ABSTRACT
To identify DNA oxidation defenses of hyperthermophilic archaea, we deleted genes encoding the putative 7,8-dihydro-8-oxoguanine (oxoG)-targeted N-glycosylase of Sulfolobus acidocaldarius (ogg; Saci_01367), the Y family DNA polymerase (dbh; Saci_0554), or both and measured the effects on cellular survival, replication accuracy, and oxoG bypass in vivo. Spontaneous G·C-to-T·A transversions were elevated in all Δogg and Δdbh constructs, and the Δogg Δdbh double mutant lost viability at a higher rate than isogenic wild-type (WT) and ogg strains. The distribution of G·C-to-T·A transversions within mutation detector genes suggested that reactivity of G toward oxidation and the effect on translation contribute heavily to the pattern of mutations that are recovered. An impact of the Ogg protein on the overall efficiency of bypassing oxoG in transforming DNA was evident only in the absence of Dbh, and Ogg status did not affect the accuracy of bypass. Dbh function, in contrast, dramatically influenced both the efficiency and accuracy of oxoG bypass. Thus, Ogg and Dbh were found to work independently to avoid mutagenesis by oxoG, and inactivating this simple but effective defense system by deleting both genes imposed a severe mutational burden on S. acidocaldarius cells.
IMPORTANCE Hyperthermophilic archaea are expected to have effective (and perhaps atypical) mechanisms to limit the genetic consequences of DNA damage, but few gene products have been demonstrated to have genome-preserving functions in vivo. This study confirmed by genetic criteria that the S. acidocaldarius Ogg protein avoids the characteristic mutagenesis of G oxidation. This enzyme and the bypass polymerase Dbh have similar impacts on genome stability but work independently and may comprise most of the DNA oxidation defense of S. acidocaldarius. The critical dependence of accurate oxoG bypass on the accessory DNA polymerase Dbh further argues that some form of polymerase exchange is important for accurate genome replication in Sulfolobus, and perhaps in related hyperthermophilic archaea.
KEYWORDS: 7,8-dihydro-8-oxoguanine; Archaea; base excision repair; hyperthermophile; spontaneous mutations; translesion DNA polymerase
INTRODUCTION
Extremophilic microorganisms provide a means to study molecular mechanisms that preserve cellular components under unusual physical and chemical stresses. The genome of a bacterial or archaeal cell represents a cellular component that is both critical to biological success and vulnerable to environmentally promoted damage, as this extremely long molecule must be replicated accurately and completely before each cell division. The biological importance of genome maintenance and the challenges that it faces are reflected in the multiple enzyme systems of bacteria and unicellular eukaryotes that repair DNA damage and minimize the impact of persistent DNA lesions (1).
Spontaneous oxidation of guanines in DNA to yield 7,8-dihydro-8-oxoguanine (oxoG) imposes a characteristic burden on genomes. Though sterically similar to G, oxoG has altered H-bonding properties that promote Hoogsteen base pairing with dATP in the polymerase active site. This causes most DNA polymerases to insert A in the nascent strand opposite oxoG (2, 3), which subsequent DNA repair or replication can convert into a permanent G·C-to-T·A transversion mutation. Study of bacteria and eukaryotes has identified multiple mechanisms that act to avoid this mutagenesis. Base excision repair (BER), which represents the first line of defense, involves cleaving of oxoG from the DNA backbone by oxoG-targeted N-glycosylases, examples of which are the Escherichia coli Fpg protein and the Saccharomyces cerevisiae Ogg protein (4). Subsequent processing of the resulting abasic site removes the sugar-phosphate unit and replaces the missing nucleotide, using the opposite, undamaged strand as the template. Alternatively, if a replication fork encounters oxoG that has escaped BER, translesion DNA synthesis (TLS) can avoid mutation by inserting C opposite this lesion, which provides a second line of defense. Corresponding DNA polymerases include S. cerevisiae Polη (5) and Saccharolobus (Sulfolobus) solfataricus Dpo4 (6, 7). A third oxoG coping strategy, represented by the MutY protein of E. coli and related enzymes found in some eukaryotes, uses a BER-like process to remove A from oxoG·A mispairs formed as a result of failure of the first two strategies (4). This recycling reaction temporarily avoids mutation fixation and thus improves the overall performance of repair or TLS indirectly.
The extremely high growth temperatures of hyperthermophilic archaea (HA) accelerate DNA decomposition reactions by orders of magnitude relative to mesophiles; this suggests that rates of spontaneous DNA damage should be greatly elevated in these archaea and may create a correspondingly greater need for DNA repair and mutation avoidance. Genetic assays nevertheless measure low rates of replication errors in Sulfolobus and related archaea (8–10). Although these results demonstrate that HA can achieve accurate replication, it has been difficult to confirm in vivo which gene products contribute to this accuracy. One of the few gene products that has been confirmed to support genetic fidelity in vivo is an accessory DNA polymerase that counters the impact of DNA oxidation. In vitro, the sole Y family DNA polymerase of Sulfolobus acidocaldarius, designated Dbh (DinB homolog), is extremely error prone on an undamaged template, yet it correctly inserts C opposite oxoG (11). Inactivating the corresponding gene, Saci_0554, specifically increases G·C-to-T·A transversion mutations, which are otherwise rare among spontaneous mutations in S. acidocaldarius (12). Inactivating this gene also decreases the efficiency and accuracy with which oxoG placed in the chromosome artificially is bypassed (13).
While these results demonstrate that TLS suppresses the signature mutagenesis of oxoG in S. acidocaldarius, the role of BER has not been investigated genetically. Putative oxoG-targeted N-glycosylases are well represented in the genomes of Sulfolobales and related crenarchaeotes, whereas putative A/G-specific adenine DNA glycosylases (i.e., MutY homologs) generally have not been found (14). In the present study, we constructed S. acidocaldarius strains (Table 1) lacking its putative oxoG-targeted N-glycosylase, the Dbh polymerase, or both and evaluated the impact on replication accuracy and oxoG bypass in vivo.
TABLE 1.
Strains and oligonucleotides
| Sulfolobus strain or oligonucleotide | Genotype | Source or derivation | Sequence (5′ to 3′) |
|---|---|---|---|
| Strains | |||
| MR31 | pyrE131 (bp 159–177 deleted) | Spontaneous Foar mutant (49) | |
| DG185 | Wild type | ATCC 33909 | |
| SD67 | Δogg::pyrESso pyrE131 | This study (see Materials and Methods) | |
| SD67-7 | Δogg::pyrE mutant pyrE131 | Spontaneous Foar mutant of SD67 | |
| RJ4 | Δdbh::pyrEStoV4 | This study (see Materials and Methods) | |
| RJ12 | wild-type | MR31 transformed to Pyr+ by pSAPE5a | |
| RJ13 | Δogg::pyrE mutant pyrE131 | Spontaneous Foar mutant of SD67 | |
| RJ8 | Δogg Δdbh | RJ13 transformed by PCR product of RJ4 | |
| RJ401 | Δdbh::pyrEStoV4 | Spontaneous Foar mutant of RJ4 | |
| RJ803 | Δogg Δdbh | RJ80 transformed to Pyr+ by pSAPE5a | |
| Oligonucleotidesb | |||
| 5′ anchor | ACTTACAAGCAATAAATGAGGCAAATGGAACGCCCCCAGTAACAACTCCCAATATCATAT | ||
| 3′ anchor | P-ATGTCGACTGCAGAACTAACGACGAATGAAAATATGTCAGGATGGTTGGGGAGTTTCCTT | ||
| Control insert | P-CAAATTGAGCNCCTTTGACT | ||
| oxoG insert | P-CAAATTGAGC[8oG]CCTTTGACT | ||
| Downstream scaffold | GCTCAATTTGATATGATATTG | ||
| Upstream scaffold | AGTCGACATAGTCAAAGG | ||
| delSso Sa1367f | AGTTAAGAAAAAAACCAGTAAGAAAAAGAAGAGAAAGAAAAGTGCTAAGATGTCTTAATCTCACAAAG | ||
| delSso Sa1367f | TCACAACTAATCATTTATACTCCTTTTTATACTTTTTTGTTCTATTTTACGATATGAGAGAGGTTTATC | ||
| delSto Sa0554f | CCTTAAATGCTTATACACCAAATACTAAATGTAAATGATAGTGGCAGTGGGTATTTAAAC | ||
| delSto Sa0554r | TTAAGCAAAATCCTTAACTCGTTGCAATTAAATGTCGAAGAAATCCCACTGCCTAGGTCT |
E. coli cloning plasmid containing the wild-type pyrE gene (53).
Synthetic DNAs used in strain constructions and TLS assays (see Materials and Methods).
