Abstract
Activation of the human cardiac α-actin (HCA) promoter in skeletal muscle cells requires the integrity of DNA binding sites for the serum response factor (SRF), Sp1, and the myogenic basic helix-loop-helix (bHLH) family. In this study we report that activation of the HCA correlates with formation of a muscle-specific multiprotein complex on the promoter. We provide evidence that proteins eluted from the multiprotein complex specifically react with antibodies directed against myogenin, Sp1, and SRF and that the complex can be assembled in vitro by using the HCA promoter and purified MyoD, E12, SRF, and Sp1. In vitro and in vivo assays revealed a direct association of Sp1 and myogenin-MyoD mediated by the DNA-binding domain of Sp1 and the HLH motif of myogenin. The results obtained in this study indicate that protein-protein interactions and the cooperative DNA binding of transcriptional activators are critical steps in the formation of a transcriptionally productive multiprotein complex on the HCA promoter and suggest that the same mechanisms might be utilized to regulate the transcription of muscle-specific and other genes.
Cooperative interactions of transcriptional activators are pivotal in ensuring the proper execution of the myogenic program. For instance, the cooperative binding to two adjacent E boxes on the muscle creatine kinase enhancer by MyoD is required for transcriptional activation (38). The transcriptional activators that bind muscle regulatory regions often establish direct contacts. In fact, protein-protein interactions govern functional cooperativity of myogenic basic helix-loop-helix (bHLH) with E proteins (17) and the myocyte enhancer factor 2 (MEF2) in directing muscle transcription (24). In addition to the E box, numerous muscle-specific regulatory regions contain binding sites for the MEF2 proteins, the serum response factor (SRF), and Sp1, suggesting that the combinatorial binding of these factors to muscle regulatory regions has been selected and is particularly favored for regulation of muscle-specific transcription (25, 39). Whereas both MEF2 and SRF have been shown to interact with the myogenic bHLH (24, 11), the question of whether Sp1 can also directly associate with myogenic bHLH has not been addressed. Furthermore, it remains to be determined whether muscle and ubiquitous transcription factors found on muscle-specific regulatory regions are associated as multiprotein complexes.
A region of the human cardiac α-actin (HCA) promoter spanning from nucleotides −110 to +68 is a composite response element that directs striated muscle-specific transcription (20). Electrophoretic mobility shift assay (EMSA) and footprinting experiments revealed sites for the binding of three nuclear proteins in myogenic cells, including a CArG box for the SRF (21, 5), a GC box for Sp1 (12), and an E box for one of the myogenic bHLH proteins (33). Mutations in any of these three DNA elements result in promoter inactivation (33). Furthermore, reconstitution experiments conducted in Drosophila melanogaster-derived Schneider cells have shown that exogenously supplied Sp1 and MyoD are concomitantly required for the transactivation of the human cardiac α-actin (HCA) promoter (33).
This study reports the formation and analysis of a single major multiprotein complex on the HCA promoter that correlates with muscle-specific transcriptional activation. The formation of this complex relies on the simultaneous presence of at least three factors that bind to the cis-regulatory elements CArG, GC, and E boxes. Through the use of a combined gel mobility shift-Western blot technique, we first demonstrate that cellular Sp1, SRF, and myogenin are all part of this transcriptional complex and are present on the same DNA template. To further substantiate this observation, we successfully attempted to reconstitute the multiprotein complex on the HCA promoter with recombinant MyoD, E12, SRF, and Sp1. We report that Sp1 and myogenic bHLH proteins associate both in vitro and in vivo and that the HLH domain of myogenin and a region spanning the DNA-binding motif of Sp1 mediate such interaction. Since many muscle regulatory regions have been shown to be regulated by Sp1 and the myogenic bHLH (see below), we believe that our findings are likely to be of significance for the understanding of the transcriptional regulation of a number of muscle genes and other genes that are activated by a combination of tissue-specific and ubiquitously expressed factors.
MATERIALS AND METHODS
Cell culture, transfections, and nuclear extract preparation.
C2C12 and HeLa cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 20 and 10% fetal calf serum (FCS), respectively. Differentiation of C2C12 cells was induced by switching the cells to DMEM containing 2% horse serum. C3H10T1/2 cells were grown in DMEM supplemented with 10% FCS. Transfections were performed by using the calcium phosphate precipitation protocol. The Gal-Sp1, Gal-E12, and VP16-MyoD plasmids have been described earlier (34). The UASx4-tk-LUC was kindly provided by R. Evans (Salk Institute). Nuclear extracts derived from C2C12 were prepared after culturing the cells for 4 days in differentiation medium and according to the method of Dignam et al. (7). Protein concentrations were measured by using the Bio-Rad protein assay kit, and extracts were aliquoted and stored at −80°C.
EMSA.
The DNA probes used were the wild-type and mutated HCA promoters extending from nucleotides −126 to −16 relative to the start of transcription. Synthetic oligonucleotides were used as specific competitors. Mutated bases of the CArG box, GC box, and E box are underlined. Mutations corresponding to the CArG-M and E-sm boxes were introduced in the HCA promoter by site-directed mutagenesis (QuikChange; Stratagene), and the resulting templates were sequenced as follows: CArG box GACCAAATAAGGCAAGGTGG μCArG box GACCCAGATCGATCTGGTGG CArG-M box GACCTATTATGGCAAGGTGG GC box CCGGGCCCCCACCCCTGCCCCCGGC μGC box CCGGGCCCCCAAACCTGCCCCCGGC E box TGCTCCAACTGACCC μE box TGCTTGGTCCTGACC E-sm box TGCTCTAACTAACCC
In vitro complex reconstitution was obtained by using bacterially produced and purified glutathione S-transferase (GST)-MyoD (∼20 ng), GST-E12 (∼20 ng), His-SRF (∼50 ng), and Sp1 (∼40 ng) obtained from HeLa cells infected with a recombinant vaccinia virus containing full-length Sp1 (Promega). The purified proteins were incubated in 1× binding buffer [20mM HEPES (pH 7.6), 50 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 300 ng of double-stranded poly(dI-dC)] at 37°C for 20 min, after which 0.4 ng of radiolabeled HCA (nucleotides −126 to −16) promoter was added to the reaction for an additional 15 min at room temperature. The reaction was loaded onto 4% nondenaturing polyacrylamide gel (electrophoresis buffer, 0.5× TBE; 150 V; room temperature; running time, 6 h).
