Abstract
Background
The results of clinical laboratory tests are an essential component of medical decision-making. To guide interpretation, test results are returned with reference intervals defined by the range in which the central 95% of values occur in healthy individuals. Clinical laboratories often set their own reference intervals to accommodate variation in local population and instrumentation. For some tests, reference intervals change as a function of sex, age, and self-identified race and ethnicity.
Methods
In this work, we develop a novel approach, which leverages electronic health record data, to identify healthy individuals and tests for differences in laboratory test values between populations.
Results
We found that the distributions of >50% of laboratory tests with currently fixed reference intervals differ among self-identified racial and ethnic groups (SIREs) in healthy individuals.
Conclusions
Our results confirm the known SIRE-specific differences in creatinine and suggest that more research needs to be done to determine the clinical implications of using one-size-fits-all reference intervals for other tests with SIRE-specific distributions.
Impact Statement
In this work, a data-driven approach of using EHR (electronic health record) precollected data is used to compare population-specific reference intervals. The results of this work may have an effect on the reporting and interpretation of laboratory values. Most potentially affected patients are from minority groups who were previously assumed to have the same reference intervals as the majority group.
Clinical laboratory tests contribute to medical diagnoses and interventional decisions. They also provide insight into physiological states that are not directly observable. However, differences in population demographics, geography, and laboratory instruments can alter distributions of test results (1, 2) and so each clinical laboratory is expected to define reference intervals for each laboratory test (1, 3). Clinical laboratories commonly base these reference intervals on published specifications and/or data from healthy individuals from the communities around the hospital (3, 4). The reference intervals are incorporated into their health systems and used to define normal/abnormal test results.
Reference intervals are typically defined as the interval in which 95% of test results in healthy individuals occur (5). The current gold standard approach is to collect a minimum of 120 healthy samples to estimate reference intervals of new tests, and as few as 20 samples per partition to verify existing ones (3, 6). However, reference samples are difficult to obtain, laboratories often use “easily collected” samples such as college students or internal laboratory staff (7), or rely on reference intervals from the literature or analytical instrument manufacturer product inserts (3). Moreover, there is no general agreement on how to define healthy individuals. To address the issues of diverse sample availability, approaches relying on large cross-sectional samples have been proposed, e.g., the Hoffman and Bhattacharya methods (8, 9). Although these approaches have shown benefits in some circumstances, we considered whether improvements could be made to this approach, because these methods require subjective parameters to be fit “by eye” and make strong distributional assumptions.
In this work, we develop a set of inclusion/exclusion criteria to define healthy individuals within an electronic health record (EHR)8 system, and examine the distributions of laboratory test results in these healthy individuals across self-identified races and ethnicities (SIREs). Currently, some clinical laboratory tests are well known to have racial or ethnic-specific differences, and are reported using SIRE-specific reference intervals (10). For example, mean serum creatinine level was found to be the highest in non-Hispanic African-American, lower in non-Hispanic European, and the lowest in Mexican-American for female and male individuals (11). Therefore, estimated glomerular filtration rate (eGFR), a widely used measure of kidney function, is calculated on the basis of serum creatinine, age, sex, and race. However, in this function, race is binary (African-American or European), and more granular categories and/or genetic ancestry could improve eGFR scaling (12). In other instances, differences in test distributions between SIRE are known in theory, but reference intervals are not altered in practice (13). For example, a genetic variant in the Duffy Antigen Receptor (14), common in African-Americans but rare in European-Americans, induces a 1-SD drop in the mean neutrophil count and is the basis of benign ethnic neutropenia (15). However, neutrophil counts are reported with the same reference interval for all SIREs.
In this work we explore differences between SIREs for an entire spectrum of clinical laboratory tests. To accomplish this goal, we first define a set of inclusion/exclusion criteria to identify healthy individuals' visits from EHR data. We use laboratory results from these visits to define reference intervals using data taken directly from the EHR system at the University of California, San Francisco (UCSF) Medical Center. On the basis of our findings, we estimated the effect that alternative reference intervals may have for biological discovery and for the healthcare system.
MATERIALS AND METHODS
General overview of the approach used in this study
We split patients with deidentified laboratory data into 2 overlapping cohorts: a healthy cohort and a general cohort. The healthy cohort consists of adults (age, 18–60 years) who had laboratory results from healthy encounters (see below). The general cohort includes adults (age, 18–72 years) who had any laboratory encounter (Table 1). We subsampled a single random healthy encounter for each patient having multiple healthy encounters. The healthy cohort was used to define the EHR-based reference interval, and the general cohort was used to estimate the effect of changes of reference intervals on classification of previous measurements.
Table 1.
