coinfection with clinical strains of C. albicans and E. faecalis reduces killing of C. elegans nematodes compared with infection with one or the other microbe, indicating that C. albicans also suppresses E. faecalis virulence in vivo (8). In addition to inhibiting C. albicans biofilm formation, wild-type E. faecalis inhibited hyphal morphogenesis, while mutants lacking functional GelE or SprE did not, confirming the involvement of the Fsr QS system in the interactions of these microbes (8). It was later revealed that E. faecalis secretes a bacteriocin called EntV, which is responsible for its inhibition of C. albicans hyphal morphogenesis and biofilm formation and that GelE was required for processing into its active form (9, 224). Thus, E. faecalis uses the Fsr system to sense C. albicans and produce the bacteriocin EntV, which antagonizes C. albicans biofilm formation and filamentation (Fig. 4).
INSIGHTS FROM OTHER C. ALBICANS INTERACTING PARTNERS
Mechanistic studies looking at the interactions between C. albicans and other bacteria may provide important framework for future research on C. albicans-Lactobacillus and C. albicans-E. faecalis interaction mechanisms—especially for potential experiments involving targeted gene expression analysis. For example, knowing that QSMs, such as farnesol, and lipid mediators, such as prostaglandins, are involved in some of these interkingdom interactions (225–229), it would be interesting to study the effects of those molecules on E. faecalis and Lactobacillus species as well as on host cells.
Quorum-Sensing Molecules and Prostaglandins
Interactions with Pseudomonas aeruginosa.
In addition to modulating phenotypic switching in response to cell density, the QSM farnesol promotes C. albicans survival in the presence of Pseudomonas aeruginosa. Like E. faecalis, P. aeruginosa is an opportunistic bacterial pathogen that is commonly found in mixed-species infections with C. albicans—particularly in the lungs of cystic fibrosis patients (230). However, unlike E. faecalis, it can antagonize the overall growth of C. albicans in addition to its virulence (231, 232). Specifically, P. aeruginosa uses an N-acyl homoserine lactone (AHL)-dependent QS system to attach to and kill C. albicans hyphae (225). 3-Oxododecanoyl-homoserine lactone (3-oxo-HSL) is a QSM produced by this system that mediates P. aeruginosa adherence to C. albicans hyphae by triggering the synthesis of surface adherence proteins (225). Once adhered, redox-active phenazines also produced by the QS system can directly kill C. albicans cells by generating reactive oxygen species (ROS) and reducing cAMP levels (225). In doing so, P. aeruginosa undermines C. albicans cell wall integrity and blocks hyphal morphogenesis. However, farnesol produced by C. albicans prevents the formation of hyphae and thereby protects against killing by P. aeruginosa (225, 233). During infection, both release arachidonic acid, which they can transform into eicosanoids (234). Farnesol also promotes the production of phenazines by P. aeruginosa, which are toxic to a variety of bacteria and may therefore enable C. albicans to indirectly outcompete other bacteria (229). The production of farnesol is also important for C. albicans dissemination, as it has been shown to promote the recruitment of circulating macrophages, which C. albicans can kill after being engulfed and transported into the bloodstream (229, 235).
While it remains to be determined whether such QSMs play a role in its interactions with E. faecalis and/or Lactobacillus species, QS systems are potential mechanisms by which C. albicans, E. faecalis, and lactobacilli adapt to and antagonize one another. It is likely that QSMs are at least partially responsible for the reduced virulence of C. albicans in the presence of these bacteria, as well as the reduced virulence of E. faecalis in the presence of C. albicans (Fig. 5).
FIG 5.
Model of the interplay between C. albicans, E. faecalis, and various Lactobacillus species in the human GI tract. Lactobacilli use a combination of surface and secreted molecules to reduce the growth and virulence of both C. albicans and E. faecalis. In turn, E. faecalis can antagonize lactobacilli via competitive exclusion and/or Lactobacillus-targeting bacteriocins. E. faecalis and C. albicans promote each other’s growth but reduce each other’s virulence. The bacteriocin EntV produced via the E. faecalis Fsr QS system is the only identified molecule mediating this interaction; however, E. faecalis has also been shown to suppress the C. albicans white cell phenotype, which is more virulent under physiological conditions. Additionally, given that SCFAs produced by lactobacilli have been shown to reduce C. albicans hyphal morphogenesis, it is possible that E. faecalis-derived SCFAs have a similar effect on C. albicans. The overall impact of C. albicans on the growth of lactobacilli, as well as the molecules and pathways involved in its interactions with E. faecalis and Lactobacillus species, have not yet been elucidated. Studies on the interactions of C. albicans with P. aeruginosa, S. gordonii, and S. aureus lead us to postulate that C. albicans QSMs may underlie its ability to suppress E. faecalis virulence. Another possibility is the production of immunomodulatory eicosanoids such as PGE2, which, by reshaping the immunological landscape, may help C. albicans evade the antagonistic effects of LAB and create a more conducive environment for its own growth as well as the growth of E. faecalis.
Interactions with Streptococcus gordonii.
QSMs are also implicated in the interplay between C. albicans and S. gordonii—another bacterium commonly found in C. albicans biofilms (229). The physical interaction of these microbes is mediated by the antigen I/II (AgI/II) polypeptides SspA and SspB expressed on the cell wall of S. gordonii. Following bacterial attachment to the mucosa, C. albicans binds these polypeptides and coaggregates with S. gordonii, enabling proximal cell-to-cell communication via secreted signals (236). These two microbes ultimately enhance each other’s persistence through mutualistic interactions; metabolic by-products released by S. gordonii can be used as a carbon source by C. albicans, and in turn, C. albicans biofilms provide an anoxic niche and nutrient source for S. gordonii (229, 236). Unlike E. faecalis, however, S. gordonii also promotes C. albicans virulence by abrogating farnesol-mediated inhibition of hyphal morphogenesis, likely via the protein kinase A (PKA) pathway (236).
Interactions with Staphylococcus aureus.
Similar to S. gordonii, the presence of S. aureus in C. albicans biofilms portends increased mortality compared with monoinfection with either microbe but can also enhance the resistance of both species to antimicrobial agents (229). In particular, farnesol secreted by C. albicans has been shown to increase the tolerance of S. aureus to the antibiotic vancomycin by inducing upregulation of efflux pump genes (227), although higher concentrations of farnesol may actually reduce S. aureus viability and biofilm formation (226). Furthermore, repeated exposure to farnesol results in the loss of staphyloxanthin, an important S. aureus virulence factor (237). However, other studies have noted that mixed-species infections involving these two microbes are associated with higher mortality rates due to induction of the agr QS system in S. aureus, which leads to increased hemolytic activity as well as production of alpha and delta toxin (238). In any case, interactions between C. albicans and S. aureus can dramatically alter the growth and virulence of both species in a host. A significant increase in PGE2 release by C. albicans and host cells has been observed in mixed-species infections involving C. albicans and S. aureus, with PGE2 promoting S. aureus biofilm growth in a dose-dependent manner (228).
Given these observations, it is conceivable that farnesol and C. albicans-derived PGE2 alter the activity of E. faecalis and Lactobacillus species either directly or by altering the immunological landscape (Fig. 5), although this possibility has not yet been investigated.
