ABSTRACT
The Mycobacterium tuberculosis H37Rv genome was sequenced and annotated over 20 years ago, yet roughly half of the protein-coding genes still lack a predicted function. We characterized two genes of unknown function, rv3679 and rv3680, for which inconsistent findings regarding their importance for virulence in mice have been reported. We confirmed that the rv3679 and rv3680 operon (rv3679-80) deletion mutant (Δrv3679-80) was virulent in mice and discovered that the Δrv3679-80 mutant suffered from a glycerol-dependent recovery defect on agar plates following mouse infection. Glycerol also exacerbated killing of the Δrv3679-80 mutant by nitric oxide. Rv3679 and Rv3680 have previously been shown to form a complex with ATPase activity, and we demonstrate that the ability of M. tuberculosis to cope with elevated levels of glycerol and nitric oxide requires intact ATP-binding motifs in both Rv3679 and Rv3680. Inactivation of glycerol kinase or Rv2370c, a protein of unknown function, suppressed glycerol-mediated toxicity in the Δrv3679-80 mutant. Glycerol catabolism led to increased intracellular methylglyoxal pools, and the Δrv3679-80 mutant was hypersusceptible to extracellular methylglyoxal, suggesting that glycerol toxicity in the Δrv3679-80 mutant is caused by methylglyoxal. Rv3679 and Rv3680 interacted with Rv1509, and Rv3679 had numerous additional interactors including proteins of the type II fatty acid synthase (FASII) pathway and mycolic acid-modifying enzymes linking Rv3679 to fatty acid or lipid synthesis. This work provides experimentally determined roles for Rv3679 and Rv3680 and stimulates future research on these and other proteins of unknown function.
IMPORTANCE A better understanding of the pathogenesis of tuberculosis requires a better understanding of gene function in M. tuberculosis. This work provides the first functional insight into the Rv3679/Rv3680 ATPase complex. We demonstrate that M. tuberculosis requires this complex and specifically its ATPase activity to resist glycerol and nitric oxide toxicity. We provide evidence that the glycerol-derived metabolite methylglyoxal causes toxicity in the absence of Rv3679/Rv3680. We further show that glycerol-dependent toxicity is reversed when glycerol kinase (GlpK) is inactivated. Our work uncovered other genes of unknown function that interact with Rv3679 and/or Rv3680 genetically or physically, underscoring the importance of understanding uncharacterized genes.
KEYWORDS: Mycobacterium tuberculosis, glycerol, nitric oxide, ATPase
INTRODUCTION
Tuberculosis (TB) remains one of the world’s most pressing global health threats, with 1.5 million deaths in 2018 alone (https://www.who.int/tb/publications/global_report/en/). Antibiotics that kill the causative agent, Mycobacterium tuberculosis, have existed since the 1940s, but the length and complexity of TB therapy give rise to the emergence of drug-resistant strains. In 2016, 600,000 new TB cases did not respond to the first-line drugs isoniazid and rifampin and are thus considered multidrug-resistant TB (MDR-TB). Moreover, 6.2% of these were further classified as extensively drug-resistant TB (XDR-TB) due to resistance to second-line drugs. Thus, the development of new antibiotics is crucial to combat the problem of resistance to current TB treatments.
One hurdle to finding new therapies is that only approximately 52% of M. tuberculosis protein-coding genes have a putative function (1, 2). Not only does the M. tuberculosis genome contain a large pool of genes without a predicted function, but also many existing annotations are based on sequence similarity of the hypothetical protein to proteins in other organisms. Many of these are evolutionarily remarkably different from mycobacteria, and thus many of the annotations are likely incorrect. Therefore, there are large gaps in our understanding of M. tuberculosis physiology and a lack of insights into new potential targets for drugs and vaccines, limiting the development of new interventions to treat and control TB disease.
Here, we describe the characterization of two genes of unknown function, rv3679 and rv3680, which were selected for study because previous work showed that mutants of each gene were underrepresented in transposon (Tn) mutant libraries isolated from infected mice, suggesting that rv3679 and rv3680 encode proteins required for virulence (3). Additionally, rv3679 and rv3680 were among the genes identified as differentially required in clinical isolates compared to the laboratory strain H37Rv, suggesting their importance for growth of strains derived from human infection (4). They are also conserved in other mycobacterial species, including Mycobacterium leprae, which has a “minimal” genome (5).
Genes rv3679 and rv3680 are transcribed as an operon (rv3679-80), and the two encoded proteins together share sequence similarity with the arsenite exporter component ArsA from Escherichia coli and the eukaryotic guided entry of tail-anchored proteins factor 3 (Get3), which forms homodimers and facilitates insertion of lipophilic proteins into the endoplasmic reticulum membrane (6, 7). Both ArsA and Get3 depend on ATPase activity for function (8–11). Recent work revealed the structure of the Rv3679 and Rv3680 heterodimeric complex (PDB codes 6BS3, 6BS4, and 6BS5) and demonstrated that it has ATPase activity (6). While the Rv3679/Rv3680 complex shows similarities to Get3 and ArsA, there are notable differences. Rv3679 and Rv3680 lack a hydrophobic groove that is functionally important in Get3 and lack the anion-binding site that is present in ArsA (6). Therefore, the Rv3679/Rv3680 complex likely has activities other than Get3 and ArsA. Surprisingly, the same study showed that an M. tuberculosis strain missing Rv3679 and Rv3680 was not attenuated in the TB mouse model, in contrast with the predictions that motivated our work. Hu et al. (6) named Rv3679 and Rv3680 “bacterial Get3-like proteins A and B” (BagA and BagB); however, given the lack of evidence that these proteins are functionally related to Get3 and because the function of the Rv3679/Rv3680 complex remains to be determined, we decided to keep their original annotation.
RESULTS
Rv3679 and Rv3680 protect M. tuberculosis against nitric oxide toxicity.
To investigate the function of Rv3679 and Rv3680, we created strains missing the entire rv3679-80 locus in M. tuberculosis and the homologous genes msmeg_6193 through msmeg_6195 (msmeg_6193-95) in Mycobacterium smegmatis. In M. tuberculosis, we first created a merodiploid strain by adding a copy of the rv3679-80 operon into the attL5 integration site, replaced the native rv3679-80 locus with a hygromycin cassette, and then performed replacement transformations to switch the rv3679-80-expressing plasmid in the attL5 site with a kanamycin resistance-conferring plasmid to generate the Δrv3679-80 mutant (see Fig. S1 in the supplemental material). Successful deletion of rv3679-80 in M. tuberculosis and of msmeg_6193-95 in M. smegmatis was verified by Southern blotting (see Fig. S2 in the supplemental material). Rv3679 and Rv3680 share sequence similarity with members of the ArsA protein family, which helps export arsenite and arsenate from the cell. Consistent with a previous report demonstrating that the arsenite substitute antimonite failed to enhance the ATPase activity of the Rv3679/Rv3680 complex (6), we found that the Δrv3679-80 mutant was not hypersusceptible to arsenite in culture medium (Fig. 1A). Rv3679 and Rv3680 also share sequence similarity with a eukaryotic ATPase, Get3. Mutants lacking Get3 have defects in resisting heat stress (7, 12–14); therefore, we tested the response of the Δmsmeg_6193-95 mutant to high temperatures but observed no growth defect compared to that of wild-type (WT) M. smegmatis or to a ΔclpB mutant (15), which is hypersusceptible to heat stress (see Fig. S3A in the supplemental material). Thus, Rv3679 and Rv3680 ATPases likely have distinct functions from both ArsA and Get3. We next tested if loss of Rv3679/Rv3680 affected the ability of M. tuberculosis to withstand other host relevant stresses and discovered that the Δrv3679-80 mutant was significantly more susceptible to nitric oxide (NO) than WT M. tuberculosis and the complemented strain (Fig. 1B; see also Fig. S3B).