Patients, encounters, and measurements.
| Healthy cohorta | General cohort | |
|---|---|---|
| Age, years | 18–60 | 18–72 |
| Number of patients | 11245 | 656148 |
| Number of encounters | 13817 | 20750914 |
| Number of laboratory tests | 174505 | 62846904 |
Based on ICD10 diagnosis codes (see Table 1 in the online Data Supplement).
To define the reference interval for each laboratory test among the 50 most common tests, we analyzed for males and females separately. We took the median value for each patient and laboratory test if multiple measurements were available from 1 individual healthy encounter. Finally, we removed outliers as described below. For tests with a single threshold of abnormal (e.g., HDL cholesterol >39 mg/dL) we used the 5th percentile to set the interval. For tests that have reference ranges with lower and upper thresholds (e.g., serum creatinine, 0.44–1.0 mg/dL), we computed the 2.5th and 97.5th percentiles as the EHR-based reference intervals.
Data extraction and cleaning
UCSF uses the Epic EHR system, which was launched in August 2012. Deidentified structured laboratory data, diagnosis and procedure codes, encounter data, and demographics were extracted from EHRs for encounters between August 2012 and May 2017.
Laboratory test methodologies and manufacturer assays are available online (http://labmed.ucsf.edu/labmanual/mftlng-mtzn/test/test-index.html). Siemens platforms were used for all chemistry, complete blood counts, urinalysis, and immunochemistry tests. Stago platforms were used for coagulation assays. We excluded patients <18 or >72 years. eGFR and hepatitis B surface antibody laboratory tests were excluded because >1% of measurements' reported values are of the form “>x” or “<x,” where “x” is a number. Data units of each test were converted to the same unit scale. For example, values reported in cells/μL were converted to cells ×109/L by dividing it by 1000.
Healthy outpatient encounters were selected by a list of International Classification of Diseases, 10th Revision, Clinical Modification (ICD10) codes representing no illness (see Table 1 in the Data Supplement that accompanies the online version of this article at http://www.jalm.org/content/vol3/issue3). Encounters associated with any other ICD10 code were excluded from the healthy encounter set. Patients older than 60 years were excluded from the healthy set. Patients with unknown/declined race or ethnicity (Table 2) were included in the healthy cohort, which was used to define EHR-calibrated reference intervals, but were excluded from SIRE-specific reference intervals calibration.
Table 2.
Summary statistics of patients in UCSF EHR database.
| Median | Mean | SD | |
|---|---|---|---|
| Age, years | 42 | 41.66 | 23.98 |
| Encounters per patient | 7 | 27.19 | 61.48 |
| Female | Male | Unknown/Unspecified | |
|---|---|---|---|
| Sex | 505400 | 415888 | 1308 |
| Ethnicity | Hispanic or Latino | Not Hispanic or Latino | Unknown/declined |
|---|---|---|---|
| Race | |||
| Native American or Native Alaskan | 595 | 2491 | 194 |
| Asian | 982 | 94032a | 3587 |
| Black or African-American | 1011 | 41644a | 2354 |
| Native Hawaiian or other Pacific Islander | 319 | 12882a | 1091 |
| Other | 74968a | 49592 | 8077 |
| Unknown/Declined | 8383 | 16568 | 197413 |
| White or Caucasian | 22162a | 368845a | 20306 |
Indicates major race-ethnicity group used in the current analysis.
Defining reference intervals
Outliers were detected using the Tukey method (4, 16) as proposed by Reed et al. (6). For each test and sex, we defined interquartile range (IQR) as IQR = Q3 − Q1, where Qi was the ith quartile of the data. Values below or above 1.5 × IQR of Q2 or Q3, respectively, were considered outliers and were removed.
According to current practice, a reference interval is defined by collecting a minimum of 120 healthy samples from a population. Values between the 2.5th and 97.5th percentiles become the reference interval. Several tests specify only a lower or upper threshold. In these cases, we defined the reference interval as being below the 5th percentile or above the 95th (e.g., LDL cholesterol <130 mg/dL and HDL cholesterol >39 mg/dL).
Statistical tests
To test for statistically significant differences in average values across SIREs, we performed analysis of variance (ANOVA) using 2 tests: 1-way ANOVA and the Kruskal–Wallis (KW) test (17). In the 1-way ANOVA, we added a patient's SIRE and age to the linear model, as well as the laboratory measurement as the dependent variable. The KW test does not support multiple independent variables, so we first computed residuals of the linear model, in which laboratory measurements were the dependent variables and age and SIRE were the independent variables. Residuals were then set as dependent variables in the KW test, with SIRE as the independent variable. Multiple-test correction was performed using the Benjamini–Hochberg procedure (18).
