Abstract
Pituitary gonadotropins are critical regulators of gonadal development and function. Expression and secretion of the mature hormones are regulated by gonadotropin-releasing hormone (GnRH), which is itself secreted from the hypothalamus. GnRH stimulation of gonadotropin expression and secretion occurs through the G-protein-linked phospholipase C/inositol triphosphate intracellular signaling pathway, which ultimately leads to protein kinase C (PKC) activation and increased intracellular calcium levels. Transcription factors mediating the effects of GnRH-induced signals on transcription of gonadotropin genes have not yet been identified. Recent studies have identified key factors involved in luteinizing hormone β (LHβ) gonadotropin gene transcription: the nuclear receptor SF-1, the bicoid-related homeoprotein Ptx1 (Pitx1), and the immediate-early Egr-1 gene. We now show that GnRH is a potent stimulator of Egr-1, but not Ptx1 or SF-1, expression. Further, Egr-1 activation of the LHβ promoter is specifically enhanced by PKC, in agreement with a role for Egr-1 in mediating a GnRH effect on transcription. Egr-1 interacts directly with Ptx1 and with SF-1, leading to an enhancement of Ptx1- and SF-1-induced LHβ transcription. Thus, Egr-1 is a likely transcriptional mediator of GnRH-induced signals for activation of the LHβ gene.
The gene for Egr-1 (also designated as NGFI-A, Krox-24, and Zif268) belongs to a group of transcriptional regulators that behave as immediate-early response genes. These genes are transiently activated at the cellular level by a variety of external stimuli, such as serum or growth factors, and they are thought to be important regulators of cellular proliferation and differentiation. The Egr-1 gene was first identified about 10 years ago as a gene rapidly induced during nerve growth factor-induced differentiation (9, 37). Egr-1 has since been cloned by several groups and shown to be rapidly and transiently induced, both at the transcriptional and protein levels, by a variety of mitogens, developmental or differentiation cues, tissue damage, and signals that induce neuronal excitation or apoptosis in numerous cell types (7, 31, 50; for a review, see reference 9). The Egr-1 protein is a zinc-finger-containing transcription factor of the C2H2 class that specifically binds the DNA sequence GCG(G/T)GGGCG to activate transcription of target genes (5, 7, 32, 37, 40, 52). The DNA-binding activity of Egr-1 is apparently controlled by the phosphorylation state of the protein. Indeed, Huang et al. have shown that phosphorylated Egr-1 binds DNA more efficiently than the nonphosphorylated form (16). Moreover, Egr-1 DNA binding is significantly increased by inhibitors of protein serine/threonine phosphatase 1 and 2A, suggesting that its DNA-binding activity is under the control of protein kinase(s) and/or phosphatase(s) (4). Taken together, these data suggest that Egr-1 serves as an intermediary regulatory factor in many cellular response pathways.
Egr-1 is broadly expressed in tissues throughout development and in the adult of many species. It can be found in the endothelial system, thymus, muscle, cartilage, bone, and parts of the central and peripheral nervous systems (36, 61). At a functional level, several in vitro studies initially characterized Egr-1 as having a role in the control of macrophage differentiation and T-lymphocyte proliferation (38, 41), as well as in platelet-derived growth factor-B gene expression in endothelial cells (23). However, in two independent Egr-1 targeted gene deletion experiments, these observations were not corroborated (30, 55). Rather, Egr-1−/− mice of both sexes have reduced body sizes and fertility problems due to a pituitary defect in the male and sterility due to combined pituitary and ovarian dysfunction in the female (29, 55). In the pituitary of knockout mice, the absence of Egr-1 results in a lack of the gonadotropin luteinizing hormone β (LHβ) gene expression in gonadotrope cells despite the presence of other gonadotrope markers (29, 55). These observations have suggested that Egr-1 is not involved in the differentiation of gonadotrope cells but rather in the expression of the gonadotrope-specific LHβ gene. Indeed, Lee et al. have shown that Egr-1 can bind to a conserved consensus GC-rich motif and directly activate LHβ transcription (29). Moreover, Egr-1 can act in synergy with the orphan nuclear receptor SF-1 to further enhance LHβ transcription (28, 29).
Pituitary gonadotropes synthesize and secrete two gonadotropin hormones: LH and follicle-stimulating hormone (FSH). Both hormones are heterodimeric glycoproteins composed of a common peptide, the glycoprotein hormone subunit α (αGSU), and either a specific FSHβ or LHβ polypeptide (43). Expression of the genes encoding the α and β subunits, as well as secretion of the mature hormones, is regulated by gonadotropin-releasing hormone (GnRH), which is itself secreted from the hypothalamus (10). Naturally occurring mutations in the GnRH (hpg mice) or GnRH receptor (GnRH-R) genes both lead to hypogonadism due to a lack of gonadotropin production (27, 35). GnRH binds the GnRH-R, a seven-transmembrane G-protein-coupled receptor, present at the membrane of pituitary gonadotropes: this triggers the activation of phospholipase C (PLC), which cleaves phosphatidylinositol-4,5-biphosphate (PIP2) to generate triphosphate inositol (IP3) and diacylglycerol (DAG). IP3 increases intracellular calcium levels, whereas DAG activates protein kinase C (PKC) (1, 2, 20). Activation of PKC leads to increased mitogen-activated protein kinase kinase (MAPKK or MEK) and mitogen-activated protein kinase (MAPK or ERK) activity and to increased gonadotropin mRNA levels (15, 20, 51). Direct activation of PKC by phorbol 12-myristate 13-acetate (PMA), as well as calcium mobilization by ionomycin, reproduce the profile of GnRH-induced LHβ mRNA (1, 2). Conversely, depletion of PKC activity significantly reduces the ability of GnRH to stimulate LHβ mRNA (1). Thus, GnRH-dependent activation of the PKC pathway appears to be a major step for stimulation of LHβ mRNA. However, the transcriptional mediator(s) of PKC action on the LHβ promoter is presently unknown.
Recent studies have identified three factors involved in LHβ gene transcription: the nuclear receptor SF-1, the bicoid-related homeoprotein Ptx1 (Pitx1), and Egr-1 (14, 22, 29, 56, 57). Numerous studies have demonstrated the importance of SF-1 at multiple levels of the reproductive axis, including gonadotrope function in the pituitary (39). Ptx1 is a homeobox transcription factor first isolated as a regulator of pro-opiomelanocortin (POMC) gene expression in pituitary corticotrope cells (25). Ptx1 was later shown to be present in all pituitary cells and to activate most pituitary-hormone-coding gene promoters, including LHβ (26, 57). In addition, Ptx1 contributes to lineage-restricted gene expression by synergism with cell-restricted transcription factors such as SF-1, Pit1, and NeuroD1/Pan1 (44, 53, 56, 57).
In the present study, we report the identification of Egr-1 as a downstream effector of the GnRH-induced PKC signal transduction pathway in pituitary gonadotropes. Indeed, GnRH markedly induces Egr-1 gene expression in model gonadotrope cells, αT3-1 and LβT2, and the Egr-1-dependent activation of the LHβ promoter is specifically enhanced by PKC. Also, we show that Egr-1 exerts its transcriptional effects on the LHβ promoter by physical interactions with Ptx1 and SF-1.
MATERIALS AND METHODS
Cell culture and transfection assays.
