Abstract
Two coordinate forms of transcriptional synergy mediate eukaryotic gene regulation: the greater-than-additive transcriptional response to multiple promoter-bound activators, and the sigmoidal response to increasing activator concentration. The mechanism underlying the sigmoidal response has not been elucidated but is almost certainly founded on the cooperative binding of activators and the general machinery to DNA. Here we explore that mechanism by using highly purified transcription factor preparations and a strong Epstein-Barr virus promoter, BHLF-1, regulated by the virally encoded activator ZEBRA. We demonstrate that two layers of cooperative binding govern transcription complex assembly. First, the architectural proteins HMG-1 and -2 mediate cooperative formation of an enhanceosome containing ZEBRA and cellular Sp1. This enhanceosome then recruits transcription factor IIA (TFIIA) and TFIID to the promoter to form the DA complex. The DA complex, however, stimulates assembly of the enhanceosome itself such that the entire reaction can occur in a highly concerted manner. The data reveal the importance of reciprocal cooperative interactions among activators and the general machinery in eukaryotic gene regulation.
The assembly of an RNA polymerase II (pol II) transcription complex involves a sophisticated network of interactions between multiple upstream activators and the general transcription machinery. Biochemical experiments have suggested that the complex assembles in two phases. First, activators bind either to naked DNA or to chromatin templates and assemble into a nucleoprotein complex termed an enhanceosome (7, 23). The enhanceosome then recruits the general transcription machinery either in discrete steps or in the form of a pol II-containing holoenzyme (7, 21, 55, 57). It has been proposed by us and others that the formation of the enhanceosome and the recruitment of the general machinery may occur in a concerted reaction, although details regarding the mechanism remain unclear (38). Here we recreate the concerted assembly of a transcription complex in vitro and explain how multiple layers of cooperativity provide a sensitive switch for activating a gene. We further discuss the complicated interrelationship between synergy and cooperativity.
The dynamic nature of gene regulation in mammalian cells requires that preinitiation complexes assemble rapidly, over small increases in activator concentration, and that they respond to and integrate signals from diverse stimuli. To achieve such regulation, the cell employs the principles of cooperativity and synergy. The promoter of a gene is arranged to allow the cooperative binding of multiple activators to DNA, while the general transcription machinery, in turn, is designed to be recruited by and respond synergistically to multiple activators. The requirement for multiple activators allows the cell to link related signaling pathways and control thousands of genes by using small combinations of activators. This phenomenon is often referred to as combinatorial control (7).
It is believed that the activation surface generated by the activators constituting the enhanceosome is complementary to a surface on coactivators and the general machinery (46). The multiple, complementary interactions are thought to be additive, which from an energetic standpoint should lead to an exponential increase in affinity of the general machinery for the enhanceosome versus any of its individual activators (7, 34, 46, 65). This exponential increase in affinity is the basis for the synergistic transcriptional effect of multiple activators.
The beta interferon (IFN-β) enhancer, for example, employs NF-κB, IRF-3, Jun, and ATF-1 to respond to viral infection (34, 46, 63). While each of the factors is keyed into multiple signaling pathways, it is the modest increase in concentration of each activator that leads to cooperative DNA binding and enhanceosome assembly upon viral infection. The activators present within the enhanceosome then synergistically activate transcription. It is important to realize that the synergistic response to multiple activators is not a result of cooperative DNA binding; it is a consequence of multiple activators interacting with the general machinery. However, as we will discuss throughout this paper, the sigmoidal response of the gene to increasing activator concentrations is due to cooperative binding. This cooperative binding has two components, which can be isolated biochemically and studied. The first component is cooperative assembly of the enhanceosome, and the second is reciprocal cooperative binding between the enhanceosome and the general machinery.
The assembly of an enhanceosome requires specific positioning of activators on the DNA surface. In many contexts, interactions among these bound activators require DNA bending and twisting. Such distortions require significant energetic input when occurring within the DNA persistence length (2, 56, 64). This energetic requirement, however, can be overcome with the assistance of DNA architectural proteins that can absorb the energetic cost. The IFN-β and the T-cell receptor alpha-chain (TCR-α) enhanceosomes both employ architectural proteins to mediate cooperative binding of activators. These architectural proteins include LEF-1 or its homologue TCF-1 (in the case of the TCR-α enhancer) and HMG-I (in the case of the IFN-β enhancer).
LEF-1 and HMG-I are both members of the high mobility group (HMG) of chromatin-associated proteins. LEF-1 is a member of the HMG-1,2 class, whereas HMG-I is a member of the HMG-I(Y) class (66). The two classes employ different DNA binding motifs to recognize and bend DNA. HMG-1 and -2 bind DNA nonspecifically (6, 24), and the resulting bend can have global effects on activator binding and enhancer activity. The yeast HMG-1 and -2 homologues NHP6A and -B, for example, affect activated transcription at a variety of yeast genes (53), while HMG-1 has been shown to affect both p53 and homeodomain DNA binding and transactivation (29, 67).
In attempting to understand the role of cooperativity in gene regulation, and how an enhanceosome functions to recruit the general transcription machinery, we began to study the Epstein-Barr virus (EBV) transactivator ZEBRA. Several lytic promoters have been shown to possess ZEBRA-dependent enhancer activity (33, 41, 59). Furthermore, ZEBRA participates in differential transcription of almost three dozen genes involved in the EBV lytic cycle. The wide range of transcriptional responses elicited by the lytic promoters represents an opportunity to understand how a single activator controls a regulatory hierarchy.
Our initial studies focused on model systems composed of multimerized ZEBRA sites positioned upstream of well-characterized core promoters. This system provided basic information on the mechanism of transcription complex assembly and how multiple activators elicited synergistic effects on transcription. We found, using the model system, that ZEBRA stimulates transcription synergistically as a function of the number of sites under conditions in which the ZEBRA sites were saturated. This result implied that the synergistic effect of sites was not due to cooperative binding of the activators to DNA. It was also shown that the amount of transcription in the model systems correlated with transcription complex assembly, as measured in open complex assays, and that the synergy was first manifested during recruitment of TFIID and TFIIA to the core promoter (8, 13, 14).
The model system allowed us to further explore how upstream activators communicate with the general machinery. By varying the affinity of the upstream promoter sites for ZEBRA and the affinity of the core promoter for TFIID and TFIIA, we were able to obtain evidence for what we will refer to as reciprocal cooperativity. The reciprocal cooperativity was manifested as the ability of strong core promoters to compensate for low-affinity ZEBRA sites and for high-affinity ZEBRA sites to compensate for weak core promoters in transcription assays. The data implied that the general machinery could facilitate cooperative binding of ZEBRA (38), an observation that might explain why genes are activated in a sigmoidal fashion.
In an effort to link the concepts of enhanceosome formation and reciprocal cooperativity, we began studying transcription complex assembly on natural ZEBRA-responsive templates. Natural templates, as opposed to model systems, are more likely to require architectural proteins for activator binding. Furthermore, the distribution and affinity of the sites may be designed to facilitate cooperative interactions. We provide biochemical evidence that the sigmoidal transcriptional response of a natural EBV gene to ZEBRA involves two layers of cooperativity: (i) cooperative assembly of an enhanceosome-like complex mediated by the architectural proteins HMG-1 and -2 and the cellular factor Sp1 and (ii) reciprocal cooperative interactions between the enhanceosome and the transcription factor IID (TFIID)-TFIIA (DA) complex which lead to concerted transcription complex assembly. We also present our initial efforts to study recruitment of the RNA pol II holoenzyme by the enhanceosome-stimulated DA complex.
MATERIALS AND METHODS
Transcription factor purification.