RESULTS
We examined deidentified EHR data from approximately 970000 patients seen between August 2012 and December 2017 (28 million encounters), yielding an initial set of 87 million laboratory test results. Table 2 shows the basic demographic information for this population. We selected our main cohort from the EHR based on inclusion and exclusion criteria, as follows. To capture healthy individuals, we selected outpatient encounters during which a diagnostic ICD10 code was assigned matching a predefined list reflecting healthy nonillness encounters (see Table 1 in the online Data Supplement) and excluded any encounter with ICD10 codes not on the list. Out of 28 million encounters, there were 13817 encounters covering 11254 healthy adults (age, 18–60 years). None of these encounters was a follow-up appointment to a previous one as defined by the visit type. A total of 174505 laboratory test results were recorded from these encounters (Table 1). Through discussions with laboratory staff, we ensured that laboratory instruments were not changed during the collection period.
EHR data are known not to represent the general population; many younger or healthier individuals may not interact with the health system. Although we did not have demographic data of subjects who were originally used to define the current reference interval, we did compare the body mass indices (BMI) of our healthy subject cohort with the BMI of a sample cohort from the general US population [National Health and Nutrition Examination Survey (NHANES), 2015–2016], which has previously been used to define reference intervals for laboratory tests (10). The average BMI in the general US population of age range 18–60 years is 25.6 ± 8 kg/m2 and 24.7 ± 7 kg/m2 for female and male individuals, respectively, where the average BMI of healthy individuals at UCSF is 25.3 ± 6 kg/m2 and 26.5 ± 5 kg/m2 for females and males, respectively and for the same age group of 18–60 years (average age, 42.0 ± 11 and 39.5 ± 10 years for healthy individuals at UCSF and NHANES, respectively). While the female individuals at UCSF have lower BMI (2-sided t-test, P = 0.04) and the male individuals have higher BMI (1-sided t-test, P = 1.95 × 10–32), the statistical significance is driven by the large sample size and the magnitude of the changes is small.
EHR-based laboratory reference intervals are stable
We collected data from the 50 most common laboratory tests having at least 120 healthy samples for each sex from the UCSF EHR. Outlier values were removed using the Tukey method (16; see Methods). For each of the 50 laboratory tests, we defined a data-driven reference interval to contain 95% of the healthy sample results (see Methods).
We compared EHR-based and currently-in-use reference intervals by taking the log base 2 of the ratio of interval's size for each laboratory test. We rejected the hypothesis that EHR-based reference intervals for laboratory tests with smaller healthy samples deviate from currently-in-use reference intervals more than laboratory tests with larger healthy samples (Spearman correlation = −0.05, P = 0.65) (Fig. 1 and see Fig. 1 in the online Data Supplement).
Fig. 1. EHR-calibrated reference intervals comparison.
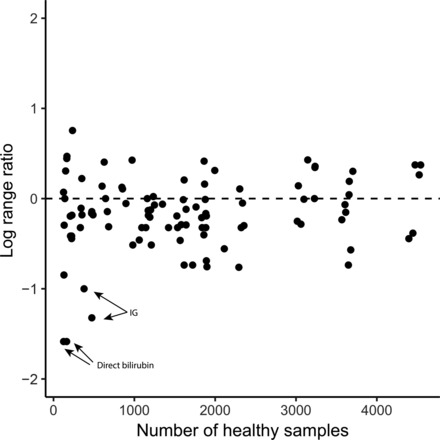
Scatter plot of the number of healthy samples vs log2 of the ratio of original reference interval size and EHR-based reference interval size. A point above the horizontal line represents a laboratory test in which the EHR-calibrated reference interval is larger than the original. For example, 2 data points on the bottom left size represent the 2 laboratory tests to measure direct bilirubin levels for male and female individuals. For these 2 laboratory tests, the reference interval is ≤0.3 mg/dL, whereas the EHR-based reference intervals were found to be ≤0.1 mg/dL.
Comparing our new calculated reference intervals with the original ones, we found that the reference intervals of only 2 laboratory tests had a >2-fold change (Fig. 1). The existing reference range for direct bilirubin is ≤0.3 mg/dL for female and male individuals at UCSF, and we defined a data-driven range of ≤0.1 mg/dL for both female and male individuals independently, a 3-fold narrowing of the reference interval. Immature granulocyte (IG) count is a marker for infection and sepsis (19). The existing reference interval for IG is ≤0.1 × 109 cells/L for both sexes in UCSF's reference interval, whereas the EHR-calibrated method gave upper threshold/95th percentile values of 0.04 and 0.05 × 109 cells/L for female and male individuals, implying a 2-fold decrease over the UCSF reference intervals.