RESULTS
Removing the putative oxoG repair N-glycosylase of S. acidocaldarius.
DNA N-glycosylases that target oxoG cluster into three families, designated OGG1, OGG2, and AGOG. Most OGG1 members occur in eukaryotes, although a subfamily is found in Gram-positive bacteria (14). The AGOG family was first discovered in Pyrobaculum aerophilum (15) and has been found so far only in a subset of HA (Table 2). The remaining family, OGG2, occurs throughout the remaining groups of bacteria and archaea. All three families of oxoG-specific BER enzymes exhibit N-glycosylase and β-lyase activity but are distinguished from each other by certain sequence motifs and functional properties, including the degree of specificity for the base opposite oxoG in duplex DNA (16).
TABLE 2.
Phylogenetic distribution of oxoG repair N-glycosylases among thermophilic archaeaa
| Family | Species | E valueb |
|---|---|---|
| OGG2 | Sulfolobus acidocaldarius | (0) |
| Sulfurisphaera tokodaii | ||
| Acidianus spp. | ||
| Saccharolobus solfataricus c | E-76 | |
| Metallosphaera spp. | ||
| Sulfodiicoccus sp. | ||
| Archaeoglobus spp. | E-39 | |
| Ferrolobus spp. | ||
| Thermoplasma spp. | ||
| Methanocaldococcus jannaschii | E-34 | |
| AGOG | Pyrobaculum aerophilum d | (0) |
| Pyrobaculum islandicum | ||
| Thermoproteus uzonensis | E-93 | |
| Caldivirga spp. | E-50 | |
| Pyrodictium spp. | ||
| Ignicoccus spp. | ||
| Pyrococcus spp. | ||
| Thermococcus spp. | ||
| Aeropyrum pernix | ||
| Hyperthermus butylicus | E-27 |
Results of BLASTP using the protein encoded by Saci_1367 (OGG2 family) or Pae2237 (AGOG family) as the query; exemplary species of cultured archaea are listed in order of decreasing similarity to the query sequence.
For reference, approximate expectation values are listed for selected genera or species (some are averaged over multiple sequences).
Reflects new genus assignment of Sulfolobus solfataricus (7).
Pyrobaculum genomes were confirmed to lack OGG2 representatives.
One open reading frame (ORF) of the S. acidocaldarius genome, Saci_1367, or ogg, has been annotated as encoding a putative oxoG-specific N-glycosylase that belongs to the OGG2 family and has no close paralog. Corresponding proteins occur in several closely related HA, but at the genus level, their distribution appears to be rather erratic and complementary to that of AGOG members (Table 2). At least three OGG2 enzymes closely related to the Saci_1367 product (those of Methanocaldococcus jannaschii, Archaeoglobus fulgidus, and Thermoplasma volcanium) have been confirmed experimentally to have oxoG-directed N-glycosylase and β-lyase activities (17–19).
Based on this evidence, we targeted Saci_1367 for deletion by replacement, using selectable cassettes consisting of heterologous pyrE genes (see Materials and Methods). Pyr+ transformants were compared to the recipient strain by PCR, and a confirmed Saci_1367 deletion, designated SD67, was retained for study (see Fig. S2 in the supplemental material). To facilitate analysis of ogg function, we constructed two isogenic deletion strains by a similar approach. In one, the dbh gene (Saci_0554) was deleted by replacement with a cassette distinct from the one used to delete Saci_1367 (see Fig. S3 in the supplemental material). The DNA of this strain then served as a template for a PCR product that transformed a pyrE mutant derivative of SD67 to pyrE+, creating the double mutant RJ8 (Δogg Δdbh). Spontaneous pyrE mutants of the ogg, dbh, and ogg dbh deletion mutants were selected; these Pyr− derivatives and the parental strain MR31 were then modified by restoring the native pyrE gene (Saci_1597) (Table 1). The resulting four isogenic strains, RJ12 (wild type [WT]), RJ13 (Δogg), RJ401 (Δdbh), and RJ803 (Δogg Δdbh), were then compared in various analyses to determine the functional impacts and epistatic interactions of the gene deletions.
Phenotypic properties.
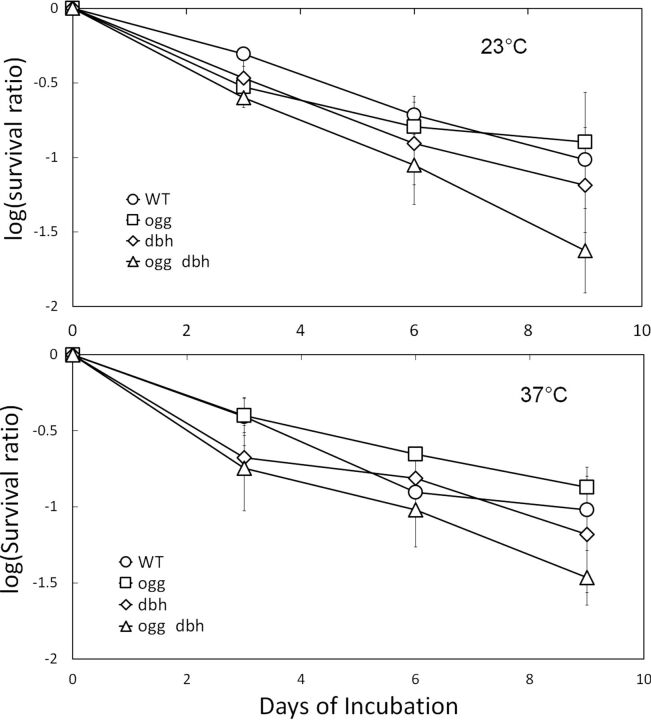
No consistent growth differences among the four isogenic strains were noted during routine cultivation, and individual cultures varied somewhat in preliminary growth curve experiments (see Table S1 in the supplemental material). When compared more systematically with respect to survival of UV and growth inhibition by chemicals that damage DNA in vivo, all the strains yielded the same MIC values for each compound tested and similar survival rates as a function of the UV dose (see Table S1). We also monitored survival under various nongrowth conditions over several days. At 4°C, rates of cell death were high and nearly identical for all four strains, whereas at slightly higher temperatures, the Δogg Δdbh double mutant died more quickly than one or more of the Dbh+ strains of the set (Fig. 1).
FIG 1.
Comparative survival of isogenic constructs. Serial dilutions of strains RJ12 (WT), RJ13 (Δogg), RJ401 (Δdbh), and RJ803 (Δogg Δdbh) in xylose-tryptone medium were incubated under nongrowth conditions (23°C with 9 h fluorescent illumination per day or 37°C in the dark). Viable counts were determined by plating at the times indicated. The data are averaged from three independent trials. The error bars indicate ±1 standard error of the mean and do not overlap for the last time points of the WT or Δogg strain versus the Δogg Δdbh strain under either condition. Similarly, the following slopes calculated by linear regression differed by the sum of twice the standard error of each regression for the following cases, indicating statistical significance (52): 23°C, WT versus Δogg Δdbh (−0.1137 versus −0.1803); 37°C, Δogg versus Δogg Δdbh (−0.1028 versus −0.1711).
Genetic properties.
A preliminary assessment of the genetic impacts of deleting ogg and dbh was made by selecting inactivation of the pyrE cassette inserted at each locus. Although cassettes with different sequence were used to delete the two genes, they exhibited similar properties relative to a cassette inserted into the trpC gene (12). Specifically, the cassettes replacing the ogg and dbh genes exhibited markedly higher frequencies of G·C-to-T·A transversions than the one replacing trpC (see Table S2 in the supplemental material). The effect was most pronounced in the Δogg Δdbh construct RJ8, in which all the identified mutations were G·C-to-T·A events.
A more systematic analysis involved constructing four isogenic strains representing the wild-type, Δogg, Δdbh, and Δogg Δdbh genotypes with the native pyrE gene restored (Table 1). For all four strains, fluctuation assays were performed on multiple sets of independent liquid cultures using 5-fluoroorotic acid (FOA) selection for loss of this native pyrE gene function (see Materials and Methods). Under these conditions, deleting either ogg or dbh had limited impact on the overall rate of forward mutation, whereas deleting both ogg and dbh increased the overall mutation rate 5-fold relative to the WT (Table 3). Thus, with respect to the rate of forming spontaneous FOA-resistant (Foar) mutations, the ogg and dbh deletions did not exhibit genetic epistasis.
