Summary
Trypanosoma brucei and other African trypanosomes are vector-borne parasites that cause substantial human suffering across sub-Saharan Africa. The T. brucei life cycle is punctuated by numerous developmental stages, each occurring in a specific environmental niche and characterized by a unique morphology, metabolism, surface protein coat, and gene expression profile. The environmental cues and signaling pathways that drive transitions between these stages remain incompletely understood. Recent studies have started to fill this gap in knowledge. Likewise, several new studies have expanded our understanding of parasite movement through specific tissues and the parasite’s ability to alter movement in response to external cues. Life cycle stage differentiation and motility are intimately integrated phenomena, as parasites must be at the right place (i.e. within a specific environmental milieu) at the right time (i.e. when they are appropriately staged and pre-adapted for perceiving and responding to signals) in order to complete their life cycle. In this review, we highlight some of the recent work that has transformed our understanding of signaling events that control parasite differentiation and motility. Increased knowledge of T. brucei environmental sensing and signal transduction advances our understanding of parasite biology and may direct prospective chemotherapeutic and transmission blockade strategies that are critical to eradication efforts.
Introduction
Throughout sub-Saharan Africa, substantial health and economic burdens are attributed to Trypanosoma brucei, the etiologic agent of human African trypanosomiasis (HAT) and nagana, a wasting disease of cattle. Tsetse fly vectors (Glossina spp.) transmit these protozoan parasites between mammalian hosts, with T.b. rhodesiense and T.b. gambiense conferring acute and chronic HAT, respectively [1]. While successful surveillance and control efforts have led to a marked reduction in cases [2], these pathogens have historically shown strong capacity for resurgence and control efforts are complicated by the potential for asymptomatic patients to serve as parasite reservoirs [3–5]. Moreover, infection of livestock promulgates economic instability in communities reliant upon agriculture [6].
T. brucei parasites navigate diverse environments within their mammalian hosts and tsetse fly vectors. Following inoculation of a mammalian host by an infected tsetse fly, T. brucei trypomastigotes traverse the circulatory system as two distinct developmental morphotypes: a proliferative slender form and a quiescent, transmission-competent stumpy form [7]. Parasites eventually escape the bloodstream and enter the central nervous system (CNS), marking onset of the lethal stage of disease [8]. Stumpy parasites are pre-adapted for survival in a new tsetse vector and they differentiate to insect-stage procyclic forms in the fly midgut [9]. Parasites then travel from the midgut lumen to the salivary glands, undergoing sequential differentiation events that are matched to the tsetse tissues encountered. The cycle is completed when infective metacyclic trypomastigotes from the tsetse salivary glands are released into a new mammalian host during telmophagous (slash-and-suck) blood feeding.
Given the diverse milieus encountered, it is paramount that trypanosomes possess signal transduction systems for sensing and responding to extracellular signals, thus enabling them to coordinate developmental cycles with movement through host tissues. This coordination, in turn, allows the parasites to maintain competence for both survival in each tissue and continuation through the life cycle. While the T. brucei life cycle is well-characterized, the identification of cues and associated signaling pathways responsible for driving developmental programs during the life cycle and controlling cell movement through host tissues remains a dynamic and exciting field of investigation. Beyond advancing basic understanding of parasite biology, elucidation of signaling pathways required for transmission and pathogenesis illuminates opportunities for therapeutic intervention.
In this review, we discuss recent seminal studies that have shifted our understanding of T. brucei environmental sensing and signal transduction systems that direct parasite development and movement. Many of these studies and their findings were made possible by innovative molecular techniques and imaging capabilities; these technical advances have propelled the field forward and will be instrumental in answering open questions in T. brucei pathogenesis and transmission biology.
Transduced environmental cues drive cellular differentiation
Stumpy induction factor (SIF) and differentiation from slender to stumpy trypomastigotes
For T. brucei trypomastigotes, existence within the mammalian host presents a paradox: proliferation and immune evasion ensure parasites are present for transmission, but high parasitemia is potentially fatal for the host [7, 10, 11]. Slender cells are well-suited for the host bloodstream, dividing by binary fission to increase parasitemia and evading the host immune system via shifting expression of variable surface glycoproteins (VSGs)[7, 10]. Slender cells develop to stumpy cells, which become growth-arrested in the G0 stage of the cell cycle and refrain from changing VSGs [9, 12]. In stumpy cells, growth arrest slows the rise in parasitemia and cells are readily cleared from circulation in the absence of VSG switching [7, 9]. Stumpy cells are also uniquely prepared for life in the tsetse fly, through distinct surface protein expression (such as PAD1) and metabolic capacity (at least partially mediated via stumpy-specific mitochondrial gene expression) [7, 13] [14]. Thus, the slender to stumpy developmental program accommodates both the needs to limit parasitemia and to prepare for transmission [9].
Stumpy formation is parasite density-dependent (i.e. quorum sensing) [13, 15] and triggered by a soluble factor or factors, termed stumpy induction factor (SIF) [13]. The identity of SIF, as well as mechanisms by which the SIF signal is perceived and transduced, remained unknown until a recent series of highly innovative studies. In a landmark study, Rojas and colleagues have now managed to identify an oligopeptide signaling pathway that promotes density-dependent stumpy formation [16]. The data indicate that they have at last identified both SIF and membrane proteins engaged in SIF signaling [16]. Trypanosomes are noted for their lack of heterotrimeric G-proteins and cognate G-protein coupled receptors (GPCRs) [17]. Nonetheless, genome searches identified a T. brucei gene with homology to the GPR89 family of “orphan GPCRs” [16], which function in diverse signaling capacities in other organisms. Through a meticulous and rigorous series of experiments, the authors established that GPR89 is an essential, slender-specific surface protein that – when ectopically expressed – accelerates stumpy formation in vitro and in mice. This phenotype requires genes previously identified as required for stumpy formation in vivo [18], demonstrating GPR89 acts via the bona fide SIF pathway. GPR89 exhibits structural homology to the POT family of oligopeptide transporters and the authors showed that a bacterial POT protein (YjdL from Escherichia coli) drives stumpy formation when expressed in T. brucei. Importantly, GPR89 transports oligopeptides in vitro and replacement of one allele with a YjdL mutant with reduced transporter activity has attenuated capacity to induce stumpy formation. As is the case for many aspects of T. brucei signaling, GPR89 from T. brucei has several variations relative to homologues in the host.
The data firmly establish GPR89 as a driver of stumpy formation. Given the oligopeptide transport activity, cell-surface location, and slender-specific expression of GPR89, Rojas and colleagues then asked whether oligopeptides can promote stumpy formation. Supplementation with oligopeptides drove stumpy formation in low-density T. brucei cultures. Moreover, the authors found that transgenic T. brucei cells expressing either of two secreted peptidases provided a paracrine signal to drive early stumpy formation in control cells during co-infection in mice. The combined data provide an elegant model in which GPR89 mediates perception of an oligopeptide SIF signal to drive stumpy formation. The source of the oligopeptides during natural infection - whether from parasite or host - and the mechanism by which they promote stumpy formation remain to be determined.
