Abstract
The recombination activating genes RAG-1 and RAG-2 are expressed in a lymphoid-cell-specific and developmentally regulated fashion. To understand the transcriptional basis for this regulation, we have cloned and characterized the murine RAG-2 promoter. The promoter was lymphoid cell specific, showing activity in various B- and T-cell lines but little activity in nonlymphoid cells. To our surprise, however, the promoter was regulated differently in B and T cells. Using nuclear extracts from B-cell lines, we found that the B-cell-specific transcription factor BSAP (Pax-5) could bind to a conserved sequence critical for promoter activity. BSAP activated the promoter in transfected cells, and the BSAP site was occupied in a tissue-specific manner in vivo. An overlapping DNA sequence binding to a distinct factor was necessary for promoter activity in T cells. Full promoter activity in T cells was also dependent on a more distal DNA sequence whose disruption had no effect on B-cell activity. The unexpected finding that a B-cell-specific factor regulates the RAG-2 promoter may explain some of the recently observed differences in the regulation of RAG transcription between B and T cells.
Antigen receptor genes are assembled during lymphocyte development by a site-specific recombination reaction known as V(D)J recombination (71). RAG-1 and RAG-2 are the essential lymphoid-cell-specific components of the V(D)J recombinase and are required for the initiation of recombination in developing B and T cells (44, 49, 59, 63). Rag-1 and Rag-2 proteins are present at significant levels only in lymphocytes, limiting V(D)J recombinase activity to lymphoid cells.
RAG expression begins at the earliest stages of lymphocyte development (22, 27, 72, 76). In developing B cells, RAG transcript levels transiently decrease after successful immunoglobulin (Ig) heavy-chain gene rearrangement and increase again during light-chain gene rearrangement in pre-B cells (22). This pattern of expression has been proposed to contribute to the allelic exclusion of heavy-chain gene rearrangement and the temporally regulated activation of light-chain gene rearrangement. RAG gene transcription persists in surface IgM-positive (sIgM+) immature bone marrow B cells but is absent in mature IgD+ peripheral B cells (22). Recent experiments have revealed two surprising aspects of RAG transcriptional regulation: (i) RAG transcription increases in immature B cells expressing an autoreactive antigen receptor leading to receptor editing, an important tolerance mechanism in B cells (19, 42, 70); and (ii) RAG transcription [and V(D)J recombination] is reactivated in germinal center B cells during an immune response (24, 25, 32, 34, 52). In developing T cells, RAG expression persists in CD4+ CD8+ CD3+ T cells until positive selection and subsequent differentiation into CD4 or CD8 single-positive T cells. CD3 cross-linking extinguishes RAG expression in thymocytes (72, 76). One recent report suggests that peripheral T cells reactivate the V(D)J recombinase in the setting of persistent negative selection (41). Thus, RAG transcription appears to be regulated differently in B and T cells.
In all species examined thus far, the genomic organization of RAG-1 and RAG-2 has been conserved, with the two genes tightly linked and convergently transcribed (60). This observation led to the hypothesis that the RAG genes derive from an ancestral transposon that integrated into an early vertebrate genome (49, 69). With one exception, the coding region of each gene is contained within a single exon (26). Basal promoters for murine and human RAG-1 and human RAG-2 have been described (5, 17, 38, 77). None of these promoters show tissue specificity, however. The promoters of the murine and human RAG-1 genes are highly conserved, supporting the idea that the mechanism of their regulation is also conserved.
Elucidating the regulation of RAG gene transcription should enhance our understanding of lymphoid lineage determination and the regulation of V(D)J recombination. In addition, defective or aberrant regulation of RAG transcription may contribute to some cases of human severe combined immunodeficiency (1) and possibly to lymphocytic leukemia. In an attempt to first understand the cis elements that control RAG expression, we have cloned and characterized the core promoter of the murine RAG-2 gene. The promoter was active in several pro-B-cell lines and two T-cell lines but showed little activity in a mature B-cell line, an immature T-cell line, and nonlymphoid cells. To our surprise, we found that different factors were responsible for the activity of the promoter in B and T cells. We have identified a conserved DNA sequence critical for promoter activity that binds the B-cell transcription factor BSAP (also known as Pax-5) in vitro and is occupied in a tissue-specific fashion in vivo. An overlapping but distinct region of the promoter is essential for its activity in T cells. We found that a more distal region of the RAG-2 promoter dispensable in pro-B cells was quantitatively important in T cells. The involvement of BSAP in the regulation of RAG-2 transcription may contribute to the differences in RAG gene expression observed in the T- and B-cell lineages.
MATERIALS AND METHODS
Cells and cell culture.
Thymocytes and splenocytes were isolated from 6- to 12-week-old BALB/c mice (National Cancer Institute). The following cell lines were used: 63-12 (63), 103 bcl2/4 (7), and 220-8 (3), transformed pro-B-cell lines; WEHI-231, M12 (21), and CH33 (28), mature B-cell lymphomas; 2017 (65), a transformed immature T-cell line; VL3-3M2, an immature T-cell line (23); Jurkat and EL4, mature T-cell lymphomas; P815, a mastocytoma cell line (13); 293T; HeLa; and NIH 3T3. The 103 bcl2/4 line was grown at 33 or 39°C in a 5% CO2 atmosphere in RPMI 1640 (Mediatech) supplemented with 10% fetal calf serum, penicillin-streptomycin, l-glutamine, and 50 μM β-mercaptoethanol. All other B- and T-cell lines, as well as P815 cells, were grown at 37°C and 5% CO2 in the same medium except with 5% fetal calf serum. 293T, NIH 3T3, and HeLa cells were grown in Dulbecco modified Eagle medium (Mediatech) with 4.5 or 1 (HeLa) g of glucose per liter, supplemented with 10% fetal calf serum, l-glutamine, penicillin-streptomycin, and 50 μM β-mercaptoethanol.
Primer extension.
Fifty nanograms of a 30-mer oligonucleotide (5′-TCCCTCTCTGAATCCTTTCGGCCAAGCCAG) was end labeled with [γ-32P]ATP (specific activity, 6,000 Ci/mmol; Amersham) and T4 polynucleotide kinase (New England Biolabs) to a specific activity of 3 × 108 cpm/μg; 105 cpm of this primer was coprecipitated with 40 μg of total RNA from thymus, 103 bcl2/4 cells grown at 33 or at 39°C for 6.5 h, CH33 cells, or yeast tRNA. Samples were resuspended in 10 μl of 10 mM Tris (pH 8.3)–90 mM KCl–1 mM EDTA, denatured for 3 min at 94°C, and then annealed 1 h at 64°C. During the last few minutes of annealing, 40 μl of prewarmed 10 mM Tris (pH 8.3)–90 mM KCl–1.25 mM MnCl2–1.25 mM dithiothreitol (DTT)–250 mM deoxynucleoside triphosphates was added to the samples, followed by 2.5 U of ΔTth DNA polymerase (Clontech), and incubated 1 h at 70°C. The samples were precipitated, and half of each sample was run on a 6% polyacrylamide–7 M urea gel alongside a DNA sequencing ladder as a size marker. The dried gel was exposed to X-Omat AR film (Kodak) at −80°C with an intensifying screen for 14 days.
S1 nuclease mapping.
A uniformly labeled probe was prepared as follows. The template for the probe was a plasmid containing an insert extending from the 3′ end of the first exon of RAG-2 to a HindIII site 402 bp upstream cloned into the XhoI and BglII sites of pGL2-Basic (Promega). Two micrograms of this plasmid was denatured with 0.2 N NaOH, neutralized, precipitated, and annealed to 5 pmol of a 25-mer oligonucleotide (5′-TCCCTCTCTGAATCCTTTCGGCCAA). The primer was extended with [α-32P]dATP for 30 min with Klenow enzyme, followed by a 20-min chase with unlabeled dATP. The extended product was linearized with XhoI digestion, and unincorporated nucleotides were separated using the Qiaquick nucleotide removal kit (Qiagen). The probe was denatured and gel purified from a 6% polyacrylamide–7 M urea gel.
Probe (5 × 104 cpm) was coprecipitated with 40 μg of total RNA from thymus, 103 bcl2/4 cells grown at 39°C for 6.5 h, CH33 cells, or yeast tRNA. Samples were resuspended in 80% formamide–0.4 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4)–0.4 M NaCl–1 mM EDTA, denatured, and hybridized overnight at 30°C. Digestion was carried out for 1 h at 37°C with 300 U of S1 nuclease (Life Technologies). Products were precipitated and loaded onto a 6% polyacrylamide–7 M urea gel alongside a DNA sequencing ladder as a size marker. The dried gel was autoradiographed.
Cloning and sequencing of the RAG-2 5′ flanking region.
Two P1 clones containing the murine RAG-2 locus were identified by screening a murine P1 phage genomic library with RAG-1- and RAG-2-specific PCR primers and obtained from Genome Systems, St. Louis, Mo. One of these clones was digested with SacI and subcloned into pBluescript SK (Stratagene). A subclone with a 17-kb insert was identified as including the region 5′ of the RAG-2 coding exon by Southern blotting using a RAG-2 cDNA probe. The region upstream and downstream of the published first exon sequence was sequenced on both strands by the dideoxy-chain termination method (Sequenase; Amersham).
Plasmid constructs.
An XmnI fragment extending from ∼−900 to +86 of the RAG-2 promoter and a blunted HindIII-XmnI fragment from −279 to +86 were cloned into the blunted HindIII site of pGL2-Basic (Promega). PCR was used to amplify a fragment from −279 to +123, which was cloned into the XhoI and BglII sites of pGL2-Basic. The NheI-PstI fragment from this construct was replaced with NheI-PstI fragments from the ∼−900 to +86 and −279 to +86 constructs described above to generate constructs containing sequences from ∼−900 to +123 and −279 to +123, respectively. The constructs extending from −71 to +123, −45 to +123, −23 to +123, −279 to +37, and −71 to −23 were made using PCR. The construct containing sequences from −279 to +9 was generated from the −279 to +123 construct by cutting with PstI and BglII, blunting, and religating. The Δ(−156/−107) construct was made by first mutating nucleotides −156 to −152 as described below, then cutting with SmaI and BsmAI, blunting, and religating.
