Abstract
The p53 tumor suppressor gene is mutated in over 50% of human cancers, resulting in inactivation of the wild-type (wt) p53 protein. The most notable biochemical feature of p53 is its ability to act as a sequence-specific transcriptional activator. Through use of the suppression subtractive hybridization differential screening technique, we identified c-fos as a target for transcriptional stimulation by p53 in cells undergoing p53-mediated apoptosis. Overexpression of wt p53 induces c-fos mRNA and protein. Moreover, in vivo induction of c-fos in the thymus following whole-body exposure to ionizing radiation is p53 dependent. p53 responsiveness does not reside in the basal c-fos promoter. Rather, a distinct region within the c-fos gene first intron binds specifically to p53 and confers upon the c-fos promoter the ability to become transcriptionally activated by wt p53. Identification of c-fos as a specific target for transcriptional activation by p53 establishes a direct link between these two pivotal regulatory proteins and raises the possibility that c-fos contributes to some of the biological effects of p53.
The p53 tumor suppressor gene plays a central role in the prevention of cancer through its ability to recruit several signaling pathways toward the regulation of cell fate (reviewed in references 4, 17, 21, 29, and 33). Mutations in the gene for p53 which inactivate its biological and biochemical functions are found in about half of all human cancers (23).
Most notable among the biochemical activities of p53 is its ability to mediate sequence-specific transactivation of genes harboring distinct p53-binding elements. Positive regulation of target genes by p53 has been implicated in the two major biological outcomes of p53 activation, growth arrest and apoptosis (21, 29, 33). p53-mediated G1 arrest is largely brought about by induction of the cyclin-dependent kinase inhibitor p21/Waf1 (13). Similarly, the p53 target BTG2 (44) and 14-3-3 ς (22) genes have been implicated in the control of the G2-M checkpoint by p53.
p53 can mediate apoptosis under a variety of physiological and pathological conditions (4, 7, 17, 48, 49). A growing number of p53-inducible genes have been suggested to be involved in this process, including those for bax (37), IGF-BP3 (6), Fas/APO1 (38), p85 (50), and PAG608 (24) and several redox-related genes (40). This diverse list suggests that p53 mediates apoptosis through several independent pathways. In addition, p53 may also utilize transcriptionally independent pathways toward induction of apoptosis (reviewed in references 4, 21, and 49).
We employed the suppression subtractive hybridization (SSH) method (10) to identify new target genes that are induced by p53 in cells undergoing p53-mediated apoptosis. Surprisingly, one strongly p53-inducible cDNA was found to correspond to the mouse c-fos proto-oncogene, suggesting that c-fos is a target for positive regulation by p53. The c-fos protein is a constituent of the AP-1 transcription factor complex (reviewed in references 1 and 26). Changes in c-fos expression have been implicated in a variety of biological processeses, including proliferation, differentiation, tumorigenesis, and apoptosis. Previously, the c-fos basal promoter was shown to be repressed in cells possessing very high levels of wild-type (wt) p53 activity (15, 28, 45). By using more physiological levels of p53 in this study, we demonstrated that, unlike the c-fos basal promoter when studied in isolation, the c-fos gene as a whole is actually positively regulated by p53, giving rise to a p53-dependent increase in both c-fos mRNA and protein. This effect is mediated through a distinct element within the first intron of the c-fos gene which binds specifically to p53. These findings establish c-fos as a new p53 target gene whose activation may contribute to the downstream effects of p53.
MATERIALS AND METHODS
Cell lines.
M1, LTR6 (51), and H1299 (36) cells were maintained routinely at 37°C in RPMI medium supplemented with 10% fetal calf serum (FCS). DA-1 (16), MCO1 (3), MCO1-cG9 (3), Clone6 (39), and F89 (normal human diploid fibroblast) cells were maintained at 37°C in Dulbecco modified Eagle medium supplemented with 10% FCS. MCO-1 cells are mouse fibrosarcoma cells devoid of p53 expression (20), while MCO1-cG9 cells are MCO1 derivatives stably transfected with temperature-sensitive (ts) mutant protein p53val135 (2).
RNA analysis.
Total RNA was extracted by using either the RNAzol or the ULTRASPEC (Biotex Laboratories) reagent. Northern blot analysis was performed as previously described (14), by using mouse c-fos and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probes. Semiquantitative reverse transcription (RT)-PCR was performed as previously described (24), by using the following primer combinations: mouse c-fos, 5′GCTGACAGATACACTCCAAGCGG3′ and 5′AGGAAGACGTGTAAGTAGTGCAG3′ or 5′GGTTTCAACGCCGACTACGAG3′ and 5′CTCCTCCGATTCCGGCACTT3′; rat c-fos, 5′GATCTGTCCGTCTCTAGTGCCAAC3′ and 5′CTCCTCCGATTCCGGCACTT3′; human c-fos, 5′CCTCACCCTTTCGGAGTCCC3′ and 5′CTCCTTCAGCAGGTTGGCAATCT3′; GAPDH, 5′CAGCAATGCATCCTGCACC3′ and 5′TGGACTGTGGTCATGAGCCC3′.
SSH.
SSH (10) was utilized to produce a cDNA library enriched for p53 target genes. The starting material consisted of two poly(A)+ RNA populations extracted from either M1 or LTR6 cells incubated for 4 h at 32°C. After removal of adapters, the cDNA clones present in the enriched library were ligated into vector pBLKS+. Positive clones were individually amplified by PCR using T3 and T7 primers. The PCR products were separated on agarose gels, transferred to membranes, and hybridized against radiolabeled population probes prepared by radiolabeling the same poly(A)+ RNA pools used as the starting material for SSH. In addition to the enriched clones, each gel also included GAPDH and PAG608 (24) cDNAs as negative and positive controls, respectively.