Purification of recombinant ZEBRA, recombinant TFIIA, and HeLa cell hemagglutinin (HA) epitope-tagged TFIID from the HeLa cell line LTRα3 were described previously (12). Recombinant human Sp1 was purchased from Promega. HMG-1 and -2 proteins were purified from calf thymus as previously described (20, 54). The mammalian RNA pol II holoenzyme was purified by affinity chromatography using glutathione S-transferase (GST)-VP16 (28a). Briefly, GST-VP16 was bound to glutathione-agarose beads at 4°C in binding buffer (20 mM HEPES [pH 7.9], 50 μM ZnCl2, 0.05% Nonidet P-40, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine) containing 50 mM KCl. HeLa nuclear extracts prepared as previously described (16) were incubated with the immobilized GST-VP16 for 1 h at 4°C, washed, and eluted in binding buffer containing 0.225 M KCl. The eluate was subjected to a second round of affinity chromatography and used in the experiments represented in Fig. 5. To ensure the integrity of the complex, the resulting eluate was subjected to Sepharose 4B and Superose 6 gel filtration chromatography. All of the general factors except TFIIA and TFIID copurified (28a). As an additional test of integrity, the complex was incubated with HA-tagged recombinant TFIIB and subjected to Superose 6 gel filtration. The HA-tagged TFIIB, although functional in vitro, did not exchange with the wild-type TFIIB present in the complex (data not shown).
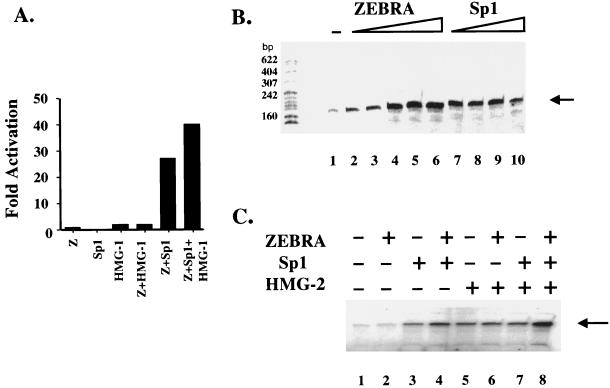
FIG. 5.
In vivo and in vitro effects on transcription by an enhanceosome. (A) In vivo transcription by an enhanceosome. Cotransfection by calcium phosphate of 1 μg of the indicated effector plasmids with 200 ng of the BHLF-1–CAT reporter into a BHK-21 parental cell line. The in vivo fold activation of transcription from the BHLF-1–CAT reporter is indicated. (B) The RNA pol II holoenzyme can respond to stimulation by ZEBRA and Sp1 from the natural BHLF-1 promoter. Basal levels of transcription are indicated in lane 1, where the holoenzyme (2 μg) and TFIID (100 ng) and TFIIA (40 ng) were incubated with the BHLF-1 promoter. Neither factor alone elicits any signal (data not shown). Lanes 2 to 6 show that threefold-increasing concentrations of ZEBRA (2.5 to 200 ng) elicit activated transcription. Similarly, lanes 7 to 10 show that increasing concentrations of Sp1 (0.1 to 3 fpu) can also activate transcription. (C) The enhanceosome responds to stimulation by a partially purified RNA pol II holoenzyme. ZEBRA (7.5 ng), Sp1 (0.3 fpu), and HMG-2 (125 ng) are able to elicit a greater overall level of transcription (lane 8) from the natural BHLF-1 promoter in a reconstituted system using the RNA pol II holoenzyme, TFIID, and TFIIA and then when any of the factors are used alone or in various paired combinations (lanes 2 to 7).
Cloning of BHLF-1 promoter and BHLF-1 promoter mutants.
PCR was used to amplify a fragment from −990 to +90 of the EBV BHLF-1 promoter from pBamW2 YFSal G, which contained a segment of the B95-8 EBV genome spanning kbp 40 to 61 (62). The primers used in the amplification reaction were HL1K (5′-GGGGGATCCGATGAAACAGGCAACTC-3′) and HLPE (5′-GACCCCGCGCCACCCGCTTCAT-3′). The DNA fragment was end repaired with Klenow fragment to remove the TA overhang and then subcloned into the HincII site of the Promega pGEM3 plasmid polylinker. The start site of transcription was oriented facing the pGEM3 T7 promoter.
A PCR-based mutagenesis technique was used to alter the ZEBRA and Sp1 binding sites in pHLGEM to establish their physiological relevance (37). ZEBRA sites located at −77 (5′-TGTGTAA-3′), −96 (5′-TGAGCAA-3′), −135 (5′-TGTGTCA-3′), and −168 (5′-TGTGTCA-3′) were changed to BsrG1, BclI, StuI, and MscI sites to generate HLΔ1ZpGEM, HLΔ2ZpGEM, HLΔ3ZpGEM, and HLΔ4ZpGEM, respectively. The Sp1 sites at −312 (5′-GGGCGG-3′) and −389 (5′-GGGCGG-3′) were also mutated to ApaI and SacII sites to generate HLΔ1Sp-1pGEM and HLΔ2Sp-1pGEM, respectively.
Pairwise binding site mutants were generated by performing a second round of mutagenesis. A primer termed HLdown was designed containing a SacI restriction site (5′-GGGGAGCTCCCGGCTGGGAGGTGTGCA-3′). HLdown in conjunction with the upstream HL1K primer was used to PCR amplify the wild-type BHLF-1 promoter. This fragment was then inserted into the EcoRI/SacI-digested E4TCAT vector (17). The 1,050-bp HL promoter regions replace the E4T promoter, leaving the chloramphenicol acetyltransferase (CAT) gene intact. All constructs were subjected to DNA sequencing to confirm their integrity.
Mutagenesis primers.
The primers used for mutagenesis were Z1-a (5′-CCTCTTTTTGGGGTCTCTGTACAATACTTTAAGGTTTGCTC-3′), Z1-b (5′-GAGCAAACCTTAAAGTATTGTACAGAGACCCCAAAAAGAGG-3′), Z2-a (5′-AAGAAGCCCCCACTCCTGATCAAACCTTAAAGTATTACA-3′), Z2-b (5′-TGTAATACTTTAAGGTTTGATCAGGAGTGGGGGCTTCTT-3′), Z3-a (5′-GGGGGCTTCTTATTGGTTAATTCAGGCCTGTCATTTTAGCCCGT-3′), Z3-b (5′-ACGGGCTAAAATGACAGGCCTGAATTAACCAATAAGAAGCCCCC-3′), Z4-a (5′-GGGTTTCATTAAGGTGTGTGGCCAGGTGGGTGGTACCT-3′), Z4-b (5′-AGGTACCACCCACCTGGCCACACACCTTAATGAAACCC-3′), Sp1-1a (5′-GGGGAGGATTGGGCTGGGCCCCGATATACCTAGTGG-3′), Sp1-1b (5′-CCACTAGGTATATCGGGGCCCAGCCCAATCCTCCCC-3′), Sp1-2a (5′-GGAGGTATCCTAAGCTCCGCGGCTATATACCAGGTGGG-3′), and Sp1-2b (5′-CCCACCTGGTATATAGCCGCGGAGCTTAGGATACCTCC-3′).
DNase I footprinting.
Plasmid pHLCAT was digested with EcoRI, 32P end labeled with polynucleotide kinase and [γ-32P]ATP, and digested again with HindIII to generate the 1,050-bp BHLF-1 promoter fragment used in the DNase I footprints. The binding reactions for DNase I footprinting were as previously described (14). The 13-μl reaction mixtures contained 5 fmol of the 32P-end-labeled probe, 100 ng of TFIID, 40 ng of TFIIA, 5 or 200 ng of ZEBRA, 6 ng of Δ161 ZEBRA (13), and 63 ng of HMG-2 or 0.3 footprint unit (fpu) of recombinant Sp1 (Promega catalog no. E3391) in binding buffer [12.5 mM HEPES (pH 7.9), 60 mM KCl, 12.5% glycerol, 5 mM MgCl2, 0.2 mM EDTA, 60 mM β-mercaptoethanol, 0.5 mg of bovine serum albumin per ml, 30 μg of poly(dG-dC) per ml]. After 60 min of incubation at 30°C, the complexes were subjected to cleavage by DNase I for 1 min, and the reactions were terminated by addition of 100 μl of stop buffer containing 0.4 M sodium acetate, 0.2% sodium dodecyl sulfate, 10 mM EDTA, 50 μg of yeast tRNA per ml, and 10 μg of proteinase K. After a 15-min incubation at 55°C, the mixtures were extracted with phenol-chloroform, and the DNA was precipitated with ethanol, resuspended in formamide dye mix, and resolved on a 6% polyacrylamide–7 M urea sequencing gel run in 1× Tris-borate-EDTA.