To this end, genes functioning in SIF signal transduction were identified in a pioneering and seminal study by Mony et al. [18]. Following an initial RNAi screen, Mony and colleagues demonstrated the identified genes participated in the natural SIF pathway through both in vitro experiments and mouse infection models. Identified genes encompass most aspects of the signaling cascade, including signal processing, signal transduction, and effector function - including control of gene expression and cell cycle exit. Moreover, beyond identifying components of the SIF quorum sensing pathway, this work set the stage for genetic interaction studies in which epistatic relationships between proposed players could be established. This has identified independent branches of the SIF pathway and allowed for nascent assembly of these branches into a comprehensive network [19, 20]. Continued genetic interaction studies [18–20] to order SIF pathway components is an important area for future work.
Among the most central SIF signaling genes identified thus far are the RNA binding protein RBP7 [18] and the dual specificity kinase, DYRK [20]. RBP7 is a global regulator of stumpy-specific gene expression, while DYRK is a master regulator in a phospho-signaling cascade that targets effectors of stumpy formation and inhibitors of slender cell maintenance. An important facet of the DYRK studies is the rigorous biochemical analyses applied, which directly demonstrated requirement for DYRK kinase activity in stumpy formation and revealed several peculiarities in T. brucei DYRK functionality as compared to DYRK proteins of mammals. Differences relative to the mammalian host is a common theme in T. brucei signaling and offers opportunity for exploitation in therapeutic development.
Parasites exist in a complicated environmental milieu where integration of multiple signals is imperative to successful differentiation. Most T. brucei differentiation events are irreversible, so multiple confirmatory or reinforcing signals may function to avoid differentiation at an inappropriate time or place. SIF-independent pathways for stumpy formation have been identified in vitro. For example, transcriptional attenuation at the VSG expression site is correlated with differentiation into stumpy forms, although these two activities can be uncoupled [21, 22]. The metabolic regulator AMPK was identified as a promoter of stumpy formation, acting in parallel with the TOR signaling pathway [23] and providing a connection between slender to stumpy differentiation and adenylate charge. Signals that drive stumpy formation may also come through cross-species interaction or from genetically distinct strain variants during co-infection [24]. Understanding how multivariate signaling pathways are integrated is a key challenge for future studies.
Entry into the insect vector and differentiation from stumpy to procyclic forms
Transfer from the mammalian host to the tsetse fly vector is a profound change in environment that T. brucei must accommodate. The transition brings changes in temperature and tissue interfaces, different host defense mechanisms, alterations in nutrient availability, new host microbiota, and novel chemical cues. The parasite responds by transitioning from growth-arrested stumpy forms into proliferating procyclics - a change that includes surface proteome remodeling, a shift in energy production mechanisms, global changes in gene expression, altered parasite morphology, and cell cycle re-entry.
The stumpy to procyclic transition is controlled by a phosphatase cascade in response to physical and chemical cues [25–27]. In stumpy cells, unphosphorylated PIP39 activates the protein tyrosine phosphatase PTP1, which in turn maintains PIP39 in the unphosphorylated state. Upon entry into the fly, the temperature drop facilitates uptake of citrate/cis-aconitate (CCA) and this blocks PIP39 activation of PTP1, shifting the pool of PIP39 to the phosphorylated form (PIP39~P) which then accumulates in the glycosome. Perception of CCA is facilitated by the PAD family of carboxylate transporters, which are upregulated during slender to stumpy differentiation and upon temperature drop in the fly [28]. Events downstream of PIP39~P activation are unclear but involve a combination of phosphoproteome and transcriptome changes [29].
In addition to temperature and CCA, mild acid and protease treatment are robust drivers of stumpy to procyclic differentiation [30–33]. Mild acid treatment engages the PTP1-PIP39 regulatory axis, while protease treatment operates in a separate or downstream branch of the pathway [34]. Local glucose concentration has also been studied as a potential regulator of procyclic differentiation. The glucose ingested in the tsetse blood meal is rapidly consumed, thrusting the newly introduced trypomastigotes into a glucose-depleted environment [35]. Without glucose, slender trypomastigotes quickly die in culture, as they are not pre-adapted to survive in glucose-deplete environments akin to the tsetse midgut [35]. Low glucose, alone, is unlikely to promote the development of procyclics [35]. However, in combination with cold shock, CCA, acid, and protease, low glucose conditions may facilitate or reinforce differentiation of stumpy trypomastigotes to procyclic forms at physiologically relevant rates [36]. Notably, non-metabolizable glucose analogues inhibit procyclic formation, indicating that glucose acts in a signaling capacity and not merely as a metabolite [35]. The role of glucose and overarching mechanisms of signal integration are still unclear.
Recent work has revealed a new dimension to the PTP1-PIP39 regulatory scheme by demonstrating that interaction between PIP39 and PTP1 occurs within a specific subcellular location near the flagellar pocket (FP), termed the “stumpy regulatory nexus” (STuRN)[37]. Relocation of PIP39~P to the glycosome prevents access to PTP1, reinforcing the differentiation process and enabling a non-reversible committed step [29, 37]. Notably, REG9.1, recently identified as a repressor of stumpy-specific genes [38], also localizes to the STuRN [37]. REG9.1 overexpression can lead to signal-independent procyclic differentiation for a subset of cells in the population [38], hinting at interrelationships between factors controlling slender to stumpy and stumpy to procyclic transitions. These new findings emphasize that the cellular location of signaling components plays an important, yet underappreciated, role in T. brucei differentiation processes. The proximity of the STuRN to the FP is also intriguing, given that the FP is a key window for sampling the extracellular environment. The full composition and physical make-up of the STuRN, as well as the mechanisms by which proteins are targeted to or away from the STuRN, are important questions to address with future studies.
Differentiation in insect stages
Following emergence of procyclics in the tsetse gut lumen, trypanosomes embark on a fraught journey through the alimentary tract and mouthparts to reach the salivary glands, where they differentiate into mammalian-infectious forms. This journey is essential for transmission and requires parasites to resist tsetse defenses, while coordinating multiple developmental changes with progression through specific tissues. At each developmental juncture, parasites risk clearance from the tsetse if their life cycle stage is not matched to the tissue they inhabit.