Switching to EHR-based data-driven references from the current UCSF ranges affects 6.7% of all measurements: 2.4% out-of-range measurements were reclassified as normal and 4.2% normal measurements exceed the new calculated thresholds. This confirms the overall utility of our inclusion/exclusion criteria, with results for individual tests provided in Table 2 in the online Data Supplement (see Fig. S2 in the online Data Supplement). For example, UCSF's current reference interval for creatinine in female individuals of age ≥19 years is 0.44–1.0 mg/dL. Based on 3061 healthy female individuals of age 19–60 years, the new EHR-defined reference interval is 0.48–0.94 mg/dL, which reclassifies 6.6% of creatinine measurements (see Table 2 in the online Data Supplement). Similarly, the existing reference interval for white blood cell (WBC) counts for female individuals of age ≥21 years is 3.4–10.0 × 109 cells/L and the EHR-calibrated reference interval (3606 subjects) is 3.4–9.7 × 109 cells/L, which reclassifies only 1.9% of measurements.
Laboratory test distributions differ across subpopulations
The general practice for defining reference intervals requires separate categories for male and female individuals and for age groups when appropriate. Few laboratory tests have reference intervals for race and ethnic groups currently in use in the clinic (e.g., creatinine). We had male-specific data from 46 tests and female-specific data from 44 tests that also had at least 2 SIREs with ≥50 healthy individuals. For each test, we compared the distribution of healthy measurements from different SIREs adjusting for age and stratifying by sex and tested for differences using ANOVA (see Methods). Table 3 shows the 10 results with the lowest ANOVA P value (complete results in Table 3 in the online Data Supplement). We find that many laboratory test results differed between SIREs. Out of 85 laboratory tests, 48 (56%) were significantly different across different SIREs (ANOVA, P < 0.05 after Benjamini–Hochberg correction for multiple testing). The nonparametric KW test also found significant differences between SIREs in 60 (71%) laboratory tests (after Benjamini–Hochberg correction for multiple testing; see Table 3 in the online Data Supplement).
Table 3.
Laboratory tests and adjusted ANOVA P values.a
| Name of laboratory test | Sex | Mean | CI | Units | Adjusted P value | Number of samples L-O/L-W/NL-B/NL-A/NL-H/NL-Wb,c | L-O mean | L-W mean | NL-B mean | NL-A mean | NL-H mean | NL-W mean |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCH | F | 29.95 | < 0.1 | pg | 9.94E-34 | 214/92/214/978/79/1594 | 29.77 ± 0.3 | 29.85 ± 0.5 | 28.35 ± 0.4 | 29.68 ± 0.2 | 29.23 ± 0.6 | 30.39 ± 0.1 |
| MCHC | F | 33.32 | < 0.1 | g/dL | 3.79E-31 | 214/92/214/978/79/1594 | 33.36 ± 0.1 | 33.33 ± 0.2 | 32.66 ± 0.1 | 33.26 ± 0.1 | 33.10 ± 0.2 | 33.45 ± 0.0 |
| MCV | F | 89.86 | 0.2 | fL | 7.75E-25 | 214/92/214/978/79/1594 | 89.41 ± 0.7 | 89.49 ± 1.1 | 86.70 ± 1.0 | 89.12 ± 0.5 | 87.99 ± 1.6 | 90.90 ± 0.2 |
| Cholesterol HDL | F | 64.74 | 0.5 | mg/dL | 1.23E-23 | 287/103/240/1414/104/1844 | 57.79 ± 1.8 | 59.56 ± 2.9 | 61.65 ± 2.1 | 63.80 ± 0.8 | 62.28 ± 2.8 | 67.37 ± 0.8 |
| Hemoglobin | F | 13.09 | < 0.1 | g/dL | 1.21E-22 | 216/93/226/988/79/1612 | 13.06 ± 0.2 | 13.15 ± 0.2 | 12.35 ± 0.2 | 13.10 ± 0.1 | 12.95 ± 0.3 | 13.19 ± 0.0 |
| Vitamin D, 25-hydroxy | F | 27.02 | 0.6 | ng/mL | 7.42E-19 | 89/40/72/572/36/837 | 23.85 ± 2.3 | 26.20 ± 3.7 | 21.39 ± 2.9 | 24.60 ± 0.8 | 24.97 ± 4.2 | 29.61 ± 0.8 |
| Triglycerides, serum | F | 86.04 | 1.8 | mg/dL | 1.93E-16 | 287/105/240/1415/104/1852 | 101.38 ± 8.3 | 90.97 ± 10.5 | 72.28 ± 5.1 | 91.05 ± 3.4 | 114.08 ± 20.8 | 79.77 ± 2.3 |
| Cholesterol HDL ratio | F | 3.25 | < 0.1 | - | 4.41E-16 | 286/102/240/1408/104/1833 | 3.62 ± 0.1 | 3.50 ± 0.2 | 3.29 ± 0.1 | 3.29 ± 0.0 | 3.43 ± 0.2 | 3.14 ± 0.0 |
| WBC count | F | 6.22 | < 0.1 | × 109 cells/L | 1.22E-10 | 214/92/214/978/79/1595 | 6.96 ± 0.3 | 6.32 ± 0.4 | 6.26 ± 0.4 | 5.92 ± 0.1 | 6.22 ± 0.4 | 6.29 ± 0.1 |
| Hematocrit | F | 39.26 | 0.1 | % | 1.22E-10 | 216/93/217/978/79/1599 | 39.13 ± 0.5 | 39.52 ± 0.7 | 37.74 ± 0.5 | 39.35 ± 0.2 | 39.07 ± 0.8 | 39.42 ± 0.1 |
Top 10 most significant laboratory tests with differences in means across SIRE in healthy patients (see complete table in Table 3 in the online Data Supplement).