In vitro transcription and primer extension.
In vitro transcription and primer extension assays were performed as described previously (8), with the following modifications. Two micrograms of the RNA pol II holoenzyme was mixed with 100 ng of TFIID, 40 ng of TFIIA, 160 ng of TFIIB in the presence of 0.5 mM nucleotides, 12.5 ng of DNA template (pHLCAT), 5 ng of pGEM3, 7.5 mM MgCl2 and 0.2 U of RNasin; 7.4 ng of ZEBRA, 0.3 fpu of Sp1, and 125 ng of HMG2 were added as shown in Fig. 5. After incubation at 30°C for 60 min, the reactions were terminated by addition of 100 μl of stop buffer containing 10 μg of proteinase K. After a 15-min incubation at 55°C, the mixtures were extracted once each with phenol and phenol-chloroform and subsequently ethanol precipitated. The RNA pellet was then resuspended in 20 μl of hybridization buffer containing 300 mM NaCl, 20 mM Tris (pH 7.6), 2 mM EDTA, and 0.2% sodium dodecyl sulfate; 0.05 fmol of the 32P-end-labeled CAT primer (5′-CTCAAAATGTTCTTTACGATGCCATTGGGA-3′) was added, and after 2 h at 37°C the hybridization mixtures were precipitated with isopropyl alcohol, washed in 70% ethanol, resuspended in 10 μl of 10 mM Tris (pH 8.3), and subjected to primer extension as previously described (8).
Cotransfection and CAT assays.
Transcription was measured in triplicate by lipofectin (Life Technology Laboratories, Gaithersburg, Md.)- or calcium phosphate-mediated transient transfection assays (1) using 0.2 to 1 μg of the BHLF-1–CAT wild-type and mutant reporters shown in Fig. 1. Effector plasmids expressing ZEBRA (1 ng to 1 μg) and HMG-1 (125 to 1,000 ng) driven by the simian virus 40 (SV40) promoter or Sp1 (1 μg) (a kind gift from N. Tanese) driven by the cytomegalovirus promoter were cotransfected along with reporter templates and equivalent β-galactosidase (β-Gal)-expressing plasmids into a baby hamster kidney cell line (BHK-21) and harvested 24 h posttransfection as previously described (38). Calcium phosphate transfections were done with larger amounts of reporter and effector DNA than the more efficient Lipofectin reagent. Whole-cell extracts were prepared by freeze-thawing the cells three times, and transfection efficiency was normalized by β-Gal expression. Typical CAT assay mixtures contained 25 to 50 μl of whole-cell extract, 0.01 μCi of [14C]chloramphenicol, and 15 μg of acetyl coenzyme A in 0.25 M Tris (pH 7.5). The mixtures were fractionated by thin-layer chromatography, and the resulting thin-layer chromatography plate was exposed to a Molecular Dynamics PhosphorImager screen, scanned, and quantitated with ImageQuant software.
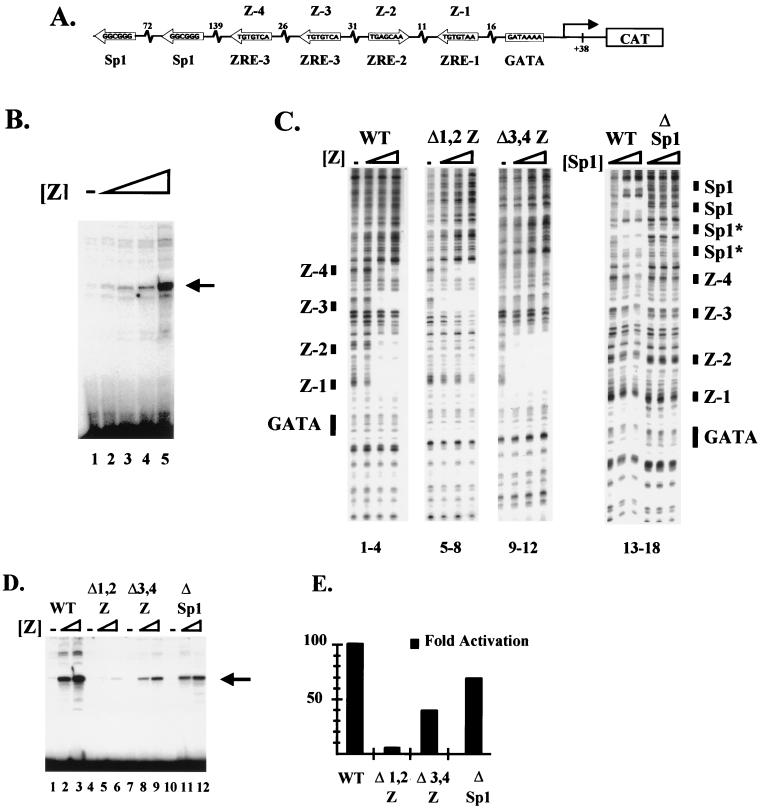
FIG. 1.
ZEBRA binding sites are required for transcriptional activation by the BHLF-1 promoter in vitro and in vivo. (A) Schematic of the BHLF-1 EBV promoter. A 1,050-bp fragment of the BHLF-1 promoter region was subcloned upstream of a CAT reporter gene. The ZEBRA and Sp1 binding sites are depicted along with their relative spacing. (B) Sigmoidal dose response of BHLF-1 to ZEBRA. Fifty nanograms of BHLF-1 was incubated in a HeLa extract with threefold-increasing steps of ZEBRA from 7.5 to 200 ng. Transcription was measured by primer extension using a primer in the CAT coding region. (C) DNase I footprint of ZEBRA on the BHLF-1 EBV wild-type promoter (lanes 1 to 4) or three mutant promoters, Δ1,2Z (lanes 5 to 8), Δ3,4Z (lanes 9 to 12), and ΔSp1 (lanes 13 to 18) (see Materials and Methods for details of construction). In the first three panels, threefold-increasing concentrations of recombinant ZEBRA (2.5 to 22.5 ng) were incubated with the BHLF-1 wild-type (WT) (lanes 1 to 4) or mutant (lanes 5 to 12) promoters and digested with DNase I. Similarly, in the last panel, threefold-increasing concentrations of Sp1 (0.1 to 1 fpu) were incubated with the wild-type promoter (lanes 13 to 15) or the Sp1 mutant promoter (lanes 16 to 18) and subjected to DNase I cleavage. The cleavage ladders are shown with the four ZEBRA binding sites numbered Z-1 to Z-4, where Z-4 is the most distal from the core promoter GATAA box. The nonconsensus Sp1 sites that were not mutated are indicated (*). (D) In vitro transcription and primer extension of the wild-type, Δ1,2Z, Δ3,4Z, and ΔSp1 BHLF-1 promoters. The promoters were incubated in HeLa nuclear extracts with threefold-increasing concentrations of recombinant ZEBRA (7.5 to 200 ng), and transcription was measured by primer extension using a primer in the CAT coding region. (E) Transient transfection assays of the wild-type, Δ1,2Z, Δ3,4Z, and ΔSp1 BHLF-1 promoters. One microgram of effector plasmid encoding ZEBRA expressed from the SV40 promoter and 200 ng of the BHLF-1 wild-type or mutant promoters fused to the CAT gene were cotransfected by calcium phosphate into BHK-21 cells. The fold activation of CAT expression is shown for each of the promoters.
RESULTS
Organization of the BHLF-1 promoter.
To study transcription complex assembly by ZEBRA on natural EBV promoters, we first attempted to identify a strong viral promoter. Among the 10 different EBV regulatory regions that we tested, BHLF-1 was found to be the strongest (data not shown). BHLF-1 and BHRF-1 are divergent genes regulated by a complex intergenic enhancer (41, 59). The BHLF-1-proximal portion of the enhancer contains binding sites for ZEBRA, the cellular factor Sp1, and another EBV regulator called Rta (Fig. 1A). Although transfection studies suggest that Rta contributes to the transcription of BHLF-1 under certain conditions, its requirement can be bypassed (18, 25, 42, 43).