Protein analysis.
Nuclear extracts were prepared from M1, LTR6, or Clone6 cells incubated at either 37 or 32°C as previously described (53). Extract proteins (100 μg per lane) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blotted, and reacted with a c-fos antibody (SC-52; Santa-Cruz). Blots were developed with the SuperSignal enhanced-chemiluminescence system (Pierce).
Reporter plasmids, transfections, and gel mobility shift assays.
A genomic fragment containing the murine c-fos promoter (550 bp), exon 1, and intron 1 was obtained from mouse genomic DNA by PCR amplification using primers 5′GGGGTACCAAAAAAAGTTCCAGATTGCTGGAC3′ and 5′GGGGATAAAGTTGGCACTAGAGA3′. The PCR product was digested with KpnI and BglII and ligated upstream of the gene for luciferase in pGL3-basic (Promega) to yield reporter plasmid FL:PEI. Subsequent deletion mutants were generated through digestion of FL:PEI with various restriction enzymes (see Fig. 3 and 4). To create FL:PEI-X, FL:PEI was digested with XhoI and BglII, and the vector containing the remaining c-fos sequences was filled in and self-ligated. To create FL-PEI-B, FL-PEI was digested with BsmI, blunt ended, and then redigested with KpnI; the excised c-fos genomic DNA fragment was ligated into pGL3-basic digested with KpnI and SmaI. To create FL-PEI-A, FL-PEI was digested with ApaLI; the desired 2-kb fragment was blunt ended and digested with KpnI, and the excised c-fos DNA fragment was ligated into pGL3-basic digested with KpnI and SmaI. To create FL-PE, FL-PEI-X was digested with KpnI and BglII, and the c-fos genomic DNA fragment was gel extracted and partially digested with HincII; the resultant DNA fragment containing the promoter and exon sequences of c-fos was ligated into pGL3-basic digested with KpnI and SmaI. To create FL-P, FL-PEI was digested with AccI, blunt ended, and digested with KpnI; the desired c-fos fragment was ligated into pGL3-basic digested with KpnI and SmaI. FL:PEI-X(mut) was created by PCR amplification of an FL:PEI-X template using primers 5′GGGGTACCAAAAAAAGTTCCAGATTGCTGGAC3′ and 5′ACAAGTGTGCACGCGCTCAGAGAATTCCTGGGTTCC3′. The 3′ primer contains a double mutation of the wt sequence (see Fig. 4b) that introduces a unique EcoRI site. The PCR product was digested with KpnI (restriction site within the 5′ primer) and ApaLI (restriction site within the 3′ primer). To restore the entire FL:PEI-X(mut) sequence, the PCR product was ligated to the 120-bp ApaLI-XhoI fragment present downstream of the ApaLI site in FL:PEI-X in a triple ligation that also included pGL3-basic DNA digested with KpnI and XhoI. The presence of the correct mutations was confirmed by sequencing the entire PCR-amplified region.
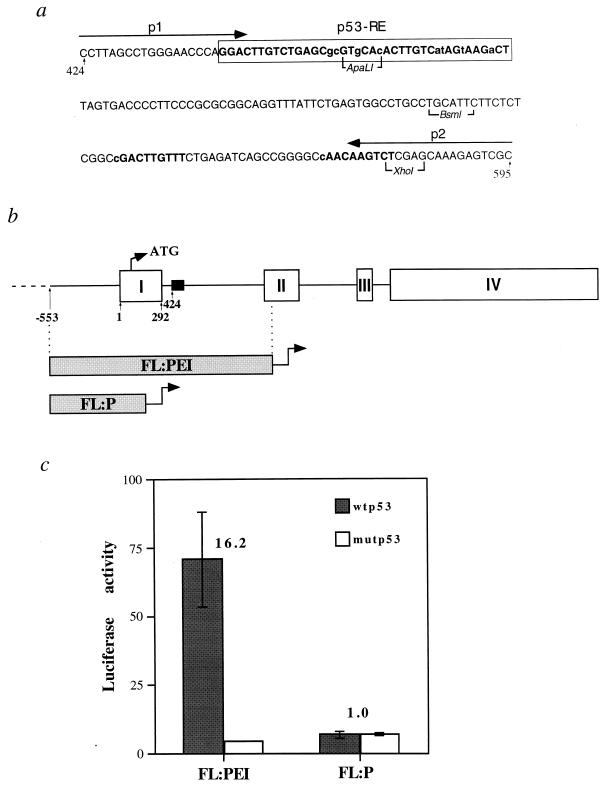
FIG. 3.
