ABSTRACT
Bacterial carboxyl-terminal processing proteases (CTPs) are widely conserved and have been linked to important processes, including signal transduction, cell wall metabolism, and virulence. However, the features that target proteins for CTP-dependent cleavage are unclear. Studies of the Escherichia coli CTP Prc suggested that it cleaves proteins with nonpolar and/or structurally unconstrained C termini, but it is not clear if this applies broadly. Pseudomonas aeruginosa has a divergent CTP, CtpA, which is required for virulence. CtpA works in complex with the outer membrane lipoprotein LbcA to degrade cell wall hydrolases. In this study, we investigated if the C termini of two nonhomologous CtpA substrates are important for their degradation. We determined that these substrates have extended C termini compared to those of their closest E. coli homologs. Removing 7 amino acids from these extensions was sufficient to reduce their degradation by CtpA both in vivo and in vitro. Degradation of one truncated substrate was restored by adding the C terminus from the other but not by adding an unrelated sequence. However, modification of the C termini of nonsubstrates, by adding the C-terminal amino acids from a substrate, did not cause their degradation by CtpA. Therefore, the C termini of CtpA substrates are required but not sufficient for their efficient degradation. Although C-terminal truncated substrates were protected from degradation, they still associated with the LbcA-CtpA complex in vivo. Therefore, degradation of a protein by CtpA requires a C terminus-independent interaction with the LbcA-CtpA complex, followed by C terminus-dependent degradation, perhaps because CtpA normally initiates cleavage at a C-terminal site.
IMPORTANCE Carboxyl-terminal processing proteases (CTPs) are found in all three domains of life, but exactly how they work is poorly understood, including how they recognize substrates. Bacterial CTPs have been associated with virulence, including CtpA of Pseudomonas aeruginosa, which works in complex with the outer membrane lipoprotein LbcA to degrade potentially dangerous peptidoglycan hydrolases. We report an important advance by revealing that efficient degradation by CtpA requires at least two separable phenomena and that one of them depends on information encoded in the substrate C terminus. A C terminus-independent association with the LbcA-CtpA complex is followed by C terminus-dependent cleavage by CtpA. Increased understanding of how CTPs target proteins is significant, due to their links to virulence, peptidoglycan remodeling, and other important processes.
KEYWORDS: Pseudomonas, cell envelope, proteases
INTRODUCTION
Pseudomonas aeruginosa is a widespread Gram-negative bacterium and a frequent cause of serious opportunistic human infections (1). Like for all other bacterial pathogens, many P. aeruginosa virulence factors are assembled in its cell envelope or must pass through the envelope on their way out of the bacterial cell. These include type II and III secretion systems, their exported substrates, pili, and the extracellular polysaccharide alginate, which plays an important role during chronic lung infections of cystic fibrosis patients (1–3). The successful production and function of these virulence factors are presumably impacted by the normal physiological functions that produce, maintain, and remodel the major components of the cell envelope (4).
Proteolysis is an important process in the bacterial cell envelope. It ranges from discrete processing events during protein export, assembly, and signal transduction to the complete degradation of proteins, which is especially important for misfolded or otherwise dangerous proteins (for examples, see references 5, to ,7). One family of proteases found in the bacterial cell envelope is the carboxyl-terminal processing proteases (CTPs), which also occur in archaea and eukaryotes. These enzymes belong to the S41 family of serine proteases, and they can cleave a substrate once for processing or degrade the substrate completely (8–11). The CTP name is derived from early findings that these proteases often cleave close to the C terminus of their substrate. However, at least one member of the CTP family in Xanthomonas campestris has now been shown to cleave close to the N terminus of its substrate (12).
Recently, it has emerged that one role played by Gram-negative bacterial CTPs is to control potentially dangerous cell wall cross-link hydrolases by degrading them (13, 14). This is likely to be a widespread phenomenon, because it occurs in the divergent species Escherichia coli and P. aeruginosa. In E. coli, the Prc protease degrades the cell wall cross-link hydrolase MepS, and in P. aeruginosa, CtpA degrades at least four predicted cell wall cross-link hydrolases (13, 14). Both of these CTPs form complexes with an outer membrane lipoprotein that is required for their proteolytic function, NlpI in E. coli and LbcA in P. aeruginosa (13, 14). Structural analysis of the E. coli NlpI-Prc complex has led to a model for protein degradation by a CTP, in which a lever-like mechanism serves to translocate the MepS substrate though a passage containing the catalytic site for successive cleavage events (10). However, despite their obvious similarities, Prc and CtpA are not orthologs. Prc is much larger than CtpA, and it is a member of the CTP-1 subfamily, whereas CtpA is in the divergent CTP-3 subfamily (15–17). Furthermore, although their lipoprotein binding partners NlpI and LbcA both contain the short, degenerate tetratricopeptide repeat motifs, they do not share obvious primary sequence similarity and are very different in size.
Exactly how CTPs recognize their substrates is unclear. In E. coli, Prc has been suggested to target proteins with nonpolar and/or structurally unconstrained C termini (10, 18–20). However, nothing is known about whether the C terminus of a substrate is important during cleavage by the CtpA protease of P. aeruginosa. The significant differences between Prc and CtpA mean that what is true for Prc cannot be assumed to be true for CtpA. In particular, they might not share similar substrate recognition requirements. Indeed, the substrates of CtpA do not have nonpolar C termini (14). Knowing the features required for substrate cleavage is an important step toward understanding how CtpA-dependent proteolysis is controlled in P. aeruginosa. Therefore, we have investigated the role of the substrate C terminus. Our results reveal that the extreme C terminus is a critical determinant for efficient degradation both in vivo and in vitro and that the C termini of two unrelated substrates are functionally interchangeable for degradation. However, the C-terminal motif is not sufficient for degradation, which suggests that it is only one of two or more checkpoints that determine if a protein will be degraded by CtpA.
RESULTS
Truncation of the C terminus protects substrates from degradation by CtpA in vivo.
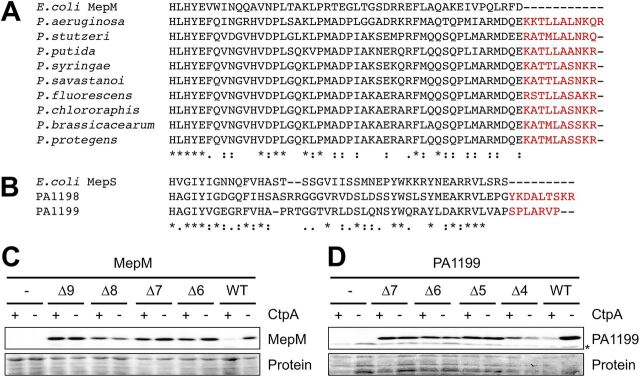
The first CtpA substrate to be discovered was PA0667, which we named MepM due to its homology with the E. coli MepM peptidoglycan cross-link hydrolase (14, 21). P. aeruginosa MepM has a short C-terminal extension that is not present in E. coli MepM but is conserved in other Pseudomonas species, all of which also have CtpA (Fig. 1A). We hypothesized that this extension might be important for CtpA-dependent degradation. To test this, we constructed araBp-mepM expression plasmids encoding MepM with C-terminal truncations and determined their steady-state levels in ctpA+ and ΔctpA strains. C-terminal truncation increased the amount of MepM in ctpA+ cells, suggesting some protection from CtpA-dependent degradation (Fig. 1C). The Δ6 truncation (C-terminal 6 amino acids removed) reproducibly had a slightly higher level in the ΔctpA strain than in the ctpA+ strain, whereas longer truncations (Δ7 to Δ9) consistently had similar levels in the two strains (Fig. 1C and data not shown). The Δ8 truncation reduced the amount of MepM even in the ΔctpA strain, perhaps due to CtpA-independent destabilization (Fig. 1C and data not shown). Therefore, from all these data we concluded that removing approximately 7 C-terminal amino acids was sufficient to protect MepM from CtpA-dependent degradation in vivo, without otherwise destabilizing the protein.