Transcription of the BHLF-1 promoter in a HeLa nuclear extract displays a strong sigmoidal response to increasing ZEBRA concentration (Fig. 1B). This response is due to binding of ZEBRA to four binding sites in the region immediately upstream of the BHLF-1 core promoter bearing the GATAA sequence in place of a consensus TATA (42, 43). Figure 1C shows the results of a DNase I footprinting experiment confirming the positions of the four sites. The sites have previously been termed ZRE-1, ZRE-2, and ZRE-3 (two copies), based on their unique sequences and their different affinities for ZEBRA (10, 26, 38, 42, 43). We will refer to them as sites Z-1 to Z-4 for clarity. In dose-response measurements, Z-1 and Z-2 become occupied at the lowest concentrations of recombinant ZEBRA, while Z-3 and Z-4 require higher concentrations.
The sites are important for ZEBRA responsiveness because pairwise point mutations of Z-1 and Z-2 (Δ1,2Z) or of Z-3 and Z-4 (Δ3,4Z) decrease ZEBRA’s affinity in a DNase I footprinting assay (Fig. 1C, lanes 5 to 12) and decrease ZEBRA’s ability to activate transcription from mutant BHLF-1 promoters in vitro in a HeLa nuclear extract (Fig. 1D). The effects of mutations in Z-1 and Z-2 were particularly severe. The two consensus Sp1 sites also contribute to the activity of the promoter in vitro, as mutants that weaken Sp1 binding in footprinting assays (Fig. 1C, lanes 13 to 18) lowered the overall levels of transcription in vitro (Fig. 1D).
The sites are also necessary for BHLF-1 promoter activity in transfection assays. A vector expressing ZEBRA from the SV40 enhancer was cotransfected into BHK-21 cells with the wild-type and mutant BHLF-1 promoters driving expression of a CAT reporter gene. The bar graph in Fig. 1E shows that the transfection results agree with the in vitro transcription and binding studies, although the effects of Sp1 site mutants were smaller in transfection assays than in vitro. Transfection into B cells elicited similar effects (data not shown).
Cooperative binding of ZEBRA and the DA complex to BHLF-1.
Our previous studies on model promoters bearing one through seven high-affinity ZEBRA binding sites had shown that ZEBRA could synergistically recruit TFIID and TFIIA to a core promoter. We interpreted this result as evidence that multiple ZEBRA molecules were simultaneously interacting with the DA complex (14). However, the model predicts that the DA complex should have a reciprocal cooperative effect on binding of ZEBRA to DNA. Our inability to observe the reciprocal effect on the model promoters was initially surprising but could be due to the fact that the ZEBRA sites were of such high affinity that the DA complex had a negligible effect on ZEBRA binding (data not shown). However, unlike the optimized model promoters, natural ZEBRA-responsive promoters frequently contain multiple medium- to low-affinity sites and rarely contain a high-affinity site. As described below, we observe strong cooperative effects of the DA complex on binding of ZEBRA to multiple sites within the BHLF-1 promoter in DNase I footprinting experiments.
Figure 2A shows the sequential binding of ZEBRA to sites Z-1 and Z-2 followed by Z-3 and then Z-4. When ZEBRA is incubated at the subsaturating concentration shown in lane 2 of Fig. 2A, it generates little protection over any of the sites (Fig. 2B, lane 4). Similarly, when subsaturating amounts of either TFIIA and TFIID are incubated with the template, little protection is observed over the GATAA motif (Fig. 2B, lane 2). However, together, ZEBRA recruits the DA complex to the GATAA box of the BHLF-1 promoter and the DA complex elicits a reciprocal effect on binding of ZEBRA such that it strongly promotes binding to sites Z-1, -2, and -3 and weakly to Z-4 (Fig. 2B, lane 3).
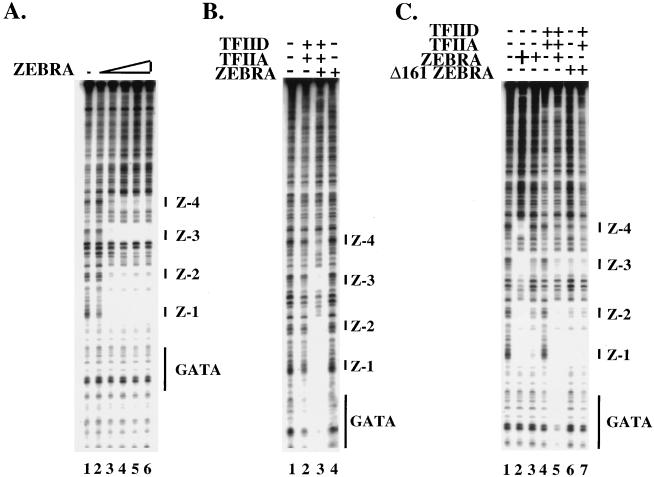
FIG. 2.
Cooperative assembly of a complex containing TFIID, TFIIA, and ZEBRA. (A) Dose-response experiment (threefold steps) using DNase I footprinting to measure ZEBRA binding to the wild-type BHLF-1 promoter. At saturating conditions (200 ng), all four ZEBRA binding sites are occupied (lane 6). (B) ZEBRA recruits the DA complex, and the DA complex has a reciprocal effect on ZEBRA binding. Lane 1 shows naked DNA. When ZEBRA is set at subsaturating concentrations (2.5 ng) as in lane 2 of panel A, none of the four ZEBRA sites are occupied (lane 4). Similarly, when DA is set at subsaturating concentrations (100 ng of TFIID and 40 ng of TFIIA), there is little to no protection of the GATA box (lane 2). When ZEBRA, TFIIA, and TFIID are present together at subsaturating concentrations, ZEBRA recruits DA to the GATA box and DA has a reciprocal effect on ZEBRA binding (lane 3). (C) Recruitment of DA by ZEBRA requires the activation domain. Lane 1 shows naked DNA. Lane 2 shows that at saturating concentrations of ZEBRA (200 ng), all four ZEBRA binding sites are filled, whereas lane 3 shows the site occupancy at subsaturating concentrations of ZEBRA (2.5 ng). Lane 4 shows the footprint over the GATAA box at subsaturating concentrations of TFIID and TFIIA. At subsaturating concentrations of ZEBRA, TFIID, and TFIIA, there is a strong protection over the GATAA box (compare lanes 4 and 5) as well as a reciprocal effect on ZEBRA binding by the DA complex (compare lanes 3 and 5). Lanes 6 and 7 demonstrate the dependence of this effect on the activation domain of ZEBRA. In lane 6, the footprint from subsaturating concentrations of an activation domain mutant of ZEBRA called Δ161 (12.5 ng) is shown. In the presence of subsaturating concentrations of TFIID and TFIIA (lane 7), Δ161 fails to recruit the DA complex to the GATAA box and the DA complex does not stimulate Δ161 binding to the upstream ZEBRA binding sites.
The cooperative recruitment required the activation domain of ZEBRA, as shown in Fig. 2C. Whereas intact ZEBRA bound cooperatively with the DA complex (Fig. 2C, lanes 3 to 5), a truncated version of ZEBRA lacking the amino-terminal nonacidic activation domain (Δ161) (13) failed to recruit the DA complex. Furthermore, the presence of TFIID and TFIIA had little effect of the binding Δ161 to the promoter (Fig. 2C, lanes 6 and 7).
Taken together, our results demonstrate that the DA complex and ZEBRA can interact at subsaturating concentrations to promote the simultaneous or concerted assembly of a transcription complex on DNA. The DA complex is a central checkpoint in transcription complex assembly, and studies using yeast have affirmed the importance of TFIID recruitment as a limiting step in gene activation (60). The ability the DA complex to simultaneously promote ZEBRA binding to multiple sites represents one mechanism for ensuring a sigmoidal response to increasing ZEBRA concentration in the cell.
HMG-1 and -2 promote binding of ZEBRA to DNA to form a simple enhanceosome.