Murine αT3-1, LβT2, and African green monkey kidney fibroblast-like CV-1 cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). CV-1 cells were transfected by the calcium phosphate method as previously described (56). LβT2 cells were plated at a density of 750,000 cells per well in a 12-well plate the evening prior to transfection and then transfected the next day by the calcium phosphate method. The following day, cells were rinsed and DMEM-FCS containing either the vehicle phosphate-buffered saline (PBS) or 100 nM GnRH was added for 15 min, followed by a 75-min incubation in regular DMEM-FCS. This pulsatile treatment (58) was repeated three more times and another four times the next day before the cells were harvested. Data are presented as the means of 3 to 10 experiments, each performed in duplicate, ± the standard error of the mean (SEM). The weak (about twofold) background activation observed with Ptx1 (and with combination of factors containing Ptx1) of mutant promoters (see, for example, Fig. 6F and G) has been observed with many negative control promoters, such as herpes simplex virus thymidine kinase promoter, minimal pituitary promoters, and Mullerian inhibiting substance promoter, that do not contain Ptx1 binding sites (56, 57); hence, we consider this to be a general nonspecific effect on reporter activity.
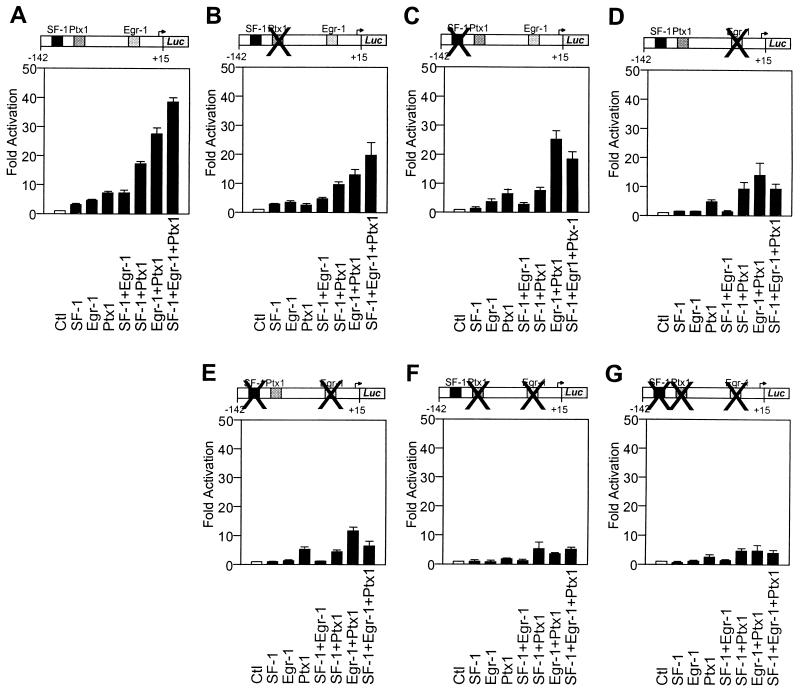
FIG. 6.
Site requirements for Egr-1–Ptx1–SF-1 cooperation. Transactivation by Egr-1, Ptx1, and SF-1 alone or in combination was tested on bp −142 bovine LHβ reporters as follows: wild-type promoter with intact binding sites for Egr-1, SF-1, and Ptx1 (A); mutated Ptx1 site (25); mutated SF-1 site (14, 22) (C); mutated Egr-1 site (29) (D); double mutation of the SF-1 and Egr-1 sites (E); double mutation of the Ptx1 and Egr-1 sites (F); and mutation of all three sites (G). Promoter constructs were cotransfected in CV-1 cells with the indicated expression plasmids. The results are shown as fold activation (± SEM).
Hormone treatment, RNA extraction, and analysis.
GnRH, forskolin, PMA, and cyclic ADP-ribose were obtained from Sigma. αT3-1 or LβT2 cells were treated with 10−5 M forskolin, 10−7 M GnRH, 5 × 10−7 M PMA, and 5 μM cyclic ADP-ribose for the times indicated. Total cellular RNA was extracted by the guanidium thiocyanate-phenol-chloroform method (6) and analyzed by Northern blot analysis as described previously (57). DNA probes used for hybridization were cDNA fragments specific for Ptx1 (25), SF-1 (34), GnRH-R (45), αGSU (11), and Egr-1 (50). As a loading control, the blots were stripped and rehybridized with a 32P-labeled oligonucleotide specific for 18S ribosomal RNA as described earlier (57).
Plasmids and oligonucleotides.
The bp −142 LHβ promoter reporter, the mutations of the SF-1 and/or Ptx1 binding sites, and the generation of Ptx1 mutants were as described elsewhere (56). The Ptx1 fragments used in the Gal4 DNA-binding domain (Gal4DBD)–Ptx1 fusions were generated by PCR with primers containing restriction sites and were subsequently subcloned in frame in the corresponding sites of a Gal4DBD vector. Mutations of the Egr-1 site and the double or triple mutants were obtained by PCR with the bp −142 bovine LHβ promoter as a template. A common reverse primer that incorporates the Egr-1 site mutation (shown in boldface) and a natural PstI site 5′-−31ACCTGCAGGCTCTAAGAACAGCAAGGCCGGGGGTGGCAGC−70-3′ was used with various forward primers that were described previously (56) to generate the mutants. The amplified fragments were subcloned back into the bp −142 LHβ reporter. All mutations and deletions were confirmed by DNA sequencing.
Recombinant protein production and pull-down assays.
Maltose-binding protein (MBP) fusion proteins (MBP–SF-1, MBP–Egr-1, MBP-Ptx1, and MBP-LacZα) were produced as described earlier (56). 35S-labeled in vitro-translated Ptx1 (wild type and mutants), SF-1, and luciferase were obtained by using the TNT-coupled transcription-translation rabbit reticulocyte lysate system (Promega). Protein-protein interaction assays were performed according to the method of Tremblay et al. (56).
RESULTS
GnRH rapidly and transiently induces Egr-1.
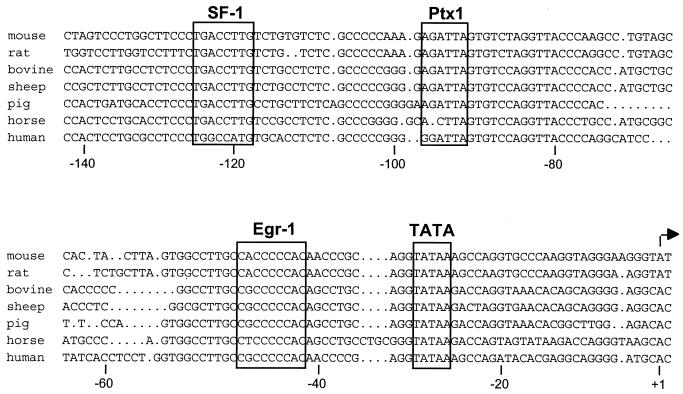
LHβ gene expression requires the concerted action of several transcription factors, some of which are involved in basal expression, whereas other inducible factors are needed to elicit a rapid response to external stimuli such as the hypothalamic hormone GnRH. As shown in Fig. 1, alignment of the LHβ promoter from several species reveals three highly conserved regions: one that binds the orphan nuclear receptor SF-1, a consensus site for binding of the homeobox transcription factor Ptx1, and a GC-rich region previously shown to bind Egr-1. All of these factors were previously shown to activate the LHβ promoter (14, 22, 29, 57).
FIG. 1.
Egr-1, Ptx1, and SF-1 binding sites are conserved across species in the LHβ promoter. Alignment of the mouse (24), rat (19), bovine (60), sheep (3), pig (8), horse (48), and human (54) LHβ promoter sequences reveals consensus SF-1, Ptx1, and Egr-1 elements (boxed) that are conserved across species.