FIG 1.
Truncation of the C termini of MepM and PA1199 protects them from degradation by CtpA in vivo. (A) CLUSTAL Omega alignment of the C termini of MepM proteins from E. coli and Pseudomonas species. (B) CLUSTAL Omega alignment of the C termini of E. coli MepS and P. aeruginosa PA1198 and PA1199. For panels A and B, amino acids in red represent C-terminal extensions in the Pseudomonas proteins. (C) MepM immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains. (D) PA1199 immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains (an asterisk indicates a protein cross-reactive with the PA1199 antiserum). For PA1199 analysis, both strains also had a Δprc mutation to eliminate any interference from degradation of mutant proteins by Prc. For panels C and D, strains contained an arabinose-inducible expression plasmid encoding wild-type MepM or PA1199 (WT), or derivatives with the indicated number of amino acids removed from the C terminus, and were grown in medium containing arabinose. MepM and PA1199 were detected with polyclonal antisera, and a Ponceau S total protein stain of the same region of the nitrocellulose membrane is shown to document loading levels. Immunoblots are single representatives of several replicate experiments.
MepM and another CtpA substrate, PA4404, are both members of the LytM/M23 peptidase family. However, the two other known substrates, PA1198 and PA1199, are in the NlpC/P60 peptidase family (14). PA1198 and PA1199 are more than 50% identical to each other and homologous to E. coli MepS. An alignment of E. coli MepS with PA1198 and PA1199 revealed that the P. aeruginosa proteins have extended C termini (Fig. 1B). Therefore, we expanded our analysis to include one of these NlpC/P60 family substrates, PA1199. However, when we began to analyze PA1199, we found that some truncation mutants had significantly reduced abundance in a ΔctpA strain compared to the wild-type protein (data not shown). P. aeruginosa has a homolog of the E. coli CTP, Prc, which has been implicated in degrading proteins with aberrant C termini (20, 22). We reasoned that P. aeruginosa Prc might cleave some truncated PA1199 proteins due to their altered C termini. In support of this, the abundance of the truncated PA1199 proteins was indistinguishable from that of full-length PA1199 in ΔctpA Δprc strains (data not shown). For this reason, the in vivo analysis of PA1199 and its derivatives in this study was done by comparing their abundances in Δprc ctpA+ and Δprc ΔctpA strains, which eliminated any interference from Prc. As with MepM, C-terminal truncations increased the amount of PA1199 in ctpA+ cells, suggesting some protection from CtpA-dependent degradation (Fig. 1D). In this case, removing five C-terminal amino acids appeared sufficient to protect PA1199 from CtpA-dependent degradation in vivo without otherwise destabilizing the protein. Together, all of these data suggest that the C termini of MepM and PA1199 contain information that is important for their efficient degradation by CtpA in vivo.
The C termini of MepM and PA1199 are interchangeable for CtpA-dependent degradation.
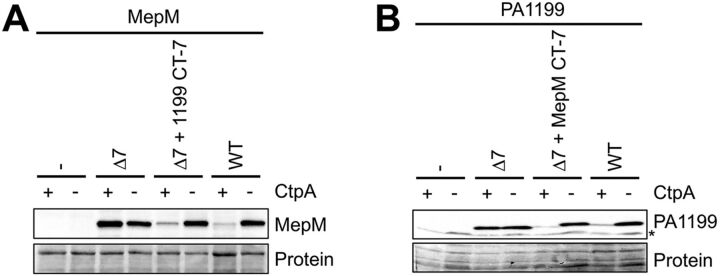
To extend our investigation, we focused on the Δ7 truncations of MepM and PA1199, because both rendered the proteins similarly abundant in ctpA+ and ΔctpA strains (Fig. 1). The amino acids removed from each protein are not obviously similar (LALNKQR for MepM and SPLARVP for PA1199 [Fig. 1]). Nevertheless, we tested our conclusion that they contain specific information important for CtpA-dependent degradation, by exchanging them.
MepM-Δ7 was similarly abundant in ctpA+ and ΔctpA strains, as seen previously (Fig. 1 and 2). However, when the C-terminal 7 amino acids of PA1199 were added onto the C terminus of MepM-Δ7, it behaved indistinguishably from wild-type MepM, suggesting that its degradation by CtpA was restored (Fig. 2A). Similarly, addition of the C-terminal 7 amino acids of MepM onto the end of PA1199-Δ7 made it behave similarly to wild-type PA1199 (Fig. 2B). These experiments further support the conclusion that the C termini of CtpA substrates are important for their degradation in vivo and also show that they can be exchanged between two different substrates.
FIG 2.
Substrate C termini are interchangeable for degradation by CtpA. (A) MepM immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains. Strains contained an arabinose-inducible expression plasmid encoding wild-type MepM or derivatives with 7 amino acids removed from the C terminus (Δ7) or with the C-terminal 7 amino acids replaced by those from PA1199 (Δ7 + 1199 CT-7). (B) PA1199 immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains (an asterisk indicates a protein cross-reactive with the PA1199 antiserum). For PA1199 analysis, both strains also had a Δprc mutation to eliminate any interference from degradation of mutant proteins by Prc. Strains contained an arabinose-inducible expression plasmid encoding wild-type PA1199 or derivatives with 7 amino acids removed from the C terminus or with the C-terminal 7 amino acids replaced by those from MepM (Δ7 + MepM CT-7). For both panels, strains were grown in medium containing arabinose. MepM and PA1199 were detected with polyclonal antisera, and a Ponceau S total protein stain of the same region of the nitrocellulose membrane is shown to document loading levels. Immunoblots are single representatives of several replicate experiments.
Restoring the length of truncated substrates is not sufficient for CtpA-dependent degradation.