Architectural proteins have been shown to play a key role in cooperative binding of activators to the upstream promoter region (7, 24). Previous studies had shown that HMG-I(Y) and LEF-1 could assist in the cooperative assembly of enhanceosomes on the IFN-β and TCR-α enhancers (19, 63). DNase I footprinting of HMG-I and LEF-1 revealed that they did bind the BHLF-1 promoter at nonconsensus sites but their binding had little effect on binding by ZEBRA (data not shown). Although it is likely that other sequence-specific DNA bending proteins exist in the cell, we attempted to assess the effects of the ubiquitous HMG-1 and -2 proteins on binding of ZEBRA. HMG-1 and -2 are 215 and 209 amino acids in size, respectively. Although the two proteins are encoded by separate genes, they share greater than 82% amino acid identity (6). Previous studies had shown that the yeast HMG-1 and -2 homologues, NHP6A and -B, elicited global effects on transcription from a wide array of promoters (53). Furthermore, biochemical studies had shown that HMG-1 could promote binding of some transcription factors to individual sites or pairs of sites (29, 48, 67). We therefore investigated the ability of the HMG-1 and -2 to mediate cooperative binding of ZEBRA.
We were unable to observe specific binding of HMG-1 or -2 to the promoter alone (Fig. 3A, lane 6). Increasing concentrations of ZEBRA led to a gradual but differential filling of Z-1 through Z-4 (Fig. 3A, lanes 2 to 5). However, as shown in Fig. 3A, when increasing concentrations of ZEBRA were added together with a fixed concentration of HMG-2 (62.5 ng), we observed cooperative ZEBRA binding to all four sites identified in the promoter mutagenesis and DNase I footprinting experiments of Fig. 1 (Fig. 3A; compare lanes 3 to 5 and 7 to 9). An identical effect was observed with HMG-1 (data not shown). There were two measurable consequences of the cooperativity. First, ZEBRA bound at eightfold-lower concentrations in the presence than in the absence of HMG-1 or -2. Second, ZEBRA occupied all four sites simultaneously, whereas the sites normally differed in affinity four- to eightfold. An interesting aspect of the footprint on sites Z-3 and Z-4 is the additional DNase I protection observed between the sites in the presence of HMG-2 (compare lanes 2 and 7). This footprint may represent HMG-2 binding.
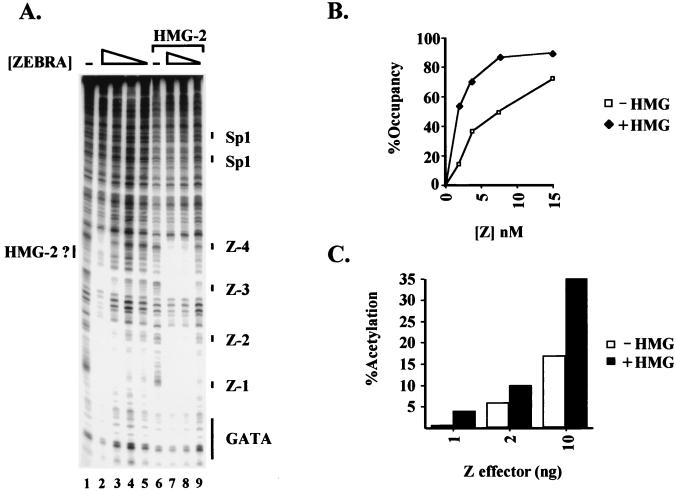
FIG. 3.
HMG-1 and -2 affect ZEBRA binding to the BHLF-1 promoter in vitro and in vivo. (A) DNase I footprint of the BHLF-1 promoter. Lane 1 shows the cleavage ladder of BHLF-1 in the absence of any proteins. Lane 2 shows ZEBRA binding at saturating concentrations (200 ng). A twofold titration of ZEBRA (1.25 to 5 ng) binding is shown in lanes 3 to 5. Lanes 7 to 9 show the effect of HMG-2 (62.5 ng) on the binding of ZEBRA (compare lanes 3 to 5 and 7 to 9). The ZEBRA binding sites are indicated as Z-1 to Z-4. (B) Graph of the results of a gel shift assay showing that HMG-2 helps ZEBRA to bind to the BHLF-1 promoter. In the presence of 62.5 ng of HMG-2, 56% of the BHLF-1 probe is bound by ZEBRA, whereas at the same concentration (1.9 nM) of ZEBRA alone, only 15% of the probe is shifted. (C) HMG-1 affects transcriptional activation by ZEBRA in vivo. ZEBRA (1 to 10 ng) and HMG-1 (800 ng) effector plasmids driven from the SV40 promoter were cotransfected by lipofection with 25 ng of the wild-type BHLF-1 CAT reporter construct and assayed for transcription as a function of CAT activity. One nanogram of ZEBRA alone gives a 0.5-fold stimulation of transcription, and the presence of HMG-1 increases the activation to 4-fold.
The sigmoidal response characteristic of cooperative binding was clearly evident in gel shift experiments. We titrated ZEBRA in the presence and absence of a fixed concentration of HMG-2. At low concentrations of ZEBRA, four distinct shifted complexes were observed. By quantitating the amount of radioactivity in each complex and in the unbound probe, we could calculate the percentage occupancy of the four sites. The occupancy was then plotted as a function of ZEBRA concentration (Fig. 3B). The addition of HMG-2 led to a mild supershift of the complex (data not shown) but, more importantly, strongly stimulated site occupancy such that even at the lowest ZEBRA concentrations tested, most of the sites in the probe were bound (compare upper and lower curves). The stimulatory effect was most apparent at the lowest ZEBRA concentrations because higher concentrations led to probe saturation in the absence of HMG-2. Similar effects were observed with HMG-1 (data not shown).
The stimulatory effect of HMG-1 was also observed in a transfection experiment into BHK-21 cells as measured by a CAT assay. Cotransfection of a vector expressing HMG-1 from the SV40 enhancer-promoter with increasing amounts of the ZEBRA expression vector resulted in a significant additional stimulation by ZEBRA of the BHLF-1 promoter (Fig. 3C). Again, the stimulatory effect was most evident at low concentrations of ZEBRA. Despite the consistent stimulatory effect of HMG-1 in transfection experiments, the effect might be viewed as surprising since there are millions of molecules of HMG-1 and -2 in a typical mammalian cell. However, it is possible that HMG-1 and -2 are sequestered into chromatin complexes in vivo and are unavailable to the transfected DNA. Alternatively, the amount of transfected DNA may simply exceed the pool of free HMG-1 and -2.
Taken together, the data reveal cooperative binding of multiple activators to a promoter is mediated by architectural proteins, which bind DNA nonspecifically. We will refer to the final structure as a simple enhanceosome, as opposed to a more complex enhanceosome (i.e., TCR-α) containing several different activators engaged in combinatorial interactions. We imagine that the cooperativity involves direct protein-protein interactions among bound ZEBRA molecules. We have not been able to observe strong interactions between ZEBRA and HMG-1 or -2 in affinity binding experiments off the DNA but can not exclude that ZEBRA and HMG-1 or -2 interact on the DNA.
Sp1, ZEBRA, and HMG-2 assemble into an enhanceosome that recruits the DA complex.
Sequence analysis of the BHLF-1 promoter revealed that it contained two consensus binding sites for the cellular factor Sp1 upstream of the ZEBRA sites (Fig. 1A). Sp1 consensus sites are found in many lytic promoters, and mutagenesis of those sites in BHLF-1 decreased transcription in vivo and in vitro (Fig. 1C, D, and E). We therefore investigated the effect of Sp1 on both enhanceosome formation and DA complex recruitment (Fig. 4). Sp1 alone or with HMG-2 generated a series of protections spanning over 100 bp immediately upstream of the four ZEBRA binding sites (Fig. 4A, lane 6, and data not shown). Indeed, four separate Sp1 footprints were observed, suggesting the possibility that Sp1 was binding cooperatively to the two consensus sites and two or more adjacent nonconsensus sites (indicated by asterisks). When subsaturating amounts of ZEBRA (lane 2) were incubated with Sp1, we observed strong binding of ZEBRA to sites Z-1 to Z-3 (Fig. 4A, lane 3). In dose-response experiments, we observed an 8- to 16-fold stimulation of ZEBRA binding by Sp1 (data not shown). Addition of HMG-2 further stimulated binding to Z-4 (Fig. 4A, lane 4). Indeed, in the presence of Sp1, we observed ZEBRA and HMG-2 DNase I protections spanning nearly all 300 bp of the proximal BHLF-1 promoter. There were several intriguing DNase I enhancements and protections between the sites, possibly indicative of DNA looping between ZEBRA and Sp1. However, we have not observed strong Sp1-ZEBRA interactions off the DNA. Nevertheless, the strong degree of cooperativity in the presence of Sp1, ZEBRA, and HMG-2 indicates the assembly of a sophisticated nucleoprotein complex at the promoter. We will refer to this structure as an enhanceosome, although we have not formally shown that the proteins directly interact.