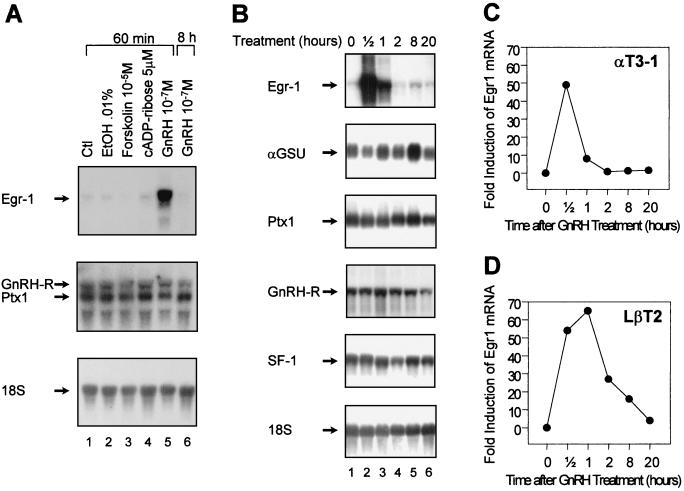
In order to identify the factor(s) responsible for GnRH-dependent activation of the LHβ promoter, we tested whether GnRH and other second messenger inducers could stimulate Ptx1, SF-1, and/or Egr-1 gene expression in the αT3-1 gonadotrope cell line. As shown in Fig. 2A, treatment of αT3-1 cells with forskolin (an inducer of protein kinase A [PKA]) or cyclic ADP-ribose (a calcium ionophore) had no significant effect on Ptx1 and Egr-1 mRNA levels. Interestingly, 1 h after treatment with 10−7 M GnRH, Egr-1, but not Ptx1, mRNA levels were dramatically increased. Egr-1 mRNA returned to normal levels by 8 h after initiation of GnRH treatment. Egr-1 induction was transient, since mRNA levels were back to basal levels by 2 h of GnRH treatment (Fig. 2B). Densitometric analysis revealed that Egr-1 mRNA levels were 50 times higher in GnRH-treated cells compared to vehicle-treated cells after only 30 min (Fig. 2B and C). Consistent with a previous report by Windle et al. (62), αGSU mRNA levels were slightly increased after 8 h of GnRH treatment (Fig. 2B). GnRH treatment did not significantly affect GnRH-R mRNA levels nor those of the two other transcription factors known to be involved in LHβ gene expression, Ptx1 and SF-1 (Fig. 2B). The effect of GnRH on Egr-1 mRNA levels was also ascertained in another model gonadotrope cell line, the LβT2 cells, and a similar transient increase was observed on Egr-1 mRNA (Fig. 2D), but not on αGSU, Ptx1, SF-1, or GnRH-R mRNAs (data not shown). Taken together, these results suggest that Egr-1 may be a downstream mediator of the GnRH-induced signal transduction pathway in pituitary gonadotropes.
FIG. 2.
GnRH rapidly induces Egr-1 gene expression. (A) αT3-1 cells were treated as indicated for 60 min or 8 h, and total RNA was isolated for use in Northern blot analysis of Egr-1, GnRH-R, and Ptx1 mRNA. The blot was subsequently probed with an 18S rRNA probe to ensure integrity and loading of the RNA. (B) Time course analysis of GnRH effect on αT3-1 cells. Total RNA was isolated at the indicated time after GnRH treatment (10−7 M) and analyzed by Northern blot as in panel A. (C) The Egr-1 mRNA levels from B were quantified by densitometry. (D) Time course analysis of GnRH effect on Egr-1 mRNA levels in LβT2 cells.
PKC enhances Egr-1-dependent LHβ promoter activation.
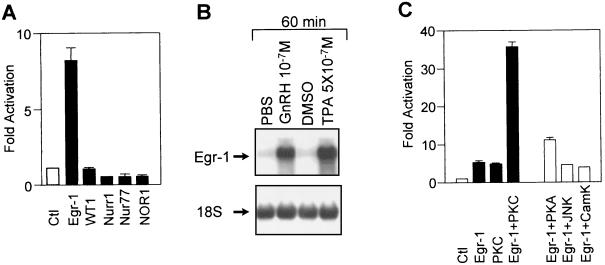
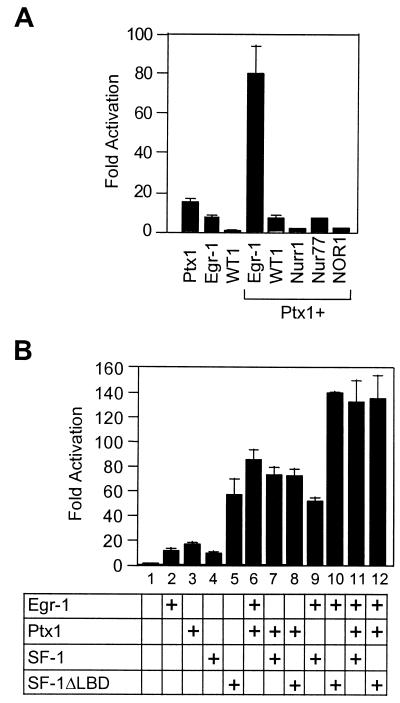
Egr-1 has been shown recently to be involved in LHβ gene expression (28, 29, 55). As shown in Fig. 3A, LHβ promoter activation by Egr-1 (NGFI-A) was specific, since the closely related factor WT-1 or the products of three other immediate-early genes, Nur77 (NGFI-B), Nurr1, and NOR-1, failed to activate the LHβ promoter. GnRH stimulation of gonadotropin expression and secretion occurs through activation of PKC and increased intracellular calcium levels (1, 2, 20); in turn, PKC is thought to activate a downstream transcription factor(s) that controls LHβ gene expression. Consistent with this model, we showed that the PKC activator PMA is as efficient as GnRH in inducing Egr-1 mRNA levels in αT3-1 cells (Fig. 3B). The activity of Egr-1 may also be enhanced by phosphorylation (4, 16) and, consequently, we tested various protein kinase catalytic subunits for the enhancement of Egr-1-dependent LHβ promoter activation. As shown in Fig. 3C, PKC potentiated the ability of Egr-1 to activate the LHβ promoter. This potentiation was specific for PKC since none of the other kinases tested, including PKA, Jun kinase (JNK), and calmodulin kinase (CamK) markedly enhanced basal (not shown) or Egr-1-induced LHβ promoter activation (Fig. 3C). These results suggest that induction of Egr-1 gene expression and phosphorylation of Egr-1, both events specifically mediated through the PKC pathway, constitute part of the intracellular signaling cascade induced by GnRH in gonadotropes.
FIG. 3.
Involvement of PKC pathway in Egr-1-dependent activation of LHβ promoter. (A) The effect of Egr-1 or the related factor WT-1 and of the products of other immediate-early genes (Nurr1, Nur77, and NOR-1) was assessed on the bp −776 bovine LHβ promoter. The LHβ-luciferase reporter was cotransfected in CV-1 cells together with a control plasmid (empty expression vector, open bar) or with expression vectors for Egr-1, WT-1, Nurr1, Nur77, or NOR-1 as indicated (solid bars). (B) Effect of PMA treatment on Egr-1 mRNA levels. αT3-1 cells were treated with 5 × 10−7 M PMA for 60 min before harvest and RNA isolation. Egr-1 mRNA was revealed by Northern blot. (C) Enhancement of Egr-1-dependent stimulation of LHβ promoter activity by PKC, but not other kinases. CV-1 cells were cotransfected with the bp −776 LHβ reporter and either a control empty expression vector (Ctl) or with expression vectors for Egr-1 with or without PKC, PKA, JNK, or CamK. Results are shown as fold activation (± SEM).