Number of samples for each SIRE.
Abbreviations: L, Hispanic or Latin;, NL, Not Hispanic or Latino; A, Asian; B, Black or African American; H, Native Hawaiian or Other Pacific Islander; O, Other; W, White or Caucasian.
Distribution of clinical measurements within healthy individuals
Results for several laboratory tests are known to have distribution differences between SIREs. Serum creatinine is an example (11), and we evaluated this test in detail as a correctly discovered true-positive finding. Healthy non-Hispanic African-Americans had the highest mean serum creatinine level, followed by non-Hispanic Europeans and Hispanic Europeans, with non-Hispanic Asians having the lowest mean serum creatinine level (Fig. 2A), consistent with previous publications (10, 11). The difference in creatinine levels between female SIRE was significant (ANOVA, P = 2.26 × 10–8; KW, P = 8.56 × 10–49; see Table 3 in the online Data Supplement), and it remained significant after excluding the more extreme non-Hispanic, African-American measurements (P = 4.21 × 10–8). The difference in males was not significant by ANOVA (P = 0.33) but was according to KW test (P = 1.7 × 10–10).
Fig. 2. Difference in laboratory test measurements among healthy individuals in different SIRE groups.
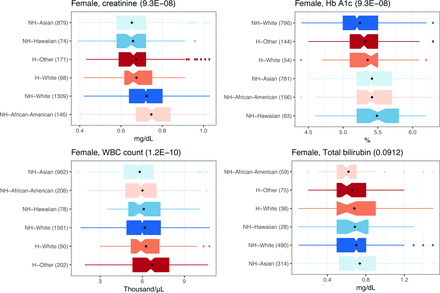
Distributions of different laboratory tests in healthy individuals for (A) creatinine, (B) Hb A1c, (C) WBC count, and (D) total bilirubin. Black diamonds: average values. Parentheses in the titles show Benjamini–Hochberg-adjusted ANOVA P values. Parentheses next to each SIRE, show the number of healthy individuals. Colors match across panels. SIRE sorted by mean. H, Hispanic or Latino; NH, Not Hispanic or Latino.
Previous studies have suggested differences in total bilirubin levels between SIREs (20). We tested this possibility and found no difference (ANOVA, P value = 0.09 and 0.54 for female and male individuals, respectively; Fig. 2D and see Table S3 in the online Data Supplement). Separately, we found that healthy African-Americans had a lower average WBC count than healthy Europeans (t-test, P = 0.02), as has been shown (14, 21). We also found a lower WBC count in non-Hispanic Asians compared with Europeans and Hispanics (t-test, P = 7.5 × 10–11; Fig. 2 and see Table 3 in the online Data Supplement). HbA1c levels in our data were higher in African-Americans compared with those in Europeans in accordance with previous work (22, 23). To the best of our knowledge, only a few laboratory tests were previously shown to differ across SIRE groups. Serum creatinine is the only test that is currently adjusted for SIRE during clinical interpretations of the test. In total, we found that >50% of common laboratory tests had significant difference across SIRE groups.
Reference intervals stratified by population demographics
We next examined the effect sizes of SIRE-specific reference intervals. For each laboratory test, sex, and SIRE, we repeated the procedure above to defined SIRE-specific EHR-based reference intervals as central 95% of values of matching healthy individuals. The number of measurements in the low/normal/high group for each reference intervals considered was calculated for each SIRE and laboratory test. Among laboratory tests with at least 120 healthy samples, 543601 measurements (2.5%) from the general data set that were originally considered abnormal, would be reclassified as normal, and 980673 (4.5%) originally normal measurements would be considered abnormal under new EHR-driven SIRE-specific reference intervals (see Table 4 in the online Data Supplement).
