ABSTRACT
The bacterial flagellum is an organelle responsible for motility and has a rotary motor comprising the rotor and the stator. Flagellar biogenesis is initiated by the assembly of the MS-ring, a supramolecular complex embedded in the cytoplasmic membrane. The MS-ring consists of a few dozen copies of the transmembrane FliF protein and is an essential core structure that is a part of the rotor. The number and locations of the flagella are controlled by the FlhF and FlhG proteins in some species. However, there is no clarity on the factors initiating MS-ring assembly or on the contributions of FlhF/FlhG to this process. Here, we show that FlhF and a C-ring component, FliG, facilitate Vibrio MS-ring formation. When Vibrio FliF alone was expressed in Escherichia coli cells, MS-ring formation rarely occurred, indicating a requirement of other factors for MS-ring assembly. Consequently, we investigated if FlhF aided FliF in MS-ring assembly. We found that FlhF allowed green fluorescent protein (GFP)-fused FliF to localize at the cell pole in a Vibrio cell, suggesting that it increases local concentration of FliF at the pole. When FliF was coexpressed with FlhF in E. coli cells, the MS-ring was effectively formed, indicating that FlhF somehow contributes to MS-ring formation. The isolated MS-ring structure was similar to that of the MS-ring formed by Salmonella FliF. Interestingly, FliG facilitates MS-ring formation, suggesting that FliF and FliG assist in each other’s assembly into the MS-ring and C-ring. This study aids in understanding the mechanism behind MS-ring assembly using appropriate spatial/temporal regulations.
IMPORTANCE Flagellar formation is initiated by the assembly of the FliF protein into the MS-ring complex, which is embedded in the cytoplasmic membrane. The appropriate spatial/temporal control of MS-ring formation is important for the morphogenesis of the bacterial flagellum. Here, we focus on the assembly mechanism of Vibrio FliF into the MS-ring. FlhF, a positive regulator of the number and location of flagella, recruits the FliF molecules at the cell pole and facilitates MS-ring formation. FliG also facilitates MS-ring formation. Our study showed that these factors control flagellar biogenesis in Vibrio by initiating the MS-ring assembly. Furthermore, it also implies that flagellar biogenesis is a sophisticated system linked with the expression of certain genes, protein localization, and a supramolecular complex assembly.
KEYWORDS: bacterial flagellum, MS-ring, supramolecular complex, FliF, FlhF, bacterial flagellum
INTRODUCTION
Bacteria can swim in a liquid environment or on a wet surface using flagella, which elongate outwards from the cell surface. The bacterial flagellum, which functions like a screw propeller of a ship, is a filamentous supramolecular assembly composed of more than 20 different proteins (1, 2) (Fig. 1A). The flagellum is rotated by a nanoscale rotary motor, which is comprised of the rotor and stator in the cytoplasmic membrane and the rod as a driving shaft connected to the rotor and extended into the periplasmic space. The motor is energized by the influx of ions, typically H+ or Na+, which drive flagellar rotation (3).
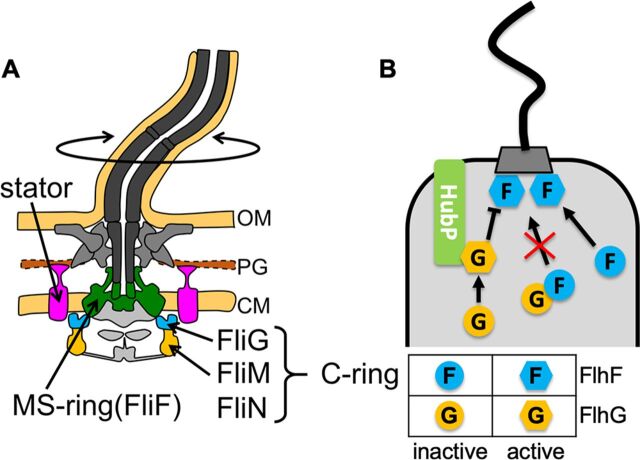
FIG 1.
(A) Schematic drawing of the flagellum in Vibrio. The basal part of the flagellar motor consists of the transmembrane MS-ring and the cytoplasmic C-ring. The MS-ring is shown in green. The C-ring component FliG is shown in cyan, and FliM/FliN is in orange. The stator complexes are shown in pink. CM, cytoplasmic membrane; PG, peptidoglycan layer; OM, outer membrane. (B) Schematic drawing of flagellar number and location control by FlhF and FlhG in Vibrio. An “F” in circles or hexagons represents inactive and active FlhF, respectively. A “G” in circles or hexagons represents inactive and active FlhG, respectively. HubP is shown in light green.
The bacterial flagellum is an interesting research topic for energy transduction by ion flow. Motor rotation is produced by the rotor-stator interaction coupled with ion influx via ion channels in the stator unit (4–8). The rotor is composed of the transmembrane MS-ring and the cytoplasmic C-ring, which is attached to the cytoplasmic region of the MS-ring (9, 10). The stator is composed of two kinds of transmembrane proteins, the A subunit and B subunit, which form complexes of 4 and 2 molecules, respectively. They form complexes of 4 PomAs and 2 PomBs in Vibrio and 4 MotAs and 2 MotBs in Escherichia coli (11, 12). A dozen stator complexes surround the rotor (13, 14). The torque for rotation is produced by the interaction between FliG in the C-ring and the A subunit in the stator. Since FliG binds to the cytoplasmic tail of FliF, which is an MS-ring component, the MS-ring assembly is responsible for the C-ring assembly (15). Therefore, the MS-ring acts as a base structure of the motor structure for flagellar rotation. The flagellum possesses a flagellar protein-specific translocator called the export apparatus, which is classified as a type III secretion system (T3SS) (16). It translocates the flagellar axial structure proteins and their cap proteins across the cell membrane into the extracellular space. Since the export apparatus is located inside the MS-ring, MS-ring formation is essential in the initiation of flagellar construction (17, 18). FliF possesses two transmembrane (TM) segments in the N-terminal and C-terminal regions and a large periplasmic domain containing ca. 400 residues between the TM segments (see Fig. S1 in the supplemental material) (19). However, the mechanism behind the insertion of FliF molecules into the cytoplasmic membrane and their assembly into a ring-shaped structure is unclear.
The number and positions of flagella are various in different bacterial species. For example, Salmonella and E. coli cells possess peritrichous flagella, whereas Vibrio and Pseudomonas cells possess a single flagellum at one of the cell poles (monotrichous flagellum). What is the difference between these species? Previous studies have shown that the flagellar gene expression in Salmonella affects the number of flagella. The overexpression of the master regulators FlhD/FlhC, which initiate transcription of the flagellar gene regulons, or the defect of negative regulators against FlhD/FlhC, FliT, or ClpX/ClpP allowed an increase of the flagellar number (20, 21). In Vibrio, FlaK (or FlrA) functions as the master regulator, instead of FlhD/FlhC (22, 23). Moreover, additional regulators, namely, FlhF and FlhG, are involved in controlling the number and position of the single flagellum in Vibrio and Pseudomonas (24–31). They ensure that only one flagellum is formed at one of the cell poles (Fig. 1B). Genes encoding these proteins do not exist in E. coli and Salmonella. FlhF and FlhG serve as positive and negative factors, respectively, in the regulation of flagellar number. In Vibrio spp., overexpression of FlhF or a defect of FlhG induces an increase in the number of flagella, whereas overexpression of FlhG or a defect of FlhF results in the loss of flagellar formation (24, 25). A double defect of FlhF and FlhG induces the development of a nonflagellate phenotype. However, in a few cases, an flhF-flhG null mutant cell produces peritrichous flagella, illustrating the importance of FlhF in the determination of flagellar location, as well as in the flagellar formation (24, 25, 31). Since green fluorescent protein (GFP)-fused FlhF is intrinsically localized at the cell pole, FlhF seems to determine the site at which flagellar formation is initiated (in this case, at the cell pole) (25). FlhG reduces the polar localization of FlhF and suppresses the function of FlhF at the cell pole, controlling the flagellar number and location to construct a single flagellum at one of the poles (32, 33) (Fig. 1B). FlhF and FlhG are a GTPase and an ATPase, respectively, and the GTPase activity of FlhF is enhanced by interaction with FlhG (33–39). Flagellar formation was not affected by a mutation in the catalytic residue of FlhF, which abolishes the GTPase activity, whereas a mutation in the GTP-binding residues prevented flagellar formation in Vibrio spp. (34, 35). These results suggest that nucleotide binding to FlhF is necessary for flagellar formation and that GTP hydrolysis is involved in removing FlhF from the cell pole. FlhG also localizes at the cell pole. This localization is important for FlhG function as a suppressor of flagellar biogenesis (25, 33). The precise roles of FlhF and FlhG are, however, still obscure. Recently, it has been reported that a gene, hubP, is involved in the regulation of the flagellar number in Vibrio (40, 41). HubP seems to assist the function of FlhG in suppressing FlhF activity at the cell pole (Fig. 1B).