Detection of the protein components present in the gel-retarded complex.
Protein-DNA complexes were resolved by gel mobility shift assay. The wet gel was autoradiographed to locate the protein-DNA complexes, and the proteins present in the shifted complex (see Fig. 1) were eluted with 0.4 M acetic acid–1 M NaCl at 65°C for 40 min, precipitated for 30 min with 10% cold CCl3COOH, washed in acetone and in cold 100% ethanol, vacuum dried, resuspended, denatured in sodium dodecyl sulfate (SDS) buffer, and subjected to SDS–10% polyacrylamide gel electrophoresis. The Western blot technique was performed by using ECL Western blotting detection reagents kit according to the manufacturer’s instructions (Amersham). The polyclonal antiserum against SRF was the gift of R. Prywes (Columbia University), and the monoclonal antibody against myogenin (FD5) was kindly supplied by W. Wright (University of Texas Southwestern Medical Center).
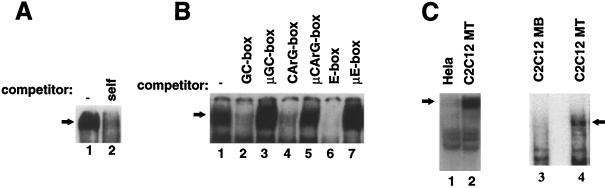
FIG. 1.
Formation of a muscle-specific protein complex on HCA promoter. (A) EMSA analysis for binding of nuclear factors. Samples (10 μg) of C2C12 myotube nuclear extracts were employed with radiolabeled HCA promoter fragment in the absence of specific competitor (lane 1) and in the presence of a 50-fold molar excess of the unlabeled fragment (lane 2). (B) Competition analysis. Nuclear extracts from C2C12 myotubes were assayed in the absence of specific competitor (lane 1) and in the presence of a 100-fold molar excess of unlabeled synthetic double-stranded oligonucleotides containing the following sequences: the normal and mutated GC boxes (lanes 2 and 3), the normal and mutated CArG boxes (lanes 4 and 5), the normal and mutated E boxes (lanes 6 and 7). (C) The shifted protein complex is cell type specific and differentiation dependent. An EMSA of the radiolabeled HCA promoter incubated with equal amounts of nuclear extracts from HeLa (lane 1) and from C2C12 myotubes (lane 2) cells is shown. Equivalent amounts of nuclear extracts derived from either undifferentiated C2C12 myoblasts (MB, lane 3) or differentiated myotubes (MT, lane 4) were analyzed by EMSA with the radiolabeled HCA promoter.
In vitro transcription and translation, GST and His fusion proteins, and protein complex precipitation.
The pBS-Sp1 plasmids (14) were linearized with EcoRI. In vitro transcription was induced with T3 RNA polymerase. The resulting synthetic RNAs were translated in vitro with a rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine according to the manufacturer’s instructions. The pGEX-Sp1C168 construct was obtained by cloning a PCR-generated DNA fragment encoding the last C-terminal 168 amino acids of Sp1 in the pGEX-2TK vector. Preparation of the GST-Sp1C168, GST-myogenin, and GST-E12 proteins was performed as previously described (32). The His-SRF protein was prepared from the pET11d-SRF plasmid (42) as suggested by the manufacturer (Novagen).
Immunoprecipitation of the Sp1-myogenin complex from radiolabeled C2 cells.
C2C12 cells were first incubated in 10 ml of methionine-free DMEM supplemented with 5% dialyzed FCS for 30 min. Then, 5 ml of prewarmed methionine-free DMEM–5% dialyzed FCS containing 1.020 mCi of [35S]methionine was added to the cells followed by incubation for 3 h. Cells were lysed in nuclear lysis buffer (20 mM HEPES, pH 7.7; 20% glycerol; 100 mM NaCl; 1.5 mM MgCl2; 0.2 mM EDTA; 0.1% Triton X-100; 1 mM dithiothreitol; 1 mM phenylmethylsulfonyl fluoride; 10 μg of pepstatin per ml; 100 μg of aprotinin per ml), rocked for 1 h at 4°C and centrifuged at 12,000 × g for 5 min at 4°C. The supernatant was recovered and approximately 3.2 × 107 cpm was incubated in 400 μl of a buffer containing 10 mM HEPES (pH 7.7), 250 mM NaCl, 0.25% Nonidet P-40, and 5 mM EDTA. Sequential immunoprecipitation with the myogenin monoclonal antibody FD5 and the Sp1 antiserum (Santa Cruz Biotechnology) was performed as previously described (34). For blocking experiments, the Sp1 antiserum (5 μg) was incubated with 25 μg of a peptide corresponding to amino acids 520 to 538 of Sp1 (Santa Cruz Biotechnology).
RESULTS
Formation of a muscle-specific multiprotein complex on the HCA promoter.