The RAG-1 promoter construct was made by cloning an EcoRV-PstI fragment extending from −243 to +72 (5) into the polylinker of pGL2-Basic. The simian virus 40 (SV40) promoter construct was pGL2-Promoter (Promega).
Oligonucleotide-directed mutagenesis was used to make five different 5-bp changes in the −279 to +123 construct, using the P-Select/P-Alter system (Promega). The −143 to −139 sequence was changed to GCCCC.
The eukaryotic BSAP expression construct was created by cloning the NotI-linkered murine BSAP cDNA (78) into the EFBneo expression vector (a modified version of pEF-BOS [43]). The six-His-tagged prokaryotic BSAP expression construct was created by cloning the same NotI fragment into the expression vector pET22b (Invitrogen) at its NotI site and then correcting the reading frame of the fusion protein by modifying its polylinker. BSAP expression was induced in Escherichia coli BL21 harboring this plasmid by using 1 mM isopropyl-β-d-thiogalactopyranoside for 2 h at 30°C. Recombinant BSAP (rBSAP) was purified from bacterial lysates under mildly denaturing conditions (5 M urea) and renatured while bound to His-Bind resin (Novagen). The eluted protein (>90% pure as assessed by gel electrophoresis) was dialyzed against an excess of 20 mM Tris (pH 8.0)–100 mM NaCl–0.1 mM EDTA–1 mM DTT–0.4 mM phenylmethylsulfonyl fluoride–20% glycerol and stored frozen at −80°C.
Transient transfection reporter assays.
220-8, 103 bcl2/4, 63-12, WEHI-231, and EL4 cells were transfected by the DEAE-dextran method. First, 107 cells were washed twice in STBS (25 mM Tris [pH 7.4], 137 mM NaCl, 5 mM KCl, 0.6 mM Na2HPO4, 0.7 mM CaCl2, 0.5 mM MgCl2) and resuspended in 1 ml of STBS with 10 μg of reporter plasmid, 0.5 μg of pCMV-β-gal (Promega), and 250 μg of DEAE-dextran per ml for 20 min at room temperature. Cells were washed with medium, resuspended in medium, and grown for approximately 48 h before harvest. 2017, VL3-3M2, Jurkat, M12, P815, and HeLa cells were transfected with Superfect reagent (Qiagen) according to the manufacturer’s instructions. In some experiments, DEAE-dextran was used for M12 and 2017.
293T and NIH 3T3 cells were transfected by the calcium phosphate-HEPES method as described elsewhere (4). Cells were grown for approximately 48 h in the presence of the precipitate before harvest. Ten micrograms of reporter plasmid and 100 ng of pCMV-β-gal were used for each 10-cm-diameter dish.
Cells were harvested as follows. A cell scraper was used to transfer the adherent cell lines to microcentrifuge tubes. Cells were washed twice with phosphate-buffered saline (PBS) and lysed for 15 min in reporter lysis buffer (Promega), vortexed for 15 s, and spun for 2 min at 4°C in a microcentrifuge. Supernatant was combined with luciferase substrate (Promega), and activity was measured with a luminometer (Analytical Luminescence Laboratories). β-Galactosidase assays were performed using the Galacto-Light Plus kit (Tropix) according to the manufacturer’s instructions.
For cotransfection experiments, 10 μg of the −279/+123 reporter and 0.5 μg of empty EFBneo vector or EFBneoBSAP was used. 293T cells were transfected as described above. Other cell lines were transfected with Superfect.
Luciferase activity for each sample was normalized to the β-galactosidase control. Unless otherwise noted, all experiments were performed in duplicate or triplicate and repeated two to three times.
EMSA.
For electrophoretic mobility shift assay, (EMSA), the wild-type and mutant versions of the −103 to −9 RAG-2 promoter probe were prepared by PCR from plasmid DNA templates by using the primers 5′-CAACCATCACAGGGGTGCAG (top strand) and 5′-GCCTACAGATGTTCCAGTGAG (bottom strand). PCR conditions were 30 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min 15 s, followed by a 10-min final extension at 72°C. Products were purified from agarose gels with a Qiaex II gel extraction kit (Qiagen). Approximately 15 ng of probe fragment was end labeled with [γ-32P]ATP for 30 min at 37°C by using T4 polynucleotide kinase (New England Biolabs). Unincorporated nucleotides were removed by using a spin column (Qiagen).
Nuclear extracts were prepared as described elsewhere (61). The integrity and relative concentration of each extract were confirmed by EMSA with a probe that binds the ubiquitous Oct-1 transcription factor (62).
Each 20-μl EMSA binding reaction mixture consisted of 10 mM Tris (pH 7.5), 1 mM EDTA, 1 mM DTT, 5% glycerol, 1 mg of bovine serum albumin per ml, 100 μg of poly(dI-dC) (Pharmacia) per ml, approximately 100 pg of probe DNA, 5 μM ZnSO4, and 5 μl of nuclear extract (7 to 15 μg, depending on the source). Recombinant BSAP (∼25 ng) was substituted for the nuclear extract in the experiment shown in Fig. 7, lanes 19 to 21. The final salt concentration was 100 mM NaCl. For competition, approximately 100-fold molar excess specific or nonspecific competitor DNA was added to the reaction. For competition with wild-type or mutant BSAP and NF-κB double-stranded oligonucleotides, a 50-, 500-, or 5,000-fold molar excess was used. The CD19 BSAP, κ 3′ enhancer wild-type and mutant BSAP, and NF-κB probes have been described elsewhere (36, 57, 62). For antibody supershift experiments, 1 μl of a 1:10 dilution in PBS of anti-BSAP(189-391) affinity-purified rabbit polyclonal antiserum (reconstituted in 100 μl of PBS from 100 μl of lyophilized antiserum) or normal rabbit serum was added to the reaction. Binding mixtures were incubated at room temperature for 20 to 30 min and then loaded onto 0.25× Tris-borate-EDTA–5% polyacrylamide gels that had been prerun for 2 h at 170 V. Gels were run for 3.5 to 4 h at 170 V, dried, and exposed to PhosphorImager screens (Molecular Dynamics) overnight.
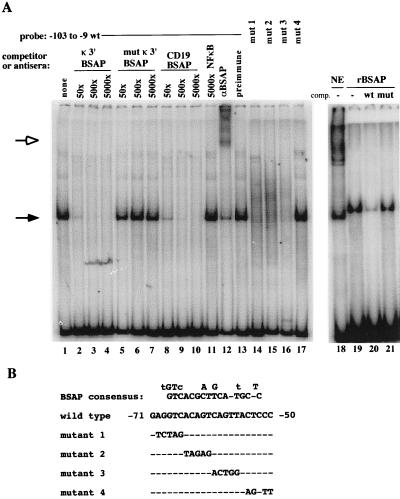
FIG. 7.
BSAP binds to a conserved element in the RAG-2 promoter. (A) Competition, antibody supershift, and mutational analysis of the B-cell-specific complex. EMSA was performed as for Fig. 5, using either 220-8 nuclear extract (NE) (lanes 1 to 18) or purified rBSAP (lanes 19 to 21) and either the wild-type (wt) −103 to −9 probe (lanes 1 to 13 and 18 to 21) or probes containing the mutations (mut) shown in panel B (lanes 14 to 17). Binding reactions contained no competitor (comp.) DNA (lanes 1 and 12 to 19) or the indicated molar excesses of unlabeled DNA containing previously characterized binding sites for BSAP from the Igκ 3′ enhancer (lanes 2 to 4) and the CD19 promoter (lanes 8 to 10), a mutant version of the κ enhancer BSAP site that no longer binds BSAP (lanes 5 to 7), the wild-type RAG-2 promoter (lane 20), the RAG-2 promoter with a BSAP site mutation (lane 21), or an NF-κB site (lane 11). Antibody supershift experiments were performed by adding either affinity-purified rabbit polyclonal anti-BSAP antiserum (lane 12) or preimmune rabbit serum (lane 13) to the binding reactions. Solid arrow, B-cell-specific complex; open arrow, supershifted complexes. (B) Comparison of the RAG-2 promoter BSAP site with the BSAP consensus sequence. The sequence shown is the complementary strand of the published consensus (9). Nucleotides above the consensus represent alternative nucleotides at each position, with lowercase letters representing nucleotides found in that position at a frequency of less than 30%. Also indicated are mutations analyzed by EMSA in panel A and by transfection in Fig. 8A.
Methylation interference.
Probes from −103 to −9 labeled on only one strand were prepared by PCR as described above, using end-labeled top- or bottom-strand primers. Dimethyl sulfate (DMS) treatment was performed as described elsewhere (4). Binding reactions were performed as above for EMSA, except scaled up to 80 μl, with 450,000 cpm of each probe and 20 μl of 220-8 nuclear extract. Bound and free probe were excised from the gel, electroeluted, and cleaved with piperidine as described above. Equivalent counts per minute of bound and free probes were loaded on a 6% polyacrylamide–7 M urea sequencing gel alongside a sequencing ladder. The dried gel was exposed to a PhosphorImager screen as well as X-ray film, and ImageQuant software (Molecular Dynamics) was used to confirm differences in band intensity.
In vivo DMS footprinting.
In vivo and in vitro DMS treatment, piperidine cleavage, and DNA purification were performed as described elsewhere (62). We subjected 0.5 to 1.5 μg of piperidine-treated DNA to ligation-mediated PCR. Primer extension was performed with the primer listed below as follows: 94°C for 5 min, 48°C for 30 min, and 76°C for 10 min. Double-stranded linker (45) was ligated to the extension products overnight at 16°C with T4 DNA ligase (New England Biolabs). Amplification of ligated products was performed with the extension primer (see below) and the linker primer. Samples were heated to 95°C for 1 to 2 min before Vent polymerase (New England Biolabs) was added. PCR cycles were as follows: 95°C for 3 min, 56°C for 2 min, and 76°C for 3 min for 1 cycle; 16 cycles of 95°C for 1 min, 56°C for 2 min, 76°C for 3 min 5 s/cycle; final extension for 10 min at 76°C. 5′-end-labeled primer was added for three cycles of 95°C for 1 min, 59°C for 2 min, and 76°C for 10 min. DNA was extracted, precipitated, and resuspended in loading dye. Samples were heated to 90°C for 5 min and run on 6% polyacrylamide–7 M urea sequencing gels alongside a sequencing ladder. Gels were dried and exposed to PhosphorImager screens. Differences in band intensity were analyzed with ImageQuant software. The following locus-specific primers were used: for primer extension, 5′-CCGTCGAGAAGCTTAAGACAGTCATTTT; for amplification, 5′-CAAATCGAGTTTCAATTAGACCTTGTCT; for end labeling, 5′-GACCTTGTCTTTACAAATGACTGGTATC.