Transactivation of a c-fos reporter by p53. (a) Sequence of a 5′ segment of c-fos intron 1 containing putative p53-binding elements. The numbers at both ends of the sequence correspond to nucleotide positions within the c-fos gene, where 1 corresponds to the transcription start site. The arrows represent the two primers (p1 and p2) used to PCR amplify the region for gel mobility analysis (see Fig. 5). The box depicts the putative p53-RE as deduced from subsequent experiments (see Fig. 4 and the text). Boldface letters denote the four tandem putative 10-mer p53-binding consensus half sites and lowercase identifies nucleotides which deviate from the consensus. The indicated restriction enzymes were used to produce luciferase constructs with deletions (Fig. 4). (b) Schematic diagram of the c-fos gene. Boxes represent the four exons. The first in-frame ATG is indicated. The position of the sequence presented in panel a is indicated by the filled box. Also shown are relative positions of the c-fos genomic DNA fragments used to construct reporter plasmids FL:PEI and FL:P (see panel c). (c) Luciferase activity of the reporter plasmids depicted in panel b. H1299 cells (4 × 105 per 60-mm-diameter dish) were transfected in triplicate with a combination of the indicated reporter plasmid (1 μg/dish) and either pCMVp53wt (wtp53, encoding murine wt p53) or pCMVp53m (mutp53, encoding the murine p53gly168ile234 mutant protein) at 25 ng/dish. Extracts were prepared 36 h later and assayed for luciferase activity. Values are presented in arbitrary machine units. The numbers above the bars represent fold of activation of wtp53 over mutp53. Transfections were done in triplicate, and standard errors are shown.
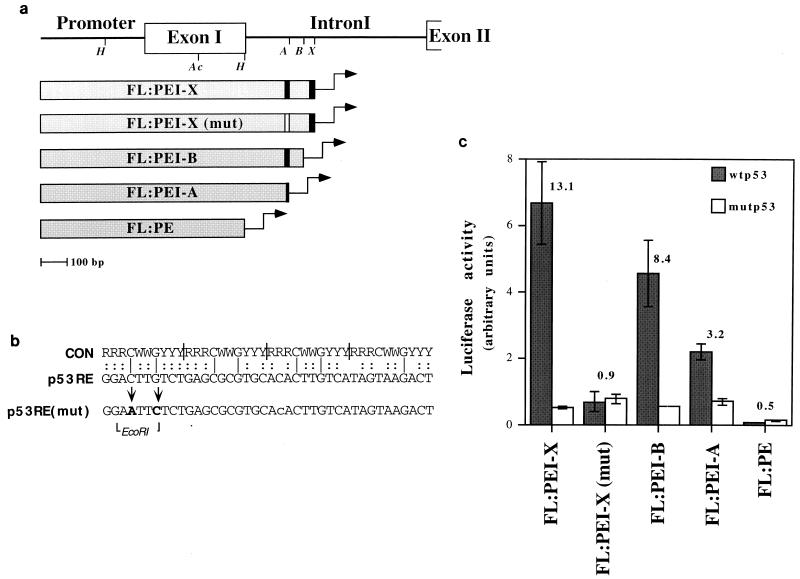
FIG. 4.
Deletion analysis mapping of the region responsible for p53-mediated c-fos transactivation. (a) Schematic map of the reporter plasmids used for transactivation analysis. Each plasmid contains the indicated region of the c-fos gene placed upstream of the luciferase gene in a promoterless luciferase construct (pGL3basic). Italics identify the positions of restriction enzymes used to create the various deletion mutants: H, HincII; Ac, AccI; A, ApaLI; B, BsmI; X, XhoI. Construct FL:PE was created through partial digestion of FL:PEI with HincII. The black vertical bars indicate the two clusters of p53 consensus half sites (Fig. 3a). The white bar represents the mutated p53-RE. (b) Alignment of the consensus (CON) p53-binding site (12) and the 40-bp p53-RE of the mouse c-fos gene. Vertical bars within the consensus sequence indicate the four half-site decamers. The positions of the site-directed mutations in the p53-RE, as well as that of the unique EcoRI site created during mutagenesis, are shown at the bottom. (c) Luciferase activity of the reporter plasmids depicted in panel a. For details, see the legend to Fig. 3c.
H1299 cells were transfected transiently by the calcium phosphate method (53). Transfections were performed in triplicate by using 1 μg of each reporter plasmid and 25 ng of either pCMVp53wt (encoding wt mouse p53) or pCMVp53m (encoding the murine mutant protein p53gly168ile234). Luciferase assays were performed by standard procedures with the aid of a Turner Designs luminometer.
Gel mobility shift assays were performed as previously described (53).
RESULTS
p53 overexpression induces c-fos.
The SSH method (10) was employed to identify p53 target genes. The starting material consisted of two mRNA populations, one extracted from p53-null M1 mouse myeloid leukemia cells and the other from LTR6 cells, derived by stable transfection of M1 cells with ts mutant p53 protein p53Val135 (51, 52). Prior to RNA extraction, cells were incubated for 4 h at 32°C; in LTR6, this restores wt p53 activity, leading to apoptosis (51, 52). cDNA was synthesized off each mRNA population, and the two cDNA pools were used for SSH. Clones in which LTR6 cDNA is overrepresented relative to M1 cDNA included the p53 target genes for GLN-LTR (53) and EI24 (32) (data not shown). Unexpectedly, c-fos transcripts were also enriched in p53-activated LTR6 cDNA (data not shown).
Induction of c-fos mRNA in LTR6 cells following p53 activation at 32°C was confirmed by Northern blot analysis (Fig. 1a, lanes 3 and 4). No induction was seen in M1 cells (lanes 1 and 2), ruling out a nonspecific temperature effect. Semiquantitative RT-PCR revealed a prominent increase in c-fos mRNA within 80 min after the temperature shift (Fig. 1b). This resembles the rate of induction in this system of p21Waf1, mdm2, gadd45, bax, and the gene for GLN-LTR, well-established p53 target genes (18, 34, 53). A corresponding increase in c-Fos protein was evident in LTR6, but not M1, cells 4 h after the temperature shift (Fig. 1d, lanes 1 to 4).