Early procyclic stage parasites in the lumen of the gut and proventriculus are characterized by a surface coat of EP and GPEET procyclins [39–41]. Eventually, late procyclic forms with repressed GPEET expression emerge, concomitant with parasite crossing of the peritrophic matrix (PM) [39–41] – a multilayered physical and biochemical barrier composed of chitin, glycoproteins, and glycosaminoglycans [42–44]. Upon crossing the PM, parasites arrive in the ectoperitrophic space, where three morphotypes – late procyclics, long procyclics, and mesocyclics – are observed [39–41]. Parasites then cross the PM a second time, moving into the proventriculus lumen [42, 44, 45] [46], where they develop into long, dividing epimastigotes and subsequently, via an asymmetric division, into short, non-dividing epimastigotes [42, 45]. Short epimastigotes ultimately arrive in the salivary glands, where they attach to the gland epithelium via their flagellar membrane [7, 45, 47]. Attached epimastigotes initiate metacyclogenesis, re-entering the cell cycle and undergoing another asymmetric division to complete the final step of differentiation into mammalian-infectious metacyclic trypomastigotes [47–49].
The complex and sophisticated developmental program observed within the tsetse fly is presumed to be orchestrated by spatially and temporally restricted, tissue-specific cues. However, the identity of these cues and the signaling systems that transduce them are almost completely unknown. There are several chemical and physical changes noted within the tsetse alimentary tract (e.g. variations in pH, reactive oxygen species, and osmolarity), but their potential as triggers of T. brucei differentiation remain to be tested. Likewise, little is known regarding downstream signaling events, although in vitro studies demonstrate that repression of GPEET during differentiation into late procyclics can be hastened in hypoxic conditions and stalled by exogenous glycerol [50]. This glycerol-dependent control is mediated by the GPEET 3’ UTR [50]. Proteome and transcriptome studies have identified several stage-specific markers that present potential effectors and targets in pathways for early to late procyclic transformation [51, 52], as well as for developmental changes in the proventriculus and salivary glands [53, 54]. Paramount among these is the discovery that RNA binding protein 6 (RBP6), when overexpressed, induces metacyclogenesis in vitro [53]. As such, RBP6 is a key effector of metacyclic differentiation, with the Q109K mutant demonstrating accelerated differentiation to metacyclic forms capable of further development to bloodstream forms in vitro [55]. Given the use of polycistronic gene expression and lack of transcriptional regulation in trypanosomes, many signaling pathways culminate in post-transcriptional regulation via RNA binding proteins [56]. Candidates for perception of signals directing metacyclogenesis include two adenylate cyclases (ACs) whose transcripts are upregulated in salivary gland parasites [54]. These are part of a large family of stage-specific ACs that localize to the flagellum [57–60] and exhibit receptor-like domain architecture, making them good candidates for initiators of second messenger signaling [61]. The final stage of the life cycle is development from metacyclic trypomastigotes into proliferating slender forms in the mammalian host, following a fly bite. Signaling events directing this transformation are completely unknown.
Metabolic reprogramming
As T. brucei parasites progress through their life cycle, they encounter environments of varying nutrient abundance and reprogram their metabolism accordingly [62, 63]. While metabolic reprogramming may merely be an adaptive maneuver to meet energy requirements, a question remains as to whether metabolites act as signals to reinforce stage differentiation [25, 36, 64], as illustrated by examples below.
Upregulation of genes involved in fat metabolism is correlated with differentiation into “adipose tissue form” (ATF) trypanosomes in the mammalian host [3]. Metabolic diversity provided by the glycosome is proposed to be important in driving differentiation events – particularly that of stumpy to procyclic forms [62]. In the glucose-depleted environment of the tsetse digestive tract, proline serves as the primary carbon source for procyclic trypanosomes [64]. RNAi knockdown of P5CDH, a component of the proline degradation pathway, results in parasites unable to metabolize proline to satisfy their energy needs. P5CDH knockdown parasites struggle to colonize the tsetse midgut and show marked growth defects in low glucose conditions in vitro [64]. Glucose is also suggested to be an important negative regulator of procyclic growth [35].
Gluconeogenesis and accumulation of reactive oxygen species (ROS) are also critical for development of infective metacyclic parasites from epimastigotes [65, 66]. Parasites with deleted fructose-1,6-bisphosphatase (Δfbpase) are unable to colonize the salivary glands [65]. When metacyclogenesis is triggered by RBP6 overexpression in vitro [53], Δfbpase parasites stall as epimastigotes and are unable to differentiate into metacyclics [65]. A recent multi-omics study has also demonstrated that depletion of ROS via expression of exogenous C. fasciculata catalase blocks progression though RBP6-induced metacyclogenesis, suggesting that ROS act as key developmental signals upon the mitochondria[66]. These studies are perhaps the best evidence that metabolic reprogramming drives stage differentiation, rather than merely being a coincident event.
Environmental cues and cell motility
Chemotaxis and role of signaling in movement of insect stage parasites
Beyond differentiation, signaling in response to environmental stimuli canonically controls cell movement [67, 68] and it is reasonable to expect this is the case for T. brucei. As discussed above, T. brucei movement to specific tissues is required for transmission and is a defining step in disease pathogenesis. Historically, external factors influencing motility of trypanosomes have proved enigmatic to study. Yet in recent years, the combination of in vitro social motility (SoMo) assays and improved in vivo imaging modalities have shed light on the ability of T. brucei to adjust its motility in response to external signals and has greatly advanced our understanding of parasite movement through tissues.
A series of in vitro experiments have demonstrated capacity for both negative and positive chemotaxis in procyclic forms. SoMo was first identified in procyclic T. brucei in 2010 [69], and has since been concisely reviewed [70, 71]. In brief, when placed on semisolid agarose, parasites collect in groups that migrate outward, generating evenly-spaced radial projections [69]. Parasites can move outward from any edge of these projections, but group movement is polarized in one direction and parasites at the leading edge actively avoid neighboring projections – demonstrating behavior consistent with negative chemotaxis [69]. Additional studies demonstrated that SoMo initiation is dependent on cell number and restricted to specific developmental stages [51]. This supports at least two signaling events: one to control SoMo initiation and a second to enable avoidance behavior. In a recent advance, DeMarco and colleagues provided the first evidence of positive chemotaxis [72]. Using SoMo assays with procyclic T. brucei, they identified a behavior termed ‘BacSoMo,’ in which parasites dramatically alter their movement toward bacteria [72]. This study showed that procyclic forms are attracted to a soluble factor derived from living E. coli that acts at a distance to alter parasite motility. Upon sensing this cue, individual parasite movement within projections grew more constrained, while projections accelerated movement at the leading edge and produced new outgrowths toward the bacteria [72]. Beyond demonstrating positive chemotaxis, the finding that parasites are affected by bacteria suggests an ability to respond to bacterial symbionts in the tsetse vector [73].