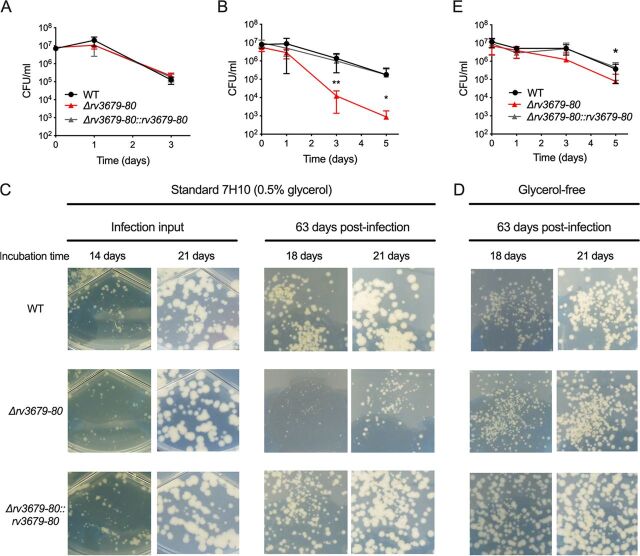
FIG 1.
The M. tuberculosis Δrv3679-80 mutant is hypersusceptible to NO and exhibits a glycerol-dependent recovery defect following mouse infection. (A) Survival in the presence of 2 mM sodium arsenite. (B) Survival of bacteria treated with diethylenetriamine NONO-ate (DETA-NO) in standard 7H9 liquid medium and cultured on standard 7H10 agar for CFU enumeration. (C) Images of bacterial colonies cultured on standard 7H10 agar (0.5% glycerol, 10% OADC) from diluted infection inoculum and from lung homogenates at day 63 postinfection. Images were taken at the indicated incubation time points. (D) Images of bacterial colonies cultured on glycerol-free 7H10 agar (0.5% glucose, 10% OADC). (E) Survival of bacteria treated with DETA-NO in glycerol-free 7H9 liquid medium and cultured on glycerol-free 7H10 agar for CFU enumeration. Data in panels B and E are means and errors from three independent experiments; error bars indicate standard deviation. Statistical significance of the Δrv3679-80 mutant relative to the WT was assessed by Student's t test; *, P < 0.05; **, P < 0.01.
Deletion of rv3679 and rv3680 results in a glycerol-dependent recovery defect after mouse infection.
Transposon mutant screens predicted that Rv3679 and Rv3680 are required for virulence in the mouse model (3, 16). The hypersusceptibility of the Δrv3679-80 mutant to NO also suggested that these proteins might contribute to virulence. In contrast, a recent study reported that in single-strain mouse infections an rv3679-80 mutant was not attenuated (6). To gain further insight, we infected mice by aerosolization with the WT, the Δrv3679-80 mutant, and the complemented mutant. The three strains replicated and persisted similarly, confirming that Rv3679 and Rv3680 are not required for virulence in the C57BL/6 mouse model (see Fig. S4 in the supplemental material). However, Δrv3679-80 yielded smaller colonies than the WT or the complemented strain, specifically after outgrowth from infected tissues (Fig. 1C). The colony sizes after 2 and 3 weeks of incubation were similar across all strains when the inoculum used to infect the mice was cultured on agar plates. However, on agar plates containing diluted organ homogenates, the Δrv3679-80 mutant colonies were smaller than those of the WT or complemented strain after 2 and 3 weeks of incubation. The catabolism of glycerol leads to the production of toxic metabolites (17), and we hypothesized that glycerol in the standard 7H10 agar might be enhancing a stressed state of the Δrv3679-80 mutant isolated from mouse organs. To test this, we cultured organ homogenates from infected mice on glycerol-free 7H10 agar plates, which contained only glucose and oleic acid from the oleic acid-albumin-dextrose-catalase (OADC) supplement as carbon sources. The Δrv3679-80 mutant colonies were similar in size to WT colonies even when isolated from mice infected for 63 days (Fig. 1D), indicating that glycerol metabolism during outgrowth on agar plates following mouse infection slows growth of Δrv3679-80. We next tested the impact of glycerol metabolism on NO susceptibility. The NO hypersusceptibility of the Δrv3679-80 mutant was largely ameliorated when glycerol was removed from both the liquid medium in which the assay was performed and the agar plates on which viability was measured (Fig. 1E; see also Fig. S3C). Together these data demonstrate that glycerol in the growth medium impairs recovery of the Δrv3679-80 mutant from the in vivo environment and synergizes with NO exposure.
Rv3679 and Rv3680 are required for resistance to elevated levels of glycerol in vitro.
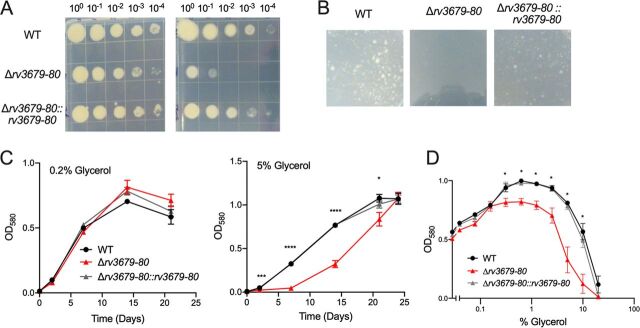
Because the Δrv3679-80 mutant was sensitive to the presence of glycerol during recovery from stress conditions, we assessed whether elevated levels of glycerol impair growth of the Δrv3679-80 mutant. We cultured the WT, the Δrv3679-80 mutant, and the complemented strain on agar plates containing 0.5% glycerol, the concentration used in standard 7H10 medium, and on plates containing 10 times more glycerol (5%). This revealed a clear growth defect of the Δrv3679-80 mutant (Fig. 2A); when the glycerol concentration was increased to 10%, the mutant failed to form colonies (Fig. 2B). In liquid growth medium containing 0.2% glycerol, WT, mutant, and complemented mutant grew similarly; however, with 5% glycerol in the medium, the Δrv3679-80 mutant had a significant growth defect (Fig. 2C). Growth analysis of the WT, the Δrv3679-80 mutant, and the complemented strain in the presence of 2-fold increasing concentrations of glycerol revealed that all three strains initially benefited from increasing amounts of glycerol, with optimal growth in the presence of 0.625% glycerol. However, the Δrv3679-80 mutant never achieved as high of a biomass as the WT or complemented strain, and its growth was more severely inhibited in the presence of 5% and 10% glycerol than that of the WT and complemented mutant (Fig. 2D). Thus, M. tuberculosis requires Rv3679 and Rv3680 for regular growth in the presence of elevated levels of glycerol. M. smegmatis lacking msmeg_6193 and msmeg_6195 was similarly sensitive to glycerol in liquid medium and on agar plates, a phenotype which was rescued by complementation with either the M. smegmatis genes or the M. tuberculosis genes, suggesting a conserved function of the encoded proteins between the species (see Fig. S5 in the supplemental material).