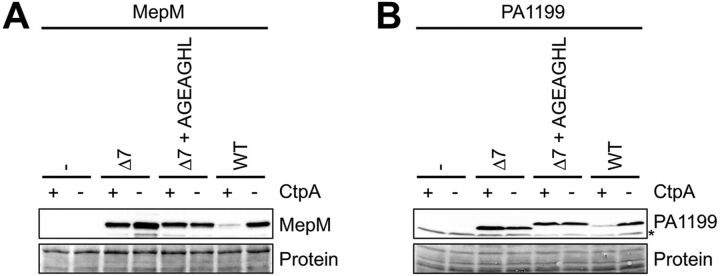
CtpA-dependent degradation of two substrates truncated by 7 amino acids was restored by adding back 7 amino acids from a different substrate (Fig. 2). This raised the question of whether it is the length of the substrates that is critical or whether their C termini contain sequence-dependent information. To distinguish between these possibilities, we generated a 7-amino-acid sequence randomly, AGEAGHL, and added it to the C termini of the MepM-Δ7 and PA1199-Δ7 proteins. In contrast to the C-terminal swaps, adding these random 7 amino acids to the truncated MepM and PA1199 proteins did not reduce their levels in a ctpA+ strain compared to those in a ΔctpA strain (Fig. 3). Therefore, the C-terminal sequences of MepM and PA1199 contain sequence-specific information important for degradation by CtpA.
FIG 3.
Restoring the length of truncated substrates is not sufficient for CtpA-dependent degradation. (A) MepM immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains. Strains contained an arabinose-inducible expression plasmid encoding wild-type MepM or derivatives with 7 amino acids removed from the C terminus or with the C-terminal 7 amino acids replaced by AGEAGHL. (B) PA1199 immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains (an asterisk indicates a protein cross-reactive with the PA1199 antiserum). For PA1199 analysis, both strains also had a Δprc mutation to eliminate any interference from degradation of mutant proteins by Prc. Strains contained an arabinose-inducible expression plasmid encoding wild-type PA1199 or derivatives with 7 amino acids removed from the C terminus or with the C-terminal 7 amino acids replaced by AGEAGHL. For both panels, strains were grown in medium containing arabinose. MepM and PA1199 were detected with polyclonal antisera, and a Ponceau S total protein stain of the same region of the nitrocellulose membrane is shown to document loading levels. Immunoblots are single representatives of several replicate experiments.
The C-terminal motif of a CtpA substrate is not sufficient for CtpA-dependent degradation.
It was reported that for E. coli, adding the C-terminal 5 amino acids of a Prc substrate onto the C terminus of a nonsubstrate rendered it cleavable by Prc in vivo and in vitro (23). Our findings suggested that the C-terminal amino acids of CtpA substrates are also important for degradation, but they did not address if they are sufficient for degradation. Therefore, we tested this possibility next.
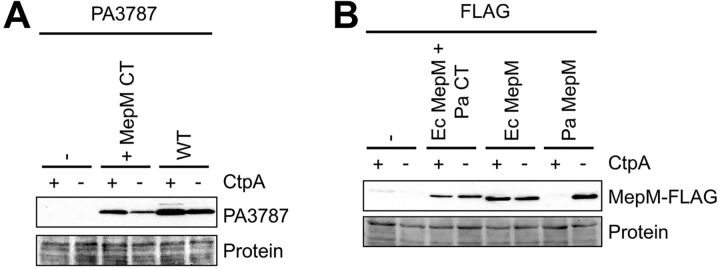
Besides the CtpA substrates MepM and PA4404, P. aeruginosa has a third member of the LytM/M23 peptidase family in its cell envelope that is predicted to be catalytically active, PA3787. However, PA3787 is not a CtpA substrate in vivo or in vitro (14). To test if the C-terminal amino acids of a substrate are sufficient for CtpA-dependent degradation, we added the C-terminal 11 amino acids of MepM onto the C terminus of PA3787. This hybrid protein was slightly less abundant than wild-type PA3787 in both ctpA+ and ΔctpA cells (Fig. 4A). However, it did not accumulate in a ΔctpA strain compared to a ctpA+ strain. Therefore, the addition of the C-terminal 11 amino acids from MepM did not make the related PA3787 a CtpA substrate in vivo.
FIG 4.
The C-terminal motif of a CtpA substrate is not sufficient for CtpA-dependent degradation. (A) PA3787 immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains. Strains contained an arabinose-inducible expression plasmid encoding wild-type PA3787 or a derivative with the 11 C-terminal amino acids from MepM added to its C terminus. (B) Anti-FLAG immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains. Strains contained an arabinose-inducible expression plasmid encoding P. aeruginosa MepM (Pa), E. coli MepM (Ec), or E. coli MepM with the 11 C-terminal amino acids from P. aeruginosa MepM added to its C terminus (+ Pa CT). The C termini of all proteins terminated with the FLAG tag sequence. For both panels, strains were grown in medium containing arabinose. Proteins were detected with ant-FLAG monoclonal antibodies, and a Ponceau S total protein stain of the same region of the nitrocellulose membrane is shown to document loading levels. Immunoblots are single representatives of several replicate experiments.
PA3787 lacks the LysM peptidoglycan-binding domain that is found in MepM, making it a shorter protein with only 18% overall identity to MepM. Therefore, to test our conclusion more rigorously, we extended our experiments to include E. coli MepM, which is a closer homolog of P. aeruginosa MepM than is PA3787. P. aeruginosa and E. coli MepM proteins are similar in length, have the same domain organization, and are approximately 30% identical (data not shown). Both have LysM peptidoglycan-binding and LytM/M23 peptidase domains, and their homology is distributed throughout their lengths (data not shown). However, the C-terminal KKTKLALNKQR motif of P. aeruginosa MepM is absent from E. coli MepM (Fig. 1A). To further test if the C-terminal amino acids are sufficient for degradation by CtpA, we added the entire KKTKLALNKQR sequence onto the C terminus of E. coli MepM. This was followed by a FLAG tag sequence, because we did not have a reagent to detect E. coli MepM and because a C-terminal FLAG tag does not prevent the degradation of any CtpA substrate, including MepM (14). As expected, P. aeruginosa MepM-FLAG was undetectable in a ctpA+ strain but abundant in a ΔctpA mutant, consistent with its degradation by CtpA as in our previous study (Fig. 4B) (14). In contrast, E. coli MepM-FLAG was equally abundant in both strains, suggesting that it is not degraded by CtpA. Furthermore, E. coli MepM-FLAG with the KKTKLALNKQR motif added was also equally abundant in both strains. Therefore, these experiments further support the conclusion that the C terminus of a CtpA substrate is important, but not sufficient, for degradation by CtpA in vivo.
Truncation of the C terminus protects substrates from degradation by CtpA in vitro.
It was possible that the C-terminal truncation mutations of MepM and PA1199 had an indirect effect in vivo, rather than rendering the proteins more resistant to direct degradation by CtpA. One example would be if their localization was affected so that they became separated from CtpA. Therefore, we used an in vitro proteolysis assay to test our conclusion that the C-terminal amino acids are important for direct degradation by CtpA. The CtpA-binding partner LbcA, which increases CtpA activity, CtpA itself, and an inactive CtpA-S302A control (catalytic serine changed to alanine) were purified with C-terminal His6 tags, as before (14). However, MepM, MepM-Δ7, PA1199, and PA1199-Δ7 were purified with N-terminal His6 tags, so that their C termini were unaltered and identical to the proteins we had studied in the in vivo experiments.