Since FlhF and FlhG regulate the number and location of flagella in Vibrio and the MS-ring formation serves as the start point for flagellar formation, it is suggested that these proteins should contribute to FliF localization at the cell pole and to the subsequent formation of the MS-ring. To elucidate the initiation mechanism of flagellar formation and the spatial/temporal control mechanism of MS-ring assembly in marine Vibrio species, we investigated the factors required for MS-ring formation by expressing Vibrio FliF in E. coli cells. Here, we provide an insight into the assembly mechanism of the MS-ring in Vibrio species, which regulates the number and location of the flagella.
RESULTS
Vibrio FliF rarely assembles into the MS-ring complex by itself.
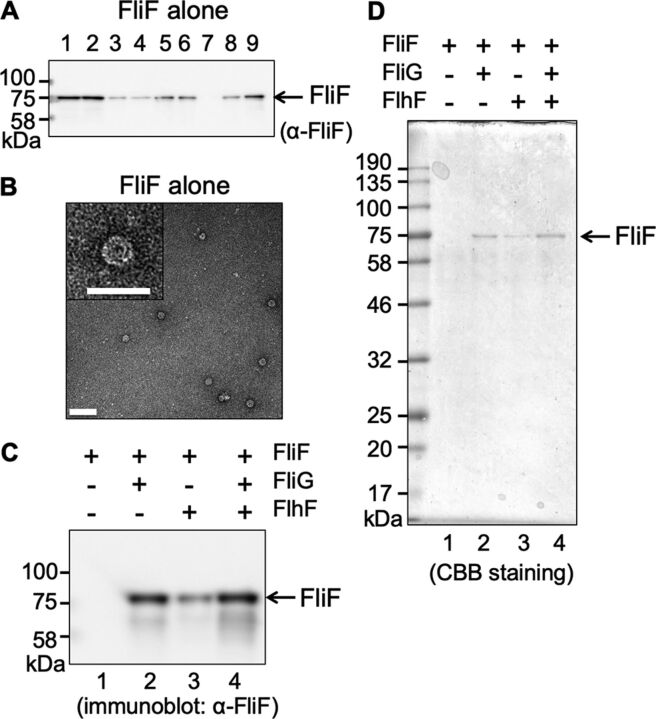
It has been shown that the overexpression of Salmonella FliF alone in E. coli cells is sufficient for its efficient assembly into the MS-ring (9, 42, 43). Salmonella FliF seems to be capable of assembling into the ring by itself, without the help of other proteins such as chaperones. First, we examined whether Vibrio FliF assembles into the MS-ring by itself in E. coli cells. To isolate the MS-rings formed by Vibrio FliF, the method used to purify the Salmonella MS-rings was slightly modified (43). The cells overproducing FliF from pColdI expression vector, pRO101, were disrupted by a French press, and the membrane fraction obtained by ultracentrifugation was solubilized using Triton X-100 (Fig. 2A, lanes 1 to 6). The suspension was treated with an alkaline solution (pH 11) to remove the insoluble outer membranes. A crude MS-ring fraction was then obtained by ultracentrifugation (Fig. 2A, lane 9). The crude MS-ring sample was separated from protein aggregates by sucrose density gradient centrifugation (see Fig. S2A in the supplemental material). The resultant fractions 5 and 6, which contained FliF, were recovered as the MS-ring fractions. Since these fractions contained impurities, they were further purified and concentrated by Ni affinity chromatography via an N-terminal His tag fused to FliF. In the isolated sample, the FliF protein was hardly detected by immunoblot. However, a small number of ring structures, with a diameter of ca. 25 nm, were infrequently observed by electron microscopy (EM); these are similar to the MS-ring of Salmonella (Fig. 2B and C, lane 1) (9, 42, 43). Although we observed other density gradient fractions using EM, we found no ring structures.
FIG 2.
MS-ring isolated from cells expressing FliF alone. (A) Immunoblot of the FliF protein during purification. Vibrio FliF was expressed in plasmid pRO101 in E. coli BL21(DE3) cells. Lane 1, suspension processed by the French press. Lane 2, supernatant from low-speed centrifugation. Lane 3, pellet from low-speed centrifugation. Lane 4, supernatant from ultracentrifugation (cytoplasmic fraction). Lane 5, pellet from ultracentrifugation (membrane fraction). Lane 6, supernatant from low-speed centrifugation following membrane solubilization by alkaline solution containing Triton X-100. Lane 7, pellet from low-speed centrifugation. Lane 8, supernatant from ultracentrifugation. Lane 9, pellet from ultracentrifugation (crude MS-ring fraction). (B) Electron microscopy (EM) image of purified MS-ring (bar, 100 nm). The right panel is an enlarged image of the MS-ring (bar, 50 nm). The grid was stained using uranyl acetate. (C) Immunoblot of the FliF protein contained in the MS-ring fraction (D) Coomassie brilliant blue-stained SDS-PAGE gel of the FliF protein contained in the MS-ring fraction. Lane 1, MS-ring fraction obtained from cells expressing Vibrio FliF alone in the pRO101 plasmid. Lane 2, MS-ring fraction obtained from cells coexpressing Vibrio FliF and FliG in the pTSK137 plasmid. Lane 3, MS-ring fraction obtained from cells coexpressing Vibrio FliF and FlhF in the pRO101 and pTSK122 plasmids, respectively. Lane 4, MS-ring fraction obtained from cells coexpressing Vibrio FliF, FliG, and FlhF in the pTSK137 and pTSK122 plasmids. The applied volume of the MS-ring samples was normalized by adjusting the culture absorbance at an optical density of 600 nm (OD600). All experiments were repeated more than twice, and the results were reproducible.
Consistent with previous results (15), the Vibrio FliF protein was overproduced, and following cell disruption, it was fractionated half-and-half into both the cytoplasm and membrane fractions (Fig. 2A, lane 4 and 5). When the membrane fraction was solubilized using Triton X-100 and fractionated into the supernatant and the precipitate by ultracentrifugation, the FliF protein was detected in both fractions (Fig. 2A, lanes 8 and 9). The crude MS-ring fraction was precipitated, whereas the supernatant solubilized from the membrane fraction seemed to contain various incomplete assemblies of the FliF protein. To confirm whether the membrane fraction contained incomplete assemblies, we solubilized it using n-decyl-β-d-maltoside (DM), and the supernatant was subjected to size exclusion chromatography (Fig. S2B). The elution profile of the FliF protein showed broad peaks. However, it also showed two significant peaks at the 9-ml and 12-ml elution volumes, suggesting that the FliF protein in the membrane was oligomerized in various stoichiometries. We were concerned that the His tag attached to the N terminus of FliF could affect membrane insertion and MS-ring formation of Vibrio FliF. However, His-tagged FliF, which attaches 16 residues derived from pColdI vector that includes a hexahistidine tag at its N terminus, showed a similar swimming ring on the soft agar plate as that of the wild-type FliF, suggesting that His-tagged FliF is fully functional compared with wild-type FliF in the fliF null mutant cell (see Fig. S3A in the supplemental material). Immunoblot analyses of His-tagged FliF using anti-FliF or anti-His-tag antibodies showed that His-tagged FliF was expressed stably in the fliF null mutant cell, suggesting that the His-tag at the N terminus does not affect the behavior of Vibrio FliF (Fig. S3B). Therefore, we concluded that Vibrio FliF is difficult to assemble properly into the MS-ring without the help of other Vibrio flagellum-related proteins and, consequently, that FliF alone does not efficiently assemble into the MS-ring.