FIG 2.
Rv3679 and Rv3680 are required for growth in the presence of elevated levels of glycerol. (A) Growth of indicated strains on agar plates with either 0.5% glycerol (left) or 5% glycerol (right) after incubation for 2 weeks. Images are representative of multiple experiments. (B) A total of 6 × 107 bacteria/plate cultured on agar plates with 10% glycerol and photographed after 4 weeks of incubation. Extended incubation of the Δrv3679-80 mutant plate also yielded no colonies. (C) Growth of the WT, the Δrv3679-80 mutant, and the Δrv3679-80::rv3679-80 strain in liquid medium containing either 0.2% glycerol (left) or 5% glycerol (right). Data points are means from three independent experiments; error bars indicate standard error of the mean (SEM). One-way ANOVA relative to WT indicated complemented strain not significant (ns) at all time points; Δrv3679-80 mutant, *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (D) Growth after 14 days of incubation in the presence of increasing concentrations of glycerol with constant 0.2% glucose. Data points are means from three independent experiments; error bars indicate SEM. One-way ANOVA at each glycerol concentration comparing strains to WT showed the complement strain was ns at all concentrations; the ∆rv3679-80 mutant was significantly different from WT where indicated. *, P < 0.05.
ATP binding is required for Rv3679 and Rv3680 function.
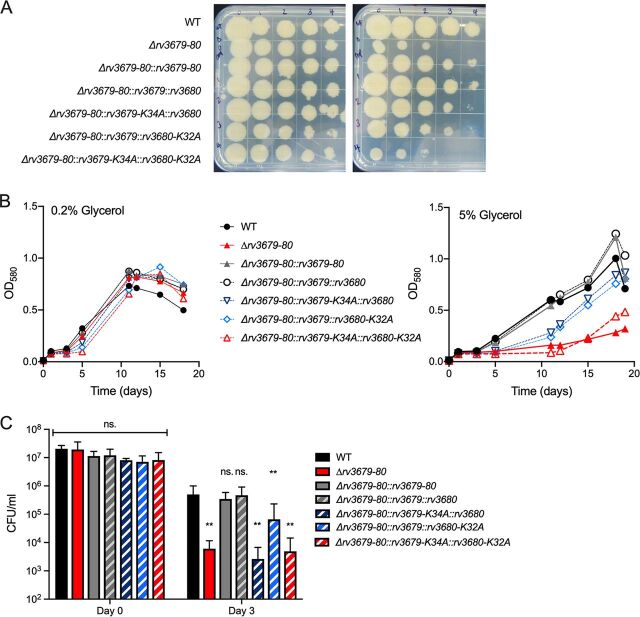
We mutated the lysine residue in the P loop of the Walker A motif, which is required for ATP binding (18), in both proteins to assess if the ATPase activity of Rv3679/Rv3680 is required to confer resistance against glycerol and NO. We also added a FLAG tag to native and mutated Rv3679 and a hemagglutinin (HA) tag to the native and mutated Rv3680, which helped demonstrate that Rv3679-K34A and Rv3680-K32A were expressed at similar levels as the nonmutated proteins (see Fig. S6 in the supplemental material). The Rv3679-FLAG- and Rv3679-K34A-FLAG-expressing plasmids integrated into the attL5 site, while the Rv3680-HA- and Rv3680-K32A-HA-expressing plasmids integrated into the Giles integration site. This enabled us to create four strains as follows: one in which both Rv3679 and Rv3680 are native, two in which either Rv3679 or Rv3680 is mutated, and one in which both Rv3679 and Rv3680 are mutated.
All strains grew similarly on standard 7H10 agar plates (0.5% glycerol); however, on agar plates containing 5% glycerol, the strain expressing the two mutated ATPases displayed a similar growth defect as that of the Δrv3679-80 mutant (Fig. 3A). The strains expressing the two native proteins with FLAG and HA tags grew like the WT, indicating that the tags did not interfere with function. Mutation of a single Walker A motif in either Rv3679 or Rv3680 resulted in intermediate phenotypes. We observed similar results when testing the tolerance of these strains to high glycerol in liquid medium (Fig. 3B). In medium containing 0.2% glycerol, all strains replicated similarly and reached similar biomass; however, in medium containing 5% glycerol, the strain expressing both mutated proteins mimicked the Δrv3679-80 mutant. The strains expressing both proteins with only one Walker A motif mutated showed intermediate growth phenotypes.
FIG 3.
Intact ATP-binding motifs are required for WT levels of resistance to elevated glycerol and NO. (A) Growth of indicated strains on agar plates containing 0.5% glycerol (left) or 5% glycerol (right). (B) Growth in liquid culture in the presence of 0.2% glycerol (left) or 5% glycerol (right). Data are representative of 3 independent experiments. (C) Survival following treatment of strains with 20 μM DETA-NO in medium with 0.2% glycerol. Data are means from 3 independent experiments; error bars indicate standard deviation (SD). One-way ANOVA with Dunnett’s multiple comparison correction at each time point to compare all strains to WT (**, P < 0.01).
We also used this set of Walker A mutant strains to assess the importance of ATP binding for NO resistance. In the presence of diethylenetriamine NONO-ate (DETA-NO) using standard glycerol-containing liquid medium and solid outgrowth agar, the strain expressing both Walker mutant proteins phenocopied the Δrv3679-80 mutant as did the strain expressing mutated Rv3679 and intact Rv3680 (Fig. 3C). Mutation of only Rv3680 resulted in an intermediate phenotype. Taken together, these data show that ATP binding is required for the Rv3679/Rv3680 complex to prevent glycerol-mediated toxicity and that optimal activity requires both Walker A motifs.
Inactivation of glycerol kinase or Rv2370c prevents glycerol-mediated toxicity.
To gain mechanistic insight into the hypersusceptibility of the Δrv3679-80 mutant to glycerol, we screened for mutations that suppress this phenotype. We generated a transposon mutant library in the Δrv3679-80 mutant and cultured it on agar plates containing 5% glycerol. We recovered 146 colonies and determined the transposon insertion sites in 21 of these mutants. Ten mutants contained transposon insertions in glpK, encoding glycerol kinase, and in two mutants the transposon inserted upstream of glpK, likely disrupting promoter activity (Table 1). The glpK gene contains a homopolymeric region that is susceptible to frameshift mutations (19, 20), and we hypothesized that the remaining transposon mutants may also have acquired glpK mutations. Indeed, sequence analysis of the glpK locus revealed spontaneous mutations in glpK leading to either an amino acid change (G36C) or extension of the previously described homopolymeric region (G191), resulting in a premature stop codon at amino acid 252. Two transposon mutants, both with interruption of rv2370c, encoding a protein of unknown function, did not carry glpK mutations.
TABLE 1.