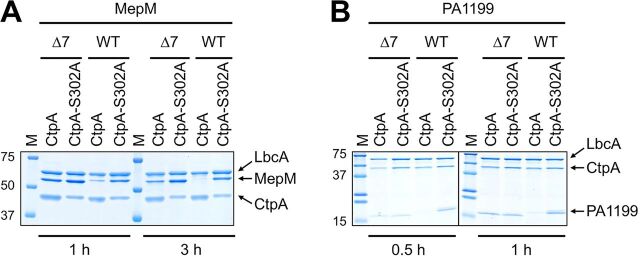
When MepM was incubated with CtpA and LbcA, it was mostly degraded after 1 h and completely degraded after 3 h (Fig. 5A). In contrast, MepM-Δ7 was resistant to degradation at both time points, with no degradation evident after 1 h and only slight degradation after 3 h. PA1199 was degraded more quickly than MepM in vitro, such that it was degraded completely after 30 min (Fig. 5B). However, the PA1199-Δ7 protein was resistant to degradation after 1 h (Fig. 5B), and we have also found that it remains resistant to degradation after 3 h (data not shown). These experiments suggest that the C-terminal 7 amino acids of MepM and PA1199 are important for their direct degradation by the LbcA-CtpA complex.
FIG 5.
Truncation of the C termini of MepM and PA1199 protects them from degradation by CtpA in vitro. (A) His6-MepM (WT) or His6–MepM-Δ7 proteins were incubated with LbcA-His6 and either active CtpA-His6 (CtpA) or inactive CtpA-S302A–His6 (CtpA-S302A) for 1 h or 3 h. Samples were separated by 10% SDS-PAGE and stained with ProtoBlue Safe (National Diagnostics). (B) His6-PA1199 (WT) or His6-PA1199-Δ7 proteins were incubated with LbcA-His6 and either active CtpA-His6 (CtpA) or inactive CtpA-S302A–His6 (CtpA-S302A) for 0.5 h or 1 h. Samples were analyzed on a single 16% SDS-PAGE gel, but the order of the left- and right-hand sides of the gel was reversed to construct the image, indicated by the vertical black line. For both panels, approximate sizes of molecular-mass marker proteins (M), in kilodaltons, are indicated on the left-hand side. Gels are single representatives of several replicate experiments.
Truncation of the C terminus does not prevent substrates from associating with the LbcA-CtpA complex in vivo.
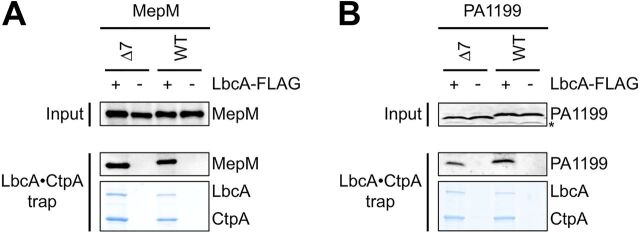
MepM was discovered as a CtpA substrate because it copurified with the proteolytically inactive LbcA–CtpA-S302A complex after in vivo formaldehyde cross-linking (14). We considered the possibility that the truncated substrates are resistant to degradation because they can no longer associate with the LbcA-CtpA complex. Alternatively, the truncated substrates might still associate with LbcA-CtpA, but their degradation is inhibited for another reason, perhaps because CtpA most efficiently initiates cleavage from a missing C-terminal motif. To distinguish between these possibilities, we repeated the procedure that discovered MepM (14), to test if the MepM-Δ7 and PA1199-Δ7 proteins were still trapped by an inactive LbcA–CtpA-S302A complex.
ΔctpA ΔmepM strains contained a plasmid encoding CtpA-S302A–His6 and a second plasmid encoding LbcA-FLAG and either MepM or MepM-Δ7. The LbcA–CtpA-S302A complex was purified using nickel-agarose first to capture CtpA-S302A–His6, followed by anti-FLAG affinity gel to capture LbcA-FLAG. Coomassie staining of the samples after SDS-PAGE revealed that LbcA and CtpA-S302A purified abundantly, as expected (Fig. 6A). Confirmation that an LbcA–CtpA-S302A complex was isolated was provided by negative-control purifications from strains that did not encode LbcA-FLAG but still encoded CtpA-S302A–His6. Neither LbcA nor CtpA-S302A purified abundantly from these controls (Fig. 6). Immunoblot analysis showed that both MepM and MepM-Δ7 were captured with the LbcA–CtpA-S302A complex but not in the negative controls (Fig. 6A). A similar experiment to analyze PA1199 revealed that both PA1199 and PA1199-Δ7 were captured by the LbcA–CtpA-S302A complex as well (Fig. 6B). These results suggest that removal of the C-terminal 7 amino acids from MepM or PA1199 does not prevent their association with the LbcA-CtpA complex in vivo. Therefore, it is likely that the C termini of these substrates are important to initiate their degradation after they have been engaged by the proteolytic complex (see Discussion).
FIG 6.
Truncation of the C terminus does not prevent substrates from associating with the LbcA-CtpA complex in vivo. (A) Proteins were purified from detergent-solubilized lysates of ΔmepM ΔctpA strains, which contained one plasmid encoding CtpA-S302A–His6 and a second plasmid encoding MepM or MepM-Δ7 and LbcA-FLAG (+) or MepM or MepM-Δ7 only (−). (B) Proteins were purified from detergent-solubilized lysates of Δ(PA1198-PA1199) ΔctpA strains, which contained one plasmid encoding CtpA-S302A–His6 and a second plasmid encoding PA1199 or PA1199-Δ7 and LbcA-FLAG (+) or PA1199 or PA1199-Δ7 only (−). For both panels, tandem affinity purification of the LbcA–CtpA-S302A complex was done with nickel-agarose followed by anti-FLAG M2–agarose resin. Input lysates (Input) and purified samples (LbcA·CtpA trap) were separated by SDS-PAGE and analyzed by anti-MepM (A) or anti-PA1199 (B) immunoblotting (an asterisk indicates a protein cross-reactive with the PA1199 antiserum). Purified samples were also separated by SDS-PAGE and stained with ProtoBlue Safe (National Diagnostics) to monitor recovery of the LbcA-CtpA complex. Immunoblots and gels are single representatives of several replicate experiments.
A conserved amino acid at the −5 position is not essential for CtpA-dependent degradation.
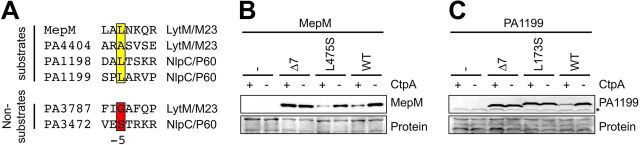
The sequences, charges, and hydrophobicities of the C-terminal amino acids of MepM and PA1199 do not have obvious similarity. However, an alignment of the C-terminal 7 amino acids of all four CtpA substrates revealed that three of them have leucine as the fifth amino acid from their C terminus (referred to as the −5 position) and the other one, PA4404, has alanine (Fig. 7A). These four CtpA substrates are LytM/M23 or NlpC/P60 peptidase family members. Two other predicted peptidoglycan cross-link hydrolase members of these families are not CtpA substrates, PA3787 and PA3472 (14). Neither of these has leucine or alanine at the −5 position (Fig. 7A). In ongoing work in our laboratory, we have identified a fifth CtpA substrate preliminarily, which also has leucine at the −5 position (D. Chakraborty, A. G. Sommerfield, and A. J. Darwin, unpublished data). This means that three or four CtpA substrates have leucine at position −5 and one has alanine, whereas neither of two related nonsubstrates share this property. Therefore, we tested if the leucine at position −5 of MepM or PA1199 was important for their degradation by CtpA.