CONCLUDING REMARKS
C. albicans and E. faecalis are two microbiologically and clinically distinct microbes that are both common gastrointestinal commensals and opportunistic pathogens (i.e., “pathobionts” [239, 240]) but have emerged as important clinical challenges in the modern acute care setting. Despite their clinical co-occurrence, their biological interactions are understudied and represent significant knowledge gaps. These gaps are unfortunate as both C. albicans and E. faecalis are poised to become yet more dangerous with invasive medical procedures becoming increasingly common. Additionally, both taxa are precariously close to antibiotic resistance crises with few options for each. Thus, an understanding of the factors that facilitate the colonization and persistence of these organisms provides an opportunity to intervene before they cause harm. Such is the potential role of various Lactobacillus species, whose colonization of mucosal surfaces antagonizes these pathobionts and augments protective mucosal immunity.
ACKNOWLEDGMENTS
The following funding sources have provided support for this project: NIH/NIAID R01AI138348 (G.B.H. and K.D.Z.) and The Nina and Jerry D. Luptak Endowment of The Mary H. Weiser Food Allergy Center at the University of Michigan (G.B.H. and K.D.Z.).
The authors have no conflicts of interest to declare.
Biographies

Karen D. Zeise is a Ph.D. candidate in the Department of Microbiology & Immunology at the University of Michigan. She received her B.S. in biology from Northeastern University in 2019. While pursuing her undergraduate degree, she held internships in the pharmacology departments at Sanofi Genzyme (Framingham, MA) and AbbVie Foundational Neuroscience Center (Cambridge, MA), as well as in in the Laboratory of Glia Biology at VIB-KU Leuven Center for Brain & Disease Research (Leuven, Belgium). She became interested in microbiome research while taking microbiology courses at Northeastern University and hopes to eventually work in industry, where her research can have a direct clinical impact. As a second-year graduate student in Huffnagle’s lab, she studies yeast-bacterial interactions in the GI tract and their impact on allergic disease.

Robert J. Woods is an Infectious Diseases Physician at the University of Michigan. Woods earned his B.S. and Ph.D. in zoology from Michigan State University and M.D. from the University of Chicago. He subsequently completed his postgraduate medical training in internal medicine and infectious diseases at the University of Michigan, where he is currently appointed Assistant Professor of Medicine. The focus of his research is the evolutionary dynamics of bacterial and viral pathogens. Current research projects largely focus on the transmission and antibiotic resistance evolution of enterococci in the acute care setting.

Gary B. Huffnagle is a Professor in the Department of Molecular Cellular & Developmental Biology and holds the Nina and Jerry D. Luptak Professorship in the Mary H. Weiser Food Allergy Center at the University of Michigan. He also holds joint faculty appointments in microbiology & immunology and pulmonary medicine at the University of Michigan. He earned his Ph.D. in immunology from the University of Texas Southwestern Medical School, received a Burroughs-Wellcome Award in pathogenic mycology, and was elected to the American Academy of Microbiology of the American Society for Microbiology. He teaches a series of courses in microbiology, immunology, and allergic diseases to students ranging from undergraduate sophomores to graduate students. His laboratory’s research interests include the role of microbiome-host interactions in disease and mucosal immunity, and his experience includes over three decades devoted to NIH-supported research in fungal-host and microbiome-host interactions in the lungs and gastrointestinal tract.
REFERENCES
- 1.Ethridge AD, Bazzi MH, Lukacs NW, Huffnagle GB. 2020. Interkingdom communication and regulation of mucosal immunity by the microbiome. J Infect Dis 223:S236–S240. 10.1093/infdis/jiaa748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, Rakoff-Nahoum S. 2021. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 591:633–638. 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV. 2016. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22:1187–1191. 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, Boushey HA, Zoratti E, Ownby D, Lukacs NW, Lynch SV. 2014. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A 111:805–810. 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. 2004. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72:4996–5003. 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. 2005. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun 73:30–38. 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann C, Hermann J, Munzel U, Rüchel R. 1999. Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 42:619–627. 10.1046/j.1439-0507.1999.00519.x. [DOI] [PubMed] [Google Scholar]
- 8.Cruz MR, Graham CE, Gagliano BC, Lorenz MC, Garsin DA. 2013. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun 81:189–200. 10.1128/IAI.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham CE, Cruz MR, Garsin DA, Lorenz MC. 2017. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc Natl Acad Sci U S A 114:4507–4512. 10.1073/pnas.1620432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB. 2013. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep 3:2191. 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason KL, Erb Downward JR, Falkowski NR, Young VB, Kao JY, Huffnagle GB. 2012. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect Immun 80:150–158. 10.1128/IAI.05162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. 2012. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 80:3371–3380. 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggimann P, Pittet D. 2014. Candida colonization index and subsequent infection in critically ill surgical patients: 20 years later. Intensive Care Med 40:1429–1448. 10.1007/s00134-014-3355-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Centre for Disease Prevention and Control. 2016. Healthcare-associated infections in intensive care units—Annual Epidemiological Report for 2016. https://www.ecdc.europa.eu/en/publications-data/healthcare-associated-infections-intensive-care-units-annual-epidemiological-0.
- 15.System N. 1999. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1990-May 1999, issued June 1999. A report from the NNIS System. Am J Infect Control 27:520–532. 10.1016/s0196-6553(99)70031-3. [DOI] [PubMed] [Google Scholar]
- 16.Lin JN, Lai CH, Chen YH, Chang LL, Lu PL, Tsai SS, Lin HL, Lin HH. 2010. Characteristics and outcomes of polymicrobial bloodstream infections in the emergency department: a matched case-control study. Acad Emerg Med 17:1072–1079. 10.1111/j.1553-2712.2010.00871.x. [DOI] [PubMed] [Google Scholar]