TABLE 3.
Effects of ogg and dbh on spontaneous mutation
| Mutation typea | Wild type |
ogg
|
dbh
|
ogg dbh
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Countb | % | Observed ratec | Relative to WT | Count | % | Observed rate | Relative to WT | Countb | % | Observed rate | Relative to WT | Count | % | Observed rate | Relative to WT | |
| Tract expansion | 46 | 42.2 | 23 | 27.4 | 29 | 26.1 | 5 | 4.9 | ||||||||
| Tract contraction | 22 | 20.2 | 10 | 11.9 | 18 | 16.2 | 0 | 0.0 | ||||||||
| Isolated base pairs inserted | 1 | 0.9 | 0 | 0.0 | 5 | 4.5 | 0 | 0.0 | ||||||||
| Isolated base pairs deleted | 2 | 1.8 | 0 | 0.0 | 1 | 0.9 | 1 | 1.0 | ||||||||
| Total indels <5 bp | 71 | 65.1 | 3.76 | 1 | 33 | 39.3 | 1.74 | 0.46 | 53 | 47.7 | 0.87 | 0.23 | 6 | 5.8 | 1.68 | 0.45 |
| G to T + C to A | 2 | 1.8 | 0.11d | 1d | 35 | 41.7 | 1.85d | 17.43d | 26 | 23.4 | 0.43d | 4.05d | 95 | 92.2 | 26.6d | 251d |
| Other transversions | 8 | 7.3 | 2 | 2.4 | 5 | 4.5 | 0 | 0.0 | ||||||||
| Transitions | 18 | 16.5 | 9 | 10.7 | 17 | 15.3 | 0 | 0.0 | ||||||||
| Total BPS | 28 | 25.7 | 1.48 | 1 | 46 | 54.8 | 2.43 | 1.64 | 48 | 43.2 | 0.79 | 0.53 | 95 | 92.2 | 26.6 | 17.9 |
| Tandem dupl >4 bp | 8 | 7.3 | 3 | 3.6 | 9 | 8.1 | 1 | 1.0 | ||||||||
| Deletions >4 bp | 2 | 1.8 | 2 | 2.4 | 1 | 0.9 | 1 | 1.0 | ||||||||
| Total indels >4 bp | 10 | 9.2 | 0.53 | 1 | 5 | 6.0 | 0.26 | 0.50 | 10 | 9.0 | 0.16 | 0.31 | 2 | 1.9 | 0.56 | 1.06 |
| All mutations | 109 | 100 | 5.77e | 1 | 84 | 100 | 4.43e | 0.77 | 111 | 100 | 1.83 | 0.31 | 103 | 100 | 28.8 | 4.99 |
Indel, insertion or deletion; dupl, duplication.
pyrE mutations in wild-type (DG185) and dbh (CS2) strains were determined by Sakofsky et al. (12); spectra for Δogg (RJ13) and Δogg Δdbh (RJ803) strains were determined in the present study, using the same procedure (see Materials and Methods).
Mutation rates for wild-type, ogg, dbh, and ogg dbh strains (RJ12, RJ13, RJ401, and RJ803, respectively) were determined in the present study (see Materials and Methods). The rate for each major type of mutation was calculated as the indicated percentage of the overall mutation rate.
Values for G-to-T transversions are provided for comparison to the aggregate categories.
The 95% confidence intervals of the mutation rates for strains RJ12 (wild type) and RJ13 (ogg) overlap.
One spontaneous Foar mutant was then picked at random from each independent culture of the Δogg construct (RJ13) and the Δogg Δdbh construct (RJ803), and the pyrE gene was analyzed by sequencing. As seen in previous studies, nearly all the independent mutants were found to contain only one mutation each, which in each case altered the predicted pyrE gene product. To estimate the impacts of ogg and dbh on particular types of mutations, we compared the proportions of different mutations in the sets of independent mutants we derived from strains RJ13 and RJ803 and the corresponding sets previously generated by the same methods from strains DG185 (WT) and CS2 (dbh) (12). Scaling the proportions of mutations observed in each set by the overall rates of spontaneous mutation in the four isogenic strains (Table 3) thus provided a basis for comparing the effects of ogg and dbh on various types of mutation.
The results suggested, for example, that inactivating either ogg or dbh had similar effects on the pattern of spontaneous mutation in S. acidocaldarius (Table 3). In both cases, the frequency of base pair substitutions (BPSs) increased relative to other spontaneous mutations, and this increase was due almost entirely to large increases in the frequency of G·C-to-T·A transversion events. The proportions of G·C-to-T·A transversions in the Δogg and Δdbh samples (42% and 23%, respectively) were markedly higher than in the wild-type sample (2%) (Table 3). In addition, the ogg and dbh strains were similar with respect to the relative proportions of the major types of mutation; corresponding chi-square tests distinguished Δogg and Δdbh strains from wild-type and double-deletion constructs but not from each other (see Table S3 in the supplemental material).
When the ogg and dbh deletions were combined in a single strain, their impact on the mutation spectrum increased dramatically. In the Δogg Δdbh double mutant, about 90% of spontaneous mutations inactivating pyrE were G·C-to-T·A transversions, and this increase accounted for the 5-fold elevation of the overall forward mutation rate versus the wild type (a 6.5-fold increase versus ogg) observed in the fluctuation assays (Table 3). The fluctuation data in the current study and previous data for wild-type and dbh strains indicate that deleting both ogg and dbh caused a 250-fold increase in the rate of G·C-to-T·A transversions (Table 3). The genetic evidence thus indicated that, although both gene products act to avoid spontaneous G·C-to-T·A transversion, they do not act in the same biochemical pathway, which is consistent with the enzymatic activities known or predicted for the two proteins.
Analysis of spontaneous transversion mutations.
The spectrum of spontaneous pyrE mutations recovered in the Δogg Δdbh strain (RJ803) is shown in Fig. 2. We observed 96 independent G·C-to-T·A events distributed over 20 of the 215 G or C nucleotides in the pyrE coding sequence. The frequency of G·C-to-T·A transversions in RJ803 varied widely among these 20 sites. The top 3 sites accounted for 56 independent events, for example, whereas 8 sites had only 1 event each (Table 4). An additional 11 G or C sites represented G·C-to-T·A events observed in other strains but not in this set of RJ803 mutants (Tables 4 and 5).
FIG 2.
Spectrum of pyrE mutations in Δogg Δdbh S. acidocaldarius. Each of 103 independent mutations inactivating the native pyrE gene of strain RJ803 is depicted above the corresponding position of the wild-type sequence. Duplication of an existing base pair is indicated by “+,” whereas a single-base deletion as indicated by “Δ”; tracts of the same base pair are overlined to indicate that they behave as a single point with respect to + or Δ events. A 26-bp interval tandemly duplicated in one mutant is overlined, and a 16-bp interval deleted from one mutant is underlined.
TABLE 4.
Distribution of observed transversions in the S. acidocaldarius pyrE genea
| Positionb | Sense strand | Countc | Motif | pre-CTd |
|---|---|---|---|---|
| 3 | G | 6 | gGATTT | ATge |
| 46 | G | 4 | gGAAAT | gGA |
| 77 | G | 1 | ||
| 187 | G | 5 | gGATTT | gGA |
| 199f | G | 20 | ||
| 202 | G | 1 | ||
| 215 | C | 1 | ||
| 250 | G | 26 | gGATAC | gGA |
| 255 | C | 1 | TAc | |
| 265 | G | 2 | gAG | |
| 334 | G | 4 | ||
| 359 | C | 1 | cAA | |
| 394 | G | 10 | gGAGGT | gGA |
| 406 | G | 2 | gAA | |
| 413 | C | 1 | ||
| 436 | G | 2 | gAA | |
| 439 | G | 5 | gGAGCT | gGA |
| 448 | G | 1 | gAA | |
| 457 | G | 1 | gAA | |
| 482 | C | 2 | TcA |
The set of 96 G·C-to-T·A events in RJ803 distributed over 20 sites in the pyrE coding sequence (Fig. 2).
Base pair number in the coding sequence.
Number of occurrences observed.
Wild-type codon sequence is shown with the affected nucleotide in lowercase.
G·C-to-T·A events at this site do not terminate translation but remove the wild-type N terminus.