Identity of glycerol-resistant transposon mutants
| Mutant code(s)d | Gene | Protein (length)a | No. of times isolated | Consequence of transposon insertion | glpK mutationc | Spontaneous GlpK mutation |
|---|---|---|---|---|---|---|
| A1, A4, A6, B3, B4, C6, D3 | rv3696c | GlpK (517 aa) | 7 | Premature stop (aa 316) | Y | N/A |
| C3, D2 | rv3696c | GlpK (517 aa) | 2 | Premature stop (aa 230) | Y | N/A |
| D4 | rv3696c | GlpK (517 aa) | 1 | Premature stop (aa 350) | Y | N/A |
| B6, C2 | Intergenic nt 4139762 | N/Ab,e | 2 | N/A | Y | N/A |
| A2 | rv2954c | Hypothetical protein (241 aa) | 1 | Premature stop (aa 142) | Y | Homopolymeric region expansion (G191) resulting in premature stop (aa 252) |
| A5 | rv0831c | Hypothetical protein (271 aa) | 1 | Frameshift (aa 211) | Y | G36C |
| B1, B2, B5 | rv2178c | AroG (462 aa) | 3 | Addition of 14 aa | Y | G36C |
| D1 | rv3273 | Probable carbonic anhydrase (764 aa) | 1 | Frameshift (aa 421) | Y | Homopolymeric region expansion (G191) resulting in premature stop (aa 252) |
| D5 | rv3049c | Probable monooxygenase (524 aa) | 1 | Premature stop (aa 492) | Y | Homopolymeric region expansion (G191) resulting in premature stop (aa 252) |
| C5, D6 | rv2370c | Hypothetical protein (437 aa) | 2 | Frameshift (aa 364) | N | N/A |
aa, amino acid(s); TSS, transcription start site.
N/A, not applicable.
Y, yes; N, no.
Mutant codes refer to the agar plates (A to D) and colonies (1 to 6) picked and analyzed.
Mutation is 6 bp upstream of the TSS of glpK (39).
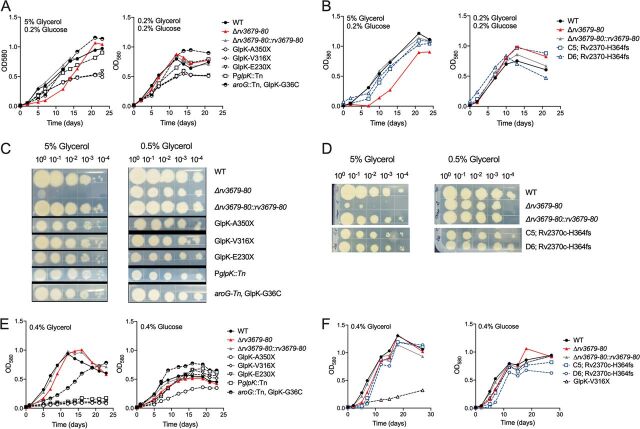
To confirm the suppressor phenotype, we tested a panel of strains carrying mutations in glpK resulting in premature termination of the protein, the mutant with transposon insertion in the glpK promoter (PglpK::Tn), a mutant containing the transposon in aroG and carrying a spontaneous glpK mutation (aroG::Tn, GlpK-G36C), and the two independently isolated mutants that contained the transposon in rv2370c and had no glpK mutation (Rv2370c-H364fs). All mutants rescued the growth defect of the Δrv3679-80 mutant in liquid cultures (Fig. 4A and B) and on agar plates (Fig. 4C and D). To determine if the ability to metabolize glycerol was affected in all mutants, we grew them with either glucose or glycerol as the single carbon source (Fig. 4E and F). As expected and as previously reported (17, 19), disruption of glpK prevented growth with glycerol as the single carbon source. The mutant carrying the transposon in the glpK promoter also failed to utilize glycerol. The mutant with a point mutation in glycerol kinase (GlpK-G36C) grew slower than the WT with glycerol as the sole carbon source, and the two mutants with disruption of Rv2370c replicated like the WT. All strains were able to utilize glucose for growth. Thus, mutations that prevent or impair glycerol metabolism suppress the glycerol-mediated growth defect of the Δrv3679-80 mutant as does interruption of rv2370c, which encodes a protein of unknown function.
FIG 4.
Inactivation of glycerol kinase or Rv2370c prevents glycerol-mediated toxicity. (A) Growth of suppressor mutants with disruption of glpK in medium with 5% or 0.2% glycerol. (B) Growth of suppressor mutants with wild-type glpK in medium with 5% or 0.2% glycerol. (C) Growth of suppressor mutants with disruption of glpK on agar with 5% or 0.5% glycerol. (D) Growth of suppressor mutants with wild-type glpK on agar with 5% or 0.5% glycerol. (E) Growth of suppressor mutants with disruption of glpK in medium with glycerol or glucose as the sole carbon source. (F) Growth of suppressor mutants with wild-type glpK in medium with glycerol or glucose. All suppressor mutants were generated in the Δrv3679-80 mutant background. All data are representative of three independent experiments.
M. tuberculosis Δrv3679-80 mutant is hypersusceptible to extracellular methylglyoxal.
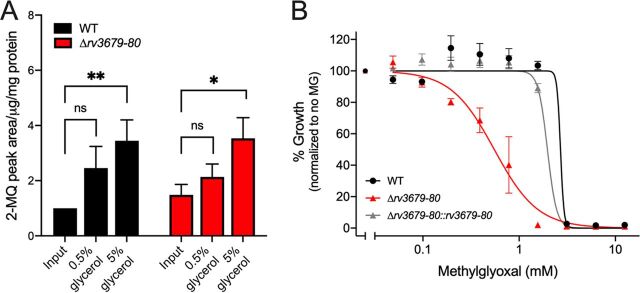
The metabolism of glycerol results in the production of triosephosphates and methylglyoxal (MG), whose accumulation can be toxic to M. tuberculosis (17, 21). To determine if the mutant accumulates more MG than the WT, we measured the intracellular MG pool in the WT and mutant after growth in the presence of 0.5 and 5% glycerol (Fig. 5A). MG pool sizes increased with increasing glycerol concentration in the medium, but there was no significant difference in pool sizes between WT and the Δrv3679-80 mutant. However, when MG was given exogenously in culture medium, the M. tuberculosis Δrv3679-80 mutant was more susceptible to extracellular MG than the WT and complemented mutant (Fig. 5B). Together these data suggest that the glycerol-dependent toxicity in the Δrv3679-80 mutant is due to heightened sensitivity to MG generation, although there is no hyperaccumulation of MG in the Δrv3679-80 mutant.
FIG 5.
The M. tuberculosis Δrv3679-80 mutant is hypersusceptible to extracellular methylglyoxal but does not accumulate it more than the WT. (A) Intrabacterial pool sizes of methylglyoxal derivative product 2-methylquinoxaline (2-MQ) in the indicated strains after 3 days of cultivation on 7H10 agar with 0.2% glucose and either 0.5% or 5% glycerol. Pool sizes are expressed as area under the curve normalized to protein content. Data from triplicate cultures are shown relative to the WT input values. *, P < 0.05; **, P < 0.01 by one-way ANOVA with Bonferroni multiple comparison. (B) Impact of methylglyoxal on growth of the indicated strains. Data are means from two independent experiments each with triplicate cultures; error bars indicate SEM.
Rv3679 interacts with Rv3680 and proteins required for mycolic acid biosynthesis.