FlhF is required for the polar localization of Vibrio FliF.
Since Vibrio FliF is not self-sufficient in forming the MS-ring in E. coli cells, there is a need for other factors to facilitate MS-ring formation. FlhF is a positive regulatory factor that controls the flagellar number and location in Vibrio cells. For Vibrio cholerae cells, it has been reported that localization of GFP-fused FliF at the cell pole requires only the FlhF protein (27). Therefore, FlhF of Vibrio alginolyticus might also be involved in the localization of FliF at the cell pole, thereby facilitating MS-ring formation. To examine this possibility, we observed the cellular localization of GFP-fused FliF (Fig. 3) in the presence or absence of FlhF in V. alginolyticus cells. First, we characterized the function and protein stability of GFP-fused FliF proteins compared with those of wild-type FliF. GFP-fused FliF was somewhat functional in the fliF null mutant cell according to the swimming assay on the soft agar plate (see Fig. S4A in the supplemental material). Immunoblot analyses of GFP-fused FliF proteins with anti-FliF antibody showed that the GFP-fused FliF proteins were certainly produced in the fliF null mutant cells, although products with lower molecular weights, probably degradation products, were detected (Fig. S4B). These showed that GFP-fused FliF proteins were expressed stably and retained the ability of flagellar assembly in the fliF null mutant cells. Plasmid-borne GFP-fused FliF was localized at the cell pole in wild-type cells (Fig. 3A, upper and lower left). Polar localization indicates the assembly of FliF into the flagellar base and is believed to result from MS-ring formation. GFP-fused FliF, however, was not localized at the cell pole in the flhF null mutant cell, indicating the involvement of FlhF in the localization of FliF at the cell pole (Fig. 3A, upper and lower right). Subsequently, we investigated the contribution of other flagellar proteins to the polar localization of FliF. Since the rpoN gene encodes the sigma factor σ54 for class II and III flagellar regulons, which encode the hook-basal body components in Vibrio, its defective mutant does not express any of the flagellar proteins except the flagellar master transcription activator FlaK (25, 44). In the rpoN mutant cell, GFP-fused FliF coexpressed with FlhF derived from plasmids was localized at the cell pole, but no localization was seen in the absence of FlhF (Fig. 3B). These results indicate that no other flagellar components are necessary for the polar localization of FliF in V. alginolyticus, as well as in V. cholerae.
FIG 3.
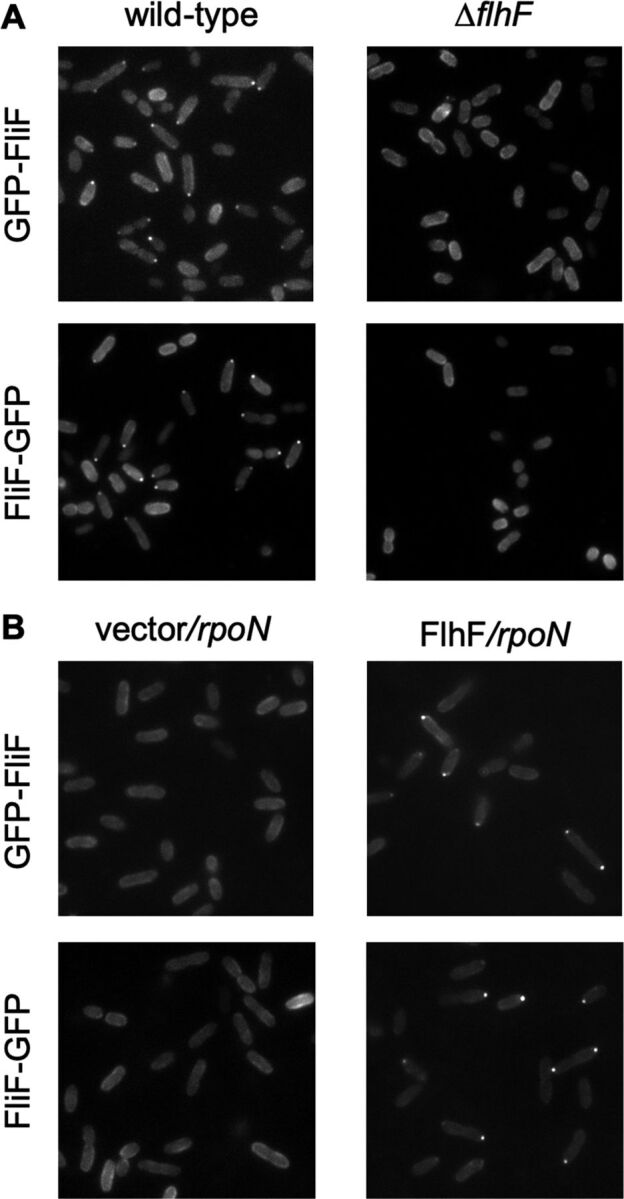
Cellular localization of green fluorescent protein (GFP)-fused FliF. GFP was fused at the N terminus of FliF (GFP-FliF) or at the C terminus of FliF (FliF-GFP). The cells expressing GFP-fused FliF were cultured in VPG broth containing 0.02% (wt/vol) arabinose for 4 h at 30°C. (A) Left upper and lower panels show the fluorescent spots of GFP-FliF expressed in the pTY701 plasmid or FliF-GFP expressed in the pTSK124 plasmid, respectively, in the wild-type strain, VIO5. Right upper and lower panels show the dispersed fluorescence of GFP-FliF expressed in the pTY701 plasmid or FliF-GFP expressed in the pTSK124 plasmid, respectively, in the ΔflhF strain, LPN1. (B) Left upper and lower panels show the dispersed fluorescence of GFP-FliF (in the pYI102 plasmid) or FliF-GFP (in the pYI101 plasmid), respectively, without FlhF (empty vector pMMB206) in the rpoN strain, YM14. Right upper and lower panels show the fluorescent spots of GFP-FliF (in the pYI102 plasmid) or FliF-GFP (in the pYI101 plasmid), respectively, coexpressed with FlhF (in the pTSK122 plasmid) in YM14.
FlhF facilitated MS-ring formation by Vibrio FliF.
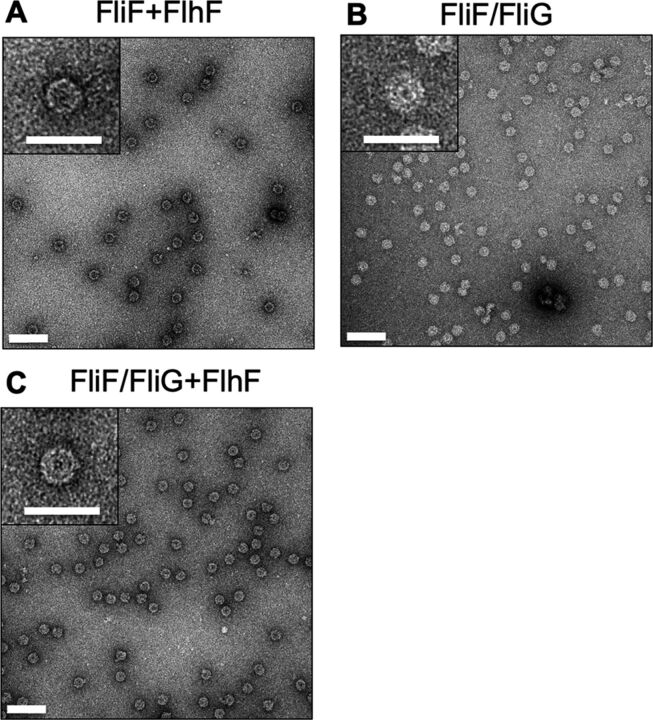
Although FlhF recruited FliF to the cell pole, it is not still clear if FlhF facilitates MS-ring formation. Therefore, we investigated the role of FlhF in MS-ring formation. We expressed Vibrio FlhF with FliF in E. coli cells and isolated the MS-ring fraction by sucrose density gradient centrifugation and Ni-affinity chromatography (see Fig. S5A and D in the supplemental material). Our immunoblot analysis revealed a larger amount of FliF protein in the MS-ring fraction isolated from cells expressing FlhF and FliF than that isolated from the cells expressing only FliF (Fig. 2C, lane 3). Moreover, we detected FliF protein in the isolated MS-ring fraction, not only by immunoblot analysis but also by Coomassie brilliant blue (CBB) staining (Fig. 2D, lane 3). Contaminants were hardly observed on the SDS-PAGE gel, indicating that the purity of the isolated MS-ring fraction was very high. As expected, we found a large number of the ring structures by EM observation (Fig. 4A). Since these ring structures are similar to that of the Salmonella MS-ring (9, 42, 43) and only the FliF proteins were contained in the isolated MS-ring fraction (Fig. 2D, lane 3), we concluded that these ring structures are MS-rings constructed by the FliF protein of Vibrio. Furthermore, this result suggests that FlhF somehow facilitates MS-ring formation.