FIG. 4.
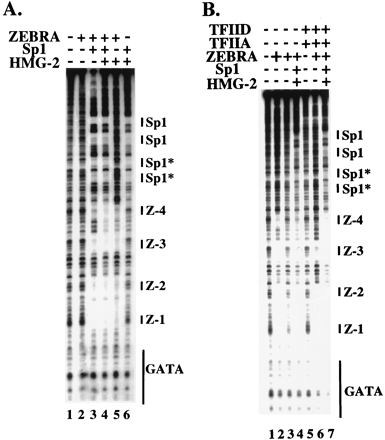
Formation of an enhanceosome. (A) Sp1 and HMG-2 help ZEBRA to bind cooperatively to the BHLF-1 EBV natural promoter. Sp1 (0.3 fpu) and HMG-2 (62.5 ng) stimulate ZEBRA (2.5 ng) binding (compare lanes 2 and 3 and lanes 2 and 5). Together, Sp1 and HMG-2 stimulate an even greater binding of ZEBRA to Z-1 through Z-4 (compare lanes 3 and 4 and lanes 4 and 5). Sp1 and HMG-2 alone exhibit no binding to or around the promoter region encompassing ZEBRA sites Z-1 through Z-4 (lane 6). (B) The enhanceosome can recruit the DA complex to the GATA box of the natural EBV BHLF-1 promoter. Lanes 2 and 3 show saturating (200 ng) and subsaturating (2.5 ng) concentrations of ZEBRA, respectively. Lane 4 shows binding to Z-1 to Z-4 in the context of the simple enhanceosome as shown in panel A, lane 4. Lane 5 shows that at subsaturating concentrations of DA (100 ng of TFIID and 40 ng of TFIIA), there is very little protection over the GATA box. Lane 6 shows the recruitment of the DA complex by ZEBRA and the reciprocal effect that DA has on ZEBRA binding to sites Z-1 to Z-4 as shown in Fig. 2. Lane 7 demonstrates that the simple enhanceosome is able to recruit the DA complex to the GATA box and that the DA complex has a reciprocal effect on ZEBRA binding to this promoter (compare lanes 6 and 7 as well as lanes 4 and 7). Sp1* indicates nonconsensus Sp1 binding sites distinguishable from the two consensus sites shown in Fig. 1A and labeled here as Sp1.
The enhanceosome was able to recruit the DA complex to the core promoter (GATAA), as shown in Fig. 4B. Subsaturating amounts of ZEBRA (lane 3) were incubated with Sp1 and HMG-2. Sp1 and HMG-2 promoted ZEBRA binding (compare lanes 3 and 4), although the sites were not entirely saturated. Addition of TFIID and TFIIA to ZEBRA strongly promoted ZEBRA binding to sites Z-1 and Z-2, and less so to Z-3, while ZEBRA recruited DA (lane 6). However, addition of TFIID, TFIIA, ZEBRA, HMG-2, and Sp1 led to even stronger recruitment of the DA complex to the core promoter and a reciprocal effect on ZEBRA binding to sites Z-3 and Z-4 (Fig. 4B; compare lanes 6 and 7).
The cooperative effects of ZEBRA and Sp1 binding were paralleled by synergistic activation in transfection assays (Fig. 5A). Cotransfection of the BHLF-1 promoter with vectors constitutively expressing Sp1 or ZEBRA alone (at subsaturating concentrations) had less than a twofold stimulatory effect on transcription from the BHLF-1 promoter. However, together the two proteins activated transcription 28-fold (Fig. 5A). HMG-1 further stimulated transcription 30%. We believe that because of the strong synergistic effects of Sp1 and ZEBRA, we observed only a small additional stimulatory effect of HMG-1. This result is somewhat analogous to the effects of HMG-1 when ZEBRA was present at saturating amounts in the transfection assay represented in Fig. 3C. We do not believe that Sp1 was inadvertently stimulating ZEBRA expression. ZEBRA and our β-Gal normalization standard are both expressed from the SV40 enhancer-promoter, and Sp1 had no effect on β-Gal activity in our transfection assays (data not shown).
We have found it difficult to precisely regulate the amount of synergy by ZEBRA, Sp1, and HMG-1 because Sp1 and HMG-1 are already present in the cell. Furthermore, different permutations of the experiment in Fig. 5A revealed differential synergistic effects as different components were made limiting. We presented this particular experiment because it demonstrated the ability of ZEBRA and Sp1 to synergize, analogous to the binding experiments represented in Fig. 4.
Interaction of a putative holoenzyme with the enhanceosome.
Recent studies have revealed that many of the general factors are assembled into a holoenzyme. Although holoenzymes isolated from yeast and mammalian extracts vary considerably in terms of composition, it remains unclear if this variability is due to purification techniques or because functionally distinct holoenzymes exist within the cell. The yeast holoenzyme isolated by Hengartner et al. (27) contains TFIIB and is complementable by TFIID and TFIIE (36). This holoenzyme has also been shown to directly interact with activators in affinity chromatography experiments (27). The unique properties of the holoenzyme suggest that it could participate in the second step of a two-step recruitment model (60). Such a model predicts that activators initially recruit a complex containing TFIID and TFIIA, which then serves as a platform for subsequent recruitment of the RNA pol II holoenzyme. Although a complex containing TFIIB and complementable solely by TFIIA and TFIID had not yet been isolated from mammalian extracts, the rationale that such a complex exists is compelling. First, it would agree with biochemical data suggesting that binding of the DA complex and recruitment of TFIIB are two biochemically separable steps. Second, most transcription occurs in multiple rounds. The first round is slow and takes longer than reinitiation (30). Biochemical data suggest that TFIID and probably TFIIA stay behind during elongation but the remaining factors dissociate from the complex (see, for example, reference 68). Thus, a TFIIB-containing holoenzyme lacking TFIID and TFIIA would make sense from a regulatory standpoint because it could support the rapid reinitiation observed in vitro. Other models have been proposed, and we will address these in Discussion.
In an effort to isolate such a holoenzyme from HeLa extracts, we used GST-VP16 and GST-ZEBRA affinity chromatography. The eluate from the first round was subjected to a second round of affinity chromatography and assayed for transcriptional activity. Although the eluate displayed a low basal activity and the ability to respond weakly to activators, that activity was greatly stimulated by TFIIA and TFIID. To determine the composition and whether the various factors constituting the putative holoenzyme existed in a complex, the affinity eluate was subjected to gel filtration. Immunoblotting revealed that TFIIB, TFIIE, TFIIF, TFIIH, pol II, SWI-SNF, p300, and other components, but not TATA binding protein (TBP), TBP-associated factors, or TFIIA, comigrated as a large (>2-MDa) complex (28a).
To determine if this holoenzyme could complement the DA complex on a natural ZEBRA-responsive promoter, we incubated it with increasing concentrations of ZEBRA in the presence of TFIID and TFIIA. The results shown in Fig. 5B and C revealed that the holoenzyme could complement the DA complex on the BHLF-1 gene and could respond strongly to ZEBRA, Sp1, and HMG-2. The synergistic effects were not nearly as dramatic as those observed in vivo, or in the intact HeLa extracts, although they present a framework for further investigation. We imagine that the differences could be due to a lack of critical coactivators, the need for chromatin templates to observe the synergistic effects in a pure system, or simply the weaker activity of the isolated holoenzyme.
DISCUSSION
Our current model for the synergistic response of a gene to multiple activators is that the general machinery has an exponentially higher affinity for multiple versus single activators. This model is based on the exponential relationship between the free energy of an interaction and the affinity (e.g., K = e−ΔG/RT). Several of our earlier studies provided support for the model by demonstrating synergistic transcription under conditions where the activator sites were saturated (8, 9). A corollary of the model is, however, that the general machinery should facilitate cooperative binding of multiple activators to DNA when the activators are present at subsaturating levels.