In vitro DMS footprinting.
The wild-type −103 to −6 RAG-2 promoter probe labeled on either strand was generated by PCR as described above, using kinase-labeled top strand or bottom strand primers. The probes were purified by electrophoresis on a native 15% polyacrylamide gel and recovered by overnight elution and precipitation. Probes (100,000 cpm) were incubated with 50 ng of rBSAP in a buffer consisting of 50 mM sodium cacodylate (pH 8.0), 50 mM NaCl, 1 mM EDTA, 200 μg of bovine serum albumin per ml, and 2 μg of poly(dI-dC) in a final volume of 30 μl. The binding reaction mixtures were incubated at room temperature for 30 min, at which time 5 μl of 10% DMS was added. Aliquots of the methylation reactions were taken after 2, 5, and 10 min and quenched by addition of 6 μl of a mixture of 1.5 M sodium acetate and 1 M β-mercaptoethanol. Carrier tRNA was added, and the methylated probes were precipitated three times with ethanol. Methylated DNA was cleaved by reaction with 1 M piperidine at 90°C for 10 min, followed by three cycles of lyophilization and resuspension in double-distilled H2O. Finally, samples were resuspended in a formamide-containing gel loading buffer, denatured by heating to 90°C, and then analyzed by electrophoresis through a denaturing 10% polyacrylamide gel alongside a series of DNA sequencing reactions using the same radiolabeled primer. Controls included unmethylated probe DNA and DNA methylated in the absence of rBSAP.
RESULTS
Mapping RAG-2 transcription initiation sites.
To obtain genomic sequences including the transcription initiation region of RAG-2, a murine P1 phage genomic library was screened by PCR using primers for RAG-1 and RAG-2 coding sequences. A positive P1 clone was further subcloned, and sequences 5′ of the RAG-2 coding exon were identified by Southern hybridization with a RAG-2 cDNA probe. Restriction mapping of this subclone revealed the most 5′ exon of RAG-2 to be approximately 5 kb upstream of the coding exon (data not shown).
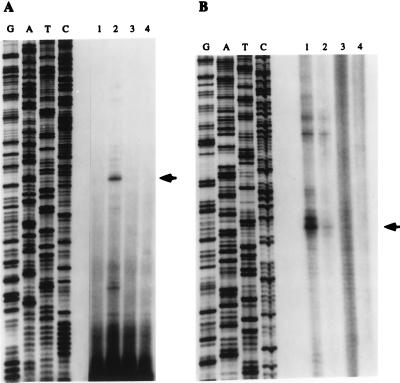
Primer extension and S1 nuclease protection assays were used to map RAG-2 transcription initiation sites (Fig. 1). Sources of RNA included 103 bcl2/4 cells cultured at 33 or 39°C, thymus, CH33 (a mature B-cell line), and yeast tRNA. 103 bcl2/4 is a conditionally transformed murine bone marrow pro-B-cell line previously shown to induce RAG-1 and RAG-2 transcription upon shift to restrictive growth conditions (7). With both assays, we observed multiple bands specific to the thymus and induced 103 bcl2/4 samples. The predominant start site, indicated by the arrow in Fig. 1, was identical in both analyses, and the patterns of start site usage were very similar between the thymus and B-cell samples.
FIG. 1.
Mapping of RAG-2 transcription initiation sites. (A) Primer extension. Total RNA from RAG-2 low-expressing 103 bcl2/4 cells grown at 33°C (lane 1), high-expressing 103 bcl2/4 cells grown at 39°C (lane 2), nonexpressing CH33 mature B cells (lane 3), or yeast tRNA (lane 4) was annealed to an end-labeled primer complementary to sequence in the first untranslated exon of RAG-2 and extended with Tth DNA polymerase. Extension products were run on a sequencing gel alongside a sequencing ladder. An autoradiograph of the dried gel is shown. Bands corresponding to multiple transcription initiation sites are seen in the high-expressing sample (lane 2). The major start site is indicated by an arrowhead. (B) S1 nuclease protection. Total RNAs from RAG-2-expressing 103 bcl2/4 cells grown at 39°C (lane 1) and thymocytes (lane 2), nonexpressing CH33 cells (lane 3), and yeast tRNA (lane 4) were hybridized to a uniformly labeled, complementary single-stranded DNA probe, followed by S1 nuclease digestion. Digested products were loaded onto a sequencing gel alongside a sequencing ladder. An autoradiograph of the dried gel is shown. Multiple transcription start sites are observed. An arrowhead indicates the major start site.
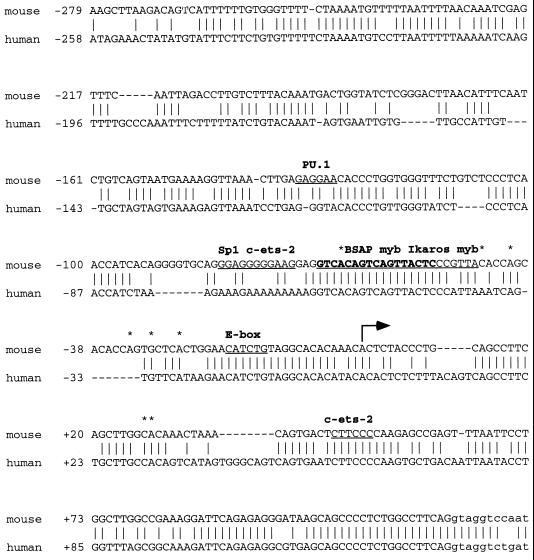
We determined the DNA sequence of the first exon and 280 bp 5′ of the major transcription initiation site (Fig. 2). There is a canonical splice donor site at the intron/exon junction. The predominant transcription start site as determined above is designated +1. There is no TATA motif upstream of any of the start sites. Several start sites (those at −65, −33, and +1) are six-of-seven matches to the initiator element consensus sequence [5′-YYA(+1)NA/TYY-3′ (14)], and the site at −46 is a perfect match. Database searching revealed a number of potential transcription factor binding sites in the sequence (29), including several that are involved in the transcription of lymphocyte-specific genes. These include an E box (consensus CANNTG) at −19, two c-ets-2 sites at −72 and +46 (consensus CTTCCC), a PU.1 site at −133 (AGAGGAACT), an Sp1 site at −82 (GGAGGGG), and an Ikaros site at −49 (YGGGAW). There are two myb sites (YAACKG) at −46 and −55. Overall there is a high degree of sequence identity (67% in the regions between −279 and +123) between the mouse and human genomic sequences (Fig. 2) (77). Of the potential transcription factor binding sites, only the Ikaros and myb sites, the E box, and the c-ets-2 site in the exon are conserved between species.
FIG. 2.
Comparison of the genomic sequence of the murine and human RAG-2 promoter regions. Sequences of the first exon and 5′ and 3′ flanking regions are shown. The human sequence has been published (77). The intronic sequence is in lowercase. The major transcription initiation site is numbered +1. Minor transcription initiation sites are indicated by asterisks. Potential transcription factor binding sites are underlined, with the BSAP site shown in boldface.
RAG-2 promoter activity is cell type specific.
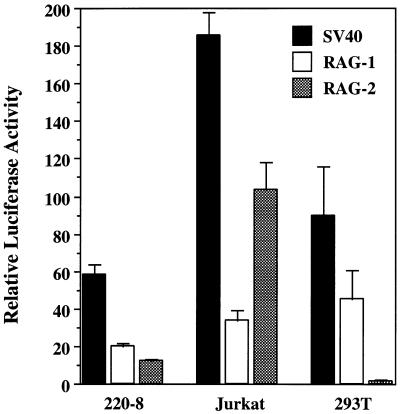
Using a luciferase reporter construct, we tested sequences 279 bp 5′ of and including the first untranslated exon of RAG-2 (−279 to +123) for functional promoter activity by transient transfection into a variety of lymphoid and nonlymphoid cell lines (Fig. 3 and Table 1). Each construct was transfected in duplicate or triplicate in each experiment, and each experiment was performed multiple times. Luciferase assay values were normalized to β-galactosidase levels from cotransfected plasmid pCMV-β-gal to control for transfection efficiency. The activity of each construct was calculated relative to that of the promoterless pGL2-Basic vector. For comparison, we also tested constructs containing the SV40 promoter and the murine RAG-1 promoter (−243/+72 [5]) (Fig. 3). The SV40 and RAG-1 promoters were active in every cell type tested, consistent with the observed lack of tissue specificity of the RAG-1 promoter (5, 38, 77). The RAG-1 promoter was more than twice as strong in 293T cells (an embryonal kidney cell line) as in several pro-B-cell lines.
FIG. 3.
Relative transcriptional activity of the SV40, RAG-1, and RAG-2 promoters in 220-8 pro-B cells, Jurkat T cells, and 293T embryonal kidney cells. Activity is expressed as fold increase in luciferase activity relative to the promoterless pGL2-Basic construct. This promoterless vector resulted in uncorrected light units in the ranges of 1,393 to 1,450 in 220-8 cells, 561 to 723 in Jurkat cells, and 109,337 to 123,208 in 293T cells. The relative values shown are adjusted for transfection efficiency by using β-galactosidase and represent averages of two or more experiments, each performed in duplicate.
TABLE 1.