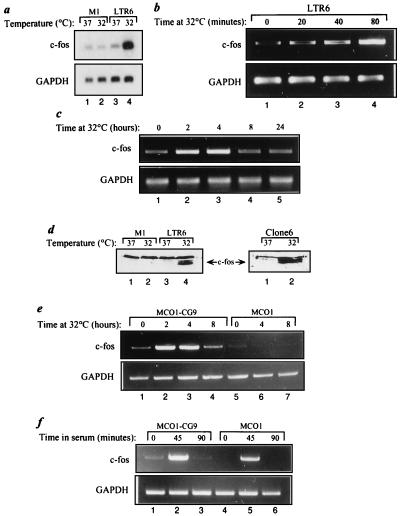
FIG. 1.
Analysis of c-fos gene expression in cell lines harboring ts mutant protein p53Val135. (a) Poly(A)+ RNA was extracted from M1 and LTR6 cells incubated at either 37 or 32°C for 4 h. Aliquots (5 μg) were separated by agarose gel electrophoresis, transferred to a nylon membrane, and probed with a mouse c-fos cDNA probe. The same membrane was subsequently reprobed for GAPDH. Total cellular RNA was extracted from LTR6 (b) or Clone6 (c) cells following incubation at 32°C for the indicated periods. An 8-μg sample of each RNA was subjected to semiquantitative RT-PCR performed with c-fos- and GAPDH-specific primers. The numbers of PCR cycles for c-fos and GAPDH were 26 and 21 (b), and 30 and 21 (c) respectively. (d) Nuclear extracts were prepared from M1, LTR6 and Clone6 cells incubated at either 37 or 32°C for 4 h. Equal amounts of protein (100 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with an anti-c-Fos antibody. Total cellular RNA was extracted from MCO1-cG9 or MCO1 cells following either incubation at 32°C for the indicated periods (e) or starvation for 24 h in 0.1% FCS, followed by incubation in 20% FCS for the indicated periods (f). A 5-μg sample of each RNA was subjected to semiquantitative RT-PCR as described above. The numbers of PCR cycles were 30 and 20 (e) and 26 and 20 (f) for c-fos and GAPDH, respectively.
Activation of ts p53 at 32°C also induced c-fos in mouse DP-16 cells transfected with p53val135 (25) (data not shown) and in the p53val135-overexpressing fibroblastic lines Clone6 (Fig. 1c) and MCO1-cG9 (Fig. 1e), with a corresponding increase in c-Fos protein (Fig. 1d). However, p53 was not required for c-fos induction by serum stimulation (Fig. 1f). In the fibroblastic cell lines Clone6 and MCO1-cG9, the induction of c-fos following p53 activation was transient, peaking within 2 h of the temperature shift and markedly declining by 8 h (Fig. 1c and e). This transient induction pattern differs from that of other p53 target genes, such as p21 (data not shown) and mdm2 (2), whose transcripts keep accumulating for at least 24 h. As observed for many other p53-responsive genes (30), the ability of p53 to elevate c-fos gene expression appears to be cell type dependent. Thus, in several situations, including HeLa cells stably transfected with p53val135, p53 activation did not result in significant induction of c-fos mRNA (data not shown).
c-fos mRNA is induced by DNA damage in a p53-dependent manner.
To determine whether c-fos is also induced upon activation of endogenous p53, c-fos expression was examined in cells exposed to ionizing radiation (IR), a potent p53 activator (27). Interleukin-3 (IL-3)-dependent DA-1 mouse lymphoma cells contain functional, IR-responsive wt p53 (16). Exposure of DA-1 cells to 3 Gy of IR, with or without IL-3, strongly elevated c-fos mRNA (Fig. 2a, lanes 3 and 4). A milder increase occurred in irradiated normal human diploid fibroblasts (Fig. 2b).
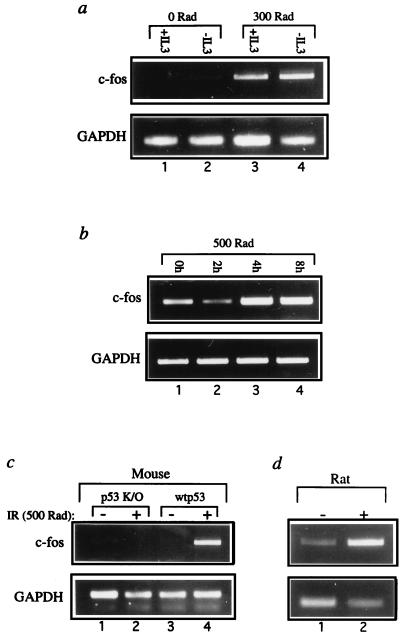
FIG. 2.
Induction of c-fos gene expression following DNA damage. (a) RNA was extracted from DA-1 cells, incubated either in the absence or in the presence of IL-3, 6 h after exposure to 3 Gy of gamma irradiation. The RNA was subjected to semiquantitative RT-PCR, as described in the legend to Fig. 1, using c-fos (28 cycles)- and GAPDH (21 cycles)-specific primers. (b) RNA was extracted from F89 normal human diploid fibroblasts at the indicated time points following exposure to 5 Gy of gamma irradiation and subjected to semiquantitative RT-PCR with c-fos (33 cycles)- and GAPDH (21 cycles)-specific primers. (c) p53 knockout mice (p53 K/O) and their wt littermates (wtp53) were obtained through a cross between two p53 +/− mice. One 40-day-old mouse of each genotype was exposed to 5 Gy of whole-body gamma irradiation (+), while another pair of mice were left untreated (−). Animals were sacrificed 4 h later, and total RNA was extracted from each thymus and subjected to semiquantitative RT-PCR with c-fos (28 cycles)- and GAPDH (23 cycles)-specific primers. (d) Two-month-old rats were either exposed to 5 Gy of whole-body gamma irradiation (+) or left untreated (−). Animals were sacrificed 4 h later, and total RNA was extracted from each thymus and subjected to semiquantitative RT-PCR with c-fos (29 cycles)- and GAPDH (21 cycles)-specific primers.