Mutagenesis of Egr-1 site affects GnRH activation of LHβ promoter.
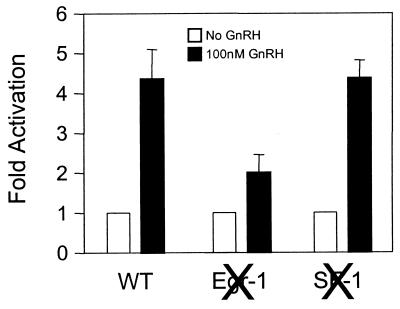
The Egr-1 and SF-1 sites of the LHβ promoter were mutated separately in order to test their involvement in responsiveness to GnRH. Pulsatile treatment of LβT2 cells with GnRH was previously shown to increase LHβ mRNA (58), and a similar approach was used to test for GnRH responsiveness of LHβ promoter constructs. This treatment increased LHβ promoter activity, and mutagenesis of the SF-1 binding site did not affect this response (Fig. 4). However, mutagenesis of the Egr-1 site significantly reduced GnRH responsiveness of the LHβ promoter. Thus, at least part of the GnRH-induced signals exert their effect on LHβ transcription through this Egr-1 site.
FIG. 4.
GnRH stimulation of LHβ promoter activity is reduced by mutation of Egr-1, but not SF-1, binding site. LβT2 cells were transfected with three different bp −142 LHβ reporters (wild type, mutated Egr-1 binding site, and mutated SF-1 binding site). Cells were subjected to four daily pulses of either vehicle (open bars) or 100 nM GnRH (solid bars) over a 2-day period as described in Materials and Methods. Results are shown as fold activation (± SEM).
Egr-1, Ptx1, and SF-1 cooperatively activate the LHβ promoter.
Ptx1 and SF-1 are both present at high levels in unstimulated pituitary gonadotropes (26, 39, 57) and in αT3-1 cells, whereas Egr-1 is not (Fig. 2). Binding sites for these three factors are conserved across species within the LHβ promoter (Fig. 1). It has already been shown that, individually, Ptx1 and SF-1 activate the LHβ promoter by binding to their cognate sites (14, 22, 57), while, together, they act synergistically (56, 57). Since Egr-1 and SF-1 have also been documented to synergize with each other (28, 29), we tested whether Ptx1 could synergistically enhance transcription with Egr-1. Both Ptx1 and Egr-1 activated the bp −776 LHβ reporter, and they also acted synergistically to enhance promoter activity (Fig. 5A). The product of another Egr-1-related gene, WT-1, or other immediate-early genes, such as Nurr1, Nur77, and NOR-1, did not synergize with Ptx1 (Fig. 5A).
FIG. 5.
Egr-1, Ptx1, and SF-1 synergize for activation of the LHβ promoter. (A) Transcriptional cooperation between Egr-1 and Ptx1. Ptx1 was tested for synergism on the bp −776 LHβ reporter with either Egr-1, WT-1, Nurr1, Nur77, or NOR-1. (B) Egr-1 has a cumulative effect on Ptx1–SF-1 synergism. CV-1 cells were cotransfected with the bp −776 LHβ reporter and the indicated expression plasmids. The SF-1ΔLBD mutant is deleted of its LBD, and it was previously shown to have constitutive transcriptional activity (47, 56). The results are shown as fold activation (± SEM).
We have recently demonstrated that Ptx1 can modulate the activity of SF-1 by bypassing the requirement for its ligand (56). Indeed, as shown in Fig. 5B, a constitutively active SF-1 mutant devoid of its ligand binding domain (ΔLBD) was as active as the synergistic activity of Ptx1–SF-1 (compare lanes 5 and 7), suggesting that Ptx1 serves to unmask SF-1 activity (56). Since Egr-1 also synergizes with SF-1 (references 28 and 33 and Fig. 5B, column 9), we tested whether Egr-1 has a similar unmasking effect on SF-1 as does Ptx1. Although Egr-1 and Ptx1 each markedly enhanced the activity of wild-type SF-1 (columns 9 and 7, respectively), only Egr-1 synergized with SF-1ΔLBD (compare lanes 10 and 8), suggesting that Egr-1 and Ptx1 have different mechanisms for synergizing with SF-1. As expected, when the three factors were combined, a cumulative effect was observed (Fig. 5B, column 11). Moreover, the cumulative activity of the three factors (column 11) was the same as that of Egr-1 with SF-1ΔLBD (column 10) or of Ptx1, Egr-1, and SF-1ΔLBD (column 12), a finding consistent with the putative role of Ptx1 in unmasking the activation domain of SF-1.
Binding site requirements for Egr-1–Ptx1–SF-1 synergism.
Previous studies have revealed that a bp −142 LHβ promoter fragment that retains the SF-1 element at bp −120, the Ptx1 binding site at bp −95, and the Egr-1 binding motif at bp −45 is sufficient for Ptx1 and SF-1 transactivation and synergy (56). This construct also allowed us to determine the binding sites required for the synergistic cooperativity between SF-1, Ptx1, and Egr-1. Like the bp −776 LHβ promoter (Fig. 5B), the three factors exhibited cumulative effects on the shorter bp −142 LHβ promoter fragment (Fig. 6A). The requirement for each promoter binding site was tested by creating mutations of each site, either individually (Fig. 6B, C, and D), in two-by-two combinations (Fig. 6E and F), or all three together (Fig. 6G).
Consistent with our previous study, mutation of the Ptx1 binding site did not affect Ptx1–SF-1 synergism (Fig. 6B and reference 56). Similarly, this same mutation did not prevent synergy between Ptx1 and Egr-1 (Fig. 6B). Thus, the cooperativity between Ptx1, SF-1, and Egr-1 appears to be independent of Ptx1 binding to DNA. In contrast, mutation of the SF-1 element abolished both Ptx1–SF-1 and SF-1–Egr-1 synergism (Fig. 6C). Finally, mutation of the Egr-1 element prevented synergy with SF-1 but did not completely abolish Egr-1 interaction with Ptx1 (Fig. 6D). Taken together, these results suggest that Egr-1–SF-1 synergism requires the binding of both factors to their cognate elements, since mutation of each site, either individually (Fig. 6C and D) or in combination (Fig. 6E) abolished synergy. Conversely, synergism between Egr-1 and Ptx1 apparently requires only one of the two elements, since promoters with single mutations still exhibited some synergism (Fig. 6B and D), although the cumulative activity was not as great in these cases as with the intact promoter (Fig. 6A). In contrast, the double Egr-1 and Ptx1 site mutation completely abrogated Egr-1–Ptx1 synergism (Fig. 6F). As expected, mutation of all three elements, blocked LHβ promoter activation by any combination of Egr-1, Ptx1, or SF-1 (Fig. 6G). The results of this mutagenesis analysis (summarized in Table 1) revealed that the synergies observed between Egr-1, SF-1, and Ptx1 on the LHβ promoter have different site requirements and, thus, are likely mediated via different molecular mechanisms.
TABLE 1.
Site requirements for synergy on the LHβ promoter
| Synergy | Site requirement(s) | Reference(s) |
|---|---|---|
| Egr-1–Ptx1 | Egr-1 or Ptx1 | This study |
| SF-1–Egr-1 | SF-1 and Egr-1 | This study |
| SF-1–Ptx1 | SF-1 only | This study and reference 56 |
Egr-1 and Ptx1 interact physically.