Relative transcriptional activity of the SV40, RAG-1, and RAG-2 promoters in B-, T-, and nonlymphoid cell lines
| Cell line | Cell type | Relative promoter activitya
|
||
|---|---|---|---|---|
| SV40 | RAG-1 | RAG-2 | ||
| 63-12 | Pro-B | 58.9 | 32.1 | 25.3 |
| 220-8 | Pro-B | 58.5 ± 5.0 | 19.6 ± 7.7 | 12.6 ± 0.1 |
| 103 bcl2/4 | ||||
| 33°C | Pro-B | 32.0 | 9.3 | 10.5 |
| 37°C | Pro-B | 25.1 | 22.6 | 33.8 |
| WEHI-231 | Immature B | 12.4 | 14.5 | 11.7 |
| M12 | Mature B | 63.6 ± 25.4 | 30.5 ± 11.3 | 4.1 ± 1.1 |
| 2017 | Pre-T | 16.1 ± 3.7 | 6.9 ± 0.6 | 1.6 ± 0.3 |
| VL3-3M2 | Immature T | 27.9 | 17.1 | 11.9 |
| Jurkat | Mature T | 185.5 ± 12.1 | 33.4 ± 5.6 | 103.3 ± 14.6 |
| EL4 | Mature T | 62.1 | 25.1 | 4.1 |
| 293T | Emb. kidney | 89.5 ± 26.5 | 45.4 ± 15.4 | 1.8 ± 0.2 |
| NIH 3T3 | Fibroblast | 184.0 | 47.3 | 4.3 |
| P815 | Mastocytoma | 110.5 ± 14.4 | 15.4 ± 2.0 | 2.0 ± 0.2 |
| HeLa | Epithelial | 81.0 | 8.8 | 2.2 |
Expressed as fold increase in luciferase activity relative to the promoterless pGL2-Basic construct. Values were adjusted for transfection efficiency by using β-galactosidase. All experiments were performed in duplicate. Where experiments were performed more than once, standard deviations are shown.
In contrast, the RAG-2 promoter was 7- to 19-fold more active in a panel of pro-B-cell lines compared with 293T cells (Table 1). Interestingly, both RAG-1 and RAG-2 promoter activity increased when transfected 103 bcl2/4 cells were cultured at the nonpermissive temperature (Table 1). The RAG-2 promoter was active in the immature B-cell line WEHI 231, which expresses the endogenous RAG-2 gene (data not shown), but showed only weak activity in M12, a mature B-cell lymphoma which lacks endogenous RAG-2 expression (Table 1), suggesting that the RAG-2 promoter may contribute to the developmental stage specificity of RAG-2 expression.
We tested the RAG-2 promoter in several T-cell lines and found it to be highly active in Jurkat and much less active in EL-4. Although both of these cell lines have been reported to lack RAG expression (49, 59), reverse transcription-PCR analysis revealed that Jurkat cells express significant amounts of endogenous RAG-2 mRNA (data not shown and reference 18). Surprisingly, the RAG-2 promoter was essentially inactive in 2017, an Abelson virus-transformed immature T-cell line reported to have detectable recombinase activity (65).
Although the RAG-2 construct had some activity in nonlymphoid cell lines, it was much less than that observed in lymphoid cell lines. To investigate the possibility of a repressor element within the promoter and first exon, 5′ and 3′ truncation constructs were also tested in 293T cells. No derepression was observed when a construct corresponding to the minimal core promoter (−71 to +37) was used (see below; data not shown). In summary, unlike the RAG-1 promoter, the RAG-2 promoter shows lymphoid-cell-specific transcriptional activity.
DNA sequences required for RAG-2 promoter activity.
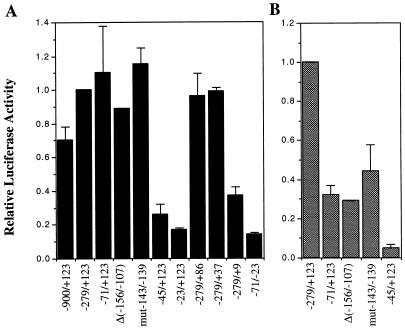
We generated an extensive series of 5′ and 3′ truncations of the RAG-2 promoter and analyzed their activity by transfection into the pro-B-cell line 220-8 and the T-cell line Jurkat (Fig. 4). Near-maximal activity in both cell types was observed with a construct extending from −279 to +123. This construct gave a signal 12-fold above the promoterless vector background in 220-8 cells and 100-fold above the background in Jurkat cells. A construct including approximately 600 bp of additional upstream sequence had somewhat less promoter activity in 220-8, eightfold above background (Fig. 4A). A 5′ deletion extending to nucleotide −71 did not affect promoter activity in 220-8 (or in other pro-B-cell lines [data not shown]) but resulted in a 70% decrease in promoter activity in Jurkat (and VL3-3M2 [data not shown]). Further deletions and point mutations localized this T-cell-specific promoter element to a region between nucleotides −156 and −107, with much of the activity dependent on the integrity of nucleotides −143 to −139 (Fig. 4B). Further truncation from the 5′ end to −45 reduced promoter activity by 75% in 220-8 and greater than 90% in Jurkat cells (and VL3-3M2 [data not shown]). Thus, a DNA sequence between nucleotides −71 and −45 is important for promoter activity in both B- and T-cell lines. From the 3′ end, constructs could be truncated to +37 without significant loss of activity. Further truncation to +9 resulted in a 64% reduction in promoter activity, however. In summary, the minimal RAG-2 promoter active in pro-B cells contains sequences between −71 and +37 while the minimal sequences required in Jurkat cells extend upstream to position −156.
FIG. 4.
Delineation of critical sequences in the RAG-2 promoter in 220-8 pro-B cells (A) and Jurkat T cells (B). Activity is expressed as fold increase in luciferase activity relative to the −279/+123 construct. The values shown are adjusted for transfection efficiency by using β-galactosidase and represent averages of two experiments, each performed in duplicate. The RAG-2 promoter sequences contained in each construct are indicated beneath each column. The Δ(−156/−107) construct is the −279/+123 construct with an internal deletion of base pairs −156 to −107. The mut −143/−139 construct is the −279/+123 construct with a 5-bp mutation from −143 to −139.
Factor binding to the RAG-2 core promoter.
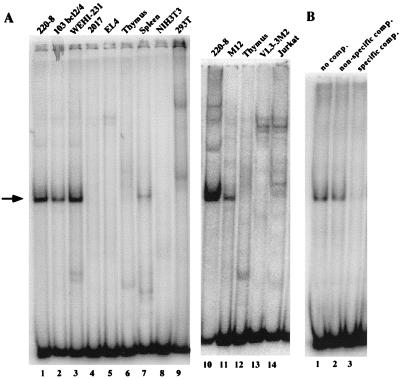
To identify nuclear factors capable of binding to RAG-2 promoter regulatory elements, EMSAs were performed with a probe which spans a portion of the functionally important pro-B-cell core promoter region (−103 to −9). As shown in Fig. 5A, a single DNA-protein complex was detected in nuclear extracts prepared from pro-B (220-8 and 103 bcl2/4)-, immature B (WEHI 231)-, and mature B (M12)-cell lines, as well as from spleen. This complex was not seen in nuclear extracts prepared from T-cell lines (2017, Jurkat, VL3-3M2, and EL4), thymus, or nonlymphoid cell lines (NIH 3T3 and 293T). All extracts demonstrated similar binding activities in assays using a control probe containing the binding site for Oct-1 (data not shown). The complex was specifically competed with excess unlabeled DNA identical in sequence to the probe but not with an equivalent molar excess of an unrelated DNA fragment (Fig. 5B). No additional complexes were observed with probes extending through the rest of the minimal core promoter region (to +37).
FIG. 5.
A B-cell-specific nuclear factor binds to the core RAG-2 promoter. (A) EMSA of a RAG-2 core promoter probe. An end-labeled probe spanning nucleotides −103 to −9 was incubated with nuclear extracts from B-cell lines (lanes 1 to 3, 10, and 11), T-cell lines (lanes 4, 5, 13, and 14), thymus (lanes 6 and 12), spleen (lane 7), and nonlymphoid cell lines (lanes 8 and 9). The binding reaction was analyzed by electrophoresis on a nondenaturing acrylamide gel, which was dried and exposed to a PhosphorImager screen. A B-cell-specific complex is indicated by the arrow. The integrity and relative concentration of the extracts were confirmed in separate assays with a probe binding the ubiquitous Oct-1 transcription factor (data not shown). (B) Specificity of the B-cell-specific complex. EMSA was performed as for panel A, using 220-8 nuclear extract and the RAG-2 promoter −103 to −9 probe. Either no competitor (comp.) DNA (lane 1) or a 100-fold molar excess of unlabeled DNA unrelated in sequence to the probe (lane 2; RAG-2 promoter nucleotides −199 to −103) or identical in sequence to the probe (lane 3; nucleotides −103 to −9) was added to the binding reaction. A PhosphorImager exposure of the dried gel is shown.
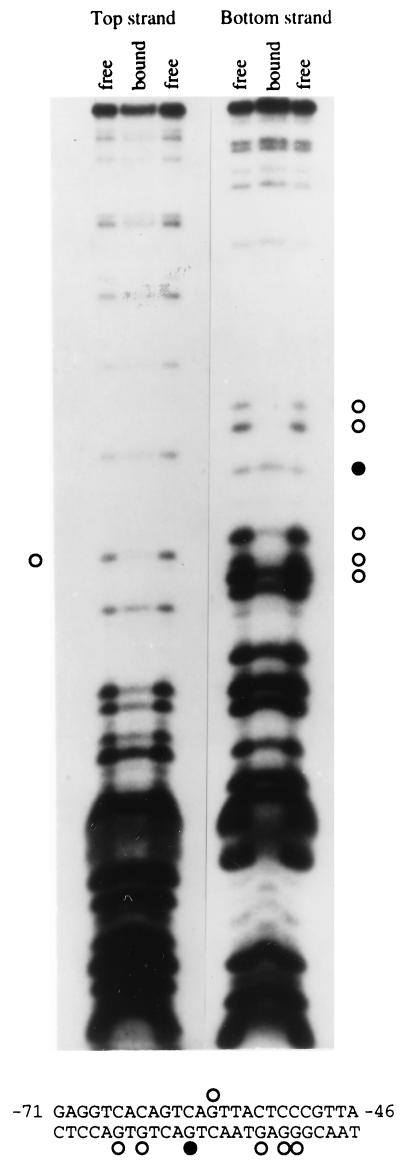
Methylation interference analysis was performed to identify essential DNA-protein contacts within the B-cell-specific complex (Fig. 6). Methylation at several G residues on the bottom strand of the sequence interfered with protein binding. Fewer contacts were made on the top strand. The binding site as discerned by this method spans at least 16 bp and is localized to a region shown to be critical for promoter activity in the transient transfection reporter construct assay (Fig. 4A; bp −66 to −51).