IR exposure in vivo activates p53 in the thymus, triggering apoptosis (9, 35). c-fos expression was strongly induced in mouse and rat thymuses (Fig. 2c and d) 4 h after whole-body irradiation. Importantly, no increase was seen in p53 knockout mice (Fig. 2c, lanes 1 and 2). Hence, c-fos is p53 responsive both in vitro and in vivo. p53-independent mechanisms may also contribute to c-fos induction by IR; however, at least in thymocytes, this is insufficient in the absence of functional p53.
The first intron of the c-fos gene contains a p53-responsive element.
The c-fos basal promoter is unlikely to mediate the observed p53 responsiveness; in fact, it can even be repressed by very high levels of p53 (15, 45). In contrast, the sequence of the c-fos first intron suggests the presence of several candidate p53-binding motifs (5). In particular, a stretch of 40 nucleotides (nt) (boxed in Fig. 3a) can be viewed as a tandem array of four 10-mer motifs exhibiting various degrees of homology to the p53 consensus half site (12) (consensus matches are indicated by uppercase letters in Fig. 3a; see also Fig. 4b). In addition, two potential half sites separated from one another by 17 bp reside further downstream in intron 1 (boldface in Fig. 3a).
A genomic fragment spanning the c-fos promoter, exon 1, intron 1, and the 5′ end of exon 2 was cloned upstream of a luciferase reporter gene (plasmid FL:PEI in Fig. 3b). When cotransfected into p53-null human H1299 cells together with small amounts of p53 expression plasmids, FL:PEI was strongly activated by wt but not tumor-derived mutant p53 (Fig. 3c). In contrast, no activation was observed with deletion mutant FL:P, comprising mainly the c-fos promoter. Thus, the c-fos gene contains a p53-responsive domain downstream of its promoter.
Deletion constructs (Fig. 4a) were next tested for retention of p53-mediated transactivation. Removal of sequences downstream of the putative p53-binding domain (plasmid FL:PEI-X) maintained full activation (Fig. 4c); the 3′ part of intron 1 is therefore dispensable. Nevertheless, FL:PEI-X did exhibit lower basal activity than FL:PEI, presumably owing to loss of a splice acceptor site.
Deletion of the promoter-distal pair of potential half sites (FL:PEI-B) had only a mild effect on p53-mediated activation (Fig. 4c). However, deletion into the promoter-proximal 40-nt stretch (FL:PEI-A) caused a substantial decrease in activation, whereas complete deletion of intron 1 (FL:PE) entirely abolished p53 inducibility. Hence, a functional p53-responsive element (p53-RE) resides within the 5′ part of intron 1.
To map more precisely the structural requirements for c-fos activation by p53, we mutated 2 nt in the 5′ part of the potential p53-RE [p53RE(mut) in Fig. 4b]. These particular nucleotides were chosen because they reside at invariant positions within a 10-mer motif perfectly matching the p53 consensus half site. As seen in Fig. 4c, these mutations [FL:PEI-X(mut)] totally eliminated the ability of the reporter to be activated by wt p53 in H1299 cells. This confirms the correct assignment of the p53-RE and defines critical residues within this element.
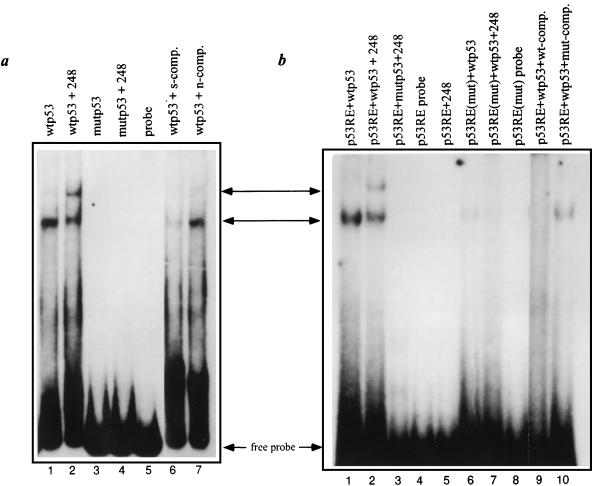
To investigate whether the c-fos intronic region binds p53 directly, the entire region was PCR amplified using primers p1 and p2 (Fig. 3a) and used as a radiolabeled probe for gel mobility shift analysis. Incubation of the probe with purified recombinant wt p53 yielded a distinct retarded band (Fig. 5a, middle arrow; compare lanes 1 and 5) partially supershifted by p53-specific monoclonal antibody PAb248 (lane 2, upper arrow). Hence, p53 can bind this c-fos intron 1-derived DNA fragment. No shift was obtained with p53cys270, a mutant protein defective in sequence-specific DNA binding (lanes 3 and 4). The specificity of the binding was confirmed by its ability to be competed efficiently by an excess of a nonlabeled probe (lane 6) but not by an unrelated DNA fragment (lane 7). A synthetic 40-bp oligonucleotide comprising the proposed p53-RE (boxed in Fig. 3a) was also shifted by wt p53 but not by p53cys270 (Fig. 5b, lanes 1 to 5), confirming that this defined DNA element is indeed sufficient for p53 binding. Importantly, a similar oligonucleotide carrying the two mutations described in Fig. 4b was severely compromised for p53 binding (Fig. 5b, lanes 6 and 7). Thus, these mutations, which abolish p53-mediated transactivation, also strongly reduce p53 binding. The residual binding seen with p53RE(mut) suggests that the mutated element retains some ability to interact with p53, albeit far less efficiently. This is consistent with the ability of excess p53RE(mut) to compete to a limited extent for p53 binding (Fig. 5b, compare lanes 1 and 10). The residual p53 binding may be mediated by the three remaining 10-mer motifs but is obviously not enough for transcriptional activation.