FIG 7.
Mutation of the −5 position affects the CtpA-dependent stability of MepM and PA1199 differently. (A) Alignment of the C-terminal 7 amino acids of predicted peptidoglycan cross-link hydrolases in the LytM/M23 or NlpC/P60 peptidase families that either are or are not CtpA substrates. The −5 positions of the substrates and nonsubstrates are highlighted in yellow and red, respectively. (B) MepM immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains. (C) PA1199 immunoblot analysis of equivalent amounts of whole-cell lysates of ctpA+ and ΔctpA strains (an asterisk indicates a protein cross-reactive with the PA1199 antiserum). For PA1199 analysis, both strains also had a Δprc mutation to eliminate any interference from degradation of mutant proteins by Prc. For panels B and C, strains were grown in medium containing arabinose and contained an arabinose-inducible expression plasmid encoding wild-type MepM or PA1199 or derivatives with the indicated amino acid substitution at the −5 position or with 7 amino acids removed from the C terminus. MepM and PA1199 were detected with polyclonal antisera, and a Ponceau S total protein stain of the same region of the nitrocellulose membrane is shown to document loading levels. Immunoblots are single representatives of several replicate experiments.
The nonsubstrate PA3472 has serine at the −5 position (Fig. 7A). Therefore, we made the conservative leucine-to-serine substitution at the −5 positions of MepM and PA1199. These substitutions had different effects on the steady-state levels of the proteins. The MepM L475S mutant behaved indistinguishably from wild-type MepM, suggesting that the leucine at position −5 is not important for its efficient degradation by CtpA (Fig. 7B). In contrast, the PA1199 L173S mutant behaved indistinguishably from the PA1199-Δ7 truncation mutant, showing that its leucine at the −5 position is important for efficient CtpA-dependent degradation. Taken together, these results suggest that while individual amino acids can influence CtpA-dependent destabilization in vivo, a conserved position-specific sequence signature for degradation is unlikely (see Discussion).
DISCUSSION
CtpA is essential for P. aeruginosa type III secretion system (T3SS) function and for virulence in a mouse model of acute pneumonia, and it affects surface attachment (14, 17). These phenotypes are probably linked to the cell wall, because CtpA degrades peptidoglycan cross-link hydrolases (14). This means that CtpA is critical for fundamental cell envelope physiology and for the ability of P. aeruginosa to cause disease. Therefore, it is important to understand all aspects of CtpA function, including the features of a protein that make it susceptible to CtpA-dependent proteolysis. The carboxyl-terminal processing protease (CTP) family was named because the first members studied were found to cleave close to the C terminus of their substrates, either as a processing event, or to initiate degradation (for examples, see references 8 to 10). This suggests that substrate C termini might contain information that is recognized by a CTP. Indeed, some CTPs, including E. coli Prc, have been proposed to target proteins with nonpolar and/or structurally unconstrained C termini (10, 18–20, 23, 24). However, the role of the substrate C terminus has not been studied for most CTPs, and the same rules are unlikely to apply to all of them. In fact, Xanthomonas campestris Prc cleaves close to the N terminus of a transmembrane substrate and cannot rely on recognition features in the C terminus, which is physically separated from Prc by the cytoplasmic membrane (12).
We have investigated if the C termini of CtpA substrates play a role in their degradation, using one substrate in the LytM/M23 peptidase family (MepM) and one in the NlpC/P60 family (PA1199) as model substrates. Despite the fact that MepM and PA1199 are not homologous, in both cases their C-terminal amino acids were important for degradation (Fig. 1 and 5). The C-terminal 7 amino acids of MepM and PA1199 could also be exchanged without affecting their CtpA-dependent degradation in vivo, whereas substitution with an unrelated 7 amino acids rendered the proteins resistant to degradation (Fig. 2 and 3). This suggests that the C termini of CtpA substrates contain specific information that is important for efficient degradation. However, the sequences of the 7 C-terminal amino acids of MepM and PA1199 are not similar, and a comparison of all four CtpA substrates also failed to reveal obvious common C-terminal features (Fig. 7A). Notably, they are not predominantly nonpolar, which is a feature that emerged from early research as something common among E. coli Prc substrates (20). We did notice that CtpA substrates have leucine or alanine at the −5 position, whereas two nonsubstrates in the LytM/M23 and NlpC/P60 peptidase families do not (Fig. 7A). However, although changing this leucine to serine protected PA1199 from CtpA-dependent degradation, it did not protect MepM (Fig. 7). From all of these observations, we conclude that the information required for efficient CtpA-dependent degradation is not a conserved C-terminal amino acid sequence motif but perhaps another property, such as a structural feature. JPred4 secondary-structure predictions of the known CtpA substrates suggests that the C-terminal 9 to 18 amino acids of PA1198, PA1199, and PA4404 are unstructured but that only the C-terminal 2 or 3 amino acids of MepM might be unstructured (25).
An early study of E. coli Prc concluded that the C-terminal 5 amino acids of a substrate were sufficient for cleavage, because when added onto the C terminus of a nonsubstrate it was cleaved by Prc (23). However, the addition of the P. aeruginosa MepM C terminus onto two other LytM/M23 peptidases did not make either of them a CtpA substrate (Fig. 4). Therefore, the C terminus of CtpA substrates is not sufficient for degradation. Later work on E. coli Prc revealed that it forms a complex with the outer membrane lipoprotein NlpI, which promotes Prc-dependent degradation of MepS in vivo and in vitro (13). Protein interaction and structure-function analysis suggested that NlpI binds to Prc and MepS independently, acting as a scaffold to bring protease and substrate together (10, 13). CtpA also has an outer membrane lipoprotein binding partner, LbcA, which promotes the degradation of all four CtpA substrates in vivo and in vitro (14). LbcA and NlpI are not homologous, but both contain tetratricopeptide repeats (TPR) that mediate the formation of multiprotein complexes (26). We have evidence that LbcA also acts as a scaffold, binding CtpA and its substrates independently (D. Chakraborty and A. J. Darwin, unpublished data). The MepM and PA1199 C-terminal truncation mutants still associated with the LbcA-CtpA complex in vivo (Fig. 6). This suggests that substrate C termini are not involved in the association with LbcA. Therefore, the C terminus of MepM is probably insufficient for degradation, because when transplanted onto a nonsubstrate it cannot provide a required association with the LbcA-CtpA complex. Interestingly, recent work indicated that cleavage of one proposed substrate of E. coli Prc, FtsI, is not helped by the NlpI binding partner of Prc in vitro (27). If something similar occurs in vivo, it would mean that NlpI promotes the cleavage of some substrates (MepS) but is not required for the cleavage of others (FtsI). That might explain why the transplantation of a Prc substrate C terminus onto at least one nonsubstrate was sufficient for its degradation (23).