- 17.Klotz SA, Chasin BS, Powell B, Gaur NK, Lipke PN. 2007. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn Microbiol Infect Dis 59:401–406. 10.1016/j.diagmicrobio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Brandt ME, Lockhart SR. 2012. Recent taxonomic developments with Candida and other opportunistic yeasts. Curr Fungal Infect Rep 6:170–177. 10.1007/s12281-012-0094-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whiteway M, Bachewich C. 2007. Morphogenesis in Candida albicans. Annu Rev Microbiol 61:529–553. 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. 2011. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10:112–122. 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gow NA, Hube B. 2012. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol 15:406–412. 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Soll DR. 1992. High-frequency switching in Candida albicans. Clin Microbiol Rev 5:183–203. 10.1128/CMR.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 48:1695–1703. 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- 24.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Candida Auris Incident Management T, Manuel R, Brown CS, Candida auris Incident Management Team. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029-17. 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarma S, Upadhyay S. 2017. Current perspective on emergence, diagnosis and drug resistance in Candida auris. Infect Drug Resist 10:155–165. 10.2147/IDR.S116229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dadar M, Tiwari R, Karthik K, Chakraborty S, Shahali Y, Dhama K. 2018. Candida albicans—biology, molecular characterization, pathogenicity, and advances in diagnosis and control—an update. Microb Pathog 117:128–138. 10.1016/j.micpath.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 27.Neville BA, d'Enfert C, Bougnoux ME. 2015. Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res 15:fov081. 10.1093/femsyr/fov081. [DOI] [PubMed] [Google Scholar]
- 28.Mallick EM, Bergeron AC, Jones SK, Jr., Newman ZR, Brothers KM, Creton R, Wheeler RT, Bennett RJ. 2016. Phenotypic plasticity regulates Candida albicans interactions and virulence in the vertebrate host. Front Microbiol 7:780. 10.3389/fmicb.2016.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pande K, Chen C, Noble SM. 2013. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet 45:1088–1091. 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soll DR. 2014. The role of phenotypic switching in the basic biology and pathogenesis of Candida albicans. J Oral Microbiol 6:10.3402/jom.v6.22993. 10.3402/jom.v6.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallick EM, Bennett RJ. 2013. Sensing of the microbial neighborhood by Candida albicans. PLoS Pathog 9:e1003661. 10.1371/journal.ppat.1003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaffin WL, López-Ribot JL, Casanova M, Gozalbo D, Martinez JP. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev 62:130–180. 10.1128/MMBR.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuoka J. 2004. Surface glycans of Candida albicans and other pathogenic fungi: physiological roles, clinical uses, and experimental challenges. Clin Microbiol Rev 17:281–310. 10.1128/CMR.17.2.281-310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ene IV, Brunke S, Brown AJ, Hube B. 2014. Metabolism in fungal pathogenesis. Cold Spring Harb Perspect Med 4:a019695. 10.1101/cshperspect.a019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgain A, Tebbji F, Khemiri I, Sellam A. 2020. Metabolic reprogramming in the opportunistic yeast Candida albicans in response to hypoxia. mSphere 5:e00913-19. 10.1128/mSphere.00913-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428, table of contents. 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E, Weindl G, Tappuni AR, Rodgers CA, Woodman AJ, Challacombe SJ, Schaller M, Hube B. 2008. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology (Reading) 154:3266–3280. 10.1099/mic.0.2008/022293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naglik JR, Rodgers CA, Shirlaw PJ, Dobbie JL, Fernandes-Naglik LL, Greenspan D, Agabian N, Challacombe SJ. 2003. Differential expression of Candida albicans secreted aspartyl proteinase and phospholipase B genes in humans correlates with active oral and vaginal infections. J Infect Dis 188:469–479. 10.1086/376536. [DOI] [PubMed] [Google Scholar]
- 39.Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miramon P, Dunker C, Windecker H, Bohovych IM, Brown AJ, Kurzai O, Hube B. 2012. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS One 7:e52850. 10.1371/journal.pone.0052850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miramon P, Kasper L, Hube B. 2013. Thriving within the host: Candida spp. interactions with phagocytic cells. Med Microbiol Immunol 202:183–195. 10.1007/s00430-013-0288-z. [DOI] [PubMed] [Google Scholar]
- 42.Ene IV, Lohse MB, Vladu AV, Morschhauser J, Johnson AD, Bennett RJ. 2016. Phenotypic profiling reveals that candida albicans opaque cells represent a metabolically specialized cell state compared to default white cells. mBio 7:e01269-16. 10.1128/mBio.01269-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng L, Kelly CJ, Colgan SP. 2015. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol 309:C350–C360. 10.1152/ajpcell.00191.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD. 2014. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol 24:2411–2416. 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopes JP, Stylianou M, Backman E, Holmberg S, Jass J, Claesson R, Urban CF. 2018. Evasion of immune surveillance in low oxygen environments enhances Candida albicans virulence. mBio 9:e02120-18. 10.1128/mBio.02120-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pradhan A, Avelar GM, Bain JM, Childers DS, Larcombe DE, Netea MG, Shekhova E, Munro CA, Brown GD, Erwig LP, Gow NAR, Brown AJP. 2018. Hypoxia promotes immune evasion by triggering beta-glucan masking on the Candida albicans cell surface via mitochondrial and cAMP-protein kinase A signaling. mBio 9:e01318-18. 10.1128/mBio.01318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huffnagle GB, Noverr MC. 2013. The emerging world of the fungal microbiome. Trends Microbiol 21:334–341. 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF. 2017. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5:153. 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angebault C, Djossou F, Abélanet S, Permal E, Ben Soltana M, Diancourt L, Bouchier C, Woerther P-L, Catzeflis F, Andremont A, d'Enfert C, Bougnoux M-E. 2013. Candida albicans is not always the preferential yeast colonizing humans: a study in Wayampi Amerindians. J Infect Dis 208:1705–1716. 10.1093/infdis/jit389. [DOI] [PubMed] [Google Scholar]
- 51.Parmeland L, Gazon M, Guerin C, Argaud L, Lehot JJ, Bastien O, Allaouchiche B, Michallet M, Picot S, Bienvenu AL, Study Group. 2013. Candida albicans and non-Candida albicans fungemia in an institutional hospital during a decade. Med Mycol 51:33–37. 10.3109/13693786.2012.686673. [DOI] [PubMed] [Google Scholar]
- 52.Soll DR, Galask R, Schmid J, Hanna C, Mac K, Morrow B. 1991. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J Clin Microbiol 29:1702–1710. 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, Auchtung JM, Ajami NJ, Petrosino JF. 2018. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 3:e00092-18. 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. 2013. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eckstein MT, Moreno-Velasquez SD, Perez JC. 2020. Gut bacteria shape intestinal microhabitats occupied by the fungus Candida albicans. Curr Biol 30:4799–4807.e4. 10.1016/j.cub.2020.09.027. [DOI] [PubMed] [Google Scholar]
- 57.Koh AY. 2013. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot Cell 12:1416–1422. 10.1128/EC.00196-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Charlet R, Bortolus C, Sendid B, Jawhara S. 2020. Bacteroides thetaiotaomicron and Lactobacillus johnsonii modulate intestinal inflammation and eliminate fungi via enzymatic hydrolysis of the fungal cell wall. Sci Rep 10:11510. 10.1038/s41598-020-68214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prieto D, Pla J. 2015. Distinct stages during colonization of the mouse gastrointestinal tract by Candida albicans. Front Microbiol 6:792. 10.3389/fmicb.2015.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Repentigny L, Phaneuf M, Mathieu LG. 1992. Gastrointestinal colonization and systemic dissemination by Candida albicans and Candida tropicalis in intact and immunocompromised mice. Infect Immun 60:4907–4914. 10.1128/iai.60.11.4907-4914.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prieto D, Román E, Correia I, Pla J. 2014. The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS One 9:e87128. 10.1371/journal.pone.0087128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrutis KA, Riggle PJ, Kumamoto CA, Tzipori S. 2000. Intestinal lesions associated with disseminated candidiasis in an experimental animal model. J Clin Microbiol 38:2317–2323. 10.1128/.38.6.2317-2323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoeflinger JL, Coleman DA, Oh SH, Miller MJ, Hoyer LL. 2014. A piglet model for studying Candida albicans colonization of the human oro-gastrointestinal tract. FEMS Microbiol Lett 357:10–15. 10.1111/1574-6968.12500. [DOI] [PubMed] [Google Scholar]
