Abstract
In recent years, the prevalence of resistance to aminoglycosides among clinical isolates of Pseudomonas aeruginosa is increasing. The aim of this study was to investigate the role of aminoglycoside-modifying enzymes (AMEs) in resistance to aminoglycosides in clinical isolates of P. aeruginosa. The clinical isolates were collected from different hospitals. Disk agar diffusion test was used to determine the antimicrobial resistance pattern of the clinical isolates, and the minimum inhibitory concentration of aminoglycosides was detected by microbroth dilution method. The PCR was performed for discovery of aminoglycoside-modifying enzyme-encoding genes. Among 100 screened isolates, 43 (43%) isolates were resistant to at least one tested aminoglycosides. However, 13 (13%) isolates were resistant to all tested aminoglycosides and 37 isolates were detected as multidrug resistant (MDR). The resistance rates of P. aeruginosa isolates against tested antibiotics were as follows: ciprofloxacin (41%), piperacillin-tazobactam (12%), cefepime (32%), piperacillin (26%), and imipenem (31%). However, according to the MIC method, 13%, 32%, 33%, and 37% of the isolates were resistant to amikacin, gentamicin, tobramycin, and netilmicin, respectively. The PCR results showed that AAC(6′)-Ib was the most commonly (26/43, 60.4%) identified AME-encoding gene followed by AAC(6′)-IIa (41.86%), APH(3′)-IIb (34.8%), ANT(3″)-Ia (18.6), ANT(2″)-Ia (13.95%), and APH(3″)-Ib (2.32%). However, APH(3′)-Ib was not found in any of the studied isolates. The high prevalence of AME-encoding genes among aminoglycoside-resistant P. aeruginosa isolates in this area indicated the important role of AMEs in resistance to these antibiotics similar to most studies worldwide. Due to the transmission possibility of these genes between the Gram-negative bacteria, we need to control the prescription of aminoglycosides in hospitals.
1. Introduction
Pseudomonas aeruginosa, as an opportunist pathogen, is responsible for several nosocomial infections such as bacteremia; urinary tract, blood, respiratory, burn, and soft tissue infections; external otitis; and endocarditis in clinical settings, which are often difficult to treat [1, 2]. Aminoglycosides, fluoroquinolones, and β-lactams are clinically effective antibiotics in the treatment of infections caused by P. aeruginosa, while carbapenems are the last-line option before colistin [3–5]. Increasing resistance to fluoroquinolones and β-lactams has led to attentiveness in clinical applications of aminoglycosides against Gram-negative bacteria [6]. However, P. aeruginosa can survive in hospital environments for a long time due to the high-level resistance against biocides and can acquire and/or spread the antibiotic resistance genes [7]. Transfer of these genes, especially in hospital environments, can further complicate the treatment of the infected patients [8]. Some of the most important resistance genes transferable between the Gram-negative bacteria, especially in P. aeruginosa, are the aminoglycoside-modifying enzyme- (AME-) encoding genes [6]. Enzymatic modification is one of the important mechanisms leading to resistance against aminoglycosides, while these antibiotics are counting as the most potent drugs of choice for the treatment of life-threatening infections caused by P. aeruginosa [8]. The AMEs are classified as acetyltransferases (AACs), nucleotidyltransferases (ANTs), and phosphotransferases (APH) [9]. These enzymes started N-acetylation, O-nucleotidylation, and/or O-phosphorylation of the aminoglycosides resulting in inactivation of the drugs and therapeutic failure [9, 10]. AAC(6′)-I, AAC (6′)-II, ANT(2″)-I, and APH(3′) are the most common AME-encoding genes in P. aeruginosa [11]. The mobile genetic elements such as integrons, plasmids, and transposons can help in the rapid spreading of these clinically important genes [12]. Recently, Thirumalmuthu et al. found that the presence of AME genes was the main cause for resistance to aminoglycosides in multidrug-resistant (MDR) ocular isolates of P. aeruginosa [13]. Therefore, understanding the prevalence of these genes is important for the practical control of antibiotic resistance. This study was aimed at evaluating the role of aminoglycoside-modifying enzyme-encoding genes in resistance to these antibiotics in the clinical isolates of P. aeruginosa collected from hospitalized patients in the north of Iran.
2. Materials and Methods
2.1. Sample Collection and Identification of the Bacteria
One hundred consecutive nonduplicated clinical isolates of P. aeruginosa were collected during 2018-2019 from 5 different teaching and therapeutic hospitals in the north of Iran. The isolates were obtained from different sources including sputum, urine, wound, catheter, blood, stool, and eye. After identification of the isolates by different conventional microbiological and biochemical methods [14], the pure overnight isolates were cultured in trypticase soy broth (Merck, Germany) containing 10% glycerol and were stocked at -20°C for the next investigations. The isolates which were detected as P. aeruginosa were included in the study and others were rejected.
2.2. Antibiotic Susceptibility Testing
The antimicrobial susceptibility pattern of the clinical P. aeruginosa isolates was determined by the Kirby-Bauer disk agar diffusion method on Mueller–Hinton agar (Merck, Germany), according to the Clinical and Laboratory Standards Institute (CLSI) recommendations against 4 different antibiotics [15]. The following antibiotics were exploited in this study: ciprofloxacin (5 μg), cefepime (30 μg), piperacillin (100 μg), piperacillin/tazobactam (100/10 μg), and imipenem (10 μg) (Rosco, Denmark). However, the minimum inhibitory concentrations (MICs) of amikacin, gentamicin, tobramycin, and netilmicin (Sigma, Germany) were detected using the microbroth dilution method conferring to the CLSI guidelines [15], and the lowest concentration of the antibiotics that inhibited the bacterial growth was reported as the MIC. P. aeruginosa ATCC 27853 was used as the control strain in antimicrobial susceptibility testing.