We undertook the present study to test the prediction and to explore other mechanisms contributing to cooperative activator binding. We chose to perform our study on a natural EBV promoter where nuances of the mechanism might be revealed and where the findings might be expanded to study other genes in the EBV regulatory switch. Our study showed that recruitment of TFIIA and TFIID can cooperatively facilitate ZEBRA binding to the upstream promoter. In addition to this form of cooperativity, we found that the architectural proteins HMG-1, HMG-2, and Sp1 could facilitate cooperative binding of ZEBRA to form an enhanceosome. The two forms of cooperativity, when superimposed, would provide a plausible mechanism for explaining the sensitive sigmoidal response to increasing activator concentration.
A model for transcription complex assembly at BHLF-1.
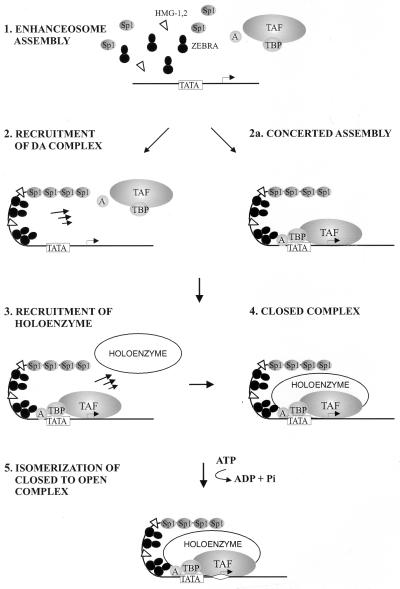
The data suggest a simple biochemical model for assembly of a transcription complex (Fig. 6), namely, that a ZEBRA-containing enhanceosome and the DA complex are recruited to the promoter in a concerted reaction. The final complex contains all or most of the information for specificity and, subsequently (or possibly concurrently), recruits the holoenzyme. We imagine that the holoenzyme can be continually rerecruited during multiple rounds of reinitiation.
FIG. 6.
A model for gene activation by the ZEBRA enhanceosome. The model depicts enhanceosome assembly and DA recruitment (steps 1 and 2) either as distinct steps or as a concerted reaction involving reciprocal cooperative interactions among ZEBRA and the general machinery. Previous data have supported a stepwise assembly of the complex, although our study revealed the assembly can occur in a concerted fashion. The RNA pol II holoenzyme is then recruited to the promoter by the enhanceosome-DA complex (step 3) to form the closed preinitiation complex (step 4). This step is then followed by ATP-dependent start site melting (step 5) and eventually elongation. TAF, TBP-associated factor.
The holoenzyme preparation which we isolated in our study apparently contains the coactivator and histone acetylase p300/CBP and components of the SWI-SNF complex involved in chromatin remodeling (35, 61). We have shown that these activities are active in the context of the holoenzyme (28a). On chromatin templates these activities may be necessary to remove nucleosomes encompassing the core promoter during initiation or to remove downstream nucleosomes during elongation (3).
Although our model incorporates the concept of a holoenzyme that can be complemented by the DA complex, other forms of the holoenzyme have been isolated (11, 15, 44, 45, 47, 51). Some preparations contain a full complement of general transcription factors along with myriad other functionally significant proteins (45, 50, 51), whereas others contain only a subset of the general factors, coactivators, and chromatin remodeling factors such as SWI-SNF (11, 44, 47). The discrepancies in the actual composition of the RNA pol II holoenzymes may be a result of different purification procedures or the existence of functionally discrete versions of the holoenzyme, or perhaps some of these factors are only loosely associated with the holoenzyme at any given time within the cell.
Given the observation of intact holoenzymes, it is plausible that the initial event in transcription of a gene includes recruitment of a holoenzyme containing TFIID and all of the general factors. Current models for elongation suggest, however, that after the first round of initiation the holoenzyme falls apart. Given the observation that TFIID has been shown to remain behind after elongation (68, 69), the extreme stability of TFIID bound to the DNA (reviewed in reference 5), and the slow kinetics of TFIID binding, it is unlikely that new TFIID-containing holoenzymes are recruited in each round.
Enhanceosome assembly.
The BHLF-1 enhanceosome consist minimally of ZEBRA and HMG-1 or -2. Sp1 strongly cooperates with ZEBRA in binding DNA. Although mutagenesis and transfection data suggest Sp1 sites are important, we have no direct evidence that Sp1 mediates a response under physiological conditions. One other potential candidate for binding to the GC-rich Sp1 sites is EGR-1. However, EGR-1 showed no stimulatory effects in cotransfection assays (data not shown). The strong cooperative effects and the DNase I-hypersensitive sites interspersed among the Sp1 and ZEBRA footprints suggest bona fide protein-protein interactions between the two proteins. ZEBRA, like Sp1, contains several glutamine-rich stretches which might engage in cooperative interactions with Sp1 similar to those seen among Sp1 protomers (52). The appearance of a new footprint between the Z-3 and Z-4 sites in the promoter suggests, however, that HMG-2 may bind there stably in the presence of ZEBRA. This issue is under investigation.
There are tremendous differences in the range and action of enhancers, and it is likely that the dynamics of enhanceosome assembly will change from case to case. There will be some common features. The principles of cooperativity and synergy, for example, are hallmarks of an enhanceosome, and they are likely to be employed in potent, broadly active enhancers like the SV40 enhancer (49) as well as in regulated enhanceosomes such as the cell-specific TCR-α enhancer or the signal-dependent IFN-β enhancer. However, it is important to consider that there will be differences. Enhancers that are designed to respond to particular signals may be tuned more sensitively to changes in helical phasing or the arrangements of activator binding sites. For example, such changes in promoter architecture are more deleterious to the IFN-β enhancer (63) than to the strong constitutive SV40 enhancer with its redundancy in activator binding sites (49, 63).
To what extent can the role of architectural proteins in assembly of other enhanceosomes be generalized? Architectural proteins are not necessary for cooperative binding of λ repressor in prokaryotes or of GAL4 and androgen receptor in eukaryotes (28, 28a, 32). The case of λ repressor is particularly interesting because the ability to interact was dependent on helical phasing even though two molecules of λ repressor stably interacted as far as 60 bp away and the DNA was clearly looping (22). Therefore, the strength of the cooperative interactions absorbed the energetic cost of DNA bending but was unable to absorb the excess cost of DNA twisting. It is not yet clear whether similar scenarios will be observed in mammalian systems, although numerous cooperative interactions have been reported for gene activators in binding reactions lacking architectural proteins.
In instances where architectural proteins are involved, the requirement for sequence specificity may vary. In the case of the IFN-β enhancer, the role of HMG-I is to reverse an intrinsic bend to allow NF-κB to bind. Similarly, LEF-1 (or TCF-1) promotes distal cooperative interactions on TCR-α but also displays a context-dependent activation domain, recently shown to interact with a coactivator called ALY (4). However, the abundance of HMG-1 in cells, and its ability to singularly twist and bend the DNA in a manner largely independent of sequence (6), suggests that it could be used by virtually any enhancer or promoter to facilitate protein-protein interactions. One can envision a scenario where different numbers of HMG-1 or -2 molecules would be required on different enhanceosomes. The number and positioning of HMG molecules would be based not on sequence but on the final free energy of the nucleoprotein structure.
Recruitment of the general machinery.
Until recently, most studies using the EBV transactivator ZEBRA had been performed in model systems with highly defined templates bearing multimerized binding sites upstream of a core promoter such as adenovirus E4. Early kinetic experiments in this system used permanganate-sensitive open complexes as an endpoint for transcription complex assembly. These studies established that complexes containing ZEBRA, crude TFIID (contaminated with upstream stimulatory activity [USA] coactivators) and crude TFIIA formed a rate-limiting intermediate in open complex assembly (13). The addition of TFIIB enhanced the stability of the complexes in these experiments but was not a limiting intermediate. Later studies using homogenous TFIID and TFIIA established that ZEBRA could directly recruit the DA complex in gel shift and DNase I footprinting experiments (14, 40).