Structural analysis of the E. coli NlpI-Prc complex, and a docking model of the C-terminal 12-amino-acid peptide of its MepS substrate, suggests that the substrate C terminus is bound by the PDZ domain of Prc (10). The Prc PDZ domain was proposed to recognize substrate C termini with low specificity, because Prc degrades MepS with or without a C-terminal His6 tag and will also degrade lysozyme in vitro if a disulfide bond in its C terminus is broken by reduction (10). In contrast, our analysis suggests that there is specific recognition of the C termini of CtpA substrates. However, all four CtpA substrates are still degraded in vivo when a FLAG tag is added onto their C termini and in vitro with C-terminal His6 tags (14). This can still be reconciled with our conclusion that substrate C termini are recognized specifically. We hypothesize that the native C-terminal amino acids of CtpA substrates can still make a required specific interaction with CtpA even if nonnative amino acids have been added onto them.
In summary, this work has shown that the C termini of P. aeruginosa CtpA substrates contain specific information that is important, but not sufficient, for degradation. We hypothesize that the cleavage of a protein by CtpA requires at least two phenomena: (i) association of the substrate with the LbcA-CtpA complex (most likely with LbcA) and (ii) a specific recognition of the substrate C terminus by CtpA. However, any interaction with LbcA might have to occur in a specific way, perhaps with one or more specific TPR motifs, of the 11 that are present in LbcA. This is because we have evidence that LbcA might associate with some proteins that are not cleaved by CtpA, including PA3787 (14; Chakraborty and Darwin, unpublished). The NlpI partner of E. coli Prc has also been proposed to interact with some non-Prc substrates (28). Perhaps CtpA substrates and nonsubstrates interact differently with LbcA, possibly engaging different TPR motifs. Regardless, specific engagement of a substrate by LbcA, and then recognition of its C terminus by CtpA, would be followed by the first cleavage event. Degradation might then proceed in a nonspecific manner that does not require specific recognition of the new C terminus generated after this first cleavage event. This might occur similarly to the lever-like mechanism proposed for E. coli Prc, which feeds the substrate into the Prc active site in a C- to N-terminal direction after each cleavage event (10). However, CtpA and Prc are in different CTP subfamilies, and their lipoprotein partners are not homologous. Therefore, the details of the organization and function of the LbcA-CtpA complex are likely to diverge from those of E. coli NlpI-CtpA. The goal of future work will be to uncover exactly what those details are.
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine growth.
Strains and plasmids are listed in Table 1. Bacteria were grown routinely in Luria-Bertani (LB) broth, composed of 1% (wt/vol) tryptone, 0.5% (wt/vol) yeast extract, and 1% (wt/vol) NaCl, or on LB agar, at 30°C or 37°C. P. aeruginosa was occasionally grown on Vogel-Bonner minimal agar (29).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or features | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAKa | Wild type | 31 |
| AJDP730 | ΔctpA | 17 |
| AJDP1228 | ΔmepM | 14 |
| AJDP1229 | ΔctpA ΔmepM | 14 |
| AJDP1264 | Δprc | This study |
| AJDP1265 | ΔctpA Δprc | This study |
| AJDP1385 | Δprc Δ(PA1198-PA1199) | This study |
| AJDP1386 | ΔctpA Δprc Δ(PA1198-PA1199) | This study |
| AJDP1390 | ΔctpA Δ(PA1198-PA1199) | This study |
| Plasmids | ||
| pHERD20T | Ampr, pMB1 ori, araBp expression vector | 32 |
| pET-24b(+) | Kanr, pMB1 ori, T7p expression vector | Novagen |
| pEX18Ap | Ampr, pMB1 ori, oriT, sacB+ | 33 |
| pQE-30 | Ampr, Col E1 ori, T5p expression vector | Qiagen |
| pAJD2290 | T7p-´ctpA-his6 in pET-24b(+) | 17 |
| pAJD2653 | T7p-´lbcA-his6 in pET-24b(+) | 14 |
| pAJD2655 | T7p-´ctpA-S302A-his6 in pET-24b(+) | 14 |
| pAJD2799 | araBp-mepM-FLAG in pHERD20T | 14 |
| pAJD2805 | araBp-mepM in pHERD20T | This study |
| pAJD2897 | araBp-PA1199 in pHERD20T | This study |
| pAJD2909 | araBp-mepM-Δ6 in pHERD20T | This study |
| pAJD2912 | araBp-PA1199-Δ7 in pHERD20T | This study |
| pAJD2929 | araBp-mepM-Δ9 in pHERD20T | This study |
| pAJD2930 | araBp-mepM-Δ8 in pHERD20T | This study |
| pAJD2931 | araBp-mepM-Δ7 in pHERD20T | This study |
| pAJD2935 | araBp-PA3787 in pHERD20T | This study |
| pAJD2936 | araBp-PA3787-MepMCT11 in pHERD20Tb | This study |
| pAJD2939 | araBp-PA1199-Δ6 in pHERD20T | This study |
| pAJD2940 | araBp-PA1199-Δ5 in pHERD20T | This study |
| pAJD2941 | araBp-PA1199-Δ4 in pHERD20T | This study |
| pAJD2946 | T5p-his6-′mepM in pQE-30 | This study |
| pAJD2947 | T5p-his6-′mepM-Δ7 in pQE-30 | This study |
| pAJD2951 | T5p-his6-′PA1199 in pQE-30 | This study |
| pAJD2952 | T5p-his6-′PA1199-Δ7 in pQE-30 | This study |
| pAJD2953 | araBp-PA1199-Δ7-MepMCT7 in pHERD20Tc | This study |
| pAJD2955 | araBp-mepM-Δ7-PA1199CT7 in pHERD20Td | This study |
| pAJD2971 | araBp-PA1199-Δ7-AGEAGHL in pHERD20T | This study |
| pAJD2972 | araBp-mepM-Δ7-AGEAGHL in pHERD20T | This study |
| pAJD2982 | araBp-mepM lbcA-FLAG in pHERD20T | This study |
| pAJD2983 | araBp-mepM-Δ7 lbcA-FLAG in pHERD20T | This study |
| pAJD2984 | araBp-mepMEC-FLAG in pHERD20Te | This study |
| pAJD2985 | araBp-mepMEC-MepMCT11-FLAG in pHERD20Tf | This study |
| pAJD2992 | araBp-PA1199 lbcA-FLAG in pHERD20T | This study |
| pAJD2993 | araBp-PA1199-Δ7 lbcA-FLAG in pHERD20T | This study |
All P. aeruginosa strains are derivatives of strain PAK.
PA3787 with the C-terminal 11 amino acids of MepM added to its C terminus.
PA1199 with its C-terminal 7 amino acids replaced by those of MepM.
MepM with its C-terminal 7 amino acids replaced by those of PA1199.
mepMEC is the E. coli mepM gene.
E. coli MepM with the 11 C-terminal amino acids of P. aeruginosa MepM added to its C terminus.
Strain constructions.