Oligonucleotides spanning the CArG box, GC box, and E box of the HCA promoter interact autonomously with their cognate binding factors in C2C12 cell nuclear extracts. Nevertheless, the affinity of the binding proteins for their individual DNA sites is relatively low since most of the DNA probe remains in an unbound state (12, 33). Because these three binding sites are each required for expression from the HCA promoter, we have attempted to determine whether or not the proteins bind simultaneously to the promoter and whether their binding is independent or interactive. After the HCA promoter fragment encompassing nucleotides −126 to −16 was incubated with C2C12 myotube nuclear extract, an EMSA revealed a slowly migrating complex (Fig. 1A, lane 1). The complex appears to be specific since its formation is inhibited by excess copies of unlabeled promoter (lane 2). To begin to identify the proteins present in the complex, we competed for binding with a series of oligonucleotides that bind either purified Sp1, MyoD, or SRF (see Materials and Methods). Surprisingly, the inclusion in the binding reactions of an oligonucleotide carrying a single GC-box, CArG-box, or E-box sequence eliminated the formation of the retarded complex (Fig. 1B, lanes 2, 4, and 6). The DNA-binding specificity of the complex was tested by using mutated CArG-box, GC-box, and E-box oligonucleotides in competition experiments (Fig. 1B, lanes 3, 5, and 7). Unlike the wild-type oligonucleotides, the mutated oligonucleotides (see Materials and Methods) μCArG box, μGC box, and μE box fail to compete for complex formation. The finding that oligonucleotides bearing any one of three solitary binding sites can disrupt the complex suggests that at least three proteins are engaged in its formation and that the removal of any one of the three destabilizes the complex. The HCA promoter fragment employed in this study is sufficient to direct tissue-specific transcription in skeletal muscle cells but is inactive in nonmuscle cells. To investigate whether the protein complex formed on the HCA promoter is cell type specific, we employed nuclear extracts from muscle and nonmuscle cells by EMSA and found that the retarded complex is generated with C2C12 muscle cells but not with HeLa cell nuclear extracts (Fig. 1C, lanes 1 and 2). Since HCA is expressed exclusively in differentiated myotubes and not in undifferentiated myoblasts, we analyzed whether formation of the shifted complex was differentially regulated in C2C12 myoblasts and myotubes. Our results indicate that the presence of the complex correlates with the activation of the endogenous HCA gene in differentiated myotubes (Fig. 1C, lanes 3 and 4).
Identification of Sp1, SRF, and myogenin as components of the multiprotein complex.
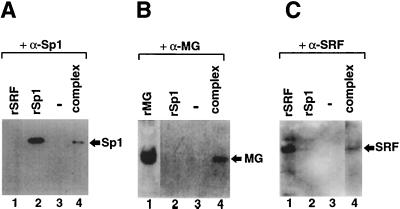
The EMSA results imply, but do not directly demonstrate, that the multiprotein complex contains Sp1, myogenic bHLH, and SRF proteins. To directly identify components of the multiprotein complex formed on the HCA promoter, we performed a preparative EMSA and subjected the proteins eluted from the shifted complex to Western blot analysis. As shown in Fig. 2, antibodies against Sp1 (A), myogenin (B), and SRF (C), but not unrelated antibodies (data not shown), reacted with proteins eluted from the complex that migrated with the same mobility as the corresponding in vitro-synthesized proteins. To eliminate the possibility of coincidental protein migration into the preparative gels that was not dependent on specific interactions with the DNA probe, a control experiment was performed in which nuclear proteins were electrophoresed under similar conditions but in the absence of the DNA probe. Under these experimental conditions we failed to detect any specific reaction between eluted proteins and the antibodies against Sp1, SRF, and myogenin (data not shown). These results are the first direct evidence that Sp1, SRF, and myogenin are all part of the multiprotein complex that occupies the HCA promoter.
FIG. 2.
Identification of the protein components of the shifted complex. (A) Proteins eluted from the shifted complex (Fig. 1, lane 1) were subjected to Western blotting with a polyclonal rabbit antiserum raised against Sp1 protein. Lanes: 1, in vitro-synthesized SRF protein; 2, recombinant Sp1 protein; 3, blank lane; and 4, proteins eluted from the shifted complex. (B) Complex proteins were subjected to Western blotting with a monoclonal antibody against myogenin (FD5). Lanes: 1, in vitro-synthesized myogenin; 2, recombinant Sp1 protein; 3, blank lane; and 4, proteins eluted from the shifted complex. (C) Complex proteins were subjected to Western blotting with polyclonal rabbit antiserum raised against SRF protein. Lanes: 1, in vitro-synthesized SRF protein; 2, recombinant Sp1 protein; 3, blank lane; and 4, proteins eluted from the complex.
In vitro assembly of a protein complex on the HCA promoter by purified MyoD, E12, SRF, and Sp1.