2.3. DNA Extraction and Amplification of AME-Encoding Genes
The alkaline lysis method based on sodium dodecyl sulfate (SDS) (Sigma) and NaOH (Sigma) was used for DNA extraction [16]. Briefly, we solved 0.5 g SDS and 0.4 g NaOH in 200 ml distilled water and suspended the bacterial colony in 20 μl of this extraction buffer. Then, the suspension was warmed at 95°C for 10 min and centrifuged in13000 × gfor 3 min. Next, 180 μl distilled water was added to it and saved at -20°C as extracted DNA until use. All isolates which were resistant to at least one aminoglycoside were examined by the polymerase chain reaction (PCR) method, and the presence of aminoglycoside-modifying enzyme-encoding genes was detected using specific primers (Table 1). The detection of ANT(3″)-Ia, ANT (2″)-Ia, APH(3′)-IIb, AAC(6′)-Ib, APH (3′)-Ib, APH (3″)-Ib, and AAC (6′)-IIa genes was performed by the PCR method. The PCRs were done in a final volume of 15 μl reaction mixtures containing 7.5 μl of Master Mix (Ampliqon, Denmark), 300 ng of the extracted DNA, and 10 pmol of each primer and added distilled water to the final volume. The thermal cycler (Bio-Rad, USA) running condition was as follows: an initial denaturation for 5 min at 94°C and 35 cycles including denaturation at 94°C for 60 seconds, annealing for 1 min at specific primer set temperatures (Table 1), and extension at 72°C for 2 min, and after the cycles, a final extension of the amplicons happened at 72°C for 10 min. Finally, the amplified DNAs were visualized by electrophoresis on 1% agarose gel (Sigma, Germany) containing a 3% safe stain (SinaClon, Iran). Distilled water was used as the negative control in PCR.
Table 1.
Primers used for detection of aminoglycoside-modifying enzyme-encoding genes by PCR.
| Target genes | Primer sequences (5′ to 3′) | Amplicon size (bp) | Annealing temperature (°C) | References |
|---|---|---|---|---|
| ANT(3 ″ )-Ia | Pri F: TGT AGA AGT CAC CAT TGT TG | 152 | 51 | This study |
| Pri R: TCA GCA AGA TAG CCA GAT | ||||
| ANT(2 ″ )-Ia | Pri F: GCA GGT CAC ATT GAT ACA C | 225 | 54 | This study |
| Pri R: TCC GCT AAG AAT CCA TAG TC | ||||
| AAC(6′)-Ib | Pri F: GAC CAA CAG CAA CGA TTC | 375 | 57 | This study |
| Pri R: AAC AGC AAC TCA ACC AGA | ||||
| AAC(6′)-IIa | Pri F: CCA TAA CTC TTC GCC TCA T | 442 | 48 | This study |
| Pri R: AAT CCT GCC TTC TCA TAG C | ||||
| APH(3′)-IIb | Pri F: TTC GTC AAG CAG GAA GTC | 662 | 50 | This study |
| Pri R: TAG AAG AAC TCG TCC AAT AGC | ||||
| APH(3′)-Ib | Pri F: TTG TTG TTG ACG CAT TGG | 284 | 56 | This study |
| Pri R: GCC GAC TAC CTT ACC TTC | ||||
| APH(3 ″ )-Ib | Pri F: GGT GAT AAC GGC AAT TCC | 548 | 56 | This study |
| Pri R: GGT CCA ATC GCA GAT AGA |
2.4. Ethical Approval Statements
We received the clinical samples without names from the laboratories of the hospitals affiliated to the Mazandaran University of Medical Sciences. This study was conducted in accordance with the Declaration of Helsinki; however, written informed consent form was provided by the patients or a close relative before hospitalization, and classifying information of each sample was kept secret. Also, this study was approved by the Iran National Committee for Ethics in Biomedical Research with the national ethical code (consent ref number) IR.MAZUMS.REC.1398.980.
3. Results
3.1. Patients and Samples
A total of 100 P. aeruginosa clinical isolates, collected from 60 males and 40 females with an average age of 46 years, were analyzed in the present study. The isolates were collected from five educational-therapeutic hospitals including Zare (burn center, 11 isolates), Razi (infectious center, 22 isolates), Bu-Ali Sina (pediatric center, 17 isolates), Fatemeh Zahra (heart center, 10 isolates), and Imam Khomeini (general center, 40 isolates). However, the isolates were collected from different hospital wards counting intensive care unit (ICU) (48 isolates), burn (9 isolates), internal medicine (4 isolates), operation room (3 isolates), men (3 isolates), women (2 isolates), emergency (13 isolates), surgery (3 isolates), oncology (1 isolate), cardiac care unit (CCU) (6 isolates), neurology (2 isolates), and pediatric (6 isolates). In terms of sample type, the isolates were obtained from sputum (n = 37), urine (n = 26), wound (n = 20), catheter (n = 8), blood (n = 5), stool (n = 2), and eye (n = 2).