- 64.Achkar JM, Fries BC. 2010. Candida infections of the genitourinary tract. Clin Microbiol Rev 23:253–273. 10.1128/CMR.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Repentigny L, Lewandowski D, Jolicoeur P. 2004. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin Microbiol Rev 17:729–759, table of contents. 10.1128/CMR.17.4.729-759.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janus MM, Willems HM, Krom BP. 2016. Candida albicans in multispecies oral communities; a keystone commensal? Adv Exp Med Biol 931:13–20. 10.1007/5584_2016_5. [DOI] [PubMed] [Google Scholar]
- 67.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713. 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barousse MM, Van Der Pol BJ, Fortenberry D, Orr D, Fidel PL, Jr.. 2004. Vaginal yeast colonisation, prevalence of vaginitis, and associated local immunity in adolescents. Sex Transm Infect 80:48–53. 10.1136/sti.2002.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fidel PL, Jr. 2002. Distinct protective host defenses against oral and vaginal candidiasis. Med Mycol 40:359–375. 10.1080/714031126. [DOI] [PubMed] [Google Scholar]
- 70.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108:4680–4687. 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chee WJY, Chew SY, Than LTL. 2020. Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microb Cell Fact 19:203. 10.1186/s12934-020-01464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Logan C, Martin-Loeches I, Bicanic T. 2020. Invasive candidiasis in critical care: challenges and future directions. Intensive Care Med 46:2001–2014. 10.1007/s00134-020-06240-x. [DOI] [PubMed] [Google Scholar]
- 73.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a Prospective Nationwide Surveillance Study. Clin Infect Dis 39:309–317. 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 75.Ricotta EE, Lai YL, Babiker A, Strich JR, Kadri SS, Lionakis MS, Prevots DR, Adjemian J. 2021. Invasive candidiasis species distribution and trends, United States, 2009–2017. J Infect Dis 223:1295–1302. 10.1093/infdis/jiaa502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wenzel RP, Gennings C. 2005. Bloodstream infections due to Candida species in the intensive care unit: identifying especially high-risk patients to determine prevention strategies. Clin Infect Dis 41:S389–S393. 10.1086/430923. [DOI] [PubMed] [Google Scholar]
- 77.Mellado E, Cuenca-Estrella M, Regadera J, González M, Díaz-Guerra TM, Rodríguez-Tudela JL, 2000. Sustained gastrointestinal colonization and systemic dissemination by Candida albicans, Candida tropicalis and Candida parapsilosis in adult mice. Diagn Microbiol Infect Dis 38:21–28. 10.1016/S0732-8893(00)00165-6. [DOI] [PubMed] [Google Scholar]
- 78.Savage DC, Dubos RJ. 1967. Localization of indigenous yeast in the murine stomach. J Bacteriol 94:1811–1816. 10.1128/jb.94.6.1811-1816.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Teoh F, Pavelka N. 2016. How chemotherapy increases the risk of systemic candidiasis in cancer patients: current paradigm and future directions. Pathogens 5:6. 10.3390/pathogens5010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol 56:397–415. 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- 81.Mukaremera L, Lee KK, Mora-Montes HM, Gow NAR. 2017. Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front Immunol 8:629. 10.3389/fimmu.2017.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, Jacobs L, Buurman ET, Gijzen K, Williams DL, Torensma R, McKinnon A, MacCallum DM, Odds FC, Van der Meer JW, Brown AJ, Kullberg BJ. 2006. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest 116:1642–1650. 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. 2002. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis 185:1483–1489. 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- 84.Rubin-Bejerano I, Abeijon C, Magnelli P, Grisafi P, Fink GR. 2007. Phagocytosis by human neutrophils is stimulated by a unique fungal cell wall component. Cell Host Microbe 2:55–67. 10.1016/j.chom.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cottier F, Hall RA. 2019. Face/Off: the interchangeable side of Candida albicans. Front Cell Infect Microbiol 9:471. 10.3389/fcimb.2019.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wagener J, Malireddi RK, Lenardon MD, Köberle M, Vautier S, MacCallum DM, Biedermann T, Schaller M, Netea MG, Kanneganti TD, Brown GD, Brown AJ, Gow NA. 2014. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog 10:e1004050. 10.1371/journal.ppat.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee KK, Maccallum DM, Jacobsen MD, Walker LA, Odds FC, Gow NA, Munro CA. 2012. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob Agents Chemother 56:208–217. 10.1128/AAC.00683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence 4:119–128. 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng G, Wozniak K, Wallig MA, Fidel PL, Jr., Trupin SR, Hoyer LL. 2005. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect Immun 73:1656–1663. 10.1128/IAI.73.3.1656-1663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murciano C, Moyes DL, Runglall M, Tobouti P, Islam A, Hoyer LL, Naglik JR. 2012. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS One 7:e33362. 10.1371/journal.pone.0033362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sundstrom P, Balish E, Allen CM. 2002. Essential role of the Candida albicans transglutaminase substrate, hyphal wall protein 1, in lethal oroesophageal candidiasis in immunodeficient mice. J Infect Dis 185:521–530. 10.1086/338836. [DOI] [PubMed] [Google Scholar]
- 92.Noverr MC, Huffnagle GB. 2004. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect Immun 72:6206–6210. 10.1128/IAI.72.11.6206-6210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lo H-J, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 94.Kumamoto CA, Gresnigt MS, Hube B. 2020. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr Opin Microbiol 56:7–15. 10.1016/j.mib.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tso GHW, Reales-Calderon JA, Tan ASM, Sem X, Le GTT, Tan TG, Lai GC, Srinivasan KG, Yurieva M, Liao W, Poidinger M, Zolezzi F, Rancati G, Pavelka N. 2018. Experimental evolution of a fungal pathogen into a gut symbiont. Science 362:589–595. 10.1126/science.aat0537. [DOI] [PubMed] [Google Scholar]
- 96.Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP, Noble SM. 2019. Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host Microbe 25:432–443.e6. 10.1016/j.chom.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Noverr MC, Erb-Downward JR, Huffnagle GB. 2003. Production of eicosanoids and other oxylipins by pathogenic eukaryotic microbes. Clin Microbiol Rev 16:517–533. 10.1128/CMR.16.3.517-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ells R, Kemp G, Albertyn J, Kock JL, Pohl CH. 2013. Phenothiazine is a potent inhibitor of prostaglandin E2 production by Candida albicans biofilms. FEMS Yeast Res 13:849–855. 10.1111/1567-1364.12093. [DOI] [PubMed] [Google Scholar]