FIG 4.
MS-rings purified from cells coexpressing FliF with FlhF and/or FliG. (A to C) EM image of MS-rings isolated and purified from E. coli BL21(DE3) cells coexpressing FliF and FlhF in pRO101 and pTSK122 plasmids (A), coexpressing FliF with FliG in pTSK137 plasmid (B), and coexpressing FliF with FliG and FlhF in pTSK137 and pTSK122 plasmids, respectively (C). Bars, 100 nm. The embedded upper left panel in each image is an enlarged image of the MS-ring (bars, 50 nm). The grid was stained using uranyl acetate.
FliG facilitated MS-ring formation by Vibrio FliF.
We examined the contribution of factors other than FlhF to MS-ring formation. Previous studies in Salmonella and E. coli have shown that FliG, a C-ring component, is important for FliF assembly in the flagellar base under chromosome-level gene expression, as monitored by fluorescence foci of fluorescent protein-fused FliF. (45, 46). Another study showed that FliF promoted the multimerization of FliG (47). These studies suggested that FliF and FliG affect each other for their assemblies. To investigate the function of FliG as a facilitator of MS-ring formation, we coexpressed Vibrio FliG with FliF in E. coli cells and isolated the MS-ring. As expected, we detected the FliF protein by immunoblot and CBB staining (lane 2 in Fig. 2C and D; see also Fig. S5C and F). Although FliG interacts with FliF in vivo, the FliG protein was not found in the isolated MS-ring fraction, and it probably dissociated from the MS-ring during its isolation procedures (Fig. 2D, lane 2). We observed many MS-rings in the MS-ring fraction by EM observation (Fig. 4C). Therefore, MS-ring formation was facilitated by the coexpression of FliG with FliF.
Subsequently, we examined how FlhF and FliG contribute to MS-ring formation. We coexpressed FliF with both FliG and FlhF in E. coli cells and purified MS-rings (lane 4 in Fig. 2C and D; see also Fig. S5B and E). EM observation revealed that many MS-rings were formed in the presence of both FlhF and FliG (Fig. 4B). On the other hand, the amount of FliF protein in the purified MS-ring fraction with FliG and FlhF was changed a little in comparison with the amount when only FliG were coexpressed with FliF (lanes 2 to 4 in Fig. 2C and D). These results suggest that FliG and FlhF do not synergistically work to assist MS-ring formation by FliF.
Purified Vibrio MS-ring was properly assembled.
Although Vibrio MS-rings were assembled in the E. coli cells producing FliF, FliG, and FlhF, it is unclear whether the purified MS-rings had the same structural assembly as the native MS-rings with the basal body. We therefore averaged the EM images of MS-rings observed by negative stain and compared them to that of the MS-ring in the hook-basal body (HBB) purified from Vibrio cells (see Fig. S6 in the supplemental material). The three-dimensional (3D) reconstruction of the HBB showed inadequate MS-ring density because the transmembrane segment of FliF was disordered in the HBB. Moreover, the purified Vibrio MS-rings were observed mostly in the top view, and side views were not obtained. Therefore, we could not reconstruct the 3D image to compare the purified Vibrio MS-ring with that of the HBB. We therefore examined the MS-ring structure by using high-speed atomic force microscopy (HS-AFM) (Fig. 5). Based on the AFM images, MS-rings had a rivet shape consisting of a large base with a diameter of ca. 30 nm and a smaller cylinder with a diameter of ca. 18 nm and a height of ca. 20 nm. The cylinder had a hole at the center, which appeared to be the translocation pathway for the flagellar axial protein (Fig. 5A). The shape of Vibrio MS-rings, as observed by HS-AFM, was similar to the 3D image of Salmonella MS-rings reconstructed by cryo-EM observation (48). From the results of HS-AFM observation, we concluded that the Vibrio FliF protein was properly folded and that it assembled into MS-rings during exogenous expression in E. coli cells.
FIG 5.
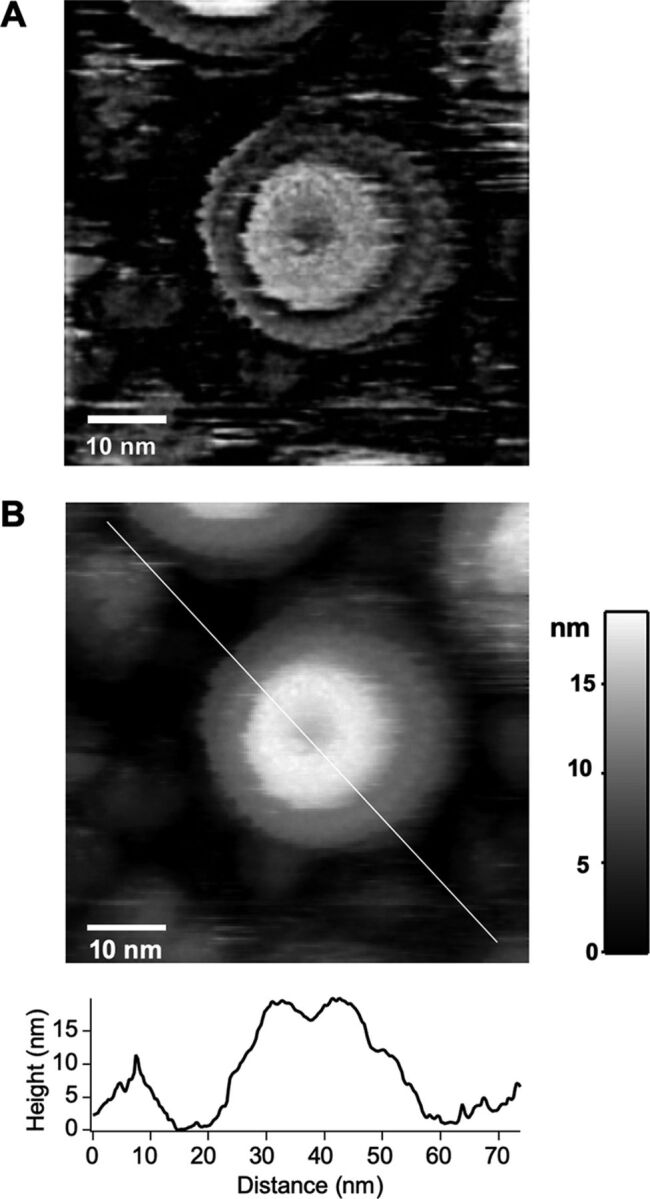
AFM image of the purified Vibrio MS-ring. (A) A filtered AFM image of the purified Vibrio MS-ring. A raw AFM image is shown in panel B. To obtain the filtered AFM image, a raw AFM image was filtered by a band-pass filter with high and low cutoff frequencies of 1/12.0 nm−1 and 1/1.2 nm−1, respectively. (B) A raw AFM image of the purified Vibrio MS-ring. A cross-section profile along the white line in the AFM image is shown in the bottom panel.
FlhG did not affect MS-ring formation facilitated by FlhF.