3.2. Antimicrobial Susceptibility Pattern of the Isolates
The resistance rates against all tested antibiotics are shown in Figure 1. According to the antimicrobial susceptibility testing, 43 (43%) isolates were resistant to at least one tested aminoglycoside. However, 13 (13%) isolates were resistant to all tested aminoglycosides in the present study. The highest aminoglycoside resistance rate was observed against gentamicin (n = 41), while 39 and 28 isolates were resistant towards tobramycin and amikacin, respectively. Piperacillin-tazobactam was the most effective antibiotic in this study, while the highest resistance rate was shown against gentamicin and ciprofloxacin. Also, 37 isolates were detected as MDR in the present study.
Figure 1.
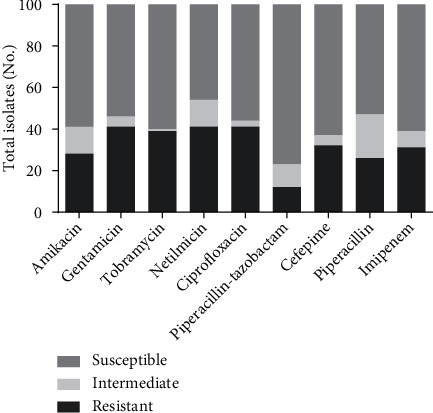
Antibiotic resistance pattern of 100 Pseudomonas aeruginosa clinical isolates.
The microbroth dilution results indicated that 6, 23, 12, and 2 isolates were high-level resistant (MIC ≥ 512 μg/ml) to amikacin, gentamicin, tobramycin, and netilmicin, respectively (Table 2).
Table 2.
The aminoglycoside MICs against 43 aminoglycoside-resistant Pseudomonas aeruginosa.
| MIC ranges (μg/ml) | Amikacin | Gentamicin | Tobramycin | Netilmicin | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤16 | 32 | 64-128 | ≥256 | ≤4 | 8 | 16-128 | ≥256 | ≤4 | 8 | 16-128 | ≥256 | ≤8 | 16 | 32-128 | ≥256 | |
| No. of isolates | 25 | 5 | 7 | 6 | 10 | 1 | 6 | 26 | 9 | 1 | 17 | 16 | 4 | 2 | 30 | 7 |
3.3. Genotypic Detection of AME-Encoding Genes
The PCR amplification exhibited that 34/43 (79%) of aminoglycoside-resistant isolates were positive for AME-encoding genes (Table 3). AAC(6′)-Ib was the most commonly (26/43, 60.4%) identified gene followed by AAC(6′)-IIa (41.86%), APH(3′)-IIb (34.8%), ANT(3″)-Ia (18.6%), ANT(2″)-Ia (13.95%), and APH(3″)-Ib (2.32%). APH(3′)-Ib was not found in any of the studied isolates. Among the 34 isolates that contained at least one of the studied genes, 12 (35.29%) isolates were carrying only one AME-encoding gene; however, 12 (35.29%) isolates were positive for two genes, 4 (11.76%) isolates had 3 genes, 4 (11.76%) others contained 4 AME-encoding genes, and 2 (5.88%) isolates had 5 AME genes. We did not detect any isolates containing all studied genes in the present study (Table 3). The simultaneous presence of the genes encoding aminoglycoside-modifying enzymes had a significant effect on the increase of the MICs of gentamicin and tobramycin (p < 0.05), whereas the combination of the two genes had less effect on the increasing of amikacin and netilmicin MICs. Interestingly, among 26 ICU isolates in this study, 22 (84.61%) of them have contained at least one AME-encoding gene, while this rate among burn isolates was 6/8 (75%). Also, Table 4 exhibits the relation between the aminoglycoside resistance phenotype and the simultaneous presence of AME-encoding genes in the studied isolates. According to this table, most aminoglycoside resistance phenotype (29/43, 67.44%) was the simultaneous resistance against gentamicin and tobramycin. However, different resistance phenotypes in the present study showed similar resistance gene profile.
Table 3.
The correlation between the AME gene profile of P. aeruginosa clinical isolates and the MIC ranges of aminoglycosides.