Evidence of clear chemotactic responses and a tractable in vitro assay by which to study underlying mechanisms provide a critical step in defining signaling mechanisms that direct trypanosome movement. To this end, further work has shown that the SoMo signaling machinery relies upon spatial and temporal regulation of cAMP [70, 74, 75]. cAMP levels are titrated through oppositional action of cAMP-generating ACs and cAMP-hydrolyzing phosphodiesterases (PDEs) [76]. Upon RNAi knockdown or mutational inactivation of procyclic-specific AC6 and AC1/AC2, parasites exhibit a hypersocial phenotype apparently due to decreased cAMP levels [74]. Correspondingly, chemical or genetic inhibition of the phosphodiesterase PDEB1, inhibits SoMo. SoMo inhibition correlates directly with increased intracellular cAMP in live parasites, and the PDEB1 knockdown phenotype is phenocopied by addition of non-hydrolyzable cAMP analogues [75]. Therefore, SoMo is regulated by flux through the cAMP signaling pathway and inhibition is due to cAMP directly, rather than downstream metabolites.
In light of robust data on chemotaxis and regulated cell motility generated through in vitro SoMo assays, a key question emerges: does in vitro SoMo reflect similar activities in the tsetse? Importantly, SoMo is restricted to early procyclics, leading to the hypothesis that this behavior is linked to successful circumnavigation to the ectoperitrophic space [51, 70]. Further work established a relationship between factors affecting SoMo and those required for fly infection, as RTF1 knockout parasites present with defective SoMo and difficulties establishing a midgut infection [70, 77]. Significantly, Shaw and DeMarco et al. demonstrated that ablation of SoMo in vitro through PDEB1 knockout correlated with a block in parasite movement from the midgut lumen into the ectoperitrophic space [42]. PDEB1 knockouts were able to establish a midgut infection, exhibited normal stage differentiation in the tsetse, and showed processive motility in vitro [42]. The combined data indicate that the inability to move beyond the midgut lumen stems from a defect in control of motility direction, rather than a defect in motility itself [42]. Therefore, cAMP signaling, in addition to being required for in vitro SoMo, is critical for traversing the PM in vivo [42], consistent with early studies suggesting correlation between AC activity and movement through the fly [78, 79]. Whether the impact on motility manifests in the tsetse as it does in in vitro SoMo remains to be determined, but it is clear that SoMo assays are an excellent tool for dissecting chemotaxis and underlying signaling pathways relevant for progression through the tsetse fly.
Despite great strides in establishing the essentiality of cAMP signaling for control of parasite motility and for completion of the transmission cycle, the extracellular cue(s) and putative receptor(s) guiding these events remain undefined. It has been posited that flagellar ACs operate as receptors, transducing extracellular cues via cAMP response proteins (CARPs) that mediate downstream responses [80]; however, rigorous biochemical analysis is needed to further interrogate this idea. Notably, T. brucei ACs are a large gene family whose sequences differ primarily in the extracellular, putative ligand-binding domain [61, 80]. Moreover, while some ACs function in SoMo, others do not [74], indicating different functions for different ACs. Consistent with this idea, different ACs exhibit distinct expression profiles as parasites progress through various life cycle stages [54, 60, 77]. Differences compared to the mammalian host make the T. brucei cAMP signaling pathway an attractive therapeutic target [80, 81]. Extracellular cues that direct parasite movement could come from neighboring parasites, from host tissues, or from microbiota. Given that the PDEB1-knockdown SoMo phenotype is trans-complemented upon co-culture with wild-type parasites [75], it stands to reason that the signal initiating SoMo can be parasite-derived [82]. Parasite-produced exosomes have been suggested to confer the negative chemotactic cues instigating avoidance behavior in SoMo [83]. From our limited understanding of the tsetse alimentary tract, there are a number of candidate signals that are worth exploring, such as cooler temperatures (20°C, versus 37°C in the mammalian bloodstream), changes in nutrient availability, and a pH gradient distributed from the posterior midgut (pH 7.9±0.4) to the proventriculus (pH 10.6±0.6) [64, 84, 85]. Parasites also encounter gram-negative microbial endosymbionts in the tsetse, namely Sodalis glossinidus, Wigglesworthia glossinidia, and Wolbachia spp. [73]. As parasites demonstrate positive chemotaxis toward gram-negative E. coli in BacSoMo [72], the possibility that factors derived from endogenous tsetse microbiota impact T. brucei movement should be explored.
As the new developments highlighted above provide insight into signaling mechanisms that control T. brucei motility, improvements in imaging and microscopy have enabled better characterization of trypanosome movement within tsetse tissues [44, 86–88]. In a groundbreaking study, Schuster and colleagues implemented multicolor light sheet fluorescence microscopy and high-speed fluorescence microscopy to meticulously trace the migration of T. brucei parasites through the tsetse alimentary tract [86]. These experiments i) generated a high-resolution, three-dimensional model of the tsetse alimentary tract; ii) defined structural and morphological parameters dictating parasite movement across life-cycle stages – including flagellar waveforms; and iii) demarcated areas in which parasites exhibit solitary or collective swimming behaviors [86]. In some areas, procyclic parasites adjacent to the PM exhibited collective motility, in which flagellar oscillations in groups of parasites were synchronized and produced tissue deforming force [86]. The ability of adjacent parasites to synchronize flagellar beating indicates capacity for mechanical signaling, perhaps similar to that enabling synchronized ciliary beating in ciliates and mammalian airway epithelial cells [89]. This study also showed a population of parasites congregating near a doughnut-shaped structure at the entrance of the proventriculus – an area where specialized tsetse cells actively synthesize the PM [86]. The authors posited that this could be a possible location of parasite PM crossing, rather than the traditional model of parasites penetrating the PM in the anterior midgut [39, 86]. This hypothesis was further bolstered by Rose et al. 2020, wherein studies combining confocal laser scanning microscopy, transmission electron microscopy (EM), scanning EM, and serial block-face scanning EM indicate that procyclic forms enter the ectoperitrophic space by traversing an immature and more fluid PM, near the proventriculus [39]. These advances in imaging trypanosome movement in vivo now set the stage to more specifically demarcate the steps of the transmission cycle that are blocked in parasite signaling mutants.
In order to control T. brucei motility, signaling cascades must ultimately target the flagellum. As such, it is noteworthy that all ACs examined thus far are located in the flagellum [59, 60], as are PDEs [90, 91]. Additionally, there are important spatial relationships within the flagellum, with some ACs restricted to the flagellar tip and others located along the entire flagellum length [60]. Based on localization of AC6 to the flagellum tip, a model is posited in which SoMo is regulated by local cAMP levels at the flagellum tip rather than bulk cellular cAMP [74, 75]. The importance of flagellar cAMP signaling pathways to parasite motility and transmission [42, 74, 75] also demonstrates that the concept of the eukaryotic flagellum as a platform for sensing and signal transduction [92, 93] extends to pathogenic protozoa. These findings further emphasize the emerging concept that the eukaryotic flagellum is composed of distinct domains, each having specialized composition and function [60, 74]. The importance for cellular location of signaling components is thus an emerging pattern in T. brucei signaling, being observed here and, as discussed above, with the STuRN as a nexus for regulation of stumpy to procyclic development [37]. As a sensory organelle, the T. brucei flagellum is ripe for future studies, particularly for elucidating connections between extracellular signaling and control of parasite motility.