In marine Vibrio species, the FlhF proteins localized at the cell pole, serve to initiate the MS-ring formation, while the FlhG protein regulates the temporal and spatial localization of FlhF, to control the number and location of the flagella. It is thought that FlhG controls the function of FlhF through direct interaction (25, 30, 33, 37, 39). Furthermore, HubP mediates the recruitment of FlhG to the cell pole in order to inhibit the FlhF activity at the cell pole (40, 41). Thus, the defect of HubP gives rise to a multiple-flagellum phenotype similar to that of an flhG mutant. We therefore investigated if FlhG affects MS-ring formation during exogenous Vibrio FliF and FlhF expression in E. coli cells. FliF was coexpressed with FlhF and FlhG in E. coli cells, and the MS-rings were purified using Ni-affinity chromatography. Ring structures were observed in the MS-ring fraction by EM (see Fig. S7A and B in the supplemental material). The amount of FliF in the isolated MS-ring fractions did not change due to FlhG coexpression (Fig. S7C). These results suggest that the assembly of FliF into MS-rings is not affected by disturbing the FlhF function with FlhG only.
DISCUSSION
FliF has two putative transmembrane segments, TM1 and TM2, in the N-terminal and C-terminal regions, respectively, and a large periplasmic domain (see Fig. S1 in the supplemental material). The FliF proteins assemble into the MS-ring complex in the cytoplasmic membrane. Overexpression of Salmonella FliF allowed the spontaneous formation of the MS-ring complex (9, 43, 48). On the other hand, such an effect was not observed when Vibrio FliF was overexpressed in E. coli cells. Therefore, it is unclear if Vibrio FliF forms the MS-ring spontaneously or if other flagellar components are required for its formation (15). In this study, we improved the MS-ring purification protocol and found a few MS-rings by EM observation in the MS-ring fraction isolated from cells overproducing FliF alone, suggesting that the assembling efficiency of FliF alone is low. We presume the following two reasons for the difficulty in MS-ring formation of Vibrio FliF alone: (i) FliF does not translocate into the cytoplasmic membrane without the aid of helper proteins (Fig. 6A) and (ii) FliF is not folded and/or assembled into MS-rings without the help of other flagellar proteins (Fig. 6B). On the other hand, coexpression of FliF with FlhF and/or FliG enhanced the formation of MS-rings composed of Vibrio FliF. Therefore, FlhF and FliG somehow play a role in the assembly of FliF into the MS-ring. We propose that MS-ring formation is facilitated by FlhF and FliG in Vibrio cells, and the mechanism is described below.
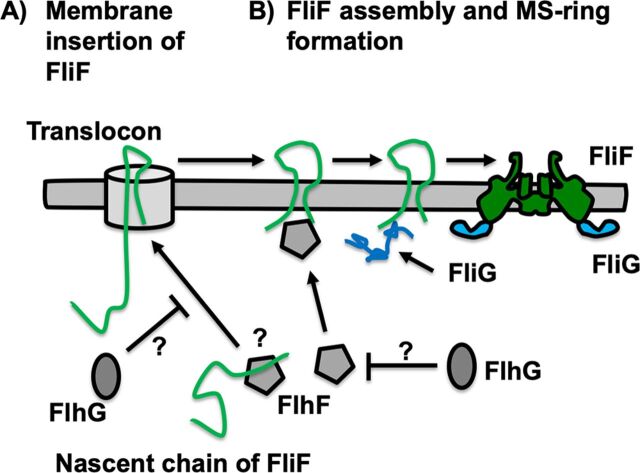
FIG 6.
Working model of MS-ring assembly regulated by the FliG, FlhF, and FlhG proteins. (A) FlhF may interact with a nascent FliF polypeptide chain and conduct the nascent chain to the Sec translocon machinery to insert FliF into the cytoplasmic membrane with appropriate topology. However, we have no evidence to prove that the FliF protein is inserted into the cytoplasmic membrane via the Sec translocon. FlhG may affect FlhF function and/or interact with FliF to inhibit the insertion. (B) FlhF may directly interact with an FliF molecule to recruit it to the cell pole and facilitate MS-ring formation at the cell pole. FliG interacts with the C-terminal region of FliF, and FliF and FliG form a complex. FliF and FliG assist in each other’s assembly into the MS-ring and C-ring. FlhG may indirectly affect FlhF function.
FlhF recruited Vibrio FliF to the cell pole. We propose that FlhF increases the local concentration of FliF at the cell pole, which facilitates MS-ring formation. We speculate that Vibrio FliF spontaneously assembles into the MS-ring if its local concentration on the membrane is very high. As another possibility, FlhF may contribute to membrane insertion of a nascent chain of FliF. FlhF is homologous to the signal recognition particle (SRP), Ffh, and the SRP receptor, FtsY (37, 49, 50). The SRP binds to the signal sequence of a nascent polypeptide translated by the ribosome and mediates the nascent polypeptide to dock with the Sec translocon to translocate it to the periplasmic space or the outer membrane (51). Considering that FlhF is a homologue of Ffh and FtsY, the TM1 region of FliF may be targeted by FlhF and inserted into the membrane surrounding the cell pole. A nascent polypeptide binds to Ffh and is transferred to the Sec translocon through recognition by Ffh-FtsY heterodimer formation. Since FlhF forms a dimer in the presence of GTP (35, 37), FlhF binding to the nascent FliF polypeptide may target other free FlhF localized at the cell pole. Consequently, FliF is released into the membrane surrounding the cell pole through the FlhF homodimer (Fig. 6A). However, we have not been able to assess this possibility yet.
FliG interacts with the C-terminal region of FliF and forms a part of the C-ring. Since the fliG null mutant of Salmonella did not allow yellow fluorescent protein (YFP)-FliF to form fluorescent puncta in the cytoplasmic membrane, as monitored by fluorescent spots, FliG was thought to play an important role in the formation of MS-rings in Salmonella cells (46). When Vibrio FliF was coexpressed with FliG, MS-rings were formed with high efficiency, suggesting that FliG contributes to the assembly of FliF into the MS-ring and that the interaction between FliF and FliG helps MS-ring and C-ring assembly (Fig. 6B). Thus, the defect in the interaction induces a defective phenotype for flagellar formation (15, 47, 52–54). The periplasmic ring structure of the MS-ring is structurally homologous to the injectisome IM-ring of enteropathogenic bacteria and to a sporulation channel in Bacillus subtilis. The IM-ring and sporulation channel display a closely packed oligomerization ring through a canonical ring-building motif (RBM) fold (55). The TM segments and cytoplasmic region seems to be a distorted arrangement compared with that of the extended and well-packed periplasmic ring because the TM segments of the MS-ring or the injectisome IM-ring are disordered (56, 57). We speculate that FliG stabilizes the TM segments of FliF by binding to the C-terminal α-helix of FliF, thereby allowing FliF and FliG to coassemble into the MS-ring and C-ring, respectively.
We showed that overexpression of FliF with FlhF and FliG in E. coli cells led to the formation of MS-rings. Nevertheless, it is still obscure how FliF, under chromosomal expression levels, forms MS-rings in Vibrio cells. If Vibrio FliF from the genome is expressed at a very low level and MS-ring formation is linked to local concentration of the FliF protein, FlhF gathers FliF around the cell pole and MS-ring formation may be initiated by interaction with FliG and/or FlhF. We inferred that not only the gene expression or the amount of protein but also the efficiency of MS-ring formation or the assembly of FliF are necessary to form a single flagellum. The function of FliG and FlhF for MS-ring formation will contribute to strictly determining the number of flagella for a single flagellum and the position of flagellar formation in a Vibrio cell.
It has been shown by bacterial two-hybrid interaction assay that Pseudomonas FlhF interacts with FliG but does not interact with FliF (58). Thus, it has been proposed that the interaction between FlhF and FliG stops the motor rotation by disengaging or jamming the interaction between stator A subunit and rotor FliG when a Pseudomonas cell is attached to a surface in the environment. We had the result that FliG and FlhF did not synergistically work to assist MS-ring formation. This may suggest that the interaction between FlhF and FliG does not assist the MS-ring assembly and that FlhF may have plural functions to regulate motor rotation to determine flagellar number and location. We would like to detect the interaction between FlhF and FliG, as well as the interaction between FlhF and FliF.