| Genotypes | No. (%) of MDRs | No. (%) of isolates with different MICs (μg/ml) against amikacin | No. (%) of isolates with different MICs (μg/ml) against gentamicin | No. (%) of isolates with different MICs (μg/ml) against tobramycin | No. (%) of isolates with different MICs (μg/ml) against netilmicin | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤16 | 32 | 64-128 | ≥256 | ≤4 | 8 | 16-128 | ≥256 | ≤4 | 8 | 16-128 | ≥256 | ≤8 | 16 | 32-128 | ≥256 | ||
| ANT(3″)-Ia (n = 8) | 7 (87.5) | 5 (62.5) | 1 (12.5) | 0 | 2 (25) | 1 (12.5) | 0 | 2 (25) | 5 (62.5) | 1 (12.5) | 0 | 6 (75) | 1 (12.5) | 1 (12.5) | 0 (0) | 7 (87.5) | 0 |
| ANT(2″)-Ia (n = 6) | 5 (83.3) | 2 (33.3) | 2 (33.3) | 2 (33.3) | 0 | 0 | 0 | 2 (33.3) | 4 (66.6) | 0 | 0 | 4 (66.6) | 2 (33.3) | 1 (16.6) | 1 (16.6) | 3 (50) | 1 (16.6) |
| AAC(6′)-Ib (n = 26) | 24 (92.3) | 11 (42.3) | 4 (15.3) | 7 (26.9) | 4 (15.3) | 1 (3.8) | 1 (3.8) | 3 (11.5) | 21 (80.7) | 1 (3.8) | 0 | 12 (46.1) | 13 (50) | 3 (11.5) | 0 | 18 (69.2) | 5 (19.2) |
| AAC(6′)-IIa (n = 18) | 16 (88.8) | 10 (55.5) | 3 (16.6) | 3 (16.6) | 2 (11.1) | 2 (11.1) | 1 (5.5) | 4 (22.2) | 11 (61.1) | 2 (11.1) | 0 | 10 (55.5) | 6 (33.3) | 4 (22.2) | 0 | 12 (66.6) | 2 (11.1) |
| APH(3′)-IIb (n = 15) | 13 (86.6) | 7 (46.6) | 3 (20) | 3 (20) | 2 (13.3) | 1 (6.6) | 0 | 4 (26.6) | 10 (66.6) | 1 (6.6) | 0 | 9 (60) | 5 (33.3) | 2 (13.3) | 1 (6.6) | 10 (66.6) | 2 (13.3) |
| APH(3″)-Ib (n = 1) | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| ANT(3″)-Ia+ ANT(2″)Ia (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
| ANT(3″)-Ia+ AAC(6′)-Ib (n = 6) | 5 (83.3) | 4 (66.6) | 1 (16.6) | 0 | 1 (16.6) | 0 | 0 | 2 (33.3) | 4 (66.6) | 0 | 0 | 5 (83.3) | 1 (16.6) | 1 (16.6) | 0 | 5 (83.3) | 0 |
| ANT(3″)-Ia+ AAC(6′)-IIa (n = 3) | 2 (66.6) | 2 (66.6) | 1 (33.3) | 0 | 0 | 0 | 0 | 1 (33.3) | 2 (66.6) | 0 | 0 | 2 (66.6) | 1 (33.3) | 1 (33.3) | 0 | 2 (66.6) | 0 |
| ANT(3″)-Ia+ APH(3′)-IIb (n = 4) | 3 (75) | 2 (50) | 1 (25) | 0 | 1 (25) | 0 | 0 | 1 (25) | 3 (75) | 0 | 0 | 3 (75) | 1 (25) | 1 (25) | 0 | 3 (75) | 0 |
| ANT(2″)-Ia+ AAC(6′)-Ib (n = 5) | 5 (100) | 1 (20) | 2 (40) | 2 (40) | 0 | 0 | 0 | 1 (20) | 4 (80) | 0 | 0 | 2 (40) | 3 (60) | 1 (20) | 0 | 3 (60) | 1 (20) |
| ANT(2″)-Ia+ AAC(6′)-IIa (n = 5) | 5 (100) | 1 (20) | 2 (40) | 2 (40) | 0 | 0 | 0 | 1 (20) | 4 (80) | 0 | 0 | 2 (40) | 3 (60) | 1 (20) | 0 | 3 (60) | 1 (20) |
| ANT(2″)-Ia+ APH(3′)-IIb (n = 6) | 5 (83.3) | 2 (33.3) | 2 (33.3) | 2 (33.3) | 0 | 0 | 0 | 2 (33.3) | 4 (66.6) | 0 | 0 | 3 (50) | 3 (50) | 1 (16.6) | 1 (16.6) | 3 (50) | 1 (16.6) |
| ANT(2″)-Ia+ APH(3″)-Ib (n = 1) | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| AAC(6′)-Ib+ AAC(6′)-IIa (n = 14) | 12 (85.7) | 7 (50) | 2 (14.2) | 3 (21.4) | 2 (14.2) | 0 | 1 (7.1) | 2 (14.2) | 11 (78.5) | 0 | 0 | 8 (57.1) | 6 (42.8) | 3 (21.4) | 0 | 9 (64.2) | 2 (14.2) |
| AAC(6′)-Ib+ APH(3′)-IIb (n = 9) | 8 (88.8) | 3 (33.3) | 2 (22.2) | 3 (33.3) | 1 (11.1) | 0 | 0 | 1 (11.1) | 8 (88.8) | 0 | 0 | 5 (55.5) | 4 (44.4) | 1 (11.1) | 0 | 6 (66.6) | 2 (22.2) |
| AAC(6′)-IIa+ APH(3′)-IIb (n = 12) | 11 (91.66) | 5 (41.6) | 3 (25) | 3 (25) | 1 (8.3) | 1 (8.3) | 0 | 3 (25) | 8 (66.6) | 1 (8.3) | 0 | 7 (58.3) | 4 (33.3) | 2 (16.6) | 0 | 8 (66.6) | 2 (16.6) |
| APH(3′)-IIb+ APH(3′′)-Ib (n = 1) | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| ANT(3″)-Ia+ ANT(2″)-Ia+ AAC(6′)-Ib (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
| ANT(3″)-Ia+ ANT(2″)-Ia+ AAC(6′)-IIa (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
| ANT(3″)-Ia+ ANT(2″)-Ia+ APH(3)-IIb (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
| ANT(3″)-Ia+ APH(3)-IIb+ AAC(6′)-Ib (n = 3) | 2 (66.6) | 2 (66.6) | 1 (33.3) | 0 | 0 | 0 | 0 | 1 (33.3) | 2 (66.6) | 0 | 0 | 0 | 0 | 1 (33.3) | 0 | 2 (66.6) | 0 |
| ANT(3″)-Ia+ APH(3′)-IIb+ AAC(6′)-IIa (n = 3) | 2 (66.6) | 2 (66.6) | 1 (33.3) | 0 | 0 | 0 | 0 | 1 (33.3) | 2 (66.6) | 0 | 0 | 2 (66.6) | 1 (33.3) | 1 (33.3) | 0 | 2 (66.6) | 0 |
| ANT(3″)-Ia+ AAC(6′)-Ib+ AAC(6′)-IIa (n = 3) | 2 (66.6) | 2 (66.6) | 1 (33.3) | 0 | 0 | 0 | 0 | 1 (33.3) | 2 (66.6) | 0 | 0 | 2 (66.6) | 1 (33.3) | 1 (33.3) | 0 | 2 (66.6) | 0 |