FIG. 6.
Methylation interference analysis of the EMSA complex. Top- and bottom-strand end-labeled versions of the −103 to −9 EMSA probe were treated with DMS, purified, and incubated with 220-8 nuclear extracts. The bound and free probes were excised from a nondenaturing gel, electroeluted, and cleaved at methylated G residues with piperidine. Single-stranded, end-labeled cleavage products were analyzed by electrophoresis on a sequencing gel alongside a sequencing ladder (not shown). An autoradiograph of the dried gel is shown. Differences in band intensity greater than sample variability in loading were confirmed by using ImageQuant software. Open circles, G residues whose methylation interferes with protein binding; filled circles, G residues whose methylation enhances protein binding. The pattern of interaction between the protein and its binding site is summarized at the bottom.
BSAP binds to the RAG-2 core promoter and is critical for its function.
Because the promoter binding activity was B cell specific and encompassed a rather large sequence, we asked whether it might contain BSAP (Pax-5), a transcription factor expressed only in B cells, the central nervous system, and testis (2). As shown in Fig. 7A, formation of the EMSA complex was competed by well-characterized BSAP sites from the Igκ 3′ enhancer (57) and the CD19 promoter (36) but not by large molar excesses (>5,000 fold) of a κ 3′ enhancer site mutant that no longer binds BSAP (57) or an irrelevant NF-κB binding site. In addition, polyclonal rabbit anti-BSAP antiserum, but not preimmune rabbit serum, caused a supershift of the complex (compare lanes 12 and 13). Taken together, the expression pattern, competition, and supershift data demonstrate that BSAP binds to a critical element in the RAG-2 promoter.
Our gel shift probe contains potential binding sites for other factors in addition to BSAP (Fig. 2). To determine whether the BSAP-containing gel shift complex described above contains other factors as well, we compared its mobility to that of a complex generated by using His-tagged rBSAP purified from E. coli. The bacterially produced BSAP generated a specific DNA-protein complex (Fig. 7A, lanes 19 to 21). This complex migrated slightly more slowly than a complex generated on the same probe, using 220-8 pro-B-cell nuclear extract (lane 18). This mobility difference can be accounted for by the additional amino acids introduced into the rBSAP by the expression vector (see Materials and Methods). Thus, we conclude that BSAP is the only protein within this specific pro-B-cell gel shift complex.
The BSAP DNA binding consensus consists of two half-sites, which contribute independently to binding affinity (10). No naturally occurring BSAP site completely conforms to the consensus (9). The RAG-2 promoter binding site contains a TAC trinucleotide in the 3′ half-site instead of the usual TGC, but the presence of an A in this position has been shown to preserve the ability of a site to bind BSAP (57, 66). There is an exact match to the GTCAC in the 5′ half-site consensus (Fig. 6B). Further evidence for the significance of this BSAP site is our observation that the murine and human RAG-2 promoter sequences are identical from positions −70 to −50 (Fig. 2).
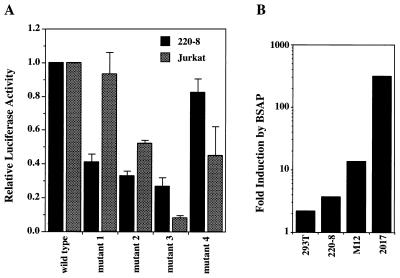
To further characterize the functional importance of the BSAP site, we generated a series of 5-bp substitution mutations across this interval (Fig. 7B). Mutation of base pairs −70 to −66, −65 to −61, or −60 to −56 abolished the binding of BSAP to the RAG-2 promoter probe, whereas binding was unaffected by mutation of bp −54 to −50 (Fig. 7A). These same mutations were introduced into the −279/+123 RAG-2 reporter construct and assayed by transient transfection into 220-8 cells. The mutations that abolished BSAP binding in the gel shift decreased promoter activity by 60 to 75%, whereas the mutation that preserved binding had little effect on promoter activity (Fig. 8A). When we tested each of these mutants by transfection into Jurkat T cells, we observed a different pattern of activity (Fig. 8A). While mutant 1 decreased promoter activity in 220-8 by approximately 60%, it had no effect in Jurkat. Conversely, mutant 4 had little effect on transcription in 220-8 but resulted in a 50% decrease in promoter activity in Jurkat. The latter mutation disrupts both the Ikaros site and one of the Myb sites in this region (Fig. 2), suggesting that these factors may contribute to core promoter activity in T cells but not B cells.
FIG. 8.
BSAP is a critical and potent activator of the RAG-2 promoter. (A) Transient transfection reporter assay comparing the four mutants described in Fig. 7B to the wild-type promoter. The mutations were introduced into the −279/+123 promoter construct, and the experiments were performed in 220-8 cells and Jurkat T cells. Activities of the mutants are depicted relative to the wild-type activity, which is assigned a value of 1. The results shown are the average of three (220-8) or five (Jurkat) experiments, each performed in duplicate and adjusted for transfection efficiency by using β-galactosidase. (B) Effect of BSAP expression vector cotransfection on RAG-2 promoter activity. The cell lines indicated were cotransfected with the −279/+123 reporter construct and either empty expression vector or BSAP expression vector. The results are depicted as the fold increase in luciferase activity by BSAP compared to empty vector. Data shown are from one representative experiment performed in duplicate and adjusted for transfection efficiency.
Because BSAP has been shown to have either transcriptional activating or repressing functions, depending on context, we performed cotransfection experiments to determine whether BSAP could transactivate the RAG-2 promoter. Cell lines were cotransfected with the −279/+123 RAG-2 promoter reporter construct and either empty expression vector or a murine BSAP expression vector. Figure 8B shows the fold increase in reporter activity over empty vector conferred by BSAP expression. The effects of BSAP expression varied depending on the recipient cell line. In 2017 cells, which had very low levels of activity with the RAG-2 promoter reporter in the absence of exogenous BSAP (Table 1), BSAP expression caused a 300-fold increase in promoter activity. M12 cells, which also had low activity in the absence of exogenous BSAP, showed a 14-fold induction by BSAP. Expression of BSAP in 220-8 and 293T cells had little effect. Interestingly, the effect of BSAP cotransfection in lymphoid cell lines appears to be inversely proportional to the amount of BSAP present in the untransfected cell line. 220-8 cells have severalfold more BSAP than M12 cells (data not shown), while 2017 cells have no BSAP. 293T cells also lack BSAP, however, and the failure of BSAP cotransfection to induce the RAG-2 promoter construct must be due to other causes (see Discussion). We believe that these experiments represent direct effects of BSAP on the RAG-2 promoter since BSAP-site mutant reporter constructs show significantly less stimulation in these cotransfection experiments (data not shown).
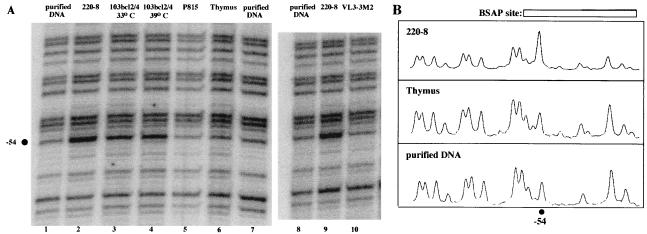
To determine whether BSAP might regulate the endogenous RAG-2 promoter in vivo, we performed in vivo DMS footprinting on 220-8 and 103 bcl2/4 pro-B cells, thymocytes, the T-cell line VL3-3M2, and the nonlymphoid mastocytoma cell line P815. As shown in Fig. 9, the G at position −54 within the BSAP site on the bottom strand exhibits enhanced DMS reactivity relative to in vitro-treated DNA specifically in pro-B cells but not in thymocytes, VL3-3M2, or P815 cells. No footprint was detected on the top strand. Interestingly, the in vivo BSAP site footprint does not change when 103 bcl2/4 cells are cultured under restrictive conditions. Thus, the RAG-2 promoter BSAP site is occupied in vivo in pro-B cells.
FIG. 9.
The RAG-2 promoter BSAP site is occupied in vivo in pro-B cells. (A) In vivo DMS footprinting of the RAG-2 promoter. Intact 220-8 or 103 bcl2/4 pro-B cells, P815 mastocytoma cells, thymocytes, VL3-3M2 T cells, or purified DNA isolated from 220-8 cells or thymocytes was treated with DMS. The purified DNA was subjected to piperidine cleavage at methylated G residues, linker ligation, and PCR amplification with nested locus-specific primers and a linker-specific primer. Labeled extension products were analyzed on a sequencing gel alongside a sequencing ladder, and the dried gel was exposed to a PhosphorImager screen. The bottom strand of the promoter is analyzed in the experiments shown. Lanes 1 to 7 and 8 to 10 represent two separate experiments. The black circle indicates the G at −54 which is hypersensitive in vivo to DMS in the 220-8 and 103 bcl2/4 cells. (B) Histograms of lanes 2, 6, and 7. ImageQuant software was used to quantify the peak intensities of the footprint ladders shown in panel A. The absolute intensities of the peaks differ among the samples, but significant differences in relative peak intensities are indicated. The G residue at −54 is hypersensitive in 220-8 cells in comparison to thymocytes and purified DNA.