Previous studies have revealed that FlhG affects FlhF activity (39). FlhG disturbs FlhF localization at the cell pole, and overexpression of FlhG, or FlhF knockout, prevents flagellar formation (24–30, 59). However, in this study, FlhG coexpressed with FliF and FlhF does not affect MS-ring formation, suggesting that FlhG is not directly involved in MS-ring formation facilitated by FlhF. Rather than being involved in MS-ring formation, FlhG seems to regulate the polar localization of FlhF to initiate flagellar formation at the appropriate position and time. Moreover, FlhG may affect the function of FlhF as a transcriptional activator because FlhF activates the transcription of class III genes in V. cholerae (26). Additionally, it has been shown that FlhG affects FleQ, which is the master regulator of the flagellar gene clusters in Pseudomonas and is an orthologue of FlaK, the Vibrio master regulator, and it thereby inhibits flagellar gene expression (60, 61). Bange and colleagues showed that FlhG mediates the interaction between FliG and FliM/FliY in an ATP- and lipid-dependent manner (62). FlhG, FliM, FliY, and FliG form a quaternary complex and contribute to C-ring assembly during flagellar biogenesis. As described above, FlhG interacts with many other proteins during flagellum biogenesis, and it achieves fine-tuned control over the number and location of the flagella. Thus, we propose that the FlhG protein is a global regulator of flagellum biogenesis that is involved in flagellar gene expression and the flagellar assembly process (Fig. 6).
The ability to form flagella with appropriate temporal and spatial control is important to survive in different environments. However, the factors involved in initiating flagellar formation in marine Vibrio species have not been clear. We think that FlhF recruits FliF to the cell pole to increase the local concentration of FliF and that FlhF and FliG facilitate MS-ring formation. We want to know how marine Vibrio species regulate the number and location of the MS-rings with appropriate temporal and spatial control by way of coordinated action between FliG, FlhF, FlhG, HubP, and so on.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Vibrio cells were cultured in VC broth (0.5% [wt/vol] HiPolypeptone, 0.5% [wt/vol] yeast extract, 3% [wt/vol] NaCl, 0.4% [wt/vol] K2HPO4, and 0.2% [wt/vol] glucose) and VPG broth (1% [wt/vol] HiPolypeptone, 3% [wt/vol] NaCl, 0.4% [wt/vol] K2HPO4, and 0.5% [wt/vol] glycerol), and E. coli was cultured in LB broth (1% [wt/vol] Bacto tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) or SB broth (1.2% [wt/vol)] Bacto tryptone, 2.4% [wt/vol] yeast extract, 1.25% [wt/vol] K2HPO4, 0.38% [wt/vol] KH2PO4, and 0.5% [vol/vol] glycerol). Chloramphenicol was added to final concentrations of 2.5 μg/ml for Vibrio and 25 μg/ml for E. coli. Ampicillin was added to a final concentration of 50 μg/ml for E. coli. Kanamycin was added to a final concentration of 100 μg/ml for Vibrio.
Purification of Vibrio FliF protein.
Vibrio FliF was overproduced in the E. coli BL21(DE3) strain from the cold shock expression plasmid pRO101. An overnight culture was inoculated in 1.5 liters of SB broth at 1/100 dilution and incubated at 37°C until an optical density at 600 nm (OD600) of 0.4 to 0.5. The culture was cooled on ice for 30 min to induce transcription from the cold shock promoter. Following cold shock treatment, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM and incubated at 16°C overnight. The cells were then harvested and suspended in 50 ml of TN solution (20 mM Tris-HCl [pH 8.0] and 150 mM NaCl) with 100 μg/ml lysozyme. Cell disruption was carried out by sonication. Following cell debris removal, the cytoplasmic membrane was precipitated by ultracentrifugation at 100,000 × g for 1 h. The membrane fraction was resuspended in 50 ml TN solution, and n-decyl-β-d-maltoside (DM) was added to a final concentration of 1% (wt/vol). The membrane fraction was stirred on ice for 1 h to solubilize the cytoplasmic membrane. Following the removal of inclusion bodies, the solubilized FliF was applied onto a HisTrap column (GE Healthcare). The column was washed using TN solution with 5 mM imidazole and 0.1% (wt/vol) DM, and the bound FliF proteins were eluted by a linear gradient of up to 500 mM imidazole. Eluted proteins were subjected to the Superdex 200 Increase column (GE Healthcare) in TN solution with 0.1% (wt/vol) DM.
Purification of Vibrio MS-ring from E. coli cells.
Purification of MS-rings formed by Vibrio FliF was carried out as described previously with several modifications (43). An overnight culture was inoculated in 2 liters of LB broth at 1/100 dilution and incubated at 37°C until an OD600 of 0.5. The culture was cooled in ice for 30 min to induce transcription from the cold shock promoter. Following cold shock treatment, IPTG (for the pCold1 and pMMB206 vectors) and arabinose (for the pBAD33 vector) were added to final concentrations of 0.5 mM and 0.02% (wt/vol), respectively, and incubated at 16°C for 16 to 24 h. The cells were then harvested, suspended in 40 ml of TEN solution (50 mM Tris-HCl [pH 8.0], 5 mM EDTA-NaOH [pH 8.0], and 50 mM NaCl) and passed through a French press cell disrupter (Otake, Inc.) under 10,000 lb/in2 pressure to disrupt the cells. Following cell debris removal, the cytoplasmic membrane was precipitated by ultracentrifugation at 90,000 × g for 1 h. The membrane fraction was suspended in 40 ml of an alkaline solution (50 mM CAPS-NaOH [pH 11.0], 5 mM EDTA-NaOH [pH 11.0], 50 mM NaCl, and 1% [wt/vol] Triton X-100) and left to stand at 4°C for 1 h to solubilize the cytoplasmic membrane. Following the removal of inclusion bodies, the crude MS-ring fraction was precipitated by ultracentrifugation at 90,000 × g for 1 h. The crude MS-ring fraction was resuspended in 3 ml S solution (25 mM Tris-HCl [pH 8.0], 1 mM EDTA-NaOH [pH 8.0], 50 mM NaCl, and 0.1% [wt/vol] Triton X-100) and separated using sucrose density gradient centrifugation (6 ml each 40% [wt/wt]/30% [wt/wt]/25% [wt/wt]/20% [wt/wt]/20% [wt/wt] sucrose stepwise gradient solution dissolved in C solution [10 mM Tris-HCl {pH 8.0}, 5 mM EDTA-NaOH {pH 8.0}, and 1% {wt/vol} Triton X-100]) at 49,100 × g (P28S rotor; Hitachi Koki) for 13 h. The sucrose solution was fractionated into 2-ml aliquots. Fractions 5, 6, and 7, which contained the MS-ring, were diluted 20 times in His-A solution (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 0.1% [wt/vol] Triton X-100) and loaded onto a Ni-nitrilotriacetic acid (Ni-NTA)–agarose column (Qiagen). The MS-ring bound to the column was washed using His-B solution (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1% [wt/vol] Triton X-100, and 10 mM imidazole). MS-rings were eluted in His-C solution (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1% [wt/vol] Triton X-100, and 500 mM imidazole) and precipitated by ultracentrifugation at 90,000 × g for 1 h. The precipitated MS-ring was resuspended in 30 μl of S solution and observed by EM.
MS-ring purification from cells where FlhG was coexpressed with FliF and FlhF was carried out as described above with further slight modifications. The cells were harvested, suspended in 45 ml of TN solution (50 mM Tris-HCl [pH 8.0] and 50 mM NaCl) and passed through a French press cell disrupter (Otake, Inc.) under 10,000 lb/in2 pressure to disrupt the cells. Following cell debris removal, the cytoplasmic membrane was precipitated by ultracentrifugation at 90,000 × g for 1 h. The membrane fraction was resuspended in 45 ml TN solution and solubilized using n-dodecyl-β-d-maltoside (DDM) at a final concentration of 1% (wt/vol). Following the removal of undissolved particles, the crude MS-ring fraction was precipitated by ultracentrifugation at 90,000 × g for 1 h. The crude MS-ring fraction was resuspended in 10 ml TND solution (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, and 0.05% [wt/vol] DDM]. The suspension was loaded onto a Ni-NTA–agarose column (Qiagen). The MS-ring bound to the column was washed using His-D solution (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.05% [wt/vol] DDM, and 50 mM imidazole). MS-rings were eluted in His-E solution (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.05% [wt/vol] DDM, and 300 mM imidazole) and precipitated by ultracentrifugation at 90,000 × g for 1 h. The precipitated MS-rings were resuspended in 50 μl of the remaining supernatant and observed by EM.