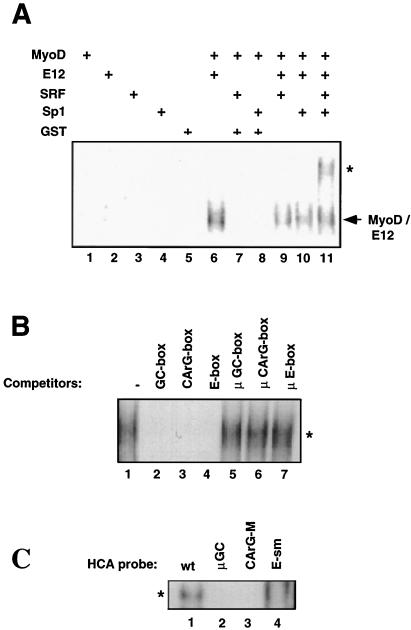
The data reported in Fig. 1 and 2 indicate that the HCA promoter is occupied by a protein complex containing myogenic bHLH, SRF, and Sp1 and that the presence of each individual protein is required for the integrity of the complex. We attempted to reconstitute the multiprotein complex by assembling in vitro the individual, purified proteins on the HCA promoter. Since the myogenic bHLH increase their DNA binding ability upon interaction with E proteins and require heterodimerization with E12 for functional activity (17), E12 was included in the experiments described below. At relatively low protein concentrations, MyoD, E12, SRF, or Sp1 is incapable of individually binding to the HCA promoter (Fig. 3A, lanes 1 to 4). Similarly, the MyoD/Sp1 and MyoD-SRF combinations fail to interact with the HCA probe (lanes 7 and 8), whereas heterodimers of MyoD and E12 are readily observed (lane 6). Only when the four individual proteins were present in the binding reaction was the formation of a slowly migrating complex evident (lane 11). This complex was absent in any of the reactions containing other protein combinations (lanes 5 to 9). The results of these experiments suggest that myogenic bHLH (MyoD), E12, SRF, and Sp1 can form a protein complex on the HCA if they are simultaneously present. As indicated by the competition experiments shown in Fig. 1B, preventing binding of any of the three factors to the HCA results in abolishment of the complex. To investigate whether the multiprotein complex described in Fig. 1 and that described in this paragraph (Fig. 3A) share similar properties, competition experiments were performed. After assembling MyoD, E12, SRF, and Sp1 on the HCA promoter, molar excess of oligonucleotides spanning the GC box (Sp1 binding), CArG box (SRF binding), and E box (MyoD/E12 binding) of the HCA promoter were added to the binding reactions (Fig. 3B). Inclusion of any of the three oligonucleotides interfered with the formation of the shifted complex (lanes 2 to 4), whereas mutant oligonucleotides that no longer interact with their respective binding proteins (μGC box, μCArG box, and μE box) left the complex unaltered (lanes 5 to 7). Similar experiments were conducted with HCA promoter fragments bearing subtle mutations in the GC, CArG, or E box (see below) and purified MyoD, E12, Sp1, and SRF proteins. Mutations of any of the three binding sites interfered with complex assembly (Fig. 3C, lanes 2 to 4), confirming our previous findings (Fig. 1) that their integrity is a prerequisite for complex formation. The experiments illustrated in Fig. 3 suggest that the affinity of the individual transcriptional activators for the HCA promoter is weak and show that a protein complex that displays higher affinity is assembled solely in the presence of a defined protein combination.
FIG. 3.
In vitro assembly of a protein complex on the HCA promoter by purified MyoD, E12, SRF, and Sp1. (A) EMSA was performed with the radiolabeled HCA promoter with different combinations of GST-MyoD, E12, His-SRF, and Sp1 purified proteins. The arrow points to a shifted complex containing MyoD-E12 heterodimers. The asterisk indicates a low-mobility complex observed exclusively in the presence of MyoD, E12, SRF, and Sp1. (B) Competition analysis of the low-mobility complex described in panel A. Oligonucleotides containing the GC, CArG, or E box but not their respective mutated sequences compete for the formation of the low-mobility complex. (C) EMSA was performed with radiolabeled HCA promoter fragments derived from the HCA wild type and the HCA-μGC, HCA-CArGM, and HCA-E-sm constructs (see Materials and Methods) and purified MyoD, E12, SRF, and Sp1 proteins. The low-mobility complex described in panel A and indicated by the asterisk is observed when the HCA wild type (lane 1) but not the HCA mutants (lanes 2 to 4) are employed.
Refining the CArG- and E-box mutations associated with HCA promoter inactivation.
The mutations introduced in the CArG and E boxes and investigated in the present study, as well as in previous studies, extend outside the minimal binding sites required for SRF and MyoD-myogenin. This raises the question of whether promoter inactivation might be the consequence of lack of binding of additional, yet uncharacterized proteins that might recognize sequences surrounding the CArG and E boxes. To stringently evaluate the individual contributions of the single trans-acting factors, finer mutations were created. The CArG-box sequence of the HCA promoter was altered to create the CArG-M motif (see Materials and Methods). The CArG-M sequence is recognized by the yeast SRF homolog MCM1 but not by mammalian SRF (13). Similarly, the E box of the HCA was slightly modified (CAACTG > TAACTA) to generate the E-sm box. The HCA promoter bearing either the CArG-M (HCA-CArG-M) or the E-sm (HCA-E-sm) boxes was tested in a transient-transfection assay and found to be inactive in muscle cells, thus confirming the findings that SRF and myogenic bHLH proteins are essential for promoter activation (Table 1). Oligonucleotides for the CArG-M and E-sm boxes were tested by EMSA and found to be unable to bind purified SRF and MyoD-myogenin, respectively. They were also not able to compete for the formation of the muscle-specific multiprotein complex analyzed in the experiments reported in Fig. 1 (data not shown).
TABLE 1.
Fine mutations of the CArG and E boxes inactivate tissue-specific activation of the HCA promotera
| Construct | Luciferase units ± SD |
|---|---|
| HCA-LUC wild type | 1,450 ± 250 |
| HCA-CArG-M-LUC | 75 ± 24 |
| HCA-E-sm-LUC | 158 ± 56 |
C2C12 cells were transiently transfected with 100 ng of the HCA-LUC constructs, 50 ng of the CMV-lacZ construct, and 10 μg of the pUC19 plasmid. Luciferase and β-galactosidase activities were determined 72 h after transfection by using the chemiluminescent Dual-Light kit (Tropix). Luciferase units, corrected for β-galacosidase activity, are indicated. Every experiment was performed with triplicated samples and was repeated at least twice.
Lack of binding by Sp1 and myogenic bHLH proteins correlates with the absence of the multiprotein complex and with inactivation of the HCA promoter.