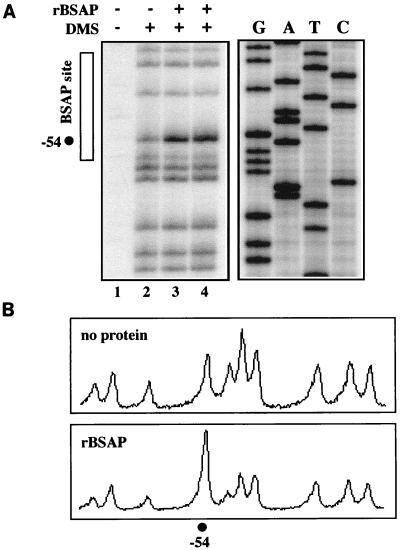
While this in vivo footprinting analysis revealed a correlation between DMS hyperreactivity at nucleotide −54 of the RAG-2 promoter, the presence of BSAP, and the activity of the RAG-2 promoter reporter construct in pro-B-cell lines, this experiment does not allow us to conclude that BSAP binding is responsible for the in vivo footprint. To determine whether the DMS hyperreactivity that we observed was caused by BSAP binding, we performed in vitro DMS footprinting on a radiolabeled RAG-2 promoter probe (−103 to −9), using purified recombinant BSAP (Fig. 10). This analysis revealed that the same nucleotide within the RAG-2 promoter which was hypersensitive to DMS within pro-B cells in vivo (−54) was also rendered hypersensitive upon interaction with rBSAP in vitro (compare lane 2 with lanes 3 and 4). We failed to observe any additional enhanced or diminished bands on either strand of this probe (Fig. 10 and data not shown). Thus, we conclude that BSAP is bound to a critical region of the RAG-2 promoter in progenitor B-cell lines in vivo.
FIG. 10.
In vitro DMS footprinting of the RAG-2 promoter. (A) Double-stranded DNA consisting of the −103 to −9 region of the RAG-2 promoter was labeled on the bottom strand by PCR using a kinase-labeled primer and incubated in vitro with buffer (lane 2) or with purified rBSAP (lanes 3 and 4). The DNA-protein complexes were then treated with DMS for either 2 min (lanes 2 and 3) or 5 min (lane 4), repurified, and finally reacted with piperidine to cleave methylated G residues. The samples were then analyzed by denaturing polyacrylamide gel electrophoresis alongside a DNA sequencing ladder of the same region generated by using the same kinase-labeled primer (lanes labeled G, A, T, and C). A phosphorimage of the gel is shown. Lane 1 contains radiolabeled probe which was not reacted with DMS. The location of the BSAP binding site is indicated along with the location of the hypersensitive site observed in vivo at −54 (Fig. 9). Note that the orientation of this direct analysis of the bottom strand of the promoter is the reverse of that generated by the indirect technique of in vivo footprinting. (B) ImageQuant-generated tracings of lanes 2 and 3. The position of the hypersensitive nucleotide indicated.
DISCUSSION
Understanding the transcriptional regulation of the RAG genes is likely to provide important insights into the control of lymphocyte development and differentiation. As a step toward this goal, we have cloned and characterized the promoter of the murine RAG-2 gene. Using a transient transfection assay, we defined a 193-bp core RAG-2 promoter, within which three regions (−156 to −107, −71 to −45, and +9 to +37) contribute most of the activity (Fig. 4). We found the −156 to −107 region to be dispensable for promoter activity in B cells but important in T cells. The −71 to −45 region was important for transcription in both cell types but apparently interacts with distinct factors in B and T cells. Our finding that the region between +9 and +37 contributes to promoter activity was of interest since sequences in the RAG-1 untranslated exon have also been shown to be critical for basal promoter function (5).
The core RAG-2 promoter demonstrated tissue specificity, with greatest activity in pro-B-cell lines and two T-cell lines and little or no activity in a mature B-cell line and several nonlymphoid cell lines. Surprisingly, the promoter was inactive in an immature T-cell line, 2017, which expresses the endogenous RAG-2 gene. Unlike most available T-cell lines, however, 2017 was transformed by the Abelson leukemia virus and thus might display unusual transcriptional regulatory properties (35, 65).
We found that a region within this promoter which is essential for its function binds to the B-cell transcription factor BSAP (Pax-5). Disruption of the BSAP binding site by site-directed mutagenesis significantly diminished promoter activity in pro-B cells (Fig. 8). Although the same general region was also important for promoter activity in T cells, analysis of a panel of specific mutants revealed the critical sequences to differ between B and T cells. Cotransfection of a BSAP expression vector into immature T-cell and mature B-cell lines resulted in dramatic increases in promoter activity, whereas BSAP cotransfection did not activate the promoter in nonlymphoid cells. The biological significance of these observations was heightened by the results of footprinting analyses which indicated that this BSAP site was occupied in vivo within B-lineage cells expressing RAG-2 and that the in vivo footprint was a consequence of BSAP binding (Fig. 9 and 10).
RAG-2 transcription initiation.
RAG-2 transcription initiates from multiple sites clustered within a 90- to 100-bp region. In this respect the murine RAG-2 gene differs from the human RAG-2 gene, which initiates transcription from one predominant site (77). The difference between the two species is curious because there is a high level of sequence identity over the region from which transcription initiates, which suggests similar transcriptional control mechanisms. The major start site that we observed does correspond closely to the major human start site, however. The RAG-2 promoter lacks a TATA sequence, but several of the initiation sites are close matches to the consensus sequence for the initiator element. Multiple start sites and absence of a TATA box are characteristics shared by several other early lymphocyte-specific promoters, including the RAG-1 (5, 17, 38, 77), VpreB (50), λ5 (37), B29 (30, 68), blk (79), and CD19 (36) promoters.
Tissue specificity of the RAG-2 promoter.
To assess its potential contribution to the tissue specificity of RAG-2 expression, we tested the activity of the RAG-2 promoter construct in various B- and T-cell lines, including 220-8, 63-12, and 103 bcl2/4 (pro-B), VL3-3M2 and 2017 (immature T), Jurkat and EL-4 (mature T), WEHI 231 (immature B), M12 (mature B), 293T (embryonal kidney), P815 (mastocytoma), HeLa (cervical carcinoma), and NIH 3T3 (fibroblast). The promoter showed extremely low but reproducible activity in each of the nonlymphoid cell lines, severalfold lower than that observed in pro-B cells. In addition, it was inactive in mature B- and T-cell lines lacking in RAG expression. These differences become even more dramatic when compared to transcription driven by the SV40 and RAG-1 promoters in these same cell lines. As others have shown, the RAG-1 promoter has significant activity in nonlymphoid cells and in a mature B-cell line (5, 17, 38, 77). Despite the coordinate and equally specific transcription of these two genes, the RAG-2 promoter confers a degree of tissue and developmental stage specificity that the RAG-1 promoter does not.
While the RAG-2 promoter was active in both pro-B- and T-cell lines, we found that different elements within the promoter were important in these two lineages. An upstream region extending from nucleotides −156 to −107 was important in T cells and had no apparent effect in pro-B cells. We have thus far been unable to identify the factors that may interact with this region of the promoter in T cells. It is possible that this region of the promoter will prove to be important for RAG-2 expression in primary B-cell progenitors but that some aspect of cell transformation interferes with its activity. In this regard, it is worth noting that RAG-2 promoter activity in Jurkat T cells is 5- to 10-fold greater than in various pro-B- and immature B-cell lines. Finally, it remains possible that additional DNA sequences important for promoter activity in vivo are not revealed by these assays using transformed cell lines.
We found that the region between −71 and −45 is important for RAG-2 promoter activity in both lymphoid lineages. This region contains potential binding sites for BSAP, Ikaros, and myb. As described below, we demonstrated that BSAP is critical for promoter activity in transfected pro-B-cell lines but not in T-lineage cells (BSAP is not expressed in T cells). Ikaros and myb, however, are essential factors expressed at multiple stages of B- and T-cell development (20, 40). It is possible that these latter factors drive constitutive promoter activity at multiple stages of B- and T-cell development, but that BSAP is involved in the B-cell-specific regulation of RAG-2 transcription (see below).
A BSAP site in the core RAG-2 promoter is critical for its activity in B cells.
We found that an essential region of the RAG-2 promoter specifically interacts with the well-characterized transcription factor BSAP. BSAP, a member of the Pax family of transcription factors, is expressed primarily in B cells, the central nervous system, and testis (6). BSAP is found in all stages of B-cell development except plasma cells. Several B-cell-specific genes have been implicated as targets for BSAP activation, including CD19 (36), blk (79), VpreB, and λ5 (51), mb-1 (16), and XBP-1 (54), as well as the germ line Ig heavy-chain Cɛ promoter (58). Interestingly, many of these genes have multiple transcription start sites and lack a TATA box. This may be coincidence, or BSAP may play a role in recruiting the basal transcription machinery to TATA-less promoters in B cells. In addition BSAP has been shown or suggested to act as a repressor at the heavy-chain 3′α enhancer (47, 64), the kappa light-chain 3′ enhancer (57, 62), and the J-chain gene promoter (55).
BSAP and the regulation of the core RAG-2 promoter.
The expression of BSAP DNA binding activity among the group of B-cell lines used in this study parallels the activity of the RAG-2 promoter in these cells. Our observation that cotransfection of a BSAP expression vector led to a dramatic increase of RAG-2 promoter activity in the immature T- and mature B-cell lines verifies BSAP’s role as a transactivator of RAG-2 transcription in lymphocytes. Interestingly, cotransfection of the BSAP expression vector into 293T cells failed to activate the RAG-2 promoter construct. We can envision several potential mechanisms for this tissue-specific effect: (i) another factor(s) present in B and T cells is required to cooperate with BSAP; (ii) BSAP is posttranslationally modified in a lymphoid-cell-specific fashion, enabling it to transactivate; or (iii) there is a repressor or silencer element within the minimal core promoter region that is active in nonlymphoid cells. Others have observed the failure of cotransfected BSAP to activate a reporter construct in nonlymphoid cell lines (2, 56). However, cotransfected BSAP was able to activate a construct with two BSAP sites upstream of a TATA-containing minimal promoter in HeLa cells, making possibility ii above unlikely and suggesting that more stringent demands are placed upon BSAP by TATA-less promoters (56).
Mice lacking BSAP have been generated using homologous recombination in embryonic stem cells (73). These mice exhibit a complete lack of detectable B-cell progenitors in the fetal liver, whereas bone marrow B cells develop to the early pro-B stage (B220+ CD43+ λ5+ VpreB+ HSA+ BP1−) (48). These pro-B cells express normal levels of RAG-1 and RAG-2 and contain numerous D-to-JH rearranged Ig heavy-chain loci. Thus, it appears from this result that BSAP is not required for B-cell-specific RAG-2 expression in vivo. There are several ways to reconcile this report with the data presented here. First, BSAP may be important but not essential for bone marrow RAG-2 expression. Second, the BSAP-deficient mice may compensate for the loss of BSAP expression through the activity of another Pax family member. Pax-2 and Pax-8, which are most closely related to BSAP and bind to the same consensus sequence, are not expressed in the spleen (12, 15, 53), but there are no data in the literature regarding their expression in bone marrow B cells. It is even possible that a currently unknown Pax family member is present in B cells and capable of substituting for BSAP at the RAG-2 promoter. Finally, it is possible that BSAP is necessary for regulated RAG-2 expression only during the later stages of B-cell development.