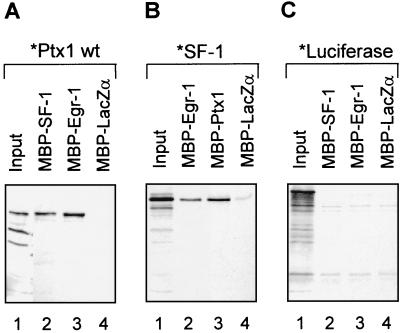
Cooperativity between Ptx1, SF-1, and Egr-1 for activation of LHβ promoter suggests that the proteins may interact directly. We have, in fact, recently demonstrated that Ptx1 and SF-1 interact in vitro and in vivo (56). As shown in Fig. 7A, both SF-1 and Egr-1 immobilized on beads also interacted with in vitro-synthesized Ptx1. These interactions were specific since no binding was observed with immobilized MBP-LacZα (Fig. 7A, lane 4) and labeled luciferase did not bind any immobilized protein (Fig. 7C). Similarly, both Egr-1 and Ptx1 interacted with labeled SF-1 protein (Fig. 7B). Thus, the Ptx1, SF-1, and Egr-1 cooperative effects are likely to occur through direct protein-protein interactions.
FIG. 7.
Egr-1 directly interacts with Ptx1 and SF-1. Pull-down assays were performed with immobilized, bacterially produced MBP fusion proteins (MBP–SF-1, MBP–Egr-1, MBP-Ptx1, and MBP-LacZα as a control) with 35S-labeled Ptx1 (A), SF-1 (B), or luciferase (C). Complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and visualized by autoradiography. The input protein (lanes 1) corresponds to 20% of the labeled protein used in the assay.
Egr-1–Ptx1 synergism maps to a C-terminal domain of Ptx1.
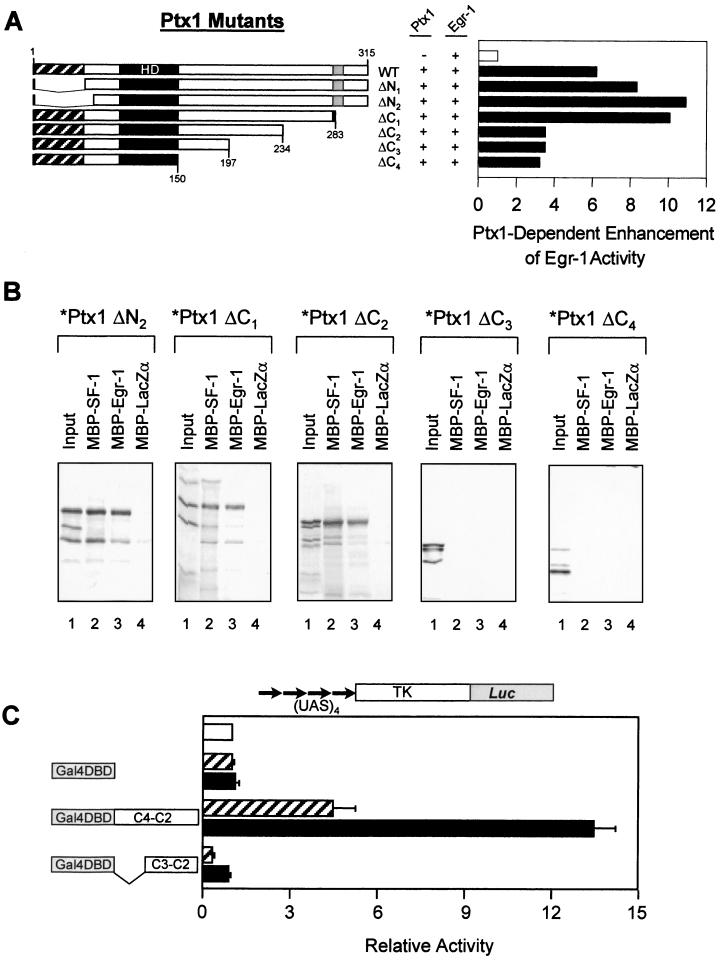
In order to identify the domain of Ptx1 involved in the synergistic and physical interactions with Egr-1, a series of Ptx1 mutants was tested in transfection and pull-down assays. The expression level, nuclear localization, and transcriptional properties of the Ptx1 mutant proteins have been assessed previously (56). Deletion of the N-terminal domain of Ptx1 (mutants ΔN1 and ΔN2) did not affect its ability to synergize with Egr-1 (Fig. 8A). However, deletion of a 49-amino-acid region in the C-terminal domain of Ptx1 (amino acids 234 to 283), which deletes an activation domain (56), led to a significant decrease in synergy with Egr-1 (Fig. 8A, compare mutant ΔC2 with mutant ΔC1). Pull-down assays were then used to identify the region involved in the physical interaction with Egr-1 (Fig. 8B). Consistent with the transactivation data, the N-terminal region of Ptx1 was not required for interaction with Egr-1 (Fig. 8B, mutant ΔN2). The Egr-1 interacting domain mapped to a 37-amino-acid region located between residues 197 (mutant ΔC3) and 234 (mutant ΔC2) of Ptx1. Interestingly, this same Ptx1 region was recently shown to be the domain involved in the physical interaction with SF-1 (Fig. 8B and reference 56). We also used an in vivo system to ascertain the direct interaction between Ptx1 and Egr-1 documented in vitro by using the pull-down assay. For this purpose, C-terminal fragments of Ptx1 that have little (between endpoints C4 and C2 defined in Fig. 8A) or no (endpoints C3 to C2) transcriptional activity were fused to the Gal4DBD. These fusion proteins did not show marked transcriptional activity when expressed with an upstream activating sequence (UAS)-containing reporter; however, coexpression of Egr-1 significantly enhanced the activity of the chimeras but not of Gal4DBD (Fig. 8C). Egr-1 alone had no effect on this reporter, and Gal4DBD fusions containing other Ptx1 fragments were not affected by Egr-1 (data not shown). Thus, it appears that an activation domain of Ptx1 (localized between residues 234 and 283) is required for its transcriptional synergism with both Egr-1 and SF-1, while physical interactions involve a contiguous region of Ptx1 (between residues 197 and 234).
FIG. 8.
The C-terminal domain of Ptx1 is required for physical interaction and transcriptional cooperation with Egr-1. (A) CV-1 cells were cotransfected with the bovine bp −142 LHβ reporter, along with expression vectors for Egr-1 (open bar) or for Egr-1 together with Ptx1 mutants (solid bars). The Ptx1 mutants were as described previously (56). (B) The indicated Ptx1 mutant proteins were labeled by in vitro translation and tested for binding to MBP–SF-1 (lanes 2), MBP–Egr-1 (lanes 3), or MBP-LacZα (lanes 4) as a control. Bound complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and visualized by autoradiography. The input proteins (lanes 1) correspond to 20% of the labeled protein used in the assay. (C) In vivo hybrid assay for interaction between Egr-1 and fragments of Ptx1 fused to Gal4DBD. A UAS-containing thymidine kinase promoter-luciferase reporter plasmid was cotransfected in CV-1 cells either alone (open bar) or with a Gal4DBD fused (or not) to Ptx1 C-terminal fragments C4-C2 (amino acids 150 to 234) or C3-C2 (amino acids 197 to 234) in the absence (hatched bars) or the presence (solid bars) of Egr-1.