| ANT(2″)-Ia+ APH(3′)-IIb+ AAC(6′)-Ib (n = 5) | 5 (100) | 1 (20) | 2 (40) | 2 (40) | 0 | 0 | 0 | 1 (20) | 4 (80) | 0 | 0 | 3 (60) | 2 (40) | 1 (20) | 0 | 3 (60) | 1 (20) |
| ANT(2″)-Ia+ AAC(6′)-Ib+ AAC(6′)-IIa (n = 5) | 5 (100) | 1 (20) | 2 (40) | 2 (40) | 0 | 0 | 0 | 1 (20) | 4 (80) | 0 | 0 | 3 (60) | 2 (40) | 1 (20) | 0 | 3 (60) | 1 (20) |
| APH(3′)-IIb+ AAC(6′)-Ib+ AAC(6′)-IIa (n = 9) | 8 (88.8) | 3 (33.3) | 2 (22.2) | 3 (33.3) | 1 (11.1) | 0 | 0 | 1 (11.1) | 8 (88.8) | 0 | 0 | 5 (55.5) | 4 (44.4) | 1 (11.1) | 0 | 6 (66.6) | 2 (22.2) |
| ANT(2″)-Ia+ APH(3′)-IIb+ APH(3″)-Ib (n = 1) | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| ANT(3″)-Ia+ ANT(2″)-Ia+ APH(3′)-IIb+ AAC(6′)-Ib (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
| ANT(2″)-Ia+ APH(3′)-IIb+ AAC(6′)-Ib+ AAC(6′)-IIa (n = 5) | 5 (100) | 1 (20) | 2 (40) | 2 (40) | 0 | 0 | 0 | 1 (20) | 4 (80) | 0 | 0 | 3 (60) | 2 (40) | 1 (20) | 0 | 3 (60) | 1 (20) |
| ANT(3″)-Ia+ APH(3′)-IIb+ AAC(6′)-Ib+ AAC(6′)-IIa (n = 3) | 2 (66.6) | 2 (66.6) | 1 (33.3) | 0 | 0 | 0 | 0 | 1 (33.3) | 2 (66.6) | 0 | 0 | 2 (66.6) | 1 (33.3) | 1 (33.3) | 0 | 2 (66.6) | 0 |
| ANT(3″)-Ia+ ANT(2″)-Ia+ AAC(6′)-Ib+ AAC(6′)-IIa (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
| ANT(3″)-Ia+ ANT(2″)-Ia+ APH(3′)-IIb+ AAC(6′)-IIa (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
| ANT(3″)-Ia+ ANT(2″)-Ia+ APH(3′)-IIb+ AAC(6′)-Ib+ AAC(6′)-IIa (n = 2) | 2 (100) | 1 (50) | 1 (50) | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 0 | 1 (50) | 0 |
Table 4.
Relation between the aminoglycoside resistance phenotypes and the presence of AME genes.
| Resistance phenotypes | AME-encoding gene profiles | No. of isolates |
|---|---|---|
| GM | AAC(6′)-Ib + AAC(6′)-IIa | 1 |
| NM | AAC(6′)-IIa + ANT(3″)-Ia | 2 |
| AK+GM | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa | 12 |
| AK+TOB | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa | 12 |
| AK+NM | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa | 12 |
| GM+TOB | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa + APH(3″)-Ib | 29 |
| GM+NM | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa + APH(3″)-Ib | 27 |
| TOB+NM | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa + APH(3″)-Ib | 27 |
| AK+GM+TOB | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa | 12 |
| AK+GM+NM | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa | 12 |
| GM+TOB+NM | ANT(3″)-Ia + ANT(2″)-Ia + APH(3′)-IIb + AAC(6′)-Ib + AAC(6′)-IIa + APH(3″)-Ib | 27 |
Abbreviations: GM: gentamicin; NM: netilmicin; AK: amikacin; TOB: tobramycin.
4. Discussion
The aminoglycosides as the broad-spectrum antibiotics have remained useful as antipseudomonal choice agents for the treatment of life-threatening infections [17]. Therefore, the continuous increase of aminoglycoside resistance levels among the clinical isolates of Gram-negative bacteria will become a growing clinical concern in the future [17, 18]. The lowest resistance to aminoglycosides in the present study was seen against amikacin (13%) with the MICs ranging from 0.25 to 2048 μg/ml, whereas 37% of the P. aeruginosa isolates were resistant to netilmicin with the MICs ranging from 4 μg/ml to 2048 μg/ml. Moreover, Kashfi et al. reported that amikacin was more effective than gentamicin against P. aeruginosa isolated from burned patients [19], while their MICs ranged from 2 μg/ml to 256 μg/ml. These lowest resistances against amikacin may be due to the lower prescription of this aminoglycoside in hospitals of Iran. On the other hand, the rate of resistance to aminoglycosides is different in various regions and countries, even in different hospitals of the same region in a similar country. These variabilities may be due to the different causes such as overuse of these drugs in hospitals, arbitrary use of the drugs by people without a prescription, geographical and cultural differences, countries' health levels, and hygienic condition [20].