Transversions at this site affected the first G in GGGGCG.
TABLE 5.
Sites of transversions not recovered in strain RJ803a
| Positionb | Sense strand | Observed in other strainsc | Not observed in other strainsd |
|---|---|---|---|
| 65 | C | TcA | |
| 87 | C | TAc | |
| 98 | G | gGAAAC | |
| 106 | C | gGAGCC | |
| 107 | C | P → H | |
| 128 | C | TcA | |
| 200 | G | G → S | |
| 203 | G | G → V | |
| 218 | C | TcA | |
| 286 | G | gAA | |
| 297 | G | gGAAGC | |
| 298 | G | gAA | |
| 337 | G | D → Y | |
| 376 | G | gAG | |
| 378 | G | gGAGGT | |
| 379 | G | gAG | |
| 431 | G | R → I | |
| 433 | G | gAA | |
| 499 | G | gAA | |
| 524 | C | TcA | |
| 538 | G | gAG | |
| 544 | G | gAA | |
| 583 | G | gGA |
G·C-to-T·A transversions recovered in various ogg+ dbh+ S. acidocaldarius strains (all are single occurrences).
Base pair number in the pyrE coding sequence.
Nonsense mutations are shown as the wild-type codon, with the affected nucleotide in lowercase, and missense mutations are indicated by the resulting amino acid substitution.
Examples of the GGANNY motif are listed where they occur; all four of the indicated occurrences generate amino acid substitutions via G-to-T transversion at the lowercase g. The remaining sites do not coincide with GGANNY, but transversion of the nucleotide shown in lowercase generates a nonsense mutation.
The nucleotide compositions of S. acidocaldarius genes are skewed by a low G+C content of the genome overall and by enrichment of purine nucleotides in the sense (coding) strand of genes; the latter bias has been noted in many thermophile genomes (20). Consistent with both biases, the pyrE coding strand has 139 G versus 76 C (ratio, 1.83), and we found evidence of strand bias in several parameters of G·C-to-T·A transversion. For example, the 20 sites at which transversions were observed in RJ803 exhibit a 3:1 skew in that only 5 have C in the sense strand (Fig. 2). Similarly, the eight most-used sites in RJ803, representing 79 independent events (83%), are all oriented with G in the sense (nontranscribed) strand of pyrE (Table 4). As a result, the observed transversion events are more strongly skewed than would be predicted solely by base composition, with 86 G-to-T versus 6 C-to-A events in the sense strand, for a ratio of 14.3.
To investigate the influence of the local sequence context on the observed distribution of events, 31 pyrE sites where transversions had been observed in any S. acidocaldarius strain were compared to each other by aligning a 21-nucleotide (nt) window centered on the affected G. The analysis identified GGANNY as the sequence motif correlating most consistently with G·C-to-T·A transversions within this sample set of known mutations (Fig. 3). The motif was distributed within pyrE with a 9:1 sense/antisense strand bias. Only six of the 10 copies of the motif had mutated in RJ803, however, accounting for 56/95 (59%) G·C-to-T·A events observed (Table 4). The six are also the only GGANNY sites in pyrE at which G·C-to-T·A changes the length of the encoded protein; at the remaining four sites (where no mutations were observed), G·C-to-T·A. events can produce only amino acid substitutions (Table 5).
FIG 3.
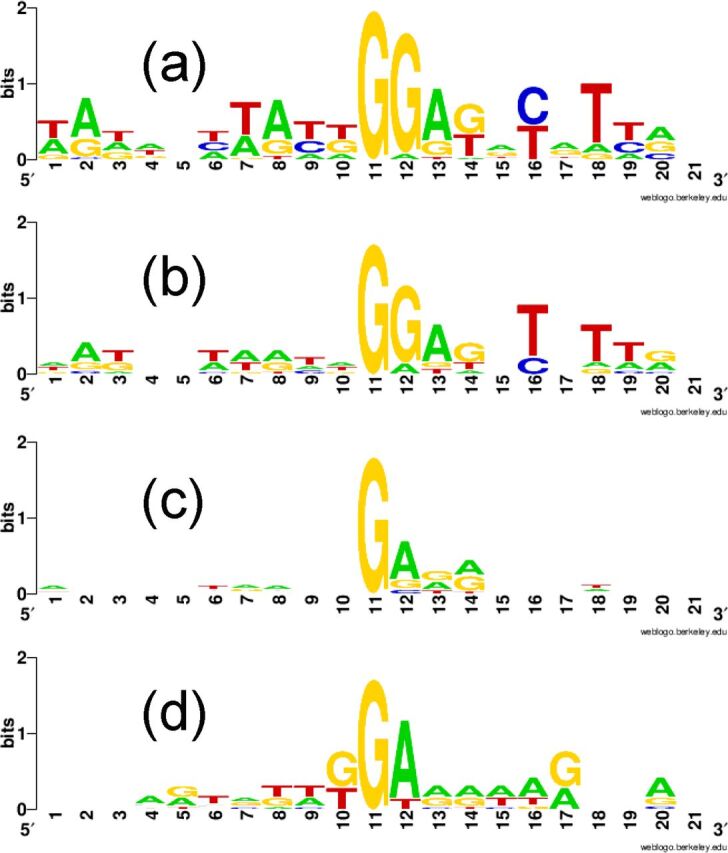
Effect of local context on the transversion rate. Nucleotide overrepresentation at sites of G·C-to-T·A transversion in the pyrE gene is depicted by letter size (see Materials and Methods). In weighted (proportional) analyses, each site was represented as many times in the alignment as in the corresponding set of mutants. The four panels represent different analyses of hot spots (a and b) and cold spots (c and d), as follows: the top eight transversion hot spots (unweighted) (a), the same eight sites weighted according to event frequency (see above) (b), sites of G·C-to-T·A transversion recovered in the mutation spectra of ogg+ dbh+ strains but not observed in RJ803 (unweighted; 11 total) (c), and sites at which G·C-to-T·A transversion creates a stop codon which nevertheless has not been observed in any S. acidocaldarius pyrE mutation analysis (unweighted; eight total) (d).
To clarify the possible role of translational consequences in detection efficiency, we identified BPSs in the pyrE gene that generated selectable (i.e., Foar) mutants in other experiments using the methods of the present study (8, 21). Of 86 independent BPS events recovered in various ogg+ dbh+ strains, 33 (38%), were G·C-to-T·A transversions, 31 (36%) were chain-terminating (CT) (i.e., nonsense) mutations, and 18 (21%) fell into both categories. Among dbh+ ogg+ strains, therefore, 18 of 33 (54%) G·C-to-T·A events created chain-terminating mutations compared to 52/89 (58%) in the Δogg Δdbh strain RJ803. Thus, similar results were obtained for two extreme cases, i.e., normal cells in which transversions are rare versus those in which loss of defense enzymes elevate G·C-to-T·A events about 250-fold. This result argued that the efficiency of detecting G·C-to-T·A transversions in pyrE by FOA selection remains relatively constant over a wide range of replication accuracy. The data also imply that nonsense mutations are overrepresented among the mutations detected by this selection, based on the following reasoning. At only 27 of the 215 G or C sites in pyrE does G·C-to-T·A transversion create a stop codon. Thus, if transversion events were distributed randomly, about 13% of them would be expected to create nonsense mutations, and the observed value is higher than this (54% to 58%). This overrepresentation of the chain-terminating sites among all transversion events that inactivate pyrE indicates that a corresponding proportion of these events (and, by extension, other BPSs) do not inactivate the gene product. This property is predicted by degeneracy of the genetic code and is typical of protein-encoding genes (8).
In considering the relative contributions of nucleotide reactivity and sequence context to the observed mutagenesis, we noted that 19 of the 27 sites (70%) where G·C-to-T·A events created CT mutations (which we designated “pre-CT” sites) appeared in spontaneous-mutation spectra of various Dbh+ Ogg+ S. acidocaldarius strains, whereas the corresponding representation of the GGANNY motif is 5/10 (50%) (Table 5). Similarly, in RJ803, 62 of 95 G·C-to-T·A events (65%) generated stop codons, whereas 56 (59%) coincided with the GGANNY motif (Table 4). In both normal and Δogg Δdbh backgrounds, therefore, creation of a stop codon was a slightly better predictor of where spontaneous G·C-to-T·A events will be recovered than was the consensus motif deduced from alignments of sequences surrounding observed G·C-to-T·A events. We also noted that S. acidocaldarius pyrE contains eight transversion “cold spots” where G·C-to-T·A events have never been observed in experiments despite the fact that they create stop codons and therefore should be detectable (Table 5). We compared the sequence contexts of these cold spots with the eight hot spots identified in RJ803 (Fig. 3c and d). Both sequence motifs contained the GG dinucleotide, but in the hot-spot motif, the chain-terminating transversion occurred on the 5′ G, whereas in the cold-spot motif, it was positioned at the 3′ G. The experimentally observed transversions thus parallel the reactivity of G toward oxidation, which is high in the 5′ member of short tracts (22).