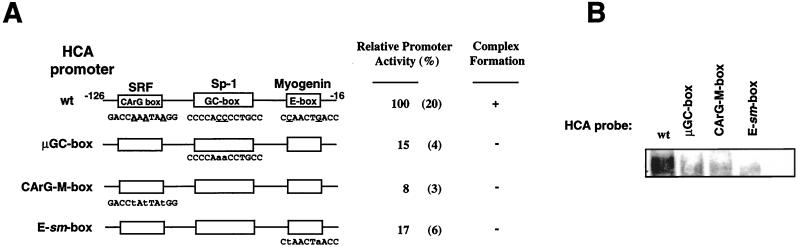
The importance of the CArG box for the activation of the cardiac α-actin promoters (20, 23; the present study) for muscle-specific expression and for expression in general has been extensively demonstrated (reference 30 and references therein), and the recent finding that SRF interacts with myogenic bHLH proteins might provide a molecular explanation for this phenomenon (11). Nevertheless, our data (Fig. 1) (33) indicate that the presence of the CArG and E boxes is insufficient for promoter activity and complex formation. In fact, deletion or mutation of the GC box is also deleterious for HCA promoter function and the results shown in Fig. 2 clearly demonstrate that Sp1 is part of the multiprotein complex. To further address the role of Sp1, SRF, and the myogenic bHLH in controlling the activation of the HCA promoter, we investigated whether formation of the multiprotein complex correlates with HCA promoter activity. The HCA promoter constructs described in Table 1 and bearing an individually mutated GC, CArG, or E box were employed in both functional and DNA binding assays. Mutations of the GC, CArG, or E box extinguish the transcriptional activity of the HCA promoter in muscle cells (Fig. 4A). Consistent with this observation, EMSA conducted with C2C12 nuclear extracts indicates that the multiprotein complex is not formed when HCA promoter probes containing a mutated GC, CArG, or E-box are used in EMSA (Fig. 4B). Our findings strongly correlate proper function of the HCA promoter with the presence of the multiprotein complex and the binding of Sp1, SRF, and myogenic bHLH proteins.
FIG. 4.
The presence of the protein complex correlates with the HCA promoter activity in muscle cells. (A) Schematic representation of the HCA promoter fragments used in transfection and EMSA assays. Mutations introduced in the GC, CArG, and E boxes are in lowercase letters. The relative promoter activity is expressed as a percentage and refers to the luciferase activities generated by the different HCA constructs when transiently transfected in C2C12 cells. The numbers in parenthesis represent standard deviations. (B) Gel retardation analysis for binding of nuclear factors from C2C12 cells with different HCA promoter fragments. Nuclear extract from C2 myotubes was employed with radiolabeled fragment of the wild type (wt) or HCA promoter containing mutated GC box (μGC), CarG box (CArG-M), or E box (E-sm).
Evidence for specific protein-protein interactions: myogenic bHLH proteins form heterodimeric complexes with Sp1 both in vitro and in vivo.
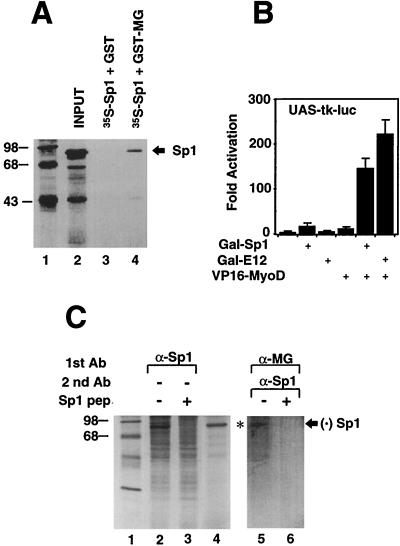
The preceding data raise the possibility that Sp1 and the myogenic bHLH might directly interact. To test for such protein-protein contacts, GST-myogenin immobilized on agarose beads was reacted with in vitro-synthesized Sp1. As shown in Fig. 5A, GST-myogenin, but not the GST alone, retained Sp1 (lanes 3 and 4). We proceeded to investigate whether the in vitro interaction between Sp1 and the myogenic bHLH proteins could be recapitulated in the cell. To this end, we performed a two-hybrid system assay employing the Sp1 coding regions fused to the DNA binding domain of Gal4 (Gal-Sp1) and MyoD grafted to the viral activator VP16 (VP16-MyoD). C3H10T1/2 fibroblasts were transiently transfected with an indicator plasmid bearing multimerized copies of Gal4 binding sites driving expression of the luciferase gene (UASx4-tk-LUC). As shown in Fig. 5B, cotransfection of the Gal-Sp1 and VP16-MyoD plasmids caused a transcriptional activation of the indicator construct that was approximately 10-fold stronger than that provoked by the individual Gal-Sp1 or VP16-MyoD plasmid and of similar magnitude to that displayed by Gal-E12 and VP16-MyoD. This indicates that a physical association between Sp1 and MyoD takes place within the cell.
FIG. 5.
Sp1 interact with two myogenic bHLH proteins, myogenin and MyoD, in vitro and in vivo. (A) The GST protein (lane 3) or the GST-myogenin (lane 4) were mixed with 10 μl of reticulocyte lysate programmed by Sp1-encoding RNA and supplemented with [35S]methionine and then processed and resolved on a 10% denaturing polyacrylamide gel. Lane 2 shows the input radiolabeled Sp1. (B) C3H10T1/2 cells were transiently transfected with the UASx4-tk-LUC indicator plasmid and the Gal-Sp1, Gal-E12, and VP16-MyoD activators. To correct for transfection efficiency, the CMV-lacZ plasmid was added to the transfection reaction. After 48 h, cells were processed and luciferase and β-galactosidase assays were performed on an automated microtiter plate luminometer (MLX; Dynex Technology). Bars indicate standard deviations. (C) Nuclear extracts derived from metabolically radiolabeled C2C12 cells were incubated with unblocked (lane 2) or blocked (lane 3) Sp1 antiserum in low-stringency conditions. Double immunoprecipitation with α-myogenin antibody, followed by unblocked (lane 5) or blocked (lane 6) Sp1 antiserum in high-stringency conditions, with radiolabeled C2C12 nuclear extracts reveals that the protein associated with myogenin is bona fide Sp1. Lane 4 shows radiolabeled in vitro-synthesized Sp1.