- 99.Ells R, Kock JL, Albertyn J, Kemp G, Pohl CH. 2011. Effect of inhibitors of arachidonic acid metabolism on prostaglandin E(2) production by Candida albicans and Candida dubliniensis biofilms. Med Microbiol Immunol 200:23–28. 10.1007/s00430-010-0169-7. [DOI] [PubMed] [Google Scholar]
- 100.Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. 2001. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun 69:2957–2963. 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Erb-Downward JR, Noverr MC. 2007. Characterization of prostaglandin E2 production by Candida albicans. Infect Immun 75:3498–3505. 10.1128/IAI.00232-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Nunez G, Shibuya A. 2014. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2). Cell Host Microbe 15:95–102. 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamaguchi N, Sugita R, Miki A, Takemura N, Kawabata J, Watanabe J, Sonoyama K. 2006. Gastrointestinal Candida colonisation promotes sensitisation against food antigens by affecting the mucosal barrier in mice. Gut 55:954–960. 10.1136/gut.2005.084954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Renga G, Moretti S, Oikonomou V, Borghi M, Zelante T, Paolicelli G, Costantini C, De Zuani M, Villella VR, Raia V, Del Sordo R, Bartoli A, Baldoni M, Renauld JC, Sidoni A, Garaci E, Maiuri L, Pucillo C, Romani L. 2018. IL-9 and mast cells are key players of Candida albicans commensalism and pathogenesis in the gut. Cell Rep 23:1767–1778. 10.1016/j.celrep.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jawhara S, Thuru X, Standaert-Vitse A, Jouault T, Mordon S, Sendid B, Desreumaux P, Poulain D. 2008. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J Infect Dis 197:972–980. 10.1086/528990. [DOI] [PubMed] [Google Scholar]
- 106.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical Practice Guideline for the Management of Candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Denning DW, Kneale M, Sobel JD, Rautemaa-Richardson R. 2018. Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect Dis 18:e339–e347. 10.1016/S1473-3099(18)30103-8. [DOI] [PubMed] [Google Scholar]
- 108.Patil S, Majumdar B, Sarode SC, Sarode GS, Awan KH. 2018. Oropharyngeal candidosis in HIV-infected patients—an update. Front Microbiol 9:980. 10.3389/fmicb.2018.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saunus JM, Kazoullis A, Farah CS. 2008. Cellular and molecular mechanisms of resistance to oral Candida albicans infections. Front Biosci 13:5345–5358. 10.2741/3085. [DOI] [PubMed] [Google Scholar]
- 110.Fraser VJ, Jones M, Dunkel J, Storfer S, Medoff G, Dunagan WC. 1992. Candidemia in a tertiary care hospital: epidemiology, risk factors, and predictors of mortality. Clin Infect Dis 15:414–421. 10.1093/clind/15.3.414. [DOI] [PubMed] [Google Scholar]
- 111.Poissy J, Damonti L, Bignon A, Khanna N, Von Kietzell M, Boggian K, Neofytos D, Vuotto F, Coiteux V, Artru F, Zimmerli S, Pagani J-L, Calandra T, Sendid B, Poulain D, van Delden C, Lamoth F, Marchetti O, Bochud P-Y, Allfun French Study Groups. 2020. Risk factors for candidemia: a prospective matched case-control study. Crit Care 24:109. 10.1186/s13054-020-2766-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Timsit JF, Azoulay E, Schwebel C, Charles PE, Cornet M, Souweine B, Klouche K, Jaber S, Trouillet JL, Bruneel F, Argaud L, Cousson J, Meziani F, Gruson D, Paris A, Darmon M, Garrouste-Orgeas M, Navellou JC, Foucrier A, Allaouchiche B, Das V, Gangneux JP, Ruckly S, Maubon D, Jullien V, Wolff M, EMPIRICUS Trial Group. 2016. Empirical micafungin treatment and survival without invasive fungal infection in adults with ICU-acquired sepsis, Candida colonization, and multiple organ failure: the EMPIRICUS Randomized Clinical Trial. JAMA 316:1555–1564. 10.1001/jama.2016.14655. [DOI] [PubMed] [Google Scholar]
- 113.Baddley JW, Benjamin DK, Jr., Patel M, Miro J, Athan E, Barsic B, Bouza E, Clara L, Elliott T, Kanafani Z, Klein J, Lerakis S, Levine D, Spelman D, Rubinstein E, Tornos P, Morris AJ, Pappas P, Fowler VG, Jr., Chu VH, Cabell C, International Collaboration on Endocarditis-Prospective Cohort Study Group. 2008. Candida infective endocarditis. Eur J Clin Microbiol Infect Dis 27:519–529. 10.1007/s10096-008-0466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sankar NP, Thakarar K, Rokas KE. 2020. Candida infective endocarditis during the infectious diseases and substance use disorder syndemic: a six-year case series. Open Forum Infect Dis 7:ofaa142. 10.1093/ofid/ofaa142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haron E, Vartivarian S, Anaissie E, Dekmezian R, Bodey GP. 1993. Primary Candida pneumonia. Experience at a large cancer center and review of the literature. Medicine 72:137–142. 10.1097/00005792-199372030-00001. [DOI] [PubMed] [Google Scholar]
- 116.Moellering RC, Jr. 1992. Emergence of Enterococcus as a significant pathogen. Clin Infect Dis 14:1173–1176. 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 117.Pendleton KM, Dickson RP, Newton DW, Hoffman TC, Yanik GA, Huffnagle GB. 2018. Respiratory tract colonization by Candida species portends worse outcomes in immunocompromised patients. Clin Pulm Med 25:197–201. 10.1097/CPM.0000000000000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pendleton KM, Huffnagle GB, Dickson RP. 2017. The significance of Candida in the human respiratory tract: our evolving understanding. Pathog Dis 75:ftx029. 10.1093/femspd/ftx029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.George F, Daniel C, Thomas M, Singer E, Guilbaud A, Tessier FJ, Revol-Junelles AM, Borges F, Foligne B. 2018. Occurrence and dynamism of lactic acid bacteria in distinct ecological niches: a multifaceted functional health perspective. Front Microbiol 9:2899. 10.3389/fmicb.2018.02899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Holzapfel WH, Wood BJB. 2014. Lactic acid bacteria: biodiversity and taxonomy. Wiley Blackwell, Chichester, UK. [Google Scholar]
- 122.De Angelis M, Gobbetti M. 1999. Lactobacillus sanfranciscensis CB1: manganese, oxygen, superoxide dismutase and metabolism. Appl Microbiol Biotechnol 51:358–363. 10.1007/s002530051402. [DOI] [PubMed] [Google Scholar]
- 123.Salvatore S, Hauser B, Devreker T, Vieira MC, Luini C, Arrigo S, Nespoli L, Vandenplas Y. 2007. Probiotics and zinc in acute infectious gastroenteritis in children: are they effective? Nutrition 23:498–506. 10.1016/j.nut.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 124.Calomme M, Hu J, Van den Branden K, Vanden Berghe DA. 1995. Seleno-lactobacillus. An organic selenium source. Biol Trace Elem Res 47:379–383. 10.1007/BF02790140. [DOI] [PubMed] [Google Scholar]
- 125.Khalid K. 2011. An overview of lactic acid bacteria. Int J Biosci 1:1–13. [Google Scholar]
- 126.Carr FJ, Chill D, Maida N. 2002. The lactic acid bacteria: a literature survey. Crit Rev Microbiol 28:281–370. 10.1080/1040-840291046759. [DOI] [PubMed] [Google Scholar]
- 127.Mokoena MP. 2017. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules 22:1255. 10.3390/molecules22081255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Heeney DD, Gareau MG, Marco ML. 2018. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr Opin Biotechnol 49:140–147. 10.1016/j.copbio.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tannock GW. 1995. Normal microflora: an introduction to microbes inhabiting the human body, 1st ed. Chapman & Hall, London, UK. [Google Scholar]