Although we analyzed the native S. acidocaldarius pyrE gene in greatest detail, spontaneous mutations were also selected in a modified cassette (see Fig. S3) that encodes a different amino acid sequence and has a higher G+C content and a lower purine/pyrimidine strand bias. In the Δogg Δdbh strain RJ8, this engineered cassette yielded 12 independent mutations, all G·C-to-T·A transversions, distributed over eight sites. In contrast to the native pyrE gene, none of the transversions created stop codons. Frequency analysis of the surrounding nucleotides (see Fig. S4 in the supplemental material), identified them as the consensus MNNNNGGM, in which the first G is converted to T in the recovered mutants. The strand bias of this motif (15 sense versus 6 antisense occurrences) was markedly weaker than that of the mutated G·C sites in the target gene (7:1) or that of independent selected events (11:1). The consensus motif inferred from the engineered cassette nevertheless overlapped the consensus observed in the native pyrE gene, sharing GGA in common.
Functional impact on TLS.
As demonstrated in yeast, various bacteria, and S. acidocaldarius, oligonucleotide-mediated transformation (OMT) can be used to select clones in which DNA lesions have been inserted into a specific site in the recipient genome by recombination and bypassed by TLS (13, 23, 24). To increase the versatility of this method in S. acidocaldarius, we designed a system that replaces an 18-bp pyrE deletion present in many of our laboratory constructs (see Fig. S1 in the supplemental material) by transformation with synthetic DNA. The transforming DNA is synthesized to place the DNA lesion at bp 165 of the native pyrE gene, which is a synonymous position within the deleted interval. Thus, after the single-stranded DNA (ssDNA) is incorporated into the recipient chromosome (see Fig. S1), insertion of any nucleotide opposite the lesion restores the wild-type amino acid sequence and generates a pyrE+ transformant. Since the recipient genome lacks the selected interval, it does not provide the template needed for either repair or accurate bypass via strand exchange processes before the transforming DNA is replicated (see Fig. S1). The resulting dependence on TLS events makes this assay complementary to that of spontaneous mutation, in that it should not be affected by the efficiency of G oxidation or the impact of transversion on the selection.
Although predicted to be specific for TLS, the transformation assay could be used to detect oxoG-specific BER indirectly by comparing the transformation efficiencies of the isogenic S. acidocaldarius constructs by intact versus oxoG-containing DNA. The results (Table 6) indicated that the impact of Ogg function on transformation by oxoG-containing DNA was conditional. Although oxoG decreased the transformation of normal (Ogg+ Dbh+) cells by the DNA sequence, the penalty associated with the oxoG lesion was not relieved by deleting only ogg (Table 6). Deleting only dbh had a dramatic effect, however, causing a further 10-fold decrease in the relative transforming ability of oxoG-containing DNA (Table 6). This result thus reinforced other genetic evidence that Dbh plays a central role in bypassing oxoG in S. acidocaldarius (12, 13). The fourth strain we evaluated, the Δogg Δdbh double mutant, exhibited an intermediate efficiency of transformation by oxoG-containing DNA (Table 6); thus, deleting ogg affected transformation of dbh mutant cells, but not dbh+ cells, by lesion-containing DNA. The observed 2.4-fold increase in the double mutant relative to the Δdbh strain is consistent with the double mutant avoiding oxoG-specific destruction of the transforming DNA by the Ogg protein. All of the observed differences in transformation efficiency between strains were found to have statistical support (see Table S3).
TABLE 6.
Transformation assay of TLSa
| Recipient strain | Genotype | Control DNAb |
oxoG DNA |
Transformation efficiency |
No. inserted at oxoGe |
||||
|---|---|---|---|---|---|---|---|---|---|
| No. of EPc | No. of transformants | No. of EP | No. of transformants | Relative to controld | Relative to WT | C | A | ||
| MR31 | ogg + dbh + | 7 | 162 | 10 | 44 | 0.181 | 1 | 27 | 0 |
| SD67-7 | Δogg | 7 | 72 | 37 | 61 | 0.160 | 0.89 | 31 | 0 |
| RJ4 | Δdbh | 30 | 219 | 185 | 23 | 0.017 | 0.094 | 1 | 18 |
| RJ8 | Δogg Δdbh | 11 | 63 | 127 | 30 | 0.041 | 0.23 | 1 | 27 |
Based on OMT, as described in Materials and Methods.
Control indicates an intact (normal) oligonucleotide, used for normalization.
Number of electroporations (EP) performed (770 pmol DNA each).
Ratio of transformants per pmol oxoG-containing DNA to transformants per pmol control DNA.
The numbers represent transformants of each type generated by oxoG-containing DNA.
Finally, we measured the accuracy of individual oxoG bypass events in these experiments by PCR and restriction scoring (see Materials and Methods). All 58 events scored in dbh+ transformants, where OMT by the lesion-containing oligonucleotide was relatively efficient, inserted C (Table 6). Conversely, in dbh mutant cells, where OMT by oxoG-containing oligonucleotides was inefficient, TLS was consistently inaccurate, with 45/47 (96%) events inserting A. Furthermore, deleting ogg in a Δdbh background, which increased transformation by oxoG-containing DNA, did not increase the accuracy of TLS (Table 6). These assays thus confirmed that a functional dbh gene is necessary for accurate TLS past oxoG in the S. acidocaldarius chromosome, regardless of the functional status of ogg.
DISCUSSION
Although the physiological and phylogenetic divergence from model organisms suggests that HA may have unusual mechanisms of genome maintenance, it also complicates the process of identifying such mechanisms experimentally. For example, the fact that HA do not encode homologs of the MutS-MutL system that is broadly conserved in mesophiles (including other archaea [25]) makes it difficult to predict a priori what proteins, if any, may correct unforced errors of DNA replication in these organisms. Conversely, whereas HA do encode homologs of highly conserved, well-studied homologous-recombination proteins, they apparently do not tolerate inactivation of these homologs (26, 27), unlike the bacteria and eukaryotes in which the biological functions of the proteins have been analyzed. Finally, the genes of HA that are implicated in genome maintenance by sequence similarity or biochemical activity, and that can be deleted, often yield phenotypes that do not support the hypothesized gene function. To cite recent examples, Sulfolobus and related HA have RPA homologs considered to be fundamental to their molecular biology (28, 29), yet deleting the sole RPA homolog of S. acidocaldarius had minimal impact on growth, arguing that it is not the major functional ssDNA-binding protein (30). Similarly, a novel mismatch-specific endonuclease could be demonstrated to suppress spontaneous mutation in corynebacteria (31), but deleting the only homologous gene in Sulfolobus had no apparent effect on mutation (32).
In this context, the present study confirms, by functional criteria, genes that control a well-defined and frequent type of spontaneous mutation in S. acidocaldarius. The phenotypes of S. acidocaldarius mutants demonstrate that ORFs Saci_1367 and Saci_0554 encode functionally complementary defenses against oxoG-promoted mutation, consistent with the biochemical properties of the encoded proteins or corresponding proteins of related HA. Specifically, deleting Saci_1367 (ogg) increased severalfold the rate of only one class of spontaneous mutations, namely, G·C-to-T·A transversion, consistent with its predicted function as an oxoG-specific N-glycosylase. Although the Ogg− phenotype resembled the phenotype caused by inactivating the sole Y family DNA polymerase of S. acidocaldarius, Dbh (12), ogg and dbh deletions were not epistatic, as demonstrated by the exaggerated defect that resulted from combining them in a single strain.