While strongly suggesting that the overexpressed Sp1 and myogenic bHLH can interact in the cell, the two-hybrid system experiments do not prove that endogenous Sp1 and myogenic bHLH proteins are naturally associated in muscle cells. To address this point, we performed a double immunoprecipitation experiment with metabolically radiolabeled C2C12 myotube nuclear extracts and antibodies directed against myogenin and Sp1 (Fig. 5C). Under low-salt conditions, the Sp1 antiserum immunoprecipitates a major band migrating at ∼95 kDa and several other minor bands (lane 2). The Sp1 antiserum failed to immunoprecipitate the ∼95-kDa band, but not the other bands, when the reaction was blocked with the immunogen, a peptide spanning amino acids 520 to 538 of Sp1 (lane 3). Furthermore, Sp1 synthesized in vitro comigrates with the major band precipitated by the Sp1 antiserum (lane 4). Altogether, these data indicate that the ∼95-kDa band represents a polypeptide immunologically indistinguishible from and of the same molecular size as Sp1. C2C12 nuclear extracts were sequentially immunoprecipitated first with an antimyogenin antibody in low-salt conditions and in a second, more stringent step, with an Sp1 antiserum to determine whether Sp1 is associated with the immunoprecipitated myogenin. Indeed, the ∼95-kDa band corresponding to bona fide Sp1 was detected according to the sequential immunoprecipitation protocol (lane 5). Blocking the Sp1 antiserum with the immunogenic peptide (lane 6) or substituting the myogenin antibody with a preimmune serum (data not shown) prevented immunoprecipitation of the ∼95-kDa band. These results indicate that the protein immunoprecipitated by the combination of myogenin and Sp1 antibodies is Sp1 and provide evidence that endogenous Sp1 and myogenin directly associate in C2C12 muscle cells.
A region encompassing the DNA-binding domain of Sp1 and the HLH domain of myogenin mediate protein-protein interactions.
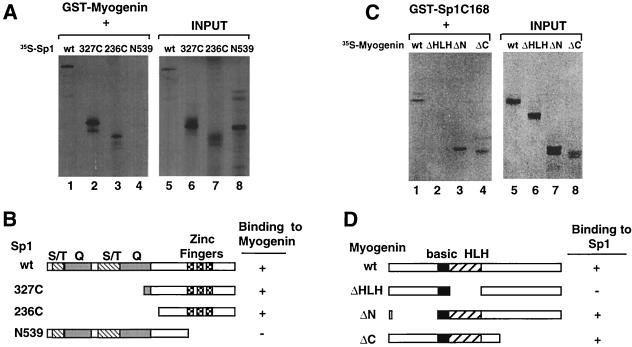
Sp1 specifically interacts with DNA regions containing defined GC boxes to activate transcription (8). Distinct regions of this protein have been shown to direct DNA binding and transcriptional activation. In particular, the C-terminal 168 amino acids of Sp1 contain three zinc-finger structures required for binding to the GC box (13) (see Fig. 6B). Two major transactivation domains rich in serine-threonine and glutamine residues, respectively, are located within the N-terminal 478 amino acids of Sp1 (15). To define the regions of Sp1 involved in contacting a myogenic bHLH protein, truncated versions of Sp1 lacking either the transactivation or the DNA binding domains were employed in affinity selection with myogenin (Fig. 6A). A total of 236 amino acids located at the C terminus and containing the DNA binding domain of Sp1 were efficiently retained on GST-myogenin-coated agarose beads (lane 3). In contrast, the N-terminal 539 amino acids spanning both transactivation domains of Sp1 failed to interact with GST-myogenin (lane 4). These results indicate that the regions of Sp1 located at the C terminus and containing the three DNA-recognizing zinc fingers are necessary and sufficient for binding to myogenin.
FIG. 6.
A region of Sp1 spanning the DNA-binding domain and the HLH domain of myogenin mediate protein interactions. (A) Wild-type and different truncated version of radiolabeled Sp1 were affinity purified on GST-myogenin-coated agarose beads. The right panel shows input proteins. (B) Schematic representation of the Sp1 polypeptides employed in (A). Regions rich in serine and threonine (S/T), glutamic acid (Q), and the zinc finger motif are indicated at the top. (C) The C-terminal 168 amino acids of Sp1 fused to GST were reacted with several versions of radiolabeled myogenin. The left panel indicates the input proteins. The myogenin ΔN and ΔC proteins were synthesized by using the myogenin DM4-79 and DM158-224 constructs described elsewhere (35). (D) Schematic representation of the myogenin deletions employed in the experiments reported in panel C.
To confirm further that Sp1 interacts with myogenin and to define the regions of myogenin involved in contacting Sp1, the C-terminal 168 amino acids of Sp1 were fused to GST (GST-Sp1C168) and used in affinity selection experiments with different truncated versions of radiolabeled myogenin. The data reported in Fig. 6C show that GST-Sp1C168 interacts with myogenin. Removal of the HLH domain of myogenin prevented interaction with GST-Sp1C168, whereas deletions of either the N or the C terminus had no effect, indicating that the HLH domain of myogenin is engaged in contacting the carboxyl terminus of Sp1.