ZEBRA was also able to directly recruit TFIIB in these experiments, reinforcing the notion that components of the DAB complex were targets of ZEBRA-mediated transactivation. Indeed coincubation of ZEBRA with TFIIA, TFIIB, TFIID, and the USA coactivator was necessary to form a stable complex resistant to challenge with the detergent Sarkosyl (39). Taken together, the data support the idea that DAB and coactivators are all needed for the final complex stability. TFIIB may also act as a potential secondary target used during reinitiation to continually rerecruit holoenzymes to the DA complex.
Our present study demonstrates that the BHLF-1 enhanceosome can recruit the DA complex and that DA can have a reciprocal effect on binding of ZEBRA. Originally we were unable to observe this reciprocal cooperativity on the model templates by DNase I footprinting, probably because the affinity of the sites for ZEBRA was so high (8). However, such reciprocal cooperativity is predicted from a thermodynamic point of view. Indeed functional in vitro transcription studies on the model templates in which high-affinity binding sites were placed upstream of low-affinity core promoters, and vice versa, established an energetic link between the two types of regulatory elements (38). Although this previous study did not provide direct binding evidence, it suggested that reciprocal cooperativity existed and provided the foundation for the direct effect observed here.
The existence and biological importance of reciprocity in gene regulation was first alluded to by the studies on λ repressor (31). Although repressor bound at the high-affinity OR1 strongly enhanced affinity for repressor to OR2, the repressor at OR2 elicited a modest reciprocal effect on repressor bound at OR1. Ironically, the initial experiments on TFIID, rather than showing an effect of the major late transcription factor (USF) activator on TFIID recruitment, showed a strong stabilizing effect of TFIID on activator binding (58). Our studies have further refined the reciprocity model, demonstrating that cooperativity could be manifested under conditions where ZEBRA, TFIID, and TFIIA were limiting in concentration. The ability of enhanceosome-DA complex assembly to occur in a concerted reaction would lead to enhanced specificity in the transcriptional response.
Many of the issues discussed have recently been established for the IFN-β enhanceosome. This structure interacts with DAB complex and the USA coactivators to form a Sarkosyl-resistant complex. The reciprocal effect of the general machinery was shown by the ability of the factors to stabilize the activators constituting the upstream enhanceosome from challenge by competitor binding site oligonucleotides (34).
Kim and Maniatis (34) and Merika et al. (46) have presented an argument that the stereospecific arrangement of activators in the IFN-β enhanceosome is important for its function, possibly by recruitment of CBP-containing coactivators. It is not clear that such a relationship exists on the EBV promoters. We have altered the ZEBRA site phasing relationships for one less potent EBV promoter (BALF-2) and did not observe an effect, although we have not yet confirmed this observation for BHLF-1. In fact the number and arrangement of the ZEBRA sites vary significantly among the three dozen or so different ZEBRA-responsive viral promoters, and there may be no strict rules for site alignment. We hope that studying enhanceosome formation on select ZEBRA-responsive promoters will reveal new principles for how nucleoprotein promoter-enhancer complexes function.
ACKNOWLEDGMENTS
We thank Naoko Tanese for her generous gift of Sp1 expression plasmids, T. K. Kim for HMG-I/Y protein, and Rudolph Grosschedl for providing LEF-1 protein.
This work was supported by grant GM057283 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F M, et al., editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 3.Brown S A, Kingston R E. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 7.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 8.Carey M, Kolman J, Katz D A, Gradoville L, Barberis L, Miller G. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J Virol. 1992;66:4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey M, Lin Y S, Green M R, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 10.Chang Y N, Dong D L, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 12.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 13.Chi T, Carey M. The ZEBRA activation domain: modular organization and mechanism of action. Mol Cell Biol. 1993;13:7045–7055. doi: 10.1128/mcb.13.11.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 15.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 17.Emami K H, Carey M. A synergistic increase in potency of a multimerized VP16 transcriptional activation domain. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin G H, Sanders C, Johns E W. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38:14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- 21.Greenblatt J. RNA polymerase II holoenzyme and transcriptional regulation. Curr Opin Cell Biol. 1997;9:310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 22.Griffith J, Hochschild A, Ptashne M. DNA loops induced by cooperative binding of lambda repressor. Nature. 1986;322:750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- 23.Grosschedl R. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 24.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 25.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayward S D, Hardwick J M. Herpes virus transcription and its regulation. Boca Raton, Fla: CRC Press; 1991. [Google Scholar]
- 27.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 28.Hochschild A, Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986;44:681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- 28a.Huang, W., and M. Carey. Unpublished data.
- 29.Jayaraman L, Moorthy N C, Murthy K G, Manley J L, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, Gralla J D. Uncoupling of initiation and reinitiation rates during HeLa RNA polymerase II transcription in vitro. Mol Cell Biol. 1993;13:4572–4577. doi: 10.1128/mcb.13.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson A, Meyer B J, Ptashne M. Mechanism of action of the cro protein of bacteriophage lambda. Proc Natl Acad Sci USA. 1978;75:1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang T, Martins T, Sadowski I. Wild type GAL4 binds cooperatively to the GAL1-10 UASG in vitro. J Biol Chem. 1993;268:9629–9635. [PubMed] [Google Scholar]
- 33.Kenney S, Holley-Guthrie E, Mar E C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 35.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 36.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 37.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehman A M, Ellwood K B, Middleton B E, Carey M. Compensatory energetic relationships between upstream activators and the RNA polymerase II general transcription machinery. J Biol Chem. 1998;273:932–939. doi: 10.1074/jbc.273.2.932. [DOI] [PubMed] [Google Scholar]
- 39.Lieberman P. Identification of functional targets of the Zta transcriptional activator by formation of stable preinitiation complex intermediates. Mol Cell Biol. 1994;14:8365–8375. doi: 10.1128/mcb.14.12.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA—promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 41.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lieberman P M, O’Hare P, Hayward G S, Hayward S D. Promiscuous trans activation of gene expression by an Epstein-Barr virus-encoded early nuclear protein. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maldonado E, Drapkin R, Reinberg D. Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 45.McCraken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 46.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 47.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onate S A, Prendergast P, Wagner J P, Nissen M, Reeves R, Pettijohn D E, Edwards D P. The DNA-bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences. Mol Cell Biol. 1994;14:3376–3391. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ondek B, Gloss L, Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988;333:40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- 50.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 51.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 52.Pascal E, Tjian R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 53.Paull T T, Carey M, Johnson R C. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 54.Paull T T, Haykinson M J, Johnson R C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 55.Reinberg D, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 56.Rippe K, von Hippel P H, Langowski J. Action at a distance: DNA-looping and initiation of transcription. Trends Biochem Sci. 1995;20:500–506. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 57.Roeder R G. Nuclear RNA polymerases: role of general initiation factors and cofactors in eukaryotic transcription. Methods Enzymol. 1996;273:165–171. doi: 10.1016/s0076-6879(96)73016-1. [DOI] [PubMed] [Google Scholar]
- 58.Sawadogo M, Roeder R G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 59.Shimizu N, Sakuma S, Ono Y, Takada K. Identification of an enhancer-type sequence that is responsive to Z and R trans-activators of Epstein-Barr virus. Virology. 1989;172:655–658. doi: 10.1016/0042-6822(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 60.Stargell L A, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 61.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 62.Sugden B, Yates J, Mark W. Transforming functions associated with Epstein-Barr virus. J Investig Dermatol. 1984;83:82s–87s. doi: 10.1111/1523-1747.ep12281491. [DOI] [PubMed] [Google Scholar]
- 63.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 64.Wang J C, Giaever G N. Action at a distance along a DNA. Science. 1988;240:300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- 65.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 66.Werner M H, Burley S K. Architectural transcription factors: proteins that remodel DNA. Cell. 1997;88:733–739. doi: 10.1016/s0092-8674(00)81917-0. [DOI] [PubMed] [Google Scholar]
- 67.Zappavigna V, Falciola L, Citterich M H, Mavilio F, Bianchi M E. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 68.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 69.Zawel L, Lu H, Cisek L J, Corden J L, Reinberg D. The cycling of RNA polymerase II during transcription. Cold Spring Harbor Symp Quant Biol. 1993;58:187–198. doi: 10.1101/sqb.1993.058.01.023. [DOI] [PubMed] [Google Scholar]