Fluorescence microscopy.
An overnight culture of Vibrio cells was diluted (1:100 dilution) in VPG broth containing antibiotics and 0.02% (wt/vol) arabinose to induce the expression of GFP-FliF. The cells were incubated further at 30°C for 4 h. To express FlhF (Fig. 3B), IPTG was added to the culture at a final concentration of 0.1 mM. GFP-FliF was expressed in plasmids pTY701 (for Fig. 3A) or pYI102 (for Fig. 3B). FlhF was expressed in plasmid pTSK122 (Fig. 3B). The cells were collected and suspended in V buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, and 5 mM MgCl2). The cells were added to a slide coated with poly-l-lysine, washed using V buffer, and observed under a BX-50 microscope (Olympus). Fluorescent images were captured using a digital camera (ORCA-Flash4.0; Hamamatsu Photonics), and the images were processed using the imaging software HSR (Hamamatsu Photonics).
Negative staining electron microscopy for MS-rings.
MS-ring solutions were applied to carbon-coated copper grids and negatively stained with 3% (wt/vol) uranyl acetate. EM images were observed with a JEM-1010 or JEM-2010 transmission electron microscope (JEOL, Japan) operating at 100 kV using a Bioscan model 792 charge-coupled device (CCD camera) or at 200 kV using an Orius SC200D model 833 CCD camera (Gatan, USA).
Purification of the hook-basal body from Vibrio cells for electron microscope images.
Purification of the hook-basal body from Vibrio cells was carried out as described previously, with several modifications (17). An overnight culture was inoculated in 1 liter of VC broth at 1/100 dilution and incubated at 30°C until an OD600 of 0.7. The cells were collected by centrifugation at 4,600 × g for 10 min. The pellet was resuspended in 46 ml of an ice-cold buffer containing 0.5 M sucrose and 50 mM Tris-HCl (pH 8.0). EDTA and lysozyme were added to final concentrations of 10 mM and 0.1 mg/ml, respectively. Subsequently, the solution was stirred for 30 min, and spheroplasts were lysed using Triton X-100 and MgSO4 at final concentrations of 1% (wt/vol) and 10 mM, respectively. The solution was stirred again on ice for 2 h. Subsequently, EDTA-NaOH (pH 11.0) was added to a final concentration of 10 mM, and the unlysed cells and cell debris were removed by centrifugation at 15,000 × g for 20 min. The pH of the supernatant was adjusted to 10.5 by adding 5 M NaOH. Following cell debris removal, the lysate was spun down by centrifugation at 60,000 × g for 60 min. The pellet was resuspended in buffer C (10 mM Tris-HCl [pH 8.0], 5 mM EDTA-NaOH [pH 8.0], and 1% [wt/vol] Triton X-100). HBB was collected from a 20 to 50% (wt/wt) fraction of sucrose density gradient centrifugation, in buffer C, at 60,000 × g for 14 h. Following the dilution of the sucrose solution-containing HBB in buffer C, HBB was collected by centrifugation at 60,000 × g for 60 min, and the pellet was resuspended in buffer C. Sample solutions were applied to carbon-coated copper grids and negatively stained with 2% (wt/vol) uranyl acetate. EM images were obtained with a JEM-1011 transmission electron microscope (JEOL, Japan) operating at 100 kV using a TemCam-F415 CCD camera (TVIPS, Germany).
3D reconstruction of electron microscope images.
The 1,881-particle images were extracted from the 43 micrographs. Once 2D classification was performed, the best particles were selected from them. The initial model was built from the selected particles. The final 3D structure was reconstructed using 1,356 particles with 34-fold rotational symmetry. All image processing was carried out with RELION3.0.
High speed-atomic force microscope observation and image analysis.
HS-AFM imaging was carried out using a laboratory-built high-speed atomic force microscope (63). The imaging mode in AFM was set as tapping mode, in which a small cantilever with a spring constant of ∼0.2 N/m and a resonant frequency of ∼800 kHz (BLAC7; Olympus) oscillated close to the resonant frequency. The change in the oscillation amplitude was detected by an optimum optical lever deflection method. For feedback control in HS-AFM imaging, the cantilever’s free oscillation amplitude of approximately 2 nm was reduced to 1.5 nm. For HS-AFM imaging, MS-rings were deposited on a mica surface treated with 3-aminopropyl-triethoxysilane (AP-mica). In brief, following cleavage of the mica substrate, 0.1% 3-aminopropyl-triethoxysilane solution was diluted with pure water, deposited on the cleaved mica surface, and incubated for 3 min. The mica surface was then thoroughly washed with pure water, and a solution of MS-rings were deposited. About 10 min postincubation, residual MS-rings were washed off using buffer C. The HS-AFM observation was carried out in buffer C. To observe the detailed structure of the MS-ring (as shown in Fig. 5A), a raw AFM image was filtered by a band-pass filter with high and low cutoff frequencies of 1/12.0 nm−1 and 1/1.2 nm−1, respectively.
ACKNOWLEDGMENTS
We thank K. Maki, Y. Kawase, and A. Abe in our laboratory for technical support in this research and T. Yorimitsu for the construction of plasmid pTY701.
This work was supported in part by JSPS KAKENHI grants JP18K07108 (to H.T.), JP16H04774 and JP18K19293 (to S.K.), JP19H05389 and JP18H01837 (to T.U.), JP25000013 (to K.N.), and JP18K06155 (to T.K.), by the Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) from AMED, under grant JP19am0101117 (to K.N.), by Cyclic Innovation for Clinical Empowerment (CiCLE) grant JP17pc0101020 from AMED (to K.N.), and by the JEOL YOKOGUSHI Research Alliance Laboratories of Osaka University (K.N.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol 270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto YV, Minamino T. 2014. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 4:217–234. doi: 10.3390/biom4010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li N, Kojima S, Homma M. 2011. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 16:985–999. doi: 10.1111/j.1365-2443.2011.01545.x. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd SA, Blair DF. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol 266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 5.Zhou JD, Blair DF. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol 273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 6.Zhou JD, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci U S A 95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yorimitsu T, Sowa Y, Ishijima A, Yakushi T, Homma M. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J Mol Biol 320:403–413. doi: 10.1016/S0022-2836(02)00426-6. [DOI] [PubMed] [Google Scholar]
- 8.Takekawa N, Kojima S, Homma M. 2014. Contribution of many charged residues at the stator-rotor interface of the Na+-driven flagellar motor to torque generation in Vibrio alginolyticus. J Bacteriol 196:1377–1385. doi: 10.1128/JB.01392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno T, Oosawa K, Aizawa S. 1992. M-ring, S-ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol 227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 10.Francis NR, Sosinsky GE, Thomas D, Derosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 11.Sato K, Homma M. 2000. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem 275:20223–20228. doi: 10.1074/jbc.M002236200. [DOI] [PubMed] [Google Scholar]
- 12.Kojima S, Blair DF. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 13.Block SM, Berg HC. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 14.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogawa R, Abe-Yoshizumi R, Kishi T, Homma M, Kojima S. 2015. Interaction of the C-terminal tail of FliF with FliG from the Na+-driven flagellar motor of Vibrio alginolyticus. J Bacteriol 197:63–72. doi: 10.1128/JB.02271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minamino T, Imada K, Namba K. 2008. Mechanisms of type III protein export for bacterial flagellar assembly. Mol Biosyst 4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamoto A, Morimoto YV, Miyata T, Minamino T, Hughes KT, Kato T, Namba K. 2013. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep 3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno T, Oosawa K, Aizawa S. 1994. Domain structures of the MS ring component protein (FliF) of the flagellar basal body of Salmonella typhimurium. J Mol Biol 236:546–555. doi: 10.1006/jmbi.1994.1164. [DOI] [PubMed] [Google Scholar]