- 130.Lievin-Le Moal V, Servin AL. 2014. Anti-infective activities of lactobacillus strains in the human intestinal microbiota: from probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin Microbiol Rev 27:167–199. 10.1128/CMR.00080-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vuik F, Dicksved J, Lam SY, Fuhler GM, van der Laan L, van de Winkel A, Konstantinov SR, Spaander M, Peppelenbosch MP, Engstrand L, Kuipers EJ. 2019. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United Eur Gastroent J 7:897–907. 10.1177/2050640619852255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Seekatz AM, Schnizlein MK, Koenigsknecht MJ, Baker JR, Hasler WL, Bleske BE, Young VB, Sun D. 2019. Spatial and temporal analysis of the stomach and small-intestinal microbiota in fasted healthy humans. mSphere 4:e00126-19. 10.1128/mSphere.00126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kashket ER. 1987. Bioenergetics of lactic acid bacteria: cytoplasmic pH and osmotolerance. FEMS Microbiology Rev 46:233–244. 10.1111/j.1574-6968.1987.tb02463.x. [DOI] [Google Scholar]
- 134.Corcoran BM, Stanton C, Fitzgerald GF, Ross RP. 2005. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl Environ Microbiol 71:3060–3067. 10.1128/AEM.71.6.3060-3067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.O’Flaherty S, Briner Crawley A, Theriot CM, Barrangou R. 2018. The lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere 3:e00140-18. 10.1128/mSphere.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Elkins CA, Moser SA, Savage DC. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology (Reading) 147:3403–3412. 10.1099/00221287-147-12-3403. [DOI] [PubMed] [Google Scholar]
- 137.Hammes WP, Vogel RF. 1995. The genus Lactobacillus, p 19–54. In Wood BJB, Holzapfel WH (ed), The genera of lactic acid bacteria, vol 2. Springer, Boston, MA. [Google Scholar]
- 138.Walter J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 74:4985–4996. 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ryan KA, Jayaraman T, Daly P, Canchaya C, Curran S, Fang F, Quigley EM, O'Toole PW. 2008. Isolation of lactobacilli with probiotic properties from the human stomach. Lett Appl Microbiol 47:269–274. 10.1111/j.1472-765x.2008.02416.x. [DOI] [PubMed] [Google Scholar]
- 140.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 103:732–737. 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roos S, Engstrand L, Jonsson H. 2005. Lactobacillus gastricus sp. nov., Lactobacillus antri sp. nov., Lactobacillus kalixensis sp. nov. and Lactobacillus ultunensis sp. nov., isolated from human stomach mucosa. Int J Syst Evol Microbiol 55:77–82. 10.1099/ijs.0.63083-0. [DOI] [PubMed] [Google Scholar]
- 142.Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol 54:1093–1101. 10.1099/jmm.0.45935-0. [DOI] [PubMed] [Google Scholar]
- 143.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104:13780–13785. 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tannock GW. 1992. The lactic microflora of pigs, mice, and rats, p 21–48. In Wood BJB (ed), The lactic acid bacteria, vol 1. Springer, Boston, MA. [Google Scholar]
- 145.Aguirre M, Collins MD. 1993. Lactic acid bacteria and human clinical infection. J Appl Bacteriol 75:95–107. 10.1111/j.1365-2672.1993.tb02753.x. [DOI] [PubMed] [Google Scholar]
- 146.Food and Agriculture Organization of the United Nations, World Health Organization. 2006. Probiotics in food: health and nutritional properties and guidelines for evaluation. Food and Agriculture Organization of the United Nations, World Health Organization, Rome, Italy. [Google Scholar]
- 147.Shreiner A, Huffnagle GB, Noverr MC. 2008. The “microflora hypothesis” of allergic disease. Adv Exp Med Biol 635:113–134. 10.1007/978-0-387-09550-9_10. [DOI] [PubMed] [Google Scholar]
- 148.Ford AC, Quigley EM, Lacy BE, Lembo AJ, Saito YA, Schiller LR, Soffer EE, Spiegel BM, Moayyedi P. 2014. Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis. Am J Gastroenterol 109:1547–1561. 10.1038/ajg.2014.202. [DOI] [PubMed] [Google Scholar]
- 149.Ganji-Arjenaki M, Rafieian-Kopaei M. 2018. Probiotics are a good choice in remission of inflammatory bowel diseases: a meta analysis and systematic review. J Cell Physiol 233:2091–2103. 10.1002/jcp.25911. [DOI] [PubMed] [Google Scholar]
- 150.Reid G, Jass J, Sebulsky MT, McCormick JK. 2003. Potential uses of probiotics in clinical practice. Clin Microbiol Rev 16:658–672. 10.1128/CMR.16.4.658-672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li J, Sung CY, Lee N, Ni Y, Pihlajamaki J, Panagiotou G, El-Nezami H. 2016. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci U S A 113:E1306–E1315. 10.1073/pnas.1518189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. 2017. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact 16:79. 10.1186/s12934-017-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pessione E. 2012. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol 2:86. 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 155.Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. 2000. The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology 118:724–734. 10.1016/S0016-5085(00)70142-9. [DOI] [PubMed] [Google Scholar]
- 156.Garcia-Solache M, Rice LB. 2019. The enterococcus: a model of adaptability to its environment. Clin Microbiol Rev 32:e00058-18. 10.1128/CMR.00058-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fisher K, Phillips C. 2009. The ecology, epidemiology and virulence of Enterococcus. Microbiology (Reading) 155:1749–1757. 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 158.Lebreton F, Willems RJL, Gilmore MS. 2014. Enterococcus diversity, origins in nature, and gut colonization. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 159.Banla LI, Salzman NH, Kristich CJ. 2019. Colonization of the mammalian intestinal tract by enterococci. Curr Opin Microbiol 47:26–31. 10.1016/j.mib.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33:e00181-19. 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Noskin GA, Peterson LR, Warren JR. 1995. Enterococcus faecium and Enterococcus faecalis bacteremia: acquisition and outcome. Clin Infect Dis 20:296–301. 10.1093/clinids/20.2.296. [DOI] [PubMed] [Google Scholar]
- 162.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hayakawa K, Marchaim D, Martin ET, Tiwari N, Yousuf A, Sunkara B, Pulluru H, Kotra H, Hasan A, Bheemreddy S, Sheth P, Lee DW, Kamatam S, Bathina P, Nanjireddy P, Chalana IK, Patel S, Kumar S, Vahia A, Ku K, Yee V, Swan J, Pogue JM, Lephart PR, Rybak MJ, Kaye KS. 2012. Comparison of the clinical characteristics and outcomes associated with vancomycin-resistant Enterococcus faecalis and vancomycin-resistant E. faecium bacteremia. Antimicrob Agents Chemother 56:2452–2458. 10.1128/AAC.06299-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ghanem G, Hachem R, Jiang Y, Chemaly RF, Raad I. 2007. Outcomes for and risk factors associated with vancomycin-resistant Enterococcus faecalis and vancomycin-resistant Enterococcus faecium bacteremia in cancer patients. Infect Control Hosp Epidemiol 28:1054–1059. 10.1086/519932. [DOI] [PubMed] [Google Scholar]