For example, 26303 non-Hispanic Europeans and 5219 non-Hispanic African-Americans had HbA1c measurements. Using the first measurement per subject, the HbA1c levels in 7887 (30%) Europeans and 2725 (52%) African-Americans exceeded 5.6%, which is the upper threshold of the existing reference interval. Alternatively, the HbA1c levels in only 3783 (14%) non-Hispanic Europeans and 1499 (29%) non-Hispanic African-Americans exceeded on using an EHR-calibrated threshold of 6.1% (see Table 2 in the online Data Supplement). When a SIRE-specific threshold (see Table 4 in the online Data Supplement) was used, the HbA1c levels was higher than the threshold in 4792 (18%) and 3064 (12%) of non-Hispanic Europeans and non-Hispanic African-Americans, respectively. These findings demonstrate that a substantial number of individuals are newly categorized to abnormal from normal or vice versa if SIRE-specific reference intervals are constructed.
DISCUSSION
In this work we found that for more than half of the commonly obtained laboratory tests, there is a difference in measurements across SIREs in our cohort of healthy individuals. Some environmental or genetic factors that affect laboratory test results are known, and these may differ across race or ethnicity (14, 24). For example, the variant rs2814778 causes benign ethnic neutropenia in African-Americans. However, our finding that WBC levels were lower in normal non-Hispanic Asians cannot be explained by this variant, because the allele associated with low neutrophil levels has a frequency of 0.82 in African-Americans and <0.001 in Asians as well as in Europeans (25).
Different subpopulations may seek care for different diseases because of genetic and environmental factors. This fact may drive differences in the distribution of test results, even if the healthy reference interval of these distributions is identical across populations. Although we found that >50% of tests varied by SIRE, only 1 of these, creatinine, is adjusted for SIRE in current clinical practice. Even in this case, we found additional heterogeneity beyond the current version of African-American vs non-African American, particularly the lower levels of creatinine among healthy Asian-Americans. The risk for mortality from stroke is 2-fold greater among African-Americans than Europeans (26), and the prevalence of hypertension is increased in the African-Americans (27). In this case, setting a different reference interval just for African-American patients may not be comprehensive enough. To get a more precise reference interval, one might also consider intra- and interindividual variations (28), but this type of information is not always available, because healthy individuals tend not to need many blood tests.
We also described an approach to defining reference intervals for clinical laboratory test results from existing clinical laboratory test measurement data and showed that the distributions of laboratory test results do not substantially differ, with the exception of direct bilirubin and IG, which we found to be 2- or 3-fold smaller. However, a study from 2003 (29) found the 95th percentile of IG values to be 0.03 × 109 cells/L for both female and male individuals, thus supporting our finding. However, we note that this result does not imply that patients with IG values between 0.04 and 0.1 × 109 cells/L have an infection.
EHR-based research has multiple advantages over traditional clinical studies (30). For example, EHR-based studies use data acquired under routine conditions, whereas large studies use measurements acquired by following research protocols. Furthermore, the adjustment of EHR-based reference interval will permit direct examination and accommodation of test distribution differences between SIREs. EHR-calibrated reference intervals may provide more accurate estimates of “real” normal values because they are based on a larger number of samples than the 120 required by the current gold standard methods (see Fig. 1 in the online Data Supplement for comparison between different sample sizes). However, there are limitations in the use of EHR data including the following: (a) abnormal laboratory values obtained during encounters that are misclassified as healthy encounters, (b) sample sizes for rarely used tests, and (c), missing knowledge of an evolving illness, which may have affected the distribution of the results. Going forward, it will be interesting to explore alternative data-driven approaches to defining a healthy distribution (18).
Despite these limitations, our method does at least as well as the standard method for sample collection, which lacks a standard for defining healthy volunteers (31). In addition, some laboratory test results change dramatically in different normal conditions. For example, calcium and phosphate levels have a circadian rhythm (32), creatinine is influenced by hydration (33), and glucose by food intake. None of these conditions were controlled for here, but they are not considered in the current scheme of determining reference intervals either.
In this work, we treated SIRE as marker of identity, as has been done previously in clinical contexts (11). First, false reporting of SIRE is common (34), and many patients identify with 2 or more racial/ethnic categories. An extension of the current work will define reference intervals for more population categories (or even a continuous space of patients) and will also include patient genome information. Discussions about the overall nature of race and ethnicity are beyond the scope of this work and widely discussed elsewhere (35, 36).
Our findings suggest that reference intervals can be calculated based on healthy baselines determined for each subpopulation. However, it is not necessarily the case that a group-specific reference interval conveys more diagnostic information, even if the healthy group-specific distribution is different from healthy distributions in other populations. For example, healthy individuals from one population may have higher LDL cholesterol or non-HDL cholesterol levels owing to genetic and environmental factors such as diet. However, this does not necessarily mean that the risk of a cardiovascular event at a given level of cholesterol differs between SIREs (37, 38). We do suggest that our technique be used to periodically compare EHR-calibrated reference intervals with the standard reference intervals to understand local population differences that may have a clinical impact.