These results reveal both parallels and differences with respect to other microorganisms. In S. cerevisiae, two studies using different detector genes found that G·C-to-T·A transversions were increased 3.3- or 24–fold by removing the oxoG-specific DNA glycosylase (Δogg1), 0.7- or 2-fold by removing the Y family TLS polymerase Polη (Δrad30), and 10- or 148-fold by removing both enzymes (33, 34). The exaggerated impact when both BER and TLS genes of yeast were inactivated resembles the S. acidocaldarius result. In contrast, the negligible effect of inactivating only the relevant TLS polymerase of yeast (Polη), despite the ability of Polη to bypass oxoG accurately in vitro (5), differed from the S. acidocaldarius results. Furthermore, in this respect E. coli resembles yeast rather than Sulfolobus: inactivating TLS polymerases II, IV, and V singly or in combination did not affect the efficiency or accuracy of oxoG bypass in M13 DNA (35). Thus, Sulfolobus differs from both yeast and E. coli in relying on TLS to avoid considerable oxoG-promoted mutation. It is not clear whether this typifies archaea generally, however. At least one hyperthermophilic species, Thermococcus gammatolerans, lacks a Y family DNA polymerase naturally and exhibits highly erroneous bypass of oxoG in vitro (36). Although the organism is a strict anaerobe and thus might be expected to avoid DNA oxidation, it has been reported to encode at least one oxoG-specific N-glycosylase and to repair artificially introduced oxoG in vivo (37).
In quantitative terms, the similar effects of deleting either ogg or dbh individually indicated that each gene product normally prevents a similar amount of oxidative mutagenesis in S. acidocaldarius (Table 3; see Table S2). Similarly, the 17-fold increase in the G·C-to-T·A transversion rate caused by deleting ogg in a dbh+ S. acidocaldarius strain demonstrates that the normal level of Dbh activity is not sufficient to accommodate all the oxoG that forms under normal growth conditions. The much (about 250-fold) larger increase that resulted from deleting both ogg and dbh further suggests that, outside of these two gene products, S. acidocaldarius probably has little capacity for removing oxoG or bypassing it accurately. Consistent with this possibility, the Δogg Δdbh double mutants also exhibited increased rates of cell death under certain conditions (Fig. 1).
The elevated rate and molecular specificity of spontaneous BPSs in Δogg Δdbh constructs, which represented 90% of the spontaneous mutations, enabled us to examine a defined process of spontaneous mutation in much greater detail than has been done in any other archaeon or hyperthermophile to date. Analysis of the mutations in a well-characterized target indicated potentially complex interactions among G oxidation, gene sequence, and protein structure and suggested that a frequent target of DNA oxidation in S. acidocaldarius is the first (5′) G of short G tracts. In the AT-rich Sulfolobus genomes the most common of these are GG dinucleotides, which are enriched in the sense strands of coding regions and commonly adjacent to A (the most common nucleotide in coding strands). The frequent conversion of GGA glycine codons to chain-terminating TGA, which contributed heavily to the GGANNY consensus motif for transversion observed in the native pyrE gene, thus reflects the abundance, reactivity toward oxidation, and high detection efficiency unique to GGA.
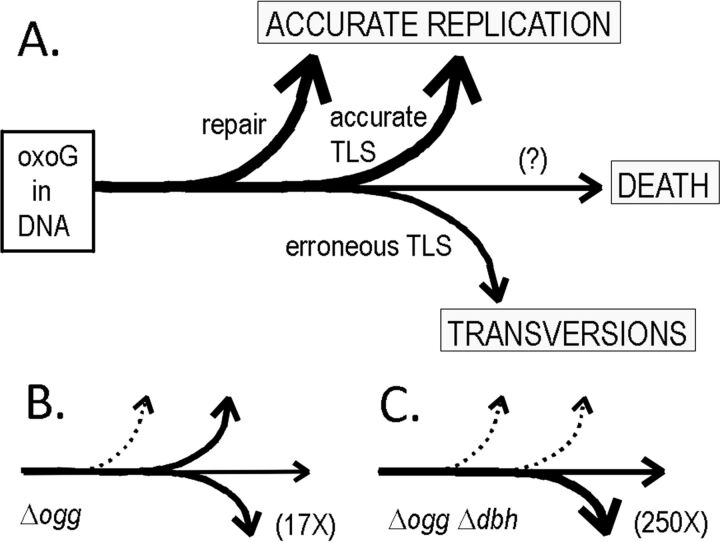
Our results combine to indicate simple relationships among DNA oxidation, biochemical defenses, and biological consequences in S. acidocaldarius (Fig. 4). The underlying principle of the scheme is that once oxoG forms in DNA, it (like other DNA lesions) ultimately leads to one of three biological fates: (i) cell death, (ii) mutagenesis, or (iii) accurate replication, where the last reflects the sum of several distinct processes that may include repair, accurate TLS, and tolerance mechanisms involving recombination (38).
FIG 4.
Fates of oxoG in vivo. The scheme depicts four alternate fates of oxoG, representing different processing pathways and biological consequences. Thus, repair avoids the deleterious consequences of oxoG if it can be completed before DNA replication, and unrepaired oxoG may be bypassed accurately by TLS. If both these options fail, bypass by the remaining polymerases has low accuracy (leading to transversion) and low efficiency (promoting cell death). (A) Predominance of repair and accurate TLS within normal S. acidocaldarius cells. (B) Cells lacking ogg activity, in which the proportion of erroneous TLS increases. (C) Consequences of losing both Ogg and Dbh activities.
Our results also provide new details regarding the role of TLS in maintenance of the Sulfolobus genome. In transformation assays, functional Dbh allowed S. acidocaldarius to replicate a chromosomal oxoG with >98% accuracy, but without Dbh, the observed accuracy was about 4%, and essentially all erroneous events insert A. Inactivating Dbh also decreased the overall success of transformation by oxoG-containing DNA, even in Δogg cells. This indicates that the remaining DNA polymerases (which may include one of the Sulfolobus primases [39]) have high intrinsic failure rates at oxoG. Combining the accuracy and efficiency data from this study and a previous one (13) suggests that in the chromosome of a normal (Dbh+) S. acidocaldarius cell, an unrepaired oxoG may be about 100 times more likely to be replicated by Dbh than by another DNA polymerase, including the highly processive PolB1. This estimate is consistent with the observation that TLS past oxoG in Dbh+ strains produced no detected insertion of A (0/58), rather than the 10% A insertion ratio predicted by combining 90% of the C-specific bypass evident in Dbh+ recipients and 10% of the A-specific bypass seen in Dbh− recipients (Table 6).
The indication that oxoG bypass accuracy is not additive in Dbh+ versus Dbh− strains is significant because it suggests that Dbh has a competitive advantage over the other DNA polymerases of S. acidocaldarius for bypass of oxoG in normal cells distinct from that of simple catalytic efficiency. In general, using TLS to avoid miscoding by DNA lesions such as oxoG requires (i) preventing the replicative complex from replicating the template lesion, (ii) giving the TLS polymerase with the appropriate specificity access to the 3′ end of the nascent strand, and (iii) excluding other DNA polymerases. Bacteria and eukaryotic cells are known to meet these fundamental requirements in different ways. Eukaryotic cells respond to perturbed replication by covalently modifying particular lysine residues of the sliding clamp (PCNA) with the peptide tags ubiquitin and SUMO (38, 40). Different perturbations induce different patterns of modification, and this allows the corrective pathway to be tailored to the nature of the replication block. Bacteria, in contrast, appear to depend largely on kinetic competition among the polymerases, assisted by colocalization with the sliding clamp, to determine which polymerase ultimately bypasses a given lesion (41–43). Archaeal DNA replication is notable in that it shares certain molecular features with eukaryotes and others with bacteria. Most archaeal replication proteins are specifically related to eukaryotic proteins, but to our knowledge, functional relevance of covalent modification for polymerase switching has not been demonstrated in archaea, and the extent to which archaea actively switch DNA polymerases remains a significant, unresolved question.
Regardless of the relative importance of kinetic competition versus active switching, Dbh orthologs appear to provide the primary alternative to the replicative polymerase in Sulfolobus and related species. Biochemical analysis of S. solfataricus indicates that its Dbh equivalent, Dpo4, has a cytoplasmic concentration about 10% that of the replicative polymerase subunit but exhibits a 10-fold-higher catalytic efficiency, as defined by the following formula: kcat/Km (44). Thus, the effective activities of the two polymerases in vivo may be comparable; in addition, both activities are much higher than those of the remaining DNA polymerases, Dpo2 and Dpo3, whose biological functions remain unclear (42, 44).