To construct Δprc and Δ(PA1198-PA1199) in-frame deletion mutants, two fragments of ∼0.5 kb each corresponding to the regions flanking the deletion site were amplified by PCR and cloned into pEX18Ap. The plasmids were integrated into the P. aeruginosa chromosome after conjugation from E. coli strain SM10 (30), and sucrose-resistant, carbenicillin-sensitive segregants were isolated on LB agar containing 10% sucrose. Deletions were verified by genomic PCR analysis.
Plasmid constructions.
araBp expression plasmids encoding MepM, PA1199, or PA3787 were constructed by amplifying the genes from P. aeruginosa chromosomal DNA using one primer that annealed ∼30 bp upstream of the start codon and a second primer that annealed immediately downstream of the stop codon. Plasmids encoding C-terminal truncated derivatives were constructed similarly, except that the downstream primers annealed within the gene and incorporated a premature stop codon. Plasmids encoding MepM or PA1199 with their C-terminal 7 amino acids exchanged, or replaced by AGEAGHL, were constructed using downstream primers that annealed 21 bp upstream of the stop codon and incorporated a region encoding the final 7 amino acids of MepM or PA1199 or the AGEAGHL sequence, followed by a stop codon. To construct a plasmid encoding PA3787 with the C-terminal 11 amino acids of MepM added, the downstream primer annealed immediately upstream of the PA3787 stop codon and incorporated a region encoding the C terminus of MepM followed by a stop codon. Plasmids encoding E. coli MepM-FLAG were constructed by amplifying mepM from E. coli strain MG1655 chromosomal DNA. The forward primer annealed immediately upstream of the start codon and incorporated the ribosome binding site from pQE-30, and the reverse primer annealed immediately upstream of the stop codon and incorporated a region encoding the FLAG tag only, or the C-terminal 11 amino acids of P. aeruginosa MepM followed by a FLAG tag, and a stop codon. In all cases, the amplified fragments were cloned into pHERD20T using restriction sites added to the fragments by the amplification primers.
For the LbcA–CtpA-S302A trap experiments, araBp expression plasmids were constructed to encode MepM or PA1199 full-length or C-terminal truncated proteins, as well as LbcA-FLAG. lbcA was amplified from P. aeruginosa chromosomal DNA using a primer that annealed ∼40 bp upstream of the start codon and a primer that annealed immediately upstream of the stop codon and incorporated a region encoding the FLAG tag followed by a stop codon. This was cloned as an XbaI-HindIII fragment (restriction sites incorporated by the amplification primers) immediately downstream of the mepM or PA1199 genes in the expression plasmids described above. pET-24b(+) derivatives used for overproduction and purification of LbcA-His6, CtpA-His6, and CtpA-S302A–His6 were described previously (14, 17). For overproduction and purification of His6-MepM or His6-PA1199 full-length and C-terminally truncated proteins, the genes were amplified without their predicted N-terminal signal sequences and cloned into pQE-30 as BamHI-HindIII fragments.
Determination of protein abundance in vivo.
Saturated cultures were diluted into 5 ml of LB broth, containing 150 μg/ml of carbenicillin and 0.02% (wt/vol) arabinose, in 18-mm-diameter test tubes so that the initial optical density (OD) at 600 nm was 0.05. The cultures were grown on a roller drum at 37°C for 5 h. Cells were collected by centrifugation and resuspended in SDS-PAGE sample buffer at equal concentrations (based on the culture OD at 600 nm) before being analyzed by immunoblotting.
Polyclonal antisera and immunoblotting.
Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane by semidry electroblotting. For analysis of total cell lysates, approximately equal loading and transfer were confirmed by total protein staining of the nitrocellulose membrane with Ponceau S (Amresco). Chemiluminescent detection followed incubation with polyclonal antiserum or monoclonal antibody and then goat anti-rabbit IgG (Sigma) or goat anti-mouse IgG (Sigma) horseradish peroxidase conjugates used at the manufacturer’s recommended dilution. The primary anti-FLAG M2 (Sigma) antibody was diluted 5,000-fold, and all polyclonal antisera used in this study have been described previously (14).
Protein purification and in vitro proteolysis assay.
LbcA-His6, CtpA-His6, and CtpA-S302A–His6 were encoded by pET-24b(+) derivatives in E. coli ER2566 (New England BioLabs [NEB]). His6-MepM and His6-PA1199 full-length and C-terminally truncated proteins were encoded by pQE-30 derivatives in E. coli M15 containing plasmid pREP4 to produce the LacI repressor (Qiagen). These strains were grown in 1 liter of LB broth at 37°C with aeration until the OD at 600 nm was 0.6 to 1.0. Protein production was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubation for 3 to 4 h at 37°C (LbcA-His6, CtpA-His6, and CtpA-S302A–His6) or at 30°C (His6-MepM and His6-PA1199), with aeration. Proteins were purified under native conditions by nitrilotriacetic acid (NTA)-agarose affinity chromatography in buffer containing 50 mM NaH2PO4 and 300 mM NaCl, as recommended by the manufacturer (Qiagen). LbcA-His6, His6-MepM, and His6-PA1199 proteins were eluted in fractions using 50 mM NaH2PO4 and 300 mM NaCl buffer containing increasing concentrations of imidazole (50 to 250 mM). Samples of each fraction were separated by SDS-PAGE and stained with ProtoBlue Safe (National Diagnostics), and 2 or 3 fractions judged to have the highest purity were combined, supplemented with 10% (wt/vol) glycerol, and stored at –70°C. The same fractions were used for His6-MepM and for His6–MepM-Δ7, and the same fractions were used for His6-PA1199 and for His6–PA1199-Δ7. CtpA-His6 and CtpA-S302A–His6 were eluted similarly, but after combining fractions the proteins were concentrated ∼10-fold using Amicon Ultra-4 centrifuge filter devices (10-kDa cutoff) and then supplemented with 50% protein-stabilizing cocktail (Thermo Fisher Scientific) before storage at –70°C. All in vitro proteolysis reaction mixtures contained approximately 2 μM LbcA and CtpA or CtpA-S302A. MepM proteins were also used at 2 μM, but the smaller PA1199 proteins were used at approximately 15 μM to facilitate visualization after staining. Reaction mixtures were incubated at 37°C for 0.5 to 3 h; reactions were terminated by adding SDS-PAGE sample buffer and boiling, followed by separation by SDS-PAGE and staining with ProtoBlue Safe (National Diagnostics).
Tandem affinity purification LbcA-FLAG–CtpA-S302A-His6 complex.