FIG. 5.
Binding of p53 to the c-fos p53-RE. (a) Equal amounts of a [32P]dATP end-labeled 180-bp probe prepared by PCR amplification (Fig. 3a) were incubated with 250 ng of either wt murine p53 (wtp53) or the murine p53cys270 mutant protein (mutp53), each purified from Sf9 cells infected with the corresponding recombinant baculovirus (53). Where indicated, reactions mixtures also included p53-specific monoclonal antibody PAb248 (lanes 2 and 4) or a 40× molar excess of a nonlabeled probe, which served as a specific competitor (s-comp., lane 6), or of the 180-bp multiple cloning site of pBluescript, which served as a nonspecific competitor (n-comp., lane 7). (b) Equal amounts of [32P]dATP end-labeled double-stranded 40-bp oligonucleotides [p53-RE and p53-RE(mut); Fig. 4b] were incubated with 250 ng of either wtp53 or p53cys270, with or without addition of PAb248 (lanes 2, 3, and 7; lane 5 contained PAb248 without p53). The reactions mixtures in lanes 9 and 10 also incubated a 150× molar excess of a nonlabeled 40-bp p53-RE oligonucleotide as a specific competitor (wt-comp., lane 9) or of p53-RE(mut) (lane 10). The arrows indicate the free probes and the shifted and supershifted bands.
Collectively, our findings identify c-fos as a new p53 target gene whose transactivation is mediated through direct binding of p53 to a distinct cognate region within intron 1.
DISCUSSION
The present study identified c-fos as a new p53 target gene. This conclusion drew on several lines of evidence: both the mRNA and protein of c-fos are strongly induced upon deliberate activation of p53, and p53-dependent induction of c-fos expression is observed in vivo following exposure to DNA damage. A region containing a putative p53-binding site resides within the first intron of the c-fos gene. This region mediates transactivation by p53 in the context of genomic c-fos DNA. Moreover, the wt, but not the mutant, form can bind directly to this region, and base substitutions that abrogate this binding also abolish p53-dependent transcriptional activation. Thus, p53-mediated transactivation of c-fos is brought about by direct binding of p53 to a distinct p53-binding element within intron 1 of the c-fos gene. This relationship suggests that c-fos acts downstream of p53 as part of a stress response pathway.
It has been shown that the basal c-fos promoter can be repressed by large amounts of p53 (15, 28, 45). The finding that the intact c-fos gene, in its chromosomal context, is actually induced in a p53-dependent manner was therefore unexpected. Yet, unlike earlier studies, the present analysis employed much smaller amounts of p53 expression plasmid DNA and was expected to represent more physiologically relevant conditions. As the region responsible for p53-mediated induction of c-fos is located within an intron, it is obvious that constructs retaining only the c-fos promoter are not suitable for studying the regulation of this gene by p53. In fact, such constructs are indeed likely to be repressed by excessive amounts of p53, presumably owing to sequestration of essential components of the transcription machinery, giving rise to transcriptional “squelching.” It is also noteworthy that the ability of p53 to trigger c-fos expression is often cell type dependent (data not shown). This might suggest that c-fos induction requires cooperation between p53 and another transcription factor(s). The functional state of such a putative factor may thus determine whether or not c-fos and, presumably, other p53 target genes will be turned on, thereby affecting the biological outcome of p53 activation.
c-fos participates in a plethora of signaling pathways (26). In many situations (e.g., growth factor stimulation), c-fos induction occurs very rapidly and transiently (26). This induction, mediated through elements in the c-fos promoter (26), does not involve p53 (Fig. 1f). However, c-fos induction can also follow a slower course, taking hours rather than minutes and persisting for an extended period; in such situations, a role for p53 might be envisaged. It is noteworthy, that persistent c-fos induction has been associated with an apoptotic outcome (8, 19, 47). c-fos also facilitates IR-induced T-lymphocyte apoptosis (42), a largely p53-dependent process (9, 35). Combined with the fact that prominent c-fos induction procedes p53-triggered apoptosis in LTR6 cells, our data therefore raise the possibility that c-fos transactivation contributes to the proapoptotic effects of p53 under circumstances such as radiation damage, hypoxia, or oxidative stress. Alternatively, since a p53-mediated increase in c-fos expression can also be observed in cells like Clone6 and MCO-cG9 cells (Fig. 1), which undergo growth arrest rather than apoptosis in response to p53 activation, it is possible that c-fos induction is, in fact, involved in biological effects of p53 distinct from apoptosis, such as positive regulation of certain differentiation processes (43). It is noteworthy, however, that in these fibroblastic cell lines, the induction of c-fos by p53 is transient and that c-fos mRNA levels return to their ground state within a few hours (Fig. 1c and e). It is thus conceivable that such a short duration of c-fos overexpression is insufficient for delivery of an irreversible apoptotic signal. Indeed, it was earlier demonstrated that overexpressed c-fos and functional wt p53 cooperate in the efficient induction of apoptosis in a human cancer cell line (41).