- 165.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16. 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Van Tyne D, Gilmore MS. 2014. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol 68:337–356. 10.1146/annurev-micro-091213-113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Baddour LM, Wilson WR, Bayer AS, Fowler VG, Jr., Tleyjeh IM, Rybak MJ, Barsic B, Lockhart PB, Gewitz MH, Levison ME, Bolger AF, Steckelberg JM, Baltimore RS, Fink AM, O'Gara P, Taubert KA, Stroke C, American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council. 2015. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals from the American Heart Association. Circulation 132:1435–1486. 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 169.Ulrich N, Vonberg RP, Gastmeier P. 2017. Outbreaks caused by vancomycin-resistant Enterococcus faecium in hematology and oncology departments: a systematic review. Heliyon 3:e00473. 10.1016/j.heliyon.2017.e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Trastoy R, Manso T, Fernandez-Garcia L, Blasco L, Ambroa A, Perez Del Molino ML, Bou G, Garcia-Contreras R, Wood TK, Tomas M. 2018. Mechanisms of bacterial tolerance and persistence in the gastrointestinal and respiratory environments. Clin Microbiol Rev 31:e00023-18. 10.1128/CMR.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lebeer S, Verhoeven TL, Francius G, Schoofs G, Lambrichts I, Dufrene Y, Vanderleyden J, De Keersmaecker SC. 2009. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl Environ Microbiol 75:3554–3563. 10.1128/AEM.02919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Allonsius CN, van den Broek MFL, De Boeck I, Kiekens S, Oerlemans EFM, Kiekens F, Foubert K, Vandenheuvel D, Cos P, Delputte P, Lebeer S. 2017. Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb Biotechnol 10:1753–1763. 10.1111/1751-7915.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tytgat HL, Douillard FP, Reunanen J, Rasinkangas P, Hendrickx AP, Laine PK, Paulin L, Satokari R, de Vos WM. 2016. Lactobacillus rhamnosus GG outcompetes Enterococcus faecium via mucus-binding pili: evidence for a novel and heterospecific probiotic mechanism. Appl Environ Microbiol 82:5756–5762. 10.1128/AEM.01243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Markowiak-Kopeć P, Śliżewska K. 2020. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 12:1107. 10.3390/nu12041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sun Y, O'Riordan MX. 2013. Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv Appl Microbiol 85:93–118. 10.1016/B978-0-12-407672-3.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nguyen LN, Lopes LCL, Cordero RJB, Nosanchuk JD. 2011. Sodium butyrate inhibits pathogenic yeast growth and enhances the functions of macrophages. J Antimicrob Chemother 66:2573–2580. 10.1093/jac/dkr358. [DOI] [PubMed] [Google Scholar]
- 177.Bhaskaran N, Quigley C, Paw C, Butala S, Schneider E, Pandiyan P. 2018. Role of short chain fatty acids in controlling Tregs and immunopathology during mucosal infection. Front Microbiol 9:1995. 10.3389/fmicb.2018.01995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sekhavat A, Sun JM, Davie JR. 2007. Competitive inhibition of histone deacetylase activity by trichostatin A and butyrate. Biochem Cell Biol 85:751–758. 10.1139/o07-145. [DOI] [PubMed] [Google Scholar]
- 179.Davie JR. 2003. Inhibition of histone deacetylase activity by butyrate. J Nutr 133:2485s–2493s. 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 180.Zacchi LF, Schulz WL, Davis DA. 2010. HOS2 and HDA1 encode histone deacetylases with opposing roles in Candida albicans morphogenesis. PLoS One 5:e12171. 10.1371/journal.pone.0012171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Simonetti G, Passariello C, Rotili D, Mai A, Garaci E, Palamara AT. 2007. Histone deacetylase inhibitors may reduce pathogenicity and virulence in Candida albicans. FEMS Yeast Res 7:1371–1380. 10.1111/j.1567-1364.2007.00276.x. [DOI] [PubMed] [Google Scholar]
- 182.Romani L. 2011. Immunity to fungal infections. Nat Rev Immunol 11:275–288. 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 183.Whibley N, Gaffen SL. 2014. Brothers in arms: Th17 and Treg responses in Candida albicans immunity. PLoS Pathog 10:e1004456. 10.1371/journal.ppat.1004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Guinan J, Wang S, Hazbun TR, Yadav H, Thangamani S. 2019. Antibiotic-induced decreases in the levels of microbial-derived short-chain fatty acids correlate with increased gastrointestinal colonization of Candida albicans. Sci Rep 9:8872. 10.1038/s41598-019-45467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Desai JD, Banat IM. 1997. Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev 61:47–64. 10.1128/mmbr.61.1.47-64.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Velraeds MM, van der Mei HC, Reid G, Busscher HJ. 1996. Inhibition of initial adhesion of uropathogenic Enterococcus faecalis by biosurfactants from Lactobacillus isolates. Appl Environ Microbiol 62:1958–1963. 10.1128/aem.62.6.1958-1963.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ceresa C, Tessarolo F, Caola I, Nollo G, Cavallo M, Rinaldi M, Fracchia L. 2015. Inhibition of Candida albicans adhesion on medical-grade silicone by a Lactobacillus-derived biosurfactant. J Appl Microbiol 118:1116–1125. 10.1111/jam.12760. [DOI] [PubMed] [Google Scholar]
- 188.Fracchia L, Cavallo M, Allegrone G, Martinotti M. 2010. A lactobacillus-derived biosurfactant inhibits biofilm formation of human pathogenic Candida albicans biofilm producers. Curr Res Technol Educ Top Appl Microbiol Microb Biotechnol 2:827–837. [Google Scholar]
- 189.De Gregorio PR, Parolin C, Abruzzo A, Luppi B, Protti M, Mercolini L, Silva JA, Giordani B, Marangoni A, Nader-Macias MEF, Vitali B. 2020. Biosurfactant from vaginal Lactobacillus crispatus BC1 as a promising agent to interfere with Candida adhesion. Microb Cell Fact 19:133. 10.1186/s12934-020-01390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Garcia-Rubio R, de Oliveira HC, Rivera J, Trevijano-Contador N. 2019. The fungal cell wall: candida, cryptococcus, and aspergillus species. Front Microbiol 10:2993. 10.3389/fmicb.2019.02993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chattaway FW, Holmes MR, Barlow AJ. 1968. Cell wall composition of the mycelial and blastospore forms of Candida albicans. J Gen Microbiol 51:367–376. 10.1099/00221287-51-3-367. [DOI] [PubMed] [Google Scholar]
- 192.Allonsius CN, Vandenheuvel D, Oerlemans EFM, Petrova MI, Donders GGG, Cos P, Delputte P, Lebeer S. 2019. Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Sci Rep 9:2900. 10.1038/s41598-019-39625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bauer MA, Kainz K, Carmona-Gutierrez D, Madeo F. 2018. Microbial wars: competition in ecological niches and within the microbiome. Microb Cell 5:215–219. 10.15698/mic2018.05.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ch’ng J-H, Chong KKL, Lam LN, Wong JJ, Kline KA. 2019. Biofilm-associated infection by enterococci. Nat Rev Microbiol 17:82–94. 10.1038/s41579-018-0107-z. [DOI] [PubMed] [Google Scholar]
- 195.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198. 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Manley KJ, Fraenkel MB, Mayall BC, Power DA. 2007. Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med J Aust 186:454–457. 10.5694/j.1326-5377.2007.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 197.Szachta P, Ignyś I, Cichy W. 2011. An evaluation of the ability of the probiotic strain Lactobacillus rhamnosus GG to eliminate the gastrointestinal carrier state of vancomycin-resistant enterococci in colonized children. J Clin Gastroenterol 45:872–877. 10.1097/MCG.0b013e318227439f. [DOI] [PubMed] [Google Scholar]
- 198.Billington EO, Phang SH, Gregson DB, Pitout JD, Ross T, Church DL, Laupland KB, Parkins MD. 2014. Incidence, risk factors, and outcomes for Enterococcus spp. blood stream infections: a population-based study. Int J Infect Dis 26:76–82. 10.1016/j.ijid.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 199.Percy MG, Grundling A. 2014. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68:81–100. 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 200.Ryu YH, Baik JE, Yang JS, Kang SS, Im J, Yun CH, Kim DW, Lee K, Chung DK, Ju HR, Han SH. 2009. Differential immunostimulatory effects of Gram-positive bacteria due to their lipoteichoic acids. Int Immunopharmacol 9:127–133. 10.1016/j.intimp.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 201.Wang S, Liu K, Seneviratne CJ, Li X, Cheung GS, Jin L, Chu CH, Zhang C. 2015. Lipoteichoic acid from an Enterococcus faecalis clinical strain promotes TNF-alpha expression through the NF-kappaB and p38 MAPK signaling pathways in differentiated THP-1 macrophages. Biomed Rep 3:697–702. 10.3892/br.2015.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ahn KB, Baik JE, Yun CH, Han SH. 2018. Lipoteichoic acid inhibits Staphylococcus aureus biofilm formation. Front Microbiol 9:327. 10.3389/fmicb.2018.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jung S, Park OJ, Kim AR, Ahn KB, Lee D, Kum KY, Yun CH, Han SH. 2019. Lipoteichoic acids of lactobacilli inhibit Enterococcus faecalis biofilm formation and disrupt the preformed biofilm. J Microbiol 57:310–315. 10.1007/s12275-019-8538-4. [DOI] [PubMed] [Google Scholar]