Tissue tropism in the mammalian most
While the variety of tissues traversed in the tsetse vector is well-established, our understanding of the extent and role for tissue tropisms seen in the mammalian host has recently expanded. For decades T. brucei was canonically characterized as living freely in blood, invading the lymph, and – during late stage infections – escaping the bloodstream to penetrate the CNS [1]. Yet in recent years, seminal studies have demonstrated that skin and adipose tissues are important reservoirs for both proliferative and transmissible morphotypes [94] [5, 95]. Intravital imaging has provided insights into infection dynamics, supporting the view that cellular development occurs in these compartments [5, 95, 96].
Movement of parasites to extravascular compartments influences disease pathogenesis and transmission. Entry into the CNS is a defining step in HAT pathogenesis because it marks onset of the lethal stage of the disease and dictates which drugs can be used for treatment [97]. Parasites in the skin are competent for transmission through the tsetse fly and field studies in endemic areas identified parasites in the dermis of people for whom no parasites could be detected in blood, lymph, or CNS [5, 94]. Therefore, in addition to influencing transmission dynamics, skin-dwelling trypanosomes present a hitherto unrealized reservoir in asymptomatic patients, posing challenges for surveillance in the effort to eradicate HAT [5, 94, 98]. The impact of adipose trypanosomes on disease pathogenesis is unclear, but they are suggested to contribute to weight loss [3] via differentiation into ATFs that have adapted their metabolic capacity for beta-oxidation of fatty acids and have potentially altered their VSG expression dynamics [3]. Importantly, while some parasites may obtain access to dermal tissues during a tsetse blood meal [96], they are also able to access the skin from the bloodstream [95]. Likewise, parasites find their way to adipose tissue remote from the infection site [3]. Although it remains to be tested, it is also possible that parasites in skin and adipose might return to the bloodstream – as has been seen for CNS parasites [99] – thus presenting reservoirs for relapse infection.
Mechanisms underlying parasite movement to and from different tissues in the mammalian host are completely unknown. Future studies should explore if these movements are directed by chemotactic signaling in response to tissue-specific cues, particularly given the demonstration of chemotaxis in insect stages [72]. Notably, parasites in the dermis were observed burrowing their anterior flagellum into dermal adipocytes [95], suggesting a possible homing mechanism localized to the flagellar tip. In addition to the potential need for chemotaxis signaling pathways, cellular differentiation observed in extracellular tissues [3, 5] likely depends on signaling pathways responsive to tissue-specific cues.
Conclusion
Environmental sensing and signaling in T. brucei are imperative for disease transmission and pathogenesis. Recent work has advanced our understanding of signal transduction systems that drive cellular differentiation and control cell movement in these organisms. In many cases, these advances have been driven by innovative technologies and conceptual approaches, establishing diverse tools for future discovery. While numerous questions still persist, the divergence of T. brucei signaling systems – as compared to those of its mammalian hosts – emerges as a common theme. Further studies must interrogate if these differences can be exploited for development of novel therapeutics or transmission blocking agents. Within the field, appreciation is also growing for the role of tissue tropism in the manifestation of disease and sustained potential for transmission from asymptomatic individuals. Looking forward, studies investigating factors that underlie tissue tropism and the means by which external signals influence parasite motility are warranted. Here again, beyond advancing fundamental biology, such studies offer potential to illuminate novel strategies for intervention.
Figure 2.
Major questions to address regarding environmental sensing and signal transduction in T. brucei.
Acknowledgments
We are grateful to members of the Hill laboratory for thoughtful comments on the manuscript. Breanna Walsh is supported by the UCLA MSTP T32 GM008042 training grant. Work in the Hill laboratory is supported by NIH grants, AI052348, AI133166, AI142544. Figure 1 and the image of the graphical abstract were created with BioRender.com.
Figure 1. T. brucei development is coordinated with movement through host tissues.
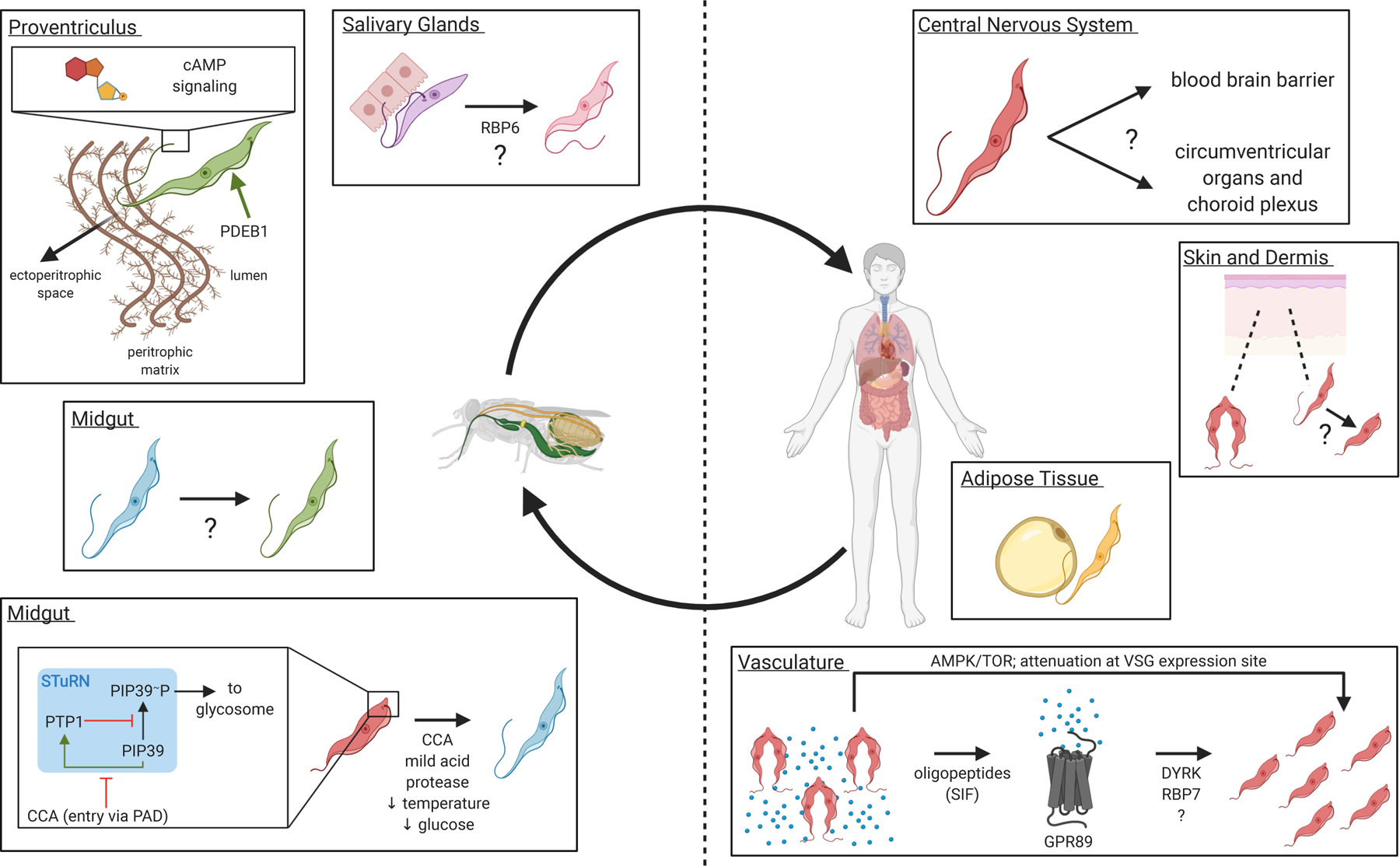
Individual panels show developmental changes and tissue tropisms observed in the T. brucei life cycle, with major components of the underlying signaling pathways shown where these are known. In the mammalian host, entry to the central nervous system may occur through the blood brain barrier or circumventricular organs/choroid plexus, but factors involved are not understood. Dermal and adipose tissues represent physiologically important reservoirs. Differentiation to transmission competent forms in the dermis (red, stumpy) and to adipose tissue forms (yellow) in adipose indicate response to local cues. Factors controlling movement between these sites and the vasculature are unknown. Within the vasculature, differentiation from proliferating slender forms (red, slender) to growth-arrested stumpy forms (red, stumpy) is driven by a quorum sensing system in which oligopeptides (blue dots) accumulate with increasing parasite density and these are perceived by the GPR89 oligopeptide transporter. Differentiation from stumpy forms (red, stumpy) to early procyclic forms (blue) in the tsetse fly midgut is controlled by an autoregulatory phosphatase pathway, with key regulatory proteins localized to a specialized “Stumpy Regulatory Nexus” (STuRN) near the base of the flagellum. In the midgut, early procyclic forms differentiate into late procyclic forms (green). Within the proventriculus, procyclic forms depend on cAMP signaling pathways in the flagellum to penetrate the peritrophic matrix and enter the ectoperitrophic space. Parasites ultimately travel to the salivary glands, where proliferating epimastigotes (purple) attach via their flagellum to the gland epithelium and differentiate into growth-arrested metacyclic trypomastigotes (pink) that are readied for the bloodstream of the mammalian host. Some steps of the life cycle are not shown (e.g. differentiation to mesocyclics and long and short epimastigotes in the proventriculus and differentiation of metacyclic trypomastigotes into slender forms in the bloodstream).