Finally, one cannot rule out the possibility that c-fos induction even plays a protective role, e.g., by facilitating recovery from DNA damage (11, 46). In fact, p53 itself has also been shown to exert an apparent antiapoptotic effect in primary fibroblasts under conditions of relatively mild stress (31). Irrespective of the exact contribution of c-fos to the p53 pathway, our data support the existence of a direct link between these two pivotal cell fate regulators.
ACKNOWLEDGMENTS
We thank S. Benchimol for the gift of DP-16 cells, Y. Shilo for the gift of F89 cells, M. Uzan for excellent technical assistance, E. Gottlieb for helpful suggestions, and A. Rosen (QBI Enterprises, Inc.) for advice on SSH.
This work was supported in part by grant RO1CA40099 from the National Cancer Institute, by the Israel-USA Binational Science Foundation, by the Leo and Julia Forchheimer Center for Molecular Genetics, and by a fellowship grant from the Israel Cancer Research Fund.
REFERENCES
- 1.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 2.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak Y, Oren M. Enhanced binding of a 95 kDa protein to p53 in cells undergoing p53-mediated growth arrest. EMBO J. 1992;11:2115–2121. doi: 10.1002/j.1460-2075.1992.tb05270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates S, Vousden K H. p53 in signaling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:12–18. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 5.Bourdon J C, Deguinchambon V, Lelong J C, Dessen P, May P, Debuire B, May E. Further characterisation of the p53 responsive element—identification of new candidate genes for trans-activation by p53. Oncogene. 1997;14:85–94. doi: 10.1038/sj.onc.1200804. [DOI] [PubMed] [Google Scholar]
- 6.Buckbinder L, Talbott R, Velasco M S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 7.Canman C E, Kastan M B. Role of p53 in apoptosis. Adv Pharmacol. 1997;41:429–460. doi: 10.1016/s1054-3589(08)61068-6. [DOI] [PubMed] [Google Scholar]
- 8.Chen S C, Curran T, Morgan J I. Apoptosis in the nervous system: new revelations. J Clin Pathol. 1995;48:7–12. doi: 10.1136/jcp.48.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wyllie A H. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 10.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dosch J, Kaina B. Induction of c-fos, c-jun, junB and junD mRNA and AP-1 by alkylating mutagens in cells deficient and proficient for the DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT) and its relationship to cell death, mutation induction and chromosomal instability. Oncogene. 1996;13:1927–1935. [PubMed] [Google Scholar]
- 12.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Definition of a consensus binding site for p53. Nat Gene. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 14.Elkeles A, Devos K M, Graur D, Zizi M, Breiman A. Multiple cDNAs of wheat voltage-dependent anion channels (VDAC): isolation, differential expression, mapping and evolution. Plant Mol Biol. 1995;29:109–124. doi: 10.1007/BF00019123. [DOI] [PubMed] [Google Scholar]
- 15.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can downmodulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottlieb E, Haffner R, Von Ruden T, Wagner E F, Oren M. Down-regulation of wild-type p53 activity interferes with apoptosis of IL-3-dependent hematopoietic cells following IL-3 withdrawal. EMBO J. 1994;13:1368–1374. doi: 10.1002/j.1460-2075.1994.tb06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 18.Guillouf C, Grana X, Selvakumaran M, Deluca A, Giordano A, Hoffman B, Liebermann D A. Dissection of the genetic programs of p53-mediated G1 growth arrest and apoptosis: blocking p53-induced apoptosis unmasks G1 arrest. Blood. 1995;85:2691–2698. [PubMed] [Google Scholar]
- 19.Hafezi F, Steinbach J P, Marti A, Munz K, Wang Z Q, Wagner E F, Aguzzi A, Reme C E. The absence of c-fos prevents light-induced apoptotic cell death of photoreceptors in retinal degeneration in vivo. Nat Med. 1997;3:346–349. doi: 10.1038/nm0397-346. [DOI] [PubMed] [Google Scholar]
- 20.Halevy O, Rodel J, Peled A, Oren M. Frequent p53 mutations in chemically-induced murine fibrosarcoma. Oncogene. 1991;6:1593–1600. [PubMed] [Google Scholar]
- 21.Hansen R, Oren M. p53; from inductive signal to cellular effect. Curr Opin Genet Dev. 1997;7:46–51. doi: 10.1016/s0959-437x(97)80108-6. [DOI] [PubMed] [Google Scholar]
- 22.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3 ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 23.Hollstein M, Soussi T, Thomas G, von-Brevern M, Bartsch H. p53 gene alterations in human tumors: perspectives for cancer control. Recent Results Cancer Res. 1997;143:369–389. doi: 10.1007/978-3-642-60393-8_26. [DOI] [PubMed] [Google Scholar]