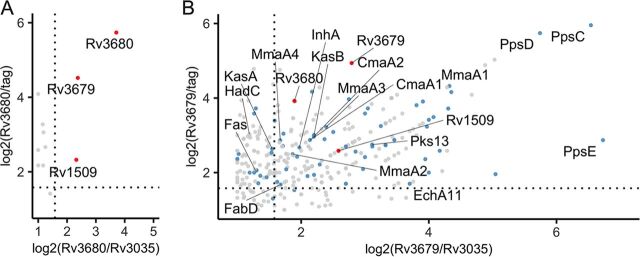
Recent structural work showed that Rv3679 and Rv3680 interact physically when coexpressed in E. coli (6). We validated this interaction in M. tuberculosis by expressing Rv3679-FLAG and Rv3680-HA in the Δrv3679-80 mutant and performing pulldown experiments (see Fig. S7 in the supplemental material). To identify additional putative interaction partners of Rv3679 and Rv3680, we generated single-deletion strains and expressed Rv3679-FLAG in the Δrv3679 mutant and Rv3680-HA in the Δrv3680 mutant. We immunoprecipitated the tagged proteins and identified putative interactors using mass spectrometry (Fig. 6). As controls, we used a strain that expressed just the FLAG tag and a strain that expressed a tagged protein of similar size and predicted cytosolic location (Rv3035-HA, 37.3 kDa). Rv3035, like Rv3679 and Rv3680, is a protein of unknown function conserved across mycobacterial species (22) and lacks a secretion signal and predicted transmembrane helix (23). We considered proteins that were 3-fold more abundant in the experimental samples than in both control samples as putative interactors. Only two proteins, Rv1509 and Rv3679, met these criteria with Rv3680 (Fig. 6A; see also Table S1 in the supplemental material). In contrast, we identified a large number of putative interaction partners of Rv3679 (Fig. 6B; see also Table S1). In addition to Rv3680 and Rv1509, the Rv3679 putative interactors were enriched for proteins with functions in lipid metabolism making up approximately 25% of the interactors (Fig. 6B). Among the strongest were PpsC, PpsD, and PpsE, which are required for biosynthesis of the virulence lipid phthiocerol dimycocerosate (PDIM). Proteins involved in the FASII system (Pks13, KasB, EchA11, InhA, FabD) and in mycolic acid modification, such as the methoxy mycolic acid synthases (MmaA1 to MmaA4) and the cyclopropane-fatty-acyl-phospholipid synthases 1 and 2 (CmaA1 and CmaA2) were also prominent among the Rv3679 putative interactors.
FIG 6.
Rv3679 interacts with Rv3680 and proteins required for mycolic acid biosynthesis. Scatterplots of proteins found bound to Rv3680-HA (A) or Rv3679-FLAG (B) by mass spectrometry. Data are from two independent experiments; proteins plotted if total spectrum count in both replicates is >3. Points indicate log2-adjusted ratios of total spectrum counts of proteins in the sample versus either a protein control of similar size (Rv3035) or a tag-alone control. Dashed lines indicate when the ratio of sample/control is equal to 3. Proteins labeled with red dots are found enriched in both the Rv3679 and Rv3680 samples. Proteins labeled with blue dots are annotated as functioning in “lipid metabolism” in Mycobrowser.
DISCUSSION
Although the ATPases Rv3679 and Rv3680 share sequence similarity with E. coli ArsA and eukaryotic Get3, previous work demonstrated significant differences from ArsA and Get3 and suggested that these proteins are unlikely to be involved in anion transport (6). This work reveals that Rv3679 and Rv3680 physically interact in M. tuberculosis and mediate resistance against nitric oxide and elevated levels of glycerol. We found that glycerol in the culture medium can synergize with other forms of stress; it exacerbated NO toxicity and impaired the recovery of the Δrv3679-80 mutant from infected mouse organs. Rv3679/Rv3680 likely depends on ATPase activity because the ATP-binding motifs in Rv3679 and Rv3680 were essential for restoring normal growth of the Δrv3679-80 mutant in the presence of elevated glycerol and for protection from NO toxicity. Mutations in glycerol kinase that prevent glycerol dissimilation restored both phenotypes, suggesting that a glycerol-derived metabolite is responsible. Of note, inactivation of glycerol kinase has previously been shown to rescue the NO hypersusceptibility of a mycobacterial proteasome ATPase mutant (24), but the underlying mechanism was not explored. Glycerol metabolism has also been reported to mediate the toxicity of pyrimidine imidazoles, which were completely inactive against M. tuberculosis in glycerol-free medium or during mouse infection (17). In that case, toxicity was associated with the accumulation of glycerol phosphate. In M. tuberculosis lacking triosephosphate isomerase, the enzyme that catalyzes the interconversion of glycerol-derived sugar phosphates, dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G3P), the intracellularly accumulated toxic by-product methylglyoxal (MG) correlated with a growth defect in glycerol-containing medium (21). We also observed that MG pool sizes increased in the WT and the Δrv3679-80 mutant in response to elevated extracellular glycerol, and although the intracellular MG pool size in the Δrv3679-80 mutant was not different from that in WT M. tuberculosis, the mutant was hypersusceptible to extracellular MG, suggesting that this glycerol-derived metabolite caused toxicity in the Δrv3679-80 mutant when grown with excess glycerol.
Frameshift mutations in a homopolymeric region of glpK have recently been reported to promote drug tolerance in M. tuberculosis and have been found in clinical isolates (19, 20). Similarly, glycerol kinase transposon mutants showed increased resistance to vancomycin and ethambutol, suggesting that disrupting glycerol metabolism can mediate drug tolerance (25). Bellerose et al. speculated that the abundance of the intracellular triose phosphate pool or a derivative might be enhancing drug efficacy (19). Given the reactivity and toxicity of methylglyoxal, it is plausible that blocking the generation or accumulation of this metabolite might contribute to drug tolerance. Although glycerol metabolism is not required for growth of M. tuberculosis in the mouse model, glpK frameshift mutations develop during human and mouse infection, and glycerol has been measured in mouse lung tissue (17, 19, 20).
Our focus on Rv3679 and Rv3680 was motivated by the finding that transposon mutants in these genes appeared to be attenuated during mouse infection (3). However, when mice were infected with single strains, M. tuberculosis lacking Rv3679 and Rv3680 was fully virulent (6; this work). The underrepresentation of Rv3679/Rv3680 transposon mutants in libraries isolated from mice is likely due to the recovery defect of M. tuberculosis lacking these genes on glycerol-containing agar plates. Hu et al. (6) also noted this slowed growth after mouse infection but ascribed it to a spontaneous ppsE mutation found in their mutant and complemented strains. Whole-genome sequencing (data not shown) revealed that the rv3679-rv3680 deletion mutant that we generated does not carry a ppsE mutation, and moreover, the glycerol-dependent outgrowth defect was restored in the complemented mutant, indicating that it was caused by the deletion of rv3679-rv3680.