Footnotes
The authors have no conflict of interest.
REFERENCES
- 1.Kennedy PGE, Human African trypanosomiasis of the CNS: current issues and challenges. The Journal of clinical investigation, 2004. 113(4): p. 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franco JR, et al. , Monitoring the elimination of human African trypanosomiasis: Update to 2016. PLOS Neglected Tropical Diseases, 2018. 12(12): p. e0006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trindade S, et al. , Trypanosoma brucei Parasites Occupy and Functionally Adapt to the Adipose Tissue in Mice. Cell Host Microbe, 2016. 19(6): p. 837–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy PGE and Rodgers J, Clinical and Neuropathogenetic Aspects of Human African Trypanosomiasis. Frontiers in immunology, 2019. 10: p. 39–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capewell P, et al. , The skin is a significant but overlooked anatomical reservoir for vector-borne African trypanosomes. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandela WR, et al. , Prevalence and control implications of bovine trypanosomes in endemic areas of northern Uganda. Trop Anim Health Prod, 2020. [DOI] [PubMed]
- 7.Silvester E, McWilliam KR, and Matthews KR, The Cytological Events and Molecular Control of Life Cycle Development of Trypanosoma brucei in the Mammalian Bloodstream. Pathogens, 2017. 6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodgers J, Trypanosomiasis and the brain. Parasitology, 2010. 137(14): p. 1995–2006. [DOI] [PubMed] [Google Scholar]
- 9.MacGregor P, et al. , Trypanosomal immune evasion, chronicity and transmission: an elegant balancing act. Nat Rev Microbiol, 2012. 10(6): p. 431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vickerman K, Antigenic variation in trypanosomes. Nature, 1978. 273(5664): p. 613–7. [DOI] [PubMed] [Google Scholar]
- 11.MacGregor P, et al. , Transmission stages dominate trypanosome within-host dynamics during chronic infections. Cell host & microbe, 2011. 9(4): p. 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro SZ, et al. , Analysis by flow cytometry of DNA synthesis during the life cycle of African trypanosomes. Acta Trop, 1984. 41(4): p. 313–23. [PubMed] [Google Scholar]
- 13.Vassella E, et al. , Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci, 1997. 110 (Pt 21): p. 2661–71. [DOI] [PubMed] [Google Scholar]
- 14.Dewar CE, et al. , Mitochondrial DNA is critical for longevity and metabolism of transmission stage Trypanosoma brucei. PLoS Pathog, 2018. 14(7): p. e1007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuner B, et al. , Cell density triggers slender to stumpy differentiation of Trypanosoma brucei bloodstream forms in culture. Mol Biochem Parasitol, 1997. 90(1): p. 269–80. [DOI] [PubMed] [Google Scholar]
- 16.Rojas F, et al. , Oligopeptide Signaling through TbGPR89 Drives Trypanosome Quorum Sensing. Cell, 2019. 176(1–2): p. 306–317.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford W, et al. , Eukaryotic G protein signaling evolved to require G protein-coupled receptors for activation. Sci Signal, 2013. 6(276): p. ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mony BM, et al. , Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature, 2014. 505(7485): p. 681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald L, et al. , Non-linear hierarchy of the quorum sensing signalling pathway in bloodstream form African trypanosomes. PLoS pathogens, 2018. 14(6): p. e1007145–e1007145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cayla M, et al. , An atypical DYRK kinase connects quorum-sensing with posttranscriptional gene regulation in Trypanosoma brucei. Elife, 2020. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McWilliam KR, et al. , Developmental competence and antigen switch frequency can be uncoupled in Trypanosoma brucei. Proc Natl Acad Sci U S A, 2019. 116(45): p. 22774–22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann H, et al. , A quorum sensing-independent path to stumpy development in Trypanosoma brucei. PLoS Pathog, 2017. 13(4): p. e1006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saldivia M, et al. , The AMPKalpha1 Pathway Positively Regulates the Developmental Transition from Proliferation to Quiescence in Trypanosoma brucei. Cell Rep, 2016. 17(3): p. 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silvester E, et al. , Interspecies quorum sensing in co-infections can manipulate trypanosome transmission potential. Nat Microbiol, 2017. 2(11): p. 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szoor B, Silvester E, and Matthews KR, A Leap Into the Unknown - Early Events in African Trypanosome Transmission. Trends Parasitol, 2020. 36(3): p. 266–278. [DOI] [PubMed] [Google Scholar]
- 26.Szoor B, et al. , Protein tyrosine phosphatase TbPTP1: A molecular switch controlling life cycle differentiation in trypanosomes. J Cell Biol, 2006. 175(2): p. 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szoor B, et al. , A novel phosphatase cascade regulates differentiation in Trypanosoma brucei via a glycosomal signaling pathway. Genes Dev, 2010. 24(12): p. 1306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dean S, et al. , A surface transporter family conveys the trypanosome differentiation signal. Nature, 2009. 459(7244): p. 213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domingo-Sananes MR, et al. , Molecular control of irreversible bistability during trypanosome developmental commitment. J Cell Biol, 2015. 211(2): p. 455–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolin S, et al. , Mild acid stress as a differentiation trigger in Trypanosoma brucei. Mol Biochem Parasitol, 1998. 93(2): p. 251–62. [DOI] [PubMed] [Google Scholar]
- 31.Hunt M, Brun R, and Kohler P, Studies on compounds promoting the in vitro transformation of Trypanosoma brucei from bloodstream to procyclic forms. Parasitol Res, 1994. 80(7): p. 600–6. [DOI] [PubMed] [Google Scholar]
- 32.Sbicego S, et al. , The use of transgenic Trypanosoma brucei to identify compounds inducing the differentiation of bloodstream forms to procyclic forms. Mol Biochem Parasitol, 1999. 104(2): p. 311–22. [DOI] [PubMed] [Google Scholar]