- 20.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aldridge C, Poonchareon K, Saini S, Ewen T, Soloyva A, Rao CV, Imada K, Minamino T, Aldridge PD. 2010. The interaction dynamics of a negative feedback loop regulates flagellar number in Salmonella enterica serovar Typhimurium. Mol Microbiol 78:1416–1430. doi: 10.1111/j.1365-2958.2010.07415.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim YK, McCarter LL. 2004. Cross-regulation in Vibrio parahaemolyticus: compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK. J Bacteriol 186:4014–4018. doi: 10.1128/JB.186.12.4014-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klose KE, Mekalanos JJ. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol 180:303–316. doi: 10.1128/JB.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusumoto A, Kamisaka K, Yakushi T, Terashima H, Shinohara A, Homma M. 2006. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem) 139:113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
- 25.Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390–1399. doi: 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 26.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green JC, Kahramanoglou C, Rahman A, Pender AM, Charbonnel N, Fraser GM. 2009. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J Mol Biol 391:679–690. doi: 10.1016/j.jmb.2009.05.075. [DOI] [PubMed] [Google Scholar]
- 28.Dasgupta N, Arora SK, Ramphal R. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 182:357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray TS, Kazmierczak BI. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol 188:6995–7004. doi: 10.1128/JB.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandza S, Baetens M, Park CH, Au T, Keyhan M, Matin A. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol Microbiol 36:414–423. doi: 10.1046/j.1365-2958.2000.01859.x. [DOI] [PubMed] [Google Scholar]
- 31.Kojima M, Nishioka N, Kusumoto A, Yagasaki J, Fukuda T, Homma M. 2011. Conversion of mono-polar to peritrichous flagellation in Vibrio alginolyticus. Microbiol Immunol 55:76–83. doi: 10.1111/j.1348-0421.2010.00290.x. [DOI] [PubMed] [Google Scholar]
- 32.Kusumoto A, Nishioka N, Kojima S, Homma M. 2009. Mutational analysis of the GTP-binding motif of FlhF which regulates the number and placement of the polar flagellum in Vibrio alginolyticus. J Biochem 146:653–660. [DOI] [PubMed] [Google Scholar]
- 33.Ono H, Takashima A, Hirata H, Homma M, Kojima S. 2015. The MinD homolog FlhG regulates the synthesis of the single polar flagellum of Vibrio alginolyticus. Mol Microbiol 98:130–141. doi: 10.1111/mmi.13109. [DOI] [PubMed] [Google Scholar]
- 34.Kondo S, Homma M, Kojima S. 2017. Analysis of the GTPase motif of FlhF in the control of the number and location of polar flagella in Vibrio alginolyticus. Biophys Physicobiol 14:173–181. doi: 10.2142/biophysico.14.0_173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo S, Imura Y, Mizuno A, Homma M, Kojima S. 2018. Biochemical analysis of GTPase FlhF which controls the number and position of flagellar formation in marine Vibrio. Sci Rep 8:12115. doi: 10.1038/s41598-018-30531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bange G, Petzold G, Wild K, Parlitz RO, Sinning I. 2007. The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proc Natl Acad Sci U S A 104:13621–13625. doi: 10.1073/pnas.0702570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bange G, Kummerer N, Grudnik P, Lindner R, Petzold G, Kressler D, Hurt E, Wild K, Sinning I. 2011. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat Struct Mol Biol 18:1376–1380. doi: 10.1038/nsmb.2141. [DOI] [PubMed] [Google Scholar]
- 38.Schniederberend M, Abdurachim K, Murray TS, Kazmierczak BI. 2013. The GTPase activity of FlhF is dispensable for flagellar localization, but not motility, in Pseudomonas aeruginosa. J Bacteriol 195:1051–1060. doi: 10.1128/JB.02013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulbronson CJ, Ribardo DA, Balaban M, Knauer C, Bange G, Hendrixson DR. 2016. FlhG employs diverse intrinsic domains and influences FlhF GTPase activity to numerically regulate polar flagellar biogenesis in Campylobacter jejuni. Mol Microbiol 99:291–306. doi: 10.1111/mmi.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takekawa N, Kwon S, Nishioka N, Kojima S, Homma M. 2016. HubP, a polar landmark protein, regulates flagellar number by assisting in the proper polar localization of FlhG in Vibrio alginolyticus. J Bacteriol 198:3091–3098. doi: 10.1128/JB.00462-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK. 2012. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki H, Yonekura K, Murata K, Hirai T, Oosawa K, Namba K. 1998. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J Struct Biol 124:104–114. doi: 10.1006/jsbi.1998.4048. [DOI] [PubMed] [Google Scholar]
- 43.Kawamoto A, Namba K. 2017. Structural study of the bacterial flagellar basal body by electron cryomicroscopy and image analysis. Meth Mol Biol 1593:119–131. doi: 10.1007/978-1-4939-6927-2_9. [DOI] [PubMed] [Google Scholar]
- 44.Kawagishi I, Nakada M, Nishioka N, Homma M. 1997. Cloning of a Vibrio alginolyticus rpoN gene that is required for polar flagellar formation. J Bacteriol 179:6851–6854. doi: 10.1128/jb.179.21.6851-6854.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli. Mol Microbiol 80:886–899. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- 46.Morimoto YV, Ito M, Hiraoka KD, Che YS, Bai F, Kami-Ike N, Namba K, Minamino T. 2014. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol 91:1214–1226. doi: 10.1111/mmi.12529. [DOI] [PubMed] [Google Scholar]
- 47.Kim EA, Panushka J, Meyer T, Ide N, Carlisle R, Baker S, Blair DF. 2017. Biogenesis of the flagellar switch complex in Escherichia coli: formation of sub-complexes independently of the basal-body MS-ring. J Mol Biol 429:2353–2359. doi: 10.1016/j.jmb.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki H, Yonekura K, Namba K. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol 337:105–113. doi: 10.1016/j.jmb.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 49.Leipe DD, Wolf YO, Koonin EV, Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 50.Bange G, Sinning I. 2013. SIMIBI twins in protein targeting and localization. Nat Struct Mol Biol 20:776–780. doi: 10.1038/nsmb.2605. [DOI] [PubMed] [Google Scholar]
- 51.du Plessis DJF, Nouwen N, Driessen AJM. 2011. The Sec translocase. Biochim Biophys Acta 1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Khan S, Guo TW, Misra S. 2018. A coevolution-guided model for the rotor of the bacterial flagellar motor. Sci Rep 8:11754. doi: 10.1038/s41598-018-30293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue C, Lam KH, Zhang H, Sun K, Lee SH, Chen X, Au S. 2018. Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum. J Biol Chem 293:2066–2078. doi: 10.1074/jbc.M117.797936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim EA, Panushka J, Meyer T, Carlisle R, Baker S, Ide N, Lynch M, Crane BR, Blair DF. 2017. Architecture of the flagellar switch complex of Escherichia coli: conformational plasticity of FliG and implications for adaptive remodeling. J Mol Biol 429:1305–1320. doi: 10.1016/j.jmb.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeytuni N, Strynadka N. 2019. A hybrid secretion system facilitates bacterial sporulation: a structural perspective. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0013-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worrall LJ, Hong C, Vuckovic M, Deng W, Bergeron JR, Majewski DD, Huang RK, Spreter T, Finlay BB, Yu Z, Strynadka NC. 2016. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 540:597–601. doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- 57.Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM. 2020. Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Nature Microbiol doi: 10.1038/s41564-020-0703-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schniederberend M, Williams JF, Shine E, Shen C, Jain R, Emonet T, Kazmierczak BI. 2019. Modulation of flagellar rotation in surface-attached bacteria: a pathway for rapid surface-sensing after flagellar attachment. PLoS Pathog 15:e1008149. doi: 10.1371/journal.ppat.1008149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balaban M, Hendrixson DR. 2011. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog 7:e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dasgupta N, Ferrell EP, Kanack KJ, West SE, Ramphal R. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J Bacteriol 184:5240–5250. doi: 10.1128/jb.184.19.5240-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuhmacher JS, Rossmann F, Dempwolff F, Knauer C, Altegoer F, Steinchen W, Dorrich AK, Klingl A, Stephan M, Linne U, Thormann KM, Bange G. 2015. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci U S A 112:3092–3097. doi: 10.1073/pnas.1419388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uchihashi T, Kodera N, Ando T. 2012. Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat Protoc 7:1193–1206. doi: 10.1038/nprot.2012.047. [DOI] [PubMed] [Google Scholar]