We validated that Rv3679 and Rv3680 form a complex in M. tuberculosis and identified one shared putative interaction partner, Rv1509, a predicted S-adenosylmethionine-dependent methyltransferase of unknown substrate specificity according to both InterPro and the Conserved Domain Database (https://www.ebi.ac.uk/interpro/; https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). In addition, Rv3679 interacts with a large number of proteins, which were not immunoprecipitated with Rv3680. These may include indirect interactors. For example, members of the fatty acid synthase II (FASII) system have been shown to interact with each other, with the mycolic acid modifying MmaA enzymes, and with Pks13, catalyzing the final Claisen condensation of mycolic acids (26). The interaction with one of these proteins could have resulted in precipitation of the whole complex. The prevalence of proteins that participate in lipid metabolism and cell envelope biogenesis suggests that Rv3679/Rv3680 participates in these processes, although we do not have direct experimental evidence for this. Potential functional redundancy could explain the lack of phenotype one would expect for deficiencies in mycolic acid biosynthesis, such as attenuation during infection. Our attempts to gain mechanistic insight into the activities mediated by Rv3679/Rv3680 by screening for suppressor mutations identified—besides mutations that interrupt glycerol metabolism—Rv2370c and thus stimulate future research on this and other proteins of unknown function. Similarly, future work on Rv1509, the only shared putative interacting protein of Rv3679 and Rv3680, may also lead to the mechanism by which Rv3679 and Rv3680 protect against glycerol and NO toxicity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 2. M. smegmatis mc2155 and derivative strains were cultured in Middlebrook 7H9 medium (BD Difco) containing 0.2% glycerol, 0.05% Tween 80, 0.2% dextrose, 0.5% bovine serum albumin (BSA) (Roche), and 0.085% NaCl. M. tuberculosis H37Rv and derivative strains were cultured in Middlebrook 7H9 medium (BD Difco) containing 0.2% glycerol, 0.05% Tween 80, and 10% OADC supplement (BD). For both species, CFU were cultured on Middlebrook 7H10 agar (BD Difco) containing 0.5% glycerol and 10% OADC supplement (BD). Strains were incubated at 37°C with 5% CO2. Antibiotics (concentrations) were used as follows: hygromycin (50 μg/ml), kanamycin (25 μg/ml), streptomycin (20 μg/ml), and zeocin (25 μg/ml).
TABLE 2.
Strains used in this work
| Species | Strain | Source or reference | Additional information |
|---|---|---|---|
| M. tuberculosis | Wild-type H37Rv | Gift from C. Sassetti, University of Massachusetts | |
| M. tuberculosis | Δrv3679-80 mutant | This work | WT background, hygromycin resistance cassette replaces rv3679 and rv3680, kanamycin resistance (Kanr) cassette at attL5 site |
| M. tuberculosis | Δrv3679-80::rv3679-80 mutant | This work | Δrv3679-80 mutant background, Phsp60-rv3679-rv3680 with kanamycin resistance at attL5 site |
| M. smegmatis | mc2155 | ATCC | |
| M. smegmatis | Δmsmeg_6193-95 mutant | This work | mc2155 background, hygromycin resistance cassette replaces msmeg_6193 and msmeg_6195 |
| M. smegmatis | Δmsmeg_6193-95::msmeg_6193_95 mutant | This work | Δmsmeg_6193-95 mutant background, Phsp60-msmeg_6193-95 |
| M. smegmatis | Δmsmeg_6193-95::rv3679-80 mutant | This work | Δmsmeg_6193-95 mutant background, Phsp60-rv3679-FLAG, phsp60-rv3680-HA |
| M. smegmatis | ΔclpB mutant | 15 | MsmΔmsmeg_0732::hyg, courtesy of Julien Vaubourgeix and Carl Nathan |
| M. tuberculosis | Δrv3679-80:: rv3679-FLAG, rv3680-HA mutant | This work | Δrv3679-80 mutant background, phsp60-rv3679-FLAG at attL5 site, phsp60-rv3680-HA at Giles site |
| M. tuberculosis | Δrv3679-80:: rv3679-K34A-FLAG, rv3680-HA mutant | This work | Δrv3679-80 background, Phsp60-rv3679-K34A-FLAG at attL5 site, Phsp60-rv3680-HA at Giles site |
| M. tuberculosis | Δrv3679-80:: rv3679-FLAG, rv3680-K32A-HA mutant | This work | Δrv3679-80 background, Phsp60-rv3679-FLAG at attL5 site, Phsp60-rv3680-K32A-HA at Giles site |
Construction of mutants and complementation strains.
The M. tuberculosis Δrv3679-80 mutant strain was generated by recombineering (27, 28) in a merodiploid background as outlined in Fig. S1 in the supplemental material. Gateway cloning technology (Life Technologies) was used to generate all plasmids. First, a plasmid expressing rv3679-80 from the hsp60 promoter with a zeocin resistance marker was inserted into the attL5 integration site in the M. tuberculosis recombineering strain. The native locus spanning rv3679 and rv3680 was then replaced with a hygromycin resistance marker utilizing a plasmid carrying the Hygr marker flanked by the upstream and downstream regions of rv3679-80. Transformants were selected with hygromycin and zeocin and subsequently cured of the recombinase plasmid as previously described (29). After deletion at the native locus, we performed replacement transformations at the attL5 site with either the kanamycin resistance cassette alone to generate the Δrv3679-80 mutant or the full operon under the control of the hsp60 promoter also with a kanamycin resistance marker to generate the Δrv3679-80::rv3679-80 strain.
The M. smegmatis Δmsmeg_6193-95 strain was generated by transformation of the M. smegmatis recombineering strain with a plasmid carrying Hygr flanked by the upstream and downstream regions of msmeg_6193-95. The recombinase plasmid was cured as above. Successful deletion of the native locus in both M. tuberculosis and M. smegmatis was confirmed by Southern blotting using an Amersham ECL direct nucleic acid labeling system kit (GE Healthcare). Complementation of the Δmsmeg_6193-95 mutant was achieved via expression of msmeg_6193-95 under the control of the hsp60 promoter with a streptomycin resistance marker.
To generate the M. tuberculosis Δrv3679 and M. tuberculosis Δrv3680 mutants, which were used in immunoprecipitation experiments, the native locus of either rv3679 or rv3680 was replaced with a hygromycin cassette by recombineering as previously described (27, 28) and confirmed by Southern blotting (see Fig. S8 in the supplemental material). The Δrv3679 and Δrv3680 mutants were transformed with an episomal plasmid expressing rv3679-FLAG or rv3680-HA.
Mouse infections.
We used mid-log-phase M. tuberculosis cultures to infect female C57BL/6 mice (Jackson Laboratory) with approximately 200 bacilli/mouse using an inhalation exposure system (Glas-Col). At each time point, animals were euthanized, and lungs and spleen were isolated and homogenized in phosphate-buffered saline (PBS), serially diluted, and plated for enumeration on either standard 7H10 (0.5% glycerol, 10% OADC) or glycerol-free 7H10 (0.5% glucose, 10% OADC). Plates were photographed to track colony size over time. Images depict equivalent areas of standard 100- by 15-mm petri plates. All animal experiments were performed following National Institutes of Health guidelines for the housing and care of laboratory animals and performed in accordance with institutional regulations after protocol review and approval by the Institutional Animal Care and Use Committee of Weill Cornell Medicine (protocol number 0601441A).
In vitro stress assays.
Susceptibility of strains to arsenite was performed as previously described (30, 31). Briefly, single-cell suspensions of M. tuberculosis strains were adjusted to an optical density at 580 nm (OD580) of 0.02 and grown in 96-well plates with 2 mM sodium arsenite (S7400; Sigma-Aldrich). At the indicated time points, aliquots of culture were removed and serially diluted for enumeration on 7H10 plates.