DISCUSSION
The regulation of gonadotropin synthesis and secretion by GnRH is well established. The lack of LHβ expression in the hpg mouse, which harbors mutations in the GnRH gene (35), and naturally occurring mutations in the GnRH receptor in humans (27), have corroborated the importance of this hypothalamic hormone for control of gonadotropin function. Although it is clear that GnRH is essential for gonadotropin gene expression, the transcription factors that, ultimately, are targets of GnRH action remain unknown. In the present study, we have identified the immediate-early response Egr-1 gene as a potential effector of GnRH-elicited responses in pituitary gonadotropes. Moreover, we propose a model in which Egr-1 physically and functionally cooperates with two other transcription factors, Ptx1 and SF-1, to elicit a rapid increase in LHβ gene expression in response to GnRH.
Regulation of Egr-1 activity.
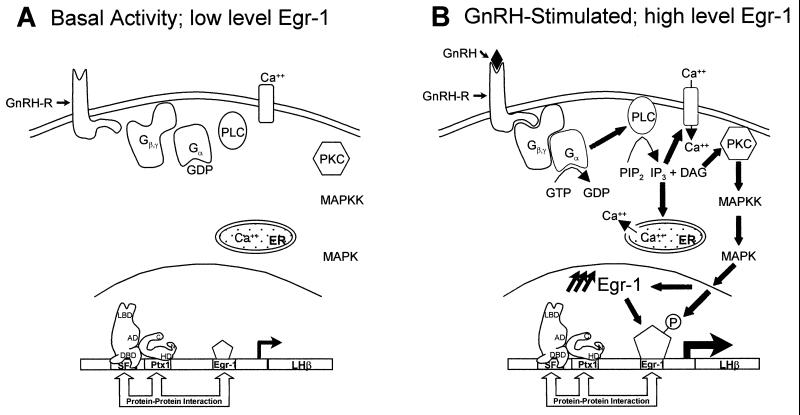
The rapid and strong induction of Egr-1 mRNA in response to GnRH (Fig. 2) and to PMA (Fig. 3B) suggested that this early response transcription factor may mediate some of the effects of the hypothalamic hormone (Fig. 9). It was previously shown that GnRH binding to its receptor elevates intracellular Ca2+ and activates the PKC cascade (1, 2, 20). We now show that PKC activation by PMA elevates Egr-1 mRNA levels (Fig. 3B) and that PKC enhances Egr-1-dependent transcription (Fig. 3C). Although these data do not exclude the putative involvement of other signaling events, they suggest that Egr-1 may be a transcriptional effector of GnRH action. This would be achieved by two complementary mechanisms: (i) stimulation of Egr-1 expression and (ii) direct enhancement by PKC-elicited modification (phosphorylation?) of Egr-1 transcriptional potency. This model (Fig. 9) is entirely consistent with the presence of a conserved Egr-1 target site in the LHβ promoter of many species (Fig. 1) and with its conserved position in relation to binding sites for Ptx1 and SF-1 which synergistically (Fig. 5, 6, and 8A) and physically (Fig. 7 and 8B and C) interact with Egr-1.
FIG. 9.
Model for control of LHβ gene expression by Egr-1, Ptx1, and SF-1. (A) In the absence of GnRH, Egr-1 is expressed at a low level. The LHβ gene is activated by Ptx1, SF-1, and low levels of Egr-1. (B) When GnRH is secreted from the hypothalamus, it binds to its receptor (GnRH-R), leading to activation of G-protein-linked PLC and IP3 intracellular signaling pathways (20). PLC cleaves PIP2 to generate IP3 and DAG. IP3 increases intracellular calcium levels (by L-type voltage-sensitive channel and release by the endoplasmic reticulum [ER]), whereas DAG activates PKC. Activation of PKC leads to increased mitogen-activated protein kinase kinase (MAPKK) and mitogen-activated protein kinase (MAPK) activity, leading to phosphorylation of Egr-1 (4, 16). Egr-1 mRNA levels are rapidly increased by GnRH via the PKC pathway, rather than by PKA or calcium (Ca2+). Egr-1 synergizes with Ptx1 and SF-1 to rapidly increase LHβ gene expression.
Mechanism of Egr-1, Ptx1, and SF-1 cooperation.
We have recently shown that synergistic cooperation between two transcription factors, Ptx1 and SF-1, contributes to LHβ gene expression. This synergism is achieved through a Ptx1–SF-1 physical interaction that mimics the activity of a constitutively active form of SF-1 (LBD deletion) and thus appears to bypass the need for an SF-1 ligand (56). We now show that Egr-1 also cooperates with these two factors, Ptx1 and SF-1, to activate LHβ promoter activity. However, the molecular mechanism of the Egr-1 synergism appears to be different (Table 1). Interestingly, the cumulative effects of these factors may serve to confer hormone responsiveness, since Egr-1 activity is greatly stimulated by GnRH, whereas Ptx1 and SF-1 mRNA levels are not hormone regulated. In resting cells, low-level expression of Egr-1 may contribute only slightly to LHβ expression (Fig. 9A); after GnRH stimulation, increased levels of Egr-1, as well as enhancement of Egr-1 transcriptional potency by PKC (for example, by phosphorylation), is likely to contribute to stimulation of LHβ gene transcription (Fig. 9B).
Egr-1 as a downstream effector of GnRH.
None of the transcription factors so far implicated in regulation of LHβ gene expression act as a downstream effector of GnRH action. The orphan nuclear receptor SF-1 was initially thought to play such a role since gonadectomy, which is known to increase hypothalamic GnRH secretion, led to a threefold increase in SF-1 mRNA levels in the pituitary (12, 59) and because SF-1 directly regulates LHβ promoter activity (14, 22, 56, 57). Moreover, targeted ablation of the SF-1 gene resulted in a severe decrease in LHβ mRNA levels (18, 49). However, the recovery of normal LHβ mRNA levels by GnRH injection in SF-1 knockout mice has unambiguously eliminated SF-1 as a mediator of GnRH action (17). The absence of LHβ mRNA in the SF-1−/− animals was later proven to be the result of a blockade of GnRH secretion (17, 49). Our results are also consistent with this interpretation since GnRH did not affect SF-1 mRNA levels in αT3-1 cells (Fig. 2B).
Ptx1 is unlikely to be a mediator of GnRH action since Ptx1 gene expression was unaffected by GnRH treatment (Fig. 2). In contrast, the dramatic upregulation of the Egr-1 mRNA by GnRH (nearly 50-fold [Fig. 2C and D]), taken together with the dependence on an intact Egr-1 binding site for GnRH responsiveness of the LHβ promoter (Fig. 4), strongly suggest that Egr-1 is an effector of GnRH action in pituitary gonadotropes. It was recently shown (13) that the proximal LHβ promoter contains a second, weaker Egr-1 binding site (ca. bp −105); this site may account for the weak GnRH responsiveness of the LHβ promoter mutated at the major (bp −45) Egr-1 site (Fig. 4). This weak GnRH responsiveness could also be the result of a direct protein-protein interaction between Egr-1 and DNA-bound Ptx1, since the Ptx1 binding site can be sufficient for some Egr-1–Ptx1 synergy (Fig. 6D and E and Table 1). Binding of GnRH to its cognate receptor activates the G-protein-linked PLC-IP3 intracellular signaling pathway, leading to PKC activation and increased intracellular calcium levels (1, 2, 20). Since Egr-1 mRNA levels are similarly induced by PMA, a PKC activator, and GnRH (Fig. 3B) but not by cyclic ADP-ribose (Fig. 2A), a calcium ionophore, it appears that GnRH-induced stimulation of Egr-1 gene expression is primarily mediated by PKC. This observation is consistent with recent work showing preferential activation of LHβ, but not αGSU, promoter activity by the PKC pathway (46). The involvement of Egr-1, together with Ptx1 and SF-1, in mediating GnRH action is also compatible with recent LHβ promoter mapping data (21). Our working model (Fig. 9) is strongly supported by the recent characterization of Egr-1 knockout mice that have undetectable LHβ expression despite the presence of other gonadotrope markers (FSHβ and αGSU) and normal GnRH secretion; further, gonadectomy, which increases GnRH secretion, induced FSHβ in these mice as in wild-type animals, but not LHβ (29, 55).