Aminoglycoside resistance in P. aeruginosa is often related to the production of various aminoglycoside-modifying enzymes [19], so the more prevalence of these enzymes is an important problem. In this study, the aminoglycoside resistance rate was 43%, while 79% of the resistant isolates carried AME-encoding genes. In total, 34 AME patterns (28 combinations and 6 single-gene forms) were identified, which showed different levels of aminoglycoside resistance. As an important result of the present study, we found that the simultaneous presence of AME genes was the cause of increasing the MIC ranges of gentamicin and tobramycin. This shows that these genes were the most effective factor in resistance to tested aminoglycosides, while this effect was lower about amikacin and netilmicin. According to the study conducted by Panahi et al. in 2020, AMEs were highly prevalent (96.2%) among the aminoglycoside nonsusceptible P. aeruginosa isolates [21]. However, Odumosu et al. in Nigeria reported that the AME-encoding genes AAC(6′)–I and ANT(2″)–I were found only in 22.22% of their isolates [8].
In the present study, AAC(6′)-Ib, AAC(6′)-IIa, and APH(3′)-IIb were more prevalent than the ANT(3″)-Ia, ANT(2″)-Ia, and APH(3″)-Ib genes among P. aeruginosa clinical isolates. Also, APH(3′)-Ib was not found in any of the isolates. The more prevalence of AAC(6′)-Ib in our study was similar to the previous reports from Iran [22, 23], while it was significantly higher compared with another research conducted by Vaziri et al. in Iran which reported a 7% prevalence of this gene [17]. Our results confirmed that the presence of AAC(6′)-Ib gene may be more effective in resistance of P. aeruginosa towards tested aminoglycosides, although probably other mechanisms contribute to this type of resistance, too [24]. However, the high prevalence of this gene in P. aeruginosa clinical isolates collected from Iranian patients demonstrates its key role in resistance to aminoglycosides and its high distribution in Iran [21–23]. While the high prevalence of the AAC(6′)-I gene can result in a higher resistance level to amikacin [10], our MIC results showed that 42.3% of AAC(6′)-Ib-positive isolates were resistant to this antibiotic.
The frequency of AAC(6′)-IIa in similar studies was 1.9% in France [25], 18.5% in Nigeria [8], and 10% in another study carried out in Iran [19]. However, 18% of our all clinical isolates were AAC(6′)-IIa positive, indicating the increasing frequency of this gene in our region. Furthermore, AAC(6′)-IIa was known as the most prevalent aminoglycoside resistance gene in Europe [26]. On the other hand, this gene can spread by integrons, causing the high prevalence of this resistance gene which plays an effective role in resistance to aminoglycosides [27]. AAC(6′)-II has a key role in resistance towards gentamicin, tobramycin, netilmicin, and kanamycin [27]. However, among the AAC(6′)-IIa-positive isolates of this study, 83%, 88%, and 77% of the isolates were resistant to gentamicin, tobramycin, and netilmicin, respectively.
It seems that ANT(2″)-Ia has a relatively important role in resistance against gentamicin and tobramycin in P. aeruginosa isolates [28]. Interestingly, all ANT(2″)-Ia-positive isolates in the present study were resistant to these aminoglycosides. However, 13.9% of our isolates were positive for this gene that was comparable with another study conducted in Iran [22]. On the other hand, other studies conducted in Iran and South Korea reported the ANT(2″)-Ia gene as the most common aminoglycoside resistance gene [19, 29]. Moreover, Michalska et al. in Poland and Odumosu et al. in Nigeria detected 36% and 16.6% of this gene in their P. aeruginosa clinical isolates, respectively [8, 30].
The results of our study revealed that 18.6% of the P. aeruginosa clinical isolates were carrying ANT(3″)-Ia gene similar to the study of Aghazadeh et al. in Iran [22], while 87.5% of other research in Tehran contained this gene [19]. This significant difference may be due to the different sources of the isolates, as 38% of the bacteria in the first study were isolated from the sputum of the cystic fibrosis patients and 62% of their isolates were collected from burned patients [22], while all isolates in another mentioned study [19] were collected from burned patients. Generally, burn patients are at high risk for bacterial infections, so isolates collected from burn injuries may show a higher resistance level [31].
Although APH(3″)-Ib is responsible for streptomycin resistance in Enterobacteriaceae [10], 2.32% of our isolates were positive for this gene. This was comparable to Michalska et al.'s study in Poland from which 8% of their isolates were carrying this gene [30]. On the other hand, besides the fact that we found that 34.8% of our isolates were APH(3′)-IIb positive, this was lower than the similar studies carried out in Iran and Thailand with the rate of 61.8% and 57%, respectively [22, 32]. Besides, we detected any isolates with the presence of the APH(3′)-Ib gene, while in other studies conducted in Iran, the frequency of this gene was 60% and 46%, respectively [19, 22].