- 24.Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, Telerman A, Oren M. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson P, Chung S, Benchimol S. Growth suppression of Friend virus-transformed erythroleukemia cells by p53 protein is accompanied by hemoglobin production and is sensitive to erythropoietin. Mol Cell Biol. 1993;13:1456–1463. doi: 10.1128/mcb.13.3.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karin M, Liu Z g, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 27.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 28.Kley N, Chung R Y, Fay S, Loeffler J P, Seizinger B R. Repression of the basal c-fos promoter by wild-type p53. Nucleic Acids Res. 1992;20:4083–4087. doi: 10.1093/nar/20.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 30.Komarova E A, Diatchenko L, Rokhlin O W, Hill J E, Wang Z J, Krivokrysenko V I, Feinstein E, Gudkov A V. Stress-induced secretion of growth inhibitors: a novel tumor suppressor function of p53. Oncogene. 1998;17:1089–1096. doi: 10.1038/sj.onc.1202303. [DOI] [PubMed] [Google Scholar]
- 31.Lassus P, Ferlin M, Piette J, Hibner U. Anti-apoptotic activity of low levels of wild-type p53. EMBO J. 1996;15:4566–4573. [PMC free article] [PubMed] [Google Scholar]
- 32.Lehar S M, Nacht M, Jacks T, Vater C A, Chittenden T, Guild B C. Identification and cloning of EI24, a gene induced by p53 in etoposide-treated cells. Oncogene. 1996;12:1181–1187. [PubMed] [Google Scholar]
- 33.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 34.Levy N, Yonish-Rouach E, Oren M, Kimchi A. Complementation by wild-type p53 of interleukin-6 effects on M1 cells induction of cell cycle exit and cooperativity with c-myc suppression. Mol Cell Biol. 1993;13:7942–7952. doi: 10.1128/mcb.13.12.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 36.Mitsudomi T, Steinberg S M, Nau M M, Carbone D, Damico D, Bodner S, Oie H K, Linnoila R I, Mulshine J L, Minna J D, Gazdar A F. p53 gene mutations is non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–180. [PubMed] [Google Scholar]
- 37.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 38.Owen-Schaub L B, Zhang W, Cusack J C, Angelo L S, Santee S M, Fujiwara T, Roth J A, Deisseroth A B, Zhang W-W, Kruzel E, Radinsky R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinhasi-Kimhi O, Michalovitz D, Oren M. Specific interaction between the p53 cellular tumor antigen and major heat-shock proteins. Nature. 1986;320:182–185. doi: 10.1038/320182a0. [DOI] [PubMed] [Google Scholar]
- 40.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 41.Preston G A, Lyon T T, Yin Y, Lang J E, Solomon G, Annab L, Srinivasan D G, Alcorta D A, Barrett J C. Induction of apoptosis by c-Fos protein. Mol Cell Biol. 1996;16:211–218. doi: 10.1128/mcb.16.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruschy M, Shi Y Q, Crompton N E A, Steinbach J, Aguzzi A, Glanzmann C, Bodis S. The proto-oncogene c-fos mediates apoptosis in murine T-lymphocytes induced by ionizing radiation and dexamethasone. Biochem Biophys Res Commun. 1997;241:519–524. doi: 10.1006/bbrc.1997.7846. [DOI] [PubMed] [Google Scholar]
- 43.Rotter V, Aloni-Grinstein R, Schwartz D, Elkind N B, Simons A, Wolkowicz R, Lavigne M, Beserman P, Kapon A, Goldfinger N. Does wild-type p53 play a role in normal cell differentiation? Semin Cancer Biol. 1994;5:229–236. [PubMed] [Google Scholar]
- 44.Rouault J P, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyretlalle C, Savatier P, Pain B, Shaw P, Berger R, Samarut J, Magaud J P, Ozturk M, Samarut C, Puisieux A. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 45.Santhanam U, Ray A, Sehgal P B. Repression of the interleukin 6 gene promoter by p53 and the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1991;88:7605–7609. doi: 10.1073/pnas.88.17.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreiber M, Baumann B, Cotten M, Angel P, Wagner E F. Fos is an essential component of the mammalian UV response. EMBO J. 1995;14:5338–5349. doi: 10.1002/j.1460-2075.1995.tb00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smeyne R J, Vendrell M, Hayward M, Baker S J, Miao G G, Schilling K, Robertson L M, Curran T, Morgan J I. Continuous c-fos expression precedes programmed cell death in vivo. Nature. 1993;363:166–169. doi: 10.1038/363166a0. [DOI] [PubMed] [Google Scholar]
- 48.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 49.Wyllie A. Apoptosis—clues in the p53 murder mystery. Nature. 1997;389:237–238. doi: 10.1038/38405. [DOI] [PubMed] [Google Scholar]
- 50.Yin Y X, Terauchi Y, Solomon G G, Aizawa S, Rangarajan P N, Yazaki Y, Kadowaki T, Barrett J C. Involvement of p85 in p53-dependent apoptotic response to oxidative stress. Nature. 1998;391:707–710. doi: 10.1038/35648. [DOI] [PubMed] [Google Scholar]
- 51.Yonish-Rouach E, Grunwald D, Wilder S, Kimchi A, May E, Lawrence J J, May P, Oren M. p53-mediated cell death: relationship to cell cycle control. Mol Cell Biol. 1993;13:1415–1423. doi: 10.1128/mcb.13.3.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 53.Zauberman A, Barak Y, Ragimov N, Levy N, Oren M. Sequence-specific DNA binding by p53: identification of target sites and lack of binding to p53-MDM2 complexes. EMBO J. 1993;12:2799–2808. doi: 10.1002/j.1460-2075.1993.tb05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]