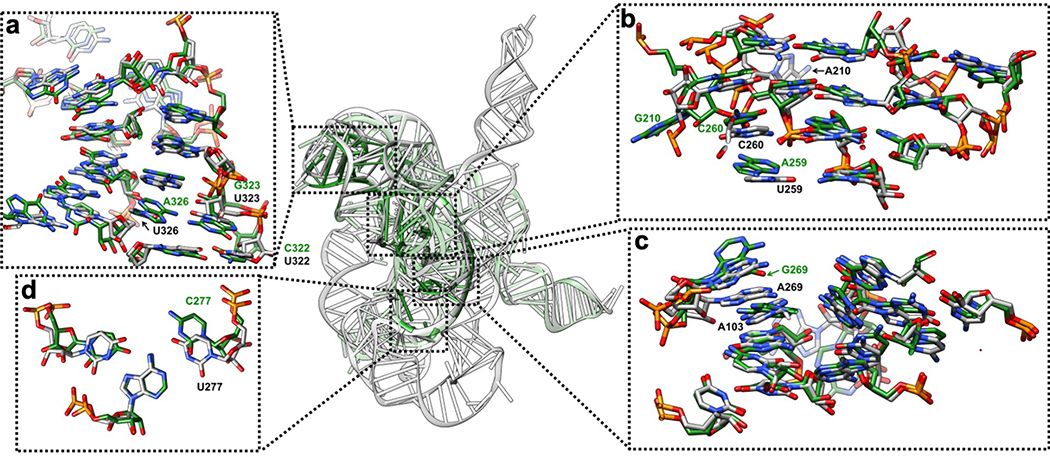
Extended Data Figure 5 related to Extended Data Figure 4. Comparison between the previous 3.8 Å crystal structure of the mutated Tetrahymena ribozyme catalytic core (green) and the cryo-EM structure of the wild type apo Tetrahymena ribozyme L-21 construct (grey) shows minor differences.
The overall RMSD for the catalytic core region (stem P3-P9) is 6.6 Å. (a) The same view of P5-J5/5a-P9 region as in Extended Data Figure 4a. The nucleotide conformations generally agree well between two models, three mutations U322C, U323G and U326A are not involved in tertiary interactions. The RMSD for this region is 4.9 Å. (b) The same view of P4-P5a-J6/7-J8/7 region as in Extended Data Figure 4b. In the crystal structure, U259A is slightly moved away from the G108-C213 base pair and disrupts this base triple interaction. A210G is moved far away from the wild type position of A210, because there is no A46 in stem P2 to interact with in the crystal structure. The very top base quartet is much more compact in the cryo-EM structure compared to the crystal structure, likely due to the presence of peripheral domain that wraps around the catalytic core to make it more compact. The RMSD for this region is 5.7 Å. (c) The same view of P3-J3/4-P6-J7/3 region as in Extended Data Figure 4c. The overall nucleotide conformations agree very well between two models, except for A269G and A270 in the crystal structure are completely moved away and disrupt their interactions with A103, which is observed in the cryo-EM structure. The RMSD for this region is 1.7 Å. (d) The same view as in Extended Data Figure 4d. U277C disrupts the U277-A97-U300 base triple. The RMSD for this base triple is 2.7 Å. See also Supplementary Table 1.