Susceptibility of strains to heat stress was performed as previously described (32). Briefly, M. smegmatis strains were adjusted to equivalent optical density and incubated at either 37°C or 53°C for 1 h prior to serial dilution and spotting onto 7H10 agar plates, which were all incubated at 37°C.
Susceptibility of strains to nitric oxide via exposure to diethylenetriamine NONO-ate (DETA-NO) (Cayman Chemicals) was performed as previously described (33). Strains were adjusted to equivalent OD and incubated with 200 μM of either DETA-NO or the control compound diethylenetriamine (DETA) (D93856; Sigma-Aldrich). At the indicated time points, cells were serially diluted for CFU enumeration on 7H10 agar with 10% OADC supplement and either 0.5% glycerol or 0% glycerol.
Western blotting.
Rv3679 and Rv3680 protein levels were assessed by Western blotting as previously described (34). Briefly, whole-cell lysates were prepared by mechanical lysis and incubated with 1% dodecyl maltoside. Thirty micrograms of protein per strain were separated by SDS-PAGE and transferred to a nitrocellulose membrane using the iBlot 2 dry blotting system (Invitrogen) in duplicate and in parallel. One blot was incubated with mouse monoclonal anti-FLAG M2 primary antibody (Sigma), while the other was incubated with mouse HA tag monoclonal antibody (2-2.2.14; Thermo Fisher Scientific). Where indicated, blots were also incubated with rabbit antiserum against proteasome beta subunit (PrcB) as a loading control. Both blots were washed and incubated with appropriate LI-COR IRDye secondary antibodies (goat anti-mouse antibody, 800 nm; donkey anti-rabbit antibody, 680 nm). After final washing steps, proteins were visualized using the Odyssey infrared imaging system (LI-COR Biosciences).
Detection of intracellular methylglyoxal levels.
To measure intracellular levels of methylglyoxal, we followed a derivatization method originally devised by Randell et al. (35) and adapted to M. tuberculosis by Trujillo et al. (21) with modifications. M. tuberculosis strains were grown for 5 days on filters laid on 7H10 agar medium with 10% OADC and 0.5% glycerol. Next, filters were split into three groups as follows: one was immediately quenched and frozen at −80°C as an input control, the second was moved onto new 7H10 agar plates with 10% OADC and 0.5% glycerol, and the third was moved onto 7H10 agar plates with 10% OADC and 5% glycerol. After 3 days, bacteria were metabolically quenched by transferring filters into tubes containing precooled acetonitrile/methanol/H2O (2:2:1) supplemented with 1% formic acid, 2.5 μM 2,3-hexanedione, and 125 μM O-phenylenediamine. Bacteria were immediately lysed by bead beating, clarified by centrifugation, and incubated for 18 to 20 h at 4°C to allow for derivatization. Methylglyoxal derivatization products were analyzed by liquid chromatography-mass spectrometry (LC-MS) using the method described by Trujillo et al. (21). Derivatized metabolite identities were established by comparing with a methylglyoxal standard curve prepared in triplicate in acetonitrile-methanol-H2O and subjected to identical bead beating, centrifugation, and derivatization as described for samples.
Suppressor screen and sequencing of isolated mutants.
To screen for suppressor mutants, we generated a transposon (Tn) mutant library in the Δrv3679-80 mutant utilizing Himar1 mutagenesis as previously described (25, 36). We cultured approximately 105 bacteria/plate of the Δrv3679-80 Tn mutant library onto 5% glycerol 7H10 plates with 10% OADC. We recovered 146 colonies from four plates. We isolated 6 colonies per plate for a total of 24 mutants, of which 21 could be successfully grown for genomic DNA extraction and subsequent sequencing of the Tn insertion site (see Table 1). Localization and sequencing of the Tn insertion site was done as previously described (37).
Immunoprecipitation and mass spectrometry.
For immunoprecipitation with Rv3679-FLAG as the “bait,” whole-cell lysates were prepared, incubated with anti-FLAG M2 affinity beads (A2220; Sigma), washed, and proteins eluted from beads as previously described (34). For immunoprecipitation with Rv3680-HA as “bait,” the procedure was modified to use EZview red anti-HA affinity gel beads (E6779; Sigma) for incubation, and proteins were eluted in Laemmli sample buffer (Bio-Rad) by boiling for 10 min and centrifuging for 30 s at 8,200 × g to pellet affinity gel beads. For immunoprecipitation using the Rv3679-FLAG and Rv3680-HA coexpression strain, protein elution was followed by Western blotting, as described above. For immunoprecipitation using the Δrv3679::rv3679-FLAG, Δrv3680::rv3680-HA, or WT::rv3035-HA strain, peptides present in eluates from two independent experiments were analyzed by mass spectrometry. Results were analyzed using Scaffold 4 (Proteome Software). Proteins required a minimum of 3 total spectrum counts in each replicate to be included in analysis. The identities of hits were assigned utilizing publicly available databases PATRIC (https://www.patricbrc.org/) and FLUTE (http://orca2.tamu.edu/U19/). Hits in each sample were compared to those of two different controls as follows: WT::Phsp60-FLAG (data courtesy of Ruojun Wang) and WT::rv3035-HA. To account for instances when a hit occurred in a sample but a control had zero total spectrum counts, creating divide-by-zero errors, a count of 1 was added to the value for all proteins analyzed. Putative functional classes of hits were obtained from Mycobrowser release 3 (5 June 2018) (38).
Statistical analysis.
Raw numerical data were recorded using Microsoft Excel. Statistical analyses were performed using GraphPad Prism version 8.3.1. When two groups are analyzed, Student's t test was utilized to compare groups. When three or more groups are analyzed, one-way analysis of variance (ANOVA) with either Dunnett or Bonferroni multiple-comparison correction was utilized. Graphics were generated in GraphPad Prism version 8.3.1. Scatterplots of coimmunoprecipitation results were prepared using R packages ggplot2 and ggrepel.