- 33.Yabu Y and Takayanagi T, Trypsin-stimulated transformation of Trypanosoma brucei gambiense bloodstream forms to procyclic forms in vitro. Parasitol Res, 1988. 74(6): p. 501–6. [DOI] [PubMed] [Google Scholar]
- 34.Szoor B, et al. , Independent pathways can transduce the life-cycle differentiation signal in Trypanosoma brucei. PLoS Pathog, 2013. 9(10): p. e1003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu Y, et al. , Glucose Signaling Is Important for Nutrient Adaptation during Differentiation of Pleomorphic African Trypanosomes. mSphere, 2018. 3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clayton C, Novel Observations Concerning Differentiation of Bloodstream-Form Trypanosomes to the Form That Is Adapted for Growth in Tsetse Flies. mSphere, 2018. 3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szöőr B, et al. , Positional Dynamics and Glycosomal Recruitment of Developmental Regulators during Trypanosome Differentiation. mBio, 2019. 10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rico E, et al. , Genome-wide RNAi selection identifies a regulator of transmission stage-enriched gene families and cell-type differentiation in Trypanosoma brucei. PLoS Pathog, 2017. 13(3): p. e1006279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose C, et al. , Trypanosoma brucei colonizes the tsetse gut via an immature peritrophic matrix in the proventriculus. Nat Microbiol, 2020. [DOI] [PubMed]
- 40.Acosta-Serrano A, et al. , The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci U S A, 2001. 98(4): p. 1513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vassella E, et al. , Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J Mol Biol, 2001. 312(4): p. 597–607. [DOI] [PubMed] [Google Scholar]
- 42.Shaw S, et al. , Flagellar cAMP signaling controls trypanosome progression through host tissues. Nat Commun, 2019. 10(1): p. 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehane MJ, Peritrophic matrix structure and function. Annu Rev Entomol, 1997. 42: p. 525–50. [DOI] [PubMed] [Google Scholar]
- 44.Rose C, et al. , An investigation into the protein composition of the teneral Glossina morsitans morsitans peritrophic matrix. PLoS Negl Trop Dis, 2014. 8(4): p. e2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rotureau B and Van Den Abbeele J, Through the dark continent: African trypanosome development in the tsetse fly. Front Cell Infect Microbiol, 2013. 3: p. 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigneron A, et al. , Single-cell RNA sequencing of Trypanosoma brucei from tsetse salivary glands unveils metacyclogenesis and identifies potential transmission blocking antigens. Proc Natl Acad Sci U S A, 2020. 117(5): p. 2613–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rotureau B, et al. , A new asymmetric division contributes to the continuous production of infective trypanosomes in the tsetse fly. Development, 2012. 139(10): p. 1842–50. [DOI] [PubMed] [Google Scholar]
- 48.Vickerman K, On the surface coat and flagellar adhesion in trypanosomes. J Cell Sci, 1969. 5(1): p. 163–93. [DOI] [PubMed] [Google Scholar]
- 49.Vickerman K, Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull, 1985. 41(2): p. 105–14. [DOI] [PubMed] [Google Scholar]
- 50.Vassella E, et al. , A major surface glycoprotein of trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes & development, 2000. 14(5): p. 615–626. [PMC free article] [PubMed] [Google Scholar]
- 51.Imhof S, et al. , Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission. PLOS Pathogens, 2014. 10(10): p. e1004493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naguleswaran A, Doiron N, and Roditi I, RNA-Seq analysis validates the use of culture-derived Trypanosoma brucei and provides new markers for mammalian and insect life-cycle stages. BMC Genomics, 2018. 19(1): p. 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolev NG, et al. , Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science, 2012. 338(6112): p. 1352–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savage AF, et al. , Transcriptome Profiling of Trypanosoma brucei Development in the Tsetse Fly Vector Glossina morsitans. PLoS One, 2016. 11(12): p. e0168877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi H, Butler K, and Tschudi C, A single-point mutation in the RNA-binding protein 6 generates Trypanosoma brucei metacyclics that are able to progress to bloodstream forms in vitro. Mol Biochem Parasitol, 2018. 224: p. 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trenaman A, et al. , A post-transcriptional respiratome regulon in trypanosomes. Nucleic Acids Res, 2019. 47(13): p. 7063–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexandre S, et al. , Families of adenylate cyclase genes in Trypanosoma brucei. Mol Biochem Parasitol, 1996. 77(2): p. 173–82. [DOI] [PubMed] [Google Scholar]
- 58.Alexandre S, et al. , Differential expression of a family of putative adenylate/guanylate cyclase genes in Trypanosoma brucei. Molecular and Biochemical Parasitology, 1990. 43(2): p. 279–88. [DOI] [PubMed] [Google Scholar]
- 59.Paindavoine P, et al. , A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei. Mol Cell Biol, 1992. 12(3): p. 1218–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saada EA, et al. , Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei Flagellar membrane. Eukaryot Cell, 2014. 13(8): p. 1064–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seebeck T, et al. , cAMP signalling in Trypanosoma brucei. International Journal for Parasitology, 2001. 31(5–6): p. 491–8. [DOI] [PubMed] [Google Scholar]
- 62.Szoor B, et al. , Evolution, dynamics and specialized functions of glycosomes in metabolism and development of trypanosomatids. Curr Opin Microbiol, 2014. 22: p. 79–87. [DOI] [PubMed] [Google Scholar]
- 63.Smith TK, et al. , Metabolic reprogramming during the Trypanosoma brucei life cycle. F1000Res, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mantilla BS, et al. , Proline Metabolism is Essential for Trypanosoma brucei brucei Survival in the Tsetse Vector. PLoS pathogens, 2017. 13(1): p. e1006158–e1006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wargnies M, et al. , Gluconeogenesis is essential for trypanosome development in the tsetse fly vector. PLOS Pathogens, 2018. 14(12): p. e1007502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dolezelova E, et al. , Cell-based and multi-omics profiling reveals dynamic metabolic repurposing of mitochondria to drive developmental progression of Trypanosoma brucei. PLoS Biol, 2020. 18(6): p. e3000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Haastert PJ and Devreotes PN, Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol, 2004. 5(8): p. 626–34. [DOI] [PubMed] [Google Scholar]