DISCUSSION
In this study we describe the formation of a single major DNA-binding complex on the HCA. This muscle-specific complex contains myogenic bHLH, Sp1, and SRF. Their simultaneous presence is essential for both complex formation and transcriptional activation of the promoter in muscle cells. In fact, competition experiments conducted with individual DNA-binding sites specific for the transcriptional activators indicate that the absence of any of the three proteins (i.e., MyoD-myogenin, Sp1, and SRF) is incompatible with promoter occupancy and therefore with transcriptional activation. When the HCA promoter DNA segments bearing single site mutations were used as a probe in EMSA experiments, we could not detect any complexes of intermediate mobility. If any of the three transcription factors binds to the HCA promoter independently or pairwise, we would have expected to observe complexes of intermediate electrophoretic mobility with two rather than three transcription factors. Thus, this protein complex appears to be formed in an all-or-none fashion with the binding of SRF, Sp1, and myogenic bHLH proteins to the HCA promoter being mutually dependent. Our data do not exclude the likelihood that other proteins, including E proteins, are involved in the formation of the multiprotein complex. Indeed, we have assembled a protein complex on the HCA promoter by using highly purified MyoD, E12, SRF, and Sp1.
E proteins are most likely a component of multiprotein complex observed with nuclear extracts since the majority of endogenous MyoD is complexed with E12-E47 in muscle cell extracts (17). Even though we have not formally demonstrated that the multiprotein complex formed with muscle cell extract corresponds to the complex assembled by recombinant MyoD, E12, SRF, and Sp1, they both share several properties. Both complexes are formed in an all-or-none fashion by myogenic bHLH, SRF, and Sp1 on the HCA promoter. Furthermore, competition with individual DNA-binding sites for the transcriptional activators engaged in the complex specifically abolishes the formation of both complexes. Other transcriptional activators and coactivators may also be present, including MEF2 that can interact with MyoD (16, 24) and the coactivators p300 (9) and PCAF (40), whose interactions with MyoD are required for MyoD transcriptional activity (32). The HCA gene product is present exclusively in differentiated myotubes despite the fact that SRF, Sp1, and MyoD are already synthesized in undifferentiated myoblasts. This raises the question of how the prevention of premature muscle gene expression is achieved. Our study indicates that assembly of multiprotein complexes might be the switch for HCA activation in particular and for muscle gene expression in general. In fact, a similar finding has been reported for the MCK gene (26). The presence of a negative regulator of DNA binding such as Id (3) or the absence of a hypothetical assembly factor might mechanistically explain the lack of complex formation in myoblasts.
The potential involvement of protein-protein interactions in the genesis of transcription complexes present on muscle-specific promoters was also suggested by the observation that SRF and myogenin can interact (11). While the interaction of SRF and myogenin might well be involved in the formation of the multiprotein complex observed on the HCA promoter, it was evident from previous data that additional proteins must be involved. We now show that Sp1 interacts with the myogenic bHLH proteins in vitro and in a mammalian two-hybrid system. Furthermore, endogenous Sp1 is coimmunoprecipitated with MyoD from extracts derived from muscle cells. Physical interaction of Sp1 and myogenic bHLH might help to explain the results obtained in Drosophila melanogaster-derived Schneider cells, where Sp1 and MyoD synergistically activated the HCA promoter (33) in the presence of endogenous SRF-like molecules (28).
Our results, combined with the observation that GC and E boxes are often juxtaposed in numerous muscle-specific enhancers, suggest the possibility that protein-protein interactions between Sp1 and the myogenic bHLH play a key role in the formation of transcriptional complexes on muscle-specific regulatory regions. It is likely that the functional synergism and physical interaction properties of the proteins described here operate also for the regulation of additional muscle genes controlled by the same set of proteins. We note that several muscle-specific genes are positively regulated by a combination of Sp1 and myogenic bHLH proteins, including the regulatory regions of HCA (33), troponin I (19), α and δ subunits of the acetylcholine receptor (4, 37), muscle sarcoplasmic reticulum Ca2+-ATPase gene (2), and muscle phosphofructokinase P2 (18). Our findings support the hypothesis that the regulation operated by Sp1 and myogenic bHLH on the HCA promoter is a general mechanism that applies to many genes expressed in skeletal muscle cells.
ACKNOWLEDGMENTS
Sarah Buranen and Terry Saluna provided excellent technical assistance, and R. Evans, E. Olson, R. Prywes, R. Tjian, and W. Wright generously provided critical biological materials.
Part of this work was done while E.B. was a Research Fellow of the American Heart Association, Greater Los Angeles Affiliate. This work was supported in parts by grants from the NIH (to L.K.) and the American Heart Association, Greater Los Angeles Affiliate (to L.K. and to V.S.). Y.H. was supported by an Initial Investigatorship Award (1104-FI1) from the American Heart Association, Greater Los Angeles Affiliate.