There are three circumstances where there might be a role for a B-cell-specific RAG-2 regulator such as BSAP. First, early pre-B cells contain very little RAG-1 or RAG-2 mRNA (22). RAG expression is reactivated, however, in small resting pre-B cells. This modulation of RAG expression has been suggested to play a role in heavy-chain gene allelic exclusion. Second, immature B cells in the bone marrow expressing an autoreactive antigen receptor can undergo receptor editing mediated by increased recombinase expression (19, 42, 70). Finally, germinal center B cells have also recently been shown to reactivate RAG expression and functional recombinase activity upon antigen stimulation (24, 25, 31, 32, 52). Interestingly, these cells also reactivate transcription of λ5 (24), which has been suggested to be a target for regulation by BSAP. Each of these circumstances occurs after the block in B-cell development observed in mice lacking BSAP. Thus, the persistence of RAG expression in BSAP-deficient pro-B cells does not rule out a later role for BSAP. Other experimental approaches will be required to test this hypothesis.
Since the initial submission of this paper, a report supporting a role for BSAP in the regulation of RAG expression in vivo has been published. Verkoczy and Berinstein used differential display to compare gene expression between two clonally related variants of a mature B-cell line, one of which expressed approximately 10-fold more RAG-1 and RAG-2 than the other; BSAP was cloned as a transcript whose expression correlated with high levels of RAG transcription (74). Aside from providing evidence that BSAP may regulate the endogenous RAG-2 gene, it is interesting that the cell line used in the study is a mature B-cell line that undergoes spontaneous secondary light-chain rearrangements in culture, again supporting a role for BSAP in receptor editing or germinal center RAG reexpression.
There is considerable evidence for a role for BSAP in several events during later stages of B-cell development (6). Overexpression of BSAP in plasma cell lines or resting splenic B cells induced proliferation, whereas down-regulation of BSAP by specific antisense oligonucleotides greatly diminished proliferation mediated by a variety of stimuli and also decreased lipopolysaccharide (LPS)- and interleukin-4 (IL-4)-induced Ig class switching (75). BSAP has been shown by similar approaches to repress the activity of the Ig heavy-chain 3′α enhancer and the J-chain promoter during B-cell development up to the plasma cell stage, when termination of BSAP expression relieves the repression (46, 47, 55, 64). Several groups have reported that a conserved BSAP site in the murine and human germ line Igɛ promoters is required for the LPS and IL-4 inducibility of these promoters (39, 58, 67), but this finding has been challenged (11). The parallels between Igɛ germ line transcription and class switching and RAG transcription are intriguing: both can be induced by LPS and IL-4, and both take place in the germinal center. BSAP may be an essential factor for both events.
Further support for a role for BSAP in RAG-2 reactivation in the periphery comes from studies of IL-7 signaling. IL-7 was recently shown to function as a cofactor for RAG reactivation in cultured IgD+ splenic B cells, and administration of antibody specific to the alpha chain of the IL-7 receptor (IL-7Rα) blocked the reactivation of the RAG genes in germinal center B cells of immunized mice (33). Mice lacking the IL-7Rα have a block in B-cell development at the same stage as BSAP-deficient mice (V-to-DJ rearrangement), and BSAP transcript levels are reduced in IL-7Rα mutant animals (8). The functional consequences of this reduction were demonstrated by reduced expression of the CD19 mRNA and protein. These data suggest that BSAP functions downstream of an IL-7 signaling pathway. RAG-2 reexpression in germinal center B cells may depend on an IL-7-mediated increase in BSAP levels.
Receptor editing in the bone marrow and germinal center reexpression of the RAG genes are two cases where the regulation of RAG transcription in B cells does not closely parallel that in T cells. RAG transcription during the earliest stages of B- and T-cell development might be regulated identically, with B-cell-specific factors coming into play only at these later stages of RAG expression. Alternatively, distinct mechanisms may regulate the RAG genes in B and T cells throughout their development, producing parallel expression in the two lineages during early stages of development but diverging later on. In addition, it is likely that other regulatory sequences besides the promoter are important for controlling RAG-2 expression. Only the elucidation of the complete repertoire of cis elements and transcription factors regulating the RAG genes will allow us to distinguish among these possibilities.
ACKNOWLEDGMENTS
We thank Eugenia Spanopoulou for providing us with 293T cells and the pEF-BOS expression vector, Meinrad Busslinger for sharing with us anti-BSAP antisera, and Steven Desiderio and Patty Zwollo for providing the BSAP cDNA. We thank Cynthia Guidos, Antony Rosen, David Schatz, Robert Siliciano, and Stephen Smale for gifts of various cell lines. We also thank Chi Dang for access to his luminometer and Qian-Fei Wang for performing gel mobility shift analyses using recombinant BSAP. This paper was improved by criticisms offered by Chi Dang and Kees Murre, by various members of the Schlissel lab, and by anonymous reviewers.
M.S.S. acknowledges the support of NIH grant HL48702, the W. W. Smith Foundation, and the Leukemia Society of America. J.L. is a Howard Hughes Medical Institute predoctoral fellow and a member of the Biochemistry, Cellular and Molecular Biology Training Program at Johns Hopkins.
REFERENCES
- 1.Abe T, Tsuge I, Kamachi Y, Torii S, Utsumi K, Akahori Y, Ichihara Y, Kurosawa Y, Matsuoka H. Evidence for defects in V(D)J rearrangements in patients with severe combined immunodeficiency. J Immunol. 1994;152:5504–5513. [PubMed] [Google Scholar]
- 2.Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 3.Alt F, Yancopoulos G, Blackwell T, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 5.Brown S, Miranda G, Galic Z, Hartman I, Lyon C, Aguilera R. Regulation of the RAG-1 promoter by the NF-Y transcription factor. J Immunol. 1997;158:5071–5074. [PubMed] [Google Scholar]
- 6.Busslinger M, Urbanek P. The role of BSAP (Pax 5) in B-cell development. Curr Opin Genet Dev. 1995;5:595–601. doi: 10.1016/0959-437x(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y-Y, Wang L C, Huang M S, Rosenberg N. An active v-abl protein tyrosine kinase blocks immunoglobulin light chain gene rearrangement. Genes Dev. 1994;8:688–697. doi: 10.1101/gad.8.6.688. [DOI] [PubMed] [Google Scholar]
- 8.Corcoran A, Riddell A, Krooshoop D, Venkitaraman A. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 9.Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 11.Delphin S, Stavnezer J. Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline ɛ promoter: regulation by NF-IL-4, a C/EBP member and NFκB/p50. J Exp Med. 1995;181:181–192. doi: 10.1084/jem.181.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dressler G, Deutsch U, Chowdhury K, Nornes H, Gruss P. Pax2, a new murine paired-box-containing gene and its expression in the developing excretory system. Development. 1990;109:787–795. doi: 10.1242/dev.109.4.787. [DOI] [PubMed] [Google Scholar]
- 13.Dunn T B, Potter M. A transplantable mast-cell neoplasm in the mouse. J Natl Cancer Inst. 1957;18:587–595. [PubMed] [Google Scholar]
- 14.Ernst P, Smale S. Combinatorial regulation of transcription I: general aspects of transcriptional control. Immunity. 1995;2:311–319. doi: 10.1016/1074-7613(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 15.Fickenscher H, Chalepakis G, Gruss P. Murine Pax-2 protein is a sequence-specific trans-activator with expression in the genital system. DNA Cell Biol. 1993;12:381–391. doi: 10.1089/dna.1993.12.381. [DOI] [PubMed] [Google Scholar]
- 16.Fitzsimmons D, Hodsdon W, Wheat W, Maira S-M, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 17.Fuller K, Storb U. Identification and characterization of the murine Rag 1 promoter. Mol Immunol. 1997;34:939–954. doi: 10.1016/s0161-5890(97)00000-x. [DOI] [PubMed] [Google Scholar]
- 18.Gaffney P M, Lund J, Miller J S. FLT-3 ligand and bone marrow stroma-derived factors promote CD3 gamma, CD3 delta, CD3 zeta, and RAG-2 gene expression in primary human CD34+LIN-DR- marrow progenitors. Blood. 1998;91:1662–1670. [PubMed] [Google Scholar]
- 19.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 21.Glimcher L, Hamano T, Asofsky R, Heber-Katz E, Hedrick S, Schwartz R H, Paul W E. I region-restricted antigen presentation by B cell-B lymphoma hybridomas. Nature. 1982;298:283–286. doi: 10.1038/298283a0. [DOI] [PubMed] [Google Scholar]
- 22.Grawunder U, Leu T M J, Schatz D G, Werner A, Rolink A G, Melchers F, Winkler T H. Down-regulation of RAG1 and RAG2 gene expression in pre-B cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 23.Groves T, Katis P, Madden Z, Manickam K, Ramsden D, Wu G, Guidos C. In vitro maturation of clonal CD4+ CD8+ cell lines in response to TCR engagement. J Immunol. 1995;154:5011–5022. [PubMed] [Google Scholar]
- 24.Han S, Dillon S, Zheng B, Shimoda M, Schlissel M, Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–305. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- 25.Han S, Zheng B, Schatz D, Spanopoulou E, Kelsoe G. Neoteny in lymphocytes: Rag1 and Rag 2 expression in germinal center B cells. Science. 1996;274:2094–2097. doi: 10.1126/science.274.5295.2094. [DOI] [PubMed] [Google Scholar]
- 26.Hansen J, Kaattari S. The recombination activating gene 1 (RAG1) of rainbow trout (Oncorhyncus mykiss): cloning, expression, and phylogenetic analysis. Immunogenetics. 1995;42:188–195. doi: 10.1007/BF00191224. [DOI] [PubMed] [Google Scholar]