5. Conclusions
The aminoglycoside-modifying enzyme-encoding genes are highly prevalent among Pseudomonas aeruginosa clinical isolates worldwide, especially in Iran. The inappropriate and indiscriminate prescription of aminoglycosides was probably one of the main reasons for the high prevalence of some aminoglycoside resistance genes in this study. Due to the high ability of P. aeruginosa in the distribution of these genes, an appropriate antibiotic stewardship policy is required for the prevention of AME gene spreading and to decrease the aminoglycoside resistance rates.
Acknowledgments
We thank the laboratory staff of Zare, Razi, Bu-Ali Sina, Fatemeh Zahra, and Imam Khomeini hospitals for providing patients' information and the collection of the clinical isolates. This study is a report of a database from MSc student thesis registered and carried out in Molecular and Cell Biology Research Center in Mazandaran University of Medical Sciences, Sari, Iran, with grant no. 1293 and the national ethical code IR.MAZUMS.REC.1398.980.
Data Availability
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
Study concept and design were performed by Hamid Reza Goli. Acquisition of data was performed by Leila Ahmadian, Zahra Norouzi Bazgir, and Hamid Reza Goli. Analysis and interpretation of data were performed by Mohammad Ahanjan and Reza Valadan. Drafting of the manuscript was performed by Leila Ahmadian. Review of the article was performed by Hamid Reza Goli.
References
- 1.Richards M. J., Edwards J. R., Culver D. H., Gaynes P. R. Nosocomial infections in medical intensive care units in the United States. Critical Care Medicine. 1999;27(5):887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 2.Dimatatac E. L., Alejandria M. M., Montalban C., Pineda C., Ang C., Delino R. Clinical outcomes and costs of care of antibiotic resistant Pseudomonas aeruginosa infections. Philippine Journal of Microbiology and Infectious Diseases. 2003;32:159–167. [Google Scholar]
- 3.Vardakas K. Z., Tansarli G. S., Bliziotis I. A., Falagas M. E. β-Lactam plus aminoglycoside or fluoroquinolone combination versus β-lactam monotherapy for _Pseudomonas aeruginosa_ infections: A meta-analysis. International Journal of Antimicrobial Agents. 2013;41(4):301–310. doi: 10.1016/j.ijantimicag.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Akhi M. T., Khalili Y., Ghotaslou R., et al. Carbapenem inactivation: a very affordable and highly specific method for phenotypic detection of carbapenemase-producing Pseudomonas aeruginosa isolates compared with other methods. Journal of Chemotherapy. 2017;29(3):144–149. doi: 10.1080/1120009X.2016.1199506. [DOI] [PubMed] [Google Scholar]
- 5.Goli H. R., Nahaei M. R., Ahangarzadeh Rezaee M., Hasani A., Samadi Kafil H., Aghazadeh M. Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iranian journal of microbiology. 2016;8(1):62–69. [PMC free article] [PubMed] [Google Scholar]
- 6.Belaynehe K. M., Shin S. W., Hong-Tae P., Yoo H. S. Occurrence of aminoglycoside-modifying enzymes among isolates of Escherichia coli exhibiting high levels of aminoglycoside resistance isolated from Korean cattle farms. FEMS Microbiology Letters. 2017;364(14) doi: 10.1093/femsle/fnx129. [DOI] [PubMed] [Google Scholar]
- 7.Hasani A., Sheikhalizadeh V., Ahangarzadeh Rezaee M., et al. Frequency of aminoglycoside-modifying enzymes and ArmA among different sequence groups of Acinetobacter baumannii in Iran. Microbial Drug Resistance. 2016;22(5):347–353. doi: 10.1089/mdr.2015.0254. [DOI] [PubMed] [Google Scholar]
- 8.Odumosu B. T., Adeniyi B. A., Chandra R. Occurrence of aminoglycoside-modifying enzymes genes (aac(6')-I and ant(2″)-I) in clinical isolates of Pseudomonas aeruginosa from Southwest Nigeria. African Health Sciences. 2015;15(4):1277–1281. doi: 10.4314/ahs.v15i4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2005;49(2):479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghotaslou R., Yeganeh Sefidan F., Akhi M. T., Asgharzadeh M., Mohammadzadeh Asl Y. Dissemination of genes encoding aminoglycoside-modifying enzymes and armA among Enterobacteriaceae isolates in Northwest Iran. Microbial Drug Resistance. 2017;23(7):826–832. doi: 10.1089/mdr.2016.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asghar A. H., Ahmed O. B. Prevalence of aminoglycoside resistance genes in Pseudomonas aeruginosa isolated from a tertiary care hospital in Makkah. KSA. Clinical Practice. 2018;15(2):541–547. [Google Scholar]
- 12.Wright G. D. Aminoglycoside-modifying enzymes. Current Opinion in Microbiology. 1999;2(5):499–503. doi: 10.1016/S1369-5274(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 13.Thirumalmuthu K., Devarajan B., Prajna L., Mohankumar V. Mechanisms of fluoroquinolone and aminoglycoside resistance in Keratitis-Associated Pseudomonas aeruginosa. Microbial Drug Resistance. 2019;25(6):813–823. doi: 10.1089/mdr.2018.0218. [DOI] [PubMed] [Google Scholar]
- 14.Sahm D. F., Weissfeld A., Trevino E. Baily and Scott’s Diagnostic Microbiology. St Louis: Mosby; 2002. [Google Scholar]