The transcriptional signaling of GnRH action through activation of Egr-1 (NGFI-A) is reminiscent of our recent identification of Nur77 (NGFI-B) as a mediator of corticotropin-releasing hormone stimulation of POMC gene transcription (42). Thus, two different pituitary lineages utilize immediate-early response genes as transcriptional effectors to mediate the effects of its trophic hypothalamic hormone.
ACKNOWLEDGMENTS
The Egr-1 expression vector and the bovine bp −776 LHβ luciferase reporter were kindly provided by Vikas Sukhatme and John Nilson, respectively. We are grateful to Keith Parker for the SF-1 expression plasmid, to Jerry Pelletier for the WT-1 expression plasmid, to Pamela Mellon for the αT3-1 and LβT2 cell lines, to Thomas Perlmann for the Gal4 plasmids, to Robert Viger for providing some of the reagents, and to Cynthia Goodyer for critical reading of the manuscript. The efficient secretarial assistance of Lise Laroche was much appreciated.
This work was funded by the National Cancer Institute of Canada supported with funds provided by the Canadian Cancer Society.
REFERENCES
- 1.Andrews W V, Maurer R A, Conn P M. Stimulation of rat luteinizing hormone-beta messenger RNA levels by gonadotropin releasing hormone. Apparent role for protein kinase C. J Biol Chem. 1988;263:13755–13761. [PubMed] [Google Scholar]
- 2.Ben-Menahem D, Naor Z. Regulation of gonadotropin mRNA levels in cultured rat pituitary cells by gonadotropin-releasing hormone (GnRH): role for Ca2+ and protein kinase C. Biochemistry. 1994;33:3698–3704. doi: 10.1021/bi00178a029. [DOI] [PubMed] [Google Scholar]
- 3.Brown P, McNeilly J R, Wallace R M, McNeilly A S, Clark A J. Characterization of the ovine LH beta-subunit gene: the promoter directs gonadotrope-specific expression in transgenic mice. Mol Cell Endocrinol. 1993;93:157–165. doi: 10.1016/0303-7207(93)90119-5. [DOI] [PubMed] [Google Scholar]
- 4.Cao X, Mahendran R, Guy G R, Tan Y H. Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 5.Cao X M, Koski R A, Gashler A, McKiernan M, Morris C F, Gaffney R, Hay R V, Sukhatme V P. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol Cell Biol. 1990;10:1931–1939. doi: 10.1128/mcb.10.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thyocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Christy B A, Lau L F, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezashi T, Hirai T, Kato T, Wakabayashi K, Kato Y. The gene for the beta subunit of porcine LH: clusters of GC boxes and CACCC elements. J Mol Endocrinol. 1990;5:137–146. doi: 10.1677/jme.0.0050137. [DOI] [PubMed] [Google Scholar]
- 9.Gashler A, Sukhatme V P. Early growth response protein 1 (Egr-1): prototype of a zinc- finger family of transcription factors. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 10.Gharib S D, Wierman M E, Shupnik M A, Chin W W. Molecular biology of the pituitary gonadotropins. Endocr Rev. 1990;11:177–199. doi: 10.1210/edrv-11-1-177. [DOI] [PubMed] [Google Scholar]
- 11.Gordon D F, Wood W M, Ridgway E C. Organization and nucleotide sequence of the mouse alpha-subunit gene of the pituitary glycoprotein hormones. DNA. 1988;7:679–690. doi: 10.1089/dna.1988.7.679. [DOI] [PubMed] [Google Scholar]
- 12.Haisenleder D J, Yasin M, Dalkin A C, Gilrain J, Marshall J C. GnRH regulates steroidogenic factor-1 (SF-1) gene expression in the rat pituitary. Endocrinology. 1996;137:5719–5722. doi: 10.1210/endo.137.12.8940405. [DOI] [PubMed] [Google Scholar]
- 13.Halvorson L M, Ito M, Jameson J L, Chin W W. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone beta-subunit gene expression. J Biol Chem. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 14.Halvorson L M, Kaiser U B, Chin W W. Stimulation of luteinizing hormone beta gene promoter activity by the orphan nuclear receptor, steroidogenic factor-1. J Biol Chem. 1996;271:6645–6650. doi: 10.1074/jbc.271.12.6645. [DOI] [PubMed] [Google Scholar]
- 15.Horn F, Bilezikjian L M, Perrin M H, Bosma M M, Windle J J, Huber K S, Blount A L, Hille B, Vale W, Mellon P L. Intracellular responses to gonadotropin-releasing hormone in a clonal cell line of the gonadotrope lineage. Mol Endocrinol. 1991;5:347–355. doi: 10.1210/mend-5-3-347. [DOI] [PubMed] [Google Scholar]
- 16.Huang R P, Adamson E D. The phosphorylated forms of the transcription factor, Egr-1, bind to DNA more efficiently than nonphosphorylated. Biochem Biophys Res Commun. 1994;200:1271–1276. doi: 10.1006/bbrc.1994.1588. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda Y, Luo X, Abbud R, Nilson J H, Parker K L. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol Endocrinol. 1995;9:478–486. doi: 10.1210/mend.9.4.7659091. [DOI] [PubMed] [Google Scholar]
- 18.Ingraham H A, Lala D S, Ikeda Y, Luo X, Shen W H, Nachtigal M W, Abbud R, Nilson J H, Parker K L. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2302–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 19.Jameson L, Chin W W, Hollenberg A N, Chang A S, Habener J F. The gene encoding the beta-subunit of rat luteinizing hormone. Analysis of gene structure and evolution of nucleotide sequence. J Biol Chem. 1984;259:15474–15480. [PubMed] [Google Scholar]
- 20.Kaiser U B, Conn P M, Chin W W. Studies of gonadotropin-releasing hormone (GnRH) action using GnRH receptor-expressing pituitary cell lines. Endocr Rev. 1997;18:46–70. doi: 10.1210/edrv.18.1.0289. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser U B, Sabbagh E, Saunders B D, Chin W W. Identification of cis-acting deoxyribonucleic acid element that mediate gonadotropin-releasing hormone stimulation of the rat luteinizing hormone β-subunit gene. Endocrinology. 1998;139:2443–2451. doi: 10.1210/endo.139.5.6003. [DOI] [PubMed] [Google Scholar]
- 22.Keri R A, Nilson J H. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone beta subunit promoter in gonadotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785. doi: 10.1074/jbc.271.18.10782. [DOI] [PubMed] [Google Scholar]
- 23.Khachigian L M, Lindner V, Williams A J, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 24.Kumar T R, Matzuk M M. Cloning of the mouse gonadotropin beta-subunit-encoding genes, II. Structure of the luteinizing hormone beta-subunit-encoding genes. Gene. 1995;166:335–336. doi: 10.1016/0378-1119(96)81753-7. [DOI] [PubMed] [Google Scholar]
- 25.Lamonerie T, Tremblay J J, Lanctôt C, Therrien M, Gauthier Y, Drouin J. PTX1, a bicoid-related homeobox transcription factor involved in transcription of pro-opiomelanocortin (POMC) gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 26.Lanctôt, C., Y. Gauthier, and J. Drouin. Pituitary homeobox 1 (Ptx1) is differentially expressed during pituitary development. Endocrinology, in press. [DOI] [PubMed]