Similarly, regardless of the mechanistic details of DNA polymerase exchange in Sulfolobus, replisome stalling seems to represent a plausible candidate for initiating the process. Although oxoG is not generally considered to block DNA replication, replicative polymerases vary considerably in their performances at this template lesion. Mammalian DNA polymerase δ, for example, inserts C opposite oxoG at a higher rate than it inserts A, but the overall accuracy of bypass is decreased by preferential extension of the oxoG·A mispair versus oxoG·C (45). Bacteriophage T7 polymerase replicates oxoG in vitro with about 25% A insertion and exhibits no discrimination by proofreading or extension kinetics (2). In contrast to both of these cases, the catalytic subunit of the S. solfataricus replicative polymerase, Dpo1, stalls at oxoG, due to efficient removal of the 3′-terminal nucleotide (i.e., proofreading) opposite this lesion (46). Archaeal replicative polymerases are also known to stall at other template lesions in vitro. The best-studied example, stalling at template dU, is the result of a conserved feature specific to archaeal DNA polymerases, namely, a specific dU-binding site (47). In Sulfolobus, therefore, multiple DNA lesions are known that interact with different features of the replicative polymerase to yield the same predicted result in vivo, i.e., a stalled replisome. The apparent convergence on this potentially deleterious common intermediate raises questions as to whether stalling may play a central role in multiple DNA damage responses of HA and what reconfigurations of the replication fork these responses may require (48).
MATERIALS AND METHODS
Strains and growth conditions.
The S. acidocaldarius constructs used in the study are listed in Table 1. Deletion mutants were constructed by electroporating corresponding pyrE131 mutants (49) with PCR products in which a functional but heterologous pyrE gene (the “cassette”) was flanked by sequences that flank the targeted gene in the S. acidocaldarius chromosome, as previously described (21). The Pyr+ strain RJ12 was constructed by corresponding transformation by a plasmid containing the cloned S. acidocaldarius pyrE gene. In all cases, Pyr+ transformants were selected by plating on xylose-tryptone medium solidified with gellan gum. Pyr− strains were propagated in XT medium supplemented with uracil (20 μg/ml); all cultures were incubated aerobically at 78°C.
Sensitivity assays.
To determine MICs, test compounds were diluted serially at a 1:3 ratio in liquid XT-plus-uracil medium and inoculated to an initial density of approximately 106 cells/ml. The growth of each culture in the dilution series was scored after 3 days of incubation; all the strains were incubated in parallel, and the results were averaged from three or more independent experiments.
Time courses of survival were determined by diluting freshly grown cultures in growth medium. Each dilution series was incubated for several days under various conditions that did not support growth. At regular intervals, each cell suspension was mixed, and an aliquot (5 μl) was spotted onto the surface of the plate. After 5 to 7 days of incubation, colonies were counted to measure the concentration of viable cells.
TLS assays.
Replication past oxoG in vivo was detected genetically by OMT (23, 50); all recipient strains bore the pyrE131 allele, which is an 18-bp internal deletion (49). As a means of increasing the transformation efficiency of the synthetic DNAs, the intervals flanking the internal deletion were extended by ligating corresponding anchor oligonucleotides to both ends of the central insert. Ligation of the three modules was assisted by scaffolding oligonucleotides (Table 1) and confirmed by agarose gel electrophoresis. The insert oligonucleotide containing oxoG was synthesized by Biosynthesis (Louisville, TX); all other oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Pyr+ transformants generated by oxoG-containing DNAs were scored by restriction. After clonal purification on solid XT medium, genomic DNA was extracted from liquid cultures of the resulting clones, and the pyrE gene was amplified by PCR (13). Amplicons were incubated with the restriction endonuclease BanI, HaeII, or SacI, followed by agarose gel electrophoresis. This scoring scheme (see Fig. S1 in the supplemental material) was validated in mock analyses in which cells were transformed by a control insert DNA containing a mixture of the four natural bases at the query position.
Analysis of spontaneous mutation.
Forward, i.e., loss-of-function, pyrE mutations were selected by plating cultures on XT-plus-uracil medium supplemented with 50 μg FOA per ml. Sets of 12 to 20 independent cultures were made by inoculating 0.2 ml XT-plus-uracil medium in wells of a microdilution plate, each with a different isolated colony, using sterile toothpicks. The cultures were grown to a final density of 5 × 108 to 10 × 108 CFU/ml, and the number of viable cells for the cultures in the set was measured by serial dilution and plating of two to four cultures on nonselective medium. The number of FOA-resistant cells in each of the remaining cultures was determined by spreading each in its entirety on solid XT-uracil-FOA medium and incubating for 7 days. The genic mutation rate (μ) and its 95% confidence interval were calculated from the numbers of viable and FOA-resistant cells in the cultures using the “bz” mutation rate calculator of Gillet-Markowska et al. (51) (http://www.lcqb.upmc.fr/bzrates).
Sequence analyses.
To sample the spectrum of mutations inactivating the pyrE gene in each construct, one Foar clone was picked randomly from each independent culture. Each independent mutant was purified and processed as described above for Pyr+ transformants, and the PCR product was analyzed by dye terminator sequencing. To investigate the sequence specificity of spontaneous-mutation sites, 21-nt windows centered around the G of an affected G·C base pair were compiled from mutation spectra. Overrepresented nucleotides were identified using the Web Logo server (https://weblogo.berkeley.edu/logo.cgi).
ACKNOWLEDGMENTS
This work was supported by the Department of Biological Sciences, University of Cincinnati. R.J. acknowledges a Wieman-Wendel-Benedict award, and S.D. acknowledges support by STEM funds for undergraduate research.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2006. DNA repair and mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 2.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. 2004. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J 23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delaney S, Jarem DA, Volle CB, Yennie CJ. 2012. Chemical and biological consequences of oxidatively damaged guanine in DNA. Free Radic Res 46:420–441. doi: 10.3109/10715762.2011.653968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David SS, O'Shea VL, Kundu S. 2007. Base-excision repair of oxidative DNA damage. Nature 447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. 2000. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet 25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 6.Kokoska RJ, McCulloch SD, Kunkel TA. 2003. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J Biol Chem 278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 7.Sakai HD, Kurosawa N. 2018. Saccharolobus caldissimus gen. nov., sp. nov., a facultatively anaerobic iron-reducing hyperthermophilic archaeon isolated from an acidic terrestrial hot spring, and reclassification of Sulfolobus solfataricus as Saccharolobus solfataricus comb. nov. and Sulfolobus shibatae as Saccharolobus shibatae comb. nov. Int J Syst Evol Microbiol 68:1271–1278. doi: 10.1099/ijsem.0.002665. [DOI] [PubMed] [Google Scholar]
- 8.Grogan DW, Carver GT, Drake JW. 2001. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci U S A 98:7928–7933. doi: 10.1073/pnas.141113098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkner S, Lipps G. 2008. Mutation and reversion frequencies of different Sulfolobus species and strains. Extremophiles 12:263–270. doi: 10.1007/s00792-007-0125-7. [DOI] [PubMed] [Google Scholar]
- 10.Mao D, Grogan DW. 2017. How a genetically stable extremophile evolves: modes of genome diversification in the archaeon Sulfolobus acidocaldarius. J Bacteriol 199:e00177-17. doi: 10.1128/JB.00177-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potapova O, Grindley ND, Joyce CM. 2002. The mutational specificity of the Dbh lesion bypass polymerase and its implications. J Biol Chem 277:28157–28166. doi: 10.1074/jbc.M202607200. [DOI] [PubMed] [Google Scholar]