- 15.In C. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute (CLSI); 2018. [Google Scholar]
- 16.Šipošová N. S., Liptáková V., Kvasnová S., Kosorínová P., Pristaš P. Genetic diversity of Acinetobacter spp. adapted to heavy metal polluted environments. Nova Biotechnologica et Chimica. 2017;16(1):42–47. doi: 10.1515/nbec-2017-0006. [DOI] [Google Scholar]
- 17.Vaziri F., Peerayeh S. N., Nejad Q. B., Farhadian A. The prevalence of aminoglycoside-modifying enzyme genes (aac(6′) − I, aac (6′) − II, ant (2") − I, aph (3′) − VI) in Pseudomonas aeruginosa. Clinics. 2011;66(9):1519–1522. doi: 10.1590/s1807-59322011000900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashemi A., Fallah F., Erfanimanesh S., Chirani A. S., Dadashi M. Detection of antibiotic resistance genes among Pseudomonas aeruginosa strains isolated from burn patients in Iran. British Microbiology Research Journal. 2016;12(4):1–6. doi: 10.9734/BMRJ/2016/23268. [DOI] [Google Scholar]
- 19.Kashfi M., Hashemi A., Eslami G., Amin M. S., Tarashi S., Taki E. The prevalence of aminoglycoside-modifying enzyme genes among Pseudomonas aeruginosa strains isolated from burn patients. Archives of Clinical Infectious Diseases. 2017;12(1) [Google Scholar]
- 20.Magnet S., Blanchard J. S. Molecular insights into aminoglycoside action and resistance. Chemical Reviews. 2005;105(2):477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 21.Panahi T., Asadpour L., Ranji N. Distribution of aminoglycoside resistance genes in clinical isolates of _Pseudomonas aeruginosa_ in north of Iran. Gene Reports. 2020;21, article 100929 doi: 10.1016/j.genrep.2020.100929. [DOI] [Google Scholar]
- 22.Aghazadeh M., Rezaee M. A., Nahaei M. R., et al. Dissemination of aminoglycoside-modifying enzymes and 16S rRNA methylases among Acinetobacter baumannii and Pseudomonas aeruginosa isolates. Microbial Drug Resistance. 2013;19(4):282–288. doi: 10.1089/mdr.2012.0223. [DOI] [PubMed] [Google Scholar]
- 23.Jafari M., Fallah F., Borhan R. S., et al. The first report of CMY, aac (6′)-Ib and 16S rRNA methylase genes among Pseudomonas aeruginosa isolates from Iran. Archives of Pediatric Infectious Diseases. 2013;1(3):109–112. [Google Scholar]
- 24.Sacha P., Jaworowska J., Ojdana D., Wieczorek P., Czaban S., Tryniszewska E. Occurrence of the aacA4 gene among multidrug resistant strains of Pseudomonas aeruginosa isolated from bronchial secretions obtained from the Intensive Therapy Unit at University Hospital in Bialystok, Poland. Folia Histochemica et Cytobiologica. 2012;50(2):322–324. doi: 10.5603/FHC.2012.0043. [DOI] [PubMed] [Google Scholar]
- 25.Dubois V., Arpin C., Dupart V., et al. Lactam and aminoglycoside resistance rates and mechanisms among Pseudomonas aeruginosa in French general practice (community and private healthcare centres) Journal of Antimicrobial Chemotherapy. 2008;62(2):316–323. doi: 10.1093/jac/dkn174. [DOI] [PubMed] [Google Scholar]
- 26.Miller G., Sabatelli F., Hare R., et al. The most frequent aminoglycoside resistance Mechanisms--Changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clinical Infectious Diseases. 1997;24(Supplement 1):S46–S62. doi: 10.1093/clinids/24.supplement_1.s46. [DOI] [PubMed] [Google Scholar]
- 27.Azucena E., Mobashery S. Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resistance Updates. 2001;4(2):106–117. doi: 10.1054/drup.2001.0197. [DOI] [PubMed] [Google Scholar]
- 28.Tam V. H., Schilling A. N., Neshat S., Poole K., Melnick D. A., Coyle E. A. Optimization of meropenem minimum concentration/MIC ratio to suppress in vitro resistance of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2005;49(12):4920–4927. doi: 10.1128/AAC.49.12.4920-4927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J. Y., Park J. Y., Kwon H. J., Han K., Kang M. W., Woo G. J. Occurrence and mechanisms of amikacin resistance and its association with β-lactamases in Pseudomonas aeruginosa: a Korean nationwide study. Journal of Antimicrobial Chemotherapy. 2008;62(3):479–483. doi: 10.1093/jac/dkn244. [DOI] [PubMed] [Google Scholar]
- 30.Michalska A. D., Sacha P. T., Ojdana D., Wieczorek A., Tryniszewska E. Prevalence of resistance to aminoglycosides and fluoroquinolones among Pseudomonas aeruginosa strains in a University Hospital in Northeastern Poland. Brazilian Journal of Microbiology. 2014;45(4):1455–1458. doi: 10.1590/S1517-83822014000400041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laupland K. B., Kibsey P. C., Gregson D. B., Galbraith J. C. Population-based laboratory assessment of the burden of community-onset bloodstream infection in Victoria, Canada. Epidemiology & Infection. 2013;141(1):174–180. doi: 10.1017/S0950268812000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poonsuk K., Tribuddharat C., Chuanchuen R. Aminoglycoside resistance mechanisms in Pseudomonas aeruginosa isolates from non-cystic fibrosis patients in Thailand. Canadian Journal of Microbiology. 2013;59(1):51–56. doi: 10.1139/cjm-2012-0465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