Strains were grown to saturation, diluted to an OD at 600 nm of 0.05 in 400 ml of LB broth containing 5 mM EGTA, and shaken at 200 rpm for 2.5 h at 37°C. The cultures were supplemented with 0.02% (wt/vol) arabinose and 1 mM IPTG and shaken at 200 rpm for a further 3 h at 37°C. Cells from the equivalent of 200 ml of culture at an OD at 600 nm of 1 were collected by centrifugation. The pellet was washed with cold 10 mM potassium phosphate buffer (pH 8.0) and resuspended to an OD at 600 nm of 5 in the same buffer. Formaldehyde (1%) was added, followed by incubation at room temperature for 30 min. Tris-HCl (0.3 M; pH 7.5) was added to quench, and the cells were collected by centrifugation. Pellets were resuspended in 3 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM imidazole), and Roche complete protease inhibitors (2× concentration), 1 μg/ml of DNase I, 1 μg/ml of RNase, and 1 mg/ml of lysozyme were added. Cells were disrupted by sonication, and then 1% n-dodecyl β-d-maltoside (DDM) was added, followed by incubation with rotation for 30 min at 4°C. Insoluble material was removed by centrifugation at 13,000 × g for 30 min at 4°C. Nickel-NTA-agarose (500 μl) in lysis buffer was added to the supernatant, followed by incubation with rotation for 50 min at 4°C. The resin was collected in a drip column and washed with 8 ml of lysis buffer and then 8 ml of lysis buffer containing 20 mM imidazole. Proteins were eluted in 1 ml of lysis buffer containing 250 mM imidazole, mixed with 40 μl of anti-FLAG M2–agarose resin (Sigma) in TBS (10 mM Tris-HCl [pH 7.5], 150 mM NaCl), and incubated with rotation for 2 h at 4°C. A 1-ml spin column (Pierce; 69725) was used to wash the resin seven times with 500 μl of TBS. Proteins were eluted by addition of 100 μl of 200-μg/ml 3×FLAG peptide (Sigma) in TBS and incubation with rotation at 4°C for 30 min.
ACKNOWLEDGMENTS
This study was supported by award number R01AI136901 from the National Institute of Allergy and Infectious Diseases (NIAID).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health.
We thank Dolonchapa Chakraborty for providing advice about the protein purification and in vitro proteolysis assays.
REFERENCES
- 1.Moore NM, Flaws ML. 2011. Epidemiology and pathogenesis of Pseudomonas aeruginosa infections. Clin Lab Sci 24:43–46. doi: 10.29074/ascls.24.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Bleves S, Viarre V, Salacha R, Michel GP, Filloux A, Voulhoux R. 2010. Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int J Med Microbiol 300:534–543. doi: 10.1016/j.ijmm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Wagner S, Königsmaier L, Lara-Tejero M, Lefebre M, Marlovits TC, Galán JE. 2010. Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci U S A 107:17745–17750. doi: 10.1073/pnas.1008053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dufresne K, Paradis-Bleau C. 2015. Biology and assembly of the bacterial envelope. Adv Exp Med Biol 883:41–76. doi: 10.1007/978-3-319-23603-2_3. [DOI] [PubMed] [Google Scholar]
- 5.Barchinger SE, Ades SE. 2013. Regulated proteolysis: control of the Escherichia coli sigma(E)-dependent cell envelope stress response. Subcell Biochem 66:129–160. doi: 10.1007/978-94-007-5940-4_6. [DOI] [PubMed] [Google Scholar]
- 6.De Geyter J, Tsirigotaki A, Orfanoudaki G, Zorzini V, Economou A, Karamanou S. 2016. Protein folding in the cell envelope of Escherichia coli. Nat Microbiol 1:16107. doi: 10.1038/nmicrobiol.2016.107. [DOI] [PubMed] [Google Scholar]
- 7.Paetzel M. 2019. Bacterial signal peptidases. Subcell Biochem 92:187–219. doi: 10.1007/978-3-030-18768-2_7. [DOI] [PubMed] [Google Scholar]
- 8.Che Y, Fu A, Hou X, McDonald K, Buchanan BB, Huang W, Luan S. 2013. C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc Natl Acad Sci U S A 110:16247–16252. doi: 10.1073/pnas.1313894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satoh K, Yamamoto Y. 2007. The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth Res 94:203–215. doi: 10.1007/s11120-007-9191-z. [DOI] [PubMed] [Google Scholar]
- 10.Su MY, Som N, Wu CY, Su SC, Kuo YT, Ke LC, Ho MR, Tzeng SR, Teng CH, Mengin-Lecreulx D, Reddy M, Chang CI. 2017. Structural basis of adaptor-mediated protein degradation by the tail-specific PDZ-protease Prc. Nat Commun 8:1516. doi: 10.1038/s41467-017-01697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng CY, Zhang H, Wu Y, Ding LL, Pan Y, Sun ST, Li YJ, Wang L, Qian W. 2018. Proteolysis of histidine kinase VgrS inhibits its autophosphorylation and promotes osmostress resistance in Xanthomonas campestris. Nat Commun 9:4791. doi: 10.1038/s41467-018-07228-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh SK, Parveen S, SaiSree L, Reddy M. 2015. Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci U S A 112:10956–10961. doi: 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava D, Seo J, Rimal B, Kim SJ, Zhen S, Darwin AJ. 2018. A proteolytic complex targets multiple cell wall hydrolases in Pseudomonas aeruginosa. mBio 9:e00972-18. doi: 10.1128/mBio.00972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings ND, Barrett AJ, Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res 38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoge R, Laschinski M, Jaeger KE, Wilhelm S, Rosenau F. 2011. The subcellular localization of a C-terminal processing protease in Pseudomonas aeruginosa. FEMS Microbiol Lett 316:23–30. doi: 10.1111/j.1574-6968.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 17.Seo J, Darwin AJ. 2013. The Pseudomonas aeruginosa periplasmic protease CtpA can affect systems that impact its ability to mount both acute and chronic infections. Infect Immun 81:4561–4570. doi: 10.1128/IAI.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. 2000. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry 39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- 19.Keiler KC, Silber KR, Downard KM, Papayannopoulos IA, Biemann K, Sauer RT. 1995. C-terminal specific protein degradation: activity and substrate specificity of the Tsp protease. Protein Sci 4:1507–1515. doi: 10.1002/pro.5560040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silber KR, Keiler KC, Sauer RT. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci U S A 89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh SK, SaiSree L, Amrutha RN, Reddy M. 2012. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 22.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 23.Keiler KC, Sauer RT. 1996. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J Biol Chem 271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 24.Kumru OS, Bunikis I, Sorokina I, Bergstrom S, Zuckert WR. 2011. Specificity and role of the Borrelia burgdorferi CtpA protease in outer membrane protein processing. J Bacteriol 193:5759–5765. doi: 10.1128/JB.05622-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blatch GL, Lassle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939. doi:. [DOI] [PubMed] [Google Scholar]
- 27.Chueh CK, Som N, Ke LC, Ho MR, Reddy M, Chang CI. 2019. Structural basis for the differential regulatory roles of the PDZ domain in C-terminal processing proteases. mBio 10:01129-19. doi: 10.1128/mBio.01129-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banzhaf M, Yau HC, Verheul J, Lodge A, Kritikos G, Mateus A, Cordier B, Hov AK, Stein F, Wartel M, Pazos M, Solovyova AS, Breukink E, van Teeffelen S, Savitski MM, den Blaauwen T, Typas A, Vollmer W. 2020. Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J 39:e102246. doi: 10.15252/embj.2019102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 30.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strom MS, Lory S. 1986. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol 165:367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]