- 12.Sakofsky CJ, Foster PL, Grogan DW. 2012. Roles of the Y-family DNA polymerase Dbh in accurate replication of the Sulfolobus genome at high temperature. DNA Repair 11:391–400. doi: 10.1016/j.dnarep.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakofsky CJ, Grogan DW. 2015. Lesion-induced mutation in the hyperthermophilic archaeon Sulfolobus acidocaldarius and its avoidance by the Y-family DNA polymerase Dbh. Genetics 201:513–523. doi: 10.1534/genetics.115.178566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denver DR, Swenson SL, Lynch M. 2003. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol 20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 15.Sartori AA, Lingaraju GM, Hunziker P, Winkler FK, Jiricny J. 2004. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic Acids Res 32:6531–6539. doi: 10.1093/nar/gkh995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faucher F, Doublie S, Jia Z. 2012. 8-oxoguanine DNA glycosylases: one lesion, three subfamilies. Int J Mol Sci 13:6711–6729. doi: 10.3390/ijms13066711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gogos A, Clarke ND. 1999. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J Biol Chem 274:30447–30450. doi: 10.1074/jbc.274.43.30447. [DOI] [PubMed] [Google Scholar]
- 18.Chung JH, Suh MJ, Park YI, Tainer JA, Han YS. 2001. Repair activities of 8-oxoguanine DNA glycosylase from Archaeoglobus fulgidus, a hyperthermophilic archaeon. Mutat Res 486:99–111. doi: 10.1016/s0921-8777(01)00081-7. [DOI] [PubMed] [Google Scholar]
- 19.Fujii M, Hata C, Ukita M, Fukushima C, Matsuura C, Kawashima-Ohya Y, Tomobe K, Kawashima T. 2016. Characterization of a thermostable 8-oxoguanine DNA glycosylase specific for GO/N mismatches from the thermoacidophilic archaeon Thermoplasma volcanium. Archaea 2016:8734894. doi: 10.1155/2016/8734894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta C, Paul S. 2012. Microbial lifestyle and genome signatures. Curr Genomics 13:153–162. doi: 10.2174/138920212799860698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakofsky CJ, Runck LA, Grogan DW. 2011. Sulfolobus mutants, generated via PCR products, which lack putative enzymes of UV photoproduct repair. Archaea 2011:864015. doi: 10.1155/2011/864015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshioka Y, Kitagawa Y, Takano Y, Yamaguchi K, Nakamura T, Saito I. 1999. Experimental and theoretical studies on the selectivity of GGG triplets toward one-electron oxidation in B-form DNA. J Am Chem Soc 121:8712–8719. doi: 10.1021/ja991032t. [DOI] [Google Scholar]
- 23.Rodriguez GP, Song JB, Crouse GF. 2013. In vivo bypass of 8-oxodG. PLoS Genet 9:e1003682. doi: 10.1371/journal.pgen.1003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss B. 2008. Removal of deoxyinosine from the Escherichia coli chromosome as studied by oligonucleotide transformation. DNA Repair 7:205–212. doi: 10.1016/j.dnarep.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White MF, Grogan DW. 2008. DNA stability and repair, p 179–188. In Robb FT, Antranikian G, Grogan DW, Driessen AJ(ed), Thermophiles: biology and technology at high temperatures. CRC Press, Boca Raton, FL. [Google Scholar]
- 26.Fujikane R, Ishino S, Ishino Y, Forterre P. 2010. Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet Syst 85:243–257. doi: 10.1266/ggs.85.243. [DOI] [PubMed] [Google Scholar]
- 27.Huang Q, Liu L, Liu J, Ni J, She Q, Shen Y. 2015. Efficient 5′-3′ DNA end resection by HerA and NurA is essential for cell viability in the crenarchaeon Sulfolobus islandicus. BMC Mol Biol 16:2. doi: 10.1186/s12867-015-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richard DJ, Bell SD, White MF. 2004. Physical and functional interaction of the archaeal single-stranded DNA-binding protein SSB with RNA polymerase. Nucleic Acids Res 32:1065–1074. doi: 10.1093/nar/gkh259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry ER, Bell SD. 2006. DNA replication in the archaea. Microbiol Mol Biol Rev 70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki S, Kurosawa N. 2019. Robust growth of archaeal cells lacking a canonical single-stranded DNA-binding protein. FEMS Microbiol Lett 366:fnz124. doi: 10.1093/femsle/fnz124. [DOI] [PubMed] [Google Scholar]
- 31.Ishino S, Skouloubris S, Kudo H, l’Hermitte-Stead C, Es-Sadik A, Lambry J-C, Ishino Y, Myllykallio H. 2018. Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res 46:6206–6217. doi: 10.1093/nar/gky460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki S, Kurosawa N. 2019. Endonucleases responsible for DNA repair of helix-distorting DNA lesions in the thermophilic crenarchaeon Sulfolobus acidocaldarius in vivo. Extremophiles 23:613–624. doi: 10.1007/s00792-019-01120-9. [DOI] [PubMed] [Google Scholar]
- 33.de Padula M, Slezak G, Auffret van Der Kemp P, Boiteux S. 2004. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res 32:5003–5010. doi: 10.1093/nar/gkh831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mudrak SV, Welz-Voegele C, Jinks-Robertson S. 2009. The polymerase eta translesion synthesis DNA polymerase acts independently of the mismatch repair system to limit mutagenesis caused by 7,8-dihydro-8-oxoguanine in yeast. Mol Cell Biol 29:5316–5326. doi: 10.1128/MCB.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. 2007. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J Biol Chem 282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 36.Killelea T, Palud A, Akcha F, Lemor M, L’haridon S, Godfroy A, Henneke G. 2019. The interplay at the replisome mitigates the impact of oxidative damage on the genetic integrity of hyperthermophilic Archaea. Elife 8:e45320. doi: 10.7554/eLife.45320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbier E, Lagorce A, Hachemi A, Dutertre M, Gorlas A, Morand L, Saint-Pierre C, Ravanat JL, Douki T, Armengaud J, Gasparutto D, Confalonieri F, Breton J. 2016. Oxidative DNA damage and repair in the radioresistant archaeon Thermococcus gammatolerans. Chem Res Toxicol 29:1796–1809. doi: 10.1021/acs.chemrestox.6b00128. [DOI] [PubMed] [Google Scholar]
- 38.Branzei D, Szakal B. 2016. DNA damage tolerance by recombination: molecular pathways and DNA structures. DNA Repair 44:68–75. doi: 10.1016/j.dnarep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jozwiakowski SK, Borazjani Gholami F, Doherty AJ. 2015. Archaeal replicative primases can perform translesion DNA synthesis. Proc Natl Acad Sci U S A 112:E633–E638. doi: 10.1073/pnas.1412982112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. 2007. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Lovett ST. 2007. Polymerase switching in DNA replication. Mol Cell 27:523–526. doi: 10.1016/j.molcel.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Trakselis MA, Cranford MT, Chu AM. 2017. Coordination and substitution of DNA polymerases in response to genomic obstacles. Chem Res Toxicol 30:1956–1971. doi: 10.1021/acs.chemrestox.7b00190. [DOI] [PubMed] [Google Scholar]
- 43.Delmas S, Matic I. 2006. Interplay between replication and recombination in Escherichia coli: impact of the alternative DNA polymerases. Proc Natl Acad Sci U S A 103:4564–4569. doi: 10.1073/pnas.0509012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi JY, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP. 2011. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J Biol Chem 286:31180–31193. doi: 10.1074/jbc.M111.258038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Einolf HJ, Guengerich FP. 2001. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J Biol Chem 276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 46.Maxwell BA, Suo Z. 2012. Kinetic basis for the differing response to an oxidative lesion by a replicative and a lesion bypass DNA polymerase from Sulfolobus solfataricus. Biochemistry 51:3485–3496. doi: 10.1021/bi300246r. [DOI] [PubMed] [Google Scholar]
- 47.Wardle J, Burgers PM, Cann IK, Darley K, Heslop P, Johansson E, Lin LJ, McGlynn P, Sanvoisin J, Stith CM, Connolly BA. 2008. Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res 36:705–711. doi: 10.1093/nar/gkm1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grogan DW. 2015. Understanding DNA repair in hyperthermophilic archaea: persistent gaps and other reasons to focus on the fork. Archaea 2015:942605. doi: 10.1155/2015/942605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reilly MS, Grogan DW. 2001. Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J Bacteriol 183:2943–2946. doi: 10.1128/JB.183.9.2943-2946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grogan DW, Stengel KR. 2008. Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli/lambdaRed and Sulfolobus acidocaldarius recombination systems. Mol Microbiol 69:1255–1265. doi: 10.1111/j.1365-2958.2008.06356.x. [DOI] [PubMed] [Google Scholar]
- 51.Gillet-Markowska A, Louvel G, Fischer G. 2015. bz-rates: a Web tool to estimate mutation rates from fluctuation analysis. G3 (Bethesda) 5:2323–2327. doi: 10.1534/g3.115.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonald JH. 2014. Handbook of biological statistics, 3rd ed. Sparky House Publishing, Baltimore, MD. [Google Scholar]
- 53.Kurosawa N, Grogan DW. 2005. Homologous recombination of exogenous DNA with the Sulfolobus acidocaldarius genome: properties and uses. FEMS Microbiol Lett 253:141–149. doi: 10.1016/j.femsle.2005.09.031. [DOI] [PubMed] [Google Scholar]