The factors affecting the differences between SIREs might be genetic, environmental, or sociological in origin. Our findings call for more exploration of the underlying biology that might be leading to these differences and the potential clinical impact of these differences.
Supplementary Material
Acknowledgments
The authors would like to thank Alan Wu and Matthew Kan for fruitful discussions, Alan Wu for reviewing the manuscript, Debajyoti Datta and Merav Heshin-Bekenstein for clinic-related advices, and Dana Ludwig and the UCSF Information Technology Services Academic Research Systems team for Clinical Data Research Consultations and data extraction. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
8 Nonstandard abbreviations
- EHR
electronic health records
- SIRE
self-identified racial and ethnic group
- eGFR
estimated glomerular filtration rate
- UCSF
University of California, San Francisco
- ICD10
International Classification of Diseases, 10th Revision, Clinical Modification
- IQR
interquartile range
- KW
Kruskal–Wallis
- ANOVA
analysis of variance
- BMI
body mass index
- NHANES
National Health and Nutrition Examination Survey
- IG
immature granulocyte
- WBC
white blood cell.
Footnotes
Author Declaration: A version of this paper was previously posted as a preprint on bioRxiv as https://www.biorxiv.org/content/early/2017/11/04/213892.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Authors' Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Research Funding: National Institute of General Medical Sciences of the National Institutes of Health R01GM079719.
Expert Testimony: None declared.
Patents: None declared.
Role of Sponsor: The sponsor played a role in the design of study, review and interpretation of data, preparation and final approval of manuscript.
References
- 1. Wu AH. Tietz clinical guide to laboratory tests. 4th ed. St. Louis: Saunders/Elsevier; 2006. 1798 p. [Google Scholar]
- 2. Lewis SM. Reference ranges and normal values. In: Lewis SM, Bain BJ, Bates I, editors. Dacie and Lewis Practical Haematology. 10th ed. Philadelphia (PA): Churchill Livingstone; 2006. p. 11–24. [Google Scholar]
- 3. Horowitz GL, Altaie S, Boyd JC. Defining, establishing, and verifying reference intervals in the clinical laboratory; approved guideline. Wayne (PA); CLSI; 2010. CLSI document EP28-A3C. [Google Scholar]
- 4. Burtis CA, Ashwood ER, Bruns DE, Tietz NW. Tietz textbook of clinical chemistry and molecular diagnostics. 5th ed. St. Louis: Saunders; 2013. 2238 p. [Google Scholar]
- 5. Marshall WJ, Bangert SK. Clinical biochemistry: metabolic and clinical aspects. 2nd ed. New York: Churchill Livingstone; 1995. 996 p. [Google Scholar]
- 6. Reed AH, Henry RJ, Mason WB. Influence of statistical method used on the resulting estimate of normal range. Clin Chem 1971;17:275–84. [PubMed] [Google Scholar]
- 7. Horn PS, Pesce AJ, Copeland BE. A robust approach to reference interval estimation and evaluation. Clin Chem 1998;44:622–31. [PubMed] [Google Scholar]
- 8. Bhattacharya C. A simple method of resolution of a distribution into gaussian components. Biometrics 1967:115–35. [PubMed] [Google Scholar]
- 9. Hoffmann RG. Statistics in the practice of medicine. JAMA 1963;185:864–73. [DOI] [PubMed] [Google Scholar]
- 10. Lim E, Miyamura J, Chen JJ. Racial/ethnic-specific reference intervals for common laboratory tests: a comparison among Asians, Blacks, Hispanics, and White. Hawaii J Med Public Health 2015;74:302–10. [PMC free article] [PubMed] [Google Scholar]
- 11. Jones CA, McQuillan GM, Kusek JW, Eberhardt MS, Herman WH, Coresh J, et al. Serum creatinine levels in the us population: third National Health and Nutrition Examination Survey. Am J Kidney Dis 1998;32:992–9. [DOI] [PubMed] [Google Scholar]
- 12. Udler MS, Nadkarni GN, Belbin G, Lotay V, Wyatt C, Gottesman O, et al. Effect of genetic African ancestry on eGFR and kidney disease. J Am Soc Nephrol 2015;26:1682–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McPherson K, Healy MJ, Flynn FV, Piper KA, Garcia-Webb P. The effect of age, sex and other factors on blood chemistry in health. Clin Chim Acta 1978;84:373–97. [DOI] [PubMed] [Google Scholar]
- 14. Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet 2009;5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haddy TB, Rana SR, Castro O. Benign ethnic neutropenia: what is a normal absolute neutrophil count? J Lab Clin Med 1999;133:15–22. [DOI] [PubMed] [Google Scholar]