REFERENCES
- 1.Andres V, Fisher S, Wearsch P, Walsh K. Regulation of Gax homeobox gene transcription by a combination of positive factors including myocyte-specific enhancer factor 2. Mol Cell Biol. 1995;15:4272–4281. doi: 10.1128/mcb.15.8.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker D L, Dave V, Reed T, Periasamy M. Multiple Sp1 binding sites in the cardiac/slow twitch muscle sarcoplasmic reticulum Ca2+-ATPase gene promoter are required for expression in Sol8 muscle cells. J Biol Chem. 1996;271:5921–5928. doi: 10.1074/jbc.271.10.5921. [DOI] [PubMed] [Google Scholar]
- 3.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 4.Bessereau J L, Mendelzon D, LePoupon C, Fiszman M, Changeux J P, Piette J. Muscle-specific expression of the acetylcholine receptor alpha-subunit gene requires both positive and negative interactions between myogenic factors, Sp1 and GBF factors. EMBO J. 1993;12:443–449. doi: 10.1002/j.1460-2075.1993.tb05676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boxer L M, Prywes R, Roeder R G, Kedes L. The sarcomeric actin CArG-binding factor is indistinguishable from the c-fos serum response factor. Mol Cell Biol. 1989;9:515–522. doi: 10.1128/mcb.9.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan T J, Olson E N. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 1990;4:582–595. doi: 10.1101/gad.4.4.582. [DOI] [PubMed] [Google Scholar]
- 7.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 8.Dynan W S, Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983;32:669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- 9.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 10.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 11.Groisman R, Masutani H, Leibovitch M P, Robin P, Soudant I, Trouche D, Harel B A. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J Biol Chem. 1996;271:5258–5264. doi: 10.1074/jbc.271.9.5258. [DOI] [PubMed] [Google Scholar]
- 12.Gustafson T A, Kedes L. Identification of multiple proteins that interact with functional regions of the human cardiac alpha-actin promoter. Mol Cell Biol. 1989;9:3269–3283. doi: 10.1128/mcb.9.8.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill C S, Marais R, John S, Wynne J, Dalton S, Treisman R. Functional analysis of a growth factor-responsive transcription factor complex. Cell. 1993;73:395–406. doi: 10.1016/0092-8674(93)90238-l. [DOI] [PubMed] [Google Scholar]
- 14.Johnson J L, McLachlan A. Novel clustering of Sp1 transcription factor binding sites at the transcription initiation site of the human muscle phosphofructokinase P1 promoter. Nucleic Acids Res. 1994;22:5085–5092. doi: 10.1093/nar/22.23.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadonaga J T, Courey A J, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 16.Kaushal S, Schneider J W, Nadal-Ginard B, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 17.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 18.Le H B, Vaisanen P A, Johnson J L, Raney A K, McLachlan A. Regulation of transcription from the human muscle phosphofructokinase P2 promoter by the Sp1 transcription factor. DNA Cell Biol. 1994;13:473–485. doi: 10.1089/dna.1994.13.473. [DOI] [PubMed] [Google Scholar]
- 19.Lin H, Yutzey K E, Konieczny S F. Muscle-specific expression of the troponin I gene requires interactions between helix-loop-helix muscle regulatory factors and ubiquitous transcription factors. Mol Cell Biol. 1991;11:267–280. doi: 10.1128/mcb.11.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minty A, Kedes L. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol Cell Biol. 1986;6:2125–2136. doi: 10.1128/mcb.6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miwa T, Kedes L. Duplicated CArG box domains have positive and mutually dependent regulatory roles in expression of the human alpha-cardiac actin gene. Mol Cell Biol. 1987;7:2803–2813. doi: 10.1128/mcb.7.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miwa T, Boxer L M, Kedes L. CArG boxes in the human cardiac alpha-actin gene are core binding sites for positive trans-acting regulatory factors. Proc Natl Acad Sci USA. 1987;84:6702–6706. doi: 10.1073/pnas.84.19.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohun T J, Taylor M V, Garrett N, Gurdon J B. The CArG promoter sequence is necessary for muscle-specific transcription of the cardiac actin gene in Xenopus embryos. EMBO J. 1989;8:1153–1161. doi: 10.1002/j.1460-2075.1989.tb03486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 25.Molkentin J D, Olson E N. Defining the regulatory networks for muscle development. Curr Opin Genet Dev. 1996;6:445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- 26.Mueller P R, Wold B. In vivo footprinting of a muscle-specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 27.Mutero A, Camp S, Taylor P. Promoter elements of the mouse acetylcholinesterase gene. Transcriptional regulation during muscle differentiation. J Biol Chem. 1995;270:1866–1872. [PubMed] [Google Scholar]
- 28.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 29.Olson E N, Perry M, Schulz R A. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 30.Phan-Dinh-Tuy F, Tuil D, Schweighoffer F, Pinset C, Kahn A, Minty A. The “CC.Ar.GG” box. A protein-binding site common to transcription-regulatory regions of the cardiac actin, c-fos and interleukin-2 receptor genes. Eur J Biochem. 1988;173:507–515. doi: 10.1111/j.1432-1033.1988.tb14027.x. [DOI] [PubMed] [Google Scholar]
- 31.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puri P L, Sartorelli V, Yang X-J, Hamamori Y, Kedes L, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 33.Sartorelli V, Webster K A, Kedes L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 1990;4:1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- 34.Sartorelli V, Huang J, Hamamori H, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz J J, Chakraborty T, Martin J, Zhou J M, Olson E N. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol Cell Biol. 1992;12:266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor M, Treisman R, Garrett N, Mohun T. Muscle-specific (CArG) and serum-responsive (SRE) promoter elements are functionally interchangeable in Xenopus embryos and mouse fibroblasts. Development. 1989;106:67–78. doi: 10.1242/dev.106.1.67. [DOI] [PubMed] [Google Scholar]
- 37.Walke W, Xiao G, Goldman D. Identification and characterization of a 47 base pair activity-dependent enhancer of the rat nicotinic acetylcholine receptor delta-subunit promoter. J Neurosci. 1996;16:3641–3651. doi: 10.1523/JNEUROSCI.16-11-03641.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weintraub H, Davis R, Lockshon D, Lassar A B. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 40.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 41.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 42.Zhu H, Joliot V, Prywes R. Role of TFIIF in SRF-activated transcription. J Biol Chem. 1994;269:3489–3497. [PubMed] [Google Scholar]