- 68.Wadhams GH and Armitage JP, Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol, 2004. 5(12): p. 1024–37. [DOI] [PubMed] [Google Scholar]
- 69.Oberholzer M, et al. , Social Motility in African Trypanosomes. PLOS Pathogens, 2010. 6(1): p. e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saada EA, et al. , “With a Little Help from My Friends”-Social Motility in Trypanosoma brucei. PLoS pathogens, 2015. 11(12): p. e1005272–e1005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roditi I, Schumann G, and Naguleswaran A, Environmental sensing by African trypanosomes. Curr Opin Microbiol, 2016. 32: p. 26–30. [DOI] [PubMed] [Google Scholar]
- 72.DeMarco SF, et al. , Identification of Positive Chemotaxis in the Protozoan Pathogen Trypanosoma brucei. mSphere, 2020. 5(4): p. e00685–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J, Weiss B, and Aksoy S, Tsetse fly microbiota: form and function. Frontiers in Cellular and Infection Microbiology, 2013. 3(69). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopez MA, Saada EA, and Hill KL, Insect stage-specific adenylate cyclases regulate social motility in african trypanosomes. Eukaryot Cell, 2015. 14(1): p. 104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oberholzer M, Saada EA, and Hill KL, Cyclic AMP Regulates Social Behavior in African Trypanosomes. MBio, 2015. 6(3): p. e01954–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gould MK and de Koning HP, Cyclic-nucleotide signalling in protozoa. FEMS Microbiol Rev, 2011. 35(3): p. 515–41. [DOI] [PubMed] [Google Scholar]
- 77.Imhof S, et al. , A Glycosylation Mutant of Trypanosoma brucei Links Social Motility Defects In Vitro to Impaired Colonization of Tsetse Flies In Vivo. Eukaryot Cell, 2015. 14(6): p. 588–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rolin S, et al. , Transient adenylate cyclase activation accompanies differentiation of Trypanosoma brucei from bloodstream to procyclic forms. Mol Biochem Parasitol, 1993. 61(1): p. 115–25. [DOI] [PubMed] [Google Scholar]
- 79.van den Abbeele J, et al. , Trypanosoma brucei: stimulation of adenylate cyclase by proventriculus and esophagus tissue of the tsetse fly, Glossina morsitans morsitans. Exp Parasitol, 1995. 81(4): p. 618–20. [DOI] [PubMed] [Google Scholar]
- 80.Tagoe DN, Kalejaiye TD, and de Koning HP, The ever unfolding story of cAMP signaling in trypanosomatids: vive la difference! Front Pharmacol, 2015. 6: p. 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Koning HP, et al. , Pharmacological validation of Trypanosoma brucei phosphodiesterases as novel drug targets. Journal of Infectious Diseases, 2012. 206(2): p. 229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salmon D, Adenylate Cyclases of Trypanosoma brucei, Environmental Sensors and Controllers of Host Innate Immune Response. Pathogens (Basel, Switzerland), 2018. 7(2): p. 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eliaz D, et al. , Exosome secretion affects social motility in Trypanosoma brucei. PLoS Pathog, 2017. 13(3): p. e1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liniger M, et al. , Cleavage of trypanosome surface glycoproteins by alkaline trypsin-like enzyme(s) in the midgut of Glossina morsitans. International Journal for Parasitology, 2003. 33(12): p. 1319–1328. [DOI] [PubMed] [Google Scholar]
- 85.Engstler M and Boshart M, Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes & development, 2004. 18(22): p. 2798–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schuster S, et al. , Developmental adaptations of trypanosome motility to the tsetse fly host environments unravel a multifaceted in vivo microswimmer system. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gibson W and Bailey M, The development of Trypanosoma brucei within the tsetse fly midgut observed using green fluorescent trypanosomes. Kinetoplastid Biol Dis, 2003. 2(1): p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vigneron A, et al. , A fine-tuned vector-parasite dialogue in tsetse’s cardia determines peritrophic matrix integrity and trypanosome transmission success. PLoS Pathog, 2018. 14(4): p. e1006972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamilton E, et al. , Motile cilia hydrodynamics: entrainment versus synchronization when coupling through flow. Philos Trans R Soc Lond B Biol Sci, 2020. 375(1792): p. 20190152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dean S, Sunter JD, and Wheeler RJ, TrypTag.org: A Trypanosome Genome-wide Protein Localisation Resource. Trends Parasitol, 2017. 33(2): p. 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oberholzer M, et al. , The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. Faseb J, 2007. 21(3): p. 720–31. [DOI] [PubMed] [Google Scholar]
- 92.Anvarian Z, et al. , Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol, 2019. 15(4): p. 199–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oberholzer M, et al. , Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol Cell Proteomics, 2011. 10(10): p. M111.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Camara M, et al. , Extravascular dermal trypanosomes in suspected and confirmed cases of gambiense Human African Trypanosomiasis. Clin Infect Dis, 2020. [DOI] [PMC free article] [PubMed]
- 95.Caljon G, et al. , The Dermis as a Delivery Site of Trypanosoma brucei for Tsetse Flies. PLoS Pathog, 2016. 12(7): p. e1005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.De Niz M, et al. , Intravital imaging of host-parasite interactions in skin and adipose tissues. Cell Microbiol, 2019. 21(5): p. e13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Babokhov P, et al. , A current analysis of chemotherapy strategies for the treatment of human African trypanosomiasis. Pathog Glob Health, 2013. 107(5): p. 242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Informal Expert Group on Gambiense, H.A.T.R., et al. , Do Cryptic Reservoirs Threaten Gambiense-Sleeping Sickness Elimination? Trends Parasitol, 2018. 34(3): p. 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gray GD, Jennings FW, and Hajduk SL, Relapse of monomorphic and pleomorphic Trypanosoma brucei infections in the mouse after chemotherapy. Z Parasitenkd, 1982. 67(2): p. 137–45. [DOI] [PubMed] [Google Scholar]