- 16. Hoaglin DC, John W. Tukey and data analysis. Statist Sci 2003:311–8. [Google Scholar]
- 17. Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 1952;47:583–621. [Google Scholar]
- 18. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995:289–300. [Google Scholar]
- 19. Ansari-Lari MA, Kickler TS, Borowitz MJ. Immature granulocyte measurement using the Sysmex XE-2100. Relationship to infection and sepsis. Am J Clin Pathol 2003;120:795–9. [DOI] [PubMed] [Google Scholar]
- 20. Carmel R, Wong ET, Weiner JM, Johnson CS. Racial differences in serum total bilirubin levels in health and in disease (pernicious anemia). JAMA 1985;253:3416–8. [PubMed] [Google Scholar]
- 21. Hsieh MM, Everhart JE, Byrd-Holt DD, Tisdale JF, Rodgers GP. Prevalence of neutropenia in the U.S. population: age, sex, smoking status, and ethnic differences. Ann Intern Med 2007;146:486–92. [DOI] [PubMed] [Google Scholar]
- 22. Menke A, Rust KF, Savage PJ, Cowie CC. Hemoglobin A1c, fasting plasma glucose, and 2-hour plasma glucose distributions in U.S. population subgroups: NHANES 2005–2010. Ann Epidemiol 2014;24:83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Selvin E, Steffes MW, Zhu H, Matsushita K, Wagenknecht L, Pankow J, et al. Glycated hemoglobin, diabetes, and cardiovascular risk in nondiabetic adults. N Engl J Med 2010;362:800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldberg DM, Handyside AJ, Winfield DA. Influence of demographic factors on serum concentrations of seven chemical constituents in healthy human subjects. Clin Chem 1973;19:395–402. [PubMed] [Google Scholar]
- 25. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lackland DT, Bachman DL, Carter TD, Barker DL, Timms S, Kohli H. The geographic variation in stroke incidence in two areas of the southeastern stroke belt: the Anderson and Pee Dee Stroke Study. Stroke 1998;29:2061–8. [DOI] [PubMed] [Google Scholar]
- 27. Ford ES. Trends in mortality from all causes and cardiovascular disease among hypertensive and nonhypertensive adults in the United States. Circulation 2011;123:1737–44. [DOI] [PubMed] [Google Scholar]
- 28. Harris EK. Effects of intra- and interindividual variation on the appropriate use of normal ranges. Clin Chem 1974;20:1535–42. [PubMed] [Google Scholar]
- 29. Bruegel M, Fiedler G, Matthes G, Thiery J. Reference values for immature granulocytes in healthy blood donors generated on the Sysmex XE-2100 automated hematology analyser. Sysmex J Int 2004;14:5–7. [Google Scholar]
- 30. Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, et al. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet 2017;49:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harris EK, Boyd JC. Statistical bases of reference values in laboratory medicine. Boca Raton: CRC Press; 1995. 384 p. [Google Scholar]
- 32. Carruthers BM, Copp DH, McIntosh HW. Diurnal variation in urinary excretion of calcium and phosphate and its relation to blood levels. J Lab Clin Med 1964;63:959–68. [PubMed] [Google Scholar]
- 33. Ship JA, Fischer DJ. The relationship between dehydration and parotid salivary gland function in young and older healthy adults. J Gerontol A Biol Sci Med Sci 1997;52:M310–9. [DOI] [PubMed] [Google Scholar]
- 34. Smith TW. Measuring race by observation and self-identification. GSS Methodological Report 89. Chicago: National Opinion Research Center, University of Chicago; 1997. [Google Scholar]
- 35. Cross M. Race and ethnicity. In: Thornley A, editor. The crisis of London. 2nd ed. London (UK): Routledge; 2003. [Google Scholar]
- 36. Das Nair R, Thomas S. Race and ethnicity. Wiley Online Library. https://onlinelibrary.wiley.com/doi/abs/10.1002/9781119967613.ch3 (Accessed May 2018). [Google Scholar]
- 37. D'Agostino RB Sr, Grundy S, Sullivan LM, Wilson P, Group CHDRP. Validation of the Framingham coronary heart disease prediction scores: results of a multiple ethnic groups investigation. JAMA 2001;286:180–7. [DOI] [PubMed] [Google Scholar]
- 38. Zaitlen N, Pasaniuc B, Sankararaman S, Bhatia G, Zhang J, Gusev A, et al. Leveraging population admixture to characterize the heritability of complex traits. Nat Genet 2014;46:1356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