- 204.Kim AR, Kang M, Yoo YJ, Yun CH, Perinpanayagam H, Kum KY, Han SH. 2020. Lactobacillus plantarum lipoteichoic acid disrupts mature Enterococcus faecalis biofilm. J Microbiol 58:314–319. 10.1007/s12275-020-9518-4. [DOI] [PubMed] [Google Scholar]
- 205.Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, Heuser J, Dodson KW, Caparon MG, Hultgren SJ. 2009. Contribution of autolysin and sortase a during Enterococcus faecalis DNA-dependent biofilm development. Infect Immun 77:3626–3638. 10.1128/IAI.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penadés JR, Lasa I. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol 67:4538–4545. 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Alvarez-Sieiro P, Montalban-Lopez M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 72:728–764, table of contents. 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Prabhurajeshwar C, Chandrakanth RK. 2017. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: an in vitro validation for the production of inhibitory substances. Biomed J 40:270–283. 10.1016/j.bj.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A 104:7617–7621. 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Messaoudi S, Manai M, Kergourlay G, Prevost H, Connil N, Chobert JM, Dousset X. 2013. Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol 36:296–304. 10.1016/j.fm.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 212.Ocana VS, Pesce De Ruiz Holgado AA, Nader-Macias ME. 1999. Characterization of a bacteriocin-like substance produced by a vaginal Lactobacillus salivarius strain. Appl Environ Microbiol 65:5631–5635. 10.1128/AEM.65.12.5631-5635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sterling AJ, Snelling WJ, Naughton PJ, Ternan NG, Dooley JSG. 2020. Competent but complex communication: the phenomena of pheromone-responsive plasmids. PLoS Pathog 16:e1008310. 10.1371/journal.ppat.1008310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tomita H, Fujimoto S, Tanimoto K, Ike Y. 1997. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J Bacteriol 179:7843–7855. 10.1128/jb.179.24.7843-7855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526:719–722. 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Balla E, Dicks LM, Du Toit M, Van Der Merwe MJ, Holzapfel WH. 2000. Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Appl Environ Microbiol 66:1298–1304. 10.1128/AEM.66.4.1298-1304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nigutova K, Morovsky M, Pristas P, Teather RM, Holo H, Javorsky P. 2007. Production of enterolysin A by rumen Enterococcus faecalis strain and occurrence of enlA homologues among ruminal Gram-positive cocci. J Appl Microbiol 102:563–569. 10.1111/j.1365-2672.2006.03068.x. [DOI] [PubMed] [Google Scholar]
- 218.Liu Y, Defourny KAY, Smid EJ, Abee T. 2018. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol 9:1502. 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Molina-Tijeras JA, Galvez J, Rodriguez-Cabezas ME. 2019. The immunomodulatory properties of extracellular vesicles derived from probiotics: a novel approach for the management of gastrointestinal diseases. Nutrients 11:1038. 10.3390/nu11051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B. 2017. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol 17:66. 10.1186/s12866-017-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dean SN, Rimmer MA, Turner KB, Phillips DA, Caruana JC, Hervey WJ, Leary DH, Walper SA. 2020. Lactobacillus acidophilus membrane vesicles as a vehicle of bacteriocin delivery. Front Microbiol 11:710. 10.3389/fmicb.2020.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Morschhäuser J. 2010. Regulation of white-opaque switching in Candida albicans. Med Microbiol Immunol 199:165–172. 10.1007/s00430-010-0147-0. [DOI] [PubMed] [Google Scholar]
- 223.Hancock LE, Perego M. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol 186:5629–5639. 10.1128/JB.186.17.5629-5639.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Brown AO, Graham CE, Cruz MR, Singh KV, Murray BE, Lorenz MC, Garsin DA. 2019. Antifungal activity of the Enterococcus faecalis peptide EntV requires protease cleavage and disulfide bond formation. mBio 10:e01334-19. 10.1128/mBio.01334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Fourie R, Ells R, Swart CW, Sebolai OM, Albertyn J, Pohl CH. 2016. Candida albicans and Pseudomonas aeruginosa interaction, with focus on the role of eicosanoids. Front Physiol 7:64. 10.3389/fphys.2016.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. 2006. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother 50:1463–1469. 10.1128/AAC.50.4.1463-1469.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kong EF, Tsui C, Kucharikova S, Van Dijck P, Jabra-Rizk MA. 2017. Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol. Antimicrob Agents Chemother 61:e01573-17. 10.1128/AAC.01573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Krause J, Geginat G, Tammer I. 2015. Prostaglandin E2 from Candida albicans stimulates the growth of Staphylococcus aureus in mixed biofilms. PLoS One 10:e0135404. 10.1371/journal.pone.0135404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Morales DK, Hogan DA. 2010. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog 6:e1000886. 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabé M, Viscogliosi E. 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313. 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kaleli I, Cevahir N, Demir M, Yildirim U, Sahin R. 2007. Anticandidal activity of Pseudomonas aeruginosa strains isolated from clinical specimens. Mycoses 50:74–78. 10.1111/j.1439-0507.2006.01322.x. [DOI] [PubMed] [Google Scholar]
- 232.Hogan DA, Vik A, Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 54:1212–1223. 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 233.Hogan DA, Kolter R. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229–2232. 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 234.Fourie R, Ells R, Kemp G, Sebolai OM, Albertyn J, Pohl CH. 2017. Pseudomonas aeruginosa produces aspirin insensitive eicosanoids and contributes to the eicosanoid profile of polymicrobial biofilms with Candida albicans. Prostaglandins Leukot Essent Fatty Acids 117:36–46. 10.1016/j.plefa.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 235.Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 67:2982–2992. 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bamford CV, d'Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77:3696–3704. 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Carolus H, Van Dyck K, Van Dijck P. 2019. Candida albicans and Staphylococcus Species: a threatening twosome. Front Microbiol 10:2162. 10.3389/fmicb.2019.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Todd OA, Fidel PL, Jr., Harro JM, Hilliard JJ, Tkaczyk C, Sellman BR, Noverr MC, Peters BM. 2019. Candida albicans augments Staphylococcus aureus virulence by engaging the staphylococcal agr quorum sensing system. mBio 10:e00910-19. 10.1128/mBio.00910-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chow J, Mazmanian SK. 2010. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7:265–276. 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jochum L, Stecher B. 2020. Label or concept—what is a pathobiont? Trends Microbiol 28:789–792. 10.1016/j.tim.2020.04.011. [DOI] [PubMed] [Google Scholar]