- 27.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haughton G, Arnold L W, Bishop G A, Mercolino T J. The CH series of murine B cell lymphomas: neoplastic analogues of Ly-1+ normal B cells. Immunol Rev. 1986;93:35–51. doi: 10.1111/j.1600-065x.1986.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 29.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel A, Kel O, Ignatieva E, Ananko E, Podkolodnaya O, Kolpakov F, Podkolodny N, Kolchanov N. Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:364–370. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermanson G, Briskin M, Sigman D, Wall R. Immunoglobulin enhancer and promoter motifs 5′ of the B29 B-cell-specific gene. Proc Natl Acad Sci USA. 1989;86:7341–7345. doi: 10.1073/pnas.86.19.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hikida M, Mori M, Kawabata T, Takai T, Ohmori H. Characterization of B cells expressing recombination activating genes in germinal centers of immunized mouse lymph nodes. J Immunol. 1997;158:2509–2512. [PubMed] [Google Scholar]
- 32.Hikida M, Mori M, Takai T, Tomochika K, Hamatani K, Ohmori H. Reexpression of RAG-1 and RAG-2 genes in activated mature mouse B cells. Science. 1996;274:2092–2094. doi: 10.1126/science.274.5295.2092. [DOI] [PubMed] [Google Scholar]
- 33.Hikida M, Nakayama Y, Yamashita Y, Kumuzawa Y, Nishikawa S, Ohmori H. Expression of recombination activating genes in germinal center B cells: involvement of interleukin 7 (IL-7) and the IL-7 receptor. J Exp Med. 1998;188:365–372. doi: 10.1084/jem.188.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hikida M, Ohmori H. Rearrangement of λ light chains in mature B cells in vitro and in vivo. Function of reexpressed recombination-activating gene (RAG) products. J Exp Med. 1998;187:795–799. doi: 10.1084/jem.187.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klug C A, Gerety S J, Shah P C, Chen Y-Y, Rice N R, Rosenberg N. The v-abl tyrosine kinase negatively regulates NF-kB/Rel factors and blocks k transcription in pre-B lymphocytes. Genes Dev. 1994;8:678–687. doi: 10.1101/gad.8.6.678. [DOI] [PubMed] [Google Scholar]
- 36.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol Cell Biol. 1992;12:2662–2672. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudo A, Sakaguchi N, Melchers F. Organization of the murine Ig-related λ5 gene transcribed selectively in pre-B lymphocytes. EMBO J. 1987;6:103–107. doi: 10.1002/j.1460-2075.1987.tb04725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurioka H, Kishi H, Isshiki H, Tagoh H, Mori K, Kitagawa T, Nagata T, Dohi K, Muragachi A. Isolation and characterization of a TATA-less promoter for the human RAG-1 gene. Mol Immunol. 1996;33:1059–1066. doi: 10.1016/s0161-5890(96)00062-4. [DOI] [PubMed] [Google Scholar]
- 39.Liao F, Birshtein B, Busslinger M, Rothman P. The transcription factor BSAP (NF-HB) is essential for immunoglobulin germ-line ɛ transcription. J Immunol. 1994;152:2904–2911. [PubMed] [Google Scholar]
- 40.Luscher B, Eisenman R. New light on Myc and Myb. Part II. Myb Genes Dev. 1990;4:2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- 41.McMahan C J, Fink P J. RAG reexpression and DNA recombination at T cell receptor loci in peripheral CD4+ T cells. Immunity. 1998;9:637–647. doi: 10.1016/s1074-7613(00)80661-5. [DOI] [PubMed] [Google Scholar]
- 42.Melamed D, Benschop R, Cambier J, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell. 1998;92:173–182. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. RAG-1 deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 45.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 46.Neurath M, Max E, Strober W. Pax5 (BSAP) regulates the murine immunoglobulin 3′α enhancer by suppressing binding of NFαP, a protein that controls heavy chain transcription. Proc Natl Acad Sci USA. 1995;92:5336–5340. doi: 10.1073/pnas.92.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neurath M, Strober W, Wakatsuki Y. The murine Ig 3′ α enhancer is a target site with repressor function for the B cell lineage-specific transcription factor BSAP (NF-HB, Sα-BP) J Immunol. 1994;153:730–742. [PubMed] [Google Scholar]
- 48.Nutt S, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax 5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 49.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 50.Okabe T, Bauer S, Kudo A. Pre-B lymphocyte-specific transcriptional control of the mouse VpreB gene. Eur J Immunol. 1992;22:31–36. doi: 10.1002/eji.1830220106. [DOI] [PubMed] [Google Scholar]
- 51.Okabe T, Watanabe T, Kudo A. A pre-B- and B cell-specific DNA-binding protein, EBB-1, which binds to the promoter of the VpreB1 gene. Eur J Immunol. 1992;22:37–43. doi: 10.1002/eji.1830220107. [DOI] [PubMed] [Google Scholar]
- 52.Papavasiliou F, Casellas R, Suh H, Qin X-F, Besmer E, Pelanda R, Nemazee D, Rajewsky K, Nussenzweig M. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science. 1997;278:298–301. doi: 10.1126/science.278.5336.298. [DOI] [PubMed] [Google Scholar]
- 53.Plachov D, Chowdhury K, Walther K, Simon D, Guenet J-L, Gruss P. Pax8, a murine paired box gene expressed in the developing excretory system and thyroid gland. Development. 1991;110:643–651. doi: 10.1242/dev.110.2.643. [DOI] [PubMed] [Google Scholar]
- 54.Reinold A, Donath P, Li Y-S, Hardy R, David C, Strominger J, Glimcher L. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinkenberger J, Wallin J, Johnson K, Koshland M. An interleukin-2 signal relieves BSAP (Pax5)-mediated repression of the immunoglobulin J chain gene. Immunity. 1996;5:377–386. doi: 10.1016/s1074-7613(00)80263-0. [DOI] [PubMed] [Google Scholar]
- 56.Riva A, Wilson G, Kehrl J. In vivo footprinting and mutational analysis of the proximal CD19 promoter reveal important roles for an SP1/Egr-1 binding site and a novel site termed the PyG box. J Immunol. 1997;159:1284–1292. [PubMed] [Google Scholar]
- 57.Roque M, Smith P, Blasquez V. A developmentally modulated chromatin structure at the mouse immunoglobulin κ 3′ enhancer. Mol Cell Biol. 1996;16:3138–3155. doi: 10.1128/mcb.16.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rothman P, Li S, Gorham B, Glimcher L, Alt F, Boothby M. Identification of a conserved lipopolysaccharide-plus-interleukin-4-responsive element located at the promoter of germ line ɛ transcripts. Mol Cell Biol. 1991;11:5551–5561. doi: 10.1128/mcb.11.11.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 60.Schatz D G, Oettinger M A, Schlissel M S. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 61.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaffer A, Peng A, Schlissel M. In vivo occupancy of the κ light chain enhancers in primary pro- and pre-B cells: a model for κ locus activation. Immunity. 1997;6:131–143. doi: 10.1016/s1074-7613(00)80420-3. [DOI] [PubMed] [Google Scholar]
- 63.Shinkai Y, Rathbun G, Lam K, Oltz E, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. RAG-2 deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 64.Singh M, Birshtein B. NF-HB (BSAP) is a repressor of the murine immunoglobulin heavy-chain 3′α enhancer at early stages of B-cell differentiation. Mol Cell Biol. 1993;13:3611–3622. doi: 10.1128/mcb.13.6.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spolski R, Miescher G, Erard R, Risser R, MacDonald H R, Mak T W. Regulation of expression of T cell gamma chain, L3T4 and Ly-2 messages in Abelson/Moloney virus-transformed T cell lines. Eur J Immunol. 1988;18:295–300. doi: 10.1002/eji.1830180218. [DOI] [PubMed] [Google Scholar]
- 66.Stuart E, Haffner R, Oren M, Gruss P. Loss of p53 function through PAX-mediated transcriptional repression. EMBO J. 1995;14:5638–5645. doi: 10.1002/j.1460-2075.1995.tb00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thienes C, De Monte L, Monticelli S, Busslinger M, Gould H, Vercelli D. The transcription factor B cell-specific activator protein (BSAP) enhances both the IL-4- and CD40-mediated activation of the human ɛ germline promoter. J Immunol. 1997;158:5874–5882. [PubMed] [Google Scholar]
- 68.Thompson A, Wood W, Gilly M, Damore M, Omori S, Wall R. The promoter and 5′ flanking sequences controlling human B29 gene expression. Blood. 1996;87:666–673. [PubMed] [Google Scholar]
- 69.Thompson C. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 70.Tiegs S, Russell D, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 72.Turka L A, Schatz D G, Oettinger M A, Chun J J M, Gorka C, Lee K, McCormack W T, Thompson C B. Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science. 1991;253:778–781. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- 73.Urbanek P, Wang Z-Q, Fetka I, Wagner E, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 74.Verkoczy L, Berinstein N. Isolation of genes negatively or positively co-expressed with human recombination activating gene 1 (RAG1) by differential display PCR (DD RT-PCR) Nucleic Acids Res. 1998;26:4497–4507. doi: 10.1093/nar/26.19.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakatsuki Y, Neurath M, Max E, Strober W. The B cell-specific transcription factor BSAP regulates B cell proliferation. J Exp Med. 1994;179:1099–1108. doi: 10.1084/jem.179.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson A, Held W, MacDonald H R. Two waves of recombinase gene expression in developing thymocytes. J Exp Med. 1994;179:1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zarrin A, Fong I, Malkin L, Marsden P, Berinstein N. Cloning and characterization of the Human Recombination Activating Gene 1 (RAG1) and RAG2 promoter regions. J Immunol. 1997;159:4382–4394. [PubMed] [Google Scholar]
- 78.Zwollo P, Arrieta H, Ede K, Molinder K, Desiderio S, Pollock R. The Pax-5 gene is alternatively spliced during B-cell development. J Biol Chem. 1997;272:10160–10168. doi: 10.1074/jbc.272.15.10160. [DOI] [PubMed] [Google Scholar]
- 79.Zwollo P, Desiderio S. Specific recognition of the blk promoter by the B-lymphoid transcription factor B-cell-specific activator protein. J Biol Chem. 1994;269:15310–15317. [PubMed] [Google Scholar]