- 27.Layman L C, Cohen D P, Jin M, Xie J, Li Z, Reindollar R H, Bolbolan S, Bick D P, Sherins R R, Duck L W, Musgrove L C, Sellers J C, Neill J D. Mutations in gonadotropin-releasing hormone receptor gene cause hypogonadotropic hypogonadism. Nat Genet. 1998;18:14–15. doi: 10.1038/ng0198-14. [DOI] [PubMed] [Google Scholar]
- 28.Le Drean Y, Liu D, Xiong F, Hew C L. Presence of distinct cis-acting elements on gonadotropin gene promoters in diverse species dictates the selective recruitment of different transcription factors by steroidogenic factor-1. Mol Cell Endocrinol. 1997;135:31–40. doi: 10.1016/s0303-7207(97)00184-6. [DOI] [PubMed] [Google Scholar]
- 29.Lee S L, Sadovsky Y, Swirnoff A H, Polish J A, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 30.Lee S L, Wang Y, Milbrandt J. Unimpaired macrophage differentiation and activation in mice lacking the zinc finger transplantation factor NGFI-A (EGR1) Mol Cell Biol. 1996;16:4566–4572. doi: 10.1128/mcb.16.8.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire P, Revelant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci USA. 1988;85:4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemaire P, Vesque C, Schmitt J, Stunnenberg H, Frank R, Charnay P. The serum-inducible mouse gene Krox-24 encodes a sequence-specific transcriptional activator. Mol Cell Biol. 1990;10:3456–3467. doi: 10.1128/mcb.10.7.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Stark M R, Johnson A D, Wolberger C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 34.Luo X, Ikeda Y, Schlosser D A, Parker K L. Steroidogenic factor 1 is the essential transcript of the mouse ftz-f1 gene. Mol Endocrinol. 1995;9:1233–1239. doi: 10.1210/mend.9.9.7491115. [DOI] [PubMed] [Google Scholar]
- 35.Mason A J, Hayflick J S, Zoeller R T, Young W S, Phillips H S, Nikolics K, Seeburg P H. A deletion truncating the gonadotropin-releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science. 1986;234:1366–1371. doi: 10.1126/science.3024317. [DOI] [PubMed] [Google Scholar]
- 36.McMahon A P, Champion J E, McMahon J A, Sukhatme V P. Developmental expression of the putative transcription factor Egr-1 suggests that Egr-1 and c-fos are coregulated in some tissues. Development. 1990;108:281–287. doi: 10.1242/dev.108.2.281. [DOI] [PubMed] [Google Scholar]
- 37.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen H Q, Hoffman-Liebermann B, Liebermann D A. The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- 39.Parker K L, Schimmer B P. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 40.Pavletich N P, Pabo C O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Castillo A, Pipaon C, Garcia I, Alemany S. NGFI-A gene expression is necessary for T lymphocyte proliferation. J Biol Chem. 1993;268:19445–19450. [PubMed] [Google Scholar]
- 42.Philips A, Lesage S, Gingras R, Maira M H, Gauthier Y, Hugo P, Drouin J. Novel dimeric Nur77 signaling mechanisms in endocrine and lymphoid cells. Mol Cell Biol. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce J G, Parsons T F. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- 44.Poulin G, Turgeon B, Drouin J. NeuroD1/BETA2 contributes to cell-specific transcription of the POMC gene. Mol Cell Biol. 1997;17:6673–6682. doi: 10.1128/mcb.17.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinhart J, Mertz L M, Catt K J. Molecular cloning and expression of cdna encoding the murine gonadotropin-releasing hormone receptor. J Biol Chem. 1992;267:21281–21284. [PubMed] [Google Scholar]
- 46.Saunders B D, Sabbagh E, Chin W W, Kaiser U B. Differential use of signal transduction pathways in the gonadotropin-releasing hormone-mediated regulation of gonadotropin subunit gene expression. Endocrinology. 1998;139:1835–1843. doi: 10.1210/endo.139.4.5972. [DOI] [PubMed] [Google Scholar]
- 47.Shen W H, Moore C C, Ikeda Y, Parker K L, Ingraham H A. Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 48.Sherman G B, Wolfe M W, Farmerie T A, Clay C M, Threadgill D S, Sharp D C, Nilson J H. A single gene encodes the beta-subunits of equine luteinizing hormone and chorionic gonadotropin. Mol Endocrinol. 1992;6:951–959. doi: 10.1210/mend.6.6.1379674. [DOI] [PubMed] [Google Scholar]
- 49.Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M, Shiba H, Sasaki H, Osawa Y, Ninomiya Y, Niwa O, et al. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev Dynamics. 1995;204:22–29. doi: 10.1002/aja.1002040104. [DOI] [PubMed] [Google Scholar]
- 50.Sukhatme V P, Cao X, Chang L C, Tsai-Morris C H, Stamenkobich D, Ferreira P C P, Cohen D R, Edwards S A, Shows T B, Curran T, Le Beau M M, Adamson E D. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 51.Sundaresan S, Colin I M, Pestell R G, Jameson J L. Stimulation of mitogen-activated protein kinase by gonadotropin-releasing hormone: evidence for the involvement of protein kinase. Endocrinology. 1996;137:304–311. doi: 10.1210/endo.137.1.8536629. [DOI] [PubMed] [Google Scholar]
- 52.Swirnoff A H, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szeto D P, Ryan A K, O’Connell S M, Rosenfeld M G. P-OTX: a PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talmadge K, Vamvakopoulos N C, Fiddes J C. Evolution of the genes for the beta subunits of human chorionic gonadotropin and luteinizing hormone. Nature. 1984;307:37–40. doi: 10.1038/307037a0. [DOI] [PubMed] [Google Scholar]
- 55.Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt M A, Rao C V, Charnay P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- 56.Tremblay, J. J., Y. Gauthier, and J. Drouin. Ptx1 regulates SF-1 activity by an interaction that bypasses the need for ligand. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 57.Tremblay J J, Lanctôt C, Drouin J. The Pan-pituitary activator of transcription, Ptx-1 (pituitary homeobox1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 58.Turgeon J L, Kimura Y, Waring D W, Mellon P L. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol Endocrinol. 1996;10:439–450. doi: 10.1210/mend.10.4.8721988. [DOI] [PubMed] [Google Scholar]
- 59.Turzillo A M, Quirk C C, Juengel J L, Nett T M, Clay C M. Effects of ovariectomy and hypothalamic-pituitary disconnection on amounts of steroidogenic factor-1 mRNA in the ovine anterior pituitary gland. Endocrine. 1997;6:251–256. doi: 10.1007/BF02820500. [DOI] [PubMed] [Google Scholar]
- 60.Virgin J B, Silver B J, Thomason A R, Nilson J H. The gene for the beta subunit of bovine luteinizing hormone encodes a gonadotropin mRNA with an unusually short 5′-untranslated region. J Biol Chem. 1985;260:7072–7077. [PubMed] [Google Scholar]
- 61.Watson M A, Milbrandt J. Expression of the nerve growth factor-regulated NGFI-A and NGFI-B genes in the developing rat. Development. 1990;110:173–183. doi: 10.1242/dev.110.1.173. [DOI] [PubMed] [Google Scholar]
- 62.Windle J J, Weiner R I, Mellon P L. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]