ACKNOWLEDGMENTS
We thank J. Vaubourgeix and C. Nathan for sharing the M. smegmatis ΔclpB strain and B. P. Rosen (Florida International University) for helpful advice. We acknowledge K. G. Papavinasasundaram (University of Massachusetts) and S. A. Shaffer (University of Massachusetts, Mass Spectrometry Facility) for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mazandu GK, Mulder NJ. 2012. Function prediction and analysis of Mycobacterium tuberculosis hypothetical proteins. Int J Mol Sci 13:7283–7302. doi: 10.3390/ijms13067283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lew JM, Kapopoulou A, Jones LM, Cole ST. 2011. TubercuList—10 years after. Tuberculosis (Edinb) 91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. 2013. Tryptophan biosynthesis protects Mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey AF, Rock JM, Krieger IV, Chase MR, Fernandez-Suarez M, Gagneux S, Sacchettini JC, Ioerger TR, Fortune SM. 2018. TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog 14:e1006939. doi: 10.1371/journal.ppat.1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vissa VD, Brennan PJ. 2001. The genome of Mycobacterium leprae: a minimal mycobacterial gene set. Genome Biol 2:reviews1023.1. doi: 10.1186/gb-2001-2-8-reviews1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu K, Jordan AT, Zhang S, Dhabaria A, Kovach A, Rangel MV, Ueberheide B, Li H, Darwin KH. 2019. Characterization of guided entry of tail-anchored proteins 3 homologues in Mycobacterium tuberculosis. J Bacteriol 201:e00159-19. doi: 10.1128/JB.00159-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karkaria CE, Chen CM, Rosen BP. 1990. Mutagenesis of a nucleotide-binding site of an anion-translocating ATPase. J Biol Chem 265:7832–7836. [PubMed] [Google Scholar]
- 9.Kaur P, Rosen BP. 1992. Mutagenesis of the C-terminal nucleotide-binding site of an anion-translocating ATPase. J Biol Chem 267:19272–19277. [PubMed] [Google Scholar]
- 10.Mateja A, Paduch M, Chang H-Y, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. 2015. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rome ME, Rao M, Clemons WM, Shan S-O. 2013. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc Natl Acad Sci U S A 110:7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Hsu C-M, Kang B-K, Rosen BP, Bhattacharjee H. 2003. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals 16:369–378. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- 13.Metz J, Wächter A, Schmidt B, Bujnicki JM, Schwappach B. 2006. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J Biol Chem 281:410–417. doi: 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- 14.Powis K, Schrul B, Tienson H, Gostimskaya I, Breker M, High S, Schuldiner M, Jakob U, Schwappach B. 2013. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. J Cell Sci 126:473–483. doi: 10.1242/jcs.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaubourgeix J, Lin G, Dhar N, Chenouard N, Jiang X, Botella H, Lupoli T, Mariani O, Yang G, Ouerfelli O, Unser M, Schnappinger D, McKinney J, Nathan C. 2015. Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe 17:178–190. doi: 10.1016/j.chom.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pethe K, Sequeira PC, Agarwalla S, Rhee K, Kuhen K, Phong WY, Patel V, Beer D, Walker JR, Duraiswamy J, Jiricek J, Keller TH, Chatterjee A, Tan MP, Ujjini M, Rao SPS, Camacho L, Bifani P, Mak PA, Ma I, Barnes SW, Chen Z, Plouffe D, Thayalan P, Ng SH, Au M, Lee BH, Tan BH, Ravindran S, Nanjundappa M, Lin X, Goh A, Lakshminarayana SB, Shoen C, Cynamon M, Kreiswirth B, Dartois V, Peters EC, Glynne R, Brenner S, Dick T. 2010. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun 1:57. doi: 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson PI, Whiteheart SW. 2005. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 19.Bellerose MM, Baek S-H, Huang C-C, Moss CE, Koh E-I, Proulx MK, Smith CM, Baker RE, Lee JS, Eum S, Shin SJ, Cho S-N, Murray M, Sassetti CM. 2019. Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. mBio 10:e00663-19. doi: 10.1128/mBio.00663-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safi H, Gopal P, Lingaraju S, Ma S, Levine C, Dartois V, Yee M, Li L, Blanc L, Ho Liang H-P, Husain S, Hoque M, Soteropoulos P, Rustad T, Sherman DR, Dick T, Alland D. 2019. Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc Natl Acad Sci U S A 116:19665–19674. doi: 10.1073/pnas.1907631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trujillo C, Blumenthal A, Marrero J, Rhee KY, Schnappinger D, Ehrt S. 2014. Triosephosphate isomerase is dispensable in vitro yet essential for Mycobacterium tuberculosis to establish infection. mBio 5:e00085-14. doi: 10.1128/mBio.00085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marmiesse M, Brodin P, Buchrieser C, Gutierrez C, Simoes N, Vincent V, Glaser P, Cole ST, Brosch R. 2004. Macro-array and bioinformatic analyses reveal mycobacterial “core” genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483–496. doi: 10.1099/mic.0.26662-0. [DOI] [PubMed] [Google Scholar]
- 23.Krogh A, Larsson B, Heijne von G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 24.Samanovic MI, Tu S, Novák O, Iyer LM, McAllister FE, Aravind L, Gygi SP, Hubbard SR, Strnad M, Darwin KH. 2015. Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol Cell 57:984–994. doi: 10.1016/j.molcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Dejesus MA, Rücker N, Engelhart CA, Wright MG, Healy C, Lin K, Wang R, Park SW, Ioerger TR, Schnappinger D, Ehrt S. 2017. Chemical genetic interaction profiling reveals determinants of intrinsic antibiotic resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e01334-17. doi: 10.1128/AAC.01334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veyron-Churlet R, Bigot S, Guerrini O, Verdoux S, Malaga W, Daffé M, Zerbib D. 2005. The biosynthesis of mycolic acids in Mycobacterium tuberculosis relies on multiple specialized elongation complexes interconnected by specific protein-protein interactions. J Mol Biol 353:847–858. doi: 10.1016/j.jmb.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei J-R, Dhiman RK, Crick DC, Rubin EJ, Sassetti CM, Alber T. 2012. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal 5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy KC, Papavinasasundaram K, Sassetti CM. 2015. Mycobacterial recombineering. Methods Mol Biol 1285:177–199. doi: 10.1007/978-1-4939-2450-9_10. [DOI] [PubMed] [Google Scholar]
- 29.Ganapathy U, Marrero J, Calhoun S, Eoh H, de Carvalho LPS, Rhee K, Ehrt S. 2015. Two enzymes with redundant fructose bisphosphatase activity sustain gluconeogenesis and virulence in Mycobacterium tuberculosis. Nat Commun 6:7912. doi: 10.1038/ncomms8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlin A, Shi W, Dey S, Rosen BP. 1995. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol 177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wysocki R, Bobrowicz P, Ułaszewski S. 1997. The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J Biol Chem 272:30061–30066. doi: 10.1074/jbc.272.48.30061. [DOI] [PubMed] [Google Scholar]
- 32.Fay A, Glickman MS. 2014. An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genet 10:e1004516. doi: 10.1371/journal.pgen.1004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. 2009. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol 191:625–631. doi: 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Kreutzfeldt K, Botella H, Vaubourgeix J, Schnappinger D, Ehrt S. 2019. Persistent Mycobacterium tuberculosis infection in mice requires PerM for successful cell division. Elife 8:e49570. doi: 10.7554/eLife.49570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randell EW, Vasdev S, Gill V. 2005. Measurement of methylglyoxal in rat tissues by electrospray ionization mass spectrometry and liquid chromatography. J Pharmacol Toxicol Methods 51:153–157. doi: 10.1016/j.vascn.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Long JE, DeJesus M, Ward D, Baker RE, Ioerger T, Sassetti CM. 2015. Identifying essential genes in Mycobacterium tuberculosis by global phenotypic profiling. Methods Mol Biol 1279:79–95. doi: 10.1007/978-1-4939-2398-4_6. [DOI] [PubMed] [Google Scholar]
- 37.Alexander DC, Jones JRW, Tan T, Chen JM, Liu J. 2004. PimF, a mannosyltransferase of mycobacteria, is involved in the biosynthesis of phosphatidylinositol mannosides and lipoarabinomannan. J Biol Chem 279:18824–18833. doi: 10.1074/jbc.M400791200. [DOI] [PubMed] [Google Scholar]
- 38.Kapopoulou A, Lew JM, Cole ST. 2011. The Mycobrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT. 2015. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet 11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]